

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122169.1 + phase: 0 /pseudo
(459 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
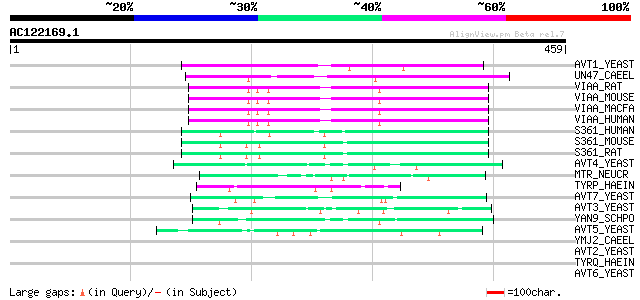
Score E
Sequences producing significant alignments: (bits) Value
AVT1_YEAST (P47082) Vacuolar amino acid transporter 1 125 3e-28
UN47_CAEEL (P34579) Vesicular GABA transporter (Uncoordinated pr... 87 1e-16
VIAA_RAT (O35458) Vesicular inhibitory amino acid transporter (G... 76 2e-13
VIAA_MOUSE (O35633) Vesicular inhibitory amino acid transporter ... 76 2e-13
VIAA_MACFA (Q95KE2) Vesicular inhibitory amino acid transporter ... 76 2e-13
VIAA_HUMAN (Q9H598) Vesicular inhibitory amino acid transporter ... 76 2e-13
S361_HUMAN (Q7Z2H8) Proton-coupled amino acid transporter 1 (Pro... 67 1e-10
S361_MOUSE (Q8K4D3) Proton-coupled amino acid transporter 1 (Pro... 65 5e-10
S361_RAT (Q924A5) Proton-coupled amino acid transporter 1 (Proto... 64 1e-09
AVT4_YEAST (P50944) Vacuolar amino acid transporter 4 59 3e-08
MTR_NEUCR (P38680) N amino acid transport system protein (Methyl... 47 8e-05
TYRP_HAEIN (P44727) Tyrosine-specific transport protein 1 (Tyros... 46 2e-04
AVT7_YEAST (P40501) Vacuolar amino acid transporter 7 46 2e-04
AVT3_YEAST (P36062) Vacuolar amino acid transporter 3 45 3e-04
YAN9_SCHPO (Q10074) Hypothetical protein C3H1.09c in chromosome I 45 4e-04
AVT5_YEAST (P38176) Vacuolar amino acid transporter 5 44 7e-04
YMJ2_CAEEL (P34479) Hypothetical protein F59B2.2 in chromosome III 43 0.002
AVT2_YEAST (P39981) Vacuolar amino acid transporter 2 43 0.002
TYRQ_HAEIN (P44747) Tyrosine-specific transport protein 2 (Tyros... 42 0.003
AVT6_YEAST (P40074) Vacuolar amino acid transporter 6 41 0.006
>AVT1_YEAST (P47082) Vacuolar amino acid transporter 1
Length = 602
Score = 125 bits (313), Expect = 3e-28
Identities = 83/257 (32%), Positives = 120/257 (46%), Gaps = 17/257 (6%)
Query: 143 TFTQTIFNGLNVLAGVGLLSAPDTVKQAGWA-SLLVIVVFAVVCFYTAELMRHCFQSREG 201
T QTIFN +NVL G+GLL+ P +K AGW L ++ +FA+ F TAEL+ C +
Sbjct: 209 TAPQTIFNSINVLIGIGLLALPLGLKYAGWVIGLTMLAIFALATFCTAELLSRCLDTDPT 268
Query: 202 IISYPDIGEAAFGKYGRVFISIVLYTELYSYCVEFIIMEGDNLSGLFPGTSLHWGSLNLD 261
+ISY D+G AAFG GR IS + +L V +I+ GD+L+ LFP S +
Sbjct: 269 LISYADLGYAAFGTKGRALISALFTLDLLGSGVSLVILFGDSLNALFPQYSTTF------ 322
Query: 262 GKHLFAILAALIILPTVW--LKDLRFVSYLSAGGVVGTALVGACVYAVGTRKDVGFHHTA 319
F I++ I+ P V+ L L +S L GT LV C + +
Sbjct: 323 ----FKIVSFFIVTPPVFIPLSVLSNISLLGILSTTGTVLVICCCGLYKSSSPGSLVNPM 378
Query: 320 PLVNW----SGVPFAFGIYGFCFAGHSVFPNIYQSMANKKDFTKAMLICFVLPVFLYGSV 375
W + + G+ C+ GH+VFPN+ M + F + + +
Sbjct: 379 ETSMWPIDLKHLCLSIGLLSACWGGHAVFPNLKTDMRHPDKFKDCLKTTYKITSVTDIGT 438
Query: 376 GAAGFLMFGERTSSQIT 392
GFLMFG +IT
Sbjct: 439 AVIGFLMFGNLVKDEIT 455
>UN47_CAEEL (P34579) Vesicular GABA transporter (Uncoordinated
protein 47) (Protein unc-47)
Length = 486
Score = 86.7 bits (213), Expect = 1e-16
Identities = 68/275 (24%), Positives = 113/275 (40%), Gaps = 21/275 (7%)
Query: 146 QTIFNGLNVLAGVGLLSAPDTVKQAGWASLLVIVVFAVVCFYTAELMRHCF-----QSRE 200
