

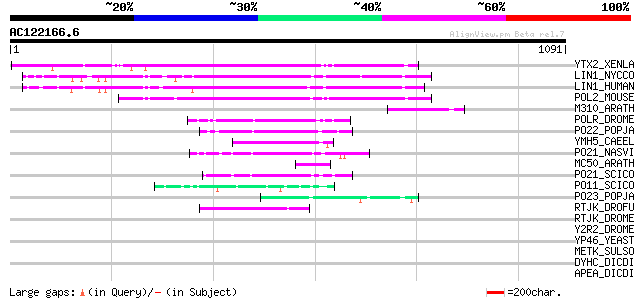
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122166.6 - phase: 0 /pseudo
(1091 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein ... 158 9e-38
LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog 150 2e-35
LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog 145 6e-34
POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains... 135 6e-31
M310_ARATH (P93295) Hypothetical mitochondrial protein AtMg00310... 87 2e-16
POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type... 77 3e-13
PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type... 76 6e-13
YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III 68 2e-10
PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type... 66 4e-10
MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250... 58 1e-07
PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type... 54 2e-06
PO11_SCICO (Q03277) Retrovirus-related Pol polyprotein from type... 54 3e-06
PO23_POPJA (Q05118) Retrovirus-related Pol polyprotein from type... 52 7e-06
RTJK_DROFU (P21329) RNA-directed DNA polymerase from mobile elem... 46 6e-04
RTJK_DROME (P21328) RNA-directed DNA polymerase from mobile elem... 41 0.015
Y2R2_DROME (P16425) Hypothetical 115 kDa protein in type I retro... 34 2.5
YP46_YEAST (Q12080) Hypothetical protein YPL146C 33 3.2
METK_SULSO (Q980S9) S-adenosylmethionine synthetase (EC 2.5.1.6)... 33 4.2
DYHC_DICDI (P34036) Dynein heavy chain, cytosolic (DYHC) 32 7.1
APEA_DICDI (P51173) DNA-(apurinic or apyrimidinic site) lyase (E... 32 9.3
>YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein
(ORF 2)
Length = 1308
Score = 158 bits (399), Expect = 9e-38
Identities = 192/829 (23%), Positives = 353/829 (42%), Gaps = 57/829 (6%)
Query: 4 SRGMVTIW-DRVEVEVWSTVRGVHFLLIHGRFLKSNEEFYIFNMYAPCDRRAKQVLWNSL 62
S G+VT++ D + EV S + L+H R +S + + N+YAP + + SL
Sbjct: 66 SCGVVTLFSDSFQPEVLSATSVIPGRLLHLRVRESGRTYNLMNVYAPTTGPERARFFESL 125
Query: 63 SSRLLSLGSSKVCVCG-DFNTV-----RSVDERRSVRGSQVVDDCAPFNDFIEDRVLINL 116
S+ + ++ S + + G DFN R+V ++R S + + A F+ ++ N
Sbjct: 126 SAYMETIDSDEALIIGGDFNYTLDARDRNVPKKRDSSESVLRELIAHFS-LVDVWREQNP 184
Query: 117 PLCGRKFTWYNGDGRSMSTLDRFLLSEEWCMVWPTCLQVAQLRGLSDHCPIVLTVDEKNW 176
+ S S +DR +S ++ +L SDH + L +
Sbjct: 185 ETVAFTYVRVRDGHVSQSRIDRIYISSH--LMSRAQSSTIRLAPFSDHNCVSLRMSIAPS 242
Query: 177 GPRPV--RLLKCWQDMPGYSDFVREKWGSFQVSGWGGFVLKEKLKLMKAALKEWHVANTL 234
P+ + G++ VR+ W GW F +++ A L +W +
Sbjct: 243 LPKAAYWHFNNSLLEDEGFAKSVRDTW-----RGWRAF--QDEF----ATLNQWWDVGKV 291
Query: 235 NLP------SKIVSLKNRRAVLDSKGEEEELSEEELG--------ELHGISSDIHSLSRL 280
+L +K VS + + GE +L + G E + ++ +
Sbjct: 292 HLKLLCQEYTKSVSGQRNAEIEALNGEVLDLEQRLSGSEDQALQCEYLERKEALRNMEQR 351
Query: 281 NTSICWQQSRLNWLRDGDANSKFFHSVLAARRRINSLSSILV-DGVVVEGVQPVRQAVFT 339
+ +SR+ L D D S+FF+++ + ++ + DG +E + +R +
Sbjct: 352 QARGAFVRSRMQLLCDMDRGSRFFYALEKKKGNRKQITCLFAEDGTPLEDPEAIRDRARS 411
Query: 340 HSKNHFLAPIMTRPSVGHLQ--FRKLSSVERGGLIKPFHEEEIKAAVWDCDSYKIPGPDG 397
+N F ++ + L +S + L P +E+ A+ K PG DG
Sbjct: 412 FYQNLFSPDPISPDACEELWDGLPVVSERRKERLETPITLDELSQALRLMPHNKSPGLDG 471
Query: 398 INLGFIKSFWHEMKAEIVRFVTEFHRNRKLSKGINSRFIALIPKVASPQSLNEFRPISLV 457
+ + F + FW + + R +TE + +L ++L+PK + + +RP+SL+
Sbjct: 472 LTIEFFQFFWDTLGPDFHRVLTEAFKKGELPLSCRRAVLSLLPKKGDLRLIKNWRPVSLL 531
Query: 458 GSLYKILAKILANRLRQVIGSVVSEVQSAFVKNRQILDGILITNEVVDEARKSKKDLLLF 517
+ YKI+AK ++ RL+ V+ V+ QS V R I D + + +++ AR++ L
Sbjct: 532 STDYKIVAKAISLRLKSVLAEVIHPDQSYTVPGRTIFDNVFLIRDLLHFARRTGLSLAFL 591
Query: 518 KVDFEKAYDSVDWGYLDEVMGSMTFPTVW*KWIKECVGTATASVLVNGCPTDEFFLERGL 577
+D EKA+D VD YL + + +F + ++K +A V +N T RG+
