

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC123574.1 + phase: 0 /pseudo
(527 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
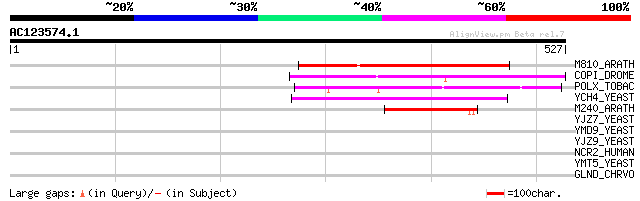
Score E
Sequences producing significant alignments: (bits) Value
M810_ARATH (P92519) Hypothetical mitochondrial protein AtMg00810... 160 8e-39
COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contain... 145 3e-34
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 142 3e-33
YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein 95 5e-19
M240_ARATH (P93290) Hypothetical mitochondrial protein AtMg00240... 82 3e-15
YJZ7_YEAST (P47098) Transposon Ty1 protein B 37 0.13
YMD9_YEAST (Q03434) Transposon Ty1 protein B 36 0.28
YJZ9_YEAST (P47100) Transposon Ty1 protein B 33 2.4
NCR2_HUMAN (Q9Y618) Nuclear receptor corepressor 2 (N-CoR2) (Sil... 33 2.4
YMT5_YEAST (Q04214) Transposon Ty1 protein B 32 3.1
GLND_CHRVO (Q7NTY6) [Protein-PII] uridylyltransferase (EC 2.7.7.... 31 9.0
>M810_ARATH (P92519) Hypothetical mitochondrial protein AtMg00810
(ORF240b)
Length = 240
Score = 160 bits (405), Expect = 8e-39
Identities = 85/201 (42%), Positives = 123/201 (60%), Gaps = 3/201 (1%)
Query: 275 LRASFKIKDLGQLKYFLGLQVAHSKQGISLCQRKYCLDLLADSGLTHSKPISTPSDPSVK 334
L ++F +KDLG + YFLG+Q+ G+ L Q KY +L ++G+ KP+STP +K
Sbjct: 27 LSSTFSMKDLGPVHYFLGIQIKTHPSGLFLSQTKYAEQILNNAGMLDCKPMSTPLP--LK 84
Query: 335 LHRD-SSPPYTDISAYRRLVGRLLYLNTTRPDITFVTQQLSQFLAKRTQVHHSAAMRVLK 393
L+ S+ Y D S +R +VG L YL TRPDI++ + Q + + T RVL+
Sbjct: 85 LNSSVSTAKYPDPSDFRSIVGALQYLTLTRPDISYAVNIVCQRMHEPTLADFDLLKRVLR 144
Query: 394 YLKTSLGKGLFFPRDSALQILGFSDADWAGCLDSRRSISGQCFFLGKSLISWRTKKQLIV 453
Y+K ++ GL+ ++S L + F D+DWAGC +RRS +G C FLG ++ISW K+Q V
Sbjct: 145 YVKGTIFHGLYIHKNSKLNVQAFCDSDWAGCTSTRRSTTGFCTFLGCNIISWSAKRQPTV 204
Query: 454 SRSSSKEEYRALAAATCELQW 474
SRSS++ EYRALA EL W
Sbjct: 205 SRSSTETEYRALALTAAELTW 225
>COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contains:
Copia VLP protein; Copia protease (EC 3.4.23.-)]
Length = 1409
Score = 145 bits (365), Expect = 3e-34
Identities = 86/266 (32%), Positives = 141/266 (52%), Gaps = 5/266 (1%)
Query: 266 SEFNNIKSVLRASFKIKDLGQLKYFLGLQVAHSKQGISLCQRKYCLDLLADSGLTHSKPI 325
+ NN K L F++ DL ++K+F+G+++ + I L Q Y +L+ + + +
Sbjct: 1099 TRMNNFKRYLMEKFRMTDLNEIKHFIGIRIEMQEDKIYLSQSAYVKKILSKFNMENCNAV 1158
