

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC123573.8 + phase: 0 /pseudo
(548 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
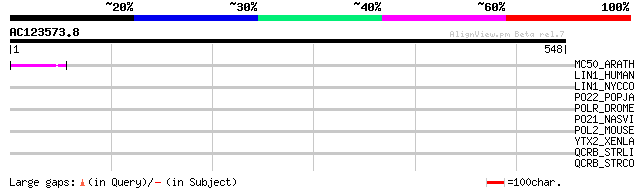
Score E
Sequences producing significant alignments: (bits) Value
MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250... 47 1e-04
LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog 43 0.002
LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog 40 0.020
PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type... 38 0.060
POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type... 34 0.86
PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type... 34 0.86
POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains... 34 1.1
YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein ... 31 9.5
QCRB_STRLI (Q9ZFB6) Ubiquinol-cytochrome c reductase cytochrome ... 31 9.5
QCRB_STRCO (Q9X806) Ubiquinol-cytochrome c reductase cytochrome ... 31 9.5
>MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250
(ORF102)
Length = 122
Score = 47.4 bits (111), Expect = 1e-04
Identities = 25/56 (44%), Positives = 31/56 (54%), Gaps = 1/56 (1%)
Query: 1 RGLRQGDPFSPFLFLLAAEGFHVLMESLSVNNFYTGYQIGINEPIVMSHLQFADDT 56
RGLRQGDP SP+LF+L E L G ++ N P + +HL FADDT
Sbjct: 26 RGLRQGDPLSPYLFILCTEVLSGLCRRAQEQGRLPGIRVSNNSPRI-NHLLFADDT 80
>LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog
Length = 1259
Score = 43.1 bits (100), Expect = 0.002
Identities = 41/152 (26%), Positives = 66/152 (42%), Gaps = 8/152 (5%)
Query: 2 GLRQGDPFSPFLFLLAAEGFHVLMESLSVNNFYTGYQIGINEPIVMSHLQFADDTLIVGE 61
G RQG P SP L + E VL ++ G Q+G E + +S FADD ++ E
Sbjct: 655 GTRQGCPLSPLLPNIVLE---VLARAIRQEKEIKGIQLG-KEEVKLS--LFADDMIVYLE 708
Query: 62 KS*ANVTGMRAALLLFETMSGLKVNFSKSLLVSVNVVGLWLSMVARVLNCRVGSLPFVYL 121
+ + + F +SG K+N KS S + L + S YL
Sbjct: 709 NPIVSAQNLLKLISNFSKVSGYKINVQKSQAFLYTNNRQTESQIMSELPFTIASKRIKYL 768
Query: 122 GMLIGGNVRCL--SF*EPIIDRIKSRLSGWKS 151
G+ + +V+ L +P+++ IK + WK+
Sbjct: 769 GIQLTRDVKDLFKENYKPLLNEIKEDTNKWKN 800
>LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog
Length = 1260
Score = 39.7 bits (91), Expect = 0.020
Identities = 30/90 (33%), Positives = 45/90 (49%), Gaps = 6/90 (6%)
Query: 2 GLRQGDPFSPFLFLLAAEGFHVLMESLSVNNFYTGYQIGINEPIVMSHLQFADDTLIVGE 61
G RQG P SP LF + E VL ++ G IG +E I +S FADD ++ E
Sbjct: 655 GTRQGCPLSPLLFNIVME---VLAIAIREEKAIKGIHIG-SEEIKLS--LFADDMIVYLE 708
Query: 62 KS*ANVTGMRAALLLFETMSGLKVNFSKSL 91
+ + T + + + +SG K+N KS+
Sbjct: 709 NTRDSTTKLLEVIKEYSNVSGYKINTHKSV 738
>PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 711
Score = 38.1 bits (87), Expect = 0.060
Identities = 32/99 (32%), Positives = 48/99 (48%), Gaps = 8/99 (8%)
Query: 1 RGLRQGDPFSPFLFLLAAEGFHVLMESLSVNNFYTGYQIGINEPIVMSHLQFADDTLIVG 60
RG++QGDP SPFLF + ++S G IG + V L FADD L++
Sbjct: 199 RGVKQGDPLSPFLFNAVLDELLCSLQSTP----GIGGTIGEEKIPV---LAFADDLLLLE 251
Query: 61 EKS*ANVTGMRAALLLFETMSGLKVNFSKSLLVSVNVVG 99
+ T + A + F + G+ +N KS+ +SV G
Sbjct: 252 DNDVLLPTTL-ATVANFFRLRGMSLNAKKSVSISVAASG 289
>POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type II
retrotransposable element R2DM [Contains: Protease (EC
3.4.23.-); Reverse transcriptase (EC 2.7.7.49);
Endonuclease]
Length = 1057
Score = 34.3 bits (77), Expect = 0.86
Identities = 31/100 (31%), Positives = 47/100 (47%), Gaps = 20/100 (20%)
Query: 1 RGLRQGDPFSPFLFLLAAEGFHVLMESLSVNNFYTGYQIGINEPIVMSHLQFADDTLIVG 60
