

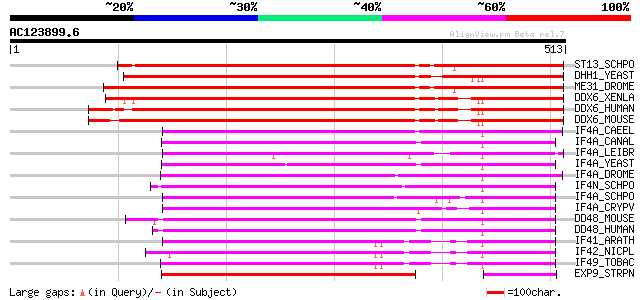
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC123899.6 + phase: 0 /pseudo
(513 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ST13_SCHPO (Q09181) Putative ATP-dependent RNA helicase ste13 486 e-137
DHH1_YEAST (P39517) Putative ATP-dependent RNA helicase DHH1 483 e-136
ME31_DROME (P23128) Putative ATP-dependent RNA helicase me31b (M... 478 e-134
DDX6_XENLA (P54824) ATP-dependent RNA helicase p54 (Xp54) 458 e-128
DDX6_HUMAN (P26196) Probable ATP-dependent RNA helicase p54 (Onc... 456 e-128
DDX6_MOUSE (P54823) Probable ATP-dependent RNA helicase p54 (Onc... 452 e-126
IF4A_CAEEL (P27639) Eukaryotic initiation factor 4A (eIF4A) (eIF... 240 7e-63
IF4A_CANAL (P87206) Eukaryotic initiation factor 4A (eIF4A) (eIF... 231 4e-60
IF4A_LEIBR (Q25225) Probable eukaryotic initiation factor 4A (eI... 230 8e-60
IF4A_YEAST (P10081) Eukaryotic initiation factor 4A (eIF4A) (eIF... 226 1e-58
IF4A_DROME (Q02748) Eukaryotic initiation factor 4A (eIF4A) (eIF... 226 1e-58
IF4N_SCHPO (Q10055) Eukaryotic initiation factor 4A-like protein... 226 1e-58
IF4A_SCHPO (P47943) Eukaryotic initiation factor 4A (eIF4A) (eIF... 225 2e-58
IF4A_CRYPV (O02494) Eukaryotic initiation factor 4A (eIF4A) (eIF... 223 7e-58
DD48_MOUSE (Q91VC3) Probable ATP-dependent helicase DDX48 (DEAD-... 214 3e-55
DD48_HUMAN (P38919) Probable ATP-dependent helicase DDX48 (DEAD-... 214 3e-55
IF41_ARATH (P41376) Eukaryotic initiation factor 4A-1 (eIF4A-1) ... 192 2e-48
IF42_NICPL (P41379) Eukaryotic initiation factor 4A-2 (eIF4A-2) ... 191 5e-48
IF49_TOBAC (Q40471) Eukaryotic initiation factor 4A-9 (eIF4A-9) ... 189 1e-47
EXP9_STRPN (P35599) Probable RNA helicase exp9 (Exported protein 9) 176 2e-43
>ST13_SCHPO (Q09181) Putative ATP-dependent RNA helicase ste13
Length = 485
Score = 486 bits (1252), Expect = e-137
Identities = 258/418 (61%), Positives = 311/418 (73%), Gaps = 12/418 (2%)
Query: 100 VEKTVQSEANDSSSQDWKARLKLPPADTRYRTEDVTATKGNEFEDYFLKRELLMGIYEKG 159
++K + ND S +K ++K P D R +TEDVT T+G EFEDY+LKRELLMGI+E G
Sbjct: 6 IQKLENANLNDRES--FKGQMKAQPVDMRPKTEDVTKTRGTEFEDYYLKRELLMGIFEAG 63
Query: 160 FERPSPIQEESIPIALTGSDILARAKNGTGKTAAFSIPALEKIDQDNNIIQVVILVPTRE 219
FERPSPIQEESIPIAL+G DILARAKNGTGKTAAF IP+LEK+D + IQ +ILVPTRE
Sbjct: 64 FERPSPIQEESIPIALSGRDILARAKNGTGKTAAFVIPSLEKVDTKKSKIQTLILVPTRE 123
Query: 220 LALQTSQVCKELGKHLQIQVMVTTGGTSLKDDIMRLYQPVHLLVGTPGRILDLAKKGVCV 279
LALQTSQVCK LGKH+ ++VMVTTGGT+L+DDI+RL VH++VGTPGR+LDLA KGV
Sbjct: 124 LALQTSQVCKTLGKHMNVKVMVTTGGTTLRDDIIRLNDTVHIVVGTPGRVLDLAGKGVAD 183
Query: 280 LKDCSMLVMDEADKLLSPEFQPSIEQLIQFLPPTRQILMFSATFPVTVKDFKDRYLRKPY 339
+C+ VMDEADKLLSPEF P IEQL+ + P RQI ++SATFP+ VK+F D++L KPY
Sbjct: 184 FSECTTFVMDEADKLLSPEFTPIIEQLLSYFPKNRQISLYSATFPLIVKNFMDKHLNKPY 243
Query: 340 IINLMDELTLKGITQFYAFVEERQKVHCLNTLFSKAANKPIHHLLQFGKSS*TPC*ENH* 399
INLMDELTL+G+TQ+YAFV+E QKVHCLNTLFSK I+ + F S T E
Sbjct: 244 EINLMDELTLRGVTQYYAFVDESQKVHCLNTLFSKL---QINQSIIFCNS--TNRVELLA 298
Query: 400 TRVFMFLYSC-----KDVARPP**SFPRLPQWCMQESRLYCLFTRGIDIQAVNVVINFDF 454
++ YSC K + F + L TRGIDIQAVNVVINFDF
Sbjct: 299 KKITELGYSCFYSHAKMLQSHRNRVFHNFRNGVCRNLVCSDLLTRGIDIQAVNVVINFDF 358
Query: 455 PKNSETYLHRVGRSGRFGHLGLAVNLITYEDRFNLYRIEQELGTEIKQIPPFIDQAIY 512
PKN+ETYLHR+GRSGRFGH GLA++ I++ DRFNLYRIE ELGTEI+ IPP ID ++Y
Sbjct: 359 PKNAETYLHRIGRSGRFGHRGLAISFISWADRFNLYRIENELGTEIQPIPPSIDPSLY 416
>DHH1_YEAST (P39517) Putative ATP-dependent RNA helicase DHH1
Length = 506
Score = 483 bits (1243), Expect = e-136
Identities = 256/419 (61%), Positives = 309/419 (73%), Gaps = 24/419 (5%)
Query: 106 SEANDSSSQDWKARLKLPPADTRYRTEDVTATKGNEFEDYFLKRELLMGIYEKGFERPSP 165
+ +N +DWK L +P DTR +T+DV TKGN FED++LKRELLMGI+E GFE+PSP
Sbjct: 12 NNSNTDLDRDWKTALNIPKKDTRPQTDDVLNTKGNTFEDFYLKRELLMGIFEAGFEKPSP 71
Query: 166 IQEESIPIALTGSDILARAKNGTGKTAAFSIPALEKIDQDNNIIQVVILVPTRELALQTS 225
IQEE+IP+A+TG DILARAKNGTGKTAAF IP LEK+ N IQ +I+VPTRELALQTS
Sbjct: 72 IQEEAIPVAITGRDILARAKNGTGKTAAFVIPTLEKVKPKLNKIQALIMVPTRELALQTS 131
Query: 226 QVCKELGKHLQIQVMVTTGGTSLKDDIMRLYQPVHLLVGTPGRILDLAKKGVCVLKDCSM 285
QV + LGKH I MVTTGGT+L+DDI+RL + VH+LVGTPGR+LDLA + V L DCS+
Sbjct: 132 QVVRTLGKHCGISCMVTTGGTNLRDDILRLNETVHILVGTPGRVLDLASRKVADLSDCSL 191
Query: 286 LVMDEADKLLSPEFQPSIEQLIQFLPPTRQILMFSATFPVTVKDFKDRYLRKPYIINLMD 345