Query: 326 STPSDPSVKLHRDSSPPYTDISAYRRLVGRLLYLNT-TRPDITFVTQQLSQFLAKRTQVH 384
STP + +S + R L+G L+Y+ TRPD+T LS++ +K
Sbjct: 1159 STPLPSKINYELLNSDEDCNTPC-RSLIGCLMYIMLCTRPDLTTAVNILSRYSSKNNSEL 1217
Query: 385 HSAAMRVLKYLKTSLGKGLFFPRDSALQ--ILGFSDADWAGCLDSRRSISGQCF-FLGKS 441
RVL+YLK ++ L F ++ A + I+G+ D+DWAG R+S +G F +
Sbjct: 1218 WQNLKRVLRYLKGTIDMKLIFKKNLAFENKIIGYVDSDWAGSEIDRKSTTGYLFKMFDFN 1277
Query: 442 LISWRTKKQLIVSRSSSKEEYRALAAATCELQWLLYLLQDLQLQTSKLHVLYCDNQSALH 501
LI W TK+Q V+ SS++ EY AL A E WL +LL + ++ +Y DNQ +
Sbjct: 1278 LICWNTKRQNSVAASSTEAEYMALFEAVREALWLKFLLTSINIKLENPIKIYEDNQGCIS 1337
Query: 502 IAANPIFHERTKHLEIDCHLVQDKLQ 527
IA NP H+R KH++I H ++++Q
Sbjct: 1338 IANNPSCHKRAKHIDIKYHFAREQVQ 1363
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 142 bits (357), Expect = 3e-33
Identities = 87/263 (33%), Positives = 142/263 (53%), Gaps = 11/263 (4%)
Query: 271 IKSVLRASFKIKDLGQLKYFLGLQVAHSKQG--ISLCQRKYCLDLLADSGLTHSKPISTP 328
+K L SF +KDLG + LG+++ + + L Q KY +L + ++KP+STP
Sbjct: 1025 LKGDLSKSFDMKDLGPAQQILGMKIVRERTSRKLWLSQEKYIERVLERFNMKNAKPVSTP 1084
Query: 329 SDPSVKLHRDSSPPYTDISA------YRRLVGRLLY-LNTTRPDITFVTQQLSQFLAKRT 381
+KL + P + Y VG L+Y + TRPDI +S+FL
Sbjct: 1085 LAGHLKLSKKMCPTTVEEKGNMAKVPYSSAVGSLMYAMVCTRPDIAHAVGVVSRFLENPG 1144
Query: 382 QVHHSAAMRVLKYLKTSLGKGLFFPRDSALQILGFSDADWAGCLDSRRSISGQCFFLGKS 441
+ H A +L+YL+ + G L F + + G++DAD AG +D+R+S +G F
Sbjct: 1145 KEHWEAVKWILRYLRGTTGDCLCFGGSDPI-LKGYTDADMAGDIDNRKSSTGYLFTFSGG 1203
Query: 442 LISWRTKKQLIVSRSSSKEEYRALAAATCELQWLLYLLQDLQLQTSKLHVLYCDNQSALH 501
ISW++K Q V+ S+++ EY A E+ WL LQ+L L K +V+YCD+QSA+
Sbjct: 1204 AISWQSKLQKCVALSTTEAEYIAATETGKEMIWLKRFLQELGLH-QKEYVVYCDSQSAID 1262
Query: 502 IAANPIFHERTKHLEIDCHLVQD 524
++ N ++H RTKH+++ H +++
Sbjct: 1263 LSKNSMYHARTKHIDVRYHWIRE 1285
>YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein
Length = 308
Score = 94.7 bits (234), Expect = 5e-19
Identities = 63/208 (30%), Positives = 105/208 (50%), Gaps = 3/208 (1%)
Query: 268 FNNIKSVLRASFKIKDLGQLKYFLGLQVAHSKQG-ISLCQRKYCLDLLADSGLTHSKPIS 326
++ +K L + +KDLG++ FLGL + S G I+L + Y ++S + K
Sbjct: 101 YDRVKQELTKLYSMKDLGKVDKFLGLNIHQSTNGDITLSLQDYIAKAASESEINTFKLTQ 160
Query: 327 TPSDPSVKLHRDSSPPYTDISAYRRLVGRLLYL-NTTRPDITFVTQQLSQFLAKRTQVHH 385
TP S L +SP DI+ Y+ +VG+LL+ NT RPDI++ LS+FL + +H
Sbjct: 161 TPLCNSKPLFETTSPHLKDITPYQSIVGQLLFCANTGRPDISYPVSLLSRFLREPRAIHL 220