Q +N N + G+ ++ P VK GW S+ +V A VC++T L+ C + R+
Sbjct: 93 QAAWNVTNAIQGMFIVGLPIAVKVGGWWSIGAMVGVAYVCYWTGVLLIECLYENGVKKRK 152
Query: 201 GIISYPDIGEAAFGKYGRVFISIVLYTELYSYCVEFIIMEGDNLSGLFPGTSLHWGSLNL 260
D + FGK+ + TEL S C+ ++++ D L FP
Sbjct: 153 TYREIADFYKPGFGKW----VLAAQLTELLSTCIIYLVLAADLLQSCFPSVD-------- 200
Query: 261 DGKHLFAILAALIILPTVWLKDLRFVSYLSAGGVVGTALVG--ACVYAVGTRKDVGFHHT 318
K + ++ + +L +L DL+ VS LS + +V +Y + F
Sbjct: 201 --KAGWMMITSASLLTCSFLDDLQIVSRLSFFNAISHLIVNLIMVLYCLSFVSQWSFSTI 258
Query: 319 APLVNWSGVPFAFGIYGFCFAGHSVFPNIYQSMANKKDFTKAMLICFVLPVFLYGSVGAA 378
+N + +P G+ F + H PN+ +M N F + + G
Sbjct: 259 TFSLNINTLPTIVGMVVFGYTSHIFLPNLEGNMKNPAQFNVMLKWSHIAAAVFKVVFGML 318
Query: 379 GFLMFGERTSSQITLDLPRDAFASKVSLWTITHML 413
GFL FGE T +I+ LP +F V+L + L
Sbjct: 319 GFLTFGELTQEEISNSLPNQSFKILVNLILVVKAL 353
>VIAA_RAT (O35458) Vesicular inhibitory amino acid transporter (GABA
and glycine transporter) (Vesicular GABA transporter)
(rGVAT)
Length = 525
Score = 76.3 bits (186), Expect = 2e-13
Identities = 59/261 (22%), Positives = 112/261 (42%), Gaps = 21/261 (8%)
Query: 149 FNGLNVLAGVGLLSAPDTVKQAGWASLLVIVVFAVVCFYTAELMRHCF--QSREGII--- 203
+N N + G+ +L P + G+ L +I+ AVVC YT +++ C ++ +G +
Sbjct: 123 WNVTNAIQGMFVLGLPYAILHGGYLGLFLIIFAAVVCCYTGKILIACLYEENEDGEVVRV 182
Query: 204 --SYPDIGEAA----FGKYGRVFISIVLYTELYSYCVEFIIMEGDNLSGLFPGTSLHWGS 257
SY I A F G +++ EL C+ ++++ G+ + FPG + S
Sbjct: 183 RDSYVAIANACCAPRFPTLGGRVVNVAQIIELVMTCILYVVVSGNLMYNSFPGLPVSQKS 242
Query: 258 LNLDGKHLFAILAALIILPTVWLKDLRFVSYLSAGGVVGTALVGACV--YAVGTRKDVGF 315
++I+A ++LP +LK+L+ VS S + ++ V Y + +D +
Sbjct: 243 --------WSIIATAVLLPCAFLKNLKAVSKFSLLCTLAHFVINILVIAYCLSRARDWAW 294
Query: 316 HHTAPLVNWSGVPFAFGIYGFCFAGHSVFPNIYQSMANKKDFTKAMLICFVLPVFLYGSV 375
++ P + GI F + P++ +M +F M + L G
Sbjct: 295 EKVKFYIDVKKFPISIGIIVFSYTSQIFLPSLEGNMQQPSEFHCMMNWTHIAACVLKGLF 354
Query: 376 GAAGFLMFGERTSSQITLDLP 396
+L + + T IT +LP
Sbjct: 355 ALVAYLTWADETKEVITDNLP 375
>VIAA_MOUSE (O35633) Vesicular inhibitory amino acid transporter
(GABA and glycine transporter) (Vesicular GABA
transporter) (mVIAAT) (mVGAT)
Length = 525
Score = 76.3 bits (186), Expect = 2e-13
Identities = 59/261 (22%), Positives = 112/261 (42%), Gaps = 21/261 (8%)
Query: 149 FNGLNVLAGVGLLSAPDTVKQAGWASLLVIVVFAVVCFYTAELMRHCF--QSREGII--- 203
+N N + G+ +L P + G+ L +I+ AVVC YT +++ C ++ +G +
Sbjct: 123 WNVTNAIQGMFVLGLPYAILHGGYLGLFLIIFAAVVCCYTGKILIACLYEENEDGEVVRV 182
Query: 204 --SYPDIGEAA----FGKYGRVFISIVLYTELYSYCVEFIIMEGDNLSGLFPGTSLHWGS 257
SY I A F G +++ EL C+ ++++ G+ + FPG + S
Sbjct: 183 RDSYVAIANACCAPRFPTLGGRVVNVAQIIELVMTCILYVVVSGNLMYNSFPGLPVSQKS 242
Query: 258 LNLDGKHLFAILAALIILPTVWLKDLRFVSYLSAGGVVGTALVGACV--YAVGTRKDVGF 315
++I+A ++LP +LK+L+ VS S + ++ V Y + +D +
Sbjct: 243 --------WSIIATAVLLPCAFLKNLKAVSKFSLLCTLAHFVINILVIAYCLSRARDWAW 294
Query: 316 HHTAPLVNWSGVPFAFGIYGFCFAGHSVFPNIYQSMANKKDFTKAMLICFVLPVFLYGSV 375
++ P + GI F + P++ +M +F M + L G
Sbjct: 295 EKVKFYIDVKKFPISIGIIVFSYTSQIFLPSLEGNMQQPSEFHCMMNWTHIAACVLKGLF 354
Query: 376 GAAGFLMFGERTSSQITLDLP 396
