

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC123975.9 + phase: 0 /pseudo
(622 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
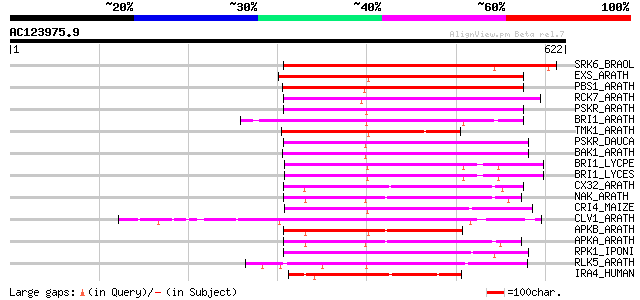
Score E
Sequences producing significant alignments: (bits) Value
SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase rec... 327 4e-89
EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase E... 219 1e-56
PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC 2.7... 215 2e-55
RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase RLC... 213 9e-55
PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (... 201 4e-51
BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC ... 197 9e-50
TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precur... 194 4e-49
PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.... 193 1e-48
BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated rec... 192 3e-48
BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.... 189 1e-47
BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precurso... 189 1e-47
CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx3... 188 4e-47
NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK ... 181 7e-45
CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4 pr... 176 2e-43
CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (... 174 5e-43
APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor ... 172 2e-42
APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor ... 169 2e-41
RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC 2... 169 3e-41
RLK5_ARATH (P47735) Receptor-like protein kinase 5 precursor (EC... 163 1e-39
IRA4_HUMAN (Q9NWZ3) Interleukin-1 receptor-associated kinase-4 (... 163 1e-39
>SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase
receptor precursor (EC 2.7.1.37) (S-receptor kinase)
(SRK)
Length = 849
Score = 327 bits (839), Expect = 4e-89
Identities = 171/321 (53%), Positives = 227/321 (70%), Gaps = 15/321 (4%)
Query: 308 NFTDSNKLGEGGFGPVYKGILSDGQEVAIKRLSICSEQGSEEFINEVMLILKLQHKNLVK 367
NF+ NKLG+GGFG VYKG L DG+E+A+KRLS S QG++EF+NEV LI +LQH NLV+
Sbjct: 527 NFSSCNKLGQGGFGIVYKGRLLDGKEIAVKRLSKTSVQGTDEFMNEVTLIARLQHINLVQ 586
Query: 368 LLGFCVDGEEKLLVYEYLPNGSLDVVLF--EQHAQLDWTKRLDIINGIARGILYLHEDSR 425
+LG C++G+EK+L+YEYL N SLD LF + ++L+W +R DI NG+ARG+LYLH+DSR
Sbjct: 587 VLGCCIEGDEKMLIYEYLENLSLDSYLFGKTRRSKLNWNERFDITNGVARGLLYLHQDSR 646
Query: 426 LQIIHRDLKASNVLLDNDMNPKISDFGMARIFAGSEGEANTTTIVGTYGYMAPEYAMEGL 485
+IIHRDLK SN+LLD +M PKISDFGMARIF E EANT +VGTYGYM+PEYAM G+
Sbjct: 647 FRIIHRDLKVSNILLDKNMIPKISDFGMARIFERDETEANTMKVVGTYGYMSPEYAMYGI 706
Query: 486 YSIKSDVFGFGVLLLEIITGIRNAGFCYSKTTPSLLAYAWHLWNDGKGLELRDPLL---- 541
+S KSDVF FGV++LEI++G +N GF LL+Y W W +G+ LE+ DP++
Sbjct: 707 FSEKSDVFSFGVIVLEIVSGKKNRGFYNLDYENDLLSYVWSRWKEGRALEIVDPVIVDSL 766
Query: 542 -----LCPGDQFLRYMNIGLLCVQEDAFDRPTMSSVVLMLMNESVMLGQPGKPPFSVGRL 596
+ + L+ + IGLLCVQE A RP MSSVV M +E+ + QP P + V R
Sbjct: 767 SSQPSIFQPQEVLKCIQIGLLCVQELAEHRPAMSSVVWMFGSEATEIPQPKPPGYCVRRS 826
Query: 597 NF-IDQN---ELDLEEEYSVN 613
+ +D + + D E ++VN
Sbjct: 827 PYELDPSSSWQCDENESWTVN 847
>EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase EXS
precursor (EC 2.7.1.37) (Extra sporogenous cells protein)
(EXCESS MICROSPOROCYTES1 protein)
Length = 1192
Score = 219 bits (559), Expect = 1e-56
Identities = 123/283 (43%), Positives = 172/283 (60%), Gaps = 9/283 (3%)