+MDEADK+LS +F+ IEQ++ FLPPT Q L+FSATFP+TVK+F ++L KPY INLM+
Sbjct: 192 FIMDEADKMLSRDFKTIIEQILSFLPPTHQSLLFSATFPLTVKEFMVKHLHKPYEINLME 251
Query: 346 ELTLKGITQFYAFVEERQKVHCLNTLFSKAANKPIHHLLQFGKSS*TPC*ENH*TRVFMF 405
ELTLKGITQ+YAFVEERQK+HCLNTLFSK I+ + F S+ RV +
Sbjct: 252 ELTLKGITQYYAFVEERQKLHCLNTLFSKL---QINQAIIFCNST---------NRVELL 299
Query: 406 LYSCKDVARPP**SFPRLPQW----CMQESR------LYC--LFTRGIDIQAVNVVINFD 453
D+ S R+ Q E R L C L TRGIDIQAVNVVINFD
Sbjct: 300 AKKITDLGYSCYYSHARMKQQERNKVFHEFRQGKVRTLVCSDLLTRGIDIQAVNVVINFD 359
Query: 454 FPKNSETYLHRVGRSGRFGHLGLAVNLITYEDRFNLYRIEQELGTEIKQIPPFIDQAIY 512
FPK +ETYLHR+GRSGRFGHLGLA+NLI + DRFNLY+IEQELGTEI IP ID+++Y
Sbjct: 360 FPKTAETYLHRIGRSGRFGHLGLAINLINWNDRFNLYKIEQELGTEIAAIPATIDKSLY 418
>ME31_DROME (P23128) Putative ATP-dependent RNA helicase me31b
(Maternal expression at 31B)
Length = 459
Score = 478 bits (1229), Expect = e-134
Identities = 253/431 (58%), Positives = 314/431 (72%), Gaps = 10/431 (2%)
Query: 87 QLGGGTDTNVVEEVEKTVQSEANDSSSQDWKARLKLPPADTRYRTEDVTATKGNEFEDYF 146
+L G + + +Q N S WK++LKLPP D R++T DVT T+GNEFE++
Sbjct: 5 KLNSGHTNLTSKGIINDLQIAGNTSDDMGWKSKLKLPPKDNRFKTTDVTDTRGNEFEEFC 64
Query: 147 LKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAKNGTGKTAAFSIPALEKIDQDN 206
LKRELLMGI+EKG+ERPSPIQE +IPIAL+G D+LARAKNGTGKT A+ IP LE+ID
Sbjct: 65 LKRELLMGIFEKGWERPSPIQEAAIPIALSGKDVLARAKNGTGKTGAYCIPVLEQIDPTK 124
Query: 207 NIIQVVILVPTRELALQTSQVCKELGKHLQIQVMVTTGGTSLKDDIMRLYQPVHLLVGTP 266
+ IQ +++VPTRELALQTSQ+C EL KHL I+VMVTTGGT LKDDI+R+YQ V L++ TP
Sbjct: 125 DYIQALVMVPTRELALQTSQICIELAKHLDIRVMVTTGGTILKDDILRIYQKVQLIIATP 184
Query: 267 GRILDLAKKGVCVLKDCSMLVMDEADKLLSPEFQPSIEQLIQFLPPTRQILMFSATFPVT 326
GRILDL K V + C +LV+DEADKLLS +FQ ++ +I LP QIL+FSATFP+T
Sbjct: 185 GRILDLMDKKVADMSHCRILVLDEADKLLSLDFQGMLDHVILKLPKDPQILLFSATFPLT 244
Query: 327 VKDFKDRYLRKPYIINLMDELTLKGITQFYAFVEERQKVHCLNTLFSKAANKPIHHLLQF 386
VK+F +++LR+PY INLM+ELTLKG+TQ+YAFV+ERQKVHCLNTLFSK I+ + F
Sbjct: 245 VKNFMEKHLREPYEINLMEELTLKGVTQYYAFVQERQKVHCLNTLFSKL---QINQSIIF 301
Query: 387 GKSS*TPC*ENH*TRVFMFLYSC-----KDVARPP**SFPRLPQWCMQESRLYCLFTRGI 441
S T E ++ Y C K F Q + LFTRGI
Sbjct: 302 CNS--TQRVELLAKKITELGYCCYYIHAKMAQAHRNRVFHDFRQGLCRNLVCSDLFTRGI 359
Query: 442 DIQAVNVVINFDFPKNSETYLHRVGRSGRFGHLGLAVNLITYEDRFNLYRIEQELGTEIK 501
D+QAVNVVINFDFP+ +ETYLHR+GRSGRFGHLG+A+NLITYEDRF+L+RIE+ELGTEIK
Sbjct: 360 DVQAVNVVINFDFPRMAETYLHRIGRSGRFGHLGIAINLITYEDRFDLHRIEKELGTEIK 419
Query: 502 QIPPFIDQAIY 512
IP ID A+Y
Sbjct: 420 PIPKVIDPALY 430
>DDX6_XENLA (P54824) ATP-dependent RNA helicase p54 (Xp54)
Length = 481
Score = 458 bits (1179), Expect = e-128
Identities = 254/452 (56%), Positives = 313/452 (69%), Gaps = 44/452 (9%)
Query: 89 GGGTDTNVVEEVEKTV------QSEANDSSS-----QDWKARLKLPPADTRYRTEDVTAT 137
GGGT T + +++ Q +A SS DWK LKLPP D R +T DVT+T
Sbjct: 33 GGGTQTQQINQLKNASTINSGSQQQAQSMSSIIKPGDDWKKTLKLPPKDLRIKTSDVTST 92
Query: 138 KGNEFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAKNGTGKTAAFSIP 197
KGNEFEDY LKRELLMGI+E G+E+PSPIQEESIPIAL+G DILARAKNGTGKT A+ IP
Sbjct: 93 KGNEFEDYCLKRELLMGIFEMGWEKPSPIQEESIPIALSGRDILARAKNGTGKTGAYLIP 152
Query: 198 ALEKIDQDNNIIQVVILVPTRELALQTSQVCKELGKHLQ-IQVMVTTGGTSLKDDIMRLY 256
LE++D + IQ +++VPTRELALQ SQ+C ++ KH+ +VM TTGGT+L+DDIMRL
Sbjct: 153 LLERLDLKKDCIQAMVIVPTRELALQVSQICIQVSKHMGGAKVMATTGGTNLRDDIMRLD 212
Query: 257 QPVHLLVGTPGRILDLAKKGVCVLKDCSMLVMDEADKLLSPEFQPSIEQLIQFLPPTRQI 316
VH+++ TPGRILDL KKGV + M+V+DEADKLLS +F +E +I LP RQI
Sbjct: 213 DTVHVVIATPGRILDLIKKGVAKVDHIQMIVLDEADKLLSQDFMQIMEDIIMTLPKNRQI 272
Query: 317 LMFSATFPVTVKDFKDRYLRKPYIINLMDELTLKGITQFYAFVEERQKVHCLNTLFSKAA 376
L++SATFP++V+ F +L+KPY INLM+ELTLKG+TQ+YA+V ERQKVHCLNTLFS+
Sbjct: 273 LLYSATFPLSVQKFMTLHLQKPYEINLMEELTLKGVTQYYAYVTERQKVHCLNTLFSRL- 331
Query: 377 NKPIHHLLQFGKSS*TPC*ENH*TRVFMFLYSCKDVARPP**SFPRLPQWCMQESR---- 432
I+ + F SS E ++ YSC + QE R
Sbjct: 332 --QINQSIIFCNSSQRV--ELLAKKISQLGYSCFYIHAK-----------MRQEHRNRVF 376
Query: 433 ----------LYC--LFTRGIDIQAVNVVINFDFPKNSETYLHRVGRSGRFGHLGLAVNL 480
L C LFTRGIDIQAVNVVINFDFPK +ETYLHR+GRSGRFGHLGLA+NL
Sbjct: 377 HDFRNGLCRNLVCTDLFTRGIDIQAVNVVINFDFPKLAETYLHRIGRSGRFGHLGLAINL 436
Query: 481 ITYEDRFNLYRIEQELGTEIKQIPPFIDQAIY 512
ITY+DRFNL IE++LGTEIK IP ID+ +Y
Sbjct: 437 ITYDDRFNLKSIEEQLGTEIKPIPSSIDKNLY 468