+L + + T IT +LP
Sbjct: 355 ALVAYLTWADETKEVITDNLP 375
>VIAA_MACFA (Q95KE2) Vesicular inhibitory amino acid transporter
(GABA and glycine transporter) (Vesicular GABA
transporter) (QccE-21148)
Length = 525
Score = 76.3 bits (186), Expect = 2e-13
Identities = 59/261 (22%), Positives = 112/261 (42%), Gaps = 21/261 (8%)
Query: 149 FNGLNVLAGVGLLSAPDTVKQAGWASLLVIVVFAVVCFYTAELMRHCF--QSREGII--- 203
+N N + G+ +L P + G+ L +I+ AVVC YT +++ C ++ +G +
Sbjct: 123 WNVTNAIQGMFVLGLPYAILHGGYLGLFLIIFAAVVCCYTGKILIACLYEENEDGEVVRV 182
Query: 204 --SYPDIGEAA----FGKYGRVFISIVLYTELYSYCVEFIIMEGDNLSGLFPGTSLHWGS 257
SY I A F G +++ EL C+ ++++ G+ + FPG + S
Sbjct: 183 RDSYVAIANACCAPRFPTLGGRVVNVAQIIELVMTCILYVVVSGNLMYNSFPGLPVSQKS 242
Query: 258 LNLDGKHLFAILAALIILPTVWLKDLRFVSYLSAGGVVGTALVGACV--YAVGTRKDVGF 315
++I+A ++LP +LK+L+ VS S + ++ V Y + +D +
Sbjct: 243 --------WSIIATAVLLPCAFLKNLKAVSKFSLLCTLAHFVINILVIAYCLSRARDWAW 294
Query: 316 HHTAPLVNWSGVPFAFGIYGFCFAGHSVFPNIYQSMANKKDFTKAMLICFVLPVFLYGSV 375
++ P + GI F + P++ +M +F M + L G
Sbjct: 295 EKVKFYIDVKKFPISIGIIVFSYTSQIFLPSLEGNMQQPSEFHCMMNWTHIAACVLKGLF 354
Query: 376 GAAGFLMFGERTSSQITLDLP 396
+L + + T IT +LP
Sbjct: 355 ALVAYLTWADETKEVITDNLP 375
>VIAA_HUMAN (Q9H598) Vesicular inhibitory amino acid transporter
(GABA and glycine transporter) (Vesicular GABA
transporter) (hVIAAT)
Length = 525
Score = 76.3 bits (186), Expect = 2e-13
Identities = 59/261 (22%), Positives = 112/261 (42%), Gaps = 21/261 (8%)
Query: 149 FNGLNVLAGVGLLSAPDTVKQAGWASLLVIVVFAVVCFYTAELMRHCF--QSREGII--- 203
+N N + G+ +L P + G+ L +I+ AVVC YT +++ C ++ +G +
Sbjct: 123 WNVTNAIQGMFVLGLPYAILHGGYLGLFLIIFAAVVCCYTGKILIACLYEENEDGEVVRV 182
Query: 204 --SYPDIGEAA----FGKYGRVFISIVLYTELYSYCVEFIIMEGDNLSGLFPGTSLHWGS 257
SY I A F G +++ EL C+ ++++ G+ + FPG + S
Sbjct: 183 RDSYVAIANACCAPRFPTLGGRVVNVAQIIELVMTCILYVVVSGNLMYNSFPGLPVSQKS 242
Query: 258 LNLDGKHLFAILAALIILPTVWLKDLRFVSYLSAGGVVGTALVGACV--YAVGTRKDVGF 315
++I+A ++LP +LK+L+ VS S + ++ V Y + +D +
Sbjct: 243 --------WSIIATAVLLPCAFLKNLKAVSKFSLLCTLAHFVINILVIAYCLSRARDWAW 294
Query: 316 HHTAPLVNWSGVPFAFGIYGFCFAGHSVFPNIYQSMANKKDFTKAMLICFVLPVFLYGSV 375
++ P + GI F + P++ +M +F M + L G
Sbjct: 295 EKVKFYIDVKKFPISIGIIVFSYTSQIFLPSLEGNMQQPSEFHCMMNWTHIAACVLKGLF 354
Query: 376 GAAGFLMFGERTSSQITLDLP 396
+L + + T IT +LP
Sbjct: 355 ALVAYLTWADETKEVITDNLP 375
>S361_HUMAN (Q7Z2H8) Proton-coupled amino acid transporter 1
(Proton/amino acid transporter 1) (Solute carrier family
36 member 1)
Length = 476
Score = 66.6 bits (161), Expect = 1e-10
Identities = 68/286 (23%), Positives = 111/286 (38%), Gaps = 38/286 (13%)
Query: 143 TFTQTIFNGLNVLAGVGLLSAPDTVKQAGWA----SLLVIVVFAVVCFYTAELMRHCFQS 198
T+ QT+ + L G GLL P VK AG SLL+I + AV C H F
Sbjct: 48 TWFQTLIHLLKGNIGTGLLGLPLAVKNAGIVMGPISLLIIGIVAVHCMGILVKCAHHFCR 107
Query: 199 REGIISYPDIGEAAF--------------GKYGRVFISIVLYTELYSYCVEFIIMEGDNL 244
R S+ D G+ +GR + L +C + + DN
Sbjct: 108 RLNK-SFVDYGDTVMYGLESSPCSWLRNHAHWGRRVVDFFLIVTQLGFCCVYFVFLADNF 166
Query: 245 SGLFPGTSLHWGSLN-------------LDGK-HLFAILAALIILPTVWLKDLRFVSYLS 290