Query: 386 SAAMRVLKYLKTSLGKGLFFPRDSALQILGFSDADWAGCLDSRRSISGQCFFLGKSLISW 445
+A RVL+YL T+ L + S + + + DA D S G L + ++W
Sbjct: 221 ESARRVLRYLYTTRSMCLKYRSGSQVALTVYCDASHGAIHDLPHSTGGYVTLLAGAPVTW 280
Query: 446 RTKK-QLIVSRSSSKEEYRALAAATCEL 472
+KK + ++ S++ EY + E+
Sbjct: 281 SSKKLKGVIPVPSTEAEYITASETVMEI 308
>M240_ARATH (P93290) Hypothetical mitochondrial protein AtMg00240
(ORF111a)
Length = 111
Score = 82.4 bits (202), Expect = 3e-15
Identities = 42/96 (43%), Positives = 61/96 (62%), Gaps = 8/96 (8%)
Query: 357 LYLNTTRPDITFVTQQLSQFLAKRTQVHHSAAMRVLKYLKTSLGKGLFFPRDSALQILGF 416
+YL TRPD+TF +LSQF + A +VL Y+K ++G+GLF+ S LQ+ F
Sbjct: 1 MYLTITRPDLTFAVNRLSQFSSASRTAQMQAVYKVLHYVKGTVGQGLFYSATSDLQLKAF 60
Query: 417 SDADWAGCLDSRRSISGQC-----FFLG---KSLIS 444
+D+DWA C D+RRS++G C +FLG KS++S
Sbjct: 61 ADSDWASCPDTRRSVTGFCSLVPLWFLGALRKSILS 96
>YJZ7_YEAST (P47098) Transposon Ty1 protein B
Length = 1755
Score = 37.0 bits (84), Expect = 0.13
Identities = 58/275 (21%), Positives = 121/275 (43%), Gaps = 40/275 (14%)
Query: 257 MILF*QEIHSEFNNIKSVLRASF--KIKDLGQ----LKY-FLGLQVAHSKQGISLCQRKY 309
MILF +++++ I + L+ + KI +LG+ ++Y LGL++ + QR
Sbjct: 1429 MILFSKDLNAN-KKIITTLKKQYDTKIINLGESDNEIQYDILGLEIKY--------QRGK 1479
Query: 310 CLDLLADSGLTHSKP------------ISTPSDPSVKLHRDSSPPYTD-----ISAYRRL 352
+ L ++ LT P +S P P + + +D D + ++L
Sbjct: 1480 YMKLGMENSLTEKIPKLNVPLNPKGRKLSAPGQPGLYIDQDELEIDEDEYKEKVHEMQKL 1539
Query: 353 VGRLLYLNTT-RPDITFVTQQLSQFLAKRTQVHHSAAMRVLKYLKTSLGKGLFF----PR 407
+G Y+ R D+ + L+Q + ++ +++++ + K L + P
Sbjct: 1540 IGLASYVGYKFRFDLLYYINTLAQHILFPSRQVLDMTYELIQFMWDTRDKQLIWHKNKPT 1599
Query: 408 DSALQILGFSDADWAGCLDSRRSISGQCFFLGKSLISWRTKKQLIVSRSSSKEEYRALAA 467
+ +++ SDA + G +S G F L +I ++ K + S+++ E A++
Sbjct: 1600 EPDNKLVAISDASY-GNQPYYKSQIGNIFLLNGKVIGGKSTKASLTCTSTTEAEIHAISE 1658
Query: 468 ATCELQWLLYLLQDLQLQTSKLHVLYCDNQSALHI 502
+ L L YL+Q+L + + L D++S + I
Sbjct: 1659 SVPLLNNLSYLIQELN-KKPIIKGLLTDSRSTISI 1692
>YMD9_YEAST (Q03434) Transposon Ty1 protein B
Length = 1328
Score = 35.8 bits (81), Expect = 0.28
Identities = 57/275 (20%), Positives = 121/275 (43%), Gaps = 40/275 (14%)
Query: 257 MILF*QEIHSEFNNIKSVLRASF--KIKDLGQ----LKY-FLGLQVAHSKQGISLCQRKY 309
MILF +++++ I + L+ + KI +LG+ ++Y LGL++ + QR
Sbjct: 1002 MILFSKDLNAN-KKIITTLKKQYDTKIINLGESDNEIQYDILGLEIKY--------QRGK 1052