>DDX6_HUMAN (P26196) Probable ATP-dependent RNA helicase p54
(Oncogene RCK) (DEAD-box protein 6)
Length = 483
Score = 456 bits (1173), Expect = e-128
Identities = 253/456 (55%), Positives = 314/456 (68%), Gaps = 40/456 (8%)
Query: 74 QQQQQQQQWLRRNQLGGGTDTNVVEEVEKTVQSEANDSSSQDWKARLKLPPADTRYRTED 133
Q QQQ Q N + GT + + T++ DWK LKLPP D R +T D
Sbjct: 37 QTQQQMNQLKNTNTINNGTQQQA-QSMTTTIKP------GDDWKKTLKLPPKDLRIKTSD 89
Query: 134 VTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAKNGTGKTAA 193
VT+TKGNEFEDY LKRELLMGI+E G+E+PSPIQEESIPIAL+G DILARAKNGTGK+ A
Sbjct: 90 VTSTKGNEFEDYCLKRELLMGIFEMGWEKPSPIQEESIPIALSGRDILARAKNGTGKSGA 149
Query: 194 FSIPALEKIDQDNNIIQVVILVPTRELALQTSQVCKELGKHLQ-IQVMVTTGGTSLKDDI 252
+ IP LE++D + IQ +++VPTRELALQ SQ+C ++ KH+ +VM TTGGT+L+DDI
Sbjct: 150 YLIPLLERLDLKKDNIQAMVIVPTRELALQVSQICIQVSKHMGGAKVMATTGGTNLRDDI 209
Query: 253 MRLYQPVHLLVGTPGRILDLAKKGVCVLKDCSMLVMDEADKLLSPEFQPSIEQLIQFLPP 312
MRL VH+++ TPGRILDL KKGV + M+V+DEADKLLS +F +E +I LP
Sbjct: 210 MRLDDTVHVVIATPGRILDLIKKGVAKVDHVQMIVLDEADKLLSQDFVQIMEDIILTLPK 269
Query: 313 TRQILMFSATFPVTVKDFKDRYLRKPYIINLMDELTLKGITQFYAFVEERQKVHCLNTLF 372
RQIL++SATFP++V+ F + +L KPY INLM+ELTLKG+TQ+YA+V ERQKVHCLNTLF
Sbjct: 270 NRQILLYSATFPLSVQKFMNSHLEKPYEINLMEELTLKGVTQYYAYVTERQKVHCLNTLF 329
Query: 373 SKAANKPIHHLLQFGKSS*TPC*ENH*TRVFMFLYSCKDVARPP**SFPRLPQWCMQESR 432
S+ I+ + F SS E ++ YSC + QE R
Sbjct: 330 SRL---QINQSIIFCNSSQRV--ELLAKKISQLGYSCFYIHAK-----------MRQEHR 373
Query: 433 --------------LYC--LFTRGIDIQAVNVVINFDFPKNSETYLHRVGRSGRFGHLGL 476
L C LFTRGIDIQAVNVVINFDFPK +ETYLHR+GRSGRFGHLGL
Sbjct: 374 NRVFHDFRNGLCRNLVCTDLFTRGIDIQAVNVVINFDFPKLAETYLHRIGRSGRFGHLGL 433
Query: 477 AVNLITYEDRFNLYRIEQELGTEIKQIPPFIDQAIY 512
A+NLITY+DRFNL IE++LGTEIK IP ID+++Y
Sbjct: 434 AINLITYDDRFNLKSIEEQLGTEIKPIPSNIDKSLY 469
>DDX6_MOUSE (P54823) Probable ATP-dependent RNA helicase p54
(Oncogene RCK homolog) (DEAD-box protein 6)
Length = 483
Score = 452 bits (1162), Expect = e-126
Identities = 251/456 (55%), Positives = 312/456 (68%), Gaps = 40/456 (8%)
Query: 74 QQQQQQQQWLRRNQLGGGTDTNVVEEVEKTVQSEANDSSSQDWKARLKLPPADTRYRTED 133
Q Q Q Q + + GT ++ A DWK LKLPP D R +T D
Sbjct: 37 QPQPQLNQLKNTSTINNGTP-------QQAQSMAATIKPGDDWKKTLKLPPKDLRIKTSD 89
Query: 134 VTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAKNGTGKTAA 193
VT+TKGNEFEDY LKRELLMGI+E G+E+PSPIQEESIPIAL+G DILARAKNGTGK+ A
Sbjct: 90 VTSTKGNEFEDYCLKRELLMGIFEMGWEKPSPIQEESIPIALSGRDILARAKNGTGKSGA 149
Query: 194 FSIPALEKIDQDNNIIQVVILVPTRELALQTSQVCKELGKHLQ-IQVMVTTGGTSLKDDI 252
+ IP LE++D + IQ +++VPTRELALQ SQ+C ++ KH+ +VM TTGGT+L+DDI
Sbjct: 150 YLIPLLERLDLKKDNIQAMVIVPTRELALQVSQICIQVSKHMGGAKVMATTGGTNLRDDI 209
Query: 253 MRLYQPVHLLVGTPGRILDLAKKGVCVLKDCSMLVMDEADKLLSPEFQPSIEQLIQFLPP 312
MRL VH+++ TPGRILDL KKGV + M+V+DEADKLLS +F +E +I LP
Sbjct: 210 MRLDDTVHVVIATPGRILDLIKKGVAKVDHVQMIVLDEADKLLSQDFVQIMEDIILTLPK 269
Query: 313 TRQILMFSATFPVTVKDFKDRYLRKPYIINLMDELTLKGITQFYAFVEERQKVHCLNTLF 372
RQIL++SATFP++V+ F + +L+KPY INLM+ELTLKG+TQ+YA+V ERQKVHCLNTLF
Sbjct: 270 NRQILLYSATFPLSVQKFMNSHLQKPYEINLMEELTLKGVTQYYAYVTERQKVHCLNTLF 329
Query: 373 SKAANKPIHHLLQFGKSS*TPC*ENH*TRVFMFLYSCKDVARPP**SFPRLPQWCMQESR 432
S+ I+ + F SS E ++ YSC + QE R
Sbjct: 330 SRL---QINQSIIFCNSSQRV--ELLAKKISQLGYSCFYIHAK-----------MRQEHR 373
Query: 433 --------------LYC--LFTRGIDIQAVNVVINFDFPKNSETYLHRVGRSGRFGHLGL 476
L C LFTRGIDIQAVNVVINFDFPK +ETYLHR+GRSGRFGHLGL
Sbjct: 374 NRVFHDFRNGLCRNLVCTDLFTRGIDIQAVNVVINFDFPKLAETYLHRIGRSGRFGHLGL 433
Query: 477 AVNLITYEDRFNLYRIEQELGTEIKQIPPFIDQAIY 512
A+NLITY+DRFNL IE++LGTEIK IP ID+++Y
Sbjct: 434 AINLITYDDRFNLKSIEEQLGTEIKPIPSNIDKSLY 469
>IF4A_CAEEL (P27639) Eukaryotic initiation factor 4A (eIF4A)
(eIF-4A)
Length = 402
Score = 240 bits (612), Expect = 7e-63
Identities = 140/375 (37%), Positives = 215/375 (57%), Gaps = 8/375 (2%)
Query: 142 FEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAKNGTGKTAAFSIPALEK 201
F+D LK ELL GIY GFE+PS IQ+ +I TG D++A+A++GTGKTA FS+ L++
Sbjct: 31 FDDMELKEELLRGIYGFGFEKPSAIQKRAIVPCTTGKDVIAQAQSGTGKTATFSVSILQR 90
Query: 202 IDQDNNIIQVVILVPTRELALQTSQVCKELGKHLQIQVMVTTGGTSLKDDIMRLYQPVHL 261
ID ++ +Q +++ PTRELA Q +V LG++L + ++ GGTS++DD +L +H+
Sbjct: 91 IDHEDPHVQALVMAPTRELAQQIQKVMSALGEYLNVNILPCIGGTSVRDDQRKLEAGIHV 150
