

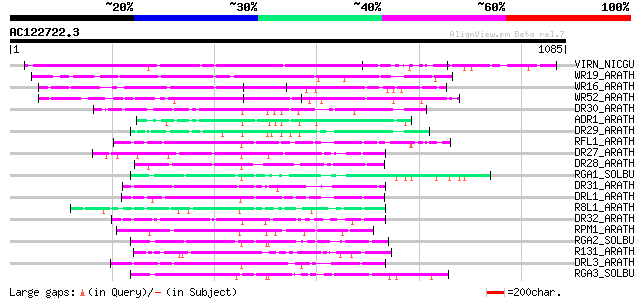
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122722.3 - phase: 0
(1085 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
VIRN_NICGU (Q40392) TMV resistance protein N 441 e-123
WR19_ARATH (Q9SZ67) Probable WRKY transcription factor 19 (WRKY ... 306 2e-82
WR16_ARATH (Q9FL92) Probable WRKY transcription factor 16 (WRKY ... 293 2e-78
WR52_ARATH (Q9FH83) Probable WRKY transcription factor 52 (WRKY ... 281 9e-75
DR30_ARATH (Q9LZ25) Probable disease resistance protein At5g04720 118 1e-25
ADR1_ARATH (Q9FW44) Disease resistance protein ADR1 (Activated d... 110 2e-23
DR29_ARATH (Q9SZA7) Probable disease resistance protein At4g33300 104 1e-21
RFL1_ARATH (Q8L3R3) Disease resistance protein RFL1 (RPS5-like p... 97 2e-19
DR27_ARATH (Q9T048) Putative disease resistance protein At4g27190 97 2e-19
DR28_ARATH (O81825) Putative disease resistance protein At4g27220 92 1e-17
RGA1_SOLBU (Q7XA42) Putative disease resistance protein RGA1 (RG... 91 1e-17
DR31_ARATH (Q9FLB4) Putative disease resistance protein At5g05400 89 6e-17
DRL1_ARATH (P60838) Probable disease resistance protein At1g12280 87 3e-16
R8L1_ARATH (O04093) Putative disease resistance protein RPP8-lik... 84 2e-15
DR32_ARATH (Q9FG91) Probable disease resistance protein At5g43730 84 3e-15
RPM1_ARATH (Q39214) Disease resistance protein RPM1 (Resistance ... 81 1e-14
RGA2_SOLBU (Q7XBQ9) Disease resistance protein RGA2 (RGA2-blb) (... 80 2e-14
R131_ARATH (Q9LRR4) Putative disease resistance RPP13-like prote... 80 3e-14
DRL3_ARATH (Q9LMP6) Probable disease resistance protein At1g15890 80 3e-14
RGA3_SOLBU (Q7XA40) Putative disease resistance protein RGA3 (RG... 75 7e-13
>VIRN_NICGU (Q40392) TMV resistance protein N
Length = 1144
Score = 441 bits (1133), Expect = e-123
Identities = 306/842 (36%), Positives = 463/842 (54%), Gaps = 78/842 (9%)
Query: 29 MASSSKSSSALVTSSKKNHYDVFVTFRGEDTRNNFTDFLFDALETKGIMVFRDVINLQKG 88
MASSS SS + YDVF++FRGEDTR FT L++ L KGI F+D L+ G
Sbjct: 1 MASSSSSS--------RWSYDVFLSFRGEDTRKTFTSHLYEVLNDKGIKTFQDDKRLEYG 52
Query: 89 ECIGPELFRAIEISQVYVAIFSKNYASSTWCLQELEKICECIKGSGKHVLPVFYDVDPSE 148
I EL +AIE SQ + +FS+NYA+S WCL EL KI EC + V+P+FYDVDPS
Sbjct: 53 ATIPGELCKAIEESQFAIVVFSENYATSRWCLNELVKIMECKTRFKQTVIPIFYDVDPSH 112
Query: 149 VRKQSGIYSEAFVKHEQRFQQDSMKVSRWREALEQVGSISG-WDLRDEPLAREIKEIVQK 207
VR Q +++AF +HE +++ D + RWR AL + ++ G D RD+ A I++IV +
Sbjct: 113 VRNQKESFAKAFEEHETKYKDDVEGIQRWRIALNEAANLKGSCDNRDKTDADCIRQIVDQ 172
Query: 208 IINILECKYS-CVSKDLVGIDSPIQALQNHLLLNSVDGVRAIGICGMGGIGKTTLATTLY 266
I + L CK S +++VGID+ ++ +++ LL ++GVR +GI GMGG+GKTT+A ++
Sbjct: 173 ISSKL-CKISLSYLQNIVGIDTHLEKIES-LLEIGINGVRIMGIWGMGGVGKTTIARAIF 230
Query: 267 GQI------SHQFSASCFIDDVTK-IYGLHDDPLDVQKQILFQTLGIEHQQICNRYHATT 319
+ S+QF +CF+ D+ + G+H +Q +L + L E N
Sbjct: 231 DTLLGRMDSSYQFDGACFLKDIKENKRGMH----SLQNALLSELLR-EKANYNNEEDGKH 285
Query: 320 LIQRKLCHERTLMILDNVDQVEQ-LEKIAVHREWLGPGSRIIIISRDEHVLKAYGVDVVY 378
+ +L ++ L++LD++D + LE +A +W G GSRIII +RD+H+++ D++Y
Sbjct: 286 QMASRLRSKKVLIVLDDIDNKDHYLEYLAGDLDWFGNGSRIIITTRDKHLIEKN--DIIY 343
Query: 379 KVSLLDWNEAHMLFCRKAFKDEKIIMSNYQNLVDQILHYAKGLPLAIKVLGSFLFGRNVT 438
+V+ L +E+ LF + AF E + N++ L ++++YAKGLPLA+KV GS L +T
Sbjct: 344 EVTALPDHESIQLFKQHAFGKE-VPNENFEKLSLEVVNYAKGLPLALKVWGSLLHNLRLT 402
Query: 439 EWKSALTRLRQSPVKDVMDVLQLSFDGLNETEKDIFLHIACFFNNDSEEDVKNILNCCGF 498
EWKSA+ ++ + ++D L++S+DGL ++++FL IACF + ++ + IL C
Sbjct: 403 EWKSAIEHMKNNSYSGIIDKLKISYDGLEPKQQEMFLDIACFLRGEEKDYILQILESCHI 462
Query: 499 HADIGLRVLIDKSLVSIS-YSIINMHSLLEELGRKIVQNSSSKEPRKWSRLWSTEQLYDV 557
A+ GLR+LIDKSLV IS Y+ + MH L++++G+ IV + K+P + SRLW +++ +V
Sbjct: 463 GAEYGLRILIDKSLVFISEYNQVQMHDLIQDMGKYIV--NFQKDPGERSRLWLAKEVEEV 520
Query: 558 MLENM-EKHVEAI-VLYYKEDEEADFEHLSKMSNLRLLFIANYISTMLGFPSCLSNKLRF 615
M N +EAI V Y + + M LR +F ST L N LR
Sbjct: 521 MSNNTGTMAMEAIWVSSYSSTLRFSNQAVKNMKRLR-VFNMGRSSTHYAI-DYLPNNLRC 578
Query: 616 VHWFRYPSKYLPSNFHPNELVELILTESNIKQLWKNKKYLPNLRTLDLRHSRNLEKIIDF 675
YP + PS F LV L L ++++ LW K+LP+LR +DL S+ L + DF
Sbjct: 579 FVCTNYPWESFPSTFELKMLVHLQLRHNSLRHLWTETKHLPSLRRIDLSWSKRLTRTPDF 638
Query: 676 GEFPNLERLDLEGCINLVELDPSIGLLRKLVYLNLKDCKSLVSIPNNIFGLSSLQYLNMC 735
PNLE ++L C NL E+ S+G K++ L L DCKSL P + SL+YL +
Sbjct: 639 TGMPNLEYVNLYQCSNLEEVHHSLGCCSKVIGLYLNDCKSLKRFP--CVNVESLEYLGLR 696
Query: 736 GCSKVFNNPRRLMKSGISSEKKQQHDIRESASHHLPGLKWIILAHDSSHMLPSLHSLCCL 795
C + P I K + I H+ G LPS
Sbjct: 697 SCDSLEKLPE------IYGRMKPEIQI------HMQG--------SGIRELPS------- 729
Query: 796 RKVDISFCYLSHVPDAIECLHWLERLNLAGNDFVTLP-SLRKLSKLVYLNLEHCKLLESL 854
F Y +HV + L W N+ + V LP S+ +L LV L++ C LESL
Sbjct: 730 ----SIFQYKTHV---TKLLLW----NM--KNLVALPSSICRLKSLVSLSVSGCSKLESL 776
Query: 855 PQ 856
P+
Sbjct: 777 PE 778
Score = 54.7 bits (130), Expect = 1e-06
Identities = 100/415 (24%), Positives = 168/415 (40%), Gaps = 79/415 (19%)
Query: 691 NLVELDPSIGLLRKLVYLNLKDCKSLVSIPNNIFGLSSLQYLNMCGCSKVFNNPRRLMKS 750
NLV L SI L+ LV L++ C L S+P I L +L+ + + + P +++
Sbjct: 748 NLVALPSSICRLKSLVSLSVSGCSKLESLPEEIGDLDNLRVFD-ASDTLILRPPSSIIRL 806
Query: 751 GISSEKKQQHDIRESASHHLPGLKWIILA----HDSSH--MLPSLHSLCCLRKVDISFCY 804
K IIL D H P L L +++S+C
Sbjct: 807 N----------------------KLIILMFRGFKDGVHFEFPPVAEGLHSLEYLNLSYCN 844
Query: 805 L--SHVPDAIECLHWLERLNLAGNDFVTLP-SLRKLSKLVYLNLEHCKLLESLPQLPFPT 861
L +P+ I L L++L+L+ N+F LP S+ +L L L+L+ C+ L LP+LP
Sbjct: 845 LIDGGLPEEIGSLSSLKKLDLSRNNFEHLPSSIAQLGALQSLDLKDCQRLTQLPELPPEL 904
Query: 862 NTGEVHREYDDYFCGAGLLIFNCPKLG----EREHCRSMTLLW----------MKQFIKA 907
N E+H + L+ KL + H +M L+ M+ I A
Sbjct: 905 N--ELHVDCHMALKFIHYLVTKRKKLHRVKLDDAHNDTMYNLFAYTMFQNISSMRHDISA 962
Query: 908 NPRSSSEIQIVNPGSE-IPSWINNQRMGYSIAIDRSPIRHDNDNNIIGIVCCAAFTMAPY 966
+ S + P E IPSW ++Q S++++ P + +G C
Sbjct: 963 SDSLSLTVFTGQPYPEKIPSWFHHQGWDSSVSVN-LPENWYIPDKFLGFAVC-------- 1013
Query: 967 REIFYSSELMNLAFKRID-SNERLLKMRVPVKLSLVTTKSSHL--WIIYL---------- 1013
YS L++ I ++++ +M + LS T+SS+ W I+
Sbjct: 1014 ----YSRSLIDTTAHLIPVCDDKMSRMTQKLALSECDTESSNYSEWDIHFFFVPFAGLWD 1069
Query: 1014 PREYPGYSCHEFGKIELKFFEVEGLEVESCGYRWVCKQDIQEFNLIMNHKNSLTP 1068
+ G + +++G I L F E + G R + K+ + L+ +NS P
Sbjct: 1070 TSKANGKTPNDYGIIRLSF----SGEEKMYGLRLLYKEGPEVNALLQMRENSNEP 1120
>WR19_ARATH (Q9SZ67) Probable WRKY transcription factor 19 (WRKY
DNA-binding protein 19)
Length = 1895
Score = 306 bits (784), Expect = 2e-82
Identities = 256/854 (29%), Positives = 421/854 (48%), Gaps = 98/854 (11%)
Query: 43 SKKNHYDVFVTFRGEDTRN-NFTDFLFDALETKGIMVFRDVINLQKGECIGPELFRAIEI 101
S YDV + + D N +F L +L +GI V+ + A+
Sbjct: 663 SSSKDYDVVIRYGRADISNEDFISHLRASLCRRGISVYEKFNEVD-----------ALPK 711
Query: 102 SQVYVAIFSKNYASSTWCLQELEKICECIKGSGKHVLPVFYDVDPSEVRKQSGIYSEAFV 161
+V + + + Y S L I E + V P+FY + P + S Y ++
Sbjct: 712 CRVLIIVLTSTYVPSN-----LLNILEHQHTEDRVVYPIFYRLSPYDFVCNSKNYERFYL 766
Query: 162 KHEQRFQQDSMKVSRWREALEQVGSISGWDLRDEPLAREIKEIVQKIINILECKYSCVSK 221
+ E + +W+ AL+++ + G+ L D+ + I EIV+ + +L S
Sbjct: 767 QDEPK---------KWQAALKEITQMPGYTLTDKSESELIDEIVRDALKVL---CSADKV 814
Query: 222 DLVGIDSPIQALQNHLLLNSVDGVRAIGICGMGGIGKTTLATTLYGQISHQFSASCFIDD 281
+++G+D ++ + + L + S+D VR+IGI G GIGKTT+A ++ +IS Q+ + D
Sbjct: 815 NMIGMDMQVEEILSLLCIESLD-VRSIGIWGTVGIGKTTIAEEIFRKISVQYETCVVLKD 873
Query: 282 VTKIYGL--HDDPLDVQKQILFQTLGIEHQQICNRYHATTLIQRKLCHERTLMILDNVDQ 339
+ K + HD V++ L + L +E I T+ ++ +L +R L+ILD+V+
Sbjct: 874 LHKEVEVKGHDA---VRENFLSEVLEVEPHVIRISDIKTSFLRSRLQRKRILVILDDVND 930
