

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124214.6 + phase: 0
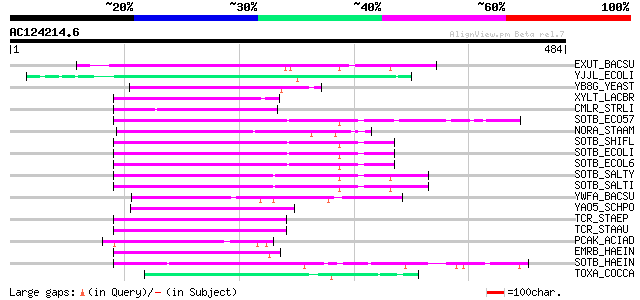
(484 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
EXUT_BACSU (O34456) Hexuronate transporter 78 4e-14
YJJL_ECOLI (P39398) Hypothetical transport protein yjjL 64 7e-10
YB8G_YEAST (P38358) Hypothetical transport protein YBR293W 63 1e-09
XYLT_LACBR (O52733) D-xylose-proton symporter (D-xylose transpor... 57 8e-08
CMLR_STRLI (P31141) Chloramphenicol resistance protein 57 8e-08
SOTB_ECO57 (P58529) Sugar efflux transporter 57 1e-07
NORA_STAAM (P21191) Quinolone resistance protein norA 56 2e-07
SOTB_SHIFL (Q7UCG7) Probable sugar efflux transporter 56 2e-07
SOTB_ECOLI (P31122) Sugar efflux transporter 55 3e-07
SOTB_ECOL6 (Q8FHE6) Probable sugar efflux transporter 55 3e-07
SOTB_SALTY (P58531) Probable sugar efflux transporter 55 5e-07
SOTB_SALTI (P58530) Probable sugar efflux transporter 55 5e-07
YWFA_BACSU (P39637) Hypothetical protein ywfA 54 9e-07
YAO5_SCHPO (Q10084) Putative transporter C11D3.05 53 2e-06
TCR_STAEP (P62967) Tetracycline resistance protein 52 4e-06
TCR_STAAU (P02983) Tetracycline resistance protein (Tetracycline... 52 4e-06
PCAK_ACIAD (Q43975) 4-hydroxybenzoate transporter 52 4e-06
EMRB_HAEIN (P44927) Multidrug resistance protein B homolog 51 6e-06
SOTB_HAEIN (P44535) Probable sugar efflux transporter 51 8e-06
TOXA_COCCA (Q00357) Putative HC-toxin efflux carrier TOXA 50 1e-05
>EXUT_BACSU (O34456) Hexuronate transporter
Length = 422
Score = 78.2 bits (191), Expect = 4e-14
Identities = 77/336 (22%), Positives = 144/336 (41%), Gaps = 42/336 (12%)
Query: 59 MLNYLDRGAIASNGVNGHRGTCTDGICKSGTGIQGDFNLTNFQDGVLSSAFMVGLLIASP 118
++NYLDR A++ + IQ D L+ Q G++ S+F +G I +
Sbjct: 18 VINYLDRSALSI----------------AAPFIQDDLTLSATQMGLIFSSFSIGYAIFNF 61
Query: 119 IFASLSKSVNPFRLIGVGLSVWTVATLCCGLSFNFWSITVCRMLVGVGEASFISLAAPFI 178
+ S + V + VW++ + L+F F S+ + R+L G+GE + +
Sbjct: 62 LGGVASDRYGAKLTLFVAMVVWSLFSGAVALAFGFVSLLIIRILFGMGEGPLSATINKMV 121
Query: 179 DDNAPASQKTVWLSIFYMCIPGGYAIGYVYGGVVGSHFGWRYAFWVEAVLMLPFAILGFV 238
++ P +Q+ + + P G AI G++ F W+ +F + ++ L +A+L F
Sbjct: 122 NNWFPPTQRASVIGVTNSGTPLGGAISGPIVGMIAVAFSWKVSFVLIMIIGLIWAVLWFK 181
Query: 239 M---KPLQ-------LKDQLSLFLK-DMKELLSDKVFVVNVLGYIAYNFVIGAYSYWGP- 286
KP + +K + S K + L K + + AYN+++ + W P
Sbjct: 182 FVKEKPQETIKEAPAIKAETSPGEKIPLTFYLKQKTVLFTAFAFFAYNYILFFFLTWFPS 241
Query: 287 ----KAGYSIYNMTNADMIFGGITIVCGILGTLAGGLVLDYMTNTLSN-----AFKLLSL 337
+ G S+ +M+ + I + G +G AGG V DY+ + + K++ +
Sbjct: 242 YLVDERGLSVESMS----VITVIPWILGFIGLAAGGFVSDYVYKKTARKGVLFSRKVVLV 297
Query: 338 TTLVGGAFCFG-AFAFKSMYGFLALFAIGELLVFAT 372
T L A G A + G + L A+ ++ T