+ + G+ N +D + ++ + L L++L V++++LR +S S
Sbjct: 167 KQVIEAAN---GTTNNCHNNETVILTPTMDSRLYMLSFLPFLVLL--VFIRNLRALSIFS 221
Query: 291 AGGVVGTALVGACVYAVGTRKDVGFHHTAPLVNWSGVPFAFGIYGFCFAGHSVFPNIYQS 350
+ + +Y ++ H + W P FG F F G + +
Sbjct: 222 LLANITMLVSLVMIYQFIVQRIPDPSHLPLVAPWKTYPLFFGTAIFSFEGIGMVLPLENK 281
Query: 351 MANKKDFTKAMLICFVLPVFLYGSVGAAGFLMFGERTSSQITLDLP 396
M + + F + + V+ LY S+G G+L FG ITL+LP
Sbjct: 282 MKDPRKFPLILYLGMVIVTILYISLGCLGYLQFGANIQGSITLNLP 327
>S361_MOUSE (Q8K4D3) Proton-coupled amino acid transporter 1
(Proton/amino acid transporter 1) (Solute carrier family
36 member 1)
Length = 475
Score = 64.7 bits (156), Expect = 5e-10
Identities = 65/282 (23%), Positives = 108/282 (38%), Gaps = 30/282 (10%)
Query: 143 TFTQTIFNGLNVLAGVGLLSAPDTVKQAGWA----SLLVIVVFAVVCFYTAELMRH--CF 196
T+ QT+ + L G GLL P VK AG SLLVI + AV C H C
Sbjct: 47 TWFQTLIHLLKGNIGTGLLGLPLAVKNAGLLLGPLSLLVIGIVAVHCMGILVKCAHHLCR 106
Query: 197 QSREGIISY-----------PDIGEAAFGKYGRVFISIVLYTELYSYCVEFIIMEGDNLS 245
+ + + Y P +GR + L +C + + DN
Sbjct: 107 RLNKPFLDYGDTVMYGLECSPSTWVRNHSHWGRRIVDFFLIVTQLGFCCVYFVFLADNFK 166
Query: 246 GLFPGTSLHWGSLN----------LDGK-HLFAILAALIILPTVWLKDLRFVSYLSAGGV 294
+ + + N +D + ++ + L L++L ++++LR +S S
Sbjct: 167 QVIEAANGTTTNCNNNVTVIPTPTMDSRLYMLSFLPFLVLLS--FIRNLRVLSIFSLLAN 224
Query: 295 VGTALVGACVYAVGTRKDVGFHHTAPLVNWSGVPFAFGIYGFCFAGHSVFPNIYQSMANK 354
+ + +Y ++ H + W P FG F F G V + M +
Sbjct: 225 ISMFVSLIMIYQFIVQRIPDPSHLPLVAPWKTYPLFFGTAIFAFEGIGVVLPLENKMKDS 284
Query: 355 KDFTKAMLICFVLPVFLYGSVGAAGFLMFGERTSSQITLDLP 396
+ F + + + LY S+G+ G+L FG ITL+LP
Sbjct: 285 QKFPLILYLGMAIITVLYISLGSLGYLQFGANIKGSITLNLP 326
>S361_RAT (Q924A5) Proton-coupled amino acid transporter 1
(Proton/amino acid transporter 1) (Solute carrier family
36 member 1) (Lysosomal amino acid transporter 1)
(LYAAT-1) (Neutral amino acid/proton symporter)
Length = 475
Score = 63.5 bits (153), Expect = 1e-09
Identities = 65/282 (23%), Positives = 107/282 (37%), Gaps = 30/282 (10%)
Query: 143 TFTQTIFNGLNVLAGVGLLSAPDTVKQAGWA----SLLVIVVFAVVCFYTAELMRH--CF 196
T+ QT+ + L G GLL P VK AG SLLVI + AV C H C
Sbjct: 47 TWFQTLIHLLKGNIGTGLLGLPLAVKNAGLLLGPLSLLVIGIVAVHCMGILVKCAHHLCR 106
Query: 197 QSREGIISY-----------PDIGEAAFGKYGRVFISIVLYTELYSYCVEFIIMEGDNLS 245
+ + + Y P +GR + L +C + + DN
Sbjct: 107 RLNKPFLDYGDTVMYGLECSPSTWIRNHSHWGRRIVDFFLVVTQLGFCCVYFVFLADNFK 166
Query: 246 GLFPGTSLHWGSLN----------LDGK-HLFAILAALIILPTVWLKDLRFVSYLSAGGV 294
+ + + N +D + ++ L L++L ++++LR +S S
Sbjct: 167 QVIEAANGTTTNCNNNETVILTPTMDSRLYMLTFLPFLVLLS--FIRNLRILSIFSLLAN 224
Query: 295 VGTALVGACVYAVGTRKDVGFHHTAPLVNWSGVPFAFGIYGFCFAGHSVFPNIYQSMANK 354
+ + +Y ++ H + W P FG F F G V + M +
Sbjct: 225 ISMFVSLIMIYQFIVQRIPDPSHLPLVAPWKTYPLFFGTAIFAFEGIGVVLPLENKMKDS 284
Query: 355 KDFTKAMLICFVLPVFLYGSVGAAGFLMFGERTSSQITLDLP 396
+ F + + + LY S+G+ G+L FG ITL+LP
Sbjct: 285 QKFPLILYLGMAIITVLYISLGSLGYLQFGADIKGSITLNLP 326
>AVT4_YEAST (P50944) Vacuolar amino acid transporter 4
Length = 713
Score = 58.5 bits (140), Expect = 3e-08
Identities = 65/291 (22%), Positives = 117/291 (39%), Gaps = 34/291 (11%)