Query: 340 VEQLEKIAVHREWLGPGSRIIIISRDEHVLKAYGVDVVYKVSLLDWNEAHMLFCRKAFKD 399
++ + GPGSRII+ SR+ V +D VY+V LD ++ +L R +
Sbjct: 931 YRDVDTFLGTLNYFGPGSRIIMTSRNRRVFVLCKIDHVYEVKPLDIPKSLLLLDRGTCQ- 989
Query: 400 EKIIMSN--YQNLVDQILHYAKGLPLAIKVLGSFLFGRNVTEWKSALTRLRQSPVKDVMD 457
I++S Y+ L +++ ++ G P ++ L S EW ++ + +
Sbjct: 990 --IVLSPEVYKTLSLELVKFSNGNPQVLQFLSSI-----DREWNKLSQEVKTTSPIYIPG 1042
Query: 458 VLQLSFDGLNETEKDIFLHIACFFNNDSEEDVKNILNCCGFHADIGLRVLIDKSLVSIS- 516
+ + S GL++ E+ IFL IACFFN +++V +L+ CGF A +G R L+DKSL++IS
Sbjct: 1043 IFEKSCCGLDDNERGIFLDIACFFNRIDKDNVAMLLDGCGFSAHVGFRGLVDKSLLTISQ 1102
Query: 517 YSIINMHSLLEELGRKIVQNSSSKEPRKWSRLWSTEQLYDVMLENM-EKHVEAIVL-YYK 574
+++++M S ++ GR+IV+ S+ P SRLW+ + + V + + +E I L
Sbjct: 1103 HNLVDMLSFIQATGREIVRQESADRPGDRSRLWNADYIRHVFINDTGTSAIEGIFLDMLN 1162
Query: 575 EDEEADFEHLSKMSNLRLLFIANYISTM-----LGFP---SCLSNKLRFVHWFRYPSKYL 626
+A+ KM NLRLL + Y S + FP L +KLR +HW YP L
Sbjct: 1163 LKFDANPNVFEKMCNLRLLKL--YCSKAEEKHGVSFPQGLEYLPSKLRLLHWEYYPLSSL 1220
Query: 627 PSNFHPNELVELILTESNIKQLWKNKK--------YLPNLRTLDLRHSRNLEKIIDFGEF 678
P +F+P LVEL L S K+LWK KK L L+ + L +S L KI
Sbjct: 1221 PKSFNPENLVELNLPSSCAKKLWKGKKARFCTTNSSLEKLKKMRLSYSDQLTKIPRLSSA 1280
Query: 679 PNLERLDLEGCINLVELDPSIGLLRKLVYLNLKDCKSLVSIPNNIFGLSSLQYLNMCGCS 738
NLE +DLEGC +L+ L SI L+KLV+LNLK C L +IP+ + L SL+ LN+ GCS
Sbjct: 1281 TNLEHIDLEGCNSLLSLSQSISYLKKLVFLNLKGCSKLENIPSMV-DLESLEVLNLSGCS 1339
Query: 739 KVFNNPRRLMKSGISSEKKQQHDIRESASHHLPGLKWIILAHDSSHMLP-SLHSLCCLRK 797
K+ N P IS P +K + + +P S+ +L L K
Sbjct: 1340 KLGNFPE------IS-----------------PNVKELYMGGTMIQEIPSSIKNLVLLEK 1376
Query: 798 VDI-SFCYLSHVPDAIECLHWLERLNLAGNDFVTL----PSLRKLSKLVYLNLEHCKLLE 852
+D+ + +L ++P +I L LE LNL+G ++L S R++ L +L+L + E
Sbjct: 1377 LDLENSRHLKNLPTSIYKLKHLETLNLSG--CISLERFPDSSRRMKCLRFLDLSRTDIKE 1434
Query: 853 SLPQLPFPTNTGEV 866
+ + T E+
Sbjct: 1435 LPSSISYLTALDEL 1448
>WR16_ARATH (Q9FL92) Probable WRKY transcription factor 16 (WRKY
DNA-binding protein 16)
Length = 1372
Score = 293 bits (750), Expect = 2e-78
Identities = 255/852 (29%), Positives = 412/852 (47%), Gaps = 115/852 (13%)
Query: 57 EDTRNNFTDFLFDALETKGIMVFRDVINLQKGECIGPELFRAIEISQVYVAIFSKNYASS 116
E+ R +F L AL+ KG+ DV + + + E +E ++V V I N S
Sbjct: 14 EEVRYSFVSHLSKALQRKGV---NDVF-IDSDDSLSNESQSMVERARVSVMILPGNRTVS 69
Query: 117 TWCLQELEKICECIKGSGKHVLPVFYDVDPSEVRKQSGIYSEAF--VKHEQRFQQDSMKV 174
L +L K+ +C K + V+PV Y V SE S + S+ F V H ++ DS V
Sbjct: 70 ---LDKLVKVLDCQKNKDQVVVPVLYGVRSSETEWLSALDSKGFSSVHHSRKECSDSQLV 126
Query: 175 SRWREALEQVGSISGWDLRDEPLAREIKEIVQKIINILECKYSCVSKDLVGIDSPIQALQ 234
++++ +K+ + + +GI S + ++
Sbjct: 127 KE-----------------------TVRDVYEKLFYM----------ERIGIYSKLLEIE 153
Query: 235 NHLLLNSVDGVRAIGICGMGGIGKTTLATTLYGQISHQFSASCFIDDVTKIYGLHDDPLD 294
+ +D +R +GI GM GIGKTTLA ++ Q+S +F A CFI+D TK
Sbjct: 154 KMINKQPLD-IRCVGIWGMPGIGKTTLAKAVFDQMSGEFDAHCFIEDYTKAIQEKGVYCL 212
Query: 295 VQKQILFQTLGIEHQQICNRYHATTLIQRKLCHERTLMILDNVDQVEQLEKIAVHREWLG 354
+++Q L + G +L++ +L ++R L++LD+V +E +W G
Sbjct: 213 LEEQFLKENAGAS-----GTVTKLSLLRDRLNNKRVLVVLDDVRSPLVVESFLGGFDWFG 267
Query: 355 PGSRIIIISRDEHVLKAYGVDVVYKVSLLDWNEAHMLFCRKAFKDEKIIMSNYQNLVDQI 414
P S III S+D+ V + V+ +Y+V L+ EA LF A D+ + N + ++
Sbjct: 268 PKSLIIITSKDKSVFRLCRVNQIYEVQGLNEKEALQLFSLCASIDD-MAEQNLHEVSMKV 326
Query: 415 LHYAKGLPLAIKVLGSFLFGRN-VTEWKSALTRLRQSPVKDVMDVLQLSFDGLNETEKDI 473
+ YA G PLA+ + G L G+ E + A +L++ P +D ++ S+D LN+ EK+I
Sbjct: 327 IKYANGHPLALNLYGRELMGKKRPPEMEIAFLKLKECPPAIFVDAIKSSYDTLNDREKNI 386
Query: 474 FLHIACFFNNDSEEDVKNILNCCGFHADIGLRVLIDKSLVSISYSIINMHSLLEELGRKI 533
FL IACFF ++ + V +L CGF +G+ VL++KSLV+IS + + MH+L++++GR+I
Sbjct: 387 FLDIACFFQGENVDYVMQLLEGCGFFPHVGIDVLVEKSLVTISENRVRMHNLIQDVGRQI 446
Query: 534 VQNSSSKEPRKWSRLW---STEQLYDVMLENMEKHVEAIVLYYKEDEEA----------- 579
+ N +++ ++ SRLW S + L + +N + + + EE
Sbjct: 447 I-NRETRQTKRRSRLWEPCSIKYLLEDKEQNENEEQKTTFERAQVPEEIEGMFLDTSNLS 505
Query: 580 -DFEHLS--KMSNLRLLFI------ANYISTML-GFPSCLSNKLRFVHWFRYPSKYLPSN 629
D +H++ M NLRL I ++++ L G S L N LR +HW YP ++LP N
Sbjct: 506 FDIKHVAFDNMLNLRLFKIYSSNPEVHHVNNFLKGSLSSLPNVLRLLHWENYPLQFLPQN 565
Query: 630 FHPNELVELILTESNIKQLWKNKKYLPNLRTLDLRHSRNLEKIIDFGEFPNLERLDLEGC 689
F P LVE+ + S +K+LW K L L+T+ L HS+ L I D + NLE +DL+GC
Sbjct: 566 FDPIHLVEINMPYSQLKKLWGGTKDLEMLKTIRLCHSQQLVDIDDLLKAQNLEVVDLQGC 625
Query: 690 INLVELDPSIGLLRKLVYLNLKDC---KSLVSIPNNIFGLSSLQYLNMCG-------CSK 739
L P+ G L L +NL C KS IP NI + LN+ G S
Sbjct: 626 TRLQSF-PATGQLLHLRVVNLSGCTEIKSFPEIPPNI------ETLNLQGTGIIELPLSI 678
Query: 740 VFNNPRRLMK--------SGISSEKKQQHDIRE-------SASHHLPGLKWIILAHDSSH 784
V N R L+ SG+S+ +Q D++ S S+ PG + +D S
Sbjct: 679 VKPNYRELLNLLAEIPGLSGVSN--LEQSDLKPLTSLMKISTSYQNPGKLSCLELNDCSR 736
Query: 785 M--LPSLHSLCCLRKVDISFCYLSHVPDAIECLHWLERLNLAGNDFVTLPSLRKLSKLVY 842
+ LP++ +L L+ +D+S C S + L+ L L G +P L + L +
Sbjct: 737 LRSLPNMVNLELLKALDLSGC--SELETIQGFPRNLKELYLVGTAVRQVPQLPQ--SLEF 792
Query: 843 LNLEHCKLLESL 854
N C L+S+
Sbjct: 793 FNAHGCVSLKSI 804
Score = 48.1 bits (113), Expect = 1e-04
Identities = 30/86 (34%), Positives = 50/86 (57%), Gaps = 2/86 (2%)
Query: 457 DVLQLSFDGLNETEKDIFLHIACFFNNDSEEDVKNIL-NCCGFHADIGLRVLIDKSLVSI 515
+VL++ + GL E K +FL+IA FN++ V ++ N GL+VL +SL+ +
Sbjct: 1049 EVLRVRYAGLQEIYKALFLYIAGLFNDEDVGLVAPLIANIIDMDVSYGLKVLAYRSLIRV 1108
Query: 516 SYS-IINMHSLLEELGRKIVQNSSSK 540
S + I MH LL ++G++I+ S K
Sbjct: 1109 SSNGEIVMHYLLRQMGKEILHTESKK 1134
>WR52_ARATH (Q9FH83) Probable WRKY transcription factor 52 (WRKY
DNA-binding protein 52) (Disease resistance protein
RRS1) (Resistance to Ralstonia solanacearum 1 protein)
Length = 1378
Score = 281 bits (718), Expect = 9e-75
Identities = 259/884 (29%), Positives = 416/884 (46%), Gaps = 117/884 (13%)
Query: 57 EDTRNNFTDFLFDALETKGIMVFRDVINLQKGECIGPELFRAIEISQVYVAIFSKNYASS 116
E+ R +F L +AL KGI V+++ + + E IE + V V + N S
Sbjct: 17 EEVRYSFVSHLSEALRRKGIN--NVVVDVDIDDLLFKESQAKIEKAGVSVMVLPGNCDPS 74
Query: 117 TWCLQELEKICECIKGSGKHVLPVFYDVDPSEVRKQSGIYSEAFVKHEQRFQQDSMKVSR 176
L + K+ EC + + + S +Y ++ ++ + + D +SR
Sbjct: 75 EVWLDKFAKVLECQRNNKDQAVV-------------SVLYGDSLLRDQWLSELDFRGLSR 121
Query: 177 WREALEQVGSISGWDLRDEPLAREIKEIVQKIINILECKYSCVSKDLVGIDSPIQALQNH 236
++ ++ +V++I+ + + V + +GI S + ++N
Sbjct: 122 IHQSRKECSD---------------SILVEEIVRDVYETHFYVGR--IGIYSKLLEIENM 164
Query: 237 LLLNSVDGVRAIGICGMGGIGKTTLATTLYGQISHQFSASCFIDDVTKIYGLHDDPLDVQ 296
+ + G+R +GI GM GIGKTTLA ++ Q+S F ASCFI+D K +
Sbjct: 165 VNKQPI-GIRCVGIWGMPGIGKTTLAKAVFDQMSSAFDASCFIEDYDK---------SIH 214
Query: 297 KQILFQTLGIEHQQICNRYHATTL----IQRKLCHERTLMILDNVDQVEQLEKIAVHREW 352
++ L+ L +Q+ AT + ++ +L +R L++LD+V E +W
Sbjct: 215 EKGLYCLL---EEQLLPGNDATIMKLSSLRDRLNSKRVLVVLDDVCNALVAESFLEGFDW 271
Query: 353 LGPGSRIIIISRDEHVLKAYGVDVVYKVSLLDWNEAHMLFCRKAFKDEKIIMSNYQNLVD 412
LGPGS III SRD+ V + G++ +Y+V L+ EA LF A E + N L
Sbjct: 272 LGPGSLIIITSRDKQVFRLCGINQIYEVQGLNEKEARQLFLLSASIMEDMGEQNLHELSV 331
Query: 413 QILHYAKGLPLAIKVLGSFLFGRN-VTEWKSALTRLRQSPVKDVMDVLQLSFDGLNETEK 471
+++ YA G PLAI V G L G+ ++E ++A +L++ P ++D + S+D L++ EK
Sbjct: 332 RVISYANGNPLAISVYGRELKGKKKLSEMETAFLKLKRRPPFKIVDAFKSSYDTLSDNEK 391
Query: 472 DIFLHIACFFNNDSEEDVKNILNCCGFHADIGLRVLIDKSLVSISYSIINMHSLLEELGR 531
+IFL IACFF ++ V +L CGF + + VL+DK LV+IS + + +H L +++GR
Sbjct: 392 NIFLDIACFFQGENVNYVIQLLEGCGFFPHVEIDVLVDKCLVTISENRVWLHKLTQDIGR 451
Query: 532 KIVQNSSSKEPRKWSRLWSTEQLYDVMLENMEK-HVEAIVLYYKEDEEADFEHL------ 584
+I+ N + + + RLW + ++ N K + E + + + E L
Sbjct: 452 EII-NGETVQIERRRRLWEPWSIKYLLEYNEHKANGEPKTTFKRAQGSEEIEGLFLDTSN 510