Query: 262 LVGTPGRILDLAKKGVCVLKDCSMLVMDEADKLLSPEFQPSIEQLIQFLPPTRQILMFSA 321
+VGTPGR+ D+ + M V+DEAD++LS F+ I ++ + +P Q+++ SA
Sbjct: 151 VVGTPGRVGDMINRNALDTSRIKMFVLDEADEMLSRGFKDQIYEVFRSMPQDVQVVLLSA 210
Query: 322 TFPVTVKDFKDRYLRKPY-IINLMDELTLKGITQFYAFVEERQ-KVHCLNTLFSKAANKP 379
T P V D +R++R P I+ DELTL+GI QFY V++ + K CL L++
Sbjct: 211 TMPSEVLDVTNRFMRNPIRILVKKDELTLEGIRQFYINVQKDEWKFDCLCDLYNVV---N 267
Query: 380 IHHLLQFGKSS*TPC*ENH*TRVFMFLYSCKDVARPP**SFPRLPQWCMQESRLYC---L 436
+ + F + F SC + ++ SR+ +
Sbjct: 268 VTQAVIFCNTRRKVDTLTEKMTENQFTVSCLHGDMDQAERDTIMREFRSGSSRVLITTDI 327
Query: 437 FTRGIDIQAVNVVINFDFPKNSETYLHRVGRSGRFGHLGLAVNLITYEDRFNLYRIEQEL 496
RGID+Q V++VIN+D P N E Y+HR+GRSGRFG G+A+N +T D L IE
Sbjct: 328 LARGIDVQQVSLVINYDLPSNRENYIHRIGRSGRFGRKGVAINFVTENDARQLKEIESYY 387
Query: 497 GTEIKQIPPFIDQAI 511
T+I+++P I I
Sbjct: 388 TTQIEEMPESIADLI 402
>IF4A_CANAL (P87206) Eukaryotic initiation factor 4A (eIF4A)
(eIF-4A)
Length = 397
Score = 231 bits (588), Expect = 4e-60
Identities = 138/369 (37%), Positives = 209/369 (56%), Gaps = 8/369 (2%)
Query: 141 EFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAKNGTGKTAAFSIPALE 200
+F+D LK ++ GI+ G+E PS IQ+ +I G D+LA+A++GTGKTA F+I AL+
Sbjct: 24 KFDDLNLKPNIVRGIFGYGYETPSAIQQRAILPITEGRDVLAQAQSGTGKTATFTISALQ 83
Query: 201 KIDQDNNIIQVVILVPTRELALQTSQVCKELGKHLQIQVMVTTGGTSLKDDIMRLYQPVH 260
+I+++ Q +IL PTRELALQ V +G +L++ V + GGTS+ DDI V
Sbjct: 84 RINENEKATQALILAPTRELALQIKNVITAIGLYLKVTVHASIGGTSMSDDIEAFRSGVQ 143
Query: 261 LLVGTPGRILDLAKKGVCVLKDCSMLVMDEADKLLSPEFQPSIEQLIQFLPPTRQILMFS 320
++VGTPGR+LD+ ++ M ++DEAD++LS F+ I + + LP T QI++ S
Sbjct: 144 IVVGTPGRVLDMIERRYFKTDKVKMFILDEADEMLSSGFKEQIYNIFRLLPETTQIVLLS 203
Query: 321 ATFPVTVKDFKDRYLRKPY-IINLMDELTLKGITQFYAFVE-ERQKVHCLNTLFSKAANK 378
AT P V + +++ P I+ DELTL+GI QFY VE E K CL L+ +
Sbjct: 204 ATMPQDVLEVTTKFMNNPVRILVKKDELTLEGIKQFYINVELEDYKFDCLCDLYDSIS-- 261
Query: 379 PIHHLLQFGKSS*TPC*ENH*TRVFMFLYSCKDVARPP**SFPRLPQWCMQESRLYC--- 435
+ + F + + R F S P + ++ SR+
Sbjct: 262 -VTQAVIFCNTRSKVEFLTNKLREQHFTVSAIHADLPQAERDTIMKEFRSGSSRILISTD 320
Query: 436 LFTRGIDIQAVNVVINFDFPKNSETYLHRVGRSGRFGHLGLAVNLITYEDRFNLYRIEQE 495
L RGID+Q V++VIN+D P N E Y+HR+GR GRFG G+A+N +T D + IE+
Sbjct: 321 LLARGIDVQQVSLVINYDLPANKENYIHRIGRGGRFGRKGVAINFVTDRDVGMMREIEKF 380
Query: 496 LGTEIKQIP 504
T+I+++P
Sbjct: 381 YSTQIEEMP 389
>IF4A_LEIBR (Q25225) Probable eukaryotic initiation factor 4A
(eIF4A) (eIF-4A)
Length = 403
Score = 230 bits (586), Expect = 8e-60
Identities = 145/389 (37%), Positives = 214/389 (54%), Gaps = 34/389 (8%)
Query: 142 FEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAKNGTGKTAAFSIPALEK 201
F+D L + LL GIY GFE+PS IQ+ +I G DI+A+A++GTGKT AFSI L++
Sbjct: 28 FDDMPLHQNLLRGIYSYGFEKPSSIQQRAIAPFTRGGDIIAQAQSGTGKTGAFSIGLLQR 87
Query: 202 IDQDNNIIQVVILVPTRELALQTSQVCKELGKHLQIQVMVT---TGGTSLKDDIMRLYQP 258
+D +N+IQ ++L PTRELALQT++V +G+ L GGT ++DD+ +L
Sbjct: 88 LDFRHNLIQGLVLSPTRELALQTAEVISRIGEFLSNSAKFCETFVGGTRVQDDLRKLQAG 147
Query: 259 VHLLVGTPGRILDLAKKGVCVLKDCSMLVMDEADKLLSPEFQPSIEQLIQFLPPTRQILM 318
V + VGTPGR+ D+ K+G + +LV+DEAD++LS F I ++ +FLP Q+ +
Sbjct: 148 VVVAVGTPGRVSDVIKRGALRTESLRVLVLDEADEMLSQGFADQIYEIFRFLPKDIQVAL 207
Query: 319 FSATFPVTVKDFKDRYLRKPY-IINLMDELTLKGITQFYAFVEERQKVHCL--------- 368
FSAT P V + +++R P I+ + LTL+GI QF+ VEE K+ L
Sbjct: 208 FSATMPEEVLELTKKFMRDPVRILVKRESLTLEGIKQFFIAVEEEHKLDTLMDLYETVSI 267
Query: 369 --NTLFSKAANKPIHHLLQFGKSS*TPC*ENH*TRVFMFLYSCKDVARPP**SFPRLPQW 426
+ +F+ K + +S+ T S P + +
Sbjct: 268 AQSVIFANTRRKVDWIAEKLNQSNHT--------------VSSMHAEMPKSDRERVMNTF 313
Query: 427 CMQESRLYC---LFTRGIDIQAVNVVINFDFPKNSETYLHRVGRSGRFGHLGLAVNLITY 483
SR+ L RGID+ VN+VINFD P N E YLHR+GR GR+G G+A+N +T
Sbjct: 314 RSGSSRVLVTTDLVARGIDVHHVNIVINFDLPTNKENYLHRIGRGGRYGVKGVAINFVTE 373
Query: 484 EDRFNLYRIEQELGTEIKQIPPFIDQAIY 512
+D L+ IE T+I ++P +D A Y
Sbjct: 374 KDVELLHEIEGHYHTQIDELP--VDFAAY 400
>IF4A_YEAST (P10081) Eukaryotic initiation factor 4A (eIF4A)
(eIF-4A) (Stimulator factor I 37 kDa component) (p37)
Length = 394
Score = 226 bits (576), Expect = 1e-58
Identities = 139/369 (37%), Positives = 205/369 (54%), Gaps = 9/369 (2%)
Query: 141 EFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAKNGTGKTAAFSIPALE 200
+F+D L LL G++ GFE PS IQ+ +I + G D+LA+A++GTGKT FSI AL+
Sbjct: 22 KFDDMELDENLLRGVFGYGFEEPSAIQQRAIMPIIEGHDVLAQAQSGTGKTGTFSIAALQ 81
