

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124215.6 - phase: 0 /pseudo
(679 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
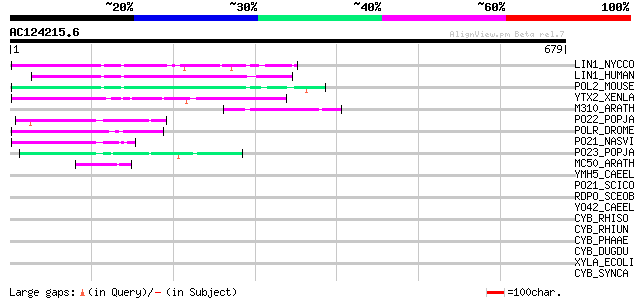
Score E
Sequences producing significant alignments: (bits) Value
LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog 91 1e-17
LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog 86 2e-16
POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains... 80 2e-14
YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein ... 75 4e-13
M310_ARATH (P93295) Hypothetical mitochondrial protein AtMg00310... 74 1e-12
PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type... 64 1e-09
POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type... 55 5e-07
PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type... 53 2e-06
PO23_POPJA (Q05118) Retrovirus-related Pol polyprotein from type... 52 4e-06
MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250... 52 7e-06
YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III 41 0.012
PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type... 37 0.13
RDPO_SCEOB (P19593) Probable reverse transcriptase (EC 2.7.7.49) 34 1.4
YO42_CAEEL (P34680) Hypothetical protein ZK757.2 in chromosome III 33 1.9
CYB_RHISO (Q9G2R8) Cytochrome b 33 1.9
CYB_RHIUN (Q96071) Cytochrome b 32 5.5
CYB_PHAAE (Q8M708) Cytochrome b 32 5.5
CYB_DUGDU (Q33401) Cytochrome b 32 7.2
XYLA_ECOLI (P00944) Xylose isomerase (EC 5.3.1.5) (D-xylulose ke... 31 9.4
CYB_SYNCA (Q9T9B9) Cytochrome b 31 9.4
>LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog
Length = 1260
Score = 90.9 bits (224), Expect = 1e-17
Identities = 96/367 (26%), Positives = 162/367 (43%), Gaps = 44/367 (11%)
Query: 3 QSAFIKGRQILDGILVANEVVDDARKRK-KDLLLFKVDFEKAYDSVD*NYLEEVMVKMGF 61
Q FI G Q I + V+ K K KD ++ +D EKA+D++ ++ + K+G
Sbjct: 562 QVGFIPGSQGWFNIRKSINVIQHINKLKNKDHMILSIDAEKAFDNIQHPFMIRTLKKIGI 621
Query: 62 PTLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVLMEAL 121
+ K I+ TA++++NG F L G RQG PLSP LF + E VL A+
Sbjct: 622 EGTFLKLIEAIYSKPTANIILNGVKLKSFPLRSGTRQGCPLSPLLFNIVME---VLAIAI 678
Query: 122 VVNNLFNGYKVGSHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGLKVNFS 181
G +GS E + FADD I+ + + + L ++ + +SG K+N
Sbjct: 679 REEKAIKGIHIGSEE---IKLSLFADDMIVYLENTRDSTTKLLEVIKEYSNVSGYKINTH 735
Query: 182 KSL-FVGVNVHGSWLAEAATVLNCNAGSIPFL-------YLGLPIGGNASRMV--FWKPL 231
KS+ F+ N + + TV + SIPF YLG+ + + + ++ L
