

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122722.5 - phase: 0
(379 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
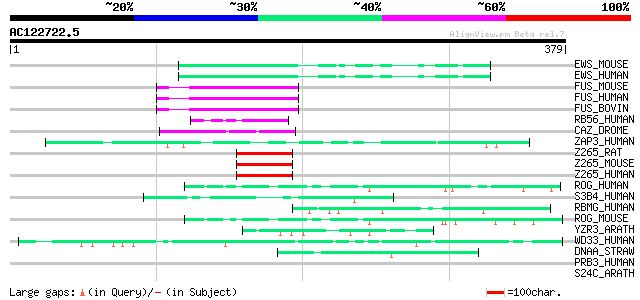
Score E
Sequences producing significant alignments: (bits) Value
EWS_MOUSE (Q61545) RNA-binding protein EWS 76 1e-13
EWS_HUMAN (Q01844) RNA-binding protein EWS (EWS oncogene) (Ewing... 76 1e-13
FUS_MOUSE (P56959) RNA-binding protein FUS (Pigpen protein) 67 6e-11
FUS_HUMAN (P35637) RNA-binding protein FUS (Oncogene FUS) (Oncog... 67 6e-11
FUS_BOVIN (Q28009) RNA-binding protein FUS (Pigpen protein) 67 6e-11
RB56_HUMAN (Q92804) TATA-binding protein associated factor 2N (R... 66 1e-10
CAZ_DROME (Q27294) RNA-binding protein cabeza (Sarcoma-associate... 61 5e-09
ZAP3_HUMAN (P49750) Nuclear protein ZAP3 (ZAP113) 58 3e-08
Z265_RAT (O35986) Zinc finger protein 265 (Zinc finger, splicing) 54 7e-07
Z265_MOUSE (Q9R020) Zinc finger protein 265 (Zinc finger, splici... 54 7e-07
Z265_HUMAN (O95218) Zinc finger protein 265 (Zinc finger, splicing) 54 7e-07
ROG_HUMAN (P38159) Heterogeneous nuclear ribonucleoprotein G (hn... 54 9e-07
S3B4_HUMAN (Q15427) Splicing factor 3B subunit 4 (Spliceosome as... 51 6e-06
RBMG_HUMAN (Q9UPN6) Putative RNA-binding protein 16 (RNA binding... 50 9e-06
ROG_MOUSE (O35479) Heterogeneous nuclear ribonucleoprotein G (hn... 49 2e-05
YZR3_ARATH (Q8GZ43) Hypothetical RanBP2-type zinc-finger protein... 47 8e-05
WD33_HUMAN (Q9C0J8) WD-repeat protein 33 (WD-repeat protein WDC146) 47 8e-05
DNAA_STRAW (Q82FD8) Chromosomal replication initiator protein dnaA 45 3e-04
PRB3_HUMAN (Q04118) Basic salivary proline-rich protein 3 precur... 42 0.002
S24C_ARATH (Q9M291) Protein transport protein Sec24-like CEF 42 0.003
>EWS_MOUSE (Q61545) RNA-binding protein EWS
Length = 655
Score = 76.3 bits (186), Expect = 1e-13
Identities = 69/213 (32%), Positives = 79/213 (36%), Gaps = 51/213 (23%)
Query: 116 PPHPYARGRPGGRPFGRAFDGPGYGHRHGRGEGLHGRNNPNVRPREGDWMCPDALCGNLN 175
P P G P GR GR D G+ R RG + NV+ R GDW CP+ CGN N
Sbjct: 473 PGGPGGPGGPMGRMGGRGGDRGGFPPRGPRGSRGNPSGGGNVQHRAGDWQCPNPGCGNQN 532
Query: 176 FARRDFCNQCKRPRPAAAGSPPRRGSPPLHAPPRRYPGPPFDRSPERPMNGYRSPPPRLM 235
FA R CNQCK P+P PP +P P DR P G R
Sbjct: 533 FAWRTECNQCKAPKPEGFLPPP-------------FPPPGGDRGRGGP-GGMRG------ 572
Query: 236 GRDGLRDYGPAAAALPPLRHEGRFPDPHLHRERMDYMDDAYRGRGKFDRPPPLDWDNRDR 295
GR GL D G + GR D RG F R
Sbjct: 573 GRGGLMDRGGPGG----MFRGGRGGD-----------------RGGFR-------GGRGM 604
Query: 296 GRDGFSNERKGFERRPLSPSAPLLPSLPPHRGG 328
R GF R+G P P PL+ + RGG
Sbjct: 605 DRGGFGGGRRG---GPGGPPGPLMEQMGGRRGG 634
>EWS_HUMAN (Q01844) RNA-binding protein EWS (EWS oncogene) (Ewing
sarcoma breakpoint region 1 protein)
Length = 656
Score = 76.3 bits (186), Expect = 1e-13
Identities = 69/213 (32%), Positives = 79/213 (36%), Gaps = 51/213 (23%)
Query: 116 PPHPYARGRPGGRPFGRAFDGPGYGHRHGRGEGLHGRNNPNVRPREGDWMCPDALCGNLN 175
P P G P GR GR D G+ R RG + NV+ R GDW CP+ CGN N
Sbjct: 474 PGGPGGPGGPMGRMGGRGGDRGGFPPRGPRGSRGNPSGGGNVQHRAGDWQCPNPGCGNQN 533
Query: 176 FARRDFCNQCKRPRPAAAGSPPRRGSPPLHAPPRRYPGPPFDRSPERPMNGYRSPPPRLM 235
FA R CNQCK P+P PP +P P DR P G R
