

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122723.7 + phase: 0 /pseudo
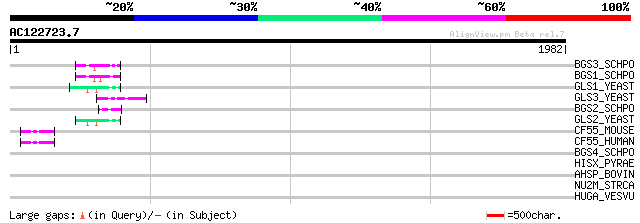
(1982 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BGS3_SCHPO (Q9P377) 1,3-beta-glucan synthase component bgs3 (EC ... 70 4e-11
BGS1_SCHPO (Q10287) 1,3-beta-glucan synthase component bgs1 (EC ... 55 3e-06
GLS1_YEAST (P38631) 1,3-beta-glucan synthase component GLS1 (EC ... 53 7e-06
GLS3_YEAST (Q04952) 1,3-beta-glucan synthase component FKS3 (EC ... 52 1e-05
BGS2_SCHPO (O13967) 1,3-beta-glucan synthase component bgs2 (EC ... 52 2e-05
GLS2_YEAST (P40989) 1,3-beta-glucan synthase component GLS2 (EC ... 47 5e-04
CF55_MOUSE (Q9CR26) Protein C6orf55 homolog 47 5e-04
CF55_HUMAN (Q9NP79) Protein C6orf55 (Dopamine responsive protein... 47 5e-04
BGS4_SCHPO (O74475) 1,3-beta-glucan synthase component bgs4 (EC ... 42 0.022
HISX_PYRAE (Q8ZY17) Histidinol dehydrogenase (EC 1.1.1.23) (HDH) 35 2.7
AHSP_BOVIN (Q865F8) Alpha-hemoglobin stabilizing protein (Erythr... 33 6.0
NU2M_STRCA (O21398) NADH-ubiquinone oxidoreductase chain 2 (EC 1... 33 7.9
HUGA_VESVU (P49370) Hyaluronoglucosaminidase (EC 3.2.1.35) (Hyal... 33 7.9
>BGS3_SCHPO (Q9P377) 1,3-beta-glucan synthase component bgs3 (EC
2.4.1.34) (1,3-beta-D-glucan-UDP glucosyltransferase)
Length = 1826
Score = 70.5 bits (171), Expect = 4e-11
Identities = 58/193 (30%), Positives = 87/193 (45%), Gaps = 49/193 (25%)
Query: 235 EDILDWLGSMFGFQKHNVANQREHLILLLANVHIRQFPNPDQQPKLDECALT---EVMKK 291
E+I L FGFQ N+ N ++L+++L + R P E LT + +
Sbjct: 163 ENIYIELAMKFGFQWDNMRNMFDYLMVMLDSRASRMTPQ--------EALLTLHADYIGG 214
Query: 292 LFKNYKKW---CK-----------------------YLDRKS--SLWLPTIQQEVQQRKL 323
N+KKW CK Y D S +LW+ + + ++
Sbjct: 215 PQSNFKKWYFACKMDQFDLKSGVLSFISRDPSTQVPYKDMSSCEALWISRMDELSNYERI 274
Query: 324 LYMGLYLLIWGEAANLRFMPECLCYIYHHMAFELYGMLAGNVSPMTGENIKPAYGGEDEA 383
+ LYLL WGEA N+RFMPECLC+IY +A++ +SP E PA +
Sbjct: 275 EQLALYLLCWGEANNVRFMPECLCFIY-KVAYDYL------ISPSFKEQKNPA---PKDY 324
Query: 384 FLRKVVTPIYNVI 396
FL +TP+YN++
Sbjct: 325 FLDNCITPLYNLM 337
>BGS1_SCHPO (Q10287) 1,3-beta-glucan synthase component bgs1 (EC
2.4.1.34) (1,3-beta-D-glucan-UDP glucosyltransferase)
Length = 1729
Score = 54.7 bits (130), Expect = 3e-06