Sbjct: 298 TCLFSSAVLIGFAGLVATTAGAVTLVALSVFFLYLT 333
>YJJL_ECOLI (P39398) Hypothetical transport protein yjjL
Length = 453
Score = 64.3 bits (155), Expect = 7e-10
Identities = 77/355 (21%), Positives = 132/355 (36%), Gaps = 43/355 (12%)
Query: 15 TSSSSTPNPVPSSVEPNMLPKSTMIPATSWFTPKRLLAIFCVINMLNYLDRGAIASNGVN 74
T SSS PVP ++ +ST I T LL F + +NYLDR +++ +
Sbjct: 15 TPSSSADIPVPPD---GLVQRSTRIKRIQ--TTAMLLLFFAAV--INYLDRSSLSVANLT 67
Query: 75 GHRGTCTDGICKSGTGIQGDFNLTNFQDGVLSSAFMVGLLIASPIFASLSKSVNPFRLIG 134
I+ + L+ + G L S F + IA L P ++G
Sbjct: 68 ----------------IREELGLSATEIGALLSVFSLAYGIAQLPCGPLLDRKGPRLMLG 111
Query: 135 VGLSVWTVATLCCGLSFNFWSITVCRMLVGVGEASFISLAAPFIDDNAPASQKTVWLSIF 194
+G+ W++ G+ NF + R+ +G+GEA I+D ++ + F
Sbjct: 112 LGMFFWSLFQAMSGMVHNFTQFVLVRIGMGIGEAPMNPCGVKVINDWFNIKERGRPMGFF 171
Query: 195 YMCIPGGYAIGYVYGGVVGSHFGWRYAFWVEAVLMLPFAILGFVMKPLQLKDQLS----- 249
G A+ + GWR F VL + AI +++ + +L+
Sbjct: 172 NAASTIGVAVSPPILAAMMLVMGWRGMFITIGVLGIFLAIGWYMLYRNREHVELTAVEQA 231
Query: 250 -------------LFLKDMKELLSDKVFVVNVLGYIAYNFVIGAYSYWGPKAGYSIYNM- 295
L + + L ++ +LG+ N+ Y W P + YN+
Sbjct: 232 YLNAGSVNARRDPLSFAEWRSLFRNRTMWGMMLGFSGINYTAWLYLAWLPGYLQTAYNLD 291
Query: 296 TNADMIFGGITIVCGILGTLAGGLVLDYMTNTLSNAFKLLSLTTLVGGAFCFGAF 350
+ + I + G G L G V D++ K + ++ G FC AF
Sbjct: 292 LKSTGLMAAIPFLFGAAGMLVNGYVTDWLVKGGMAPIKSRKI-CIIAGMFCSAAF 345
>YB8G_YEAST (P38358) Hypothetical transport protein YBR293W
Length = 474
Score = 63.2 bits (152), Expect = 1e-09
Identities = 38/170 (22%), Positives = 82/170 (47%), Gaps = 6/170 (3%)
Query: 105 LSSAFMVGLLIASPIFASLSKSVNPFRLIGVGLSVWTVATLCCGLSFNFWSITVCRMLVG 164
+++A+++ P++ S S ++ + +T+ L CG S N + ++ R L G
Sbjct: 7 ITTAYLITSTSFQPLYGSFSDALGRRNCLFFANGAFTIGCLACGFSKNIYMLSFMRALTG 66
Query: 165 VGEASFISLAAPFIDDNAPASQKTVWLSIFYMCIPGGYAIGYVYGGVVGSHFGWRYAFWV 224
+G I+L+ D P+S++ ++ + + + G G +GG + S GWR+ F +
Sbjct: 67 IGGGGLITLSTIVNSDVIPSSKRGIFQAFQNLLLGFGAICGASFGGTIASSIGWRWCFLI 126
Query: 225 EAVLMLPFAIL--GFVMKPLQLKDQLSLFLKDMKELLSDKVFVVNVLGYI 272
+ + + +IL +V + Q S ++ ++L D ++V+G I
Sbjct: 127 QVPISVISSILMNYYVPNQKEYNRQNSSIFQNPGKILRD----IDVMGSI 172
>XYLT_LACBR (O52733) D-xylose-proton symporter (D-xylose
transporter)
Length = 457
Score = 57.4 bits (137), Expect = 8e-08
Identities = 34/146 (23%), Positives = 71/146 (48%), Gaps = 4/146 (2%)
Query: 91 IQGDFNLTNFQDGVLSSAFMVGLLIASPIFASLSKSVNPFRLIGVGLSVWTVATLCCGLS 150
IQ NL ++Q G + SA ++G ++ + I S +L+ + ++ V L S
Sbjct: 34 IQKQMNLGSWQQGWVVSAVLLGAILGAAIIGPSSDRFGRRKLLLLSAIIFFVGALGSAFS 93
Query: 151 FNFWSITVCRMLVGVGEASFISLAAPFIDDNAPASQKTVWLSIFYMCIPGGYAIGYVYG- 209
FW++ + R+++G+ + +L ++ + AP+ ++ S+F + + G + Y+
Sbjct: 94 PEFWTLIISRIILGMAVGAASALIPTYLAELAPSDKRGTVSSLFQLMVMTGILLAYITNY 153