Query: 302 GDCISV--NFTDSNKLGEGGFGPVYKGILSDGQEVAIKRLSICSEQGSEEFINEVMLILK 359
GD + +F+ N +G+GGFG VYK L + VA+K+LS QG+ EF+ E+ + K
Sbjct: 908 GDIVEATDHFSKKNIIGDGGFGTVYKACLPGEKTVAVKKLSEAKTQGNREFMAEMETLGK 967
Query: 360 LQHKNLVKLLGFCVDGEEKLLVYEYLPNGSLDVVLFEQHAQ---LDWTKRLDIINGIARG 416
++H NLV LLG+C EEKLLVYEY+ NGSLD L Q LDW+KRL I G ARG
Sbjct: 968 VKHPNLVSLLGYCSFSEEKLLVYEYMVNGSLDHWLRNQTGMLEVLDWSKRLKIAVGAARG 1027
Query: 417 ILYLHEDSRLQIIHRDLKASNVLLDNDMNPKISDFGMARIFAGSEGEANTTTIVGTYGYM 476
+ +LH IIHRD+KASN+LLD D PK++DFG+AR+ + E +T I GT+GY+
Sbjct: 1028 LAFLHHGFIPHIIHRDIKASNILLDGDFEPKVADFGLARLISACESHV-STVIAGTFGYI 1086
Query: 477 APEYAMEGLYSIKSDVFGFGVLLLEIITGIRNAGFCYSKTT-PSLLAYAWHLWNDGKGLE 535
PEY + K DV+ FGV+LLE++TG G + ++ +L+ +A N GK ++
Sbjct: 1087 PPEYGQSARATTKGDVYSFGVILLELVTGKEPTGPDFKESEGGNLVGWAIQKINQGKAVD 1146
Query: 536 LRDPLLLCPG--DQFLRYMNIGLLCVQEDAFDRPTMSSVVLML 576
+ DPLL+ + LR + I +LC+ E RP M V+ L
Sbjct: 1147 VIDPLLVSVALKNSQLRLLQIAMLCLAETPAKRPNMLDVLKAL 1189
>PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC
2.7.1.37) (AvrPphB susceptible protein 1)
Length = 456
Score = 215 bits (548), Expect = 2e-55
Identities = 119/278 (42%), Positives = 175/278 (62%), Gaps = 7/278 (2%)
Query: 306 SVNFTDSNKLGEGGFGPVYKGIL-SDGQEVAIKRLSICSEQGSEEFINEVMLILKLQHKN 364
++NF LGEGGFG VYKG L S GQ VA+K+L QG+ EF+ EV+++ L H N
Sbjct: 83 TMNFHPDTFLGEGGFGRVYKGRLDSTGQVVAVKQLDRNGLQGNREFLVEVLMLSLLHHPN 142
Query: 365 LVKLLGFCVDGEEKLLVYEYLPNGSLDVVLFE---QHAQLDWTKRLDIINGIARGILYLH 421
LV L+G+C DG+++LLVYE++P GSL+ L + LDW R+ I G A+G+ +LH
Sbjct: 143 LVNLIGYCADGDQRLLVYEFMPLGSLEDHLHDLPPDKEALDWNMRMKIAAGAAKGLEFLH 202
Query: 422 EDSRLQIIHRDLKASNVLLDNDMNPKISDFGMARIFAGSEGEANTTTIVGTYGYMAPEYA 481
+ + +I+RD K+SN+LLD +PK+SDFG+A++ + +T ++GTYGY APEYA
Sbjct: 203 DKANPPVIYRDFKSSNILLDEGFHPKLSDFGLAKLGPTGDKSHVSTRVMGTYGYCAPEYA 262
Query: 482 MEGLYSIKSDVFGFGVLLLEIITGIRNAGFCYSKTTPSLLAYAWHLWND-GKGLELRDPL 540
M G ++KSDV+ FGV+ LE+ITG + +L+A+A L+ND K ++L DP
Sbjct: 263 MTGQLTVKSDVYSFGVVFLELITGRKAIDSEMPHGEQNLVAWARPLFNDRRKFIKLADPR 322
Query: 541 L--LCPGDQFLRYMNIGLLCVQEDAFDRPTMSSVVLML 576
L P + + + +C+QE A RP ++ VV L
Sbjct: 323 LKGRFPTRALYQALAVASMCIQEQAATRPLIADVVTAL 360
>RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase
RLCKVII (EC 2.7.1.37)
Length = 423
Score = 213 bits (543), Expect = 9e-55
Identities = 124/295 (42%), Positives = 173/295 (58%), Gaps = 7/295 (2%)
Query: 308 NFTDSNKLGEGGFGPVYKGILSD-GQEVAIKRLSICSEQGSEEFINEVMLILKLQHKNLV 366
NF LGEGGFG V+KG + Q VAIK+L QG EF+ EV+ + H NLV
Sbjct: 102 NFRSDCFLGEGGFGKVFKGTIEKLDQVVAIKQLDRNGVQGIREFVVEVLTLSLADHPNLV 161
Query: 367 KLLGFCVDGEEKLLVYEYLPNGSLDV---VLFEQHAQLDWTKRLDIINGIARGILYLHED 423
KL+GFC +G+++LLVYEY+P GSL+ VL LDW R+ I G ARG+ YLH+
Sbjct: 162 KLIGFCAEGDQRLLVYEYMPQGSLEDHLHVLPSGKKPLDWNTRMKIAAGAARGLEYLHDR 221
Query: 424 SRLQIIHRDLKASNVLLDNDMNPKISDFGMARIFAGSEGEANTTTIVGTYGYMAPEYAME 483
+I+RDLK SN+LL D PK+SDFG+A++ + +T ++GTYGY AP+YAM
Sbjct: 222 MTPPVIYRDLKCSNILLGEDYQPKLSDFGLAKVGPSGDKTHVSTRVMGTYGYCAPDYAMT 281
Query: 484 GLYSIKSDVFGFGVLLLEIITGIRNAGFCYSKTTPSLLAYAWHLWNDGKGL-ELRDPLL- 541
G + KSD++ FGV+LLE+ITG + ++ +L+ +A L+ D + ++ DPLL
Sbjct: 282 GQLTFKSDIYSFGVVLLELITGRKAIDNTKTRKDQNLVGWARPLFKDRRNFPKMVDPLLQ 341
Query: 542 -LCPGDQFLRYMNIGLLCVQEDAFDRPTMSSVVLMLMNESVMLGQPGKPPFSVGR 595
P + + I +CVQE RP +S VVL L + P P S G+
Sbjct: 342 GQYPVRGLYQALAISAMCVQEQPTMRPVVSDVVLALNFLASSKYDPNSPSSSSGK 396
>PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (EC
2.7.1.37) (Phytosulfokine LRR receptor kinase)
Length = 1008
Score = 201 bits (512), Expect = 4e-51
Identities = 112/274 (40%), Positives = 162/274 (58%), Gaps = 6/274 (2%)
Query: 308 NFTDSNKLGEGGFGPVYKGILSDGQEVAIKRLSICSEQGSEEFINEVMLILKLQHKNLVK 367
+F +N +G GGFG VYK L DG++VAIK+LS Q EF EV + + QH NLV
Sbjct: 733 SFDQANIIGCGGFGMVYKATLPDGKKVAIKKLSGDCGQIEREFEAEVETLSRAQHPNLVL 792
Query: 368 LLGFCVDGEEKLLVYEYLPNGSLDVVLFEQH---AQLDWTKRLDIINGIARGILYLHEDS 424
L GFC ++LL+Y Y+ NGSLD L E++ A L W RL I G A+G+LYLHE
Sbjct: 793 LRGFCFYKNDRLLIYSYMENGSLDYWLHERNDGPALLKWKTRLRIAQGAAKGLLYLHEGC 852
Query: 425 RLQIIHRDLKASNVLLDNDMNPKISDFGMARIFAGSEGEANTTTIVGTYGYMAPEYAMEG 484
I+HRD+K+SN+LLD + N ++DFG+AR+ + E +T +VGT GY+ PEY
Sbjct: 853 DPHILHRDIKSSNILLDENFNSHLADFGLARLMSPYETHV-STDLVGTLGYIPPEYGQAS 911
Query: 485 LYSIKSDVFGFGVLLLEIITGIRNAGFCYSKTTPSLLAYAWHLWNDGKGLELRDPLLLCP 544
+ + K DV+ FGV+LLE++T R C K L+++ + ++ + E+ DPL+
Sbjct: 912 VATYKGDVYSFGVVLLELLTDKRPVDMCKPKGCRDLISWVVKMKHESRASEVFDPLIYSK 971
Query: 545 GD--QFLRYMNIGLLCVQEDAFDRPTMSSVVLML 576
+ + R + I LC+ E+ RPT +V L
Sbjct: 972 ENDKEMFRVLEIACLCLSENPKQRPTTQQLVSWL 1005
>BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC
2.7.1.37) (AtBRI1) (Brassinosteroid LRR receptor kinase)
Length = 1196
Score = 197 bits (500), Expect = 9e-50
Identities = 121/327 (37%), Positives = 185/327 (56%), Gaps = 19/327 (5%)
Query: 259 NLGGKNNSSKIWMITVIAVGVGLVIIIFICYLCFLRNRQSKLFGDCISVN--FTDSNKLG 316
N G + ++ W +T G+ + I F + + F D + F + + +G
Sbjct: 837 NSGDRTANNTNWKLT------GVKEALSINLAAFEKPLRKLTFADLLQATNGFHNDSLIG 890
Query: 317 EGGFGPVYKGILSDGQEVAIKRLSICSEQGSEEFINEVMLILKLQHKNLVKLLGFCVDGE 376
GGFG VYK IL DG VAIK+L S QG EF+ E+ I K++H+NLV LLG+C G+
Sbjct: 891 SGGFGDVYKAILKDGSAVAIKKLIHVSGQGDREFMAEMETIGKIKHRNLVPLLGYCKVGD 950
Query: 377 EKLLVYEYLPNGSLDVVLFEQH---AQLDWTKRLDIINGIARGILYLHEDSRLQIIHRDL 433
E+LLVYE++ GSL+ VL + +L+W+ R I G ARG+ +LH + IIHRD+
Sbjct: 951 ERLLVYEFMKYGSLEDVLHDPKKAGVKLNWSTRRKIAIGSARGLAFLHHNCSPHIIHRDM 1010
Query: 434 KASNVLLDNDMNPKISDFGMARIFAGSEGEANTTTIVGTYGYMAPEYAMEGLYSIKSDVF 493
K+SNVLLD ++ ++SDFGMAR+ + + + +T+ GT GY+ PEY S K DV+
Sbjct: 1011 KSSNVLLDENLEARVSDFGMARLMSAMDTHLSVSTLAGTPGYVPPEYYQSFRCSTKGDVY 1070
Query: 494 GFGVLLLEIITGIR---NAGFCYSKTTPSLLAYAWHLWNDGKGLEL--RDPLLLCPGDQF 548
+GV+LLE++TG R + F + + +A +D EL DP L +
Sbjct: 1071 SYGVVLLELLTGKRPTDSPDFGDNNLVGWVKQHAKLRISDVFDPELMKEDPALEI---EL 1127
Query: 549 LRYMNIGLLCVQEDAFDRPTMSSVVLM 575
L+++ + + C+ + A+ RPTM V+ M
Sbjct: 1128 LQHLKVAVACLDDRAWRRPTMVQVMAM 1154
>TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precursor
(EC 2.7.1.-)
Length = 942
Score = 194 bits (494), Expect = 4e-49
Identities = 98/207 (47%), Positives = 141/207 (67%), Gaps = 7/207 (3%)
Query: 305 ISVNFTDSNKLGEGGFGPVYKGILSDGQEVAIKRLS--ICSEQGSEEFINEVMLILKLQH 362
++ NF+ N LG GGFG VYKG L DG ++A+KR+ + + +G EF +E+ ++ K++H
Sbjct: 584 VTNNFSSDNILGSGGFGVVYKGELHDGTKIAVKRMENGVIAGKGFAEFKSEIAVLTKVRH 643
Query: 363 KNLVKLLGFCVDGEEKLLVYEYLPNGSLDVVLFEQHAQ----LDWTKRLDIINGIARGIL 418
++LV LLG+C+DG EKLLVYEY+P G+L LFE + L W +RL + +ARG+
Sbjct: 644 RHLVTLLGYCLDGNEKLLVYEYMPQGTLSRHLFEWSEEGLKPLLWKQRLTLALDVARGVE 703
Query: 419 YLHEDSRLQIIHRDLKASNVLLDNDMNPKISDFGMARIFAGSEGEANTTTIVGTYGYMAP 478
YLH + IHRDLK SN+LL +DM K++DFG+ R+ +G T I GT+GY+AP
Sbjct: 704 YLHGLAHQSFIHRDLKPSNILLGDDMRAKVADFGLVRLAPEGKGSIE-TRIAGTFGYLAP 762
Query: 479 EYAMEGLYSIKSDVFGFGVLLLEIITG 505
