

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC123574.2 + phase: 0 /pseudo
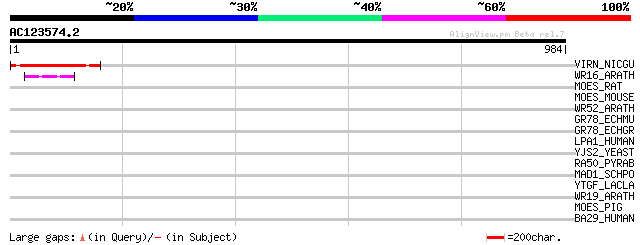
(984 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
VIRN_NICGU (Q40392) TMV resistance protein N 148 8e-35
WR16_ARATH (Q9FL92) Probable WRKY transcription factor 16 (WRKY ... 47 2e-04
MOES_RAT (O35763) Moesin (Membrane-organizing extension spike pr... 35 0.99
MOES_MOUSE (P26041) Moesin (Membrane-organizing extension spike ... 35 0.99
WR52_ARATH (Q9FH83) Probable WRKY transcription factor 52 (WRKY ... 34 1.7
GR78_ECHMU (Q24895) 78 kDa glucose-regulated protein precursor (... 34 1.7
GR78_ECHGR (Q24798) 78 kDa glucose-regulated protein precursor (... 34 1.7
LPA1_HUMAN (Q13136) Liprin-alpha 1 (Protein tyrosine phosphatase... 34 2.2
YJS2_YEAST (P46986) Hypothetical 12.5 kDa protein in SWE1-ATP12 ... 33 3.7
RA50_PYRAB (Q9UZC8) DNA double-strand break repair rad50 ATPase 33 3.7
MAD1_SCHPO (P87169) Spindle assembly checkpoint component mad1 (... 33 3.7
YTGF_LACLA (Q9CEE2) Hypothetical UPF0144 protein ytgF 32 6.4
WR19_ARATH (Q9SZ67) Probable WRKY transcription factor 19 (WRKY ... 32 6.4
MOES_PIG (P26042) Moesin (Membrane-organizing extension spike pr... 32 8.3
BA29_HUMAN (Q9UHQ4) B-cell receptor-associated protein 29 (BCR-a... 32 8.3
>VIRN_NICGU (Q40392) TMV resistance protein N
Length = 1144
Score = 148 bits (373), Expect = 8e-35
Identities = 81/160 (50%), Positives = 107/160 (66%), Gaps = 7/160 (4%)
Query: 1 MQSSSSSSSISYVFKYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEIT 60
M SSSSSS SY VFLSFRG DTR FT +LY+ L DKGI TF DD L+ G I
Sbjct: 1 MASSSSSSRWSY----DVFLSFRGEDTRKTFTSHLYEVLNDKGIKTFQDDKRLEYGATIP 56
Query: 61 PSLDNAIEESRIFIPVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHH 120
L AIEES+ I VFS NYA+S +CL+ELV I+ + + V+P+F+ VDPSHVR+
Sbjct: 57 GELCKAIEESQFAIVVFSENYATSRWCLNELVKIMECKTRFKQTVIPIFYDVDPSHVRNQ 116
Query: 121 RGSYGEALAKHEERFQHNTDHMERLQKWKIALTQAANLSG 160
+ S+ +A +HE +++ D +E +Q+W+IAL +AANL G
Sbjct: 117 KESFAKAFEEHETKYK---DDVEGIQRWRIALNEAANLKG 153
>WR16_ARATH (Q9FL92) Probable WRKY transcription factor 16 (WRKY
DNA-binding protein 16)
Length = 1372
Score = 47.4 bits (111), Expect = 2e-04
Identities = 27/90 (30%), Positives = 47/90 (52%), Gaps = 7/90 (7%)
Query: 26 DTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSLDNAIEESRIFIPVFSANYASSS 85
+ RY F +L KAL KG++ DS+ D ++ + +E +R+ + + N S
Sbjct: 15 EVRYSFVSHLSKALQRKGVNDVFIDSD----DSLSNESQSMVERARVSVMILPGNRTVS- 69
Query: 86 FCLDELVHIIHLYKQNGRLVLPVFFGVDPS 115