Query: 210 GVVGSHFGWRYAFWVEAVLMLPFAIL 235
G + GWR W+ +P A+L
Sbjct: 154 SFSGFYTGWR---WMLGFAAIPAALL 176
>CMLR_STRLI (P31141) Chloramphenicol resistance protein
Length = 392
Score = 57.4 bits (137), Expect = 8e-08
Identities = 39/144 (27%), Positives = 67/144 (46%), Gaps = 2/144 (1%)
Query: 91 IQGDFNLTNFQDGVLSSAFMVGLLIASPIFASLSKSVNPFRLIGVGLSVWTVATLCCGLS 150
I D +T G L+SAF G+++ +P+ A+L+++ P R +G + A G
Sbjct: 29 IASDLGVTVGTAGTLTSAFATGMIVGAPLVAALARTW-PRRSSLLGFILAFAAAHAVGAG 87
Query: 151 FNFWSITV-CRMLVGVGEASFISLAAPFIDDNAPASQKTVWLSIFYMCIPGGYAIGYVYG 209
+ + V CR++ + A F+++A PA ++ L++ G G
Sbjct: 88 TTSFPVLVACRVVAALANAGFLAVALTTAAALVPADKQGRALAVLLSGTTVATVAGVPGG 147
Query: 210 GVVGSHFGWRYAFWVEAVLMLPFA 233
++G+ GWR FW AV LP A
Sbjct: 148 SLLGTWLGWRATFWAVAVCCLPAA 171
>SOTB_ECO57 (P58529) Sugar efflux transporter
Length = 396
Score = 56.6 bits (135), Expect = 1e-07
Identities = 87/362 (24%), Positives = 151/362 (41%), Gaps = 23/362 (6%)
Query: 91 IQGDFNLTNFQDGVLSSAFMVGLLIASPIFASLSKSVNPFRLIGVGLSVWTVATLCCGLS 150
I F++ Q G++ + + + + S F ++ V +L+ V+ + + LS
Sbjct: 39 IAQSFHMQTAQVGIMLTIYAWVVALMSLPFMLMTSQVERRKLLICLFVVFIASHVLSFLS 98
Query: 151 FNFWSITVCRMLVGVGEASFISLAAPFIDDNAPASQKTVWLSIFYMCIPGGYAIGYVYGG 210
++F + + R+ V A F S+ A APA ++ LS+ +G G
Sbjct: 99 WSFTVLVISRIGVAFAHAIFWSITASLAIRMAPAGKRAQALSLIATGTALAMVLGLPLGR 158
Query: 211 VVGSHFGWRYAFWVEAVLMLPFAILGFVMKPLQLKDQLSLFLKDMKELLSDKVFV-VNVL 269
+VG +FGWR F+ + L + + PL L + S LK + L + + +L
Sbjct: 159 IVGQYFGWRMTFFAIGIGALITLLCLIKLLPL-LPSEHSGSLKSLPLLFRRPALMSIYLL 217
Query: 270 GYIAYNFVIGAYSYWGP----KAGYSIYNMTNADMIFGGITIVCGILGT-LAGGLVLDYM 324
+ AYSY P AG+S T ++ GG GI+G+ + G L Y
Sbjct: 218 TVVVVTAHYTAYSYIEPFVQNIAGFSANFATALLLLLGG----AGIIGSVIFGKLGNQYA 273
Query: 325 TNTLSNAFKLLSLTTLVGGAFCFGAFAFKSMYGFLALFAIGELLVFATQGPVNFVCLHCV 384
+ +S A LL LV A A + G L++F +++ V + L
Sbjct: 274 SALVSTAIALL----LVCLALLLPAANSEIHLGVLSIFWGIAMMIIGLGMQVKVLAL--- 326
Query: 385 KPSLRPLSMAMSTVAIHIFGDVPSAPLVGVVQDHINNWRTTAL-ILTTIFFPAAAIWFIG 443
P ++MA+ + +I + + LVG +W + + + T+ AA IW I
Sbjct: 327 APDATDVAMALFSGIFNI--GIGAGALVG--NQVSLHWSMSMIGYVGTVPAFAALIWSII 382
Query: 444 IF 445
IF
Sbjct: 383 IF 384
>NORA_STAAM (P21191) Quinolone resistance protein norA
Length = 388
Score = 56.2 bits (134), Expect = 2e-07
Identities = 58/230 (25%), Positives = 99/230 (42%), Gaps = 16/230 (6%)
Query: 94 DFNLTNFQDGVLSSAFMVGLLIASPIFASLSKSVNPFRLIGVGLSVWTVATLCCGLSFNF 153
D LT G+L +AF + +I SP +L+ + +I +GL +++V+ + NF
Sbjct: 32 DLGLTGSDLGLLVAAFALSQMIISPFGGTLADKLGKKLIICIGLILFSVSEFMFAVGHNF 91
Query: 154 WSITVCRMLVGVGEASFISLAAPFIDDNAPASQKTVWLSIFYMCIPGGYAIGYVYGGVVG 213
+ + R++ G+ + I D +P+ QK I G+ +G GG +
Sbjct: 92 SVLMLSRVIGGMSAGMVMPGVTGLIADISPSHQKAKNFGYMSAIINSGFILGPGIGGFM- 150