Query: 585 ----------SKMSNLRLL--FIAN-YISTMLGFPS----CLSNKLRFVHWFRYPSKYLP 627
M NLRLL + +N + ++ FP+ L N+LR +HW YP K LP
Sbjct: 511 LRFDLQPSAFKNMLNLRLLKIYCSNPEVHPVINFPTGSLHSLPNELRLLHWENYPLKSLP 570
Query: 628 SNFHPNELVELILTESNIKQLWKNKKYLPNLRTLDLRHSRNLEKIIDFGEFPNLERLDLE 687
NF P LVE+ + S +++LW K L LRT+ L HS++L I D + NLE +DL+
Sbjct: 571 QNFDPRHLVEINMPYSQLQKLWGGTKNLEMLRTIRLCHSQHLVDIDDLLKAENLEVIDLQ 630
Query: 688 GCINLVELDPSIGLLRKLVYLNLKDC---KSLVSIPNNIFGLSSLQYLNMCG--CSKVFN 742
GC L P+ G L +L +NL C KS++ IP NI L LQ + S V
Sbjct: 631 GCTRLQNF-PAAGRLLRLRVVNLSGCIKIKSVLEIPPNIEKL-HLQGTGILALPVSTVKP 688
Query: 743 NPRRLMK-----SGISSEKKQQH--DIRESASHHLPGLKWIILAHDSSHMLPSLHSLC-- 793
N R L+ G+S K + + ES S K I L L SL ++
Sbjct: 689 NHRELVNFLTEIPGLSEASKLERLTSLLESNSSCQDLGKLICLELKDCSCLQSLPNMANL 748
Query: 794 CLRKVDISFC----------------YL--SHVPDAIECLHWLERLNLAGNDFVTLPSLR 835
L +D+S C YL + + + + LE LN G+ +LP++
Sbjct: 749 DLNVLDLSGCSSLNSIQGFPRFLKQLYLGGTAIREVPQLPQSLEILNAHGSCLRSLPNMA 808
Query: 836 KLSKLVYLNLEHCKLLESLPQLPFPTNTGEVHREYDDYFCGAGL 879
L L L+L C LE++ FP N E+ YF G L
Sbjct: 809 NLEFLKVLDLSGCSELETIQ--GFPRNLKEL------YFAGTTL 844
Score = 57.4 bits (137), Expect = 2e-07
Identities = 41/115 (35%), Positives = 66/115 (56%), Gaps = 4/115 (3%)
Query: 457 DVLQLSFDGLNETEKDIFLHIACFFNNDSEEDVKNILNCCGFHADIGLRVLIDKSLVSIS 516
+VL++S+D L E +K +FL+IA FN++ + V ++ GL+VL D SL+S+S
Sbjct: 1087 EVLRVSYDDLQEMDKVLFLYIASLFNDEDVDFVAPLIAGIDLDVSSGLKVLADVSLISVS 1146
Query: 517 YS-IINMHSLLEELGRKIVQNSSSKEPRKWSRLWSTEQLYDVMLENMEKHVEAIV 570
+ I MHSL ++G++I+ S S + TE L DV + +KH E+ V
Sbjct: 1147 SNGEIVMHSLQRQMGKEILHGQSMLLSDCESSM--TENLSDVP-KKEKKHRESKV 1198
>DR30_ARATH (Q9LZ25) Probable disease resistance protein At5g04720
Length = 811
Score = 118 bits (295), Expect = 1e-25
Identities = 163/723 (22%), Positives = 294/723 (40%), Gaps = 153/723 (21%)
Query: 164 EQRFQQDSMKVSRWREALEQVGSISGWDLRDEPLAREIKEIVQKIINILECKYSCVSKDL 223
E RF + KV E L GS+ LR RE + + + ++ + +
Sbjct: 129 EFRFDRIDRKVDSLNEKL---GSMK---LRGSESLREALKTAEATVEMVTTDGADLG--- 179
Query: 224 VGIDSPIQALQNHLLLNSVDGVRAIGICGMGGIGKTTLATTLYGQISHQFSASCFIDDVT 283
VG+D + ++ +L S+DG R IGI GM G GKTTLA L ++V
Sbjct: 180 VGLDLGKRKVKE-MLFKSIDGERLIGISGMSGSGKTTLAKELARD-----------EEVR 227
Query: 284 KIYGLHDDPLDVQKQILFQTLGIEHQQICNRYHATTLIQRKLCHERTLMILDNVDQVEQL 343
+G L V + + L Y A + L R L+ILD+V E L
Sbjct: 228 GHFGNKVLFLTVSQSPNLEELRAHIWGFLTSYEAG--VGATLPESRKLVILDDVWTRESL 285
Query: 344 EKIAVHREWLGPGSRIIIISRDEHVLKAYGVDVVYKVSLLDWNEAHMLFCRKAFKDEKII 403
+++ PG+ +++SR K V Y V LL+ +EA LFC F ++K++
Sbjct: 286 DQLMFENI---PGTTTLVVSRS----KLADSRVTYDVELLNEHEATALFCLSVF-NQKLV 337
Query: 404 MSNY-QNLVDQILHYAKGLPLAIKVLGSFLFGRNVTEWKSALTRLRQSPVKD------VM 456
S + Q+LV Q++ KGLPL++KV+G+ L R W+ A+ RL + D V
Sbjct: 338 PSGFSQSLVKQVVGECKGLPLSLKVIGASLKERPEKYWEGAVERLSRGEPADETHESRVF 397
Query: 457 DVLQLSFDGLNETEKDIFLHIACFFNNDSEEDVKNILNCCGFHADI----GLRVLID--- 509
++ + + L+ +D FL + F D + + ++N D+ V++D
Sbjct: 398 AQIEATLENLDPKTRDCFLVLGA-FPEDKKIPLDVLINVLVELHDLEDATAFAVIVDLAN 456
Query: 510 KSLVSI-----------SY--SIINMHSLLEELG-------------RKIVQNSSSKEPR 543
++L+++ SY + H +L ++ R ++ S PR
Sbjct: 457 RNLLTLVKDPRFGHMYTSYYDIFVTQHDVLRDVALRLSNHGKVNNRERLLMPKRESMLPR 516
Query: 544 KWSRLWSTEQLYDVMLENME--------------KHVEAIVLYYKEDEEADFEHLSKMSN 589
+W R + ++ Y + ++ E ++L++ D+ ++KM
Sbjct: 517 EWER--NNDEPYKARVVSIHTGEMTQMDWFDMELPKAEVLILHFSSDKYVLPPFIAKMGK 574
Query: 590 LRLLFIANYISTMLGFPSCLSNKLRFVHWFRYPSKYLPSNFHPNELVELILTESNIKQLW 649
L L I N + P+ H + + +K LW
Sbjct: 575 LTALVIIN-------------------------NGMSPARLHD---FSIFTNLAKLKSLW 606
Query: 650 KNKKYLPNL--RTLDLRHSRNLEKI------------IDFGE-FPNLERLDLEGCINLVE 694
+ ++P L T+ L++ L I +D + FP L L ++ C +L+E
Sbjct: 607 LQRVHVPELSSSTVPLQNLHKLSLIFCKINTSLDQTELDIAQIFPKLSDLTIDHCDDLLE 666
Query: 695 LDPSIGLLRKLVYLNLKDCKSLVSIPNNIFGLSSLQYLNMCGCSKVFNNPRRLMKSGISS 754
L +I + L +++ +C + +P N+ L +LQ L + C ++ + P + +
Sbjct: 667 LPSTICGITSLNSISITNCPRIKELPKNLSKLKALQLLRLYACHELNSLPVEICE----- 721
Query: 755 EKKQQHDIRESASHHLPGLKWIILAH--DSSHMLPSLHSLCCLRKVDISFCYLSHVPDAI 812
LP LK++ ++ S + + + L K+D C LS +P+++
Sbjct: 722 ---------------LPRLKYVDISQCVSLSSLPEKIGKVKTLEKIDTRECSLSSIPNSV 766
Query: 813 ECL 815
L
Sbjct: 767 VLL 769
>ADR1_ARATH (Q9FW44) Disease resistance protein ADR1 (Activated
disease resistance protein 1)
Length = 787
Score = 110 bits (275), Expect = 2e-23
Identities = 141/611 (23%), Positives = 244/611 (39%), Gaps = 114/611 (18%)
Query: 249 GICGMGGIGKTTLATTLYGQISHQFSASCFIDDVTKIYGLHDDPLDVQKQILFQTLG--- 305
GI GM G GKTTLA L DDV ++ + ++LF T+
Sbjct: 190 GISGMSGSGKTTLAIELSKD-----------DDVRGLF---------KNKVLFLTVSRSP 229
Query: 306 -IEHQQICNRYHATTLIQRKLCHERTLMILDNVDQVEQLEKIAVHREWLGPGSRIIIISR 364
E+ + C R + H+R L+ILD+V E L+++ GS +++SR
Sbjct: 230 NFENLESCIREFLYDGV-----HQRKLVILDDVWTRESLDRLMSKIR----GSTTLVVSR 280
Query: 365 DEHVLKAYGVDVVYKVSLLDWNEAHMLFCRKAFKDEKIIMSNYQNLVDQILHYAKGLPLA 424
K Y V LL +EA L C AF+ + + LV Q++ KGLPL+
Sbjct: 281 S----KLADPRTTYNVELLKKDEAMSLLCLCAFEQKSPPSPFNKYLVKQVVDECKGLPLS 336
Query: 425 IKVLGSFLFGRNVTEWKSALTRLRQSPVKD------VMDVLQLSFDGLNETEKDIFLHIA 478
+KVLG+ L + W+ + RL + D V ++ S + L+ +D FL +
Sbjct: 337 LKVLGASLKNKPERYWEGVVKRLLRGEAADETHESRVFAHMEESLENLDPKIRDCFLDMG 396
Query: 479 CFFNNDSEEDVKNILNCCGFHADIGLRV-------LIDKSLVSISYS------------- 518
F D + + + + DI L DK+L++I +
Sbjct: 397 AF-PEDKKIPLDLLTSVWVERHDIDEETAFSFVLRLADKNLLTIVNNPRFGDVHIGYYDV 455
Query: 519 IINMHSLLEELG-------------RKIVQNSSSKEPRKWSRLWSTEQLYDVMLENMEK- 564
+ H +L +L R ++ + PR+W + + ++ +D + ++
Sbjct: 456 FVTQHDVLRDLALHMSNRVDVNRRERLLMPKTEPVLPREWEK--NKDEPFDAKIVSLHTG 513
Query: 565 -------------HVEAIVLYYKEDEEADFEHLSKMSNLRLLFIANY---------ISTM 602
E ++L + D + KMS LR+L I N S
Sbjct: 514 EMDEMNWFDMDLPKAEVLILNFSSDNYVLPPFIGKMSRLRVLVIINNGMSPARLHGFSIF 573
Query: 603 LGFPSCLSNKLRFVHWFRYPSKYLP-SNFHPNELVELILTESNIKQLWKNKKYLPNLRTL 661
S L+ VH S +P N H L+ + S ++ + K P+L L
Sbjct: 574 ANLAKLRSLWLKRVHVPELTSCTIPLKNLHKIHLIFCKVKNSFVQTSFDISKIFPSLSDL 633
Query: 662 DLRHSRNLEKIIDFGEFPNLERLDLEGCINLVELDPSIGLLRKLVYLNLKDCKSLVSIPN 721
+ H +L ++ +L L + C ++EL ++ ++ L L L C L+S+P
Sbjct: 634 TIDHCDDLLELKSIFGITSLNSLSITNCPRILELPKNLSNVQSLERLRLYACPELISLPV 693
Query: 722 NIFGLSSLQYLNMCGCSKVFNNPRRLMKSGISSEKKQQHDIRESASHHLP-------GLK 774
+ L L+Y+++ C + + P + K G ++ D+RE + LP L+
Sbjct: 694 EVCELPCLKYVDISQCVSLVSLPEKFGKLG----SLEKIDMRECSLLGLPSSVAALVSLR 749
Query: 775 WIILAHDSSHM 785
+I ++S M
Sbjct: 750 HVICDEETSSM 760
>DR29_ARATH (Q9SZA7) Probable disease resistance protein At4g33300
Length = 855
Score = 104 bits (260), Expect = 1e-21
Identities = 159/683 (23%), Positives = 270/683 (39%), Gaps = 155/683 (22%)
Query: 237 LLLNSVDGVRAIGICGMGGIGKTTLATTLYGQISHQFSASCFIDDVTKIYGLHDDPL-DV 295
++ S GV GI GMGG+GKTTLA L Q H+ C ++ + PL +
Sbjct: 194 MMFESQGGV--FGISGMGGVGKTTLAKEL--QRDHE--VQCHFENRILFLTVSQSPLLEE 247
Query: 296 QKQILFQTLGIEHQQICNRYHATTLIQRKLCHERTLMILDNVDQVEQLEKIAVHREWLGP 355
+++++ L C + R L+ILD+V + L+++ + P
Sbjct: 248 LRELIWGFLSG-----CEAGNPVPDCNFPFDGARKLVILDDVWTTQALDRLTSFKF---P 299
Query: 356 GSRIIIISRDEHVLKAYGVDVVYKVSLLDWNEAHMLFCRKAFKDEKIIMSNYQNLVDQ-- 413
G +++SR + + Y V +L +EA LFC AF + I + ++LV Q
Sbjct: 300 GCTTLVVSRSKLTEPKF----TYDVEVLSEDEAISLFCLCAFGQKSIPLGFCKDLVKQHN 355
Query: 414 --------------ILHYAKGLPLAIKVLGSFLFGRNVTEWKSALTRLRQSPVKD----- 454
+ + KGLPLA+KV G+ L G+ WK L RL + D
Sbjct: 356 IQSFSILRVLCLAQVANECKGLPLALKVTGASLNGKPEMYWKGVLQRLSKGEPADDSHES 415
Query: 455 -VMDVLQLSFDGLNETEKDIFLHIACFFNNDSEEDVKNILNCCGFHADI----GLRVLID 509
