

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122162.12 - phase: 0
(400 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
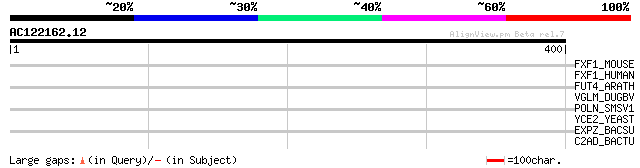
Score E
Sequences producing significant alignments: (bits) Value
FXF1_MOUSE (Q61080) Forkhead box protein F1 (Forkhead-related pr... 36 0.15
FXF1_HUMAN (Q12946) Forkhead box protein F1 (Forkhead-related pr... 36 0.15
FUT4_ARATH (Q9SJP2) Probable fucosyltransferase 4 (EC 2.4.1.-) (... 34 0.58
VGLM_DUGBV (Q02004) M polyprotein precursor [Contains: Glycoprot... 33 0.99
POLN_SMSV1 (P36286) Non-structural polyprotein [Contains: Unknow... 31 4.9
YCE2_YEAST (P25572) Hypothetical protein YCL042W 31 6.4
EXPZ_BACSU (P39115) Nucleotide binding protein expZ 31 6.4
C2AD_BACTU (Q9RMG3) Pesticidial crystal protein cry2Ad (Insectic... 30 8.4
>FXF1_MOUSE (Q61080) Forkhead box protein F1 (Forkhead-related
protein FKHL5) (Forkhead-related transcription factor 1)
(FREAC-1) (Hepatocyte nuclear factor 3 forkhead homolog
8) (HFH-8)
Length = 353
Score = 36.2 bits (82), Expect = 0.15
Identities = 30/129 (23%), Positives = 49/129 (37%), Gaps = 8/129 (6%)
Query: 58 HNILPFQFPLLK--YVPTPDSISNNNEIIDNIENTSTSFGHLSKRSFFLVKPPQEQQQQE 115
+ L +FP + Y +S+ +N + + G K ++ + P E +E
Sbjct: 50 YQFLQARFPFFRGAYQGWKNSVRHNLSLNECFIKLPKGLGRPGKGHYWTIDPASEFMFEE 109
Query: 116 TLIRRRPWLIRITQNSSGKTQLLKPPFLSLTSTAFSHLPNVVDFTKFSPQHLATDFIIDK 175
RRRP R K Q LKP + + F+HLP+ F A + + +
Sbjct: 110 GSFRRRPRGFR------RKCQALKPVYSMVNGLGFNHLPDTYGFQGSGGLSCAPNSLALE 163
Query: 176 DDLTFQNQH 184
L N H
Sbjct: 164 GGLGMMNGH 172
>FXF1_HUMAN (Q12946) Forkhead box protein F1 (Forkhead-related
protein FKHL5) (Forkhead-related transcription factor 1)
(FREAC-1) (Forkhead-related activator-1)
Length = 354
Score = 36.2 bits (82), Expect = 0.15
Identities = 26/104 (25%), Positives = 42/104 (40%), Gaps = 8/104 (7%)
Query: 58 HNILPFQFPLLK--YVPTPDSISNNNEIIDNIENTSTSFGHLSKRSFFLVKPPQEQQQQE 115
+ L +FP + Y +S+ +N + + G K ++ + P E +E
Sbjct: 50 YQFLQSRFPFFRGSYQGWKNSVRHNLSLNECFIKLPKGLGRPGKGHYWTIDPASEFMFEE 109
Query: 116 TLIRRRPWLIRITQNSSGKTQLLKPPFLSLTSTAFSHLPNVVDF 159
RRRP R K Q LKP + + F+HLP+ F
Sbjct: 110 GSFRRRPRGFR------RKCQALKPMYSMMNGLGFNHLPDTYGF 147
>FUT4_ARATH (Q9SJP2) Probable fucosyltransferase 4 (EC 2.4.1.-)
(AtFUT4)
Length = 503
Score = 34.3 bits (77), Expect = 0.58
Identities = 45/176 (25%), Positives = 71/176 (39%), Gaps = 32/176 (18%)
Query: 12 SMASVDWSELPKDIIFLISQRLDVELDLIRFRSVCSTWRSSPIPNDHNILPFQFPLLKYV 71
++ASV L D I L+ R D+ DL+ P P +LP FPL+KY
Sbjct: 129 TLASVFLYALLTDRIILVDNRKDIG-DLL----------CEPFPGTSWLLPLDFPLMKYA 177
Query: 72 PTPDSISNN--NEIIDNIENTSTSF-GHLSKRSFFLVKPPQEQ---QQQETLIRRRPWLI 125
+ +++N STSF HL + + + Q+ ++LI + PWLI
Sbjct: 178 DGYHKGYSRCYGTMLENHSINSTSFPPHLYMHNLHDSRDSDKMFFCQKDQSLIDKVPWLI 237
Query: 126 ---------RITQNSSGKTQLLKPPFLSLTSTAFSHLPNVVDFTKFSPQHLATDFI 172
+ N + +T+L K T F HL + F P++ D +
Sbjct: 238 FRANVYFVPSLWFNPTFQTELTK--LFPQKETVFHHLGRYL----FHPKNQVWDIV 287
>VGLM_DUGBV (Q02004) M polyprotein precursor [Contains: Glycoprotein