Query: 136 LPIGYGCTFTQTIFNGL-NVLAGVGLLSAPDTVKQAG-WASLLVIVVFAVVCFYTAELMR 193
LP G T T+ +F L G G+L P+ G + S+ ++ F + ++ ++
Sbjct: 290 LPSAKGTTSTKKVFLILLKSFIGTGVLFLPNAFHNGGLFFSVSMLAFFGIYSYWCYYILV 349
Query: 194 HCFQSREGIISYPDIGEAAFGKYGRVFISIVLYTELYSYCVEFIIMEGDNLSGLFPGTSL 253
+S G+ S+ DIG +G + R+ I L + ++I NL F
Sbjct: 350 QA-KSSCGVSSFGDIGLKLYGPWMRIIILFSLVITQVGFSGAYMIFTAKNLQA-FLDNVF 407
Query: 254 HWGSLNLDGKHLFAILAALIILPTVWLKDLRFVSYLSAGGVVGTALV--GACVYAVGTRK 311
H G L L + +I +P L +R +S LS ++ + G + + T K
Sbjct: 408 HVGVLPLS---YLMVFQTIIFIP---LSFIRNISKLSLPSLLANFFIMAGLVIVIIFTAK 461
Query: 312 DVGFHHTAPLVNWSGVPFAFGIYG--------------FCFAGHSVFPNIYQSMANKKDF 357
+ F + G P +YG F F G + + SM N + F
Sbjct: 462 RLFF-------DLMGTPAMGVVYGLNADRWTLFIGTAIFAFEGIGLIIPVQDSMRNPEKF 514
Query: 358 TKAMLICFVLPVFLYGSVGAAGFLMFGERTSSQITLDLPR-DAFASKVSLW 407
+ + + L+ S+ G+L +G + I L+LP+ + F + + L+
Sbjct: 515 PLVLALVILTATILFISIATLGYLAYGSNVQTVILLNLPQSNIFVNLIQLF 565
>MTR_NEUCR (P38680) N amino acid transport system protein
(Methyltryptophan resistance protein)
Length = 470
Score = 47.4 bits (111), Expect = 8e-05
Identities = 50/253 (19%), Positives = 98/253 (37%), Gaps = 30/253 (11%)
Query: 158 VGLLSAPDTVKQAGWASLLVIVV-FAVVCFYTAELMRHCFQSREGIISYPDIGEAAFGKY 216
+G LS P G +++ V ++C YTA ++ I Y D+G FG++
Sbjct: 69 LGSLSLPGAFATLGMVPGVILSVGMGLICIYTAHVIGQTKLKHPEIAHYADVGRVMFGRW 128
Query: 217 GRVFISIVLYTELYSYCVEFIIMEGDNLSGLFPGTSLHWGSLNLDGKH----LFAILAAL 272
G IS + + ++ I + G S + GT + WG++ +G +F I++A+
Sbjct: 129 GYEIIS-------FMFVLQLIFIVG---SHVLTGT-IMWGTITDNGNGTCSLVFGIVSAI 177
Query: 273 II----LPTVWLKDLRFVSYLSAGGVVGTALVGACVYAVGTRKDVGFHHTAPLVNWSGVP 328
I+ +P + ++ + Y+ + L+ + + G P W
Sbjct: 178 ILFLLAIPPSF-AEVAILGYIDFVSICAAILITMIATGIRSSHQEGGLAAVPWSCWPKED 236
Query: 329 FAFGIYGFCFAGHSVFP--------NIYQSMANKKDFTKAMLICFVLPVFLYGSVGAAGF 380
+ GF + VF + M D+ K+++ ++ +F+Y G +
Sbjct: 237 LSLA-EGFIAVSNIVFAYSFAMCQFSFMDEMHTPSDYKKSIVALGLIEIFIYTVTGGVVY 295
Query: 381 LMFGERTSSQITL 393
G S L
Sbjct: 296 AFVGPEVQSPALL 308
>TYRP_HAEIN (P44727) Tyrosine-specific transport protein 1 (Tyrosine
permease 1)
Length = 400
Score = 46.2 bits (108), Expect = 2e-04
Identities = 47/190 (24%), Positives = 86/190 (44%), Gaps = 26/190 (13%)
Query: 155 LAGVGLLSAPDTVKQAGWASLLVIVV--FAVVCFYTAELMRHCFQSREGIISYPDIGEAA 212
+ G G+L+ P T G+ LV+++ +A++ F +A L +Q+ E + E
Sbjct: 15 MIGAGMLAMPLTSAGIGFGFTLVLLLGLWALLTF-SALLFVELYQTAESDAGIGTLAEQY 73
Query: 213 FGKYGRVFISIVLYTELYSYCVEFIIMEGDNLSGLFPGT----------SLHWGSLNLDG 262
FGK GR+ + VL LY+ +I G L L P + ++ +GS + G
Sbjct: 74 FGKTGRIIATAVLIIFLYALIAAYISGGGSLLKDLLPESFGDKVSVLLFTVIFGSFIVIG 133
Query: 263 KH---------LFAILAALIILPTVWLKDLRFVSYLSAGGVVGTALVGACVYAVGTRKDV 313
H F +LAA ++ ++ L +++F + ++ + AL+ + T
Sbjct: 134 THSVDKINRVLFFVMLAAFAVVLSLMLPEIKFDNLMAT--PIDKALIISASPVFFTA--F 189
Query: 314 GFHHTAPLVN 323
GFH + P +N
Sbjct: 190 GFHGSIPSLN 199
>AVT7_YEAST (P40501) Vacuolar amino acid transporter 7
Length = 490
Score = 46.2 bits (108), Expect = 2e-04
Identities = 56/270 (20%), Positives = 108/270 (39%), Gaps = 48/270 (17%)