++ ++ S D L++T KD FL + F D + + ++N DI +L+D
Sbjct: 416 RLLRQMEASLDNLDQTTKDCFLDLGA-FPEDRKIPLDVLINIWIELHDIDEGNAFAILVD 474
Query: 510 ---------------KSLVSISYSI-INMHSLLEELG-------------RKIVQNSSSK 540
SL + Y I + H +L +L R ++
Sbjct: 475 LSHKNLLTLGKDPRLGSLYASHYDIFVTQHDVLRDLALHLSNAGKVNRRKRLLMPKRELD 534
Query: 541 EPRKWSR-----------------LWSTEQLYDVM-LENMEKH----------------- 565
P W R +ST Q++ V+ N + H
Sbjct: 535 LPGDWERNNDEHYIAQIVSIHTGKCFSTLQIFLVLSCNNHQDHNVGEMNEMQWFDMEFPK 594
Query: 566 VEAIVLYYKEDEEADFEHLSKMSNLRLLFIANYISTMLGFPSCLSNKLRFVHWFRYPSKY 625
E ++L + D+ +SKMS L++L I N + P+ L + F H
Sbjct: 595 AEILILNFSSDKYVLPPFISKMSRLKVLVIINNGMS----PAVLHDFSIFAHL------- 643
Query: 626 LPSNFHPNELVELILTESNIKQLWKNKKYLPNLRTLDL---RHSRNLEKI-IDFGE-FPN 680
++L L L ++ QL + L NL + L + +++ ++ +D + FP
Sbjct: 644 -------SKLRSLWLERVHVPQLSNSTTPLKNLHKMSLILCKINKSFDQTGLDVADIFPK 696
Query: 681 LERLDLEGCINLVELDPSIGLLRKLVYLNLKDCKSLVSIPNNIFGLSSLQYLNMCGCSKV 740
L L ++ C +LV L SI L L L++ +C L +P N+ L +L+ L + C ++
Sbjct: 697 LGDLTIDHCDDLVALPSSICGLTSLSCLSITNCPRLGELPKNLSKLQALEILRLYACPEL 756
Query: 741 FNNPRRLMKSGISSEKKQQHDIRESASHHLPGLKWIILAH--DSSHMLPSLHSLCCLRKV 798
P + + LPGLK++ ++ S + + L L K+
Sbjct: 757 KTLPGEICE--------------------LPGLKYLDISQCVSLSCLPEEIGKLKKLEKI 796
Query: 799 DISFCYLSHVPDAIECLHWLERL 821
D+ C S P + L L +
Sbjct: 797 DMRECCFSDRPSSAVSLKSLRHV 819
>RFL1_ARATH (Q8L3R3) Disease resistance protein RFL1 (RPS5-like
protein 1) (pNd13/pNd14)
Length = 885
Score = 97.4 bits (241), Expect = 2e-19
Identities = 165/702 (23%), Positives = 293/702 (41%), Gaps = 113/702 (16%)
Query: 203 EIVQKIINILECKYSCVSKDLVGIDSPIQALQNHLLLNSVDGVRAIGICGMGGIGKTTLA 262
+IV + I E + + +VG DS + + N L+ D V +G+ GMGG+GKTTL
Sbjct: 137 DIVTEAAPIAEVEELPIQSTIVGQDSMLDKVWNCLM---EDKVWIVGLYGMGGVGKTTLL 193
Query: 263 TTLYGQISHQFS--ASCFIDDVTKIYGLHDDPLDVQKQILFQTLGI--EHQQICNRYHAT 318
T QI+++FS F + + + +QK I + LG+ ++ N+
Sbjct: 194 T----QINNKFSKLGGGFDVVIWVVVSKNATVHKIQKSI-GEKLGLVGKNWDEKNKNQRA 248
Query: 319 TLIQRKLCHERTLMILDNVDQVEQLEKIAVHREWLGPGSRIIIISRDEHVLKAYGVDVVY 378
I L ++ +++LD++ + +L+ I V G ++ + + V GVD
Sbjct: 249 LDIHNVLRRKKFVLLLDDIWEKVELKVIGVPYPSGENGCKVAFTTHSKEVCGRMGVDNPM 308
Query: 379 KVSLLDWNEAHMLFCRKAFKDEKIIMSNYQNLVDQILHYAKGLPLAIKVLGSFL-FGRNV 437
++S LD A L +K ++ + L ++ GLPLA+ V+G + F R +
Sbjct: 309 EISCLDTGNAWDLLKKKVGENTLGSHPDIPQLARKVSEKCCGLPLALNVIGETMSFKRTI 368
Query: 438 TEWKSALTRLRQSP-----VKDVMDVLQLSFDGLN-ETEKDIFLHIACFFNNDSEEDVKN 491
EW+ A L + +++ +L+ S+D LN E K FL+ + F D E +
Sbjct: 369 QEWRHATEVLTSATDFSGMEDEILPILKYSYDSLNGEDAKSCFLYCS-LFPEDFEIRKEM 427
Query: 492 ILN---CCGFHADIGLRVLIDKSLVSISYSIINMHSLLEELGRKIVQNSSSKEPRKWSRL 548
++ C GF + R I +++ LLE G K S + + L
Sbjct: 428 LIEYWICEGFIKEKQGREKAFNQGYDILGTLVRSSLLLE--GAKDKDVVSMHDMVREMAL 485
Query: 549 WSTEQLYDVMLENMEKHVEAIVLY--YKEDEEADFEHLSKMSNLRLLFIANYISTMLGFP 606
W + ++ KH E ++ DE + E+ + + L+ N +LG P
Sbjct: 486 W--------IFSDLGKHKERCIVQAGIGLDELPEVENWRAVKRMSLM--NNNFEKILGSP 535
Query: 607 SCLSNKLRFVHWFRYPSKYLPSNFHPNELVELILTESNIKQLWKNKKY---LPNLRTLDL 663
C+ EL+ L L ++N K + + ++ +P+L LDL
Sbjct: 536 ECV------------------------ELITLFL-QNNYKLVDISMEFFRCMPSLAVLDL 570
Query: 664 RHSRNLEKI-IDFGEFPNLERLDLEGCINLVELDPSIGLLRKLVYLNLKDCKSLVSIPNN 722
+ +L ++ + E +L+ LDL G + L + LRKLV+L L+ + L SI +
Sbjct: 571 SENHSLSELPEEISELVSLQYLDLSGTY-IERLPHGLHELRKLVHLKLERTRRLESI-SG 628
Query: 723 IFGLSSLQYLNMCGCSKVFNNPRRLMKSGISSEKKQQHDIRESASHHLPGLKWIILAHDS 782
I LSSL+ L + + + + +G+ E + + + GL + +
Sbjct: 629 ISYLSSLRTLRL-------RDSKTTLDTGLMKELQLLEHLELITTDISSGLVGELFCYPR 681
Query: 783 -----SH-----------------MLPSLHSLCCLRKVDISFCYLSHVPDAIECLHWLER 820
H +LP++H+LC + I C++ + IE W +
Sbjct: 682 VGRCIQHIYIRDHWERPEESVGVLVLPAIHNLC---YISIWNCWMWEI--MIEKTPW--K 734
Query: 821 LNLAGNDFVTLPSLRKLSKLVYLNLEHCKLLESLPQLPFPTN 862
NL +F L ++R +E C L+ L L F N
Sbjct: 735 KNLTNPNFSNLSNVR---------IEGCDGLKDLTWLLFAPN 767
>DR27_ARATH (Q9T048) Putative disease resistance protein At4g27190
Length = 985
Score = 97.1 bits (240), Expect = 2e-19
Identities = 153/634 (24%), Positives = 268/634 (42%), Gaps = 97/634 (15%)
Query: 163 HEQRFQQDS---MKVSRWREALEQVGSIS----------GWDLRDEPLAREIKEIVQKI- 208
HE +D +K+ RW+ E+V S + G LR ++R++ +I+ ++
Sbjct: 54 HETLLTKDKPLRLKLMRWQREAEEVISKARLKLEERVSCGMSLRPR-MSRKLVKILDEVK 112
Query: 209 ---------INILECKYSCVSKDLV-GIDSPIQALQNHLLLNSVDGVRA-----IGICGM 253
+++L + + + V G+ Q + +++L DG+ + IG+ GM
Sbjct: 113 MLEKDGIEFVDMLSVESTPERVEHVPGVSVVHQTMASNMLAKIRDGLTSEKAQKIGVWGM 172
Query: 254 GGIGKTTLATTLYGQISHQFSASCFIDDVTKIYGLHDDPLDVQKQILFQTLGIEHQ---- 309
GG+GKTTL TL ++ + + F + I DP +VQKQI + L I+ Q
Sbjct: 173 GGVGKTTLVRTLNNKLREEGATQPFGLVIFVIVSKEFDPREVQKQIA-ERLDIDTQMEES 231
Query: 310 --QICNRYHATTLIQRKLCHERTLMILDNVDQVEQLEKIAVHREWLGPGSRIIIISRDEH 367
++ R + + +RK L+ILD+V + L+ + + R GS++I+ SR
Sbjct: 232 EEKLARRIYVGLMKERKF-----LLILDDVWKPIDLDLLGIPRTEENKGSKVILTSRFLE 286
Query: 368 VLKAYGVDVVYKVSLLDWNEAHMLFCRKAFKDEKIIMSNYQNLVDQILHYAKGLPLAIKV 427
V ++ D+ +V L +A LFC+ A + + + + + + GLPLAI
Sbjct: 287 VCRSMKTDLDVRVDCLLEEDAWELFCKNA--GDVVRSDHVRKIAKAVSQECGGLPLAIIT 344
Query: 428 LGSFLFG-RNVTEWKSALTRLRQSP------VKDVMDVLQLSFDGLNETEKDIFLHIACF 480
+G+ + G +NV W L++L +S + + L+LS+D L + K FL A F
Sbjct: 345 VGTAMRGKKNVKLWNHVLSKLSKSVPWIKSIEEKIFQPLKLSYDFLEDKAKFCFLLCALF 404
Query: 481 FNNDSEE--DVKNILNCCGFHADIG------------LRVLIDKSLVSIS--YSIINMHS 524
+ S E +V GF ++G + L D L+ + MH
Sbjct: 405 PEDYSIEVTEVVRYWMAEGFMEELGSQEDSMNEGITTVESLKDYCLLEDGDRRDTVKMHD 464
Query: 525 LLEELGRKIVQNSSSKEPRKWSRLWSTEQLYDVMLENMEKHVEAIVLYYKEDEE-ADFEH 583
++ + I+ SS + S + S L D+ + + + + L + E D
Sbjct: 465 VVRDFAIWIM---SSSQDDSHSLVMSGTGLQDIRQDKLAPSLRRVSLMNNKLESLPDLVE 521
Query: 584 LSKMSNLRLLFIANYI--STMLGFPSCLSNKLRFVHWFRYPSKYLPSNFHPNELVELILT 641
+ LL N++ +GF LR ++ K PS L+ L
Sbjct: 522 EFCVKTSVLLLQGNFLLKEVPIGFLQAFPT-LRILNLSGTRIKSFPS----CSLLRLF-- 574
Query: 642 ESNIKQLWKNKKYLPNLRTLDLRHSRNLEKIIDFGEFPNLERLDLEGCINLVELDPSIGL 701
+L +L LR L K+ LE LDL G +++E +
Sbjct: 575 ---------------SLHSLFLRDCFKLVKLPSLETLAKLELLDLCG-THILEFPRGLEE 618
Query: 702 LRKLVYLNLKDCKSLVSIPNNIFG-LSSLQYLNM 734
L++ +L+L L SIP + LSSL+ L+M
Sbjct: 619 LKRFRHLDLSRTLHLESIPARVVSRLSSLETLDM 652
Score = 32.7 bits (73), Expect = 5.4
Identities = 18/58 (31%), Positives = 37/58 (63%), Gaps = 3/58 (5%)
Query: 651 NKKYLPNLRTLDLRHSRNLEKIIDFGE-FPNLERLDLEGC--INLVELDPSIGLLRKL 705
++ ++PNLR L LR+ NL I ++GE + LE++++ C +N + + + G ++K+
Sbjct: 885 HQPFVPNLRVLKLRNLPNLVSICNWGEVWECLEQVEVIHCNQLNCLPISSTCGRIKKI 942
>DR28_ARATH (O81825) Putative disease resistance protein At4g27220
Length = 919
Score = 91.7 bits (226), Expect = 1e-17
Identities = 121/504 (24%), Positives = 213/504 (42%), Gaps = 61/504 (12%)
Query: 245 VRAIGICGMGGIGKTTLATTLYGQI-----SHQFSASCFIDDVTKIYGLHDDPLDVQKQI 299
V+ IG+ GMGG+GKTTL TL + + QF+ ++ V+K + L +D+ K++
Sbjct: 134 VQKIGVWGMGGVGKTTLVRTLNNDLLKYAATQQFALVIWVT-VSKDFDLKRVQMDIAKRL 192
Query: 300 LFQTLGIEHQQICNRYHATTLIQRKLCHERTLMILDNVDQVEQLEKIAVHREW-LGPGSR 358
+ + Q+ T+ +R + + L+ILD+V L+++ + S+
Sbjct: 193 GKRFTREQMNQL-----GLTICERLIDLKNFLLILDDVWHPIDLDQLGIPLALERSKDSK 247
Query: 359 IIIISRDEHVLKAYGVDVVYKVSLLDWNEAHMLFCRKAFKDEKIIMSNYQNLVDQILHYA 418
+++ SR V + + KV+ L EA LFC E N + + + H
Sbjct: 248 VVLTSRRLEVCQQMMTNENIKVACLQEKEAWELFCHNV--GEVANSDNVKPIAKDVSHEC 305
Query: 419 KGLPLAIKVLGSFLFGRNVTE-WKSALTRLRQSPV-----KDVMDVLQLSFDGLNETEKD 472
GLPLAI +G L G+ E WK L L++S + + L+LS+D L + K
Sbjct: 306 CGLPLAIITIGRTLRGKPQVEVWKHTLNLLKRSAPSIDTEEKIFGTLKLSYDFLQDNMKS 365