G2; Nonstructural protein NS-M; Glycoprotein G1]
Length = 1551
Score = 33.5 bits (75), Expect = 0.99
Identities = 19/73 (26%), Positives = 34/73 (46%)
Query: 73 TPDSISNNNEIIDNIENTSTSFGHLSKRSFFLVKPPQEQQQQETLIRRRPWLIRITQNSS 132
TP + + + + I T+T + +S + K Q ++ET+I R P ++ +S
Sbjct: 48 TPQTTTTSTAVSTTITATTTPTASWTTQSQYFNKTTQHHWREETMISRNPTVLDRQSRAS 107
Query: 133 GKTQLLKPPFLSL 145
+LL FL L
Sbjct: 108 SVRELLNTKFLML 120
>POLN_SMSV1 (P36286) Non-structural polyprotein [Contains: Unknown
protein; Helicase (2C-like protein) (P2C); 3A-like
protein; Viral genome-linked protein (VPg); Thiol
protease P3C (EC 3.4.22.-) (3C-like protease) (3C-pro);
RNA-directed RNA polymerase (EC
Length = 1879
Score = 31.2 bits (69), Expect = 4.9
Identities = 18/52 (34%), Positives = 27/52 (51%), Gaps = 2/52 (3%)
Query: 123 WLIRITQNSSGKTQLLKPPF--LSLTSTAFSHLPNVVDFTKFSPQHLATDFI 172
W +R +G+ QL+K + + TA L N +D T FS +H+ T FI
Sbjct: 928 WALRRHLKWTGQWQLIKAAYEIMCYPDTAACALRNWMDSTDFSEEHVVTQFI 979
>YCE2_YEAST (P25572) Hypothetical protein YCL042W
Length = 119
Score = 30.8 bits (68), Expect = 6.4
Identities = 18/62 (29%), Positives = 29/62 (46%), Gaps = 8/62 (12%)
Query: 62 PFQFPLLKYVPTPDSISNNNEIIDNIENTSTSFGHLSKRSFFLVKPPQEQQQQETLIRRR 121
P+++PL PTP S+ N+ + N T+T+ + S KP +E+ R
Sbjct: 50 PYKYPLSTEPPTPPSVPNSASVNHN-TTTNTTLSYTRCHSTTYTKPLRERSS-------R 101
Query: 122 PW 123
PW
Sbjct: 102 PW 103
>EXPZ_BACSU (P39115) Nucleotide binding protein expZ
Length = 547
Score = 30.8 bits (68), Expect = 6.4
Identities = 13/44 (29%), Positives = 22/44 (49%)
Query: 220 HDRDKRWTPVSNLSTAHGDICLFKGRFYVVDQSGQTVTVGPDSS 263
H KR+ V N++ A G+ LFK + + + +GP+ S
Sbjct: 285 HKTGKRFLEVQNVTKAFGERTLFKNANFTIQHGEKVAIIGPNGS 328
>C2AD_BACTU (Q9RMG3) Pesticidial crystal protein cry2Ad
(Insecticidal delta-endotoxin CryIIA(d)) (Crystaline
entomocidal protoxin) (71 kDa crystal protein)
Length = 633
Score = 30.4 bits (67), Expect = 8.4
Identities = 14/67 (20%), Positives = 32/67 (46%), Gaps = 3/67 (4%)
Query: 243 KGRFYVVDQSGQTVTVGPDSSTELAAQPLYRRCVRGENRKLLVE---NEGELLLLDIHQT 299
K Y V ++G + + P+ +T P++ V + R + E N+G+ L + T
Sbjct: 471 KNNIYAVHENGTMIHLAPEDNTGFTISPIHATQVNNQTRTFISEKFGNQGDSLRFEQSNT 530
Query: 300 FFQFSIK 306
+++++
Sbjct: 531 TARYTLR 537
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.322 0.138 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 49,253,905
Number of Sequences: 164201
Number of extensions: 2125283
Number of successful extensions: 4401
Number of sequences better than 10.0: 8
Number of HSP's better than 10.0 without gapping: 2
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 4399
Number of HSP's gapped (non-prelim): 8
length of query: 400
length of database: 59,974,054
effective HSP length: 112
effective length of query: 288
effective length of database: 41,583,542
effective search space: 11976060096
effective search space used: 11976060096
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 67 (30.4 bits)
Medicago: description of AC122162.12