Query: 201 KIDQDNNIIQVVILVPTRELALQTSQVCKELGKHLQIQVMVTTGGTSLKDDIMRLYQPVH 260
+ID Q ++L PTRELALQ +V L H+ I+V GGTS +D L +
Sbjct: 82 RIDTSVKAPQALMLAPTRELALQIQKVVMALAFHMDIKVHACIGGTSFVEDAEGL-RDAQ 140
Query: 261 LLVGTPGRILDLAKKGVCVLKDCSMLVMDEADKLLSPEFQPSIEQLIQFLPPTRQILMFS 320
++VGTPGR+ D ++ M ++DEAD++LS F+ I Q+ LPPT Q+++ S
Sbjct: 141 IVVGTPGRVFDNIQRRRFRTDKIKMFILDEADEMLSSGFKEQIYQIFTLLPPTTQVVLLS 200
Query: 321 ATFPVTVKDFKDRYLRKPY-IINLMDELTLKGITQFYAFVEERQ-KVHCLNTLFSKAANK 378
AT P V + +++R P I+ DELTL+GI QFY VEE + K CL L+ +
Sbjct: 201 ATMPNDVLEVTTKFMRNPVRILVKKDELTLEGIKQFYVNVEEEEYKYECLTDLYDSIS-- 258
Query: 379 PIHHLLQFGKSS*TPC*ENH*TRVFMFLYSCKDVARPP**SFPRLPQWCMQESRLYC--- 435
+ + F + R F S P + ++ SR+
Sbjct: 259 -VTQAVIFCNTRRKVEELTTKLRNDKFTVSAIYSDLPQQERDTIMKEFRSGSSRILISTD 317
Query: 436 LFTRGIDIQAVNVVINFDFPKNSETYLHRVGRSGRFGHLGLAVNLITYEDRFNLYRIEQE 495
L RGID+Q V++VIN+D P N E Y+HR+GR GRFG G+A+N +T ED + +E+
Sbjct: 318 LLARGIDVQQVSLVINYDLPANKENYIHRIGRGGRFGRKGVAINFVTNEDVGAMRELEKF 377
Query: 496 LGTEIKQIP 504
T+I+++P
Sbjct: 378 YSTQIEELP 386
>IF4A_DROME (Q02748) Eukaryotic initiation factor 4A (eIF4A)
(eIF-4A)
Length = 403
Score = 226 bits (576), Expect = 1e-58
Identities = 141/376 (37%), Positives = 209/376 (55%), Gaps = 6/376 (1%)
Query: 140 NEFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAKNGTGKTAAFSIPAL 199
+ F+D L+ ELL GIY GFE+PS IQ+ +I + G D++A+A++GTGKTA FSI L
Sbjct: 30 DNFDDMNLREELLRGIYGYGFEKPSAIQQRAIIPCVRGRDVIAQAQSGTGKTATFSIAIL 89
Query: 200 EKIDQDNNIIQVVILVPTRELALQTSQVCKELGKHLQIQVMVTTGGTSLKDDIMRLYQPV 259
++ID Q +IL PTRELA Q +V LG+++++ GGT++++D L
Sbjct: 90 QQIDTSIRECQALILAPTRELATQIQRVVMALGEYMKVHSHACIGGTNVREDARILESGC 149
Query: 260 HLLVGTPGRILDLAKKGVCVLKDCSMLVMDEADKLLSPEFQPSIEQLIQFLPPTRQILMF 319
H++VGTPGR+ D+ + V + + V+DEAD++LS F+ I+ + + LPP Q+++
Sbjct: 150 HVVVGTPGRVYDMINRKVLRTQYIKLFVLDEADEMLSRGFKDQIQDVFKMLPPDVQVILL 209
Query: 320 SATFPVTVKDFKDRYLRKPY-IINLMDELTLKGITQFYAFVEERQKVHCLNTLFSKAANK 378
SAT P V + ++R P I+ +ELTL+GI QFY V +Q+ L TL
Sbjct: 210 SATMPPDVLEVSRCFMRDPVSILVKKEELTLEGIKQFY--VNVKQENWKLGTLCDLYDTL 267
Query: 379 PIHHLLQFGKSS*TPC*ENH*TRVFMFLYSCKDVARPP**SFPRLPQWCMQESRLYC--- 435
I + F + + F S + Q+ SR+
Sbjct: 268 SITQSVIFCNTRRKVDQLTQEMSIHNFTVSAMHGDMEQRDREVIMKQFRSGSSRVLITTD 327
Query: 436 LFTRGIDIQAVNVVINFDFPKNSETYLHRVGRSGRFGHLGLAVNLITYEDRFNLYRIEQE 495
L RGID+Q V++VIN+D P N E Y+HR+GR GRFG G+A+N IT +DR L IEQ
Sbjct: 328 LLARGIDVQQVSLVINYDLPSNRENYIHRIGRGGRFGRKGVAINFITDDDRRILKDIEQF 387
Query: 496 LGTEIKQIPPFIDQAI 511
T I+++P I I
Sbjct: 388 YHTTIEEMPANIADLI 403
>IF4N_SCHPO (Q10055) Eukaryotic initiation factor 4A-like protein
C1F5.10
Length = 394
Score = 226 bits (575), Expect = 1e-58
Identities = 142/378 (37%), Positives = 210/378 (54%), Gaps = 8/378 (2%)
Query: 131 TEDVTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAKNGTGK 190
+EDV A + FE+ LK +LL GIY G+E PS +Q +I G D++A+A++GTGK
Sbjct: 14 SEDVNAV--SSFEEMNLKEDLLRGIYAYGYETPSAVQSRAIIQICKGRDVIAQAQSGTGK 71
Query: 191 TAAFSIPALEKIDQDNNIIQVVILVPTRELALQTSQVCKELGKHLQIQVMVTTGGTSLKD 250
TA FSI L+ ID Q +IL PTRELA+Q V LG H+ +Q GGTS+ +
Sbjct: 72 TATFSIGILQSIDLSVRDTQALILSPTRELAVQIQNVVLALGDHMNVQCHACIGGTSVGN 131
Query: 251 DIMRLYQPVHLLVGTPGRILDLAKKGVCVLKDCSMLVMDEADKLLSPEFQPSIEQLIQFL 310
DI +L H++ GTPGR+ D+ ++ ++ ML++DEAD+LL+ F+ I + ++L
Sbjct: 132 DIKKLDYGQHVVSGTPGRVTDMIRRRNLRTRNVKMLILDEADELLNQGFKEQIYDIYRYL 191
Query: 311 PPTRQILMFSATFPVTVKDFKDRYLRKPY-IINLMDELTLKGITQFYAFVEERQKVHCLN 369
PP Q+++ SAT P V + +++ P I+ DELTL+G+ Q++ VE+ + +
Sbjct: 192 PPGTQVVVVSATLPQDVLEMTNKFTTNPVRILVKRDELTLEGLKQYFIAVEKEE--WKFD 249
Query: 370 TLFSKAANKPIHHLLQFGKSS*TPC*ENH*TRVFMFLYSCKDVARPP**SFPRLPQWCMQ 429
TL I + F S R F + P + +
Sbjct: 250 TLCDLYDTLTITQAVIFCNSRRKVDWLTEKMREANFTVTSMHGEMPQKERDAIMQDFRQG 309
Query: 430 ESR-LYC--LFTRGIDIQAVNVVINFDFPKNSETYLHRVGRSGRFGHLGLAVNLITYEDR 486
SR L C ++ RGID+Q V++VIN+D P N E Y+HR+GRSGRFG G+A+N +T ED
Sbjct: 310 NSRVLICTDIWARGIDVQQVSLVINYDLPANRENYIHRIGRSGRFGRKGVAINFVTNEDV 369
Query: 487 FNLYRIEQELGTEIKQIP 504
L IEQ T I ++P
Sbjct: 370 RILRDIEQYYSTVIDEMP 387
>IF4A_SCHPO (P47943) Eukaryotic initiation factor 4A (eIF4A)
(eIF-4A)
Length = 392
Score = 225 bits (574), Expect = 2e-58
Identities = 143/371 (38%), Positives = 208/371 (55%), Gaps = 14/371 (3%)