Sbjct: 592 SLDQEKAFDRVDHQYLIGTLQAYSFGPQFVGYLKTMYASAECLVKINWSLTAPLAFGRGV 651
Query: 578 RQGDPLSPFLFLLAAEGLNVLMKALVDAGLFKGYSVGRADPVVVSHLQFADDTLLIGNKS 637
RQG PLS L+ LA E L++ + + K + VV+S +ADD +L+ +
Sbjct: 652 RQGCPLSGQLYSLAIEPFLCLLRKRLTGLVLKEPDM----RVVLS--AYADDVILVA-QD 704
Query: 638 WANVRALRAGLILLEAMSGLKVNFHKSS-LVGVNITGSWLSEAASVLGCKVGKIPFLYLG 696
++ + + A S ++N+ KSS L+ ++ +L A + + I +L +
Sbjct: 705 LVDLERAQECQEVYAAASSARINWSKSSGLLEGSLKVDFLPPAFRDISWESKIIKYLGVY 764
Query: 697 LSIGGDPRRLLFWEPVVNRIKNRLSGWQ--SRFLSFGGRLVLLKFVLTALPVYALSFFKA 754
LS P F E + + RL W+ ++ LS GR +++ ++ + Y L
Sbjct: 765 LSAEEYPVSQNFIE-LEECVLTRLGKWKGFAKVLSMRGRALVINQLVASQIWYRLICLSP 823
Query: 755 PSGIISSIESLFNKFFWGGGEDKRKISWIRWDTLSLRKEYGGLGVTRLR 803
I+ I+ F W G W+ SL + GG GV +R
Sbjct: 824 TQEFIAKIQRRLLDFLWIGKH------WVSAGVSSLPLKEGGQGVVCIR 866
>LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog
Length = 1260
Score = 150 bits (378), Expect = 2e-35
Identities = 199/845 (23%), Positives = 356/845 (41%), Gaps = 80/845 (9%)
Query: 26 HFLLIHGRFLKSNEEFYIFNMYAPCDRRAKQVLWNSLSSRLLSLGSSKVCVCGDFNTVRS 85
HF+ + G +E I N+YAP + A Q + +L+ + +L SS V GDFNT +
Sbjct: 95 HFIFVKGN--TQYDEISIINIYAP-NHNAPQFIRETLTD-MSNLISSTSIVVGDFNTPLA 150
Query: 86 VDERRSVRGSQVVDDCAPFNDFIEDRVLINLPLCGR----KFTWYNGDGRSMSTLDRFL- 140
V +R S + ++ + N I+ L ++ ++T+++ + S +D L
Sbjct: 151 VLDRSSKK--KLSKEILDLNSTIQHLDLTDIYRTFHPNKTEYTFFSSAHGTYSKIDHILG 208
Query: 141 ----LSEEWCMVWPTCLQVAQLRGLSDHCPIVLTVDE--------KNWGPRPVRLLKCW- 187
LS+ + C+ SDH I + ++ K W + L W
Sbjct: 209 HKSNLSKFKKIEIIPCI-------FSDHHGIKVELNNNRNLHTHTKTWKLNNLMLKDTWV 261
Query: 188 -----QDMPGYSDFVREKWGSFQVSGW--GGFVLKEKLKLMKAALKEWHVANTLNLPSKI 240
+++ + + + ++Q + W VL+ K ++A LK+ NL +
Sbjct: 262 IDEIKKEITKFLEQNNNQDTNYQ-NLWDTAKAVLRGKFIALQAFLKKTEREEVNNLMGHL 320
Query: 241 VSLKNRRAVLDSKGEEEELSEEELGELHGISSDIHSLSRLNTSICWQQSRLNWLRDGDAN 300
L+ +E+++ EL+ I + + ++N S W ++N + AN
Sbjct: 321 KQLEKEEHSNPKPSRRKEITKIR-AELNEIENK-RIIQQINKSKSWFFEKINKIDKPLAN 378
Query: 301 SKFFHSVLAARRRINSLSSILVDG---------VVVEGVQPVRQAVFTHSKNHFLAPIMT 351
L ++R+ SL S + +G + + + + +++H K L I
Sbjct: 379 -------LTRKKRVKSLISSIRNGNDEITTDPSEIQKILNEYYKKLYSH-KYENLKEIDQ 430
Query: 352 RPSVGHLQFRKLSSVERGGLIKPFHEEEIKAAVWDCDSYKIPGPDGINLGFIKSFWHEMK 411
HL +LS E L +P EI + + + K PGPDG F ++F E+
Sbjct: 431 YLEACHLP--RLSQKEVEMLNRPISSSEIASTIQNLPKKKSPGPDGFTSEFYQTFKEELV 488
Query: 412 AEIVRFVTEFHRNRKLSKGINSRFIALIPKVAS-PQSLNEFRPISLVGSLYKILAKILAN 470
++ + L I LIPK P +RPISL+ KIL KIL N
Sbjct: 489 PILLNLFQNIEKEGILPNTFYEANITLIPKPGKDPTRKENYRPISLMNIDAKILNKILTN 548
Query: 471 RLRQVIGSVVSEVQSAFVKNRQILDGILITNEVVDEARKSK-KDLLLFKVDFEKAYDSVD 529
R++Q I ++ Q F+ Q I + V+ K K KD ++ +D EKA+D++
Sbjct: 549 RIQQHIKKIIHHDQVGFIPGSQGWFNIRKSINVIQHINKLKNKDHMILSIDAEKAFDNIQ 608
Query: 530 WGYLDEVMGSMTFPTVW*KWIKECVGTATASVLVNGCPTDEFFLERGLRQGDPLSPFLFL 589
++ + + + K I+ TA++++NG F L G RQG PLSP LF
Sbjct: 609 HPFMIRTLKKIGIEGTFLKLIEAIYSKPTANIILNGVKLKSFPLRSGTRQGCPLSPLLFN 668
Query: 590 LAAEGLNVLMKALVDAGLFKGYSVGRADPVVVSHLQFADDTLLIGNKSWANVRALRAGLI 649
+ E VL A+ + KG +G + + FADD ++ + + L +
Sbjct: 669 IVME---VLAIAIREEKAIKGIHIGSEE---IKLSLFADDMIVYLENTRDSTTKLLEVIK 722
Query: 650 LLEAMSGLKVNFHKS-SLVGVNITGSWLSEAASVLGCKVGKIPFLYLGLSIGGDPRRLL- 707
+SG K+N HKS + + N + + S+ V K YLG+ + D + L
Sbjct: 723 EYSNVSGYKINTHKSVAFIYTNNNQAEKTVKDSIPFTVVPK-KMKYLGVYLTKDVKDLYK 781
Query: 708 -FWEPVVNRIKNRLSGWQSRFLSFGGRLVLLKFVLTALPVYALSF--FKAPSGIISSIES 764
+E + I ++ W++ S+ GR+ ++K + +Y + KAP +E
Sbjct: 782 ENYETLRKEIAEDVNKWKNIPCSWLGRINIVKMSILPKAIYNFNAIPIKAPLSYFKDLEK 841
Query: 765 LFNKFFWGGGEDKRKISWIRWDTLSLRKEYGGLGVTRLREFNLALLGKWCWRLLLEKEA- 823