Query: 473 IFLHIACFFNNDSEEDVKNILNCCGFHADIGLRVLIDKSLVSISYSIINMHSLLEEL--- 529
FL C F D ++V + + ++ +++ E++
Sbjct: 366 CFLF------------------CALFPEDYSIKVS-ELIMYWVAEGLLDGQHHYEDMMNE 406
Query: 530 GRKIVQNSSSKEPRKWSRLWSTEQLYDVMLENMEKHVEAIVLYYKEDEEADFEHLSKMSN 589
G +V+ + T +++DV V +++ + F L
Sbjct: 407 GVTLVERLKDSCLLEDGDSCDTVKMHDV--------VRDFAIWFMSSQGEGFHSLVMAGR 458
Query: 590 LRLLFIANYISTMLGFPSCLSNKLRFVHWFRYPSKYLPSNFHPNELVELILTESNIKQLW 649
+ F + + + S ++NKL R P+ + LV L+ S++K++
Sbjct: 459 GLIEFPQDKFVSSVQRVSLMANKLE-----RLPNNVIEG---VETLVLLLQGNSHVKEVP 510
Query: 650 KN-KKYLPNLRTLDLRHSRNLEKIIDFGEFPNLERLDLEGCINLVELDPSIGLLRKLVYL 708
+ PNLR LDL R F +L L L C L L PS+ L KL +L
Sbjct: 511 NGFLQAFPNLRILDLSGVRIRTLPDSFSNLHSLRSLVLRNCKKLRNL-PSLESLVKLQFL 569
Query: 709 NLKDCKSLVSIPNNIFGLSSLQYL 732
+L + ++ +P + LSSL+Y+
Sbjct: 570 DLHE-SAIRELPRGLEALSSLRYI 592
Score = 33.5 bits (75), Expect = 3.2
Identities = 53/202 (26%), Positives = 84/202 (41%), Gaps = 40/202 (19%)
Query: 671 KIIDFGEFPNLERLDLEGCINLVELDPSIGLLRKLVYLNLKDCKSLVSIPNNIFG--LSS 728
K + FP+L GC + ++L P++ L L +NL+ L N G L
Sbjct: 744 KALSIHYFPSLSLAS--GCESQLDLFPNLEEL-SLDNVNLESIGEL----NGFLGMRLQK 796
Query: 729 LQYLNMCGCSKVFNNPRRLMKSGISSEKKQQHDIRESASHHLPGLKWIILAHDSSHMLPS 788
L+ L + GC ++ +RL I + LP L+ I +
Sbjct: 797 LKLLQVSGCRQL----KRLFSDQILAGT-------------LPNLQEIKVVS-----CLR 834
Query: 789 LHSLCCLRKVDISFCYLSHVPD-AIECLHWLERLNLAGNDFVTLPSLRKLSKLVYLNLEH 847
L L V + FC S +P + L +L +L ND V L SL +L +E
Sbjct: 835 LEELFNFSSVPVDFCAESLLPKLTVIKLKYLPQLRSLCNDRVVLESLE------HLEVES 888
Query: 848 CKLLESLPQLPFPTNTGEVHRE 869
C+ L++LP + P NTG ++ +
Sbjct: 889 CESLKNLPFV--PGNTGMINEQ 908
Score = 32.3 bits (72), Expect = 7.1
Identities = 28/86 (32%), Positives = 44/86 (50%), Gaps = 6/86 (6%)
Query: 776 IILAHDSSHM--LPS--LHSLCCLRKVDISFCYLSHVPDAIECLHWLERLNLAG-NDFVT 830
++L +SH+ +P+ L + LR +D+S + +PD+ LH L L L
Sbjct: 497 VLLLQGNSHVKEVPNGFLQAFPNLRILDLSGVRIRTLPDSFSNLHSLRSLVLRNCKKLRN 556
Query: 831 LPSLRKLSKLVYLNLEHCKLLESLPQ 856
LPSL L KL +L+L H + LP+
Sbjct: 557 LPSLESLVKLQFLDL-HESAIRELPR 581
>RGA1_SOLBU (Q7XA42) Putative disease resistance protein RGA1
(RGA3-blb)
Length = 1025
Score = 91.3 bits (225), Expect = 1e-17
Identities = 172/765 (22%), Positives = 308/765 (39%), Gaps = 140/765 (18%)
Query: 237 LLLNSVDGVRAIG---ICGMGGIGKTTLATTLYG--QISHQFSASCFIDDVTKIYGLHDD 291
+L+N+ + + I GMGG+GKTTL+ ++ +++ +F KI+ D
Sbjct: 210 ILINTASDAQKLSVLPILGMGGLGKTTLSQMVFNDQRVTERF--------YPKIWICISD 261
Query: 292 PLDVQKQILFQTLGIEHQQICNRYHATTL--IQRKLCHERTLMILDNVDQVEQLEKIAVH 349
+ ++ I IE + + + A +Q L +R ++LD+V +Q K A
Sbjct: 262 DFNEKRLIKAIVESIEGKSLSDMDLAPLQKKLQELLNGKRYFLVLDDVWNEDQ-HKWANL 320
Query: 350 REWL---GPGSRIIIISRDEHVLKAYGVDVVYKVSLLDWNEAHMLFCRKAFKDEKIIMSN 406
R L G+ ++ +R E V G Y++S L + LF ++AF ++ I N
Sbjct: 321 RAVLKVGASGAFVLTTTRLEKVGSIMGTLQPYELSNLSPEDCWFLFMQRAFGHQEEINPN 380
Query: 407 YQNLVDQILHYAKGLPLAIKVLGSFL-FGRNVTEWKSALTRLRQSPV-------KDVMDV 458
+ +I+ G+PLA K LG L F R EW+ +R SP+ ++
Sbjct: 381 LMAIGKEIVKKCGGVPLAAKTLGGILRFKREEREWE----HVRDSPIWNLPQDESSILPA 436
Query: 459 LQLSFDGLNETEKDIFLHIACFFNNDSEEDVKNILNCCGFHADIGLRVLIDKSLVSISYS 518
L+LS+ L + F++ A F D++ +N++ H L+ K +
Sbjct: 437 LRLSYHHLPLDLRQCFVYCA-VFPKDTKMAKENLIAFWMAHG-----FLLSKGNLE---- 486
Query: 519 IINMHSLLEELGRKIVQNSSSKEPRKWSRLWSTEQLYDVMLENMEKHVEAIVLYYKEDEE 578
LE++G ++ W+ L+ ++ +E+ + + + L + +
Sbjct: 487 -------LEDVGNEV-----------WNELYLRSFFQEIEVESGKTYFKMHDLIH-DLAT 527
Query: 579 ADFEHLSKMSNLRLLFIANYISTM--LGFPSCLSNKLRFVHWFRYPSKYLPSNFHPNELV 636
+ F + SN+R + ANY M +GF + S++ P+ L
Sbjct: 528 SLFSANTSSSNIREI-NANYDGYMMSIGFAEVV------------------SSYSPSLLQ 568
Query: 637 ELILTESNIKQLWKNKKYLPNLRTLDLRHSRNLEKIIDFGEFPNLERLDLEGCINLVELD 696
+ + +LR L+LR+S + G+ +L LDL G + L
Sbjct: 569 KFV-----------------SLRVLNLRNSNLNQLPSSIGDLVHLRYLDLSGNFRIRNLP 611
Query: 697 PSIGLLRKLVYLNLKDCKSLVSIPNNIFGLSSLQYLNMCGCSKVFNNPRRLMKSGISS-- 754
+ L+ L L+L C SL +P L SL+ L + GCS PR + + + S
Sbjct: 612 KRLCKLQNLQTLDLHYCDSLSCLPKQTSKLGSLRNLLLDGCSLTSTPPRIGLLTCLKSLS 671
Query: 755 ----EKKQQHDIRESASHHLPG----LKWIILAHDSS------HMLPSLHSLCCLRKVDI 800
K++ H + E + +L G K + D+ +LHSLC +D
Sbjct: 672 CFVIGKRKGHQLGELKNLNLYGSISITKLDRVKKDTDAKEANLSAKANLHSLCLSWDLDG 731
Query: 801 SFCYLSHVPDAIECLHWLERLNLAGNDFVTLPS------LRKLSKLVYLNLEHCKLLESL 854
Y S V +A++ L+ L + G + LP L+ + + E+C L
Sbjct: 732 KHRYDSEVLEALKPHSNLKYLEINGFGGIRLPDWMNQSVLKNVVSIRIRGCENCSCLPPF 791
Query: 855 PQLP----FPTNTGEVHREYDDYFCGAG-------LLIFNCPKL-------GEREH--CR 894
+LP +TG EY + G L+I++ L GE++
Sbjct: 792 GELPCLESLELHTGSADVEYVEDNVHPGRFPSLRKLVIWDFSNLKGLLKMEGEKQFPVLE 851
Query: 895 SMTLLWMKQFIKANPRSSSEIQIVNPGSEIPSWINNQRMGYSIAI 939
MT W F+ S ++++ + + I+N R S+ I
Sbjct: 852 EMTFYWCPMFVIPTLSSVKTLKVIVTDATVLRSISNLRALTSLDI 896
Score = 42.7 bits (99), Expect = 0.005
Identities = 47/184 (25%), Positives = 86/184 (46%), Gaps = 12/184 (6%)
Query: 559 LENMEKHVEAIVLYYKEDEEADFEHLSKMSNLRLLFIANYISTMLGFPSCLSNK----LR 614
LE++E H + + Y ED H + +LR L I ++ S + G K L
Sbjct: 797 LESLELHTGSADVEYVEDNV----HPGRFPSLRKLVIWDF-SNLKGLLKMEGEKQFPVLE 851
Query: 615 FVHWFRYPSKYLPSNFHPNELVELILTESNIKQLWKNKKYLPNLRTLDLRHSRNLEKIID 674
+ ++ P +P+ + +++I+T++ + + N + L +L D + +L + +
Sbjct: 852 EMTFYWCPMFVIPT-LSSVKTLKVIVTDATVLRSISNLRALTSLDISDNVEATSLPEEM- 909
Query: 675 FGEFPNLERLDLEGCINLVELDPSIGLLRKLVYLNLKDCKSLVSIP-NNIFGLSSLQYLN 733
F NL+ L + NL EL S+ L L L + C +L S+P + GL+SL L+
Sbjct: 910 FKSLANLKYLKISFFRNLKELPTSLASLNALKSLKFEFCDALESLPEEGVKGLTSLTELS 969
Query: 734 MCGC 737
+ C
Sbjct: 970 VSNC 973
>DR31_ARATH (Q9FLB4) Putative disease resistance protein At5g05400
Length = 874
Score = 89.0 bits (219), Expect = 6e-17
Identities = 127/538 (23%), Positives = 227/538 (41%), Gaps = 88/538 (16%)
Query: 221 KDLVGIDSPIQALQNHLLLNSVDGVRAIGICGMGGIGKTTLATTL---YGQISHQFSASC 277
+++VG ++ +++ N ++ GV +GI GMGG+GKTTL + + + +S+ F +
Sbjct: 154 QEIVGQEAIVESTWNSMM---EVGVGLLGIYGMGGVGKTTLLSQINNKFRTVSNDFDIAI 210
Query: 278 FIDDVTKIYGLHDDPLDVQKQILFQTLGIEHQQICNRYHATTLIQRKLCHERTLMILDNV 337
++ V+K + D+ K++ G E Q+ N +T I+R L +++ +++LD++
Sbjct: 211 WVV-VSKNPTVKRIQEDIGKRLDLYNEGWE-QKTENEIAST--IKRSLENKKYMLLLDDM 266
Query: 338 DQVEQLEKIAVHREWLGPGSRIIIISRDEHVLKAYGVDVVYKVSLLDWNEAHMLFCRKAF 397
L I + GS+I SR V GVD +V+ L W++A LF R
Sbjct: 267 WTKVDLANIGIPVPKRN-GSKIAFTSRSNEVCGKMGVDKEIEVTCLMWDDAWDLFTRN-M 324
Query: 398 KDEKIIMSNYQNLVDQILHYAKGLPLAIKVLGSFLFGR-NVTEWKSALTRLRQSPVKDVM 456
K+ + I GLPLA+ V+G + + ++ EW A+ + D++
Sbjct: 325 KETLESHPKIPEVAKSIARKCNGLPLALNVIGETMARKKSIEEWHDAVG-VFSGIEADIL 383
Query: 457 DVLQLSFDGLN-ETEKDIFLHIACFFNNDSEEDVKNILNCCGFHADIGLRVLIDKSLVSI 515
+L+ S+D L E K FL A F D E +++ + G+ +L K +
Sbjct: 384 SILKFSYDDLKCEKTKSCFLFSA-LFPEDYEIGKDDLIE---YWVGQGI-ILGSKGINYK 438
Query: 516 SYSIIN------------------MHSLLEELGRKIVQNSSSKEPRKWSRLWSTEQLYDV 557
Y+II MH ++ E+ I ++ + + + QL D+
Sbjct: 439 GYTIIGTLTRAYLLKESETKEKVKMHDVVREMALWISSGCGDQKQKNVLVVEANAQLRDI 498
Query: 558 MLENMEKHVEAIVLYYKEDEEADFEHLSKMSNLRLLFIANYISTMLGFPSCLSNKLRFVH 617
+K V + L Y + EEA C S
Sbjct: 499 PKIEDQKAVRRMSLIYNQIEEA----------------------------CES------- 523
Query: 618 WFRYPSKYLPSNFHPNELVELILTESNIKQLWKN-KKYLPNLRTLDLRHSRNLEKIIDFG 676
H +L L+L ++ ++++ + ++P L LDL + NL ++ F
Sbjct: 524 ------------LHCPKLETLLLRDNRLRKISREFLSHVPILMVLDLSLNPNLIELPSFS 571
Query: 677 EFPNLERLDLEGCINLVELDPSIGLLRKLVYLNLKDCKSLVSIPNNIFGLSSLQYLNM 734
+L L+L C + L + LR L+YLNL+ L I I L +L+ L +
Sbjct: 572 PLYSLRFLNLS-CTGITSLPDGLYALRNLLYLNLEHTYMLKRI-YEIHDLPNLEVLKL 627
Score = 43.5 bits (101), Expect = 0.003