Query: 310 CLDLLADSGLTHSKP------------ISTPSDPSVKLHRDSSPPYTD-----ISAYRRL 352
+ L ++ LT P +S P P + + +D D + ++L
Sbjct: 1053 YMKLGMENSLTEKIPKLNVPLNPKGRKLSAPGQPGLYIDQDELEIDEDEYKEKVHEMQKL 1112
Query: 353 VGRLLYLNTT-RPDITFVTQQLSQFLAKRTQVHHSAAMRVLKYLKTSLGKGLFF----PR 407
+G Y+ R D+ + L+Q + ++ +++++ + K L + P
Sbjct: 1113 IGLASYVGYKFRFDLLYYINTLAQHILFPSRQVLDMTYELIQFMWDTRDKQLIWHKNKPT 1172
Query: 408 DSALQILGFSDADWAGCLDSRRSISGQCFFLGKSLISWRTKKQLIVSRSSSKEEYRALAA 467
+ +++ SDA + G +S G + L +I ++ K + S+++ E A++
Sbjct: 1173 EPDNKLVAISDASY-GNQPYYKSQIGNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAISE 1231
Query: 468 ATCELQWLLYLLQDLQLQTSKLHVLYCDNQSALHI 502
+ L L YL+Q+L + + L D++S + I
Sbjct: 1232 SVPLLNNLSYLIQELN-KKPIIKGLLTDSRSTISI 1265
>YJZ9_YEAST (P47100) Transposon Ty1 protein B
Length = 1755
Score = 32.7 bits (73), Expect = 2.4
Identities = 50/254 (19%), Positives = 110/254 (42%), Gaps = 37/254 (14%)
Query: 257 MILF*QEIHSEFNNIKSV-LRASFKIKDLGQ----LKY-FLGLQVAHSKQGISLCQRKYC 310
M+LF + ++S I+ + ++ KI +LG+ ++Y LGL++ + QR
Sbjct: 1429 MVLFSKNLNSNKRIIEKLKMQYDTKIINLGESDEEIQYDILGLEIKY--------QRGKY 1480
Query: 311 LDLLADSGLTHSKP------------ISTPSDPSVKLHRDSSPPYTD-----ISAYRRLV 353
+ L ++ LT P +S P P + + + D + ++L+
Sbjct: 1481 MKLGMENSLTEKIPKLNVPLNPKGRKLSAPGQPGLYIDQQELELEEDDYKMKVHEMQKLI 1540
Query: 354 GRLLYLNTT-RPDITFVTQQLSQFLAKRTQVHHSAAMRVLKYLKTSLGKGLFFPRDSALQ 412
G Y+ R D+ + L+Q + ++ +++++ + K L + + ++
Sbjct: 1541 GLASYVGYKFRFDLLYYINTLAQHILFPSKQVLDMTYELIQFIWNTRDKQLIWHKSKPVK 1600
Query: 413 ----ILGFSDADWAGCLDSRRSISGQCFFLGKSLISWRTKKQLIVSRSSSKEEYRALAAA 468
++ SDA + G +S G + L +I ++ K + S+++ E A++ +
Sbjct: 1601 PTNKLVVISDASY-GNQPYYKSQIGNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAISES 1659
Query: 469 TCELQWLLYLLQDL 482
L L YL+Q+L
Sbjct: 1660 VPLLNNLSYLIQEL 1673
>NCR2_HUMAN (Q9Y618) Nuclear receptor corepressor 2 (N-CoR2)
(Silencing mediator of retinoic acid and thyroid hormone
receptor) (SMRT) (SMRTe) (Thyroid-,
retinoic-acid-receptor-associated corepressor) (T3
receptor-associating factor) (TRAC) (CTG repeat pr
Length = 2517
Score = 32.7 bits (73), Expect = 2.4
Identities = 28/80 (35%), Positives = 35/80 (43%), Gaps = 11/80 (13%)
Query: 312 DLLADSGLTHS-KPISTPSDPSVKLHRDSSPPYTDISAYRRLVGRLLYLNTTRPD--ITF 368
DL D LT +P+S PS P + PP RL L N R D IT
Sbjct: 134 DLTKDRSLTGKLEPVSPPSPPHTDPELELVPP--------RLSKEELIQNMDRVDREITM 185