Sbjct: 534 FAWRTECNQCKAPKPEGFLPPP-------------FPPPGGDRGRGGP-GGMRG------ 573
Query: 236 GRDGLRDYGPAAAALPPLRHEGRFPDPHLHRERMDYMDDAYRGRGKFDRPPPLDWDNRDR 295
GR GL D G + GR D RG F R
Sbjct: 574 GRGGLMDRGGPGG----MFRGGRGGD-----------------RGGFR-------GGRGM 605
Query: 296 GRDGFSNERKGFERRPLSPSAPLLPSLPPHRGG 328
R GF R+G P P PL+ + RGG
Sbjct: 606 DRGGFGGGRRG---GPGGPPGPLMEQMGGRRGG 635
>FUS_MOUSE (P56959) RNA-binding protein FUS (Pigpen protein)
Length = 518
Score = 67.4 bits (163), Expect = 6e-11
Identities = 38/97 (39%), Positives = 44/97 (45%), Gaps = 13/97 (13%)
Query: 101 NRGFGGGRDFGRFRDPPHPYARGRPGGRPFGRAFDGPGYGHRHGRGEGLHGRNNPNVRPR 160
NRG G GR GR G P GR G G GRG G + R
Sbjct: 369 NRGGGNGRG-------------GRGRGGPMGRGGYGGGGSGGGGRGGFPSGGGGGGGQQR 415
Query: 161 EGDWMCPDALCGNLNFARRDFCNQCKRPRPAAAGSPP 197
GDW CP+ C N+NF+ R+ CNQCK P+P G P
Sbjct: 416 AGDWKCPNPTCENMNFSWRNECNQCKAPKPDGPGGGP 452
>FUS_HUMAN (P35637) RNA-binding protein FUS (Oncogene FUS) (Oncogene
TLS) (Translocated in liposarcoma protein) (POMp75) (75
kDa DNA-pairing protein)
Length = 526
Score = 67.4 bits (163), Expect = 6e-11
Identities = 38/97 (39%), Positives = 44/97 (45%), Gaps = 13/97 (13%)
Query: 101 NRGFGGGRDFGRFRDPPHPYARGRPGGRPFGRAFDGPGYGHRHGRGEGLHGRNNPNVRPR 160
NRG G GR GR G P GR G G GRG G + R
Sbjct: 376 NRGGGNGRG-------------GRGRGGPMGRGGYGGGGSGGGGRGGFPSGGGGGGGQQR 422
Query: 161 EGDWMCPDALCGNLNFARRDFCNQCKRPRPAAAGSPP 197
GDW CP+ C N+NF+ R+ CNQCK P+P G P
Sbjct: 423 AGDWKCPNPTCENMNFSWRNECNQCKAPKPDGPGGGP 459
>FUS_BOVIN (Q28009) RNA-binding protein FUS (Pigpen protein)
Length = 512
Score = 67.4 bits (163), Expect = 6e-11
Identities = 38/97 (39%), Positives = 44/97 (45%), Gaps = 13/97 (13%)
Query: 101 NRGFGGGRDFGRFRDPPHPYARGRPGGRPFGRAFDGPGYGHRHGRGEGLHGRNNPNVRPR 160
NRG G GR GR G P GR G G GRG G + R
Sbjct: 362 NRGGGNGRG-------------GRGRGGPMGRGGYGGGGSGGGGRGGFPSGGGGGGGQQR 408
Query: 161 EGDWMCPDALCGNLNFARRDFCNQCKRPRPAAAGSPP 197
GDW CP+ C N+NF+ R+ CNQCK P+P G P
Sbjct: 409 AGDWKCPNPTCENMNFSWRNECNQCKAPKPDGPGGGP 445
>RB56_HUMAN (Q92804) TATA-binding protein associated factor 2N
(RNA-binding protein 56) (TAFII68) (TAF(II)68)
Length = 592
Score = 66.2 bits (160), Expect = 1e-10
Identities = 34/67 (50%), Positives = 39/67 (57%), Gaps = 8/67 (11%)
Query: 124 RPGGRPFGRAFDGPGYGHRHGRGEGLHGRNNPNVRPREGDWMCPDALCGNLNFARRDFCN 183
R GG GR G G GRG G GR P+ GDW+CP+ CGN+NFARR+ CN
Sbjct: 326 RGGGSGGGRR----GRGGYRGRG-GFQGRGGD---PKSGDWVCPNPSCGNMNFARRNSCN 377
Query: 184 QCKRPRP 190
QC PRP
Sbjct: 378 QCNEPRP 384
Score = 30.8 bits (68), Expect = 5.9
Identities = 29/94 (30%), Positives = 37/94 (38%), Gaps = 10/94 (10%)
Query: 295 RGRDGFSNERKGFERRPLSPSAPLLPSLPPHRGGDRWSR--------DVRDRSRSPIRGG 346
RGR G+ R GF+ R P + P G ++R + R P G
Sbjct: 335 RGRGGYRG-RGGFQGRGGDPKSGDWVCPNPSCGNMNFARRNSCNQCNEPRPEDSRPSGGD 393
Query: 347 PPAKDYRRDP-VMSRGGRDDRRGGVGRDRIGGMY 379
+ Y + RGGR RGG G DR GG Y
Sbjct: 394 FRGRGYGGERGYRGRGGRGGDRGGYGGDRSGGGY 427
>CAZ_DROME (Q27294) RNA-binding protein cabeza (Sarcoma-associated
RNA-binding fly homolog) (P19)
Length = 399
Score = 60.8 bits (146), Expect = 5e-09
Identities = 40/94 (42%), Positives = 46/94 (48%), Gaps = 4/94 (4%)
Query: 103 GFGGGRD-FGRFRDPPHPYARGRPGGRPFGRAFDGPGYGHRHGRGEGLHGRNNPNVRPRE 161