Sbjct: 736 KSVAFIYTNNNQA----EKTVKD----SIPFTVVPKKMKYLGVYLTKDVKDLYKENYETL 787
Query: 232 INRINSRLSSWKSKHLSLGGRLVLLKSVLSSLPVYALSF----FKAPSGIVSSIESILNN 287
I ++ WK+ S GR+ ++K +S LP +F KAP +E I+ +
Sbjct: 788 RKEIAEDVNKWKNIPCSWLGRINIVK--MSILPKAIYNFNAIPIKAPLSYFKDLEKIILH 845
Query: 288 FFFWGGWGGGSADHRKIIWVDWNSVCRSQEVGGLEVRRIKEFNLALLGK--WCWRVLTER 345
F W K + + + GG+ + ++ + +++ K W W E
Sbjct: 846 FI----WNQKKPQIAKTL------LSNKNKAGGITLPDLRLYYKSIVIKTAWYWHKNREV 895
Query: 346 DSLWFRV 352
D +W R+
Sbjct: 896 D-VWNRI 901
>LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog
Length = 1259
Score = 86.3 bits (212), Expect = 2e-16
Identities = 75/324 (23%), Positives = 139/324 (42%), Gaps = 20/324 (6%)
Query: 27 RKRKKDLLLFKVDFEKAYDSVD*NYLEEVMVKMGFPTLWRKWIKKCVGTATASVLVNGSP 86
R + + ++ +D EKA+D + ++ + + K+G + K I+ TA++++NG
Sbjct: 587 RTKDTNHMIISIDAEKAFDKIQQPFMLKPLNKLGIDGTYLKIIRAIYDKPTANIILNGQK 646
Query: 87 TYEFSLHRGLRQGDPLSPFLFLLAAEGFHVLMEALVVNNLFNGYKVGSHEMVGVTHLQFA 146
L G RQG PLSP L + E VL A+ G ++G E V FA
Sbjct: 647 LEAPPLKTGTRQGCPLSPLLPNIVLE---VLARAIRQEKEIKGIQLGKEE---VKLSLFA 700
Query: 147 DDTIILCDKSWANIRALRAILLLFQELSGLKVNFSKSLFVGVNVHGSWLAEAATVLNCNA 206
DD I+ + + + L ++ F ++SG K+N KS + ++ + L
Sbjct: 701 DDMIVYLENPIVSAQNLLKLISNFSKVSGYKINVQKSQAFLYTNNRQTESQIMSELPFTI 760
Query: 207 GSIPFLYLGLPIGGNASRMV--FWKPLINRINSRLSSWKSKHLSLGGRLVLLKSVLSSLP 264
S YLG+ + + + +KPL+N I + WK+ S GR+ ++K +
Sbjct: 761 ASKRIKYLGIQLTRDVKDLFKENYKPLLNEIKEDTNKWKNIPCSWVGRINIVKMAILPKV 820
Query: 265 VYALSF--FKAPSGIVSSIESILNNFFFWGGWGGGSADHRKIIWVDWNSVCRSQEVGGLE 322
+Y + K P + +E F + ++K + +++ + + GG+
Sbjct: 821 IYRFNAIPIKLPMTFFTELEKTTLKFIW----------NQKRAHIAKSTLSQKNKAGGIT 870
Query: 323 VRRIKEFNLALLGKWCWRVLTERD 346
+ K + A + K W RD
Sbjct: 871 LPDFKLYYKATVTKTAWYWYQNRD 894
>POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains:
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1300
Score = 80.1 bits (196), Expect = 2e-14
Identities = 94/399 (23%), Positives = 156/399 (38%), Gaps = 39/399 (9%)
Query: 3 QSAFIKGRQILDGILVANEVVDDARKRK-KDLLLFKVDFEKAYDSVD*NYLEEVMVKMGF 61
Q FI G Q I + V+ K K K+ ++ +D EKA+D + ++ +V+ + G
Sbjct: 589 QVGFIPGMQGWFNIRKSINVIHYINKLKDKNHMIISLDAEKAFDKIQHPFMIKVLERSGI 648
Query: 62 PTLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVLMEAL 121
+ IK A++ VNG L G RQG PLSP+LF + E VL A+
Sbjct: 649 QGPYLNMIKAIYSKPVANIKVNGEKLEAIPLKSGTRQGCPLSPYLFNIVLE---VLARAI 705
Query: 122 VVNNLFNGYKVGSHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGLKVNFS 181
G ++G E V ADD I+ + R L ++ F E+ G K+N +
Sbjct: 706 RQQKEIKGIQIGKEE---VKISLLADDMIVYISDPKNSTRELLNLINSFGEVVGYKINSN 762
Query: 182 KSLFVGVNVHGSWLAEAATVLNCNAGSIPFLYLGLPIGGNASRMV--FWKPLINRINSRL 239
KS+ + E + + YLG+ + + +K L I L
Sbjct: 763 KSMAFLYTKNKQAEKEIRETTPFSIVTNNIKYLGVTLTKEVKDLYDKNFKSLKKEIKEDL 822
Query: 240 SSWKSKHLSLGGRLVLLKSVLSSLPVYALSF--FKAPSGIVSSIESILNNFFFWGGWGGG 297
WK S GR+ ++K + +Y + K P+ + +E + F W
Sbjct: 823 RRWKDLPCSWIGRINIVKMAILPKAIYRFNAIPIKIPTQFFNELEGAICKFV----WNNK 878
Query: 298 SADHRKIIWVDWNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVLTERDSLWFRVLAVR- 356
K + D + GG+ + +K + A++ K W W+R V
Sbjct: 879 KPRIAKSLLKD------KRTSGGITMPDLKLYYRAIVIKTAW--------YWYRDRQVDQ 924
Query: 357 -NRVEG--------GHLCSGGRNESMWWRNIVALHSEGW 386
NR+E GHL +++ W+ ++ W
Sbjct: 925 WNRIEDPEMNPHTYGHLIFDKGAKTIQWKKDSIFNNWCW 963
>YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein
(ORF 2)
Length = 1308
Score = 75.5 bits (184), Expect = 4e-13
Identities = 87/347 (25%), Positives = 151/347 (43%), Gaps = 25/347 (7%)
Query: 3 QSAFIKGRQILDGILVANEVVDDARKRKKDLLLFKVDFEKAYDSVD*NYLEEVMVKMGFP 62
QS + GR I D + + +++ AR+ L +D EKA+D VD YL + F
Sbjct: 558 QSYTVPGRTIFDNVFLIRDLLHFARRTGLSLAFLSLDQEKAFDRVDHQYLIGTLQAYSFG 617
Query: 63 TLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVLMEALV 122
+ ++K +A V +N S T + RG+RQG PLS L+ LA E F L+
Sbjct: 618 PQFVGYLKTMYASAECLVKINWSLTAPLAFGRGVRQGCPLSGQLYSLAIEPFLCLL---- 673
Query: 123 VNNLFNGYKVGSHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGLKVNFSK 182
G + +M V +ADD +IL + ++ + ++ S ++N+SK
Sbjct: 674 -RKRLTGLVLKEPDM-RVVLSAYADD-VILVAQDLVDLERAQECQEVYAAASSARINWSK 730
Query: 183 S--LFVGVNVHGSWLAEAATVLNCNAGSIPFL--YLG---LPIGGNASRMVFWKPLINRI 235
S L G ++ +L A ++ + I +L YL P+ N + L +
Sbjct: 731 SSGLLEG-SLKVDFLPPAFRDISWESKIIKYLGVYLSAEEYPVSQN------FIELEECV 783
Query: 236 NSRLSSWK--SKHLSLGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFFFWG- 292
+RL WK +K LS+ GR +++ +++S Y L ++ I+ L +F + G
Sbjct: 784 LTRLGKWKGFAKVLSMRGRALVINQLVASQIWYRLICLSPTQEFIAKIQRRLLDFLWIGK 843
Query: 293 GWGGGSADHRKIIWVDWNSVCRSQEVGGLEVRRIKEFNLA-LLGKWC 338
W + VC +V +++I+ + A +WC
Sbjct: 844 HWVSAGVSSLPLKEGGQGVVCIRSQVHTFRLQQIQRYLYADPSPQWC 890
>M310_ARATH (P93295) Hypothetical mitochondrial protein AtMg00310
(ORF154)
Length = 154
Score = 74.3 bits (181), Expect = 1e-12
Identities = 48/147 (32%), Positives = 71/147 (47%), Gaps = 9/147 (6%)