Query: 142 FEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAKNGTGKTAAFSIPALEK 201
F+D LK ELL GIY GFERPS IQ+ +I L D+LA+A++GTGKTA FSI L+K
Sbjct: 21 FDDMNLKPELLRGIYAYGFERPSAIQQRAIMPILGERDVLAQAQSGTGKTATFSISVLQK 80
Query: 202 IDQDNNIIQVVILVPTRELALQTSQVCKELGKHLQIQVMVTTGGTSLKDDIMRLYQPVHL 261
ID Q +IL PTRELA Q +V LG + ++ GGT ++DD+ L VH+
Sbjct: 81 IDTSLKQCQALILAPTRELAQQIQKVVVALGDLMNVECHACIGGTLVRDDMAALQAGVHV 140
Query: 262 LVGTPGRILDLAKKGVCVLKDCSMLVMDEADKLLSPEFQPSIEQLIQFLPPTRQILMFSA 321
+VGTPGR+ D+ ++ M V+DEAD++LS F+ I + Q LPPT Q+++ SA
Sbjct: 141 VVGTPGRVHDMIQRRALPTDAVQMFVLDEADEMLSRGFKDQIYDIFQLLPPTAQVVLLSA 200
Query: 322 TFPVTVKDFKDRYLRKPY-IINLMDELTLKGITQFYAFVEERQKVHCLNTLFSKAANKPI 380
T P V + +++R P I+ DELTL+GI QF +V ++ L+TL +
Sbjct: 201 TMPQDVLEVTTKFMRDPIRILVKKDELTLEGIKQF--YVAVEKEEWKLDTLCDLYETVTV 258
Query: 381 HHLLQFGKSS*TP--C*ENH*TRVFMF--LYSCKDVARPP**SFPRLPQWCMQESRLYC- 435
+ F + E R F ++ D A+ + ++ SR+
Sbjct: 259 TQAVIFCNTRRKVDWLTEQLTERDFTVSSMHGDMDQAQRD----TLMHEFRTGSSRILIT 314
Query: 436 --LFTRGIDIQAVNVVINFDFPKNSETYLHRVGRSGRFGHLGLAVNLITYEDRFNLYRIE 493
L RGID+Q V++VIN+D P N E Y+HR+GR GRFG G+++N +T +D + IE
Sbjct: 315 TDLLARGIDVQQVSLVINYDLPANRENYIHRIGRGGRFGRKGVSINFVTNDDVRMMREIE 374
Query: 494 QELGTEIKQIP 504
Q T I+++P
Sbjct: 375 QFYNTHIEEMP 385
>IF4A_CRYPV (O02494) Eukaryotic initiation factor 4A (eIF4A)
(eIF-4A)
Length = 405
Score = 223 bits (569), Expect = 7e-58
Identities = 138/378 (36%), Positives = 211/378 (55%), Gaps = 28/378 (7%)
Query: 142 FEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAKNGTGKTAAFSIPALEK 201
FE L+ +LL GI+ GFE+PS IQ+ I L G D + +A++GTGKTA F I AL+K
Sbjct: 33 FEALNLEGDLLRGIFAYGFEKPSAIQQRGIKPILDGYDTIGQAQSGTGKTATFVIAALQK 92
Query: 202 IDQDNNIIQVVILVPTRELALQTSQVCKELGKHLQIQVMVTTGGTSLKDDIMRLYQPVHL 261
ID N QV++L PTRELA Q +V LG + +++ GGTS++DD+ +L VH+
Sbjct: 93 IDYSLNACQVLLLAPTRELAQQIQKVALALGDYCELRCHACVGGTSVRDDMNKLKSGVHM 152
Query: 262 LVGTPGRILDLAKKGVCVLKDCSMLVMDEADKLLSPEFQPSIEQLIQFLPPTRQILMFSA 321
+VGTPGR+ D+ KG + + + ++DEAD++LS F+ I + + LP Q+ +FSA
Sbjct: 153 VVGTPGRVFDMLDKGYLRVDNLKLFILDEADEMLSRGFKVQIHDIFKKLPQDVQVALFSA 212
Query: 322 TFPVTVKDFKDRYLRKPY-IINLMDELTLKGITQFYAFV-EERQKVHCLNTLFSKAA--- 376
T P + +++R P I+ +ELTL+GI QFY V ++ K+ L L+
Sbjct: 213 TMPNEILHLTTQFMRDPKRILVKQEELTLEGIRQFYVGVEKDEWKMDTLIDLYETLTIVQ 272
Query: 377 -------NKPIHHLLQFGKSS*TPC*ENH*TRVFMFLYSCKDVARPP**SFPRLPQWCMQ 429
+ + L + + C H M + + R Q+
Sbjct: 273 AIIYCNTRRRVDQLTKQMRERDFTCSSMHGD---MDQKDREVIMR----------QFRSG 319
Query: 430 ESRLYC---LFTRGIDIQAVNVVINFDFPKNSETYLHRVGRSGRFGHLGLAVNLITYEDR 486
SR+ L RGID+Q V++VIN+D P + ETY+HR+GRSGRFG G+++N +T +D
Sbjct: 320 SSRVLITTDLLARGIDVQQVSLVINYDLPVSPETYIHRIGRSGRFGKKGVSINFVTDDDI 379
Query: 487 FNLYRIEQELGTEIKQIP 504
L IE+ T+I+++P
Sbjct: 380 VCLRDIERHYNTQIEEMP 397
>DD48_MOUSE (Q91VC3) Probable ATP-dependent helicase DDX48 (DEAD-box
protein 48)
Length = 411
Score = 214 bits (546), Expect = 3e-55
Identities = 137/409 (33%), Positives = 220/409 (53%), Gaps = 19/409 (4%)
Query: 108 ANDSSSQDWKARLKLPPADTRYRTE-------DVTATKGNEFEDYFLKRELLMGIYEKGF 160
AN + + AR +L + + E DVT T F+ L+ +LL GIY GF
Sbjct: 3 ANATMATSGSARKRLLKEEDMTKVEFETSEEVDVTPT----FDTMGLREDLLRGIYAYGF 58
Query: 161 ERPSPIQEESIPIALTGSDILARAKNGTGKTAAFSIPALEKIDQDNNIIQVVILVPTREL 220
E+PS IQ+ +I + G D++A++++GTGKTA FS+ L+ +D Q +IL PTREL
Sbjct: 59 EKPSAIQQRAIKQIIKGRDVIAQSQSGTGKTATFSVSVLQCLDIQVRETQALILAPTREL 118
Query: 221 ALQTSQVCKELGKHLQIQVMVTTGGTSLKDDIMRLYQPVHLLVGTPGRILDLAKKGVCVL 280
A+Q + LG ++ +Q GGT++ +DI +L H++ GTPGR+ D+ ++
Sbjct: 119 AVQIQKGLLALGDYMNVQCHACIGGTNVGEDIRKLDYGQHVVAGTPGRVFDMIRRRSLRT 178
Query: 281 KDCSMLVMDEADKLLSPEFQPSIEQLIQFLPPTRQILMFSATFPVTVKDFKDRYLRKPY- 339
+ MLV+DEAD++L+ F+ I + ++LPP Q+++ SAT P + + ++++ P
Sbjct: 179 RAIKMLVLDEADEMLNKGFKEQIYDVYRYLPPATQVVLISATLPHEILEMTNKFMTDPIR 238
Query: 340 IINLMDELTLKGITQFYAFVE-ERQKVHCLNTLFSKAANKPIHHLLQFGKSS*TPC*ENH 398
I+ DELTL+GI QF+ VE E K L L+ I + F +
Sbjct: 239 ILVKRDELTLEGIKQFFVAVEREEWKFDTLCDLYDTLT---ITQAVIFCNTKRKVDWLTE 295
Query: 399 *TRVFMFLYSCKDVARPP**SFPRLPQWCMQESRLYC---LFTRGIDIQAVNVVINFDFP 455
R F S P + ++ SR+ ++ RG+D+ V+++IN+D P
Sbjct: 296 KMREANFTVSSMHGDMPQKERESIMKEFRSGASRVLISTDVWARGLDVPQVSLIINYDLP 355
Query: 456 KNSETYLHRVGRSGRFGHLGLAVNLITYEDRFNLYRIEQELGTEIKQIP 504
N E Y+HR+GRSGR+G G+A+N + +D L IEQ T+I ++P
