

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122162.1 + phase: 0 /pseudo/partial
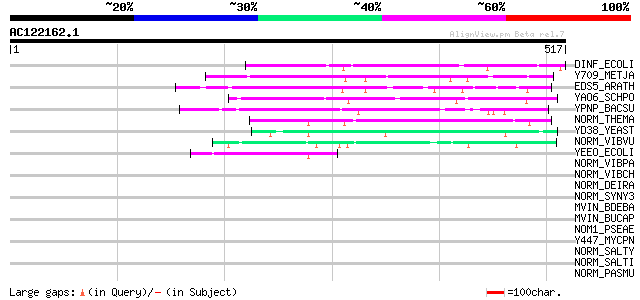
(517 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
DINF_ECOLI (P28303) DNA-damage-inducible protein F 76 2e-13
Y709_METJA (Q58119) Hypothetical protein MJ0709 72 5e-12
EDS5_ARATH (Q945F0) Enhanced disease susceptibility 5 (Eds5) (Sa... 66 2e-10
YAO6_SCHPO (Q10085) Hypothetical protein C11D3.06 in chromosome I 61 8e-09
YPNP_BACSU (P54181) Hypothetical protein ypnP 55 4e-07
NORM_THEMA (Q9WZS2) Probable multidrug resistance protein norM (... 55 6e-07
YD38_YEAST (Q05497) Hypothetical 77.8 kDa protein in MRPS28-HXT7... 51 6e-06
NORM_VIBVU (Q8D9N8) Probable multidrug resistance protein norM (... 47 1e-04
YEEO_ECOLI (P76352) Hypothetical protein yeeO 47 2e-04
NORM_VIBPA (O82855) Multidrug resistance protein norM (Na(+)/dru... 42 0.005
NORM_VIBCH (Q9KRU4) Probable multidrug resistance protein norM (... 42 0.005
NORM_DEIRA (Q9RY44) Probable multidrug resistance protein norM (... 42 0.005
NORM_SYNY3 (Q55364) Probable multidrug resistance protein norM (... 41 0.007
MVIN_BDEBA (Q8VNZ2) Virulence factor mviN homolog 41 0.007
MVIN_BUCAP (Q8K9L3) Virulence factor mviN homolog 40 0.011
NOM1_PSEAE (Q9I3Y3) Probable multidrug resistance protein norM 1... 40 0.015
Y447_MYCPN (P75130) Hypothetical protein MG447 homolog (K05_orf499) 39 0.032
NORM_SALTY (Q8ZPP2) Multidrug resistance protein norM (Na(+)/dru... 38 0.055
NORM_SALTI (Q8Z6N7) Multidrug resistance protein norM (Na(+)/dru... 38 0.055
NORM_PASMU (Q9CMZ9) Probable multidrug resistance protein norM (... 38 0.055
>DINF_ECOLI (P28303) DNA-damage-inducible protein F
Length = 459
Score = 75.9 bits (185), Expect = 2e-13
Identities = 71/312 (22%), Positives = 145/312 (45%), Gaps = 21/312 (6%)
Query: 220 MLVPAVKYLRLRALGAPAVLLSLAMQGIFRGFKDTTTPLYVIVSGYALNVAMDPLLIFYF 279
+L A ++L +R L APA L +L + G G + P+ ++V G LN+ +D L+
Sbjct: 142 VLEQARRFLEIRWLSAPASLANLVLLGWLLGVQYARAPVILLVVGNILNIVLDVWLVMGL 201
Query: 280 KLGIRGAAISHVLSQYIMATLLLFILM-KKV--------DLLPPSMKDLQIFRFLKNGGL 330
+ ++GAA++ V+++Y ATLL+ +LM +K+ ++L + + R L
Sbjct: 202 HMNVQGAALATVIAEY--ATLLIGLLMVRKILKLRGISGEMLKTAWRG-NFRRLLALNRD 258
Query: 331 LLARVIAVTFCVTLSASLAARLGPIPMAAFQTCLQVWMTSSLLADGLAVAIQAILACSFA 390
++ R + + C L ARLG +A + + ++ DG A A++A ++
Sbjct: 259 IMLRSLLLQLCFGAITVLGARLGSDIIAVNAVLMTLLTFTAYALDGFAYAVEAHSGQAYG 318
Query: 391 EKDYNKVTTAATRTLQMSFVLGVGLSLVVGGGLYFGAGVFSKNVAVIHLIRLG---LPFV 447
+D +++ + S ++ + S+V L G + + ++ + +L L +
Sbjct: 319 ARDGSQLLDVWRAACRQSGIVALLFSVVY---LLAGEHIIALLTSLTQIQQLADRYLIWQ 375
Query: 448 AATQPINSLAFVFDGVNYGASDFAYSAYSLVMVSIASVTSLFFLYKSKGFIGIWIALTIY 507
+ ++ DG+ GA+ S+ + + +L L G +W+ALT++
Sbjct: 376 VILPVVGVWCYLLDGMFIGATRATEMRNSMAVAAAGFALTLLTL-PWLGNHALWLALTVF 434
Query: 508 MSLR--MFAGVW 517
++LR A +W
Sbjct: 435 LALRGLSLAAIW 446