Query: 262 SLPVYALSFFKAPSGIVSSIESILNNFFFWGGWGGGSADHRKIIWVDWNSVCRSQE-VGG 320
+LPVYA+S F+ + + S + F W + RKI WV W +C+S+E GG
Sbjct: 2 ALPVYAMSCFRLSKLLCKKLTSAMTEF-----WWSSCENKRKISWVAWQKLCKSKEDDGG 56
Query: 321 LEVRRIKEFNLALLGKWCWRVLTERDSLWFRVLAVRNRVEGGHL-CSGGRNESMWWRNIV 379
L R + FN ALL K +R++ + +L R+L R + CS G S WR+I+
Sbjct: 57 LGFRDLGWFNQALLAKQSFRIIHQPHTLLSRLLRSRYFPHSSMMECSVGTRPSYAWRSII 116
Query: 380 ALHSEGWFQNHVSRSLGDGSSVLFWTD 406
H + R++GDG W D
Sbjct: 117 --HGRELLSRGLLRTIGDGIHTKVWLD 141
>PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 711
Score = 63.9 bits (154), Expect = 1e-09
Identities = 51/190 (26%), Positives = 92/190 (47%), Gaps = 13/190 (6%)
Query: 8 KGRQILDGILVANEVVD----DARKRKKDLLLFKVDFEKAYDSVD*NYLEEVMVKMGFPT 63
KG +DG LV + ++D R+++K + +D KA+D+V + + + ++G
Sbjct: 108 KGYARIDGTLVNSLLLDTYISSRREQRKTYNVVSLDVRKAFDTVSHSSICRALQRLGIDE 167
Query: 64 LWRKWIKKCVGTATASVLVN-GSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVLMEALV 122
+I + +T ++ V GS T + + RG++QGDPLSPFL F+ +++ L+
Sbjct: 168 GTSNYITGSLSDSTTTIRVGPGSQTRKICIRRGVKQGDPLSPFL-------FNAVLDELL 220
Query: 123 VNNLFNGYKVGSHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGLKVNFSK 182
+ G+ + L FADD ++L D L + F+ L G+ +N K
Sbjct: 221 CSLQSTPGIGGTIGEEKIPVLAFADDLLLLEDNDVLLPTTLATVANFFR-LRGMSLNAKK 279
Query: 183 SLFVGVNVHG 192
S+ + V G
Sbjct: 280 SVSISVAASG 289
>POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type II
retrotransposable element R2DM [Contains: Protease (EC
3.4.23.-); Reverse transcriptase (EC 2.7.7.49);
Endonuclease]
Length = 1057
Score = 55.5 bits (132), Expect = 5e-07
Identities = 45/186 (24%), Positives = 83/186 (44%), Gaps = 10/186 (5%)
Query: 3 QSAFIKGRQILDGILVANEVVDDARKRKKDLLLFKVDFEKAYDSVD*NYLEEVMVKMGFP 62
Q F+ D + + V+ + K + + +D KA+DS+ + + + G P
Sbjct: 446 QRGFLPTDGCADNATIVDLVLRHSHKHFRSCYIANLDVSKAFDSLSHASIYDTLRAYGAP 505
Query: 63 TLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVLMEALV 122
+ +++ S+ +G + EF RG++QGDPLSP LF L + + + +
Sbjct: 506 KGFVDYVQNTYEGGGTSLNGDGWSSEEFVPARGVKQGDPLSPILFNLVMDRLLRTLPSEI 565
Query: 123 VNNLFNGYKVGSHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGLKVNFSK 182
G KVG+ +T+ D ++L ++ ++ L L F + GLK+N K
Sbjct: 566 ------GAKVGN----AITNAAAFADDLVLFAETRMGLQVLLDKTLDFLSIVGLKLNADK 615
Query: 183 SLFVGV 188
VG+
Sbjct: 616 CFTVGI 621
>PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1025
Score = 53.1 bits (126), Expect = 2e-06