G GGGR FG R G GG F R G G + G G G G NV+PR+
Sbjct: 218 GGGGGRGGFGGRRGGGGGGGGGGGGGGRFDRGGGGGGGRYDRGGGGG-GGGGGGNVQPRD 276
Query: 162 GDWMCPDALCGNLNFARRDFCNQCKRPRPAAAGS 195
GDW C C N NFA R+ CN+CK P+ GS
Sbjct: 277 GDWKCNS--CNNTNFAWRNECNRCKTPKGDDEGS 308
>ZAP3_HUMAN (P49750) Nuclear protein ZAP3 (ZAP113)
Length = 1822
Score = 58.2 bits (139), Expect = 3e-08
Identities = 95/355 (26%), Positives = 119/355 (32%), Gaps = 76/355 (21%)
Query: 25 VVRPSLTENSPPGAAAHSNDYEPGELRRDPPPVNYSRSDRYSDDAGYRFRAGSSSPVHHR 84
+VRP + PG S D R PP +R G RAGS + R
Sbjct: 888 LVRPGSSREKVPGGLQGSQDRGAAGSRERGPPRRAGSQER-----GPLRRAGSRERIPPR 942
Query: 85 RDADHRFPSDYNHFPRNRGFGG---GRDFGRFRDPP-------HPYARGRPGGRPFGRAF 134
R R RG G GRD G FR P +PY R P P+
Sbjct: 943 RAGSRERGPPRGPGSRERGLGRSDFGRDRGPFRPEPGDGGEKMYPYHRDEPPRAPWN--- 999
Query: 135 DGPGYGHRHGRGE-GLHGRNNPNVRPREGDWMCPDALCGNLNFARRDFCNQCKRPRPAAA 193
+G G E L GRN P R R DW R + +C+R
Sbjct: 1000 ----HGEERGHEEFPLDGRNAPMERERLDDW------------DRERYWRECERDY---- 1039
Query: 194 GSPPRRGSPPLHAPPRRYPGPPFDRSPERPMNGYRSPPPRLMGRDGLRDYGPAAAALPPL 253
+ + L+ R+ P P R +G R P D RD
Sbjct: 1040 ----QDDTLELYNREDRFSAP-----PSRSHDGDRRGP---WWDDWERD----------- 1076
Query: 254 RHEGRFPDPHLHRERMDYMDDAYRGRGKFDRPPPLDWDNRDRGRDGFSNERKGFERR-PL 312
+ + + R+ +D R +DR +WD RD GR E + ER P
Sbjct: 1077 QDMDEDYNREMERDMDRDVDRISRPMDMYDRSLDNEWD-RDYGRPLDEQESQFRERDIPS 1135
Query: 313 SPSAPLLPSLPP--HRGGDRW----------SRDVRDRSRSPIRGGPPAKDYRRD 355
P P LP LPP DRW RD RDR IR P D R+
Sbjct: 1136 LPPLPPLPPLPPLDRYRDDRWREERNREHGYDRDFRDRGELRIREYPERGDTWRE 1190
Score = 31.6 bits (70), Expect = 3.5
Identities = 39/137 (28%), Positives = 53/137 (38%), Gaps = 49/137 (35%)
Query: 189 RPAAAGSPPRRGSPPLHAPPRRYPGPPFDRSPERPMNGYRSPPPRLMGRDGLRDYGPAAA 248
RP+A PP R S P+ PP P PP P P+ PPP ++ P +
Sbjct: 1332 RPSA---PPARSSVPVTRPPVPIPPPP----PPPPL----PPPPPVI--------KPQTS 1372
Query: 249 ALPPLRHEGRFPDPHLHRERMDYMDDAYRGRGKFDRPPPLDWDNRDRGRDGFSNERKGFE 308
A + +ER D +D++ G WD D G ++E K
Sbjct: 1373 A--------------VEQERWD--EDSFYGL----------WDTND--EQGLNSEFK--S 1402
Query: 309 RRPLSPSAPLLPSLPPH 325
PSAP+LP P H
Sbjct: 1403 ETAAIPSAPVLPPPPVH 1419
>Z265_RAT (O35986) Zinc finger protein 265 (Zinc finger, splicing)
Length = 332
Score = 53.9 bits (128), Expect = 7e-07
Identities = 21/38 (55%), Positives = 26/38 (68%)
Query: 156 NVRPREGDWMCPDALCGNLNFARRDFCNQCKRPRPAAA 193
N R +GDW+CPD CGN+NFARR CN+C R + A
Sbjct: 5 NFRVSDGDWICPDKKCGNVNFARRTSCNRCGREKTTEA 42
Score = 34.7 bits (78), Expect = 0.41
Identities = 14/29 (48%), Positives = 17/29 (58%), Gaps = 2/29 (6%)
Query: 163 DWMCPDALCGNLNFARRDFCNQCKRPRPA 191
DW C C N+N+ARR CN C P+ A
Sbjct: 68 DWQCKT--CSNVNWARRSECNMCNTPKYA 94
>Z265_MOUSE (Q9R020) Zinc finger protein 265 (Zinc finger, splicing)
(Fragment)
Length = 326
Score = 53.9 bits (128), Expect = 7e-07
Identities = 21/38 (55%), Positives = 26/38 (68%)
Query: 156 NVRPREGDWMCPDALCGNLNFARRDFCNQCKRPRPAAA 193
N R +GDW+CPD CGN+NFARR CN+C R + A
Sbjct: 5 NFRVSDGDWICPDKKCGNVNFARRTSCNRCGREKTTEA 42
Score = 32.7 bits (73), Expect = 1.6
Identities = 13/29 (44%), Positives = 17/29 (57%), Gaps = 2/29 (6%)
Query: 163 DWMCPDALCGNLNFARRDFCNQCKRPRPA 191