Identities = 51/188 (27%), Positives = 82/188 (43%), Gaps = 44/188 (23%)
Query: 235 EDILDWLGSMFGFQKHNVANQREHLILLLANVHIRQFPNPDQQPKLDECALT---EVMKK 291
++IL L + GFQK N+ N +++++LL + R P+ LT +V+
Sbjct: 81 QEILLDLTNKLGFQKDNMRNIFDYVMVLLDSRASRMSPS--------SALLTIHADVIGG 132
Query: 292 LFKNYKKW-----------CKYLDRKSSLWLPTIQQEVQQ------------RKLLYMGL 328
N+ KW + D S + +E +Q R ++ + L
Sbjct: 133 EHANFSKWYFASHFNDGHAIGFHDMSSPIVETMTLKEAEQAWRDQMAAFSPHRMMVQVCL 192
Query: 329 YLLIWGEAANLRFMPECLCYIYHHMAFELYGMLAGNVSPMTGENIKPAYGGEDEAFLRKV 388
Y L WGEA N+RF+PECLC+I+ A++ Y ++ E E +L V
Sbjct: 193 YFLCWGEANNVRFVPECLCFIF-ECAYDYY---------ISSEAKDVDAALPKEFYLDSV 242
Query: 389 VTPIYNVI 396
+TPIY I
Sbjct: 243 ITPIYRFI 250
>GLS1_YEAST (P38631) 1,3-beta-glucan synthase component GLS1 (EC
2.4.1.34) (1,3-beta-D-glucan-UDP glucosyltransferase)
(CND1 protein) (CWN53 protein) (FKS1 protein)
(Papulacandin B sensitivity protein 1)
Length = 1876
Score = 53.1 bits (126), Expect = 7e-06
Identities = 59/238 (24%), Positives = 93/238 (38%), Gaps = 63/238 (26%)
Query: 212 AAVYALRNTRGLPWPNDYKK----KKDEDILDWLGSMFGFQKHNVANQREHLILLLANVH 267
A AL N W D + ++ EDI L + GFQ+ ++ N +H ++LL +
Sbjct: 138 AIAMALPNEPYPAWTADSQSPVSIEQIEDIFIDLTNRLGFQRDSMRNMFDHFMVLLDSRS 197
Query: 268 IRQFPNP----------------------------DQQPKLDECALTEVMKKLFKNYKKW 299
R P+ D + +L ++ +K K KK
Sbjct: 198 SRMSPDQALLSLHADYIGGDTANYKKWYFAAQLDMDDEIGFRNMSLGKLSRKARKAKKKN 257
Query: 300 CKYL------DRKSSL---------------WLPTIQQEVQQRKLLYMGLYLLIWGEAAN 338
K + D + +L W + Q ++ ++ LYLL WGEA
Sbjct: 258 KKAMEEANPEDTEETLNKIEGDNSLEAADFRWKAKMNQLSPLERVRHIALYLLCWGEANQ 317
Query: 339 LRFMPECLCYIYHHMAFELYGMLAGNVSPMTGENIKPAYGGEDEAFLRKVVTPIYNVI 396
+RF ECLC+IY L SP+ + +P G+ FL +V+TPIY+ I
Sbjct: 318 VRFTAECLCFIYKCALDYL-------DSPLCQQRQEPMPEGD---FLNRVITPIYHFI 365
>GLS3_YEAST (Q04952) 1,3-beta-glucan synthase component FKS3 (EC
2.4.1.34) (1,3-beta-D-glucan-UDP glucosyltransferase)
Length = 1785
Score = 52.4 bits (124), Expect = 1e-05
Identities = 47/186 (25%), Positives = 79/186 (42%), Gaps = 25/186 (13%)
Query: 310 WLPTIQQEVQQRKLLYMGLYLLIWGEAANLRFMPECLCYIYHHMAFELYGMLAGNVSPMT 369
W +++ + + + LYLL WGEA +RF PECLC+I+ L ++S +
Sbjct: 170 WKLKMKKLTPENMIRQLALYLLCWGEANQVRFAPECLCFIFK-------CALDYDISTSS 222
