

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC123570.3 + phase: 0 /pseudo
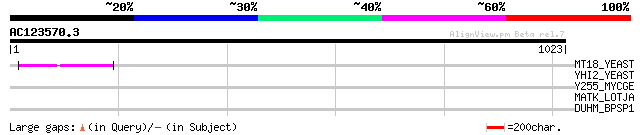
(1023 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
MT18_YEAST (P40469) DNA repair/transcription protein MET18/MMS19 67 3e-10
YHI2_YEAST (P38763) Hypothetical 29.0 kDa protein in DED81-MYO1 ... 36 0.60
Y255_MYCGE (P47497) Hypothetical protein MG255 32 6.7
MATK_LOTJA (Q9BBU2) Maturase K (Intron maturase) 32 6.7
DUHM_BPSP1 (P31654) Deoxyuridylate hydroxymethyltransferase (EC ... 32 8.7
>MT18_YEAST (P40469) DNA repair/transcription protein MET18/MMS19
Length = 1032
Score = 66.6 bits (161), Expect = 3e-10
Identities = 49/177 (27%), Positives = 81/177 (45%), Gaps = 6/177 (3%)
Query: 16 QHLHFQSLGLYDRKLCFELLDCLLEHHA--DSVASLEEDLIFGFCAAIDAERDPECLMPA 73
Q H S L+ K+ ++ D + + + V + + I F + E+DP L+ +
Sbjct: 136 QGQHLASTRLWPFKILRKIFDRFFVNGSSTEQVKRINDLFIETFLHVANGEKDPRNLLLS 195
Query: 74 FHIVESLARLYPDPSGLFASFARDVFDLLEPYFPIQFTHTTSGDAHVHRDDLSRTLMNAF 133
F + +S+ + +F D+FD+L YFPI F + DL L +A
Sbjct: 196 FALNKSITSSLQNVE----NFKEDLFDVLFCYFPITFKPPKHDPYKISNQDLKTALRSAI 251
Query: 134 SSTPLFEPFVIPLLLQKLSSSLHSAKIDSLQYLRVCSSKYGAERIAKYVGAIWSSLK 190
++TPLF LL KL++S K D+L L C K+G I + +W++LK
Sbjct: 252 TATPLFAEDAYSNLLDKLTASSPVVKNDTLLTLLECVRKFGGSSILENWTLLWNALK 308
Score = 38.9 bits (89), Expect = 0.071
Identities = 54/262 (20%), Positives = 101/262 (37%), Gaps = 48/262 (18%)
Query: 766 WIGKGLLLRGHEKIKDITKILTECLISDRNSSLPLIEGLDENNEEHKGDHLARKCAADAF 825
W+ KGL+++ + +I K + L ++ SL + F
Sbjct: 797 WLTKGLIMQNSLESSEIAKKFIDLLSNEEIGSL----------------------VSKLF 834
Query: 826 HVLMSDAEDCLNRK---FHATMRPLYKQRFFSSMMPIFLQLISRSDSSSSRYLLLRAFAR 882
V + D K ++ ++ LYKQ+FF + + + + + L A +
Sbjct: 835 EVFVMDISSLKKFKGISWNNNVKILYKQKFFGDIFQTLVSNYKNTVDMTIKCNYLTALSL 894
Query: 883 VMSVTPLIVILNDAKELISVLLDCLSMLTEDIQDKDILYGLLLVLSGMLTEKNESL---- 938
V+ TP + +L +LL L D+ D ++ L L T+K+ +L
Sbjct: 895 VLKHTPSQSVGPFINDLFPLLLQAL-----DMPDPEVRVSALETLKD-TTDKHHTLITEH 948
Query: 939 ------LFKAIQIKNDVNG*FLFLPILQLVRESCIQCLVALSKLPHVRIYPLRTQVLEAI 992
L ++ + + N + L LQL ++ + + L + Y + VL A+
Sbjct: 949 VSTIVPLLLSLSLPHKYNSVSVRLIALQL-----LEMITTVVPLNYCLSY--QDDVLSAL 1001
Query: 993 SKCLDDTKRSVRNEAVKCRQAW 1014
L D KR +R + V RQ +
Sbjct: 1002 IPVLSDKKRIIRKQCVDTRQVY 1023
>YHI2_YEAST (P38763) Hypothetical 29.0 kDa protein in DED81-MYO1
intergenic region
Length = 256
Score = 35.8 bits (81), Expect = 0.60
Identities = 39/162 (24%), Positives = 68/162 (41%), Gaps = 12/162 (7%)
Query: 179 AKYVGAIWSSLKDTINTYLMEPNFSFTLAPTDGID---FPKNEVVIEALSLLQQLIVQNS 235
AK G I+ S + + L+ N S T ID F N++ + LS L L ++ S
Sbjct: 91 AKNSGTIYESCGNFLEERLINANKS-TAQRRTSIDVQVFDTNQMEVSYLSELTTLQIRQS 149
Query: 236 SQLVSLIIDDKDVNFIINSIASYEMYDAVSVQEKKKLHVIGRILYIFAKTSIPSCNAVFQ 295
++ D +S+AS E Y + + + + I+ K + S +
Sbjct: 150 DAIILCFDSTND-----SSLASLESYICIIHHVRLECELDIPIIIACTKCDLDSERTITH 204