DW C C N+N+ARR C+ C P+ A
Sbjct: 68 DWQCKT--CSNVNWARRSECDMCNTPKYA 94
>Z265_HUMAN (O95218) Zinc finger protein 265 (Zinc finger, splicing)
Length = 337
Score = 53.9 bits (128), Expect = 7e-07
Identities = 21/38 (55%), Positives = 26/38 (68%)
Query: 156 NVRPREGDWMCPDALCGNLNFARRDFCNQCKRPRPAAA 193
N R +GDW+CPD CGN+NFARR CN+C R + A
Sbjct: 5 NFRVSDGDWICPDKKCGNVNFARRTSCNRCGREKTTEA 42
Score = 34.7 bits (78), Expect = 0.41
Identities = 14/29 (48%), Positives = 17/29 (58%), Gaps = 2/29 (6%)
Query: 163 DWMCPDALCGNLNFARRDFCNQCKRPRPA 191
DW C C N+N+ARR CN C P+ A
Sbjct: 68 DWQCKT--CSNVNWARRSECNMCNTPKYA 94
>ROG_HUMAN (P38159) Heterogeneous nuclear ribonucleoprotein G (hnRNP
G) (RNA binding motif protein, X chromosome)
(Glycoprotein p43)
Length = 391
Score = 53.5 bits (127), Expect = 9e-07
Identities = 85/283 (30%), Positives = 101/283 (35%), Gaps = 54/283 (19%)
Query: 120 YARGRPGGRPFGRAFDGPGYGHRHGRGEGLHGRNNPNVRPREGDWMCPDALCGNLNFARR 179
+ GR G P R+ GP G R GRG G G P P G M N N +
Sbjct: 89 FESGRRGPPPPPRS-RGPPRGLRGGRG-GSGGTRGP---PSRGGHMDDGGYSMNFNMS-- 141
Query: 180 DFCNQCKRPRPAAAGSPPRRGSPPLHAPPRRYPGPPFDRSPERPMNGYRSPPPRLMGRDG 239
+ P P G PPR G PP P R P P R +G P GRD
Sbjct: 142 ----SSRGPLPVKRGPPPRSGGPP---PKRSAPS-----GPVRSSSGMGGRAPVSRGRD- 188
Query: 240 LRDYG--PAAAALPPLRHEGRFPDPHLHRERMDYMDDAY-RGRGKFD-RPPPLDWDNRDR 295
YG P LP R P + + Y Y R D PPP D+ RD
Sbjct: 189 --SYGGPPRREPLPSRRDVYLSPRDDGYSTKDSYSSRDYPSSRDTRDYAPPPRDYTYRDY 246
Query: 296 G----RDGFS----NERKGFER-RPLSPSAPLLPSLPPHRGGDRWSRDVRDRSRSPIRGG 346
G RD + ++R G+ R R S PS +R D + RS P RG
Sbjct: 247 GHSSSRDDYPSRGYSDRDGYGRDRDYSDH----PSGGSYR--DSYESYGNSRSAPPTRGP 300
Query: 347 PPA-------KDYRRDPVMSRGGRDDRRG------GVGRDRIG 376
PP+ DY G RD GRDR+G
Sbjct: 301 PPSYGGSSRYDDYSSSRDGYGGSRDSYSSSRSDLYSSGRDRVG 343
>S3B4_HUMAN (Q15427) Splicing factor 3B subunit 4 (Spliceosome
associated protein 49) (SAP 49) (SF3b50) (Pre-mRNA
splicing factor SF3b 49 kDa subunit)
Length = 424
Score = 50.8 bits (120), Expect = 6e-06
Identities = 50/177 (28%), Positives = 57/177 (31%), Gaps = 37/177 (20%)
Query: 92 PSDYNHFPRNRGFGGGRDFGRFRDPPHPYARGRPGGRPFGRAFDGPGYGHRHGRGEGLHG 151
P H P + G G G PHP+ PGG P PG G HG
Sbjct: 268 PGAAGHGPPSAGTPGAGHPGHGHSHPHPFP---PGGMPH------PGMSQMQLAHHGPHG 318
Query: 152 RNNPNVRPREGDWMCPDALCGNLNFARRDFCNQCKRPRPAAAGSPPRRGSPPLHAPPRRY 211
+P+ P P RP P P G PP+ PPR
Sbjct: 319 LGHPHAGPPGSGGQPPP------------------RPPPGM----PHPGPPPMGMPPRGP 356
Query: 212 P-GPPFDRSPERPMNGYRSPPPRL-----MGRDGLRDYGPAAAALPPLRHEGRFPDP 262
P G P P +G R PPP + G YG LPP R R P P
Sbjct: 357 PFGSPMGHPGPMPPHGMRGPPPLMPPHGYTGPPRPPPYGYQRGPLPPPRPTPRPPVP 413
>RBMG_HUMAN (Q9UPN6) Putative RNA-binding protein 16 (RNA binding
motif protein 16)
Length = 1271
Score = 50.1 bits (118), Expect = 9e-06
Identities = 59/202 (29%), Positives = 74/202 (36%), Gaps = 37/202 (18%)
Query: 194 GSPPRRGSPP-------LHAPPRRYPGPPFD--RSPERP--------MNGYRSPPPRL-M 235
G PP+RG PP LH PPR P PP D PERP ++ +P R+ +
Sbjct: 945 GIPPQRGIPPPSVLDSALHPPPRG-PFPPGDIFSQPERPFLAPGRQSVDNVTNPEKRIPL 1003
Query: 236 GRDGLRDYGPAAAALPPL--RHEGRFPDPHLHRERMDYMDDAYRGRGKFDRPPPLDWDNR 293
G D ++ G PP+ R P P R+ + D G G+ PPLD
Sbjct: 1004 GNDNIQQEGDRDYRFPPIETRESISRPPPVDVRDVVGRPIDPREGPGR----PPLD---- 1055