Query: 370 GENI--KPAYGGEDEAFLRKVVTPIYNVI---AELKKAKGEGQSIHNGGTMMI*MNISGW 424
E P Y ++L V+TP+Y + K AKG + NI G+
Sbjct: 223 SEKTVKSPEY-----SYLNDVITPLYEFLRGQVYKKDAKGNWKRREKDH-----KNIIGY 272
Query: 425 PMRADADFFCLPAERVVFDKSNPCCNSNA*DDKPPNRDGWFGKV---NFVEIRSFWHLFR 481
++ ER++ + + + +D + KV + E RS+ H F
Sbjct: 273 DDINQLFWYPEGFERIILNNGERLVDKPLEERYLYFKDVAWSKVFYKTYRETRSWKHCFT 332
Query: 482 SFDRMW 487
+F+R W
Sbjct: 333 NFNRFW 338
>BGS2_SCHPO (O13967) 1,3-beta-glucan synthase component bgs2 (EC
2.4.1.34) (1,3-beta-D-glucan-UDP glucosyltransferase)
(Meiotic expression up-regulated protein 21)
Length = 1894
Score = 52.0 bits (123), Expect = 2e-05
Identities = 34/83 (40%), Positives = 45/83 (53%), Gaps = 15/83 (18%)
Query: 317 EVQQRKLLYMGLYLLIWGEAANLRFMPECLCYIYHHMAFELYGMLAGNVSPMTGENIKPA 376
E Q R+L LYLL WGEA N+RF PECLC+I+ LA + M E+ +
Sbjct: 333 ETQVRQL---ALYLLCWGEANNIRFCPECLCFIF---------KLANDF--MQSEDYAKS 378
Query: 377 YGGEDEAF-LRKVVTPIYNVIAE 398
ED+ F L V+TP+Y I +
Sbjct: 379 EPIEDDCFYLDNVITPLYEFIRD 401
>GLS2_YEAST (P40989) 1,3-beta-glucan synthase component GLS2 (EC
2.4.1.34) (1,3-beta-D-glucan-UDP glucosyltransferase)
Length = 1895
Score = 47.0 bits (110), Expect = 5e-04
Identities = 51/211 (24%), Positives = 75/211 (35%), Gaps = 59/211 (27%)
Query: 235 EDILDWLGSMFGFQKHNVANQREHLILLLANVHIRQFPNP-------------------- 274
EDI L + FGFQ+ ++ N +H + LL + R P
Sbjct: 184 EDIFIDLTNKFGFQRDSMRNMFDHFMTLLDSRSSRMSPEQALLSLHADYIGGDTANYKKW 243
Query: 275 --------DQQPKLDECALTEVMKKLFKNYKKWCKYLDRKSS------------------ 308
D + L ++ +K K KK K + S
Sbjct: 244 YFAAQLDMDDEIGFRNMKLGKLSRKARKAKKKNKKAMQEASPEDTEETLNQIEGDNSLEA 303
Query: 309 ---LWLPTIQQEVQQRKLLYMGLYLLIWGEAANLRFMPECLCYIYHHMAFELYGMLAGNV 365
W + Q + + L+LL WGEA +RF PECLC+IY + L
Sbjct: 304 ADFRWKSKMNQLSPFEMVRQIALFLLCWGEANQVRFTPECLCFIYKCASDYL-------D 356
Query: 366 SPMTGENIKPAYGGEDEAFLRKVVTPIYNVI 396
S + P G+ FL +V+TP+Y I
Sbjct: 357 SAQCQQRPDPLPEGD---FLNRVITPLYRFI 384
>CF55_MOUSE (Q9CR26) Protein C6orf55 homolog
Length = 309
Score = 47.0 bits (110), Expect = 5e-04
Identities = 36/126 (28%), Positives = 61/126 (47%), Gaps = 12/126 (9%)
Query: 38 VPSSLVEIAPILRVANEVEKTHPRVAYLCRFYAFEKAHRLDPTSSGRGVRQFKTALLQRL 97