Query: 296 SLLLRMMDSLGFSVSNIDGLKNAGILASQSVNFGFLYLCIEL 337
+L + LGFS N+D + + + +VN L+L + L
Sbjct: 205 EKVLTFIQELGFSPGNLDYFETS---SKFNVNVEDLFLAVLL 243
>Y255_MYCGE (P47497) Hypothetical protein MG255
Length = 365
Score = 32.3 bits (72), Expect = 6.7
Identities = 37/132 (28%), Positives = 58/132 (43%), Gaps = 27/132 (20%)
Query: 162 SLQYLRVCSSKYGAERIAKYVG--AIWSSLKD---------TINTYLMEPNFSFTLAPTD 210
+L +LR C K +I+K+V I S +K T+ T L P FS T+A D
Sbjct: 116 ALLWLRQCWLKLQLTKISKFVNQKGILSFIKQQWPILTTLVTVGTTLGTPVFSLTIAQQD 175
Query: 211 GIDFPKNEVVIEALSLLQQLIVQNSSQLVSLIIDDKDVNFIINSIASYEMYDAVSVQEKK 270
GI V L + + S LVS +I F+++S+ S+++KK
Sbjct: 176 GIKQNAGNDVFIFLIIFS--VFSISLGLVSSLI------FLVSSL--------FSIRQKK 219
Query: 271 KLHVIGRILYIF 282
L + ++L F
Sbjct: 220 TLDALDKVLSKF 231
>MATK_LOTJA (Q9BBU2) Maturase K (Intron maturase)
Length = 508
Score = 32.3 bits (72), Expect = 6.7
Identities = 21/82 (25%), Positives = 38/82 (45%), Gaps = 5/82 (6%)
Query: 409 KSTFENILKKFMSIIIEDFNKTILWNSTLKSLFHIGSLFQNFSESEKAMSYRSFVLDKTM 468
K+ + I+ + +II+E IL++ L S + +++ S F DK
Sbjct: 87 KNFYSQIISEGFAIIVE-----ILFSLQLSSSLEEAEIIKSYKNLRSIHSIFPFFEDKVT 141
Query: 469 ELLSLDDISLPFSLKLEVLSDI 490
L + DI +P+ + LE+L I
Sbjct: 142 YLNYISDIRVPYPIHLEILVQI 163
>DUHM_BPSP1 (P31654) Deoxyuridylate hydroxymethyltransferase (EC
2.1.2.-) (Deoxyuridylate hydroxymethylase) (dUMP
hydroxymethylase) (dUMP-HMase)
Length = 383
Score = 32.0 bits (71), Expect = 8.7
Identities = 35/169 (20%), Positives = 68/169 (39%), Gaps = 20/169 (11%)
Query: 32 FELLDCLLEHHADSVASLEEDLIFGFCAAIDAERDPECLMPAFHIVESLARLYPDPSGLF 91
+++ D L + D A + ED+ + ++ + P +VE L DP+
Sbjct: 217 YQITDSLHVYLDDYGAKITEDIQKVYGVNLETDEAP--------VVEDFEELL-DPNPRM 267
Query: 92 ASFARDVFDLLEPYFPIQFTHTTSGDAHVHRDDLSRTLMNAFSSTPLFEPFVIPLLL--- 148
+ ++ L YF I + + ++H D ++ L+N P E + LLL
Sbjct: 268 SYTMNELNQFLNEYFGIVDSLMRDDETYMHDGDCAQMLLNQIRHEP-DEYIKVTLLLMVA 326
Query: 149 -QKLSSSLHSAKIDSLQYLRVCSSKYGAERIAKYVGAIWSSLKDTINTY 196
Q L+ + ++ ++ +C K A R ++ S + IN Y
Sbjct: 327 KQALNRGMKDTVATAMNWIPLCEIKVSALRF------LYKSYPEYINQY 369
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.326 0.141 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 109,444,429
Number of Sequences: 164201
Number of extensions: 4467285
Number of successful extensions: 13785
Number of sequences better than 10.0: 5
Number of HSP's better than 10.0 without gapping: 1
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 13779
Number of HSP's gapped (non-prelim): 9
length of query: 1023
length of database: 59,974,054
effective HSP length: 120
effective length of query: 903
effective length of database: 40,269,934
effective search space: 36363750402
effective search space used: 36363750402
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 71 (32.0 bits)
Medicago: description of AC123570.3