LD+LV ++ K ++V+PV +GV S
Sbjct: 70 --LDKLVKVLDCQKNKDQVVVPVLYGVRSS 97
>MOES_RAT (O35763) Moesin (Membrane-organizing extension spike
protein)
Length = 576
Score = 35.0 bits (79), Expect = 0.99
Identities = 22/75 (29%), Positives = 40/75 (53%), Gaps = 3/75 (4%)
Query: 900 KYESEMDKALLENEWIRVELELKSFNLSEEEKNEMLRSAQMGIHVLKEKNNAEEENVIFT 959
K++ +M++ALLENE + EL K E EK E++ + + ++ A++E T
Sbjct: 312 KHQKQMERALLENEKKKRELAEKEKEKIEREKEELMEKLK---QIEEQTKKAQQELEEQT 368
Query: 960 DPYRETKSQKKRKQN 974
E + ++KR Q+
Sbjct: 369 RRALELEQERKRAQS 383
>MOES_MOUSE (P26041) Moesin (Membrane-organizing extension spike
protein)
Length = 576
Score = 35.0 bits (79), Expect = 0.99
Identities = 22/75 (29%), Positives = 40/75 (53%), Gaps = 3/75 (4%)
Query: 900 KYESEMDKALLENEWIRVELELKSFNLSEEEKNEMLRSAQMGIHVLKEKNNAEEENVIFT 959
K++ +M++ALLENE + EL K E EK E++ + + ++ A++E T
Sbjct: 312 KHQKQMERALLENEKKKRELAEKEKEKIEREKEELMEKLK---QIEEQTKKAQQELEEQT 368
Query: 960 DPYRETKSQKKRKQN 974
E + ++KR Q+
Sbjct: 369 RRALELEQERKRAQS 383
>WR52_ARATH (Q9FH83) Probable WRKY transcription factor 52 (WRKY
DNA-binding protein 52) (Disease resistance protein
RRS1) (Resistance to Ralstonia solanacearum 1 protein)
Length = 1378
Score = 34.3 bits (77), Expect = 1.7
Identities = 22/87 (25%), Positives = 41/87 (46%), Gaps = 3/87 (3%)
Query: 26 DTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSLDNAIEESRIFIPVFSANYASSS 85
+ RY F +L +AL KGI+ + D ++ D + IE++ + + V N S
Sbjct: 18 EVRYSFVSHLSEALRRKGINNVVVDVDID--DLLFKESQAKIEKAGVSVMVLPGNCDPSE 75
Query: 86 FCLDELVHIIHLYKQN-GRLVLPVFFG 111
LD+ ++ + N + V+ V +G
Sbjct: 76 VWLDKFAKVLECQRNNKDQAVVSVLYG 102
>GR78_ECHMU (Q24895) 78 kDa glucose-regulated protein precursor (GRP
78)
Length = 649
Score = 34.3 bits (77), Expect = 1.7
Identities = 21/88 (23%), Positives = 45/88 (50%), Gaps = 2/88 (2%)
Query: 894 RRIYGIKYESEMDKALLENEWIRVELELKSFNLSEEEKNEMLRSAQMGIHVLKEKNNAEE 953
R I + S+ DK + E +R +LE ++++ + K++ ++ +K +A +
Sbjct: 539 RMIQDAEKFSDQDKQVKERVEVRNDLESLAYSIKNQVKDKEKMGGKLSDDEIKTIEDAAD 598
Query: 954 ENV--IFTDPYRETKSQKKRKQNMFSIL 979
E + + +P ET KK+K N+ S++
Sbjct: 599 EAIKWMENNPQAETSDYKKQKANLESVV 626
>GR78_ECHGR (Q24798) 78 kDa glucose-regulated protein precursor (GRP
78)
Length = 651
Score = 34.3 bits (77), Expect = 1.7
Identities = 21/88 (23%), Positives = 45/88 (50%), Gaps = 2/88 (2%)
Query: 894 RRIYGIKYESEMDKALLENEWIRVELELKSFNLSEEEKNEMLRSAQMGIHVLKEKNNAEE 953
R I + S+ DK + E +R +LE ++++ + K++ ++ +K +A +
Sbjct: 539 RMIQDAEKFSDQDKQVKERVEVRNDLESLAYSIKNQVKDKEKMGGKLSDDEIKTIEDAAD 598