+P+ I LR A E +K P VAY CR YA + ++D S R+F + L+ +L
Sbjct: 10 LPAQFKSIQHHLRTAQEHDKRDPVVAYYCRLYAMQTGMKID--SKTPECRKFLSKLMDQL 67
Query: 98 ERENDPTLKGRVKKSDAREMQSFYQHYYKKYIQALQNAADKADRA-----QLTKAYQTAN 152
E LK ++ ++A + + + Y + AD DRA + K++ TA+
Sbjct: 68 E-----ALKKQLGDNEAVTQEIVGCAHLENYALKMFLYADNEDRAGRFHKNMIKSFYTAS 122
Query: 153 VLFEVL 158
+L +V+
Sbjct: 123 LLIDVI 128
>CF55_HUMAN (Q9NP79) Protein C6orf55 (Dopamine responsive protein
DRG-1) (My012 protein) (HSPC228)
Length = 307
Score = 47.0 bits (110), Expect = 5e-04
Identities = 36/126 (28%), Positives = 61/126 (47%), Gaps = 12/126 (9%)
Query: 38 VPSSLVEIAPILRVANEVEKTHPRVAYLCRFYAFEKAHRLDPTSSGRGVRQFKTALLQRL 97
+P+ I LR A E +K P VAY CR YA + ++D S R+F + L+ +L
Sbjct: 10 LPAQFKSIQHHLRTAQEHDKRDPVVAYYCRLYAMQTGMKID--SKTPECRKFLSKLMDQL 67
Query: 98 ERENDPTLKGRVKKSDAREMQSFYQHYYKKYIQALQNAADKADRA-----QLTKAYQTAN 152
E LK ++ ++A + + + Y + AD DRA + K++ TA+
Sbjct: 68 E-----ALKKQLGDNEAITQEIVGCAHLENYALKMFLYADNEDRAGRFHKNMIKSFYTAS 122
Query: 153 VLFEVL 158
+L +V+
Sbjct: 123 LLIDVI 128
>BGS4_SCHPO (O74475) 1,3-beta-glucan synthase component bgs4 (EC
2.4.1.34) (1,3-beta-D-glucan-UDP glucosyltransferase)
Length = 1955
Score = 41.6 bits (96), Expect = 0.022
Identities = 24/84 (28%), Positives = 42/84 (49%), Gaps = 10/84 (11%)
Query: 310 WLPTIQQEVQQRKLLYMGLYLLIWGEAANLRFMPECLCYIYHHMAFELYGMLAGNVSPMT 369
W ++ Q + + L+LL+WGEA N+RFMPE + +++ + A NV+
Sbjct: 353 WRSHMRSMTQFERAQQIALWLLLWGEANNVRFMPEVIAFLFKCAYDYIISPEAQNVT--- 409
Query: 370 GENIKPAYGGEDEAFLRKVVTPIY 393
E + Y +L +V+P+Y
Sbjct: 410 -EPVPEGY------YLDNIVSPLY 426
>HISX_PYRAE (Q8ZY17) Histidinol dehydrogenase (EC 1.1.1.23) (HDH)
Length = 368
Score = 34.7 bits (78), Expect = 2.7
Identities = 37/122 (30%), Positives = 51/122 (41%), Gaps = 12/122 (9%)
Query: 506 GDPTVIFHGDVFKKVLSVFITA----AILKFGQAVLGVILSWKARRSMSLYVKLRYILKV 561
GDP V+ K L + A + F +L IL WK R +LYV RYI +
Sbjct: 50 GDPEVVSAALAAAKSLEALYSRISPPAAVDFYGGILRQIL-WKPVRRAALYVPARYISTL 108
Query: 562 ISAAAWVILLSVTYAYTWDNPPGFAETIKSWFGSNSSAPSLFIVAVVVYLSPNMLA-AIF 620
+ A L V Y P G +E + + A L + AV+ P+ LA A+F
Sbjct: 109 VMLAVPARLAGVEEVYVVTPPRGVSEELL------AVAKELGVKAVLALGGPHGLAYAVF 162
Query: 621 FM 622
M
Sbjct: 163 HM 164
>AHSP_BOVIN (Q865F8) Alpha-hemoglobin stabilizing protein (Erythroid