EYA+ G + K DV+ FGV+L+E+ITG
Sbjct: 763 EYAVTGRVTTKVDVYSFGVILMELITG 789
>PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.37)
(Phytosulfokine LRR receptor kinase)
Length = 1021
Score = 193 bits (490), Expect = 1e-48
Identities = 110/279 (39%), Positives = 158/279 (56%), Gaps = 6/279 (2%)
Query: 308 NFTDSNKLGEGGFGPVYKGILSDGQEVAIKRLSICSEQGSEEFINEVMLILKLQHKNLVK 367
+F +N +G GGFG VYK L DG +VAIKRLS + Q EF EV + + QH NLV
Sbjct: 742 SFNQANIIGCGGFGLVYKATLPDGTKVAIKRLSGDTGQMDREFQAEVETLSRAQHPNLVH 801
Query: 368 LLGFCVDGEEKLLVYEYLPNGSLDVVLFEQ---HAQLDWTKRLDIINGIARGILYLHEDS 424
LLG+C +KLL+Y Y+ NGSLD L E+ LDW RL I G A G+ YLH+
Sbjct: 802 LLGYCNYKNDKLLIYSYMDNGSLDYWLHEKVDGPPSLDWKTRLRIARGAAEGLAYLHQSC 861
Query: 425 RLQIIHRDLKASNVLLDNDMNPKISDFGMARIFAGSEGEANTTTIVGTYGYMAPEYAMEG 484
I+HRD+K+SN+LL + ++DFG+AR+ + TT +VGT GY+ PEY
Sbjct: 862 EPHILHRDIKSSNILLSDTFVAHLADFGLARLILPYDTHV-TTDLVGTLGYIPPEYGQAS 920
Query: 485 LYSIKSDVFGFGVLLLEIITGIRNAGFCYSKTTPSLLAYAWHLWNDGKGLELRDPLLLCP 544
+ + K DV+ FGV+LLE++TG R C + + L+++ + + + E+ DP +
Sbjct: 921 VATYKGDVYSFGVVLLELLTGRRPMDVCKPRGSRDLISWVLQMKTEKRESEIFDPFIYDK 980
Query: 545 --GDQFLRYMNIGLLCVQEDAFDRPTMSSVVLMLMNESV 581
++ L + I C+ E+ RPT +V L N V
Sbjct: 981 DHAEEMLLVLEIACRCLGENPKTRPTTQQLVSWLENIDV 1019
>BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated
receptor kinase 1 precursor (EC 2.7.1.37)
(BRI1-associated receptor kinase 1) (Somatic
embryogenesis receptor-like kinase 3)
Length = 615
Score = 192 bits (487), Expect = 3e-48
Identities = 118/284 (41%), Positives = 162/284 (56%), Gaps = 9/284 (3%)
Query: 306 SVNFTDSNKLGEGGFGPVYKGILSDGQEVAIKRLSICSEQGSE-EFINEVMLILKLQHKN 364
S NF++ N LG GGFG VYKG L+DG VA+KRL QG E +F EV +I H+N
Sbjct: 286 SDNFSNKNILGRGGFGKVYKGRLADGTLVAVKRLKEERTQGGELQFQTEVEMISMAVHRN 345
Query: 365 LVKLLGFCVDGEEKLLVYEYLPNGSLDVVLFEQ---HAQLDWTKRLDIINGIARGILYLH 421
L++L GFC+ E+LLVY Y+ NGS+ L E+ LDW KR I G ARG+ YLH
Sbjct: 346 LLRLRGFCMTPTERLLVYPYMANGSVASCLRERPESQPPLDWPKRQRIALGSARGLAYLH 405
Query: 422 EDSRLQIIHRDLKASNVLLDNDMNPKISDFGMARIFAGSEGEANTTTIVGTYGYMAPEYA 481
+ +IIHRD+KA+N+LLD + + DFG+A++ + TT + GT G++APEY
Sbjct: 406 DHCDPKIIHRDVKAANILLDEEFEAVVGDFGLAKLMDYKDTHV-TTAVRGTIGHIAPEYL 464
Query: 482 MEGLYSIKSDVFGFGVLLLEIITGIRNAGFCYSKTTPSLLAYAW--HLWNDGKGLELRDP 539
G S K+DVFG+GV+LLE+ITG R ++ W L + K L D
Sbjct: 465 STGKSSEKTDVFGYGVMLLELITGQRAFDLARLANDDDVMLLDWVKGLLKEKKLEALVDV 524
Query: 540 LLL--CPGDQFLRYMNIGLLCVQEDAFDRPTMSSVVLMLMNESV 581
L ++ + + + LLC Q +RP MS VV ML + +
Sbjct: 525 DLQGNYKDEEVEQLIQVALLCTQSSPMERPKMSEVVRMLEGDGL 568
>BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.37)
(Brassinosteroid LRR receptor kinase)
Length = 1207
Score = 189 bits (481), Expect = 1e-47
Identities = 114/300 (38%), Positives = 168/300 (56%), Gaps = 15/300 (5%)
Query: 309 FTDSNKLGEGGFGPVYKGILSDGQEVAIKRLSICSEQGSEEFINEVMLILKLQHKNLVKL 368
F + + +G GGFG VYK L DG VAIK+L S QG EF E+ I K++H+NLV L
Sbjct: 888 FHNDSLVGSGGFGDVYKAQLKDGSVVAIKKLIHVSGQGDREFTAEMETIGKIKHRNLVPL 947
Query: 369 LGFCVDGEEKLLVYEYLPNGSLDVVLFEQHA---QLDWTKRLDIINGIARGILYLHEDSR 425
LG+C GEE+LLVYEY+ GSL+ VL ++ +L+W R I G ARG+ +LH +
Sbjct: 948 LGYCKVGEERLLVYEYMKYGSLEDVLHDRKKTGIKLNWPARRKIAIGAARGLAFLHHNCI 1007
Query: 426 LQIIHRDLKASNVLLDNDMNPKISDFGMARIFAGSEGEANTTTIVGTYGYMAPEYAMEGL 485
IIHRD+K+SNVLLD ++ ++SDFGMAR+ + + + +T+ GT GY+ PEY
Sbjct: 1008 PHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVSTLAGTPGYVPPEYYQSFR 1067