Query: 294 DRGRDGFSNERKGFERRPLSPSAPLLPSL------PPHRGGDRWSRDVRDRSRSPIRGGP 347
GRD F + P L P G R D R+ P+ GGP
Sbjct: 1056 --GRDHFGRPPVDIRENLVRPGIDHLGRRDHFGFNPEKPWGHRGDFDEREHRVLPVYGGP 1113
Query: 348 PAKDYRRDPVMSRGGRDDRRGG 369
R S R D R G
Sbjct: 1114 KGLHEERGRFRSGNYRFDPRSG 1135
>ROG_MOUSE (O35479) Heterogeneous nuclear ribonucleoprotein G (hnRNP
G) (RNA binding motif protein, X chromosome)
Length = 388
Score = 49.3 bits (116), Expect = 2e-05
Identities = 91/321 (28%), Positives = 106/321 (32%), Gaps = 88/321 (27%)
Query: 120 YARGRPGGRPFGRAFDGPGYGHRHGRGEGLHGRNNPNVRPREGDWMCPDALCGNLNFARR 179
+ GR G P R+ GP G R G G G P P G +M N N +
Sbjct: 89 FESGRRGPPPPPRS-RGPPRGLRGGSG----GTRGP---PSRGGYMDDGGYSMNFNMS-- 138
Query: 180 DFCNQCKRPRPAAAGSPPRRGSPPLHAPPRRYPGPPFDRSPERPMNGYRSPPPRLMGRDG 239
+ P P G PPR G PP P R P P R +G P GRD
Sbjct: 139 ----SSRGPLPVKRGPPPRSGGPP---PKRSTPS-----GPVRSSSGMGGRTPVSRGRD- 185
Query: 240 LRDYG--PAAAALPPLRHEGRFPDPHLHRERMDYMDDAY-RGRGKFD-RPPPLDWDNRD- 294
YG P LP R P + + Y Y R D PPP D+ RD
Sbjct: 186 --SYGGPPRREPLPSRRDVYLSPRDDGYSTKDSYSSRDYPSSRDTRDYTPPPRDYTYRDY 243
Query: 295 -----------RG---RDGFSNE------------RKGFERRPLSPSAPLLPSLPPHRGG 328
RG RDG+ + R +E S SAP PP GG
Sbjct: 244 CHSSSRDDYPSRGYGDRDGYGRDRDYSDHPSGGSYRDSYESYGNSRSAPPTRGPPPSYGG 303
Query: 329 DR-------------WSRDVRDRSRSPI------------RGGPPAKDYRRDP------V 357
SRD SRS + RG PP+ + P
Sbjct: 304 SSRYDDYSSSRDGYGGSRDSYSSSRSDLYSSGRDRVGRQERGLPPSMERGYPPPRDSYSS 363
Query: 358 MSRGG-RDDRRGGVGRDRIGG 377
SRG R RGG DR GG
Sbjct: 364 SSRGAPRGGGRGGSRSDRGGG 384
>YZR3_ARATH (Q8GZ43) Hypothetical RanBP2-type zinc-finger protein
At1g67325
Length = 288
Score = 47.0 bits (110), Expect = 8e-05
Identities = 48/157 (30%), Positives = 60/157 (37%), Gaps = 46/157 (29%)
Query: 160 REGDWMCPDALCGNLNFARRDFCN--QCKRPRPA-----AAGSPPRR------------- 199
RE DW+CP CGN+NF+ R CN C +PRPA +A P +
Sbjct: 22 REDDWICPS--CGNVNFSFRTTCNMRNCTQPRPADHNGKSAPKPMQHQQGFSSPGAYLGS 79
Query: 200 -GSPPLHAPPRRYPGPPFDRSPERPMNGYRSPP---PRLMGRDGLRDYG---PAAAALPP 252
G PP++ Y P F NG PP P G +Y PA A P
Sbjct: 80 GGPPPVYMGGSPYGSPLF--------NGSSMPPYDVPFSGGSPYHFNYNSRMPAGAHYRP 131
Query: 253 LRHEGRFPDPHLHRERMDYMDDAYRGRGKFDRPPPLD 289
L G P P+ M G + PPP+D
Sbjct: 132 LHMSG--PPPYHGGSMMG-------SGGMYGMPPPID 159
Score = 41.2 bits (95), Expect = 0.004
Identities = 47/156 (30%), Positives = 57/156 (36%), Gaps = 28/156 (17%)
Query: 54 PPPVNYSRSDRYSDDAGYRFRAGSSSPVHHRRDADHRFPSDYNHFPRNRGFGGGRDFGRF 113
PPPV Y Y G GSS P + D S Y HF N G +
Sbjct: 82 PPPV-YMGGSPY----GSPLFNGSSMPPY---DVPFSGGSPY-HFNYNSRMPAGAHYRPL 132
Query: 114 R-DPPHPYARGRP--GGRPFGRA--FDGPGYGHRHGRGEGLHGRNNPNVRP--------- 159
P PY G G +G D G G G G P P
Sbjct: 133 HMSGPPPYHGGSMMGSGGMYGMPPPIDRYGLGMAMGPGSAAAMMPRPRFYPDEKSQKRDS 192
Query: 160 -REGDWMCPDALCGNLNFARRDFCN--QCKRPRPAA 192
R+ DW CP+ CGN+NF+ R CN +C P+P +
Sbjct: 193 TRDNDWTCPN--CGNVNFSFRTVCNMRKCNTPKPGS 226
Score = 32.3 bits (72), Expect = 2.0
Identities = 16/42 (38%), Positives = 22/42 (52%), Gaps = 6/42 (14%)
Query: 161 EGDWMCPDALCGNLNFARRDFCNQ----CKRPRPAAAGSPPR 198
EG W C + CGN+N+ R CN+ +P + GSP R
Sbjct: 242 EGSWKCDN--CGNINYPFRSKCNRQNCGADKPGDRSNGSPSR 281