Query: 150 NGLNVLAGVGLLSAPDTVKQAGWASLLVIVVFAVVC-----FYTAELMRHCFQSREG--- 201
N + + G G L+ P + K G +++ + A V F ++ + R
Sbjct: 13 NLVKTIVGAGTLAIPYSFKSDGVLVGVILTLLAAVTSGLGLFVLSKCSKTLINPRNSSFF 72
Query: 202 ---IISYPDIGEAAFGKYGRVFISIVLYTELYSYCVEFIIMEGDNLSGLFPGTSLHWGSL 258
+++YP + + + + + + ++++ GD GLF G +W
Sbjct: 73 TLCMLTYPTLAP---------IFDLAMIVQCFGVGLSYLVLIGDLFPGLFGGERNYW--- 120
Query: 259 NLDGKHLFAILAALIILPTVWLKDLRFVSYLSAGGVVGTALVGACVYA-----VGT---- 309
I +A+II+P +K L + Y S G+ A + V++ +G
Sbjct: 121 --------IIASAVIIIPLCLVKKLDQLKYSSILGLFALAYISILVFSHFVFELGKGELT 172
Query: 310 ---RKDVGFHHTAPLVNWSGVPFAFGIYGFCFAGH-SVFPNIYQSMANK-KDFTKAMLIC 364
R D+ + + ++ G+ F I F F G ++FP I + N ++ T +
Sbjct: 173 NILRNDICWWK---IHDFKGLLSTFSIIIFAFTGSMNLFPMINELKDNSMENITFVINNS 229
Query: 365 FVLPVFLYGSVGAAGFLMFGERTSSQITLD 394
L L+ VG +G+L FG T + L+
Sbjct: 230 ISLSTALFLIVGLSGYLTFGNETLGNLMLN 259
>AVT3_YEAST (P36062) Vacuolar amino acid transporter 3
Length = 692
Score = 45.4 bits (106), Expect = 3e-04
Identities = 61/266 (22%), Positives = 105/266 (38%), Gaps = 37/266 (13%)
Query: 152 LNVLAGVGLLSAPDTVKQAGWASLLVIVVFAVVCFYTAELMRH-CFQS------REGIIS 204
L G G+L P GW F+ +C + L+ + CF S + G+
Sbjct: 307 LKSFVGTGVLFLPKAFHNGGWG-------FSALCLLSCALISYGCFVSLITTKDKVGVDG 359
Query: 205 YPDIGEAAFGKYGRVFISIVLYTELYSYCVEFIIMEGDNLSGLFPGTSLHW--GSLNLDG 262
Y D+G +G + I + + + + NL +F H GS++L
Sbjct: 360 YGDMGRILYGPKMKFAILSSIALSQIGFSAAYTVFTATNLQ-VFSENFFHLKPGSISL-A 417
Query: 263 KHLFAILAALIILPTVWLKDLRFVS--YLSAGGVVGTALVGACVYAV------GTRKDVG 314
++FA LI +P +++ +S L A + LV VY++ G D
Sbjct: 418 TYIFA--QVLIFVPLSLTRNIAKLSGTALIADLFILLGLVYVYVYSIYYIAVNGVASD-- 473
Query: 315 FHHTAPLVNWSGVPFAFGIYGFCFAGHSVFPNIYQSMANKKDFTKAM--LICFVLPVFLY 372
T + N + G F F G + I +SM + K F ++ ++C V +F+
Sbjct: 474 ---TMLMFNKADWSLFIGTAIFTFEGIGLLIPIQESMKHPKHFRPSLSAVMCIVAVIFI- 529
Query: 373 GSVGAAGFLMFGERTSSQITLDLPRD 398
S G + FG + + L+ P+D
Sbjct: 530 -SCGLLCYAAFGSDVKTVVLLNFPQD 554
>YAN9_SCHPO (Q10074) Hypothetical protein C3H1.09c in chromosome I
Length = 656
Score = 45.1 bits (105), Expect = 4e-04
Identities = 56/259 (21%), Positives = 98/259 (37%), Gaps = 23/259 (8%)
Query: 152 LNVLAGVGLLSAPDTVKQAGW----ASLLVIVVFAVVCFYTAELMRHCFQSREGII-SYP 206
L G G+L P K G A+LL++ V + +CF Q+R + S+
Sbjct: 285 LKSFVGTGVLFLPKAFKLGGLVFSSATLLIVGVLSHICFLLL------IQTRMKVPGSFG 338
Query: 207 DIGEAAFGKYGRVFISIVLYTELYSYCVEFIIMEGDNLSGLFPGTSLHWGSLNLDGKHLF 266
DIG +G + R I + + +I L S +L +F
Sbjct: 339 DIGGTLYGPHMRFAILASIVVSQIGFSSAYISFVASTLQACVKVISTTHREYHLA---VF 395
Query: 267 AILAALIILPTVWLKDLRFVSYLSAGGVVGTALVGACV-----YAVGTRKDVGFHHTAPL 321
+ L+ +P L +R +S LSA ++ + + + V T G A +
Sbjct: 396 IFIQFLVFVP---LSLVRKISKLSATALIADVFILLGILYLYFWDVITLATKGIADVA-M 451
Query: 322 VNWSGVPFAFGIYGFCFAGHSVFPNIYQSMANKKDFTKAMLICFVLPVFLYGSVGAAGFL 381
N + G+ F + G + I + MA K+ K + L+ S+G +
Sbjct: 452 FNKTDFSLFIGVAIFTYEGICLILPIQEQMAKPKNLPKLLTGVMAAISLLFISIGLLSYA 511
Query: 382 MFGERTSSQITLDLPRDAF 400
FG + + + L++P F
Sbjct: 512 AFGSKVKTVVILNMPESTF 530
>AVT5_YEAST (P38176) Vacuolar amino acid transporter 5