Query: 486 YSIKSDVFGFGVLLLEIITGIR---NAGFCYSKTTPSLLAYAWHLWNDGKGLELRDPLLL 542
S K DV+ +GV+LLE++TG + +A F + + +A GK ++ D LL
Sbjct: 1068 CSTKGDVYSYGVVLLELLTGKQPTDSADFGDNNLVGWVKLHA-----KGKITDVFDRELL 1122
Query: 543 CPGD----QFLRYMNIGLLCVQEDAFDRPTMSSVVLMLMNESVMLGQPGKPPFSVGRLNF 598
+ L+++ + C+ + + RPTM V+ M G +NF
Sbjct: 1123 KEDASIEIELLQHLKVACACLDDRHWKRPTMIQVMAMFKEIQAGSGMDSTSTIGADDVNF 1182
>BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precursor (EC
2.7.1.37) (tBRI1) (Altered brassinolide sensitivity 1)
(Systemin receptor SR160)
Length = 1207
Score = 189 bits (481), Expect = 1e-47
Identities = 114/300 (38%), Positives = 168/300 (56%), Gaps = 15/300 (5%)
Query: 309 FTDSNKLGEGGFGPVYKGILSDGQEVAIKRLSICSEQGSEEFINEVMLILKLQHKNLVKL 368
F + + +G GGFG VYK L DG VAIK+L S QG EF E+ I K++H+NLV L
Sbjct: 888 FHNDSLVGSGGFGDVYKAQLKDGSVVAIKKLIHVSGQGDREFTAEMETIGKIKHRNLVPL 947
Query: 369 LGFCVDGEEKLLVYEYLPNGSLDVVLFEQHA---QLDWTKRLDIINGIARGILYLHEDSR 425
LG+C GEE+LLVYEY+ GSL+ VL ++ +L+W R I G ARG+ +LH +
Sbjct: 948 LGYCKVGEERLLVYEYMKYGSLEDVLHDRKKIGIKLNWPARRKIAIGAARGLAFLHHNCI 1007
Query: 426 LQIIHRDLKASNVLLDNDMNPKISDFGMARIFAGSEGEANTTTIVGTYGYMAPEYAMEGL 485
IIHRD+K+SNVLLD ++ ++SDFGMAR+ + + + +T+ GT GY+ PEY
Sbjct: 1008 PHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVSTLAGTPGYVPPEYYQSFR 1067
Query: 486 YSIKSDVFGFGVLLLEIITGIR---NAGFCYSKTTPSLLAYAWHLWNDGKGLELRDPLLL 542
S K DV+ +GV+LLE++TG + +A F + + +A GK ++ D LL
Sbjct: 1068 CSTKGDVYSYGVVLLELLTGKQPTDSADFGDNNLVGWVKLHA-----KGKITDVFDRELL 1122
Query: 543 CPGD----QFLRYMNIGLLCVQEDAFDRPTMSSVVLMLMNESVMLGQPGKPPFSVGRLNF 598
+ L+++ + C+ + + RPTM V+ M G +NF
Sbjct: 1123 KEDASIEIELLQHLKVACACLDDRHWKRPTMIQVMAMFKEIQAGSGMDSTSTIGADDVNF 1182
>CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx32,
chloroplast precursor (EC 2.7.1.37)
Length = 419
Score = 188 bits (477), Expect = 4e-47
Identities = 116/284 (40%), Positives = 165/284 (57%), Gaps = 18/284 (6%)
Query: 308 NFTDSNKLGEGGFGPVYKGILS----------DGQEVAIKRLSICSEQGSEEFINEVMLI 357
NF + LG+GGFG VY+G + G VAIKRL+ S QG E+ +EV +
Sbjct: 85 NFKPDSMLGQGGFGKVYRGWVDATTLAPSRVGSGMIVAIKRLNSESVQGFAEWRSEVNFL 144
Query: 358 LKLQHKNLVKLLGFCVDGEEKLLVYEYLPNGSLDVVLFEQHAQLDWTKRLDIINGIARGI 417
L H+NLVKLLG+C + +E LLVYE++P GSL+ LF ++ W R+ I+ G ARG+
Sbjct: 145 GMLSHRNLVKLLGYCREDKELLLVYEFMPKGSLESHLFRRNDPFPWDLRIKIVIGAARGL 204
Query: 418 LYLHEDSRLQIIHRDLKASNVLLDNDMNPKISDFGMARIFAGSEGEANTTTIVGTYGYMA 477
+LH R ++I+RD KASN+LLD++ + K+SDFG+A++ E TT I+GTYGY A
Sbjct: 205 AFLHSLQR-EVIYRDFKASNILLDSNYDAKLSDFGLAKLGPADEKSHVTTRIMGTYGYAA 263
Query: 478 PEYAMEGLYSIKSDVFGFGVLLLEIITGIRNAGFCYSKTTPSLLAYAW-HLWNDGKGLEL 536
PEY G +KSDVF FGV+LLEI+TG+ + SL+ + L N + ++
Sbjct: 264 PEYMATGHLYVKSDVFAFGVVLLEIMTGLTAHNTKRPRGQESLVDWLRPELSNKHRVKQI 323
Query: 537 RDPLLLCPGDQFLR----YMNIGLLCVQEDAFDRPTMSSVVLML 576
D + G + I L C++ D +RP M VV +L
Sbjct: 324 MDKGI--KGQYTTKVATEMARITLSCIEPDPKNRPHMKEVVEVL 365
>NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK (EC
2.7.1.37)
Length = 389
Score = 181 bits (458), Expect = 7e-45
Identities = 112/284 (39%), Positives = 170/284 (59%), Gaps = 21/284 (7%)
Query: 308 NFTDSNKLGEGGFGPVYKGILSD----------GQEVAIKRLSICSEQGSEEFINEVMLI 357
NF + +GEGGFG V+KG + + G +A+KRL+ QG E++ E+ +
Sbjct: 67 NFRPDSVVGEGGFGCVFKGWIDESSLAPSKPGTGIVIAVKRLNQEGFQGHREWLAEINYL 126
Query: 358 LKLQHKNLVKLLGFCVDGEEKLLVYEYLPNGSLDVVLFEQ---HAQLDWTKRLDIINGIA 414