>WD33_HUMAN (Q9C0J8) WD-repeat protein 33 (WD-repeat protein WDC146)
Length = 1336
Score = 47.0 bits (110), Expect = 8e-05
Identities = 101/398 (25%), Positives = 136/398 (33%), Gaps = 61/398 (15%)
Query: 7 SQSPPSHHHHHQPLLSSLVVRPSLTENSPPGAAAHSNDYEP---GELRRDPP-------- 55
S+ PP+HH S + G H + P G+ R PP
Sbjct: 959 SRGPPNHHMGPM----------SERRHEQSGGPEHGPERGPFRGGQDCRGPPDRRGPHPD 1008
Query: 56 -PVNYSRSDRYSDDA--GYRFRA--GSSSPVHH-----RRDADHRFPSDYNHFPRNRGFG 105
P ++SR D + D G+R R G P+ R FP D+ F G
Sbjct: 1009 FPDDFSRPDDFHPDKRFGHRLREFEGRGGPLPQEEKWRRGGPGPPFPPDHREFSE----G 1064
Query: 106 GGRDFGRFRDPPHPYARGRPGGRPFGRAFDGPGYGHRHGRG--EGLHGRNNPNVRPREGD 163
GR G R PP + RPG F R + P + R G R+ PR D
Sbjct: 1065 DGR--GAARGPPGAWEGRRPGDERFPRDPEDPRFRGRREESFRRGAPPRHEGRAPPRGRD 1122
Query: 164 -WMCPDALCGNLNFARRDFCNQCKRPRPAAAGSPPRRGSPPLHAPPRRYPGPPFDRSPER 222
+ P+ NF + + + R G+ PR G L P +P R P+
Sbjct: 1123 GFPGPEDFGPEENFDASEEAARGRDLRGRGRGT-PRGGRKGLLPTPDEFPRFEGGRKPD- 1180
Query: 223 PMNGYRSPPPRLMGRDGLRDYGPAAAALPPLRHEGRFPDPHLHRERMDYMDDAY---RGR 279
+G R P P G + RD PP H+G P + MD A R R
Sbjct: 1181 SWDGNREPGP---GHEHFRD--TPRPDHPP--HDGHSPASRERSSSLQGMDMASLPPRKR 1233
Query: 280 GKFDRPPPLDWDNRDRGRDGFSNERKGFERRPLSPSAPLLPSLPPHRGGDRWSRDVRDRS 339
D P ++R+ G +E +G + R A +P D D R
Sbjct: 1234 PWHDGPGTS--EHREMEAPGGPSEDRGGKGRGGPGPAQRVPKSGRSSSLDGEHHDGYHRD 1291
Query: 340 RSPIRGGPPAKDYRRDPVMSRGGRDDRRGGVGRDRIGG 377
GGPP SRGGR G G + G
Sbjct: 1292 EP--FGGPPGSG-----TPSRGGRSGSNWGRGSNMNSG 1322
Score = 39.7 bits (91), Expect = 0.013
Identities = 72/299 (24%), Positives = 102/299 (34%), Gaps = 58/299 (19%)
Query: 99 PRNRGFGGGRDFGRFRDPPHPYA-RGRPGGRPF-GRAFDGPGYGHRHGRGEGLHGRNNPN 156
P +G G + PPHP+ +G PG + G GP G +G G
Sbjct: 735 PGTQGMQGPPGPRGMQGPPHPHGIQGGPGSQGIQGPVSQGPLMGLNPRGMQGPPG----- 789
Query: 157 VRPREGDWMCPDALCGNLNFARRDFCNQCKRPRPAAA--GSPPR-----------RGSPP 203
PRE P + + + P P G P+ +G PP
Sbjct: 790 --PRENQGPAPQGMIMGHP------PQEMRGPHPPGGLLGHGPQEMRGPQEIRGMQGPPP 841
Query: 204 ---LHAPPRRYPGPPFDRSPERPMNGYRSPPPR--LMGRDGLRDYGPAAAALPPLRHEGR 258
+ PP+ GPP +S + P G PPP+ + G G G A P +G
Sbjct: 842 QGSMLGPPQELRGPPGSQSQQGPPQGSLGPPPQGGMQGPPG--PQGQQNPARGPHPSQGP 899
Query: 259 FPDPHLHRERMDYMDDAYRG----RGKFDRPPPLDWDNRDRGRDGFSNERKGFERRPLSP 314
P +++ + D R G+ PPPL +G G PL+P
Sbjct: 900 IP---FQQQKTPLLGDGPRAPFNQEGQSTGPPPLIPGLGQQGAQG--------RIPPLNP 948
Query: 315 SAPLLPSLPPHRGGDRWSRDVRDRSRSPIRGGPPAKDYRRDPVMSRGGRD-----DRRG 368
P+ RG R GGP ++ + RGG+D DRRG
Sbjct: 949 GQGPGPNKGDSRGPPNHHMGPMSERRHEQSGGP---EHGPERGPFRGGQDCRGPPDRRG 1004
Score = 32.7 bits (73), Expect = 1.6
Identities = 70/275 (25%), Positives = 93/275 (33%), Gaps = 28/275 (10%)
Query: 116 PPHPYARGRPGGRPFGRAFDGPGYGHRHGRGEGLHGRNNPNVRPREGDWMCPDALCGNLN 175
PP P + RP G GP G +G G P P+ P + G
Sbjct: 619 PPGPQGQFRPPGPQGQMGPQGPPLHQGGGGPQGFMGPQGPQGPPQ--GLPRPQDMHGPQG 676
Query: 176 FARRDFCNQCKRPRPAAAGSPPRRGS--PPLHAPPRRYPGPPFDRSPERP--MNGYRSP- 230
R + P+ G P +GS P H P+ PGP P+ P G+ P
Sbjct: 677 MQRHPGPHGPLGPQ----GPPGPQGSSGPQGHMGPQGPPGPQSHIGPQGPPGPQGHLGPQ 732
Query: 231 -PPRLMGRDGLRDYGPAAAALPPLRHEGRFPDPHLHRERMDYMDDAYRG---RGKFDRPP 286
PP G G GP PP H G P + G RG + P
Sbjct: 733 GPPGTQGMQG--PPGPRGMQGPPHPH-GIQGGPGSQGIQGPVSQGPLMGLNPRGM--QGP 787