Query: 214 SHFGWRYAFWVEAVLMLPFAILGFVMKPLQLKDQLSLFLKDMKELLSD---KVFVVNVLG 270
+ R F+ L + I+ V+ K S F K +LL+ KVF+ V+
Sbjct: 151 AEVSHRMPFYFAGALGILAFIMSIVLIHDPKKSTTSGFQKLEPQLLTKINWKVFITPVIL 210
Query: 271 YIAYNFVIGAYS-----YWGPKAGYSIYNMTNADMIFGGITIVCGILGTL 315
+ +F + A+ Y K YS +++ A I GG GI G L
Sbjct: 211 TLVLSFGLSAFETLYSLYTADKVNYSPKDISIA--ITGG-----GIFGAL 253
>SOTB_SHIFL (Q7UCG7) Probable sugar efflux transporter
Length = 396
Score = 55.8 bits (133), Expect = 2e-07
Identities = 62/251 (24%), Positives = 107/251 (41%), Gaps = 11/251 (4%)
Query: 91 IQGDFNLTNFQDGVLSSAFMVGLLIASPIFASLSKSVNPFRLIGVGLSVWTVATLCCGLS 150
I F++ Q G++ + + + + S F ++ V +L+ V+ + + LS
Sbjct: 39 IAQSFHMQTAQVGIMLTIYAWVVALMSLPFMLMTSQVERRKLLICLFVVFIASHVLSFLS 98
Query: 151 FNFWSITVCRMLVGVGEASFISLAAPFIDDNAPASQKTVWLSIFYMCIPGGYAIGYVYGG 210
++F + + R+ V A F S+ A APA ++ LS+ +G G
Sbjct: 99 WSFTVLVISRIGVAFAHAIFWSITASLAIRMAPAGKRAQALSLIATGTALAMVLGLPLGR 158
Query: 211 VVGSHFGWRYAFWVEAVLMLPFAILGFVMKPLQLKDQLSLFLKDMKELLSDKVFV-VNVL 269
+VG +FGWR F+ + L + + PL L + S LK + L + + +L
Sbjct: 159 IVGQYFGWRMTFFAIGIGALVTLLCLIKLLPL-LPSEHSGSLKSLPLLFRRPALMSIYLL 217
Query: 270 GYIAYNFVIGAYSYWGP----KAGYSIYNMTNADMIFGGITIVCGILGT-LAGGLVLDYM 324
+ AYSY P AG+S T ++ GG GI+G+ + G L Y
Sbjct: 218 TVVVVTAHYTAYSYIEPFVQNIAGFSANFATALLLLLGG----AGIIGSVIFGKLGNQYA 273
Query: 325 TNTLSNAFKLL 335
+ +S A LL
Sbjct: 274 SALVSTAIALL 284
>SOTB_ECOLI (P31122) Sugar efflux transporter
Length = 396
Score = 55.5 bits (132), Expect = 3e-07
Identities = 62/251 (24%), Positives = 107/251 (41%), Gaps = 11/251 (4%)
Query: 91 IQGDFNLTNFQDGVLSSAFMVGLLIASPIFASLSKSVNPFRLIGVGLSVWTVATLCCGLS 150
I F++ Q G++ + + + + S F ++ V +L+ V+ + + LS
Sbjct: 39 IAQSFHMQTAQVGIMLTIYAWVVALMSLPFMLMTSQVERRKLLICLFVVFIASHVLSFLS 98
Query: 151 FNFWSITVCRMLVGVGEASFISLAAPFIDDNAPASQKTVWLSIFYMCIPGGYAIGYVYGG 210
++F + + R+ V A F S+ A APA ++ LS+ +G G
Sbjct: 99 WSFTVLVISRIGVAFAHAIFWSITASLAIRMAPAGKRAQALSLIATGTALAMVLGLPLGR 158
Query: 211 VVGSHFGWRYAFWVEAVLMLPFAILGFVMKPLQLKDQLSLFLKDMKELLSDKVFV-VNVL 269
+VG +FGWR F+ + L + + PL L + S LK + L + + +L
Sbjct: 159 IVGQYFGWRMTFFAIGIGALITLLCLIKLLPL-LPSEHSGSLKSLPLLFRRPALMSIYLL 217
Query: 270 GYIAYNFVIGAYSYWGP----KAGYSIYNMTNADMIFGGITIVCGILGT-LAGGLVLDYM 324
+ AYSY P AG+S T ++ GG GI+G+ + G L Y
Sbjct: 218 TVVVVTAHYTAYSYIEPFVQNIAGFSANFATALLLLLGG----AGIIGSVIFGKLGNQYA 273
Query: 325 TNTLSNAFKLL 335
+ +S A LL
Sbjct: 274 SALVSTAIALL 284
>SOTB_ECOL6 (Q8FHE6) Probable sugar efflux transporter
Length = 396
Score = 55.5 bits (132), Expect = 3e-07
Identities = 62/251 (24%), Positives = 107/251 (41%), Gaps = 11/251 (4%)
Query: 91 IQGDFNLTNFQDGVLSSAFMVGLLIASPIFASLSKSVNPFRLIGVGLSVWTVATLCCGLS 150