+ F W +K I LS + + GG+ + LR + +++ K W +E
Sbjct: 842 IILHFIW-----NQKKPQIAKTLLSNKNKAGGITLPDLRLYYKSIVIKTAWYWHKNREVD 896
Query: 824 LWRKV 828
+W ++
Sbjct: 897 VWNRI 901
>LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog
Length = 1259
Score = 145 bits (366), Expect = 6e-34
Identities = 185/825 (22%), Positives = 340/825 (40%), Gaps = 67/825 (8%)
Query: 26 HFLLIHGRFLKSNEEFYIFNMYAP---CDRRAKQVLWNSLSSRLLSLGSSKVCVCGDFNT 82
H++++ G EE I N+YAP R KQVL S L S + GDFNT
Sbjct: 95 HYIMVKGSI--QQEELTILNIYAPNTGAPRFIKQVL-----SDLQRDLDSHTIIMGDFNT 147
Query: 83 VRSVDERRSVRGSQVVDDCAPFNDFIEDRVLINLPLC----GRKFTWYNGDGRSMSTLDR 138
S +R + + ++ D N + LI++ ++T+++ + S D
Sbjct: 148 PLSTLDRSTRQ--KINKDIQELNSALHQADLIDIYRTLHPKSTEYTFFSAPHHTYSKTDH 205
Query: 139 FLLSEEWCMVWPTCLQVAQLRG-LSDHCPIVLTVDEKN--------WGPRPVRLLKCW-- 187
L S+ + C + + LSDH I L + K W + L W
Sbjct: 206 ILGSKT---LLSKCKRTEIITNCLSDHSAIKLELRIKKLTQNHSTTWKLNNLLLNDYWVH 262
Query: 188 ----QDMPGYSDFVREKWGSFQVSGW--GGFVLKEKLKLMKAALKEWHVANTLNLPSKIV 241
++ + + K ++Q + W V + K + A ++ + L S++
Sbjct: 263 NEMKAEIKKFFETNENKDTTYQ-NLWDTAKAVCRGKFIALNAHKRKQERSKIDTLISQLK 321
Query: 242 SLKNRRAVLDSKGEEEELSEEELGELHGISSDIHSLSRLNTSICWQQSRLNWLRDGDANS 301
L+ + +E+ + EL I + +L ++N S W ++N +
Sbjct: 322 ELEKQEQTNSKASRRQEIIKIR-AELKEIETQ-KTLQKINESRSWFFEKINKI------D 373
Query: 302 KFFHSVLAARRRINSLSSILVD-GVVVEGVQPVRQAVFTHSKNHFLAPIMTRPSVGHL-- 358
+ ++ +R N + +I D G + ++ + + K+ + + +
Sbjct: 374 RPLARLIKKKREKNQIDTIKNDRGDITTDPTEIQTTIREYYKHLYANKLENLEEMDKFLD 433
Query: 359 --QFRKLSSVERGGLIKPFHEEEIKAAVWDCDSYKIPGPDGINLGFIKSFWHEMKAEIVR 416
+L+ E L +P EI+A + + K PGP+G F + + E+ +++
Sbjct: 434 TYTLPRLNQEEVESLNRPITSSEIEAIINSLPNKKSPGPEGFTAEFYQRYKEELVPFLLK 493
Query: 417 FVTEFHRNRKLSKGINSRFIALIPKVASPQSLNE-FRPISLVGSLYKILAKILANRLRQV 475
+ L I LIPK + E FRPISL+ KIL KILAN+++Q
Sbjct: 494 LFQSIEKEGILPNSFYEASIILIPKPGRDTTKKENFRPISLMNIDAKILNKILANQIQQH 553
Query: 476 IGSVVSEVQSAFVKNRQILDGILITNEVVDEARKSKK-DLLLFKVDFEKAYDSVDWGYLD 534
I ++ Q F+ Q I + ++ ++K + ++ +D EKA+D + ++
Sbjct: 554 IKKLIHHDQVGFIPAMQGWFNIRKSINIIQHINRTKDTNHMIISIDAEKAFDKIQQPFML 613
Query: 535 EVMGSMTFPTVW*KWIKECVGTATASVLVNGCPTDEFFLERGLRQGDPLSPFLFLLAAEG 594
+ + + + K I+ TA++++NG + L+ G RQG PLSP LL
Sbjct: 614 KPLNKLGIDGTYLKIIRAIYDKPTANIILNGQKLEAPPLKTGTRQGCPLSP---LLPNIV 670
Query: 595 LNVLMKALVDAGLFKGYSVGRADPVVVSHLQFADDTLLIGNKSWANVRALRAGLILLEAM 654
L VL +A+ KG +G+ + V FADD ++ + + L + +
Sbjct: 671 LEVLARAIRQEKEIKGIQLGKEE---VKLSLFADDMIVYLENPIVSAQNLLKLISNFSKV 727
Query: 655 SGLKVNFHKSSLVGVNITGSWLSEAASVLGCKVGKIPFLYLGLSIGGDPRRLL--FWEPV 712
SG K+N KS S+ S L + YLG+ + D + L ++P+
Sbjct: 728 SGYKINVQKSQAFLYTNNRQTESQIMSELPFTIASKRIKYLGIQLTRDVKDLFKENYKPL 787
Query: 713 VNRIKNRLSGWQSRFLSFGGRLVLLKFVLTALPVYALSF--FKAPSGIISSIESLFNKFF 770
+N IK + W++ S+ GR+ ++K + +Y + K P + +E KF
Sbjct: 788 LNEIKEDTNKWKNIPCSWVGRINIVKMAILPKVIYRFNAIPIKLPMTFFTELEKTTLKFI 847
Query: 771 WGGGEDKRKISWIRWDTLSLRKEYGGLGVTRLREFNLALLGKWCW 815
W +K + I TLS + + GG+ + + + A + K W
Sbjct: 848 W-----NQKRAHIAKSTLSQKNKAGGITLPDFKLYYKATVTKTAW 887
>POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains:
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1300
Score = 135 bits (340), Expect = 6e-31
Identities = 144/624 (23%), Positives = 271/624 (43%), Gaps = 25/624 (4%)
Query: 214 LKEKLKLMKAALKEWHVANTLNLPSKIVSLKNRRAVLDSKGEEEELSEEELGELHGISSD 273
L+ KL + A+ K+ A+T +L + + +L+ + A + +E+ + GE++ + +
Sbjct: 321 LRGKLIALSASKKKRETAHTSSLTTHLKALEKKEANSPKRSRRQEIIKLR-GEINQVETR 379
Query: 274 IHSLSRLNTSICWQQSRLNWLRDGDAN-SKFFHSVLAARRRINSLSSILVDGVVVEGVQP 332
++ R+N + W ++N + A +K + + N I D E +Q
Sbjct: 380 -RTIQRINQTRSWFFEKINKIDKPLARLTKGHRDKILINKIRNEKGDITTDP---EEIQN 435