Query: 287 PLDWDNRDRGRDGFSNERKGFERRPLSPSAPLLPSLPPHRGGDRWSRDVRDRSRSPIRG- 345
P +N+ G E R P LL P G +++R P +G
Sbjct: 788 PGPRENQGPAPQGMIMGHPPQEMRGPHPPGGLLGHGPQEMRG---PQEIRGMQGPPPQGS 844
Query: 346 --GPPAKDYRRDPVMSRGGRDDRRGGVGRDRIGGM 378
GPP + R P S+ + +G +G GGM
Sbjct: 845 MLGPPQE--LRGPPGSQSQQGPPQGSLGPPPQGGM 877
>DNAA_STRAW (Q82FD8) Chromosomal replication initiator protein dnaA
Length = 653
Score = 45.1 bits (105), Expect = 3e-04
Identities = 44/143 (30%), Positives = 51/143 (34%), Gaps = 10/143 (6%)
Query: 184 QCKRPRPAAAGSPPRRGSPPLHAPPRRYPGPPFDRSPERPMN-GYRSPPPRLMGRDGLRD 242
+C RP A G PP A P PP PE P G P GR+G
Sbjct: 77 ECGRPIRIAITVDDSAGEPPASASP----APPRYEEPELPSGPGQGRDPYESRGREGYEG 132
Query: 243 YGPAAAALPPLRHEGRF---PDPHLHRERMDYMDDAYRGRGKFDRPPPLDWDNRDRGRDG 299
YG A H R P L R D + A + RP P W + G
Sbjct: 133 YGRHRADDHRQGHNDRHQGGPGDQLPPSRGDQLPTARPAYPDYQRPEPGAWPRPSQDDYG 192
Query: 300 FSNERKGF-ERRP-LSPSAPLLP 320
+ +R GF ER P SPS P
Sbjct: 193 WQQQRLGFPERDPYASPSQDYRP 215
>PRB3_HUMAN (Q04118) Basic salivary proline-rich protein 3 precursor
(Parotid salivary glycoprotein G1) (Proline-rich protein
G1)
Length = 309
Score = 42.4 bits (98), Expect = 0.002
Identities = 60/218 (27%), Positives = 77/218 (34%), Gaps = 38/218 (17%)
Query: 27 RPSLTENSPPGAAAHSNDYEPGELRRD-PPPVNYSRSD----RYSDDAGYRFRAGSSS-- 79
RP E PP S P + + PPP ++S R + G + G+ S
Sbjct: 74 RPGKPEGQPPQGGNQSQGPPPRPGKPEGPPPQGGNQSQGPPPRPGEPEGPPPQGGNQSQG 133
Query: 80 PVHHRRDADHRFPSDYNHFPRNRGFGGGRDFGRFRDPPHPYARGRPGGRPFGRAFDGPGY 139
P H + P GG + G PP P G+P G P G
Sbjct: 134 PPPHPGKPEGPPPQ-----------GGNQSQG---PPPRP---GKPEGPPPQGGNQSQGP 176
Query: 140 GHRHGRGEG---LHGRNNPNVRPREGDWMCPDALCGNLNFARRDFCNQCKRPRPAAA-GS 195
R G+ EG G + PR G P GN + Q PRP GS
Sbjct: 177 PPRPGKPEGPPPQGGNQSQGPPPRPGKPEGPPPQGGNQS--------QGPPPRPGKPEGS 228
Query: 196 PPRRGSPPLHAPPRRYPGPPFDRSPERPMNGYRSPPPR 233
P + G+ P PP +PG P P+ R PPPR
Sbjct: 229 PSQGGNKPRGPPP--HPGKPQGPPPQEGNKPQRPPPPR 264
Score = 41.2 bits (95), Expect = 0.004
Identities = 50/201 (24%), Positives = 73/201 (35%), Gaps = 37/201 (18%)
Query: 188 PRPAAAGSPPRRGSPPLHAPPRRYPGPPFDRSPERPMNGYRSPPPRLMGRDGLRDYGPAA 247
PRP PP +G PP R PG P + P + N + PPPR +G G
Sbjct: 52 PRPGKPEGPPPQGGNQSQGPPPR-PGKP-EGQPPQGGNQSQGPPPRPGKPEGPPPQGGNQ 109
Query: 248 AALPPLR------------HEGRFPDPHLHRERMDYMDDAYRGRGKFDRP-----PPLDW 290
+ PP R ++ + P PH + + +G RP PP
Sbjct: 110 SQGPPPRPGEPEGPPPQGGNQSQGPPPHPGKPEGPPPQGGNQSQGPPPRPGKPEGPPPQG 169
Query: 291 DNRDRG---RDGFSN--ERKGFERRPLSPSAPLLPSLPPHRGGDRWSRDVRDRSRSPIRG 345
N+ +G R G +G + P P P PP +GG++
Sbjct: 170 GNQSQGPPPRPGKPEGPPPQGGNQSQGPPPRPGKPEGPPPQGGNQ-------------SQ 216
Query: 346 GPPAKDYRRDPVMSRGGRDDR 366
GPP + + + S+GG R
Sbjct: 217 GPPPRPGKPEGSPSQGGNKPR 237
Score = 33.9 bits (76), Expect = 0.70
Identities = 40/131 (30%), Positives = 49/131 (36%), Gaps = 16/131 (12%)
Query: 105 GGGRDFGRFRDPPHPYARGRPGGRPFGRAFDGPGYGHRHGRGEGL--HGRNNPN-VRPRE 161
GG + G PP P G+P G P G R G+ EG G N P P
Sbjct: 190 GGNQSQG---PPPRP---GKPEGPPPQGGNQSQGPPPRPGKPEGSPSQGGNKPRGPPPHP 243
Query: 162 GDWMCPDALCGNLNFARRDFCNQCKRPR-PAAAGSPPRRGSPPLHAPPRRYPGPPFDRSP 220
G P GN + +RP+ P G P++ PP P+ P PP P