Query: 954 ENV--IFTDPYRETKSQKKRKQNMFSIL 979
E + + +P ET KK+K N+ S++
Sbjct: 599 EAIKWMENNPQAETSDYKKQKANLESVV 626
>LPA1_HUMAN (Q13136) Liprin-alpha 1 (Protein tyrosine phosphatase
receptor type f polypeptide-interacting protein alpha 1)
(PTPRF-interacting protein alpha 1) (LAR-interacting
protein 1) (LIP.1)
Length = 1202
Score = 33.9 bits (76), Expect = 2.2
Identities = 24/77 (31%), Positives = 39/77 (50%), Gaps = 9/77 (11%)
Query: 887 EEHNKRLRRIYGIKYESEMDKALLENEWIRVELELKSFNLSEEEKNEMLRSAQMGIHVLK 946
EEHNKRL +DK L E+ R++L LK + E+KN +LR + L+
Sbjct: 441 EEHNKRL--------SDTVDKLLSESNE-RLQLHLKERMAALEDKNSLLREVESAKKQLE 491
Query: 947 EKNNAEEENVIFTDPYR 963
E + +++ V+ + R
Sbjct: 492 ETQHDKDQLVLNIEALR 508
>YJS2_YEAST (P46986) Hypothetical 12.5 kDa protein in SWE1-ATP12
intergenic region
Length = 105
Score = 33.1 bits (74), Expect = 3.7
Identities = 16/37 (43%), Positives = 23/37 (61%)
Query: 590 LKYLPNSLRVLKWKGCLLESLSSSILSKASEVTSFYN 626
L+YL NSLRVL WKG + + +S + T+F+N
Sbjct: 27 LEYLHNSLRVLSWKGIITKIAASPFVIVLYFNTAFFN 63
>RA50_PYRAB (Q9UZC8) DNA double-strand break repair rad50 ATPase
Length = 880
Score = 33.1 bits (74), Expect = 3.7
Identities = 28/107 (26%), Positives = 47/107 (43%), Gaps = 4/107 (3%)
Query: 873 VPQVEEC*RVSLKLEEHNKRLRRIYGIKYESEMDKALLENEWIRVELELKSFNLSEEEKN 932
+ ++EE + KL+E K R++ G + E E LE E + E ELK+ +E
Sbjct: 279 ISELEEIVKDIPKLQEKEKEYRKLKGFRDEYESKLRRLEKELSKWESELKAIEEVIKEGE 338
Query: 933 EMLRSAQMGIHVLKEKNNAEEENVIFTDPYRETKSQKKRKQNMFSIL 979
+ A+ ++EK + E+ + PY E K+ Q L
Sbjct: 339 KKKERAE----EIREKLSEIEKRLEELKPYVEELEDAKQVQKQIERL 381
>MAD1_SCHPO (P87169) Spindle assembly checkpoint component mad1
(Mitotic arrest deficient protein 1)
Length = 689
Score = 33.1 bits (74), Expect = 3.7
Identities = 23/82 (28%), Positives = 42/82 (51%), Gaps = 5/82 (6%)
Query: 885 KLEEHNKRLRR-IYGIKYESEMDKALLENEWIRVEL-ELKSFNLSEEEKNEMLRSAQMGI 942
+L HN++L I + E++K E R+++ EL+ ++EE+ E L S +
Sbjct: 211 ELSVHNQQLEESIKQVSSSIELEKINAEQ---RLQISELEKLKAAQEERIEKLSSNNRNV 267
Query: 943 HVLKEKNNAEEENVIFTDPYRE 964
+LKE+ N E + + YR+
Sbjct: 268 EILKEEKNDLESKLYRFEEYRD 289
>YTGF_LACLA (Q9CEE2) Hypothetical UPF0144 protein ytgF
Length = 531
Score = 32.3 bits (72), Expect = 6.4
Identities = 22/96 (22%), Positives = 48/96 (49%), Gaps = 5/96 (5%)
Query: 883 SLKLEEHNKRLRRIYGIKYESEMDKALLENEWIRVELELKSFNLSEEEKNEMLRSAQMGI 942
SLK+E + Y ++ E + + +E+E+ + ELK +++ E+L +
Sbjct: 57 SLKMEAEAFKKEARYTLREEEQKQRREIEDEFKQERQELKETEKRLKQREEILDRKDDTL 116