Identities = 32/82 (39%), Positives = 45/82 (54%), Gaps = 3/82 (3%)
Query: 654 YLPNLRTLDLRHSRNLEKIIDF-GEFPNLERLDLEGCINLVELDPSIGLLRKLVYLNLKD 712
+ P L TL LR +R + +F P L LDL NL+EL PS L L +LNL
Sbjct: 525 HCPKLETLLLRDNRLRKISREFLSHVPILMVLDLSLNPNLIEL-PSFSPLYSLRFLNLS- 582
Query: 713 CKSLVSIPNNIFGLSSLQYLNM 734
C + S+P+ ++ L +L YLN+
Sbjct: 583 CTGITSLPDGLYALRNLLYLNL 604
>DRL1_ARATH (P60838) Probable disease resistance protein At1g12280
Length = 894
Score = 86.7 bits (213), Expect = 3e-16
Identities = 133/536 (24%), Positives = 225/536 (41%), Gaps = 69/536 (12%)
Query: 219 VSKDLVGIDSPIQALQNHLLLNSVDGVRAIGICGMGGIGKTTLATTLYGQISHQFSASC- 277
+ +VG ++ ++ + L + DG +G+ GMGG+GKTTL T +I+++FS C
Sbjct: 153 IQPTIVGQETMLERVWTRL---TEDGDEIVGLYGMGGVGKTTLLT----RINNKFSEKCS 205
Query: 278 -----FIDDVTKIYGLHDDPLDVQKQILFQTLGIEHQQICNRYHATTLIQRKLCHERTLM 332
V+K +H D+ K++ LG E N I L ++ ++
Sbjct: 206 GFGVVIWVVVSKSPDIHRIQGDIGKRL---DLGGEEWDNVNENQRALDIYNVLGKQKFVL 262
Query: 333 ILDNVDQVEQLEKIAVHREWLGPGSRIIIISRDEHVLKAYGVDVVYKVSLLDWNEAHMLF 392
+LD++ + LE + V G +++ +R V VD +VS L+ NEA LF
Sbjct: 263 LLDDIWEKVNLEVLGVPYPSRQNGCKVVFTTRSRDVCGRMRVDDPMEVSCLEPNEAWELF 322
Query: 393 CRKAFKDEKIIMSNYQNLVDQILHYAKGLPLAIKVLG-SFLFGRNVTEWKSALTRLRQ-- 449
K ++ + L ++ GLPLA+ V+G + R V EW++A+ L
Sbjct: 323 QMKVGENTLKGHPDIPELARKVAGKCCGLPLALNVIGETMACKRMVQEWRNAIDVLSSYA 382
Query: 450 ---SPVKDVMDVLQLSFDGLN-ETEKDIFLHIACFFNNDSEEDVKNILN---CCGFHADI 502
++ ++ +L+ S+D LN E K FL+ + F D + + +++ C GF +
Sbjct: 383 AEFPGMEQILPILKYSYDNLNKEQVKPCFLYCS-LFPEDYRMEKERLIDYWICEGFIDEN 441
Query: 503 GLRVLIDKSLVSISYSIINMHSLLEELGRKIVQNSSSKEPRKWSRLWSTEQLYDVMLENM 562
R I ++ LLEE K + + + LW L E+
Sbjct: 442 ESRERALSQGYEIIGILVRACLLLEEAINK--EQVKMHDVVREMALWIASDLG----EHK 495
Query: 563 EKHVEAIVLYYKEDEEADFEHLSKMSNLRLLFIANYISTMLGFPSCLSNKLRFVHWFRYP 622
E+ + + + +E + + S R+ + N I + G P CL
Sbjct: 496 ERCIVQVGVGLREVPKVK----NWSSVRRMSLMENEIEILSGSPECL------------- 538
Query: 623 SKYLPSNFHPNELVELILTESNIKQLWKNKKY---LPNLRTLDLRHSRNLEKIID-FGEF 678
EL L L + N L + ++ +P L LDL + +L K+ + +
Sbjct: 539 -----------ELTTLFL-QKNDSLLHISDEFFRCIPMLVVLDLSGNSSLRKLPNQISKL 586
Query: 679 PNLERLDLEGCINLVELDPSIGLLRKLVYLNLKDCKSLVSIP--NNIFGLSSLQYL 732
+L LDL + L + L+KL YL L K L SI +NI L LQ L
Sbjct: 587 VSLRYLDLSWTY-IKRLPVGLQELKKLRYLRLDYMKRLKSISGISNISSLRKLQLL 641
>R8L1_ARATH (O04093) Putative disease resistance protein RPP8-like
protein 1
Length = 821
Score = 84.3 bits (207), Expect = 2e-15
Identities = 164/657 (24%), Positives = 268/657 (39%), Gaps = 107/657 (16%)
Query: 120 LQELEKICECIKGSGKHVLPVFYDVDPSEVRKQSGIYSEAFVKHEQRFQQDSMKVSRWRE 179
LQ L K + K + V DV ++ + E+F+ +E R ++ +K R
Sbjct: 41 LQSLLKDADAKKHESERVRNFLEDV--RDIVYDAEDIIESFLLNEFRTKEKGIKKHARRL 98
Query: 180 AL-------EQVGSISGWDLRDEPLAREIKEIVQKIINILECKYSCVSKDLVGIDSPIQA 232
A E + S L++ RE KEI Q N E DLVG++ ++A
Sbjct: 99 ACFLSLGIQEIIDGASSMSLQERQ--REQKEIRQTFANSSE-------SDLVGVEQSVEA 149
Query: 233 LQNHLLLNSVDGVRAIGICGMGGIGKTTLATTLYGQISHQFSASCFIDDVTKIYGLHDDP 292
L HL+ N D ++ + I GMGGIGKTTLA Q+ H D ++
Sbjct: 150 LAGHLVEN--DNIQVVSISGMGGIGKTTLAR----QVFHHDMVQRHFDGFAWVFVSQQFT 203
Query: 293 LDVQKQILFQTLGIEHQQICNRYHATTLIQRKLCH----ERTLMILDNVDQVEQLEKIAV 348
Q ++Q L ++ I + ++Q KL R L++LD+V + E ++I
Sbjct: 204 QKHVWQRIWQELQPQNGDI--SHMDEHILQGKLFKLLETGRYLVVLDDVWKEEDWDRIKA 261
Query: 349 ----HREWLGPGSRIIIISRDEHV-LKAYGVDVVYKVSLLDWNEAHMLFCRKAF---KDE 400
R W ++++ SR+E V + A +K +L E+ L C K +DE
Sbjct: 262 VFPRKRGW-----KMLLTSRNEGVGIHADPKSFGFKTRILTPEESWKL-CEKIVFHRRDE 315
Query: 401 KIIMSNYQNLVDQILHYAKGLPLAIKVLGSFLFGRN-VTEWKSALTRL---------RQS 450
+S+ + + +++ GLPLA+KVLG L ++ V EWK +
Sbjct: 316 TGTLSDMEAMGKEMVTCCGGLPLAVKVLGGLLATKHTVPEWKRVYDNIGPHLAGRSSLDD 375
Query: 451 PVKDVMDVLQLSFDGLNETEKDIFLHIACFFNNDSEEDVKNILNCCGFHADIGLRVLIDK 510
+ + VL LS++ L K FL++A F E VK + N + A G+ D
Sbjct: 376 NLNSIYRVLSLSYENLPMCLKHCFLYLA-HFPEYYEIHVKRLFN---YLAAEGIITSSDD 431
Query: 511 SLVSISYSIINMHSLLEELGRKIVQNSSSKEPRKWSRLWSTEQLYDVMLENMEKHVEAIV 570
LEEL R+ N + + Q++D+M E
Sbjct: 432 GTTIQDKG----EDYLEELARR---NMITIDKNYMFLRKKHCQMHDMMRE---------- 474
Query: 571 LYYKEDEEADFEHLSKMS------NLRLLFIANYISTMLG--FPS---CLSNKLRFVHWF 619
+ + +E +F + K+S N R L + +S G PS ++ K+R + +F
Sbjct: 475 VCLSKAKEENFLEIFKVSTATSAINARSLSKSRRLSVHGGNALPSLGQTINKKVRSLLYF 534
Query: 620 RYPSKYLPSNFHPNELVELILTESNIKQLWKNKKYLPNLRTLDLRHSR--NLEKIIDFGE 677
+ E + ES + LP LR LDL + + G+
Sbjct: 535 AFED-------------EFCILESTTPCF----RSLPLLRVLDLSRVKFEGGKLPSSIGD 577
Query: 678 FPNLERLDLEGCINLVELDPSIGLLRKLVYLNLKDCKSLVSIPNNIFGLSSLQYLNM 734
+L L L + L S+ L+ L+YLNL +V +PN + + L+YL +
Sbjct: 578 LIHLRFLSLHRAW-ISHLPSSLRNLKLLLYLNL-GFNGMVHVPNVLKEMQELRYLQL 632
Score = 33.1 bits (74), Expect = 4.2
Identities = 20/59 (33%), Positives = 34/59 (56%), Gaps = 1/59 (1%)
Query: 788 SLHSLCCLRKVDISFCYLSHVPDAIECLHWLERLNLAGNDFVTLPS-LRKLSKLVYLNL 845
S+ L LR + + ++SH+P ++ L L LNL N V +P+ L+++ +L YL L
Sbjct: 574 SIGDLIHLRFLSLHRAWISHLPSSLRNLKLLLYLNLGFNGMVHVPNVLKEMQELRYLQL 632
>DR32_ARATH (Q9FG91) Probable disease resistance protein At5g43730
Length = 848
Score = 83.6 bits (205), Expect = 3e-15
Identities = 142/564 (25%), Positives = 234/564 (41%), Gaps = 88/564 (15%)
Query: 199 REIKEIVQKIINILECKYSCVSKDLVGIDSPIQALQNHLLLNSVDGVRAIGICGMGGIGK 258
+ + + QKII E K+ + VG+D+ + L+ D +R +G+ GMGGIGK
Sbjct: 132 KNFEVVAQKIIPKAEKKHI---QTTVGLDTMVGIAWESLI---DDEIRTLGLYGMGGIGK 185
Query: 259 TTLATTL---YGQISHQFSASCFIDDVTKIYGLHDDPLDVQKQILFQTLGIEHQQICNRY 315
TTL +L + ++ +F ++ V+K + L +Q QIL + + +
Sbjct: 186 TTLLESLNNKFVELESEFDVVIWV-VVSKDFQLE----GIQDQILGRLRPDKEWERETES 240
Query: 316 HATTLIQRKLCHERTLMILDNVDQVEQLEKIAVHREWLGPGSRIIIISRDEHVLKAYGVD 375
+LI L ++ +++LD++ L KI V GS+I+ +R + V K D
Sbjct: 241 KKASLINNNLKRKKFVLLLDDLWSEVDLIKIGVPPPSRENGSKIVFTTRSKEVCKHMKAD 300
Query: 376 VVYKVSLLDWNEAHMLFCRKAFKDEKIIMSNYQN---LVDQILHYAKGLPLAIKVLG-SF 431
KV L +EA LF R D II+ ++Q+ L + GLPLA+ V+G +
Sbjct: 301 KQIKVDCLSPDEAWELF-RLTVGD--IILRSHQDIPALARIVAAKCHGLPLALNVIGKAM 357
Query: 432 LFGRNVTEWKSALTRLRQSPVK------DVMDVLQLSFDGLNETEKDIFLHIACFFNNDS 485
+ V EW+ A+ L K ++ +L+ S+D L E + F D
Sbjct: 358 VCKETVQEWRHAINVLNSPGHKFPGMEERILPILKFSYDSLKNGEIKLCFLYCSLFPEDF 417
Query: 486 EEDVKNILN---CCGF-----HADIGLRVLIDKSLVSISYSIINMHSLLE-ELGRKIVQN 536
E + ++ C G+ + D G D I ++ H L+E EL K+ +
Sbjct: 418 EIEKDKLIEYWICEGYINPNRYEDGGTNQGYD-----IIGLLVRAHLLIECELTDKVKMH 472
Query: 537 SSSKEPRKWSRLWSTEQLYDVMLENMEKHVEAIVLYYKEDEEADFEHLSKMSNLRLLFIA 596
+E W Q + +++ HV I + +E + +MS I+
Sbjct: 473 DVIREMALWINSDFGNQQETICVKS-GAHVRLI------PNDISWEIVRQMS-----LIS 520
Query: 597 NYISTMLGFPSC--LSNKLRFVHWFRYPSKYLPSNFHPNELVELILTESNIKQLWKNKKY 654
+ + P+C LS L LP N + V L +
Sbjct: 521 TQVEKIACSPNCPNLSTLL------------LPYNKLVDISVGFFL-------------F 555
Query: 655 LPNLRTLDLRHSRNL----EKIIDFGEFPNLERLDLEGCINLVELDPSIGLLRKLVYLNL 710
+P L LDL + +L E+I + G L L L G + L + LRKL+YLNL
Sbjct: 556 MPKLVVLDLSTNWSLIELPEEISNLGSLQYL-NLSLTG---IKSLPVGLKKLRKLIYLNL 611
Query: 711 KDCKSLVSIPNNIFGLSSLQYLNM 734
+ L S+ L +LQ L +
Sbjct: 612 EFTNVLESLVGIATTLPNLQVLKL 635
Score = 41.2 bits (95), Expect = 0.015
Identities = 30/94 (31%), Positives = 49/94 (51%), Gaps = 4/94 (4%)
Query: 805 LSHVPDAIECLHWLERLNLAGNDFVTLP-SLRKLSKLVYLNLEHCKLLESLPQLPFPTNT 863
L +P+ I L L+ LNL+ +LP L+KL KL+YLNLE +LESL +
Sbjct: 570 LIELPEEISNLGSLQYLNLSLTGIKSLPVGLKKLRKLIYLNLEFTNVLESLVGIATTLPN 629
Query: 864 GEVHREYDDYFCGAGLLIFNCPKLGEREHCRSMT 897
+V + + FC +++ +L +H + +T
Sbjct: 630 LQVLKLFYSLFCVDDIIMEELQRL---KHLKILT 660