Identities = 43/153 (28%), Positives = 77/153 (50%), Gaps = 11/153 (7%)
Query: 3 QSAFIKGRQILDGILVANEVVDDARKRKKDLLLFKVDFEKAYDSVD*NYLEEVMVKMGFP 62
Q AFI + + + + ++ +AR + K L + +D +KA+DSV+ + + + + P
Sbjct: 420 QRAFIVADGVAENTSLLSAMIKEARMKIKGLYIAILDVKKAFDSVEHRSILDALRRKKLP 479
Query: 63 TLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVLMEALV 122
R +I + + V + RG+RQGDPLSP L F+ +M+A++
Sbjct: 480 LEMRNYIMWVYRNSKTRLEVVKTKGRWIRPARGVRQGDPLSPLL-------FNCVMDAVL 532
Query: 123 VNNLFN-GYKVGSHEMVGVTHLQFADDTIILCD 154
N G+ +G+ E +G L FADD ++L +
Sbjct: 533 RRLPENTGFLMGA-EKIGA--LVFADDLVLLAE 562
>PO23_POPJA (Q05118) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 606
Score = 52.4 bits (124), Expect = 4e-06
Identities = 61/282 (21%), Positives = 110/282 (38%), Gaps = 24/282 (8%)
Query: 13 LDGILVANEVVDDARKRKKDLLLFKVDFEKAYDSVD*NYLEEVMVKMGFPTLWRKWIKKC 72
L I++ + R + K + +D KA+D+V + M G + +I
Sbjct: 3 LANIIMLEHYIKLRRLKGKTYNVVSLDIRKAFDTVSHPAILRAMRAFGIDDGMQDFIMST 62
Query: 73 VGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVLMEALVVNNLFNGYKV 132
+ A +++V G T + + G++QGDPLSP L F+++++ LV N +
Sbjct: 63 ITDAYTNIVVGGRTTNKIYIRNGVKQGDPLSPVL-------FNIVLDELVTR--LNDEQP 113
Query: 133 GSH--EMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGLKVNFSKSLFV-GVN 189
G+ + L FADD ++L D+ +L F+ G+ +N K +
Sbjct: 114 GASMTPACKIASLAFADDLLLLEDRDIDVPNSLATTCAYFR-TRGMTLNPEKCASISAAT 172
Query: 190 VHGSWLAEAATVLNCN-------AGSIPFLYLGLPIGGNASRMVFWKPLINRINSRLSSW 242
V G + + + G F YLGL KP + + L +
Sbjct: 173 VSGRSVPRSKPSFTIDGRYIKPLGGINTFKYLGLTFSSTGVA----KPTVYNLTRWLRNL 228
Query: 243 KSKHLSLGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESI 284
+ L + +LK+ L Y L +GI+ + +
Sbjct: 229 EKAPLKPNQKFYILKTHLLPRLFYGLQSPGVTAGILQECDRL 270
>MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250
(ORF102)
Length = 122
Score = 51.6 bits (122), Expect = 7e-06
Identities = 29/69 (42%), Positives = 38/69 (55%), Gaps = 1/69 (1%)
Query: 81 LVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVLMEALVVNNLFNGYKVGSHEMVGV 140
++NG+P + RGLRQGDPLSP+LF+L E L G +V S+ +
Sbjct: 13 IINGAPQGLVTPSRGLRQGDPLSPYLFILCTEVLSGLCRRAQEQGRLPGIRV-SNNSPRI 71
Query: 141 THLQFADDT 149
HL FADDT
Sbjct: 72 NHLLFADDT 80
>YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III
Length = 1222
Score = 40.8 bits (94), Expect = 0.012
Identities = 41/157 (26%), Positives = 65/157 (41%), Gaps = 9/157 (5%)
Query: 3 QSAFIKGRQILDGILVANEVVDDARKRKKDLLLFKVDFEKAYDSVD*NYLEEVMVKMGFP 62