I F++ Q G++ + + + + S F ++ V +L+ V+ + + LS
Sbjct: 39 IAHSFHMQTAQVGIMLTIYAWVVALMSLPFMLMTSQVERRKLLICLFVVFIASHVLSFLS 98
Query: 151 FNFWSITVCRMLVGVGEASFISLAAPFIDDNAPASQKTVWLSIFYMCIPGGYAIGYVYGG 210
++F + + R+ V A F S+ A APA ++ LS+ +G G
Sbjct: 99 WSFTVLVISRIGVAFAHAIFWSITASLAIRMAPAGKRAQALSLIATGTALAMVLGLPLGR 158
Query: 211 VVGSHFGWRYAFWVEAVLMLPFAILGFVMKPLQLKDQLSLFLKDMKELLSDKVFV-VNVL 269
+VG +FGWR F+ + L + + PL L + S LK + L + + +L
Sbjct: 159 IVGQYFGWRMTFFAIGIGALITLLCLIKLLPL-LPSEHSGSLKSLPLLFRRPALMSIYLL 217
Query: 270 GYIAYNFVIGAYSYWGP----KAGYSIYNMTNADMIFGGITIVCGILGT-LAGGLVLDYM 324
+ AYSY P AG+S T ++ GG GI+G+ + G L Y
Sbjct: 218 TVVVVTAHYTAYSYIEPFVQNIAGFSANFATALLLLLGG----AGIIGSVIFGKLGNQYA 273
Query: 325 TNTLSNAFKLL 335
+ +S A LL
Sbjct: 274 SALVSTAIALL 284
>SOTB_SALTY (P58531) Probable sugar efflux transporter
Length = 396
Score = 54.7 bits (130), Expect = 5e-07
Identities = 67/283 (23%), Positives = 120/283 (41%), Gaps = 13/283 (4%)
Query: 91 IQGDFNLTNFQDGVLSSAFMVGLLIASPIFASLSKSVNPFRLIGVGLSVWTVATLCCGLS 150
I F++ Q G++ + + + + S F L+ + +L+ ++ + + L+
Sbjct: 39 IAESFHMQTAQVGIMLTIYAWVVAVMSLPFMLLTSQMERRKLLIYLFVLFIASHVLSFLA 98
Query: 151 FNFWSITVCRMLVGVGEASFISLAAPFIDDNAPASQKTVWLSIFYMCIPGGYAIGYVYGG 210
+NF + + R+ + A F S+ A APA ++ LS+ +G G
Sbjct: 99 WNFTVLVISRIGIAFAHAIFWSITASLAIRLAPAGKRAQALSLIATGTALAMVLGLPIGR 158
Query: 211 VVGSHFGWRYAFWVEAVLMLPFAILGFVMKPLQLKDQLSLFLKDMKELLSDKVFV-VNVL 269
VVG +FGWR F+ + L +L + +L + S LK + L + + VL
Sbjct: 159 VVGQYFGWRTTFFAIGMGAL-ITLLCLIKLLPKLPSEHSGSLKSLPLLFRRPALMSLYVL 217
Query: 270 GYIAYNFVIGAYSYWGP----KAGYSIYNMTNADMIFGGITIVCGILGTLAGGLVLDYMT 325
+ AYSY P AG S T +I GG GI+G+L G + +
Sbjct: 218 TVVVVTAHYTAYSYIEPFVQNVAGLSANFATVLLLILGG----AGIIGSLVFGKLGNRHA 273
Query: 326 NTLSN---AFKLLSLTTLVGGAFCFGAFAFKSMYGFLALFAIG 365
++L + A ++ L L+ A A S++ +A+ IG
Sbjct: 274 SSLVSIAIALLVVCLLLLLPAAESEAHLAILSIFWGIAIMVIG 316
>SOTB_SALTI (P58530) Probable sugar efflux transporter
Length = 396
Score = 54.7 bits (130), Expect = 5e-07
Identities = 67/283 (23%), Positives = 120/283 (41%), Gaps = 13/283 (4%)
Query: 91 IQGDFNLTNFQDGVLSSAFMVGLLIASPIFASLSKSVNPFRLIGVGLSVWTVATLCCGLS 150
I F++ Q G++ + + + + S F L+ + +L+ ++ + + L+
Sbjct: 39 IAESFHMQTAQVGIMLTIYAWVVAVMSLPFMLLTSQMERRKLLICLFVLFIASHVLSFLA 98
Query: 151 FNFWSITVCRMLVGVGEASFISLAAPFIDDNAPASQKTVWLSIFYMCIPGGYAIGYVYGG 210
+NF + + R+ + A F S+ A APA ++ LS+ +G G
Sbjct: 99 WNFTVLVISRIGIAFAHAIFWSITASLAIRLAPAGKRAQALSLIATGTALAMVLGLPIGR 158
Query: 211 VVGSHFGWRYAFWVEAVLMLPFAILGFVMKPLQLKDQLSLFLKDMKELLSDKVFV-VNVL 269
VVG +FGWR F+ + L +L + +L + S LK + L + + VL
Sbjct: 159 VVGQYFGWRTTFFAIGMGAL-ITLLCLIKLLPKLPSEHSGSLKSLPLLFRRPALMSLYVL 217
Query: 270 GYIAYNFVIGAYSYWGP----KAGYSIYNMTNADMIFGGITIVCGILGTLAGGLVLDYMT 325