Length = 509
Score = 44.3 bits (103), Expect = 7e-04
Identities = 67/297 (22%), Positives = 112/297 (37%), Gaps = 43/297 (14%)
Query: 122 WWEKASIQKNIPEELPIGYGCTFTQTIFNGLNVLAGVGLLSAPDTVKQAGWASLLVIVVF 181
W K S N+P + G + L+ G G+L+ P K G L+ + F
Sbjct: 41 WKRKVSSFLNMPSNVRSG--------VLTLLHTACGAGVLAMPFAFKPFGLMPGLITLTF 92
Query: 182 AVVCFYTAELMRHCFQSREGIISY-PDIGEAAFGKYGRVF---ISIVLYTELYSYC---- 233
+C L+ Q+R I Y P A+F K ++ IS+V + C
Sbjct: 93 CGICSLCGLLL----QTR--IAKYVPKSENASFAKLTQLINPSISVVFDFAIAVKCFGVG 146
Query: 234 VEFIIMEGDNLSGL-----------FPGTSLHWGSLNLDGKHLFAILAALIILPTVWLKD 282
V ++I+ GD + + G+ H + LD + ++ +I P + +
Sbjct: 147 VSYLIIVGDLVPQIVQSIFYRNDDNMSGSQEH--HMFLDRRLYITLIIVFVISPLCFKRS 204
Query: 283 LRFVSYLSAGGVVGTA-LVGACVYAVGTRKDVGFHHTAPLV----NWSGVPFA-FGIYGF 336
L + Y S +V A L G +Y R + +V + S P I+ F
Sbjct: 205 LNSLRYASMIAIVSVAYLSGLIIYHFVNRHQLERGQVYFMVPHGDSQSHSPLTTLPIFVF 264
Query: 337 CFAGHSVFPNIYQSMANK--KDFTKAMLICFVLPVFLYGSVGAAGFLMFGERTSSQI 391
+ H ++ +K K + + VL FLY +G G++ FGE I
Sbjct: 265 AYTCHHNMFSVINEQVDKSFKVIRRIPIFAIVLAYFLYIIIGGTGYMTFGENIVGNI 321
>YMJ2_CAEEL (P34479) Hypothetical protein F59B2.2 in chromosome III
Length = 460
Score = 42.7 bits (99), Expect = 0.002
Identities = 60/295 (20%), Positives = 108/295 (36%), Gaps = 38/295 (12%)
Query: 143 TFTQTIFNGLNVLAGVGLLSAPDTVKQAG-WASLLVIVVFAVVCFYTAELMRHCFQ---- 197
T ++++FN G S P K G W S ++ V A + +Y ++ Q
Sbjct: 45 TLSKSMFNA-------GCFSLPYAWKLGGLWVSFVMSFVIAGLNWYGNHILVRASQHLAK 97
Query: 198 -SREGIISYP-------DIGEAAF----GKYGRVFISIVLYTELYSYCVEFIIMEGDNLS 245
S + Y D + F K F+++ + C I+ DNL
Sbjct: 98 KSDRSALDYGHFAKKVCDYSDIRFLRNNSKAVMYFVNVTILFYQLGMCSVAILFISDNLV 157
Query: 246 GLFPGTSLHWGSLNLDGKHLFAILAALIILPTVWLKDLRFVSYLSAGGVVGTALVGACVY 305
L H G L A ++ IL T ++R VS+ + V + A +
Sbjct: 158 NLVGD---HLGGTRHQQMILMATVSLFFILLTNMFTEMRIVSFFALVSSVFFVIGAAVIM 214
Query: 306 AVGTRKDVGFHHTAPLVNWSGVPFAFGIYGFCFAGHSVFPNIYQSMANKKDF-------T 358
++ + N++G G+ + F G ++ I + N F +
Sbjct: 215 QYTVQQPNQWDKLPAATNFTGTITMIGMSMYAFEGQTMILPIENKLDNPAAFLAPFGVLS 274
Query: 359 KAMLICFVLPVFLYGSVGAAGFLMFGERTSSQITLDLPRDAFASKVSLWTITHML 413
M+IC L G G+ FG+ + IT ++P++ S V+++ + L
Sbjct: 275 TTMIICTAFMTAL----GFFGYTGFGDSIAPTITTNVPKEGLYSTVNVFLMLQSL 325
>AVT2_YEAST (P39981) Vacuolar amino acid transporter 2
Length = 480
Score = 42.7 bits (99), Expect = 0.002
Identities = 53/257 (20%), Positives = 103/257 (39%), Gaps = 11/257 (4%)
Query: 150 NGLNVLAGVGLLSAPDTVKQAG-WASLLVIVVFAVVCFYTAELMRHCFQSREGIISYPDI 208
N N + G G+++ P +K AG LL V + +T L+ + G +Y
Sbjct: 75 NLANSILGAGIITQPFAIKNAGILGGLLSYVALGFIVDWTLRLIVINL-TLAGKRTYQGT 133
Query: 209 GEAAFGKYGRVFISIVLYTELYSYCVEFIIMEGDNLSGLFPGT-SLHWGSLN--LDGKHL 265
E GK G++ I + C+ + I+ GD + + S + G+++ L +
Sbjct: 134 VEHVMGKKGKLLILFTNGLFAFGGCIGYCIIIGDTIPHVLRAIFSQNDGNVHFWLRRNVI 193
Query: 266 FAILAALIILPTVWLKDLRFVSYLSAGGVVGTALVGACVYAVGTRKDVGFH-HTAPLVNW 324
++ I P +++ +S S V+ ++ V G + H+ L ++
Sbjct: 194 IVMVTTFISFPLSMKRNIEALSKASFLAVISMIIIVLTVVIRGPMLPYDWKGHSLKLSDF 253
Query: 325 ---SGVPFAFGIYGFCFAGHSVFPNIYQSMANKK--DFTKAMLICFVLPVFLYGSVGAAG 379