Query: 333 VRQAVFTHSKNHFLAPIMTRPS-VGHLQFRKLSSVERGGLIKPFHEEEIKAAVWDCDSYK 391
++ + + L + + Q KL+ + L P +EI+A + + K
Sbjct: 436 TIRSFYKRLYSTKLENLDEMDKFLDRYQVPKLNQDQVDHLNSPISPKEIEAVINSLPTKK 495
Query: 392 IPGPDGINLGFIKSFWHEMKAEIVRFVTEFHRNRKLSKGINSRFIALIPKVAS-PQSLNE 450
PGPDG + F ++F ++ + + + L I LIPK P +
Sbjct: 496 SPGPDGFSAEFYQTFKEDLIPILHKLFHKIEVEGTLPNSFYEATITLIPKPQKDPTKIEN 555
Query: 451 FRPISLVGSLYKILAKILANRLRQVIGSVVSEVQSAFVKNRQILDGILITNEVVDEARKS 510
FRPISL+ KIL KILANR+++ I +++ Q F+ Q I + V+ K
Sbjct: 556 FRPISLMNIDAKILNKILANRIQEHIKAIIHPDQVGFIPGMQGWFNIRKSINVIHYINKL 615
Query: 511 K-KDLLLFKVDFEKAYDSVDWGYLDEVMGSMTFPTVW*KWIKECVGTATASVLVNGCPTD 569
K K+ ++ +D EKA+D + ++ +V+ + IK A++ VNG +
Sbjct: 616 KDKNHMIISLDAEKAFDKIQHPFMIKVLERSGIQGPYLNMIKAIYSKPVANIKVNGEKLE 675
Query: 570 EFFLERGLRQGDPLSPFLFLLAAEGLNVLMKALVDAGLFKGYSVGRADPVVVSHLQFADD 629
L+ G RQG PLSP+LF + L VL +A+ KG +G+ + V +S L ADD
Sbjct: 676 AIPLKSGTRQGCPLSPYLFNIV---LEVLARAIRQQKEIKGIQIGK-EEVKISLL--ADD 729
Query: 630 TLLIGNKSWANVRALRAGLILLEAMSGLKVNFHKSSLVGVNITGSWLSEAASVLGCKVGK 689
++ + + R L + + G K+N +KS E +
Sbjct: 730 MIVYISDPKNSTRELLNLINSFGEVVGYKINSNKSMAFLYTKNKQAEKEIRETTPFSIVT 789
Query: 690 IPFLYLGLSIGGDPRRLL--FWEPVVNRIKNRLSGWQSRFLSFGGRLVLLKFVLTALPVY 747
YLG+++ + + L ++ + IK L W+ S+ GR+ ++K + +Y
Sbjct: 790 NNIKYLGVTLTKEVKDLYDKNFKSLKKEIKEDLRRWKDLPCSWIGRINIVKMAILPKAIY 849
Query: 748 ALSF--FKAPSGIISSIESLFNKFFWGGGEDKRKISWIRWDTLSLRKEYGGLGVTRLREF 805
+ K P+ + +E KF W + + I L ++ GG+ + L+ +
Sbjct: 850 RFNAIPIKIPTQFFNELEGAICKFVWNNKKPR-----IAKSLLKDKRTSGGITMPDLKLY 904
Query: 806 NLALLGKWCWRLLLEKEA-LWRKV 828
A++ K W +++ W ++
Sbjct: 905 YRAIVIKTAWYWYRDRQVDQWNRI 928
>M310_ARATH (P93295) Hypothetical mitochondrial protein AtMg00310
(ORF154)
Length = 154
Score = 87.4 bits (215), Expect = 2e-16
Identities = 55/153 (35%), Positives = 76/153 (48%), Gaps = 10/153 (6%)
Query: 743 ALPVYALSFFKAPSGIISSIESLFNKFFWGGGEDKRKISWIRWDTLSLRKE-YGGLGVTR 801
ALPVYA+S F+ + + S +F+W E+KRKISW+ W L KE GGLG
Sbjct: 2 ALPVYAMSCFRLSKLLCKKLTSAMTEFWWSSCENKRKISWVAWQKLCKSKEDDGGLGFRD 61
Query: 802 LREFNLALLGKWCWRLLLEKEALWRKVLVARYGVADGGLE-DGGRSCSSWWREIVRIRDG 860
L FN ALL K +R++ + L ++L +RY +E G S WR I+ R+
Sbjct: 62 LGWFNQALLAKQSFRIIHQPHTLLSRLLRSRYFPHSSMMECSVGTRPSYAWRSIIHGREL 121
Query: 861 IGEGGEGWFGSCVRRRVGDGAETDFWWDCWCGD 893
+ G + R +GDG T W D W D
Sbjct: 122 LSRG--------LLRTIGDGIHTKVWLDRWIMD 146
>POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type II
retrotransposable element R2DM [Contains: Protease (EC
3.4.23.-); Reverse transcriptase (EC 2.7.7.49);
Endonuclease]
Length = 1057
Score = 77.0 bits (188), Expect = 3e-13
Identities = 83/324 (25%), Positives = 140/324 (42%), Gaps = 22/324 (6%)
Query: 349 IMTRPSVGHLQFRKLS---SVERGGLIKPFHEEEIKAAVWDCDSYKIPGPDGINLGFIKS 405
+MT+PS + S+ER + E++++A+ S PGPDGI KS
Sbjct: 317 VMTQPSPSSCSGEVIQMDHSLER--VWSAITEQDLRASRVSLSSS--PGPDGITP---KS 369
Query: 406 FWHEMKAEIVRFVTEFHRNRKLSKGINSRFIALIPKVASPQSLNEFRPISLVGSLYKILA 465
++R + L I IPK + + +FRPIS+ L + L
Sbjct: 370 AREVPSGIMLRIMNLILWCGNLPHSIRLARTVFIPKTVTAKRPQDFRPISVPSVLVRQLN 429
Query: 466 KILANRLRQVIGSVVSEVQSAFVKNRQILDGILITNEVVDEARKSKKDLLLFKVDFEKAY 525
ILA RL I Q F+ D I + V+ + K + + +D KA+
Sbjct: 430 AILATRLNSSINW--DPRQRGFLPTDGCADNATIVDLVLRHSHKHFRSCYIANLDVSKAF 487
Query: 526 DSVDWGYLDEVMGSMTFPTVW*KWIKECVGTATASVLVNGCPTDEFFLERGLRQGDPLSP 585
DS+ + + + + P + +++ S+ +G ++EF RG++QGDPLSP
Sbjct: 488 DSLSHASIYDTLRAYGAPKGFVDYVQNTYEGGGTSLNGDGWSSEEFVPARGVKQGDPLSP 547
Query: 586 FLFLLAAEGLNVLMKALVDAGLFKGYSVGRADPVVVSHLQFADDTLLIGNKSWANVRALR 645
LF L + L + + + G VG A + + FADD +L ++ ++ L