Q F+ R ++ + + K +K L + DF KA+D V L + + G
Sbjct: 718 QHGFLNFRSCPSSLVRSISLYHSILKNEKSLDILFFDFAKAFDKVSHPILLKKLALFGLD 777
Query: 63 TLWRKWIKKCVGTATASVLVN---GSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVLME 119
L W K+ + T SV +N S Y S G+ QG P LF+L + +E
Sbjct: 778 KLTCSWFKEFLHLRTFSVKINKFVSSNAYPIS--SGVPQGSVSGPLLFILFINDLLIDLE 835
Query: 120 ALV-VNNLFNGYKVGSHEMVGVTHLQFADDTIILCDK 155
+ V+ + K+ H + LQ + DTI+ K
Sbjct: 836 PNIHVSCFADDIKIFHH---NPSTLQNSIDTIVKWSK 869
>PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 869
Score = 37.4 bits (85), Expect = 0.13
Identities = 28/110 (25%), Positives = 53/110 (47%), Gaps = 1/110 (0%)
Query: 3 QSAFIKGRQILDGILVANEVVDDARKRKKDLLLFKVDFEKAYDSVD*NYLEEVMVKMGFP 62
Q+A++ + + + ++ +A++ +K+L + +D KA++SV + L + + + G P
Sbjct: 261 QTAYLPIDGVCINVSMLTAIIAEAKRLRKELHIAILDLVKAFNSVYHSALIDAITEAGCP 320
Query: 63 TLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAE 112
+I + G S+ G+ QGDPLS LF LA E
Sbjct: 321 PGVVDYIADMYNNVITEMQFEGKCELA-SILAGVYQGDPLSGPLFTLAYE 369
>RDPO_SCEOB (P19593) Probable reverse transcriptase (EC 2.7.7.49)
Length = 608
Score = 33.9 bits (76), Expect = 1.4
Identities = 33/147 (22%), Positives = 65/147 (43%), Gaps = 18/147 (12%)
Query: 37 KVDFEKAYDSVD*NYLEEVMVKMGFPTLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGL 96
++D + D+++ ++ +V + LW W+K ++ L PT G+
Sbjct: 196 EIDIKGCVDNINHQFISQVTPFIPKKILWA-WLKCGYIERNSNTL---QPTTT-----GV 246
Query: 97 RQGDPLSPFLFLLAAEGFHVLMEALVVNNLFNGYKVGSHEMVGVTHLQFADDTIILCDKS 156
QG +SP + L +G +++ + S + G T+ ++ADD +IL
Sbjct: 247 PQGGIISPLIMNLTLDGLEF--------HIYKKIQKSSSQSKGNTYCRYADDMVILTTTE 298
Query: 157 WANIRALRAILLLFQELSGLKVNFSKS 183
+ AL A+ F + GL+V +K+
Sbjct: 299 ETALIALPAV-KEFLAVRGLEVKLAKT 324
>YO42_CAEEL (P34680) Hypothetical protein ZK757.2 in chromosome III
Length = 294
Score = 33.5 bits (75), Expect = 1.9
Identities = 21/71 (29%), Positives = 34/71 (47%), Gaps = 2/71 (2%)
Query: 92 LHRGLRQGDPLSPFLFL--LAAEGFHVLMEALVVNNLFNGYKVGSHEMVGVTHLQFADDT 149
++R RQ + P+L++ LAA VL V NL G+++ + + V HL D+
Sbjct: 7 INREQRQCSQIRPYLYVSGLAALSPRVLSRFCVCINLIPGFRLSAPPHMKVVHLPLQDNE 66
Query: 150 IILCDKSWANI 160
WAN+
Sbjct: 67 TTDLSPHWANV 77
>CYB_RHISO (Q9G2R8) Cytochrome b
Length = 379
Score = 33.5 bits (75), Expect = 1.9
Identities = 45/185 (24%), Positives = 80/185 (42%), Gaps = 38/185 (20%)
Query: 134 SHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGL----------KVNFSKS 183
SH ++ + + F D SW N +L I L+ Q L+GL FS