Query: 943 HVLKEKNNAEEENVIFTDPYRETKSQKKRKQNMFSI 978
+E +++EEN++ R+T + KR++ + I
Sbjct: 117 TKKEENLDSKEENLV-----RKTDTLSKREEQLAHI 147
>WR19_ARATH (Q9SZ67) Probable WRKY transcription factor 19 (WRKY
DNA-binding protein 19)
Length = 1895
Score = 32.3 bits (72), Expect = 6.4
Identities = 21/52 (40%), Positives = 28/52 (53%), Gaps = 8/52 (15%)
Query: 570 FKKMTKLKTLII-----ENGH---FSNGLKYLPNSLRVLKWKGCLLESLSSS 613
F+KM L+ L + E H F GL+YLP+ LR+L W+ L SL S
Sbjct: 1172 FEKMCNLRLLKLYCSKAEEKHGVSFPQGLEYLPSKLRLLHWEYYPLSSLPKS 1223
>MOES_PIG (P26042) Moesin (Membrane-organizing extension spike
protein)
Length = 576
Score = 32.0 bits (71), Expect = 8.3
Identities = 19/56 (33%), Positives = 31/56 (54%), Gaps = 1/56 (1%)
Query: 900 KYESEMDKALLENEWIRVELELKSFNLSEEEKNEML-RSAQMGIHVLKEKNNAEEE 954
K++ +M++ALLENE + E+ K E EK E++ R Q+ K + EE+
Sbjct: 312 KHQKQMERALLENEKKKREMAEKEKEKIEREKEELMERLKQIEEQTKKAQQELEEQ 367
>BA29_HUMAN (Q9UHQ4) B-cell receptor-associated protein 29
(BCR-associated protein Bap29)
Length = 241
Score = 32.0 bits (71), Expect = 8.3
Identities = 44/184 (23%), Positives = 80/184 (42%), Gaps = 29/184 (15%)
Query: 795 VSKIATSQMNVYQYFSSGLLM*KG*TYRITSISIFFPSVLMNVTL*RSLNSIVASLLRKL 854
+ K +TS+ + Y++ L + Y I+ S+FF VL R L +++ L ++L
Sbjct: 79 IEKSSTSRPDAYEHTQMKLFRSQRNLY-ISGFSLFFWLVL------RRLVTLITQLAKEL 131
Query: 855 EGFHQILKSFPLINVNR*VPQVEEC*RVSLKLEEHNKRLRRIY-------GIKYESEMDK 907
+LK+ Q E + + K E N++L+RI E+E K
Sbjct: 132 SN-KGVLKT-----------QAENTNKAAKKFMEENEKLKRILKSHGKDEECVLEAENKK 179
Query: 908 ALLENEWIRVELELKSFNLSEEEKNEMLRSAQMGIHVLKEKNNAEEENVIFTDPYRETKS 967
+ + E ++ EL S LS+ + N+++ + KE + +E+ D R +
Sbjct: 180 LVEDQEKLKTELRKTSDALSKAQ-NDVMEMKMQSERLSKEYDQLLKEHSELQD--RLERG 236
Query: 968 QKKR 971
KKR
Sbjct: 237 NKKR 240
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.355 0.155 0.549
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 92,942,261
Number of Sequences: 164201
Number of extensions: 3344310
Number of successful extensions: 18186
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 3
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 18172
Number of HSP's gapped (non-prelim): 24
length of query: 984
length of database: 59,974,054
effective HSP length: 120
effective length of query: 864
effective length of database: 40,269,934
effective search space: 34793222976
effective search space used: 34793222976
T: 11
A: 40
X1: 14 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.6 bits)
S2: 71 (32.0 bits)
Medicago: description of AC123574.2