+ AYSY P AG S T +I GG GI+G+L G + +
Sbjct: 218 TVVVVTAHYTAYSYIEPFVQNVAGLSANFATVLLLILGG----AGIIGSLVFGKLGNRHA 273
Query: 326 NTLSN---AFKLLSLTTLVGGAFCFGAFAFKSMYGFLALFAIG 365
++L + A ++ L L+ A A S++ +A+ IG
Sbjct: 274 SSLVSIAIALLVVCLLLLLPAADSEAHLAILSIFWGIAIMVIG 316
>YWFA_BACSU (P39637) Hypothetical protein ywfA
Length = 412
Score = 53.9 bits (128), Expect = 9e-07
Identities = 53/245 (21%), Positives = 103/245 (41%), Gaps = 19/245 (7%)
Query: 107 SAFMVGLLIASPIFASLSKSVNPFRLIGVGLSVWTVATLCCGLSFNFWSITVCRMLVGVG 166
S + V + I +P+ L + + GL ++ + T+ C L+ N + + R L G+
Sbjct: 60 SIYGVMIFIGAPLLVPLGDKYSRELSLLAGLMIFIIGTVICALAQNIFFFFLGRALSGLA 119
Query: 167 EASFISLAAPFIDDNAPASQKTVWLSIFYMCIPGGYAIGYVYGGVVGSHFG----WRYAF 222
+F+ A + D P + + + + I +++ ++G +GS G WR+ F
Sbjct: 120 AGAFVPTAYAVVGDRVPYTYRGKVMGL----IVSSWSLALIFGVPLGSFIGGVLHWRWTF 175
Query: 223 WVEAVL--MLPFAILGFVMKPLQLKDQLSLFLKDMKELLSDKVFVVNVLGYIAYNF--VI 278
W+ A++ ++ IL + + Q K+ +++ D + V V YI F +I
Sbjct: 176 WIFALMGVLVVLLILLEMRRHAQHKNSGKEEIEEPAGTFRDALKVPRVPVYITITFCNMI 235
Query: 279 GAYSYWGPKAGYSIYNMTNADMIFGGITIVCGILGTLAGGLVLDYMTNTLSNAF-KLLSL 337
G Y YS D+ GG T + G + +T +++ K+ SL
Sbjct: 236 GFYGM------YSFLGTYLQDVFTGGNTAAGLFIMIYGIGFSMSVITGKIADRIGKMRSL 289
Query: 338 TTLVG 342
+G
Sbjct: 290 LIALG 294
>YAO5_SCHPO (Q10084) Putative transporter C11D3.05
Length = 546
Score = 52.8 bits (125), Expect = 2e-06
Identities = 38/144 (26%), Positives = 64/144 (44%), Gaps = 1/144 (0%)
Query: 106 SSAFMVGLLIASPIFASLSKSVNPFRLIGVGLSVWTVATLCCGLSFNFWSITVCRMLVGV 165
S F+VG + S FA LS F + V L ++T+ + G + N W++ + R GV
Sbjct: 148 SCTFLVGFGVGSLPFAPLSDIYGRFIIYFVTLLIFTIFQVGGGCAHNVWTLAIVRFFQGV 207
Query: 166 GEASFISLAAPFIDDNAPASQKTVWLSIFYMCIPGGYAIGYVYGG-VVGSHFGWRYAFWV 224
++ ++ A I D Q+T L F G IG + G + S+ WR+ FW+
Sbjct: 208 FGSTPLANAGGTISDLFTPVQRTYVLPGFCTFPYLGPIIGPIIGDFITQSYLEWRWTFWI 267
Query: 225 EAVLMLPFAILGFVMKPLQLKDQL 248
+ + F+ P +D +
Sbjct: 268 NMIWAAAVIVFVFIFFPETHEDTI 291
>TCR_STAEP (P62967) Tetracycline resistance protein
Length = 459
Score = 51.6 bits (122), Expect = 4e-06
Identities = 35/152 (23%), Positives = 69/152 (45%), Gaps = 1/152 (0%)
Query: 91 IQGDFNLTNFQDGVLSSAFMVGLLIASPIFASLSKSVNPFRLIGVGLSVWTVATLCCGLS 150
I FN T +++A+M+ I + ++ LS +N +L+ +G+S+ + +L +
Sbjct: 40 IANHFNTTPGITNWVNTAYMLTFSIGTAVYGKLSDYINIKKLLIIGISLSCLGSLIAFIG 99
Query: 151 FN-FWSITVCRMLVGVGEASFISLAAPFIDDNAPASQKTVWLSIFYMCIPGGYAIGYVYG 209
N F+ + R++ GVG A+F SL + N ++ + G +G G
Sbjct: 100 HNHFFILIFGRLVQGVGSAAFPSLIMVVVARNITRKKQGKAFGFIGSIVALGEGLGPSIG 159
Query: 210 GVVGSHFGWRYAFWVEAVLMLPFAILGFVMKP 241
G++ + W Y + + ++ L VM P
Sbjct: 160 GIIAHYIHWSYLLILPMITIVTIPFLIKVMVP 191