+ + + + F H I+ SM N+ FT+ I ++ V +G +G
Sbjct: 254 FMKATIFRSLSVISFALVCHHNTSFIFFSMRNRSVAKFTRLTHISIIISVICCALMGYSG 313
Query: 380 FLMFGERTSSQITLDLP 396
F +F E+T + P
Sbjct: 314 FAVFKEKTKGNVLNSFP 330
>TYRQ_HAEIN (P44747) Tyrosine-specific transport protein 2 (Tyrosine
permease 2)
Length = 406
Score = 42.4 bits (98), Expect = 0.003
Identities = 51/219 (23%), Positives = 94/219 (42%), Gaps = 9/219 (4%)
Query: 157 GVGLLSAPDTVKQAGWA-SLLVIVVFAVVCFYTAELMRHCFQSREGIIS-YPDIGEAAFG 214
G G+L+ P T G+ +LL++V + Y+ L +Q+ + + + E FG
Sbjct: 19 GAGMLAMPLTSAGMGFGYTLLLLVGLWALLVYSGLLFVEVYQTADQLDDGVATLAEKYFG 78
Query: 215 KYGRVFISIVLYTELYSYCVEFIIMEGDNLSGLFPGTSLHWGSLNLDGKHLFAILAALII 274
GR+F ++ L LY+ +I G LSGL + SL AI+ ++
Sbjct: 79 VPGRIFATLSLLVLLYALSAAYITGGGSLLSGLPTAFGMEAMSLKT------AIIIFTVV 132
Query: 275 LPTVWLKDLRFVSYLSAGGVVGTALVGACVYAVGTRKDVGFHHTAPLVNWSGVPFAFGIY 334
L + + + V L+ +G + A V + K + A ++++ V A I+
Sbjct: 133 LGSFVVVGTKGVDGLTRVLFIGKLIAFAFVLFMMLPKVATDNLMALPLDYAFVVSAAPIF 192
Query: 335 GFCFAGHSVFPNIYQSMANKKD-FTKAMLICFVLPVFLY 372
F H + ++ + D F +A+LI +P+ Y
Sbjct: 193 LTSFGFHVIMASVNSYLGGSVDKFRRAILIGTAIPLAAY 231
>AVT6_YEAST (P40074) Vacuolar amino acid transporter 6
Length = 448
Score = 41.2 bits (95), Expect = 0.006
Identities = 57/273 (20%), Positives = 110/273 (39%), Gaps = 45/273 (16%)
Query: 152 LNVLAGVGLLSAPDTVKQAGWASLLVIVVFAVVCFYTAELMRHCF-QSREGIISYPDIGE 210
L+ G G+L+ P K G ++++V C M+ F Q+R + Y G
Sbjct: 13 LHTACGAGILAMPYAFKPFGLIPGVIMIVLCGAC-----AMQSLFIQAR--VAKYVPQGR 65
Query: 211 AAFGKYGR-------VFISIVLYTELYSYCVEFIIMEGDNLSGLFPGTSLHWGSLNLDGK 263
A+F R + + + + + V ++I+ GD + + + + LN + +
Sbjct: 66 ASFSALTRLINPNLGIVFDLAIAIKCFGVGVSYMIVVGDLMPQIMSVWTRNAWLLNRNVQ 125
Query: 264 HLFAILAALIILPTVWLKDLRFVSYLSAGGVVGTALVGACVYAVGTRKDVGFHHTAP--- 320
+++ + P +LK L + Y S + A + CV + H+ AP
Sbjct: 126 --ISLIMLFFVAPLSFLKKLNSLRYASMVAISSVAYL--CVLVL-------LHYVAPSDE 174
Query: 321 LVNWSG--------------VPFAFGIYGFCFA-GHSVFPNIYQSMANKKDFT-KAMLIC 364
++ G V I+ F + H++F I + +++ + K LI
Sbjct: 175 ILRLKGRISYLLPPQSHDLNVLNTLPIFVFAYTCHHNMFSIINEQRSSRFEHVMKIPLIA 234
Query: 365 FVLPVFLYGSVGAAGFLMFGERTSSQITLDLPR 397
L + LY ++G AG+L FG+ I + P+
Sbjct: 235 ISLALILYIAIGCAGYLTFGDNIIGNIIMLYPQ 267
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.327 0.141 0.449
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 54,557,611
Number of Sequences: 164201
Number of extensions: 2370303
Number of successful extensions: 12990
Number of sequences better than 10.0: 140
Number of HSP's better than 10.0 without gapping: 70
Number of HSP's successfully gapped in prelim test: 74
Number of HSP's that attempted gapping in prelim test: 11079
Number of HSP's gapped (non-prelim): 960
length of query: 459
length of database: 59,974,054
effective HSP length: 114
effective length of query: 345
effective length of database: 41,255,140
effective search space: 14233023300
effective search space used: 14233023300
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 67 (30.4 bits)
Medicago: description of AC122169.1