>RPM1_ARATH (Q39214) Disease resistance protein RPM1 (Resistance to
Pseudomonas syringae protein 3)
Length = 926
Score = 81.3 bits (199), Expect = 1e-14
Identities = 132/572 (23%), Positives = 238/572 (41%), Gaps = 88/572 (15%)
Query: 210 NILECKYSCVSKDLVGIDSPIQALQNHLLLNSVDGVRAIGICGMGGIGKTTLATTLYGQI 269
NI E LVGID+P L LL + + + GMGG GKTTL+ ++
Sbjct: 159 NISESSLFFSENSLVGIDAPKGKLIGRLLSPEPQRI-VVAVVGMGGSGKTTLSANIFKSQ 217
Query: 270 SHQ----------FSASCFIDDVTKIYGLHDDPLDVQKQILFQTLGIEHQQICNRYHATT 319
S + S S I+DV + + + + QI + + ++++ +
Sbjct: 218 SVRRHFESYAWVTISKSYVIEDVFRTM-IKEFYKEADTQIPAELYSLGYRELVEK----- 271
Query: 320 LIQRKLCHERTLMILDNVDQVEQLEKIAVHREWLGPGSRIIIISRDEHVLK-AYGV-DVV 377
+ L +R +++LD+V +I++ GSR+++ +RD +V YG+
Sbjct: 272 -LVEYLQSKRYIVVLDDVWTTGLWREISIALPDGIYGSRVMMTTRDMNVASFPYGIGSTK 330
Query: 378 YKVSLLDWNEAHMLFCRKAFKD--EKIIMSNYQNLVDQILHYAKGLPLAIKVLGSFLFGR 435
+++ LL +EA +LF KAF E+ N + + +++ +GLPLAI LGS + +
Sbjct: 331 HEIELLKEDEAWVLFSNKAFPASLEQCRTQNLEPIARKLVERCQGLPLAIASLGSMMSTK 390
Query: 436 NV-TEWKSALTRL-----RQSPVKDVMDVLQLSFDGLNETEKDIFLHIACFFNNDSEEDV 489
+EWK + L +K V ++ LSF+ L K FL+ + F N +
Sbjct: 391 KFESEWKKVYSTLNWELNNNHELKIVRSIMFLSFNDLPYPLKRCFLYCSLFPVNYRMKRK 450
Query: 490 KNILNCCGFH-------------ADIGLRVLIDKSLVSISY-------SIINMHSLLEEL 529
+ I AD L L+ ++++ + MH ++ E+
Sbjct: 451 RLIRMWMAQRFVEPIRGVKAEEVADSYLNELVYRNMLQVILWNPFGRPKAFKMHDVIWEI 510
Query: 530 GRKIVQNSSSKEPRKWSRLWSTEQLYDVMLENMEKHVEAIVLYYKE-------------- 575
+ + ++ +++ + D E ME + + KE
Sbjct: 511 ALSV------SKLERFCDVYNDDSDGDDAAETMENYGSRHLCIQKEMTPDSIRATNLHSL 564
Query: 576 ----DEEADFEHLSKMSNLRLLFIANYISTMLGFPSCLSN--KLRFVHWFRYPSKYLPSN 629
+ E L ++ LR L + + S++ P CL L++++ + K LP N
Sbjct: 565 LVCSSAKHKMELLPSLNLLRALDLED--SSISKLPDCLVTMFNLKYLNLSKTQVKELPKN 622
Query: 630 FHPNELVELILTE-SNIKQL----WKNKK--YLPNLRTLDLRHSRNLEKIIDFGEFP--- 679
FH +E + T+ S I++L WK KK YL R + H N ++ P
Sbjct: 623 FHKLVNLETLNTKHSKIEELPLGMWKLKKLRYLITFRRNE-GHDSNWNYVLGTRVVPKIW 681
Query: 680 NLERLDLEGCINLV-ELDPSIGLLRKLVYLNL 710
L+ L + C N EL ++G + +L ++L
Sbjct: 682 QLKDLQVMDCFNAEDELIKNLGCMTQLTRISL 713
Score = 39.3 bits (90), Expect = 0.058
Identities = 26/81 (32%), Positives = 42/81 (51%), Gaps = 2/81 (2%)
Query: 776 IILAHDSSHMLPSLHSLCCLRKVDISFCYLSHVPDAIECLHWLERLNLAGNDFVTLP-SL 834
+++ + H + L SL LR +D+ +S +PD + + L+ LNL+ LP +
Sbjct: 564 LLVCSSAKHKMELLPSLNLLRALDLEDSSISKLPDCLVTMFNLKYLNLSKTQVKELPKNF 623
Query: 835 RKLSKLVYLNLEHCKLLESLP 855
KL L LN +H K +E LP
Sbjct: 624 HKLVNLETLNTKHSK-IEELP 643
>RGA2_SOLBU (Q7XBQ9) Disease resistance protein RGA2 (RGA2-blb)
(Blight resistance protein RPI)
Length = 970
Score = 80.5 bits (197), Expect = 2e-14
Identities = 133/546 (24%), Positives = 227/546 (41%), Gaps = 99/546 (18%)
Query: 237 LLLNSVDGVRAIG---ICGMGGIGKTTLATTLYG--QISHQFSASCFIDDVTKIYGLHDD 291
+L+N+V + + I GMGG+GKTTLA ++ +++ F + KI+ +
Sbjct: 164 ILINNVSDAQHLSVLPILGMGGLGKTTLAQMVFNDQRVTEHFHS--------KIWICVSE 215
Query: 292 PLDVQKQILFQTLGIEHQQICNRYHATTL---IQRKLCHERTLMILDNVDQVEQLEKIAV 348
D ++ I IE + + L +Q L +R L++LD+V +Q +K A
Sbjct: 216 DFDEKRLIKAIVESIEGRPLLGEMDLAPLQKKLQELLNGKRYLLVLDDVWNEDQ-QKWAN 274
Query: 349 HREWL---GPGSRIIIISRDEHVLKAYGVDVVYKVSLLDWNEAHMLFCRKAFKDEKIIMS 405
R L G+ ++ +R E V G Y++S L + +LF ++AF ++ I
Sbjct: 275 LRAVLKVGASGASVLTTTRLEKVGSIMGTLQPYELSNLSQEDCWLLFMQRAFGHQEEINP 334
Query: 406 NYQNLVDQILHYAKGLPLAIKVLGSFL-FGRNVTEWKSALTRLRQSPV-------KDVMD 457
N + +I+ + G+PLA K LG L F R W+ +R SP+ ++
Sbjct: 335 NLVAIGKEIVKKSGGVPLAAKTLGGILCFKREERAWE----HVRDSPIWNLPQDESSILP 390
Query: 458 VLQLSFDGLNETEKDIFLHIACFFNNDSEEDVKNILNCCGFHA-----------DIG--- 503
L+LS+ L K F + A F D++ + + +++ H D+G
Sbjct: 391 ALRLSYHQLPLDLKQCFAYCA-VFPKDAKMEKEKLISLWMAHGFLLSKGNMELEDVGDEV 449
Query: 504 -----LRVLIDKSLVSISYSIINMHSLLEELGRKIVQ-NSSSKEPRKWSRLWSTEQLYDV 557
LR + V + MH L+ +L + N+SS R+ ++ T +
Sbjct: 450 WKELYLRSFFQEIEVKDGKTYFKMHDLIHDLATSLFSANTSSSNIREINKHSYTHMMSIG 509
Query: 558 MLENMEKHVEAIVLYYKEDEEADFEHLSKMSNLRLLFIANYISTMLGFPSCLSN--KLRF 615
E +V +Y L K +LR+L + + ST PS + + LR+
Sbjct: 510 FAE--------VVFFY------TLPPLEKFISLRVLNLGD--STFNKLPSSIGDLVHLRY 553
Query: 616 VHWFRYPSKYLPSNFHPNELVELILTESNIKQLWKNKKYLPNLRTLDLRHSRNLEKI-ID 674
++ + + LP KQL K L NL+TLDL++ L + +
Sbjct: 554 LNLYGSGMRSLP------------------KQLCK----LQNLQTLDLQYCTKLCCLPKE 591
Query: 675 FGEFPNLERLDLEGCINLVELDPSIGLLRKLVYLNLKDCKSLVSIPNNIFGLSSLQYLNM 734
+ +L L L+G +L + P IG L LK V + L L LN+
Sbjct: 592 TSKLGSLRNLLLDGSQSLTCMPPRIGSL-----TCLKTLGQFVVGRKKGYQLGELGNLNL 646
Query: 735 CGCSKV 740
G K+
Sbjct: 647 YGSIKI 652
Score = 39.7 bits (91), Expect = 0.044
Identities = 55/225 (24%), Positives = 87/225 (38%), Gaps = 38/225 (16%)
Query: 649 WKNKKYLPNLRTLDLRHSRNLEKIIDFGEFPNLERLDLE------GCINLVELDPSIGL- 701
W N L N+ ++ + + RN + FG+ P LE L+L + V++D G
Sbjct: 723 WMNHSVLKNIVSILISNFRNCSCLPPFGDLPCLESLELHWGSADVEYVEEVDIDVHSGFP 782
Query: 702 -------LRKLVYLNLKDCKSLVSIP-NNIFGLSSLQYLNMCGCSKVFNNPRRLMKSGIS 753
LRKL + K L+ F + ++ C + +N R L I
Sbjct: 783 TRIRFPSLRKLDIWDFGSLKGLLKKEGEEQFPVLEEMIIHECPFLTLSSNLRALTSLRIC 842
Query: 754 SEKKQQHDIRESASHHLPGLKWIILA--HDSSHMLPSLHSLCCLRKVDISFCYLSHVPDA 811
K E +L LK++ ++ ++ + SL SL L+ + I C
Sbjct: 843 YNKVAT-SFPEEMFKNLANLKYLTISRCNNLKELPTSLASLNALKSLKIQLCCA------ 895
Query: 812 IECLHWLERLNLAGNDFVTLPSLRKLSKLVYLNLEHCKLLESLPQ 856
LE L G L LS L L +EHC +L+ LP+
Sbjct: 896 ------LESLPEEG--------LEGLSSLTELFVEHCNMLKCLPE 926
Score = 38.9 bits (89), Expect = 0.076
Identities = 34/109 (31%), Positives = 53/109 (48%), Gaps = 7/109 (6%)
Query: 635 LVELILTESNIKQLWKNKKYLPNLRTLDLRHSRNLEKIIDFGEFPNLERLDLEGCINLVE 694
L E+I+ E L N + L +LR + + + + + F NL+ L + C NL E
Sbjct: 816 LEEMIIHECPFLTLSSNLRALTSLRICYNKVATSFPEEM-FKNLANLKYLTISRCNNLKE 874
Query: 695 LDPSIGLLRKLVYLNLKDCKSLVSIP-NNIFGLSSL-----QYLNMCGC 737
L S+ L L L ++ C +L S+P + GLSSL ++ NM C
Sbjct: 875 LPTSLASLNALKSLKIQLCCALESLPEEGLEGLSSLTELFVEHCNMLKC 923
Score = 36.2 bits (82), Expect = 0.49
Identities = 25/74 (33%), Positives = 38/74 (50%), Gaps = 1/74 (1%)
Query: 784 HMLPSLHSLCCLRKVDISFCYLSHVPDAIECLHWLERLNLAGNDFVTLP-SLRKLSKLVY 842
+ LP L LR +++ + +P +I L L LNL G+ +LP L KL L
Sbjct: 517 YTLPPLEKFISLRVLNLGDSTFNKLPSSIGDLVHLRYLNLYGSGMRSLPKQLCKLQNLQT 576
Query: 843 LNLEHCKLLESLPQ 856
L+L++C L LP+
Sbjct: 577 LDLQYCTKLCCLPK 590
Score = 32.7 bits (73), Expect = 5.4
Identities = 32/116 (27%), Positives = 55/116 (46%), Gaps = 14/116 (12%)
Query: 641 TESNIKQLWKNKKYLPNLRTLDLRHSRNLEKIIDFGEFPNLERLDLEGCINLVE-----L 695
+ SNI+++ NK ++ ++ +++ F P LE+ +NL + L
Sbjct: 490 SSSNIREI--NKHSYTHMMSI------GFAEVVFFYTLPPLEKFISLRVLNLGDSTFNKL 541
Query: 696 DPSIGLLRKLVYLNLKDCKSLVSIPNNIFGLSSLQYLNMCGCSKVFNNPRRLMKSG 751
SIG L L YLNL + S+P + L +LQ L++ C+K+ P+ K G
Sbjct: 542 PSSIGDLVHLRYLNLYG-SGMRSLPKQLCKLQNLQTLDLQYCTKLCCLPKETSKLG 596
>R131_ARATH (Q9LRR4) Putative disease resistance RPP13-like protein
1
Length = 1054
Score = 80.1 bits (196), Expect = 3e-14
Identities = 133/535 (24%), Positives = 221/535 (40%), Gaps = 92/535 (17%)
Query: 243 DGVRAIGICGMGGIGKTTLATTLYGQISHQFSASCFIDDVTKIYGLHDDPLDVQKQILFQ 302
+G+ + I G+GG+GKTTL+ LY + Q S F TK++ + DV F+
Sbjct: 194 NGITVVAIVGIGGVGKTTLSQLLY---NDQHVRSYF---GTKVWAHVSEEFDV-----FK 242
Query: 303 TLGIEHQQICNR---YHATTLIQRKLCHERT------LMILD-----NVDQVEQLEKIAV 348
++ + +R + ++Q KL T L++LD N + L + +
Sbjct: 243 ITKKVYESVTSRPCEFTDLDVLQVKLKERLTGTGLPFLLVLDDLWNENFADWDLLRQPFI 302
Query: 349 HREWLGPGSRIIIISRDEHVLKAYGVDVVYKVSLLDWNEAHMLFCRKAF-KDEKIIMSNY 407
H GS+I++ +R + V V+ + L + LF + F E +