>TCR_STAAU (P02983) Tetracycline resistance protein (Tetracycline
efflux protein)
Length = 459
Score = 51.6 bits (122), Expect = 4e-06
Identities = 35/152 (23%), Positives = 69/152 (45%), Gaps = 1/152 (0%)
Query: 91 IQGDFNLTNFQDGVLSSAFMVGLLIASPIFASLSKSVNPFRLIGVGLSVWTVATLCCGLS 150
I FN T +++A+M+ I + ++ LS +N +L+ +G+S+ + +L +
Sbjct: 40 IANHFNTTPGITNWVNTAYMLTFSIGTAVYGKLSDYINIKKLLIIGISLSCLGSLIAFIG 99
Query: 151 FN-FWSITVCRMLVGVGEASFISLAAPFIDDNAPASQKTVWLSIFYMCIPGGYAIGYVYG 209
N F+ + R++ GVG A+F SL + N ++ + G +G G
Sbjct: 100 HNHFFILIFGRLVQGVGSAAFPSLIMVVVARNITRKKQGKAFGFIGSIVALGEGLGPSIG 159
Query: 210 GVVGSHFGWRYAFWVEAVLMLPFAILGFVMKP 241
G++ + W Y + + ++ L VM P
Sbjct: 160 GIIAHYIHWSYLLILPMITIVTIPFLIKVMVP 191
>PCAK_ACIAD (Q43975) 4-hydroxybenzoate transporter
Length = 457
Score = 51.6 bits (122), Expect = 4e-06
Identities = 41/162 (25%), Positives = 69/162 (42%), Gaps = 17/162 (10%)
Query: 82 DGICKSGTG-----IQGDFNLTNFQDGVLSSAFMVGLLIASPIFASLSKSVNPFRLIGVG 136
DGI + G + D+ + Q G + SA + G++I + + + ++ +
Sbjct: 48 DGIDTAAMGFIAPALAQDWGVDRSQLGPVMSAALGGMIIGALVSGPTADRFGRKIVLSMS 107
Query: 137 LSVWTVATLCCGLSFNFWSITVCRMLVGVGEASFISLAAPFIDDNAPASQKTVWLSIFYM 196
+ V+ TL C S N S+ + R L G+G + + A + PA + S+
Sbjct: 108 MLVFGGFTLACAYSTNLDSLVIFRFLTGIGLGAAMPNATTLFSEYCPARIR----SLLVT 163
Query: 197 CIPGGYAIGYVYGGVVGS----HFGWRYAF----WVEAVLML 230
C+ GY +G GG + S FGW F W +LML
Sbjct: 164 CMFCGYNLGMAIGGFISSWLIPAFGWHSLFLLGGWAPLILML 205
>EMRB_HAEIN (P44927) Multidrug resistance protein B homolog
Length = 510
Score = 51.2 bits (121), Expect = 6e-06
Identities = 35/151 (23%), Positives = 72/151 (47%), Gaps = 5/151 (3%)
Query: 91 IQGDFNLTNFQDGVLSSAFMVGLLIASPIFASLSKSVNPFRLIGVGLSVWTVATLCCGLS 150
I GD + Q + ++F V I+ PI L+K RL V ++ V++ CG++
Sbjct: 44 IAGDLGASFSQGTWVITSFGVANAISIPITGWLAKRFGEVRLFLVSTFLFVVSSWLCGIA 103
Query: 151 FNFWSITVCRMLVGVGEASFISLAAPFIDDNAPASQKTVWLSIFYMCIPGGYAIGYVYGG 210
+ ++ + R++ G I L+ + +N P ++ + L+ + M I G + GG
Sbjct: 104 DSLEALIIFRVIQGAVAGPVIPLSQSLLLNNYPPEKRGMALAFWSMTIVVAPIFGPILGG 163
Query: 211 VVGSHFGWRYAFWVE-----AVLMLPFAILG 236
+ + W + F++ +V+++ + ILG
Sbjct: 164 WISDNIHWGWIFFINVPIGLSVVLISWKILG 194
>SOTB_HAEIN (P44535) Probable sugar efflux transporter
Length = 396
Score = 50.8 bits (120), Expect = 8e-06
Identities = 85/376 (22%), Positives = 158/376 (41%), Gaps = 41/376 (10%)
Query: 91 IQGDFNLTNFQDGVLSSAFMVGLLIAS-PIFASLSKSVNPFRLIGVGLSVWTVATLCCGL 149
I F++ G++ + + +LI S P + LI + + ++ V + +
Sbjct: 39 IAQSFDMQTADTGLMMTVYAWTVLIMSLPAMLATGNMERKSLLIKLFI-IFIVGHILSVI 97
Query: 150 SFNFWSITVCRMLVGVGEASFISLAAPFIDDNAPASQKTVWLSIFYMCIPGGYAIGYVYG 209
++NFW + + RM + + + F S+ A + +P +KT L + + +G G
Sbjct: 98 AWNFWILLLARMCIALAHSVFWSITASLVMRISPKHKKTQALGMLAIGTALATILGLPIG 157