+L H NLVKL+G+C++ E +LLVYE++ GSL+ LF + + L W R+ + G A
Sbjct: 127 GQLDHPNLVKLIGYCLEEEHRLLVYEFMTRGSLENHLFRRGTFYQPLSWNTRVRMALGAA 186
Query: 415 RGILYLHEDSRLQIIHRDLKASNVLLDNDMNPKISDFGMARIFAGSEGEANTTTIVGTYG 474
RG+ +LH +++ Q+I+RD KASN+LLD++ N K+SDFG+AR + +T ++GT G
Sbjct: 187 RGLAFLH-NAQPQVIYRDFKASNILLDSNYNAKLSDFGLARDGPMGDNSHVSTRVMGTQG 245
Query: 475 YMAPEYAMEGLYSIKSDVFGFGVLLLEIITGIRNAGFCYSKTTPSLLAYAW-HLWNDGKG 533
Y APEY G S+KSDV+ FGV+LLE+++G R +L+ +A +L N +
Sbjct: 246 YAAPEYLATGHLSVKSDVYSFGVVLLELLSGRRAIDKNQPVGEHNLVDWARPYLTNKRRL 305
Query: 534 LELRDPLLLCPGDQFL-RYMNIGLL---CVQEDAFDRPTMSSVV 573
L + DP L G L R + I +L C+ DA RPTM+ +V
Sbjct: 306 LRVMDPRL--QGQYSLTRALKIAVLALDCISIDAKSRPTMNEIV 347
>CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4
precursor (EC 2.7.1.-)
Length = 901
Score = 176 bits (445), Expect = 2e-43
Identities = 104/287 (36%), Positives = 168/287 (58%), Gaps = 11/287 (3%)
Query: 309 FTDSNKLGEGGFGPVYKGILSDGQEVAIKRLSICSE--QGSEEFINEVMLILKLQHKNLV 366
F++ +++G+G F V+KGIL DG VA+KR S+ + S+EF NE+ L+ +L H +L+
Sbjct: 505 FSEDSQVGKGSFSCVFKGILRDGTVVAVKRAIKASDVKKSSKEFHNELDLLSRLNHAHLL 564
Query: 367 KLLGFCVDGEEKLLVYEYLPNGSLDVVLFEQHA----QLDWTKRLDIINGIARGILYLHE 422
LLG+C DG E+LLVYE++ +GSL L + +L+W +R+ I ARGI YLH
Sbjct: 565 NLLGYCEDGSERLLVYEFMAHGSLYQHLHGKDPNLKKRLNWARRVTIAVQAARGIEYLHG 624
Query: 423 DSRLQIIHRDLKASNVLLDNDMNPKISDFGMARIFAGSEGEANTTTIVGTYGYMAPEYAM 482
+ +IHRD+K+SN+L+D D N +++DFG++ + G + GT GY+ PEY
Sbjct: 625 YACPPVIHRDIKSSNILIDEDHNARVADFGLSILGPADSGTPLSELPAGTLGYLDPEYYR 684
Query: 483 EGLYSIKSDVFGFGVLLLEIITGIRNAGFCYSKTTPSLLAYAWHLWNDGKGLELRDPLLL 542
+ KSDV+ FGV+LLEI++G + + + +++ +A L G + DP+L
Sbjct: 685 LHYLTTKSDVYSFGVVLLEILSGRKAIDMQFEE--GNIVEWAVPLIKAGDIFAILDPVLS 742
Query: 543 CPGD--QFLRYMNIGLLCVQEDAFDRPTMSSVVLMLMNE-SVMLGQP 586
P D + ++ CV+ DRP+M V L + ++++G P
Sbjct: 743 PPSDLEALKKIASVACKCVRMRGKDRPSMDKVTTALEHALALLMGSP 789
>CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (EC
2.7.1.-)
Length = 980
Score = 174 bits (442), Expect = 5e-43
Identities = 150/511 (29%), Positives = 242/511 (47%), Gaps = 69/511 (13%)
Query: 123 EFCQVRYSNKNFIGSLNVNGNIGKDNVQNISEPVKFETSVNKL-------LNGLTKIASF 175
E ++++ ++ + N+ G I D++ S + + S N++ +N + + +
Sbjct: 499 EIFELKHLSRINTSANNITGGI-PDSISRCSTLISVDLSRNRINGEIPKGINNVKNLGTL 557
Query: 176 NVSANMYATGEVPFEDKTIYALVQCTRDLAANDCSRCLLSAIGDIP-GCCYASIGARVMS 234
N+S N TG +P + +L T DL+ ND S G +P G + +
Sbjct: 558 NISGNQL-TGSIPTGIGNMTSLT--TLDLSFNDLS-------GRVPLGGQFLVFNETSFA 607
Query: 235 RSCYLRYEFYPFYLGEKEQTKSSTNLGGKNNSSKIWMITVIAVGVGLVII-IFICYLCFL 293
+ YL QT S N + S+I +ITVIA GL++I + I +
Sbjct: 608 GNTYLCLPHRVSCPTRPGQT-SDHNHTALFSPSRI-VITVIAAITGLILISVAIRQMNKK 665
Query: 294 RNRQSKLF-----------GDCISVNFTDSNKLGEGGFGPVYKGILSDGQEVAIKRLSIC 342
+N++S + + + + N +G+GG G VY+G + + +VAIKRL
Sbjct: 666 KNQKSLAWKLTAFQKLDFKSEDVLECLKEENIIGKGGAGIVYRGSMPNNVDVAIKRLVGR 725
Query: 343 SEQGSEE-FINEVMLILKLQHKNLVKLLGFCVDGEEKLLVYEYLPNGSLDVVLF-EQHAQ 400
S+ F E+ + +++H+++V+LLG+ + + LL+YEY+PNGSL +L +
Sbjct: 726 GTGRSDHGFTAEIQTLGRIRHRHIVRLLGYVANKDTNLLLYEYMPNGSLGELLHGSKGGH 785
Query: 401 LDWTKRLDIINGIARGILYLHEDSRLQIIHRDLKASNVLLDNDMNPKISDFGMARIFAGS 460
L W R + A+G+ YLH D I+HRD+K++N+LLD+D ++DFG+A+
Sbjct: 786 LQWETRHRVAVEAAKGLCYLHHDCSPLILHRDVKSNNILLDSDFEAHVADFGLAKFLVDG 845
Query: 461 EGEANTTTIVGTYGYMAPEYAMEGLYSIKSDVFGFGVLLLEIITGIRNAGFCYSK----- 515
++I G+YGY+APEYA KSDV+ FGV+LLE+I G + G