Sbjct: 356 NNRELYIHRIGRSGRYGRKGVAINFVKNDDIRILRDIEQYYSTQIDEMP 404
>DD48_HUMAN (P38919) Probable ATP-dependent helicase DDX48 (DEAD-box
protein 48) (Eukaryotic initiation factor 4A-like
NUK-34) (Nuclear matrix protein 265) (hNMP 265)
(Eukaryotic translation initiation factor 4A isoform 3)
Length = 411
Score = 214 bits (546), Expect = 3e-55
Identities = 132/377 (35%), Positives = 209/377 (55%), Gaps = 12/377 (3%)
Query: 133 DVTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAKNGTGKTA 192
DVT T F+ L+ +LL GIY GFE+PS IQ+ +I + G D++A++++GTGKTA
Sbjct: 35 DVTPT----FDTMGLREDLLRGIYAYGFEKPSAIQQRAIKQIIKGRDVIAQSQSGTGKTA 90
Query: 193 AFSIPALEKIDQDNNIIQVVILVPTRELALQTSQVCKELGKHLQIQVMVTTGGTSLKDDI 252
FSI L+ +D Q +IL PTRELA+Q + LG ++ +Q GGT++ +DI
Sbjct: 91 TFSISVLQCLDIQVRETQALILAPTRELAVQIQKGLLALGDYMNVQCHACIGGTNVGEDI 150
Query: 253 MRLYQPVHLLVGTPGRILDLAKKGVCVLKDCSMLVMDEADKLLSPEFQPSIEQLIQFLPP 312
+L H++ GTPGR+ D+ ++ + MLV+DEAD++L+ F+ I + ++LPP
Sbjct: 151 RKLDYGQHVVAGTPGRVFDMIRRRSLRTRAIKMLVLDEADEMLNKGFKEQIYDVYRYLPP 210
Query: 313 TRQILMFSATFPVTVKDFKDRYLRKPY-IINLMDELTLKGITQFYAFVE-ERQKVHCLNT 370
Q+++ SAT P + + ++++ P I+ DELTL+GI QF+ VE E K L
Sbjct: 211 ATQVVLISATLPHEILEMTNKFMTDPIRILVKRDELTLEGIKQFFVAVEREEWKFDTLCD 270
Query: 371 LFSKAANKPIHHLLQFGKSS*TPC*ENH*TRVFMFLYSCKDVARPP**SFPRLPQWCMQE 430
L+ I + F + R F S P + ++
Sbjct: 271 LYDTLT---ITQAVIFCNTKRKVDWLTEKMREANFTVSSMHGDMPQKERESIMKEFRSGA 327
Query: 431 SRLYC---LFTRGIDIQAVNVVINFDFPKNSETYLHRVGRSGRFGHLGLAVNLITYEDRF 487
SR+ ++ RG+D+ V+++IN+D P N E Y+HR+GRSGR+G G+A+N + +D
Sbjct: 328 SRVLISTDVWARGLDVPQVSLIINYDLPNNRELYIHRIGRSGRYGRKGVAINFVKNDDIR 387
Query: 488 NLYRIEQELGTEIKQIP 504
L IEQ T+I ++P
Sbjct: 388 ILRDIEQYYSTQIDEMP 404
>IF41_ARATH (P41376) Eukaryotic initiation factor 4A-1 (eIF4A-1)
(eIF-4A-1)
Length = 412
Score = 192 bits (488), Expect = 2e-48
Identities = 128/393 (32%), Positives = 195/393 (49%), Gaps = 58/393 (14%)
Query: 142 FEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAKNGTGKTAAFSIPALEK 201
F+ L+ LL GIY GFE+PS IQ+ I G D++ +A++GTGKTA F L++
Sbjct: 41 FDAMGLQENLLRGIYAYGFEKPSAIQQRGIVPFCKGLDVIQQAQSGTGKTATFCSGVLQQ 100
Query: 202 IDQDNNIIQVVILVPTRELALQTSQVCKELGKHLQIQVMVTTGGTSLKDDIMRLYQPVHL 261
+D Q ++L PTRELA Q +V + LG +L ++V GGTS+++D L VH+
Sbjct: 101 LDFSLIQCQALVLAPTRELAQQIEKVMRALGDYLGVKVHACVGGTSVREDQRILQAGVHV 160
Query: 262 LVGTPGRILDLAKKGVCVLKDCSMLVMDEADKLLSPEFQPSIEQLIQFLPPTRQILMFSA 321
+VGTPGR+ D+ K+ + M V+DEAD++LS F+ I + Q LPP Q+ +FSA
Sbjct: 161 VVGTPGRVFDMLKRQSLRADNIKMFVLDEADEMLSRGFKDQIYDIFQLLPPKIQVGVFSA 220
Query: 322 TFPVTVKDFKDRYLR-----------------KPYIIN----------LMDELTLKGITQ 354
T P + +++ K + +N L D ITQ
Sbjct: 221 TMPPEALEITRKFMSKPVRILVKRDELTLEGIKQFYVNVEKEEWKLETLCDLYETLAITQ 280
Query: 355 FYAFVEERQKVHCLNTLFSKAANKPIHHLLQFGKSS*TPC*ENH*TRVFMFLYSCKDVAR 414
FV R+KV + L K ++ G + +D+
Sbjct: 281 SVIFVNTRRKV---DWLTDKMRSRDHTVSATHGDMD----------------QNTRDII- 320
Query: 415 PP**SFPRLPQWCMQESRLYC---LFTRGIDIQAVNVVINFDFPKNSETYLHRVGRSGRF 471
+ ++ SR+ L RGID+Q V++VINFD P E YLHR+GRSGRF
Sbjct: 321 --------MREFRSGSSRVLITTDLLARGIDVQQVSLVINFDLPTQPENYLHRIGRSGRF 372
Query: 472 GHLGLAVNLITYEDRFNLYRIEQELGTEIKQIP 504
G G+A+N +T +D L+ I++ ++++P
Sbjct: 373 GRKGVAINFVTRDDERMLFDIQKFYNVVVEELP 405
>IF42_NICPL (P41379) Eukaryotic initiation factor 4A-2 (eIF4A-2)
(eIF-4A-2)
Length = 413
Score = 191 bits (484), Expect = 5e-48
Identities = 133/423 (31%), Positives = 202/423 (47%), Gaps = 72/423 (17%)
Query: 126 DTRYRTEDVTATKGNEFEDYF--------------LKRELLMGIYEKGFERPSPIQEESI 171
D R +T G E E++F L+ LL GIY GFE+PS IQ+ I
Sbjct: 12 DARQFDAKMTELLGTEQEEFFTSYDEVYDSFDAMGLQENLLRGIYAYGFEKPSAIQQRGI 71
Query: 172 PIALTGSDILARAKNGTGKTAAFSIPALEKIDQDNNIIQVVILVPTRELALQTSQVCKEL 231
G D++ +A++GTGKTA F L+++D Q ++L PTRELA Q +V + L
Sbjct: 72 VPFCKGLDVIQQAQSGTGKTATFCSGVLQQLDYSLVECQALVLAPTRELAQQIEKVMRAL 131
Query: 232 GKHLQIQVMVTTGGTSLKDDIMRLYQPVHLLVGTPGRILDLAKKGVCVLKDCSMLVMDEA 291
G +L ++V GGTS+++D L VH++VGTPGR+ D+ ++ M V+DEA
Sbjct: 132 GDYLGVKVHACVGGTSVREDQRILQSGVHVVVGTPGRVFDMLRRQSLRPDHIKMFVLDEA 191
Query: 292 DKLLSPEFQPSIEQLIQFLPPTRQILMFSATFPVTVKDFKDRYLR--------------- 336
D++LS F+ I + Q LPP Q+ +FSAT P + +++
Sbjct: 192 DEMLSRGFKDQIYDIFQLLPPKIQVGVFSATMPPEALEITRKFMNKPVRILVKRDELTLE 251
Query: 337 --KPYIIN----------LMDELTLKGITQFYAFVEERQKVHCLNTLFSKAANKPIHHLL 384