RG++QGDP SP LF L + L+ +L G ++G + + FADD ++
Sbjct: 537 RGVKQGDPLSPILFNLVMDR---LLRTLPSE---IGAKVG---NAITNAAAFADDLVLFA 587
Query: 61 EKS*ANVTGMRAALLLFETMS-----GLKVNFSKSLLVSV 95
E T M +LL +T+ GLK+N K V +
Sbjct: 588 E------TRMGLQVLLDKTLDFLSIVGLKLNADKCFTVGI 621
>PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1025
Score = 34.3 bits (77), Expect = 0.86
Identities = 26/78 (33%), Positives = 40/78 (50%), Gaps = 13/78 (16%)
Query: 1 RGLRQGDPFSPFLFLLAAEGFHVLMESLSVNNFYTGYQIGINEPIVMSHLQFADDTLIVG 60
RG+RQGDP SP LF + ++ L N TG+ +G + + L FADD +++
Sbjct: 511 RGVRQGDPLSPLLFNCVMD---AVLRRLPEN---TGFLMGAEK---IGALVFADDLVLLA 561
Query: 61 EKS*ANVTGMRAALLLFE 78
E G++A+L E
Sbjct: 562 ETR----EGLQASLSRIE 575
>POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains:
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1300
Score = 33.9 bits (76), Expect = 1.1
Identities = 28/90 (31%), Positives = 43/90 (47%), Gaps = 6/90 (6%)
Query: 2 GLRQGDPFSPFLFLLAAEGFHVLMESLSVNNFYTGYQIGINEPIVMSHLQFADDTLIVGE 61
G RQG P SP+LF + E VL ++ G QIG E + +S L ADD ++
Sbjct: 682 GTRQGCPLSPYLFNIVLE---VLARAIRQQKEIKGIQIG-KEEVKISLL--ADDMIVYIS 735
Query: 62 KS*ANVTGMRAALLLFETMSGLKVNFSKSL 91
+ + + F + G K+N +KS+
Sbjct: 736 DPKNSTRELLNLINSFGEVVGYKINSNKSM 765
>YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein
(ORF 2)
Length = 1308
Score = 30.8 bits (68), Expect = 9.5
Identities = 28/93 (30%), Positives = 47/93 (50%), Gaps = 13/93 (13%)
Query: 1 RGLRQGDPFSPFLFLLAAEGFHVLMESLSVNNFYTGYQIGINEP---IVMSHLQFADDTL 57
RG+RQG P S L+ LA E F L+ TG + + EP +V+S +ADD +
Sbjct: 649 RGVRQGCPLSGQLYSLAIEPFLCLLRKR-----LTG--LVLKEPDMRVVLS--AYADDVI 699
Query: 58 IVGEKS*ANVTGMRAALLLFETMSGLKVNFSKS 90
+V + ++ + ++ S ++N+SKS
Sbjct: 700 LVAQDL-VDLERAQECQEVYAAASSARINWSKS 731
>QCRB_STRLI (Q9ZFB6) Ubiquinol-cytochrome c reductase cytochrome b
subunit
Length = 549
Score = 30.8 bits (68), Expect = 9.5
Identities = 19/71 (26%), Positives = 34/71 (47%), Gaps = 12/71 (16%)
Query: 68 TGMRAALLLFETMSGLKVNFSKSLLVSVNVVGLWLS------------MVARVLNCRVGS 115
TG L + +SG + F + ++SV +VG ++S V+R + +
Sbjct: 163 TGFTGYSLPDDLLSGTGIRFMEGAILSVPIVGTYISFFLFGGEFPGHDFVSRFYSIHILL 222
Query: 116 LPFVYLGMLIG 126
LP + LG+L+G
Sbjct: 223 LPGIMLGLLVG 233
>QCRB_STRCO (Q9X806) Ubiquinol-cytochrome c reductase cytochrome b
subunit
Length = 545
Score = 30.8 bits (68), Expect = 9.5
Identities = 19/71 (26%), Positives = 34/71 (47%), Gaps = 12/71 (16%)
Query: 68 TGMRAALLLFETMSGLKVNFSKSLLVSVNVVGLWLS------------MVARVLNCRVGS 115
TG L + +SG + F + ++SV +VG ++S V+R + +
Sbjct: 163 TGFTGYSLPDDLLSGTGIRFMEGAILSVPIVGTYISFFLFGGEFPGHDFVSRFYSIHILL 222
Query: 116 LPFVYLGMLIG 126
LP + LG+L+G
Sbjct: 223 LPGIMLGLLVG 233
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.351 0.160 0.556
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 56,856,738
Number of Sequences: 164201
Number of extensions: 2186183
Number of successful extensions: 7138
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 1
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 7130
Number of HSP's gapped (non-prelim): 12
length of query: 548
length of database: 59,974,054
effective HSP length: 115
effective length of query: 433
effective length of database: 41,090,939
effective search space: 17792376587
effective search space used: 17792376587
T: 11
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.9 bits)
S2: 68 (30.8 bits)
Medicago: description of AC123573.8