Sbjct: 846 AASECMSSIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELIAGKKPVGEFGEGVDIVR 905
Query: 516 ---------TTPSLLAYAWHLWNDGKGLELRDPLLL-CPGDQFLRYMNIGLLCVQEDAFD 565
T PS A + + DP L P + I ++CV+E+A
Sbjct: 906 WVRNTEEEITQPSDAAIV---------VAIVDPRLTGYPLTSVIHVFKIAMMCVEEEAAA 956
Query: 566 RPTMSSVVLMLMNESVMLGQPGKPPFSVGRL 596
RPTM VV ML N PP SV L
Sbjct: 957 RPTMREVVHMLTN----------PPKSVANL 977
>APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor (EC
2.7.1.-)
Length = 412
Score = 172 bits (437), Expect = 2e-42
Identities = 90/213 (42%), Positives = 137/213 (64%), Gaps = 14/213 (6%)
Query: 308 NFTDSNKLGEGGFGPVYKGILSD----------GQEVAIKRLSICSEQGSEEFINEVMLI 357
NF + LGEGGFG V+KG + + G +A+K+L+ QG +E++ EV +
Sbjct: 68 NFRPDSVLGEGGFGSVFKGWIDEQTLTASKPGTGVVIAVKKLNQDGWQGHQEWLAEVNYL 127
Query: 358 LKLQHKNLVKLLGFCVDGEEKLLVYEYLPNGSLDVVLFEQHAQ---LDWTKRLDIINGIA 414
+ H NLVKL+G+C++ E +LLVYE++P GSL+ LF + + L WT RL + G A
Sbjct: 128 GQFSHPNLVKLIGYCLEDEHRLLVYEFMPRGSLENHLFRRGSYFQPLSWTLRLKVALGAA 187
Query: 415 RGILYLHEDSRLQIIHRDLKASNVLLDNDMNPKISDFGMARIFAGSEGEANTTTIVGTYG 474
+G+ +LH ++ +I+RD K SN+LLD++ N K+SDFG+A+ + +T I+GTYG
Sbjct: 188 KGLAFLH-NAETSVIYRDFKTSNILLDSEYNAKLSDFGLAKDGPTGDKSHVSTRIMGTYG 246
Query: 475 YMAPEYAMEGLYSIKSDVFGFGVLLLEIITGIR 507
Y APEY G + KSDV+ +GV+LLE+++G R
Sbjct: 247 YAAPEYLATGHLTTKSDVYSYGVVLLEVLSGRR 279
>APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor (EC
2.7.1.-)
Length = 410
Score = 169 bits (429), Expect = 2e-41
Identities = 105/285 (36%), Positives = 163/285 (56%), Gaps = 23/285 (8%)
Query: 308 NFTDSNKLGEGGFGPVYKGILSD----------GQEVAIKRLSICSEQGSEEFINEVMLI 357
NF + LGEGGFG V+KG + + G +A+K+L+ QG +E++ EV +
Sbjct: 67 NFRPDSVLGEGGFGCVFKGWIDEKSLTASRPGTGLVIAVKKLNQDGWQGHQEWLAEVNYL 126
Query: 358 LKLQHKNLVKLLGFCVDGEEKLLVYEYLPNGSLDVVLFEQ---HAQLDWTKRLDIINGIA 414
+ H++LVKL+G+C++ E +LLVYE++P GSL+ LF + L W RL + G A
Sbjct: 127 GQFSHRHLVKLIGYCLEDEHRLLVYEFMPRGSLENHLFRRGLYFQPLSWKLRLKVALGAA 186
Query: 415 RGILYLHEDSRLQIIHRDLKASNVLLDNDMNPKISDFGMARIFAGSEGEANTTTIVGTYG 474
+G+ +LH S ++I+RD K SN+LLD++ N K+SDFG+A+ + +T ++GT+G
Sbjct: 187 KGLAFLH-SSETRVIYRDFKTSNILLDSEYNAKLSDFGLAKDGPIGDKSHVSTRVMGTHG 245
Query: 475 YMAPEYAMEGLYSIKSDVFGFGVLLLEIITGIRNAGFCYSKTTPSLLAYAW-HLWNDGKG 533
Y APEY G + KSDV+ FGV+LLE+++G R +L+ +A +L N K
Sbjct: 246 YAAPEYLATGHLTTKSDVYSFGVVLLELLSGRRAVDKNRPSGERNLVEWAKPYLVNKRKI 305
Query: 534 LELRDPLLLCPGDQF-----LRYMNIGLLCVQEDAFDRPTMSSVV 573
+ D L DQ+ + + L C+ + RP MS VV
Sbjct: 306 FRVIDNRL---QDQYSMEEACKVATLSLRCLTTEIKLRPNMSEVV 347
>RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC
2.7.1.37)
Length = 1109
Score = 169 bits (427), Expect = 3e-41
Identities = 102/284 (35%), Positives = 158/284 (54%), Gaps = 11/284 (3%)
Query: 308 NFTDSNKLGEGGFGPVYKGILSDGQEVAIKRLSICS-EQGSEEFINEVMLILKLQHKNLV 366
N D +G+G G +YK LS + A+K+L + GS + E+ I K++H+NL+
Sbjct: 815 NLNDKYVIGKGAHGTIYKATLSPDKVYAVKKLVFTGIKNGSVSMVREIETIGKVRHRNLI 874
Query: 367 KLLGFCVDGEEKLLVYEYLPNGSLDVVLFEQHAQ--LDWTKRLDIINGIARGILYLHEDS 424
KL F + E L++Y Y+ NGSL +L E + LDW+ R +I G A G+ YLH D
Sbjct: 875 KLEEFWLRKEYGLILYTYMENGSLHDILHETNPPKPLDWSTRHNIAVGTAHGLAYLHFDC 934
Query: 425 RLQIIHRDLKASNVLLDNDMNPKISDFGMARIFAGSEGEANTTTIVGTYGYMAPEYAMEG 484
I+HRD+K N+LLD+D+ P ISDFG+A++ S + T+ GT GYMAPE A
Sbjct: 935 DPAIVHRDIKPMNILLDSDLEPHISDFGIAKLLDQSATSIPSNTVQGTIGYMAPENAFTT 994
Query: 485 LYSIKSDVFGFGVLLLEIITGIRNAGFCYSKTTPSLLAYAWHLWND-GKGLELRDPLLL- 542