Sbjct: 244 GKPQGPPPQEGN----KPQRPPPPRRPQGPPPPGGNPQQPLPPPAGKPQGPPPPPQGGRP 299
Query: 221 ERPMNGYRSPP 231
RP G PP
Sbjct: 300 HRPPQG--QPP 308
Score = 30.8 bits (68), Expect = 5.9
Identities = 40/132 (30%), Positives = 53/132 (39%), Gaps = 21/132 (15%)
Query: 105 GGGRDFGRFRDPPHPYARGRPGGRPFGRAFDGPGYGHRHGRGEG---LHGRNNPNVRPRE 161
GG + G PP P G+P G P G R G+ EG G + PR
Sbjct: 169 GGNQSQG---PPPRP---GKPEGPPPQGGNQSQGPPPRPGKPEGPPPQGGNQSQGPPPRP 222
Query: 162 GDWMCPDALCGNLNFARRDFCNQCKRPRPAAAGSPPRRGSPPLHAPPRRYPG-PPFDRSP 220
G + GN + + P P G+ P+R PP PPRR G PP +P
Sbjct: 223 GKPEGSPSQGGNKPRGPPPHPGKPQGP-PPQEGNKPQR--PP---PPRRPQGPPPPGGNP 276
Query: 221 ERPMNGYRSPPP 232
++P+ PPP
Sbjct: 277 QQPL-----PPP 283
>S24C_ARATH (Q9M291) Protein transport protein Sec24-like CEF
Length = 1097
Score = 41.6 bits (96), Expect = 0.003
Identities = 82/337 (24%), Positives = 110/337 (32%), Gaps = 76/337 (22%)
Query: 8 QSPPSHHHHHQPLLSSLVVRPS-LTENSPPGAAAHSNDYEPGELRRDP--PPVNYSRSDR 64
QS P QP RPS + PP AA + P ++ + PPV
Sbjct: 68 QSFPQQQQQQQP-------RPSPMARPGPPPPAAMARPGGPPQVSQPGGFPPV------- 113
Query: 65 YSDDAGYRFRAGSSSPVHHRRDADHRFPSDYNHFPRNRGF--------------GGGRDF 110
G S+ P R + + FP+ GF G R
Sbjct: 114 -----GRPVAPPSNQPPFGGRPSTGPLVGGGSSFPQPGGFPASGPPGGVPSGPPSGARPI 168
Query: 111 GRFRDPPHPYARGR----PGGRPFGRAFDGPGYGHRHGRGEGLHGRNNPNVRPREGDWMC 166
G F PP P G P G P G +GP HG H N P G +
Sbjct: 169 G-FGSPP-PMGPGMSMPPPSGMPGGPLSNGPPPSGMHGG----HLSNGPPPSGMPGGTLS 222
Query: 167 ---PDALCGNLNFAR-RDFCNQCKRPRPAAAGSPPRRG---------SPPLHA--PPRRY 211
P + G F R F + P G PP G SPP H+ P +
Sbjct: 223 NGPPPPMMGPGAFPRGSQFTSGPMMAPPPPYGQPPNAGPFTGNSPLSSPPAHSIPAPTNF 282
Query: 212 PGPPFDRSPERPMNGYRSPPPRLMGRDG----------LRDYGPAAAALPPLRHEGRFPD 261
PG P+ R P Y +PP +L G +++ + + P + P
Sbjct: 283 PGVPYGRPPMPGGFPYGAPPQQLPSAPGTPGSIYGMGPMQNQSMTSVSSPSKIDLNQIPR 342
Query: 262 PHLHRERMDYMDDAYRGRGKFDRPPP--LDWDNRDRG 296
P + Y R K + PPP +D+ RD G
Sbjct: 343 PGSSSSPIVY---ETRVENKANPPPPTTVDYITRDTG 376
Score = 35.4 bits (80), Expect = 0.24
Identities = 31/91 (34%), Positives = 36/91 (39%), Gaps = 12/91 (13%)
Query: 172 GNLNFARRDFCN-QCKRPRPAAAGSPPRRGSPPLHAPPRRYPGPPFDRSPE-RPMNGYRS 229
GN N + N RP P GS PR SPP P+ +P + P PM
Sbjct: 31 GNPNSLAANMQNLNINRPPPPMPGSGPRP-SPPFGQSPQSFPQQQQQQQPRPSPMARPGP 89
Query: 230 PPPRLMGRDGLRDYGPAAAALPPLRHEGRFP 260
PPP M R G GP P + G FP
Sbjct: 90 PPPAAMARPG----GP-----PQVSQPGGFP 111
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.143 0.479
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 62,313,996
Number of Sequences: 164201
Number of extensions: 3602683
Number of successful extensions: 12708
Number of sequences better than 10.0: 332
Number of HSP's better than 10.0 without gapping: 43
Number of HSP's successfully gapped in prelim test: 303
Number of HSP's that attempted gapping in prelim test: 10998
Number of HSP's gapped (non-prelim): 1355
length of query: 379
length of database: 59,974,054
effective HSP length: 112
effective length of query: 267
effective length of database: 41,583,542
effective search space: 11102805714
effective search space used: 11102805714
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 67 (30.4 bits)
Medicago: description of AC122722.5