associated factor)
Length = 92
Score = 33.5 bits (75), Expect = 6.0
Identities = 17/56 (30%), Positives = 30/56 (53%), Gaps = 2/56 (3%)
Query: 84 RGVRQFKTALLQRLERENDPTLKGRVKKSDAREMQSFYQHYYKKYIQALQNAADKA 139
+G+++F L Q++ +DP + + + SFY +YYKK + Q+ DKA
Sbjct: 13 KGIKEFNILLNQQVF--SDPAISEEAMVTVVNDWVSFYINYYKKQLSGEQDEQDKA 66
>NU2M_STRCA (O21398) NADH-ubiquinone oxidoreductase chain 2 (EC
1.6.5.3)
Length = 346
Score = 33.1 bits (74), Expect = 7.9
Identities = 20/66 (30%), Positives = 35/66 (52%), Gaps = 4/66 (6%)
Query: 558 ILKVISAAAWVILLSVTYAYTWDNPPGFAETIKSWFGSNSSAPSLFIV----AVVVYLSP 613
I+ ++S L + Y T PP A +K W+ SN + S+ I+ A+++ +SP
Sbjct: 281 IIALLSLLGLFFYLRLAYYATITLPPNSANHMKQWYISNPTNTSIAILSSLSAILLPISP 340
Query: 614 NMLAAI 619
+LAA+
Sbjct: 341 MILAAL 346
>HUGA_VESVU (P49370) Hyaluronoglucosaminidase (EC 3.2.1.35)
(Hyaluronidase) (Allergen Ves v 2) (Ves v II)
Length = 331
Score = 33.1 bits (74), Expect = 7.9
Identities = 29/126 (23%), Positives = 54/126 (42%), Gaps = 23/126 (18%)
Query: 228 DYKKKKDEDILDWLGSMFGFQKHNVANQREHLILLLANVHIRQFPNPDQQPKLDECALTE 287
D + D + WL F NV LL +V++RQ PDQ+ L + + E
Sbjct: 198 DVTAMHENDKMSWL-----FNNQNV---------LLPSVYVRQELTPDQRIGLVQGRVKE 243
Query: 288 VMKKLFKNYKK--------WCKYLDRKSSLWLPTIQQEVQQRKLLYMGLYLLIWGEAANL 339
++ + N K W Y D ++ T ++ Q ++ G ++IWG ++++
Sbjct: 244 AVR-ISNNLKHSPKVLSYWWYVYQDETNTFLTETDVKKTFQEIVINGGDGIIIWGSSSDV 302
Query: 340 RFMPEC 345
+ +C
Sbjct: 303 NSLSKC 308
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.348 0.154 0.546
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 207,689,051
Number of Sequences: 164201
Number of extensions: 8206193
Number of successful extensions: 28048
Number of sequences better than 10.0: 13
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 28007
Number of HSP's gapped (non-prelim): 38
length of query: 1982
length of database: 59,974,054
effective HSP length: 125
effective length of query: 1857
effective length of database: 39,448,929
effective search space: 73256661153
effective search space used: 73256661153
T: 11
A: 40
X1: 14 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.8 bits)
S2: 74 (33.1 bits)
Medicago: description of AC122723.7