Sbjct: 7 SHPLIKIINHSFIDLPTPSNISSWWNFSSLLGICLILQILTGLFLAMHYTPDTTTAFSSV 66
Query: 184 LFVGVNVHGSWLAEAATVLNCNAGSIPFLYLGLPIGGNASRMVFWKPLINRINSRLSSWK 243
+ +V+ W+ L+ N S+ F+ L + +G + L ++ L +W
Sbjct: 67 AHICRDVNYGWMIR---YLHANGASMFFICLFIHVG---------RGLYYGSHTFLETW- 113
Query: 244 SKHLSLGGRLVLLKSVLSS------LPVYALSFFKAP--SGIVSSIESILNNFFFWGGWG 295
++G ++LL +++++ LP +SF+ A + ++S+I I N W WG
Sbjct: 114 ----NIG--IILLLTLMATAFMGYVLPWGQMSFWGATVITNLLSAIPYIGTNLVEW-IWG 166
Query: 296 GGSAD 300
G S D
Sbjct: 167 GFSVD 171
>CYB_RHIUN (Q96071) Cytochrome b
Length = 379
Score = 32.0 bits (71), Expect = 5.5
Identities = 46/185 (24%), Positives = 79/185 (41%), Gaps = 38/185 (20%)
Query: 134 SHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGL----------KVNFSKS 183
SH +V + + F D SW N +L I L+ Q L+GL FS
Sbjct: 7 SHPLVKIINHSFIDLPTPSNISSWWNFGSLLGICLILQILTGLFLAMHYTPDTTTAFSSV 66
Query: 184 LFVGVNVHGSWLAEAATVLNCNAGSIPFLYLGLPIGGNASRMVFWKPLINRINSRLSSWK 243
+ +V+ W+ L+ N S+ F+ L + +G + L + L +W
Sbjct: 67 THICRDVNYGWMIR---YLHANGASMFFICLFIHVG---------RGLYYGSYTFLETW- 113
Query: 244 SKHLSLGGRLVLLKSVLSS------LPVYALSFFKAP--SGIVSSIESILNNFFFWGGWG 295
++G ++LL +++++ LP +SF+ A + ++S+I I N W WG
Sbjct: 114 ----NIG--IILLFTLMATAFMGYVLPWGQMSFWGATVITNLLSAIPYIGTNLVEW-IWG 166
Query: 296 GGSAD 300
G S D
Sbjct: 167 GFSVD 171
>CYB_PHAAE (Q8M708) Cytochrome b
Length = 379
Score = 32.0 bits (71), Expect = 5.5
Identities = 46/185 (24%), Positives = 79/185 (41%), Gaps = 38/185 (20%)
Query: 134 SHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGL----------KVNFSKS 183
SH ++ + + F D SW N +L I L+ Q L+GL FS
Sbjct: 7 SHPLMKIVNNAFIDLPTPSNISSWWNFGSLLGICLILQILTGLFLAMHYTSDTTTAFSSV 66
Query: 184 LFVGVNVHGSWLAEAATVLNCNAGSIPFLYLGLPIGGNASRMVFWKPLINRINSRLSSWK 243
+ + +V+ W+ L+ N S+ F+ L + +G + L L +W
Sbjct: 67 VHICRDVNYGWVIR---YLHANGASMFFICLFIHVG---------RGLYYGSYMFLETW- 113
Query: 244 SKHLSLGGRLVLLKSVLSS------LPVYALSFFKAP--SGIVSSIESILNNFFFWGGWG 295
++G ++LL +V+++ LP +SF+ A + ++S+I I N W WG
Sbjct: 114 ----NIG--VILLLTVMATAFMGYVLPWGQMSFWGATVITNLLSAIPYIGTNLVEW-VWG 166
Query: 296 GGSAD 300
G S D
Sbjct: 167 GFSVD 171
>CYB_DUGDU (Q33401) Cytochrome b
Length = 379
Score = 31.6 bits (70), Expect = 7.2
Identities = 47/185 (25%), Positives = 76/185 (40%), Gaps = 38/185 (20%)
Query: 134 SHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGL----------KVNFSKS 183
SH ++ + + F D + SW N +L L+ Q L+GL FS
Sbjct: 7 SHPLIKILNNSFIDLPTPVNISSWWNFGSLLGACLIIQILTGLFLAMHYTSDTLTAFSSV 66