Sbjct: 303 H---AAQGSQILVTTRSQRVASIMCAVHVHNLQPLSDGDCWSLFMKTVFGNQEPCLNREI 359
Query: 408 QNLVDQILHYAKGLPLAIKVLGSFL-FGRNVTEWKSAL-TRLRQSPV--KDVMDVLQLSF 463
+L ++I+H +GLPLA+K LG L F V EW+ L +R+ P +++ VL++S+
Sbjct: 360 GDLAERIVHKCRGLPLAVKTLGGVLRFEGKVIEWERVLSSRIWDLPADKSNLLPVLRVSY 419
Query: 464 DGLNETEKDIFLHIACFFNNDSEEDVKNILNCCGFHADIGLRVLIDKSLVSISYSIINMH 523
L K F + + F + E K +L + + + + + S N
Sbjct: 420 YYLPAHLKRCFAYCSIFPKGHAFEKDKVVL------------LWMAEGFLQQTRSSKN-- 465
Query: 524 SLLEELGRKIVQNSSSKEPRKWSRLWSTEQLYDVMLENMEKHVEAIVLYYKEDEEADFEH 583
LEELG + S+ S L T+ Y M + + + + + FE
Sbjct: 466 --LEELGNEYFSELESR-----SLLQKTKTRY-----IMHDFINELAQFASGEFSSKFED 513
Query: 584 LSKMSNLRLLFIANYISTMLGFPSCLSNKLRFVHWFRYPSKYLPSNFHPNELVELILTES 643
K+ +Y+ P LR V + R +LP L LT S
Sbjct: 514 GCKLQVSERTRYLSYLRDNYAEPMEF-EALREVKFLR---TFLP----------LSLTNS 559
Query: 644 N----IKQLWKNK--KYLPNLRTLDLRHSR------NLEKIIDFGEFPNLERLDLEGCIN 691
+ + Q+ K L LR L L H + + K I F +L R +LE
Sbjct: 560 SRSCCLDQMVSEKLLPTLTRLRVLSLSHYKIARLPPDFFKNISHARFLDLSRTELE---- 615
Query: 692 LVELDPSIGLLRKLVYLNLKDCKSLVSIPNNIFGLSSLQYLNMCGCSKVFNNPRR 746
+L S+ + L L L C SL +P +I L +L+YL++ G +K+ PRR
Sbjct: 616 --KLPKSLCYMYNLQTLLLSYCSSLKELPTDISNLINLRYLDLIG-TKLRQMPRR 667
Score = 34.3 bits (77), Expect = 1.9
Identities = 21/67 (31%), Positives = 36/67 (53%), Gaps = 3/67 (4%)
Query: 657 NLRTLDLRHSRNLEKIIDFGEFPNLERLDLEGCINLVELDPSIGLLR---KLVYLNLKDC 713
NL+TL ++ S + F NL++L+++ C +L L+ S LR L L + DC
Sbjct: 925 NLQTLSIKSSCDTLVKFPLNHFANLDKLEVDQCTSLYSLELSNEHLRGPNALRNLRINDC 984
Query: 714 KSLVSIP 720
++L +P
Sbjct: 985 QNLQLLP 991
>DRL3_ARATH (Q9LMP6) Probable disease resistance protein At1g15890
Length = 921
Score = 80.1 bits (196), Expect = 3e-14
Identities = 131/557 (23%), Positives = 230/557 (40%), Gaps = 68/557 (12%)
Query: 197 LAREIKEIVQKIINILECKYSCVSKDLVGIDSPIQALQNHLLLNSVDGVRAIGICGMGGI 256
LA+ + E+V + I + + + + VG+D+ + N L+ D R +G+ GMGG+
Sbjct: 200 LAKGVFEVVAEKIPAPKVEKKHI-QTTVGLDAMVGRAWNSLMK---DERRTLGLYGMGGV 255
Query: 257 GKTTLATTLYGQISHQFSASCFIDDVTKIYGLHDDPLDVQKQILFQTLGIEH--QQICNR 314
GKTTL ++ + + + V L ++ +Q+QIL + LG+ +Q+ +
Sbjct: 256 GKTTLLASINNKFLEGMNGFDLVIWVVVSKDLQNE--GIQEQILGR-LGLHRGWKQVTEK 312
Query: 315 YHATTLIQRKLCHERTLMILDNVDQVEQLEKIAVHREWLGPGSRIIIISRDEHVLKAYGV 374
A+ I L ++ +++LD++ LEKI V GS+I+ +R + V + V
Sbjct: 313 EKAS-YICNILNVKKFVLLLDDLWSEVDLEKIGVPPLTRENGSKIVFTTRSKDVCRDMEV 371
Query: 375 DVVYKVSLLDWNEAHMLFCRKAFKDEKIIMSNYQNLVDQILHYAKGLPLAIKVLGSFLFG 434
D KV L +EA LF +K + L ++ GLPLA+ V+G +
Sbjct: 372 DGEMKVDCLPPDEAWELFQKKVGPIPLQSHEDIPTLARKVAEKCCGLPLALSVIGKAMAS 431
Query: 435 R-NVTEWKSALTRLRQSP------VKDVMDVLQLSFDGLNETEKDIFLHIACFFNNDSE- 486
R V EW+ + L S + ++ VL+ S+D L + + + F D E
Sbjct: 432 RETVQEWQHVIHVLNSSSHEFPSMEEKILPVLKFSYDDLKDEKVKLCFLYCSLFPEDYEV 491
Query: 487 --EDVKNILNCCGFHADIGLRVLIDKSLVSISYSIINMHSLLE-ELGRKIVQNSSSKEPR 543
E++ C GF + I S++ H L++ EL K+ + +E
Sbjct: 492 RKEELIEYWMCEGFIDGNEDEDGANNKGHDIIGSLVRAHLLMDGELTTKVKMHDVIREMA 551
Query: 544 KWSRLWSTEQLYDVMLENMEKHVEAI-----VLYYKEDEEADFEHLSKMSNLRLLFIANY 598
W + N K E + V ++ ++E L +MS L IAN
Sbjct: 552 LW------------IASNFGKQKETLCVKPGVQLCHIPKDINWESLRRMS-LMCNQIANI 598
Query: 599 ISTMLGFPSCLSNKLRFVHWFRYPSKYLPSNFHPNELVELILTESNIKQLWKNKKYLPNL 658
S S+ PN L+ + +++P L
Sbjct: 599 SS---------------------------SSNSPNLSTLLLQNNKLVHISCDFFRFMPAL 631
Query: 659 RTLDLRHSRNLEKIID-FGEFPNLERLDLEGCINLVELDPSIGLLRKLVYLNLKDCKSLV 717
LDL + +L + + + +L+ ++L + L S L+KL++LNL+ L
Sbjct: 632 VVLDLSRNSSLSSLPEAISKLGSLQYINL-STTGIKWLPVSFKELKKLIHLNLEFTDELE 690
Query: 718 SIPNNIFGLSSLQYLNM 734
SI L +LQ L +
Sbjct: 691 SIVGIATSLPNLQVLKL 707
Score = 37.4 bits (85), Expect = 0.22
Identities = 36/126 (28%), Positives = 58/126 (45%), Gaps = 14/126 (11%)
Query: 765 SASHHLPGLKWIILAH--------DSSHMLPSLHSLCCLRKVDISFCYLSHVPDAIECLH 816
S+S + P L ++L + D +P+L L R LS +P+AI L
Sbjct: 599 SSSSNSPNLSTLLLQNNKLVHISCDFFRFMPALVVLDLSRNSS-----LSSLPEAISKLG 653
Query: 817 WLERLNLAGNDFVTLP-SLRKLSKLVYLNLEHCKLLESLPQLPFPTNTGEVHREYDDYFC 875
L+ +NL+ LP S ++L KL++LNLE LES+ + +V + + C
Sbjct: 654 SLQYINLSTTGIKWLPVSFKELKKLIHLNLEFTDELESIVGIATSLPNLQVLKLFSSRVC 713
Query: 876 GAGLLI 881
G L+
Sbjct: 714 IDGSLM 719
>RGA3_SOLBU (Q7XA40) Putative disease resistance protein RGA3
(RGA1-blb) (Blight resistance protein B149)
Length = 947
Score = 75.5 bits (184), Expect = 7e-13
Identities = 162/696 (23%), Positives = 282/696 (40%), Gaps = 119/696 (17%)
Query: 237 LLLNSV---DGVRAIGICGMGGIGKTTLATTLYG--QISHQFSASCFIDDVTKIYGLHDD 291
+L+N+V + V + I GMGG+GKTTLA ++ +I+ F+ KI+ D
Sbjct: 164 ILINNVSYSEEVPVLPILGMGGLGKTTLAQMVFNDQRITEHFNL--------KIWVCVSD 215
Query: 292 PLDVQKQILFQTLGIEHQQICNRYHATTL--IQRKLCHERTLMILDNV--DQVEQLEKIA 347
D ++ I IE + + + A +Q L +R ++LD+V + E+ + +
Sbjct: 216 DFDEKRLIKAIVESIEGKSLGDMDLAPLQKKLQELLNGKRYFLVLDDVWNEDQEKWDNLR 275
Query: 348 VHREWLGPGSRIIIISRDEHVLKAYGVDVVYKVSLLDWNEAHMLFCRKAFKDEKIIMSNY 407
+ G+ I+I +R E + G +Y++S L + +LF ++AF +
Sbjct: 276 AVLKIGASGASILITTRLEKIGSIMGTLQLYQLSNLSQEDCWLLFKQRAFCHQTETSPKL 335
Query: 408 QNLVDQILHYAKGLPLAIKVLGSFL-FGRNVTEWK----SALTRLRQSPVKDVMDVLQLS 462
+ +I+ G+PLA K LG L F R +EW+ S + L Q V+ L+LS
Sbjct: 336 MEIGKEIVKKCGGVPLAAKTLGGLLRFKREESEWEHVRDSEIWNLPQDE-NSVLPALRLS 394
Query: 463 FDGLNETEKDIFLHIACFFNNDSEEDVKNILNCCGFHA-----------DIG-------- 503
+ L + F + A F D++ + + ++ H+ D+G
Sbjct: 395 YHHLPLDLRQCFAYCA-VFPKDTKIEKEYLIALWMAHSFLLSKGNMELEDVGNEVWNELY 453
Query: 504 LRVLIDKSLVSISYSIINMHSLLEELGRKIVQNSSSKEPRKWSRLWSTEQLYDVMLENME 563
LR + V + MH L+ +L + S+S + + E D+M
Sbjct: 454 LRSFFQEIEVKSGKTYFKMHDLIHDLATSMFSASASSRSIRQINVKDDE---DMMF---- 506
Query: 564 KHVEAIVLYYKEDEEADFEHLSKMSNLRLLFIANYISTMLGFPSCLSNKLRFVHWFRYPS 623
IV YK+ F + +S+ ++S LR ++
Sbjct: 507 -----IVTNYKDMMSIGFSEV--VSSYSPSLFKRFVS------------LRVLNLSNSEF 547
Query: 624 KYLPSNFHPNELVE---LILTESNIKQLWKNKKYLPNLRTLDLRHSRNLEKI-IDFGEFP 679
+ LPS+ +LV L L+ + I L K L NL+TLDL + ++L + +
Sbjct: 548 EQLPSSV--GDLVHLRYLDLSGNKICSLPKRLCKLQNLQTLDLYNCQSLSCLPKQTSKLC 605
Query: 680 NLERLDLEGCINLVELDPSIGLLRKLVYLNLKDCKSLVSIPNNIFGLSSLQYLNMCGCSK 739
+L L L+ C L + P IGLL LK V + L L+ LN+ G
Sbjct: 606 SLRNLVLDHC-PLTSMPPRIGLL-----TCLKTLGYFVVGERKGYQLGELRNLNLRGAIS 659
Query: 740 V-----FNNPRRLMKSGISSE----------------KKQQHDIRESASHHLPGLKWIIL 778
+ N ++ +S++ + ++ + E+ H P LK++ +
Sbjct: 660 ITHLERVKNDMEAKEANLSAKANLHSLSMSWDRPNRYESEEVKVLEALKPH-PNLKYLEI 718
Query: 779 AHDSSHMLPSLHSLCCLRKVD---ISFC-YLSHVP--DAIECLHWLERLN-------LAG 825
LP + L+ V IS C S +P + CL LE + +
Sbjct: 719 IDFCGFCLPDWMNHSVLKNVVSILISGCENCSCLPPFGELPCLESLELQDGSVEVEYVED 778
Query: 826 NDFVT---LPSLRKLSKLVYLNLEHCKLLESLPQLP 858
+ F+T PSLRKL + NL+ + ++ Q P
Sbjct: 779 SGFLTRRRFPSLRKLHIGGFCNLKGLQRMKGAEQFP 814
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.322 0.138 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 129,453,043
Number of Sequences: 164201
Number of extensions: 5602456
Number of successful extensions: 14988
Number of sequences better than 10.0: 177
Number of HSP's better than 10.0 without gapping: 52
Number of HSP's successfully gapped in prelim test: 125
Number of HSP's that attempted gapping in prelim test: 14341
Number of HSP's gapped (non-prelim): 528
length of query: 1085
length of database: 59,974,054
effective HSP length: 121
effective length of query: 964
effective length of database: 40,105,733
effective search space: 38661926612
effective search space used: 38661926612
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 71 (32.0 bits)
Medicago: description of AC122722.3