+ S +SDV+ +GV+LLE+IT + ++ T ++ + +W G+ ++ DP LL
Sbjct: 995 VKSRESDVYSYGVVLLELITRKKALDPSFNGET-DIVGWVRSVWTQTGEIQKIVDPSLLD 1053
Query: 543 -----CPGDQFLRYMNIGLLCVQEDAFDRPTMSSVVLMLMNESV 581
+Q +++ L C +++ RPTM VV L S+
Sbjct: 1054 ELIDSSVMEQVTEALSLALRCAEKEVDKRPTMRDVVKQLTRWSI 1097
>RLK5_ARATH (P47735) Receptor-like protein kinase 5 precursor (EC
2.7.1.37)
Length = 999
Score = 163 bits (413), Expect = 1e-39
Identities = 123/355 (34%), Positives = 182/355 (50%), Gaps = 46/355 (12%)
Query: 265 NSSKIWMITVIAVGVGLV----IIIFICYLCFLRNRQSKLFG------------------ 302
N +W++ I + GLV I++FI LR +S
Sbjct: 621 NIGYVWILLTIFLLAGLVFVVGIVMFIAKCRKLRALKSSTLAASKWRSFHKLHFSEHEIA 680
Query: 303 DCISVNFTDSNKLGEGGFGPVYKGILSDGQEVAIKRLSICSEQGSEE----------FIN 352
DC+ + N +G G G VYK L G+ VA+K+L+ + G +E F
Sbjct: 681 DCLD----EKNVIGFGSSGKVYKVELRGGEVVAVKKLNKSVKGGDDEYSSDSLNRDVFAA 736
Query: 353 EVMLILKLQHKNLVKLLGFCVDGEEKLLVYEYLPNGSLDVVLFEQH---AQLDWTKRLDI 409
EV + ++HK++V+L C G+ KLLVYEY+PNGSL VL L W +RL I
Sbjct: 737 EVETLGTIRHKSIVRLWCCCSSGDCKLLVYEYMPNGSLADVLHGDRKGGVVLGWPERLRI 796
Query: 410 INGIARGILYLHEDSRLQIIHRDLKASNVLLDNDMNPKISDFGMARI--FAGSEGEANTT 467
A G+ YLH D I+HRD+K+SN+LLD+D K++DFG+A++ +GS+ +
Sbjct: 797 ALDAAEGLSYLHHDCVPPIVHRDVKSSNILLDSDYGAKVADFGIAKVGQMSGSKTPEAMS 856
Query: 468 TIVGTYGYMAPEYAMEGLYSIKSDVFGFGVLLLEIITGIRNAGFCYSKTTPSLLAYAWHL 527
I G+ GY+APEY + KSD++ FGV+LLE++TG + S+ +A
Sbjct: 857 GIAGSCGYIAPEYVYTLRVNEKSDIYSFGVVLLELVTGKQPTD---SELGDKDMAKWVCT 913
Query: 528 WNDGKGLE-LRDPLL-LCPGDQFLRYMNIGLLCVQEDAFDRPTMSSVVLMLMNES 580
D GLE + DP L L ++ + ++IGLLC +RP+M VV+ML S
Sbjct: 914 ALDKCGLEPVIDPKLDLKFKEEISKVIHIGLLCTSPLPLNRPSMRKVVIMLQEVS 968
>IRA4_HUMAN (Q9NWZ3) Interleukin-1 receptor-associated kinase-4 (EC
2.7.1.37) (IRAK-4) (NY-REN-64 antigen)
Length = 460
Score = 163 bits (413), Expect = 1e-39
Identities = 94/200 (47%), Positives = 129/200 (64%), Gaps = 11/200 (5%)
Query: 313 NKLGEGGFGPVYKGILSDGQEVAIKRLS----ICSEQGSEEFINEVMLILKLQHKNLVKL 368
NK+GEGGFG VYKG +++ VA+K+L+ I +E+ ++F E+ ++ K QH+NLV+L
Sbjct: 190 NKMGEGGFGVVYKGYVNN-TTVAVKKLAAMVDITTEELKQQFDQEIKVMAKCQHENLVEL 248
Query: 369 LGFCVDGEEKLLVYEYLPNGSL--DVVLFEQHAQLDWTKRLDIINGIARGILYLHEDSRL 426
LGF DG++ LVY Y+PNGSL + + L W R I G A GI +LHE+
Sbjct: 249 LGFSSDGDDLCLVYVYMPNGSLLDRLSCLDGTPPLSWHMRCKIAQGAANGINFLHENHH- 307
Query: 427 QIIHRDLKASNVLLDNDMNPKISDFGMARIFAGSEGEANTTTIVGTYGYMAPEYAMEGLY 486
IHRD+K++N+LLD KISDFG+AR T+ IVGT YMAPE A+ G
Sbjct: 308 --IHRDIKSANILLDEAFTAKISDFGLARASEKFAQTVMTSRIVGTTAYMAPE-ALRGEI 364
Query: 487 SIKSDVFGFGVLLLEIITGI 506
+ KSD++ FGV+LLEIITG+
Sbjct: 365 TPKSDIYSFGVVLLEIITGL 384
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.322 0.141 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 73,622,105
Number of Sequences: 164201
Number of extensions: 3206269
Number of successful extensions: 11196
Number of sequences better than 10.0: 1635
Number of HSP's better than 10.0 without gapping: 1166
Number of HSP's successfully gapped in prelim test: 469
Number of HSP's that attempted gapping in prelim test: 7449
Number of HSP's gapped (non-prelim): 1878
length of query: 622
length of database: 59,974,054
effective HSP length: 116
effective length of query: 506
effective length of database: 40,926,738
effective search space: 20708929428
effective search space used: 20708929428
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 69 (31.2 bits)
Medicago: description of AC123975.9