Query: 369 VTQQLSQFLAKRTQVHHSAA 388
V QQ+S+ K+ Q+ AA
Sbjct: 186 VEQQISKLKKKQQQLEEEAA 205
>YMT5_YEAST (Q04214) Transposon Ty1 protein B
Length = 1328
Score = 32.3 bits (72), Expect = 3.1
Identities = 51/254 (20%), Positives = 109/254 (42%), Gaps = 37/254 (14%)
Query: 257 MILF*QEIHSEFNNI-KSVLRASFKIKDLGQ----LKY-FLGLQVAHSKQGISLCQRKYC 310
M+LF + ++S I K ++ KI +LG+ ++Y LGL++ + QR
Sbjct: 1002 MVLFSKNLNSNKRIIDKLKMQYDTKIINLGESDEEIQYDILGLEIKY--------QRGKY 1053
Query: 311 LDLLADSGLTHSKP------------ISTPSDPSVKLHRDSSPPYTD-----ISAYRRLV 353
+ L ++ LT P +S P P + + + D + ++L+
Sbjct: 1054 MKLGMENSLTEKIPKLNVPLNPKGRKLSAPGQPGLYIDQQELELEEDDYKMKVHEMQKLI 1113
Query: 354 GRLLYLNTT-RPDITFVTQQLSQFLAKRTQVHHSAAMRVLKYLKTSLGKGLFFPRDSALQ 412
G Y+ R D+ + L+Q + ++ +++++ + K L + + ++
Sbjct: 1114 GLASYVGYKFRFDLLYYINTLAQHILFPSKQVLDMTYELIQFIWNTRDKQLIWHKSKPVK 1173
Query: 413 ----ILGFSDADWAGCLDSRRSISGQCFFLGKSLISWRTKKQLIVSRSSSKEEYRALAAA 468
++ SDA + G +S G + L +I ++ K + S+++ E A++ +
Sbjct: 1174 PTNKLVVISDASY-GNQPYYKSQIGNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAISES 1232
Query: 469 TCELQWLLYLLQDL 482
L L YL+Q+L
Sbjct: 1233 VPLLNNLSYLIQEL 1246
>GLND_CHRVO (Q7NTY6) [Protein-PII] uridylyltransferase (EC 2.7.7.59)
(PII uridylyl-transferase) (Uridylyl removing enzyme)
(UTase)
Length = 856
Score = 30.8 bits (68), Expect = 9.0
Identities = 14/58 (24%), Positives = 32/58 (55%)
Query: 280 KIKDLGQLKYFLGLQVAHSKQGISLCQRKYCLDLLADSGLTHSKPISTPSDPSVKLHR 337
K+ L + +GL++ HS + + C R+ D+ ++ L ++ ++ P++P +L R
Sbjct: 96 KVSHFIGLMWDIGLEIGHSVRTLDECLREAAGDITIETNLLENRLVAGPAEPWRELMR 153
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.355 0.156 0.561
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 53,111,165
Number of Sequences: 164201
Number of extensions: 2002060
Number of successful extensions: 10282
Number of sequences better than 10.0: 11
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 10260
Number of HSP's gapped (non-prelim): 16
length of query: 527
length of database: 59,974,054
effective HSP length: 115
effective length of query: 412
effective length of database: 41,090,939
effective search space: 16929466868
effective search space used: 16929466868
T: 11
A: 40
X1: 14 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.6 bits)
S2: 68 (30.8 bits)
Medicago: description of AC123574.1