Query: 184 LFVGVNVHGSWLAEAATVLNCNAGSIPFLYLGLPIGGNASRMVFWKPLINRINSRLSSWK 243
+ +V+ W+ L+ N S+ FL L IG + P +W
Sbjct: 67 THICRDVNYGWIIR---YLHANGASMFFLCLYAHIGRGIYYGSYLYP---------ETW- 113
Query: 244 SKHLSLGGRLVLLKSVLSS------LPVYALSFFKAP--SGIVSSIESILNNFFFWGGWG 295
++G +VLL +V+++ LP +SF+ A + ++S+I I N W WG
Sbjct: 114 ----NIG--IVLLLTVMATAFMGYVLPWGQMSFWGATVITNLLSAIPYIGTNLVEW-VWG 166
Query: 296 GGSAD 300
G S D
Sbjct: 167 GFSVD 171
>XYLA_ECOLI (P00944) Xylose isomerase (EC 5.3.1.5) (D-xylulose
keto-isomerase)
Length = 440
Score = 31.2 bits (69), Expect = 9.4
Identities = 26/87 (29%), Positives = 44/87 (49%), Gaps = 9/87 (10%)
Query: 207 GSIPFLYLGLPIGGNASRMVFWKPLINRINSRLSSWKSKHLSLGGRLVLLKSVLSSLPVY 266
G++ + L L I A+RM+ L RI R S W S+ LG +++ + L+ L Y
Sbjct: 358 GAMDTMALALKI---AARMIEDGELDKRIAQRYSGWNSE---LGQQILKGQMSLADLAKY 411
Query: 267 ALSFFKAP---SGIVSSIESILNNFFF 290
A +P SG +E+++N++ F
Sbjct: 412 AQEHHLSPVHQSGRQEQLENLVNHYLF 438
>CYB_SYNCA (Q9T9B9) Cytochrome b
Length = 379
Score = 31.2 bits (69), Expect = 9.4
Identities = 45/185 (24%), Positives = 79/185 (42%), Gaps = 38/185 (20%)
Query: 134 SHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGL----------KVNFSKS 183
SH ++ + + F D SW N +L I L+ Q L+GL FS
Sbjct: 7 SHPLMKILNNAFIDLPAPSNISSWWNFGSLLGICLILQILTGLFLAMHYTSDTTTAFSSV 66
Query: 184 LFVGVNVHGSWLAEAATVLNCNAGSIPFLYLGLPIGGNASRMVFWKPLINRINSRLSSWK 243
+ +V+ W+ ++ N S+ F+ L + +G + L + L +W
Sbjct: 67 AHICXDVNYGWIIR---YMHANGASMFFICLYMHVG---------RGLYYGSYTFLETW- 113
Query: 244 SKHLSLGGRLVLLKSVLSS------LPVYALSFFKAP--SGIVSSIESILNNFFFWGGWG 295
++G ++LL +V+++ LP +SF+ A + ++S+I I N W WG
Sbjct: 114 ----NIG--VILLFTVMATAFMGYVLPWGQMSFWGATVITNLLSAIPYIGTNLVEW-IWG 166
Query: 296 GGSAD 300
G S D
Sbjct: 167 GFSVD 171
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.340 0.151 0.516
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 74,583,076
Number of Sequences: 164201
Number of extensions: 3058285
Number of successful extensions: 8831
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 8800
Number of HSP's gapped (non-prelim): 29
length of query: 679
length of database: 59,974,054
effective HSP length: 117
effective length of query: 562
effective length of database: 40,762,537
effective search space: 22908545794
effective search space used: 22908545794
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.9 bits)
S2: 69 (31.2 bits)
Medicago: description of AC124215.6