Query: 210 GVVGSHFGWRYAFWVEAVLMLPFAILGFVMKP-LQLKDQLSLFLKDM--KELLSDKVFVV 266
+VG GWR F + AVL L L + P L K+ S+ + K L ++V
Sbjct: 158 RIVGQLVGWRVTFGIIAVLALSIMFLIIRLLPNLPSKNAGSIASLPLLAKRPLLLWLYVT 217
Query: 267 NVLGYIAYNFVIGAYSYWGPKAGYSIYNMTNADMIFG-GITIVCGILGTLAGGLVLDYMT 325
+ A+ AY+Y P + ++ + D F + +V G G +A L+ + +
Sbjct: 218 TAIVISAH---FTAYTYIEP----FMIDVGHLDPNFATAVLLVFGFSG-IAASLLFNRLY 269
Query: 326 NTLSNAFKLLSLTTLVGG--AFCFGAFAFKSMYGFLALFAIGELLVFATQGPVNFVCLHC 383
F ++S++ L+ F +M+ + ++ IG + C
Sbjct: 270 RFAPTKFIVVSMSLLMFSLLLLLFSTKTIIAMFSLVFIWGIG---------------ISC 314
Query: 384 VKPSL--RPLSMA--MSTVAIHIFGDVPSAPL-VGVVQDHINNWRTTALILTTIFFPAAA 438
+ SL R L +A + VA I+ + +A + G + N TT L L I + AA
Sbjct: 315 IGLSLQMRVLKLAPDATDVATAIYSGIFNAGIGAGAL---FGNLATTYLGLNEIGYTGAA 371
Query: 439 IWFIG--IFLNSKDKF 452
+ IG IF+ + K+
Sbjct: 372 LGLIGFIIFITTHLKY 387
>TOXA_COCCA (Q00357) Putative HC-toxin efflux carrier TOXA
Length = 548
Score = 50.1 bits (118), Expect = 1e-05
Identities = 56/247 (22%), Positives = 91/247 (36%), Gaps = 20/247 (8%)
Query: 118 PIFASLSKSVNPFRLIGVGLSVWTVATLCCGLSFNFWSITVCRMLVGVGEASFISLAAPF 177
P+F L +P L L V + +L C L+ N + V R + G+G +S A
Sbjct: 102 PLFGKLYNEFSPKWLFITCLIVLQLGSLVCALARNSPTFIVGRAVAGIGAGGILSGALNI 161
Query: 178 IDDNAPASQKTVWLSIFYMCIPGGYAIGYVYGGVVGSHFGWRYAFWVEAVL-MLPFAILG 236
+ P + + + IG + GG + + GWR+ FW+ + AIL
Sbjct: 162 VALIVPLHHRAAFTGMIGALECVALIIGPIIGGAIADNIGWRWCFWINLPIGAAVCAILL 221
Query: 237 FVMKPLQLKDQLSLFLKDMKELLSDKVFVVNVLGYIAYNFVIG-------AYSYWGPKAG 289
F P + S + E+L + L YI +I A + G K
Sbjct: 222 FFFHPPRSTYSASGVPRSYSEILGN-------LDYIGAGMIISSLVCLSLALQWGGTKYK 274
Query: 290 YSIYNMTNADMIFGGITIVCGILGTLAGGLVLDYMTNTLSNAFKLLSLTTLVGGAFCFGA 349
+ + ++FG + + G L + T L LLSL CFG
Sbjct: 275 WGDGRVVALLVVFGVLFLSASGHQYWKGEKAL-FPTRLLRQRGFLLSLF----NGLCFGG 329
Query: 350 FAFKSMY 356
+ ++Y
Sbjct: 330 VQYAALY 336
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.324 0.139 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 57,748,362
Number of Sequences: 164201
Number of extensions: 2522395
Number of successful extensions: 10715
Number of sequences better than 10.0: 230
Number of HSP's better than 10.0 without gapping: 91
Number of HSP's successfully gapped in prelim test: 139
Number of HSP's that attempted gapping in prelim test: 10474
Number of HSP's gapped (non-prelim): 321
length of query: 484
length of database: 59,974,054
effective HSP length: 114
effective length of query: 370
effective length of database: 41,255,140
effective search space: 15264401800
effective search space used: 15264401800
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 68 (30.8 bits)
Medicago: description of AC124214.6