Sbjct: 548 ILFNLVMDRLLRTLPSEI------GAKVGNA---ITNAAAFADDLVLFA-ETRMGLQVLL 597
Query: 646 AGLILLEAMSGLKVNFHKSSLVGV 669
+ ++ GLK+N K VG+
Sbjct: 598 DKTLDFLSIVGLKLNADKCFTVGI 621
>PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 711
Score = 75.9 bits (185), Expect = 6e-13
Identities = 73/305 (23%), Positives = 132/305 (42%), Gaps = 23/305 (7%)
Query: 373 KPFHEEEIKAAV--WDCDSYKIPGPDGINLGFIKSFWHEMKAEIVRFVTEFHRNR-KLSK 429
+P EEI+ A+ W + PG DG+ + I + + R + H R +
Sbjct: 4 RPIAREEIQCAIKGWKPSA---PGSDGLTVQAIT------RTRLPRNFVQLHLLRGHVPT 54
Query: 430 GINSRFIALIPKVASPQSLNEFRPISLVGSLYKILAKILANRLRQVIGSVVSEVQSAFVK 489
+ LIPK ++ + +RPI++ +L ++L +ILA RL + + Q + +
Sbjct: 55 PWTAMRTTLIPKDGDLENPSNWRPITIASALQRLLHRILAKRLEAAVE--LHPAQKGYAR 112
Query: 490 NRQILDGILITNEVVDEARKSKKDLLLFKVDFEKAYDSVDWGYLDEVMGSMTFPTVW*KW 549
L L+ + + R+ +K + +D KA+D+V + + + +
Sbjct: 113 IDGTLVNSLLLDTYISSRREQRKTYNVVSLDVRKAFDTVSHSSICRALQRLGIDEGTSNY 172
Query: 550 IKECVGTATASVLVN-GCPTDEFFLERGLRQGDPLSPFLFLLAAEGLNVLMKALVDAGLF 608
I + +T ++ V G T + + RG++QGDPLSPFLF + L +++ G
Sbjct: 173 ITGSLSDSTTTIRVGPGSQTRKICIRRGVKQGDPLSPFLFNAVLDELLCSLQSTPGIG-- 230
Query: 609 KGYSVGRADPVVVSHLQFADDTLLIGNKSWANVRALRAGLILLEAMSGLKVNFHKSSLVG 668
G + L FADD LL+ + L A + + G+ +N KS +
Sbjct: 231 -----GTIGEEKIPVLAFADDLLLLEDNDVLLPTTL-ATVANFFRLRGMSLNAKKSVSIS 284
Query: 669 VNITG 673
V +G
Sbjct: 285 VAASG 289
>YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III
Length = 1222
Score = 67.8 bits (164), Expect = 2e-10
Identities = 52/204 (25%), Positives = 93/204 (45%), Gaps = 13/204 (6%)
Query: 439 IPKVASPQSLNEFRPISLVGSLYKILAKILANRLRQVIGSVVSEVQSAFVKNRQILDGIL 498
IPK +P S + +RPISL +I+ +I+ +R+R ++S Q F+ R ++
Sbjct: 673 IPKKGNPSSPSNYRPISLTDPFARIMERIICSRIRSEYSHLLSPHQHGFLNFRSCPSSLV 732
Query: 499 ITNEVVDEARKSKKDLLLFKVDFEKAYDSVDWGYLDEVMGSMTFPTVW*KWIKECVGTAT 558
+ + K++K L + DF KA+D V L + + + W KE + T
Sbjct: 733 RSISLYHSILKNEKSLDILFFDFAKAFDKVSHPILLKKLALFGLDKLTCSWFKEFLHLRT 792
Query: 559 ASVLVNG-CPTDEFFLERGLRQGDPLSPFLFLLAAEGLNVLMKALVDAGLFKGYSVGRAD 617
SV +N ++ + + G+ QG P LF+L L + ++ + F AD
Sbjct: 793 FSVKINKFVSSNAYPISSGVPQGSVSGPLLFILFINDLLIDLEPNIHVSCF-------AD 845
Query: 618 PVVVSH-----LQFADDTLLIGNK 636
+ + H LQ + DT++ +K
Sbjct: 846 DIKIFHHNPSTLQNSIDTIVKWSK 869
>PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1025
Score = 66.2 bits (160), Expect = 4e-10
Identities = 91/375 (24%), Positives = 152/375 (40%), Gaps = 40/375 (10%)
Query: 353 PSVGHLQFRKLSSVERGGLIKPFHEEEIKAAVWDCDSYKIPGPDGINLGFIKSFWHEMKA 412
P+ HL+ R ++ L +P +EIK + GPDG+ S +K+
Sbjct: 299 PTTRHLRSRMQGEIKN--LWRPISNDEIKEV--EACKRTAAGPDGMTTTAWNSIDECIKS 354
Query: 413 EIVRFVTEFHRNRKLSKGINSRFIALIPKVASPQSLNEFRPISLVGSLYKILAKILANRL 472
F + + + ++SR + LIPK FRP+S+ + +ILANR+
Sbjct: 355 ---LFNMIMYHGQCPRRYLDSRTV-LIPKEPGTMDPACFRPLSIASVALRHFHRILANRI 410
Query: 473 RQVIGSVVSEVQSAFVKNRQILDGILITNEVVDEARKSKKDLLLFKVDFEKAYDSVDWGY 532
+ ++ Q AF+ + + + + ++ EAR K L + +D +KA+DSV+
Sbjct: 411 GE--HGLLDTRQRAFIVADGVAENTSLLSAMIKEARMKIKGLYIAILDVKKAFDSVEHRS 468
Query: 533 LDEVMGSMTFPTVW*KWIKECVGTATASVLVNGCPTDEFFLERGLRQGDPLSPFLFLLAA 592
+ + + P +I + + V RG+RQGDPLSP LF
Sbjct: 469 ILDALRRKKLPLEMRNYIMWVYRNSKTRLEVVKTKGRWIRPARGVRQGDPLSPLLFNCVM 528
Query: 593 EGLNVLMKALVDAGLFKGYSVGRADPVVVSHLQFADDTLLIGNKS---WANVRALRAGL- 648
+ VL + + G G + L FADD +L+ A++ + AGL
Sbjct: 529 DA--VLRRLPENTGFLMGAE-------KIGALVFADDLVLLAETREGLQASLSRIEAGLQ 579
Query: 649 ------------ILLEAMSG----LKVNFHKSSLVG-VNITGSWLSEAASVLGCKVGKIP 691
L SG +KV HK VG IT ++ LG
Sbjct: 580 EQGLEMMPRKCHTLALVPSGKEKKIKVETHKPFTVGNQEITQLGHADQWKYLGVVYNSYG 639
Query: 692 FLYLGLSIGGDPRRL 706
+ + ++I GD +R+
Sbjct: 640 PIQVKINIAGDLQRV 654
>MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250
(ORF102)
Length = 122
Score = 58.2 bits (139), Expect = 1e-07
Identities = 31/69 (44%), Positives = 40/69 (57%), Gaps = 1/69 (1%)
Query: 562 LVNGCPTDEFFLERGLRQGDPLSPFLFLLAAEGLNVLMKALVDAGLFKGYSVGRADPVVV 621
++NG P RGLRQGDPLSP+LF+L E L+ L + + G G V P +
Sbjct: 13 IINGAPQGLVTPSRGLRQGDPLSPYLFILCTEVLSGLCRRAQEQGRLPGIRVSNNSP-RI 71
Query: 622 SHLQFADDT 630
+HL FADDT
Sbjct: 72 NHLLFADDT 80
>PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 869
Score = 53.9 bits (128), Expect = 2e-06
Identities = 75/298 (25%), Positives = 123/298 (41%), Gaps = 24/298 (8%)
Query: 379 EIKAAVWDCDSYKIPGPDGINLGFIKSFWHEMKAEIVRFVTEFHRNRKLSKGINSRFIAL 438
EIK+A + K GPDG+ + K + + R KG + F
Sbjct: 163 EIKSA--RASNEKGAGPDGVTPRSWNALDDRYKRLLYNIFVFYGRVPSPIKGSRTVFT-- 218
Query: 439 IPKVASPQSLNEFRPISLVGSLYKILAKILANRLRQVIGSVVSEVQSAFVKNRQILDGIL 498
PK+ FRP+S+ + + KILA R V E Q+A++ + +
Sbjct: 219 -PKIEGGPDPGVFRPLSICSVILREFNKILARRF--VSCYTYDERQTAYLPIDGVCINVS 275
Query: 499 ITNEVVDEARKSKKDLLLFKVDFEKAYDSVDWGYLDEVMGSMTFPTVW*KWIKECVGTAT 558
+ ++ EA++ +K+L + +D KA++SV L + + P +I +
Sbjct: 276 MLTAIIAEAKRLRKELHIAILDLVKAFNSVYHSALIDAITEAGCPPGVVDYIADMYNNVI 335
Query: 559 ASVLVNGCPTDEFFLERGLRQGDPLSPFLFLLAAEGLNVLMKALVDAGLFKGYSVGRADP 618
+ G + + G+ QGDPLS LF LA E ++AL + G F
Sbjct: 336 TEMQFEG-KCELASILAGVYQGDPLSGPLFTLAYE---KALRALNNEGRFD------IAD 385
Query: 619 VVVSHLQFADDTLLIGNKSWANVRALRAGLILLE---AMSGLKVNFHKSSLVGVNITG 673
V V+ ++DD LL+ V L+ L A GL++N KS V + +G
Sbjct: 386 VRVNASAYSDDGLLLA----MTVIGLQHNLDKFGETLAKIGLRINSRKSKTVSLVPSG 439
>PO11_SCICO (Q03277) Retrovirus-related Pol polyprotein from type I
retrotransposable element R1 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1004
Score = 53.5 bits (127), Expect = 3e-06
Identities = 88/363 (24%), Positives = 144/363 (39%), Gaps = 49/363 (13%)
Query: 285 CWQQSRLNWLRDGDANSKFFHSVLAARRRINSLSSILVDGVVVEGVQPVRQAVFTHSKNH 344
CWQ+ + + F + RR+ + S+ VDGV + + A+ N
Sbjct: 326 CWQEFVASESNENPWGRVF--KLCRGRRKPVDVCSVKVDGVYTDTWEGSVNAMM----NV 379
Query: 345 FLAPIMTRPSVGHLQFRKLSSVERGGLIKPFHEEEIKAAVWDCDSYKIPGPDGINLGFIK 404
F + S + +L ++ R L +E+ +V C K PGPDGI ++
Sbjct: 380 FFPASIDDAS----EIDRLKAIARP-LPPDLEMDEVSDSVRRCKVRKSPGPDGIVGEMVR 434
Query: 405 SFW----HEMKAEIVRFVTEFHRNRKLSKGINSRFIALIPKVASPQSLNEFRPISLVGSL 460
+ W M + + E + +K + L+ ++ S +RPI L+ +L
Sbjct: 435 AVWGAIPEYMFCLYKQCLLESYFPQKWKIASLVILLKLLDRIRSDP--GSYRPICLLDNL 492
Query: 461 YKILAKILANRLRQVIGSV-VSEVQSAFVKNRQILDGILITNEVVDEARKSKKDLLLFKV 519
K+L I+ RL Q + V VS Q AF + D V+ K ++ +
Sbjct: 493 GKVLEGIMVKRLDQKLMDVEVSPYQFAFTYGKSTEDAWRCVQRHVE--CSEMKYVIGLNI 550
Query: 520 DFEKAYDSVDW----GYLDEVMGSMTFPTVW*KWIKECVGTATASVLVNGCPTDEFFLER 575
DF+ A+D++ W LDE + + W+ G V G + + R
Sbjct: 551 DFQGAFDNLGWLSMLLKLDEAQSN-----EFGLWMSYFGGRKVYYVGKTGIVRKD--VTR 603
Query: 576 GLRQGDPLSPFLFLLAAEGLNVLMKALVDAGLFKGYSVGRADPVVVSHLQFADD-TLLIG 634
G QG P ++ L +N L+ ALV AG F + FADD T++IG
Sbjct: 604 GCPQGSKSGPAMWKLV---MNELLLALVAAGFF--------------IVAFADDGTIVIG 646
Query: 635 NKS 637
S
Sbjct: 647 ANS 649
>PO23_POPJA (Q05118) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 606
Score = 52.4 bits (124), Expect = 7e-06
Identities = 72/322 (22%), Positives = 127/322 (39%), Gaps = 31/322 (9%)
Query: 494 LDGILITNEVVDEARKSKKDLLLFKVDFEKAYDSVDWGYLDEVMGSMTFPTVW*KWIKEC 553
L I++ + R K + +D KA+D+V + M + +I
Sbjct: 3 LANIIMLEHYIKLRRLKGKTYNVVSLDIRKAFDTVSHPAILRAMRAFGIDDGMQDFIMST 62
Query: 554 VGTATASVLVNGCPTDEFFLERGLRQGDPLSPFLFLLAAEGLNVLMKALVDAGLFKGYSV 613
+ A +++V G T++ ++ G++QGDPLSP LF N+++ LV +
Sbjct: 63 ITDAYTNIVVGGRTTNKIYIRNGVKQGDPLSPVLF-------NIVLDELVTRLNDEQPGA 115