K + +N L D ITQ FV R+KV + L K ++
Sbjct: 252 GIKQFYVNVDKEEWKLETLCDLYETLAITQSVIFVNTRRKV---DWLTDKMRSRDHTVSA 308
Query: 385 QFGKSS*TPC*ENH*TRVFMFLYSCKDVARPP**SFPRLPQWCMQESRLYC---LFTRGI 441
G + +D+ + ++ SR+ L RGI
Sbjct: 309 THGDMD----------------QNTRDII---------MREFRSGSSRVLITTDLLARGI 343
Query: 442 DIQAVNVVINFDFPKNSETYLHRVGRSGRFGHLGLAVNLITYEDRFNLYRIEQELGTEIK 501
D+Q V++VIN+D P E YLHR+GRSGRFG G+A+N +T +D L+ I++ I+
Sbjct: 344 DVQQVSLVINYDLPTQPENYLHRIGRSGRFGRKGVAINSVTKDDERMLFDIQKFYNVVIE 403
Query: 502 QIP 504
++P
Sbjct: 404 ELP 406
>IF49_TOBAC (Q40471) Eukaryotic initiation factor 4A-9 (eIF4A-9)
(eIF-4A-9)
Length = 413
Score = 189 bits (480), Expect = 1e-47
Identities = 127/395 (32%), Positives = 195/395 (49%), Gaps = 58/395 (14%)
Query: 140 NEFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAKNGTGKTAAFSIPAL 199
+ F+ LK LL GIY GFE+PS IQ+ I G D++ +A++GTGKTA F L
Sbjct: 40 HSFDAMGLKENLLRGIYAYGFEKPSAIQQRGIVPFCKGLDVIQQAQSGTGKTATFCSGIL 99
Query: 200 EKIDQDNNIIQVVILVPTRELALQTSQVCKELGKHLQIQVMVTTGGTSLKDDIMRLYQPV 259
+++D + Q ++L PTRELA Q +V + LG +L ++V GGTS+++D L V
Sbjct: 100 QQLDYELLDCQALVLAPTRELAQQIEKVMRALGDYLGVKVHACVGGTSVREDQRILSSGV 159
Query: 260 HLLVGTPGRILDLAKKGVCVLKDCSMLVMDEADKLLSPEFQPSIEQLIQFLPPTRQILMF 319
H++VGTPGR+ D+ ++ M V+DEAD++LS F+ I + Q LPP Q+ +F
Sbjct: 160 HVVVGTPGRVFDMLRRQSLRPDHIKMFVLDEADEMLSRGFKDQIYDIFQLLPPKIQVGVF 219
Query: 320 SATFPVTVKDFKDRYLR-----------------KPYIIN----------LMDELTLKGI 352
SAT P + +++ K + +N L D I
Sbjct: 220 SATMPPEALEITRKFMNKPVRILVKRDELTLEGIKQFYVNVDKEEWKLETLCDLYETLAI 279
Query: 353 TQFYAFVEERQKVHCLNTLFSKAANKPIHHLLQFGKSS*TPC*ENH*TRVFMFLYSCKDV 412
TQ FV R+KV + L K ++ G + +D+
Sbjct: 280 TQSVIFVNTRRKV---DWLTDKMRSRDHTVSATHGDMD----------------QNTRDI 320
Query: 413 ARPP**SFPRLPQWCMQESRLYC---LFTRGIDIQAVNVVINFDFPKNSETYLHRVGRSG 469
+ ++ SR+ L RGID+Q V++VIN+D P E YLHR+GRSG
Sbjct: 321 I---------MREFRSGSSRVLITTDLLARGIDVQQVSLVINYDLPTQPENYLHRIGRSG 371
Query: 470 RFGHLGLAVNLITYEDRFNLYRIEQELGTEIKQIP 504
RFG G+++N +T +D L I++ I+++P
Sbjct: 372 RFGRKGVSINFVTSDDERMLSDIQRFYNVVIEELP 406
>EXP9_STRPN (P35599) Probable RNA helicase exp9 (Exported protein 9)
Length = 524
Score = 176 bits (445), Expect = 2e-43
Identities = 92/236 (38%), Positives = 144/236 (60%), Gaps = 1/236 (0%)
Query: 141 EFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAKNGTGKTAAFSIPALE 200
+F + L +LL I + GF SPIQE++IP+AL G D++ +A+ GTGKTAAF +P LE
Sbjct: 2 KFNELNLSADLLAEIEKAGFVEASPIQEQTIPLALEGKDVIGQAQTGTGKTAAFGLPTLE 61
Query: 201 KIDQDNNIIQVVILVPTRELALQTSQVCKELGKHLQIQVMVTTGGTSLKDDIMRLYQPVH 260
KI + IQ +++ PTRELA+Q+ + G+ ++V GG+S++ I L H
Sbjct: 62 KIRTEEATIQALVIAPTRELAVQSQEELFRFGRSKGVKVRSVYGGSSIEKQIKALKSGAH 121
Query: 261 LLVGTPGRILDLAKKGVCVLKDCSMLVMDEADKLLSPEFQPSIEQLIQFLPPTRQILMFS 320
++VGTPGR+LDL K+ L+D L++DEAD++L+ F IE +I +P RQ L+FS
Sbjct: 122 IVVGTPGRLLDLIKRKALKLQDIETLILDEADEMLNMGFLEDIEAIISRVPENRQTLLFS 181
Query: 321 ATFPVTVKDFKDRYLRKPYIINL-MDELTLKGITQFYAFVEERQKVHCLNTLFSKA 375
AT P +K ++++ P + + ELT + + Q+Y V+E++K + L A
Sbjct: 182 ATMPDAIKRIGVQFMKAPEHVKIAAKELTTELVDQYYIRVKEQEKFDTMTRLMDVA 237
Score = 51.6 bits (122), Expect = 5e-06
Identities = 24/67 (35%), Positives = 38/67 (55%)
Query: 439 RGIDIQAVNVVINFDFPKNSETYLHRVGRSGRFGHLGLAVNLITYEDRFNLYRIEQELGT 498
RG+DI V V N+D P++ E+Y+HR+GR+GR G G ++ + + L IE
Sbjct: 301 RGLDISGVTHVYNYDIPQDPESYVHRIGRTGRAGKSGQSITFVAPNEMGYLQIIENLTKK 360
Query: 499 EIKQIPP 505
+K + P
Sbjct: 361 RMKGLKP 367
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.325 0.140 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 61,006,150
Number of Sequences: 164201
Number of extensions: 2695861
Number of successful extensions: 50699
Number of sequences better than 10.0: 778
Number of HSP's better than 10.0 without gapping: 645
Number of HSP's successfully gapped in prelim test: 149
Number of HSP's that attempted gapping in prelim test: 26872
Number of HSP's gapped (non-prelim): 7724
length of query: 513
length of database: 59,974,054
effective HSP length: 115
effective length of query: 398
effective length of database: 41,090,939
effective search space: 16354193722
effective search space used: 16354193722
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 68 (30.8 bits)
Medicago: description of AC123899.6