Query: 614 GRADPVVVSHLQFADDTLLIGNKSWANVRALRAGLILLEAMSGLKVNFHK-SSLVGVNIT 672
++ L FADD LL+ ++ +V A G+ +N K +S+ ++
Sbjct: 116 SMTPACKIASLAFADDLLLLEDRD-IDVPNSLATTCAYFRTRGMTLNPEKCASISAATVS 174
Query: 673 GSWLSEAA---SVLGCKV----GKIPFLYLGLSIGGDPRRLLFWEPVVNRIKNRLSGWQS 725
G + + ++ G + G F YLGL+ +P V + L +
Sbjct: 175 GRSVPRSKPSFTIDGRYIKPLGGINTFKYLGLTFSSTG----VAKPTVYNLTRWLRNLEK 230
Query: 726 RFLSFGGRLVLLKFVLTALPVYALSFFKAPSGIISSIESLFNKFFWGGGEDKRKISWIRW 785
L + +LK L Y L +GI+ + L + RKI +
Sbjct: 231 APLKPNQKFYILKTHLLPRLFYGLQSPGVTAGILQECDRLARR-------TTRKIFHLNV 283
Query: 786 DT----LSLRKEYGGLGVTRLR 803
T L R GGLG+ ++R
Sbjct: 284 HTGSQFLHARIRDGGLGLVQMR 305
>RTJK_DROFU (P21329) RNA-directed DNA polymerase from mobile element
jockey (EC 2.7.7.49) (Reverse transcriptase)
Length = 916
Score = 45.8 bits (107), Expect = 6e-04
Identities = 53/218 (24%), Positives = 91/218 (41%), Gaps = 4/218 (1%)
Query: 374 PFHEEEIKAAVWDCDSYKIPGPDGINLGFIKSFWHEMKAEIVRFVTEFHRNRKLSKGINS 433
P EE+K V K PG D ++ I+ + +V R K +
Sbjct: 437 PVTLEEVKELVSKLKPKKAPGEDLLDNRTIRLLPDQALLYLVLIFNSILRVGYFPKARPT 496
Query: 434 RFIALIPKVAS-PQSLNEFRPISLVGSLYKILAKILANRL--RQVIGSVVSEVQSAFVKN 490
I +I K P ++ +RP SL+ SL K+L +++ NR+ + + + + Q F
Sbjct: 497 ASIIMILKPGKQPLDVDSYRPTSLLPSLGKMLERLILNRILTSEEVTRAIPKFQFGFRLQ 556
Query: 491 RQILDGILITNEVVDEARKSKKDLLLFKVDFEKAYDSVDWGYLDEVMGSMTFPTVW*KWI 550
+ + EA + K+ +D ++A+D V L S+ P ++ + I
Sbjct: 557 HGTPEQLHRVVNFALEALEKKEYAGSCFLDIQQAFDRVWHPGLLYKAKSLLSPQLF-QLI 615
Query: 551 KECVGTATASVLVNGCPTDEFFLERGLRQGDPLSPFLF 588
K SV +GC + F+E G+ QG L P L+
Sbjct: 616 KSFWEGRKFSVTADGCRSSVKFIEAGVPQGSVLGPTLY 653
>RTJK_DROME (P21328) RNA-directed DNA polymerase from mobile element
jockey (EC 2.7.7.49) (Reverse transcriptase)
Length = 916
Score = 41.2 bits (95), Expect = 0.015
Identities = 108/479 (22%), Positives = 185/479 (38%), Gaps = 45/479 (9%)
Query: 378 EEIKAAVWDCDSYKIPGPDGINLGFIKSFWHEMKAEIVRFVTEFHRNRKLSKGINSRFIA 437
EE+K + K PG D ++ I+ + + K S I
Sbjct: 441 EEVKNLIAKLPLKKAPGEDLLDNRTIRLLPDQALQFLALIFNSVLDVGYFPKAWKSASII 500
Query: 438 LIPKVA-SPQSLNEFRPISLVGSLYKILAKILANRLR--QVIGSVVSEVQSAFVKNRQIL 494
+I K +P ++ +RP SL+ SL KI+ +++ NRL + + + + Q F
Sbjct: 501 MIHKTGKTPTDVDSYRPTSLLPSLGKIMERLILNRLLTCKDVTKAIPKFQFGFRLQHGTP 560
Query: 495 DGILITNEVVDEARKSKKDLLLFKVDFEKAYDSVDWGYLDEVMGSMTFPTVW*KWIKECV 554
+ + EA ++K+ + +D ++A+D V W FP +K +
Sbjct: 561 EQLHRVVNFALEAMENKEYAVGAFLDIQQAFDRV-WHPGLLYKAKRLFPPQLYLVVKSFL 619
Query: 555 GTATASVLVNGCPTDEFFLERGLRQGDPLSPFLFLLAAEGLNVLMKALVDAGLFKGYSVG 614
T V V+G + + G+ QG L P L+ + A + V
Sbjct: 620 EERTFHVSVDGYKSSIKPIAAGVPQGSVLGPTLYSVFASDMPT------------HTPVT 667
Query: 615 RADPVVVSHLQFADDTLLIGNKSWANVRALRAGLILLEAMS------GLKVNFHK-SSLV 667
D V +ADDT ++ KS + + A L+A +++N K +++
Sbjct: 668 EVDEEDVLIATYADDTAVL-TKSKSILAATSGLQEYLDAFQQWAENWNVRINAEKCANVT 726
Query: 668 GVNITGSWLSEAASVLGCKV-GKIPFLYLGLSIGGDPRRLLFWEPVVN-----RIKNRLS 721
N TGS S+ G + + YLG+++ R+L F + N R K
Sbjct: 727 FANRTGS--CPGVSLNGRLIRHHQAYKYLGITL---DRKLTFSRHITNIQQAFRTKVARM 781
Query: 722 GW---QSRFLSFGGRLVLLKFVLTALPVYALSFFKAPSGIISSIESLFNKFFWGGGEDKR 778
W LS G ++ + K +L Y L + GI + +S NK + R
Sbjct: 782 SWLIAPRNKLSLGCKVNIYKSILAPCLFYGLQVY----GI--AAKSHLNKIRILQAKTLR 835
Query: 779 KISWIRWDTLSLRKEYGGLGVTRLREFNLALLGKWCWRLLLEKEALWRKVLVARYGVAD 837
+IS W + R L V +L + + K+ RL + +L RK+ A AD
Sbjct: 836 RISGAPW-YMRTRDIERDLKVPKLGDKLQNIAQKYMERLNVHPNSLARKLGTAAVVNAD 893
>Y2R2_DROME (P16425) Hypothetical 115 kDa protein in type I
retrotransposable element R1DM (ORF 2)
Length = 1021
Score = 33.9 bits (76), Expect = 2.5
Identities = 45/212 (21%), Positives = 82/212 (38%), Gaps = 8/212 (3%)
Query: 379 EIKAAVWDCDSYKIPGPDGINLGFIKSFWHEMKAEIVRFVTEFHRNRKLSKGINSRFIAL 438
E+ V S + PG DGIN K+ W + + + R +
Sbjct: 438 EVDTCVARLKSRRSPGLDGINGTICKAVWRAIPEHLASLFSRCIRLGYFPAEWKCPRVVS 497
Query: 439 IPKVASPQSL--NEFRPISLVGSLYKILAKILANRLRQVIGSVVSEVQSAFVKNRQILDG 496
+ K + +R I L+ K+L I+ NR+R+V+ Q F + R + D
Sbjct: 498 LLKGPDKDKCEPSSYRGICLLPVFGKVLEAIMVNRVREVLPE-GCRWQFGFRQGRCVEDA 556
Query: 497 ILITNEVVDEARKSKKDLLLFKVDFEKAYDSVDWGYLDEVMGSMTFPTVW*KWIKECVGT 556
V + + +L VDF+ A+D+V+W + + + + + +
Sbjct: 557 WRHVKSSV--GASAAQYVLGTFVDFKGAFDNVEWSAALSRLADLGCREM---GLWQSFFS 611
Query: 557 ATASVLVNGCPTDEFFLERGLRQGDPLSPFLF 588
+V+ + T E + RG QG PF++
Sbjct: 612 GRRAVIRSSSGTVEVPVTRGCPQGSISGPFIW 643
>YP46_YEAST (Q12080) Hypothetical protein YPL146C
Length = 455
Score = 33.5 bits (75), Expect = 3.2
Identities = 21/84 (25%), Positives = 41/84 (48%), Gaps = 3/84 (3%)
Query: 204 FQVSGWGGFVLKEKL---KLMKAALKEWHVANTLNLPSKIVSLKNRRAVLDSKGEEEELS 260
F V G +LK KL K +K LK + + + SKI +L + + + + + +S
Sbjct: 61 FHVDVEGDEILKNKLIKRKQIKKVLKSKEILDAVKTNSKIAALNHHKNSSGNPNKIQGVS 120
Query: 261 EEELGELHGISSDIHSLSRLNTSI 284
+ EL +L ++ +H S++ +
Sbjct: 121 KHELKKLMALAGRVHGESKIKNRV 144
>METK_SULSO (Q980S9) S-adenosylmethionine synthetase (EC 2.5.1.6)
(Methionine adenosyltransferase) (AdoMet synthetase)
Length = 404
Score = 33.1 bits (74), Expect = 4.2
Identities = 25/79 (31%), Positives = 41/79 (51%), Gaps = 5/79 (6%)
Query: 430 GINSRFIALIPKVASPQSLNEF---RPISLVGSLYKILAKILANRLRQVIGSV-VSEVQS 485
G +R + LI + P SL P++ VG LY +LA ++AN++ Q + V S+VQ
Sbjct: 287 GRGNRGVGLITPMR-PMSLEATAGKNPVNHVGKLYNVLANLIANKIAQEVKDVKFSQVQV 345
Query: 486 AFVKNRQILDGILITNEVV 504
R I D ++ +V+
Sbjct: 346 LGQIGRPIDDPLIANVDVI 364
>DYHC_DICDI (P34036) Dynein heavy chain, cytosolic (DYHC)
Length = 4725
Score = 32.3 bits (72), Expect = 7.1
Identities = 24/97 (24%), Positives = 47/97 (47%), Gaps = 7/97 (7%)
Query: 429 KGINSRFIALIPKVASPQSLNEFRPISLVGSLYKILAKILANRLRQVIGSVVSEVQSAFV 488
K INS FIA++ KV+ + E I + + L+ +L ++++ +G + +SAF
Sbjct: 1632 KSINSEFIAILKKVSGAPLILEVLAIERIQQTMERLSDLL-GKVQKALGEYLERQRSAFA 1690
Query: 489 KNRQILDGILITNEVVDEARKSKKDLLLFKVDFEKAY 525
+ + D L+ E + KD++ + F K +
Sbjct: 1691 RFYFVGDEDLL------EIIGNSKDIIKIQKHFRKMF 1721
>APEA_DICDI (P51173) DNA-(apurinic or apyrimidinic site) lyase (EC
4.2.99.18) (Class II
apurinic/apyrimidinic(AP)-endonuclease)
Length = 361
Score = 32.0 bits (71), Expect = 9.3
Identities = 41/159 (25%), Positives = 70/159 (43%), Gaps = 29/159 (18%)
Query: 40 EFYIFNMYAP---------CDRRAKQVLWN-SLSSRLLSLGSSKVCV-CGDFNTVRSVDE 88
+FYI N Y P D R K+ W+ + L L ++K + CGD N + +
Sbjct: 204 QFYIVNTYIPNAGTRGLQRLDYRIKE--WDVDFQAYLEKLNATKPIIWCGDLNVAHTEID 261
Query: 89 RRSVRGSQ-----VVDDCAPFNDFIEDRVL-----INLPLCGRKFTW-YNGDGRSMST-- 135
++ + ++ +++ F++F+E + N G W Y G GRS +
Sbjct: 262 LKNPKTNKKSAGFTIEERTSFSNFLEKGYVDSYRHFNPGKEGSYTFWSYLGGGRSKNVGW 321
Query: 136 -LDRFLLSEEWC-MVWPTCLQVAQLRGLSDHCPIVLTVD 172
LD F++S+ + + + G SDHCPI + VD
Sbjct: 322 RLDYFVVSKRLMDSIKISPFHRTSVMG-SDHCPIGVVVD 359
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.323 0.140 0.445
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 129,771,130
Number of Sequences: 164201
Number of extensions: 5643251
Number of successful extensions: 11902
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 11858
Number of HSP's gapped (non-prelim): 31
length of query: 1091
length of database: 59,974,054
effective HSP length: 121
effective length of query: 970
effective length of database: 40,105,733
effective search space: 38902561010
effective search space used: 38902561010
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 71 (32.0 bits)
Medicago: description of AC122166.6