Score = 30.8 bits (68), Expect = 8.8
Identities = 20/67 (29%), Positives = 30/67 (43%), Gaps = 1/67 (1%)
Query: 27 MDSLAKEILGIAFPSALAVAADPIASLIDTAFIGHL-GPVELAAAGVSIAVFNQASRITI 85
+ S K + +A P + P+ L+DTA IGHL PV L V + + +
Sbjct: 22 LTSSDKALWHLALPMIFSNITVPLLGLVDTAVIGHLDSPVYLGGVAVGATATSFLFMLLL 81
Query: 86 FPLVSIT 92
F +S T
Sbjct: 82 FLRMSTT 88
>Y709_METJA (Q58119) Hypothetical protein MJ0709
Length = 450
Score = 71.6 bits (174), Expect = 5e-12
Identities = 77/342 (22%), Positives = 145/342 (41%), Gaps = 25/342 (7%)
Query: 183 ASTALLFGTVLGLIQAATLIFAAKPLLGAMGLKYDSPMLVPAVKYLRLRALGAPAVLLSL 242
A+ A++ + G++ + L MG D L A+KY + LG +
Sbjct: 92 ANHAIILALIAGILYIIAVYPNLDTLFSLMGTYGDCKSL--AIKYSSILVLGTVIFTICD 149
Query: 243 AMQGIFRGFKDTTTPLYVIVSGYALNVAMDPLLIFYFKLGIRGAAISHVLSQYIMATLLL 302
A+ GIFRG +T + V G N+ +DP+ I+ LGI GA+ + +++ I +L
Sbjct: 150 ALYGIFRGEGNTKIVMIASVIGTLTNIILDPIFIYMLNLGISGASYATLIAIIISLLILA 209
Query: 303 FILMKKVDL-----LPPSMKDLQIFRFLKNGGL---LLARVIAVTFCVTLSASLAARLGP 354
+ L K L DL+I L G+ L+ +AV+F + S +
Sbjct: 210 YELFIKKSCYVTVKLSKFKPDLKIIADLIRVGIPSALIEITVAVSFFIMTSIIMMVG-DS 268
Query: 355 IPMAAFQTCLQVWMTSSLLADGLAVAIQAILACSFAEKDYNKVTTAATRTLQMSF---VL 411
+A + L++ + GLA +++ ++ + + K+ TA T+++ ++
Sbjct: 269 RGLAVYTGALRITEFGFIPMLGLASGATSVIGATYGARSFEKLKTAYFYTIKIGVLMEII 328
Query: 412 GVGLSLVVGGGLYF-------GAGVFSKNVAVIHLIRLGLPFVAATQPINSLAFVFDGVN 464
V L +++ L + G+ + V + ++ L L F T I + + +F G+
Sbjct: 329 IVALIMLLSPILAYLFTYTKTSMGIHEELVKALRIVPLYLLF---TPFILTTSAMFQGIG 385
Query: 465 YGASDFAYSAYSLVMVSIASVTSLFFLYKSKGFIGIWIALTI 506
G S + ++ I S LF + G GI+ L +
Sbjct: 386 KGEKSLIISIFRSLICHI-SYAYLFAVILGLGMFGIYSGLVV 426
Score = 31.2 bits (69), Expect = 6.8
Identities = 15/50 (30%), Positives = 27/50 (54%)
Query: 27 MDSLAKEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAAAGVSIAV 76
+D K ++ ++ P +A + I SL+D+ ++ LG LAA G S +
Sbjct: 8 LDDPKKAVIEVSKPIIVATFIESIYSLVDSIWVSGLGADALAAVGASFPI 57
>EDS5_ARATH (Q945F0) Enhanced disease susceptibility 5 (Eds5)
(Salicylic acid induction deficient 1) (Sid1)
Length = 543
Score = 65.9 bits (159), Expect = 2e-10
Identities = 87/369 (23%), Positives = 153/369 (40%), Gaps = 38/369 (10%)
Query: 155 NISKSSIVTNSSNKSESKPIRKKRHIASASTALLFGTVLGLIQAA-TLIFAAKPLLGAMG 213
+++ S++V S K + K + + S L G V GL+ T +F P
Sbjct: 156 SVATSNMVATSLAKQDKKEAQHQ-----ISVLLFIGLVCGLMMLLLTRLFG--PWAVTAF 208
Query: 214 LKYDSPMLVPAV-KYLRLRALGAPAVLLSLAMQGIFRGFKDTTTPLYVIVSGYALNVAMD 272
+ + +VPA KY+++R L P +L+ L Q G K++ PL + + +N D
Sbjct: 209 TRGKNIEIVPAANKYIQIRGLAWPFILVGLVAQSASLGMKNSWGPLKALAAATIINGLGD 268
Query: 273 PLLIFYFKLGIRGAAISHVLSQYIMATLLLFILMKK----VDLLPPSMKDLQIFRFLKNG 328
+L + GI GAA + SQ + A +++ L K+ PS ++L L
Sbjct: 269 TILCLFLGQGIAGAAWATTASQIVSAYMMMDSLNKEGYNAYSFAIPSPQELWKISALAAP 328
Query: 329 GL--LLARVIAVTFCVTLSASLAARLGPIPMAAFQTCLQVWMTSSLLADGLAVAIQAILA 386
+ +++ +F + + S+ + +AA Q Q + ++ + L+ Q+ +
Sbjct: 329 VFISIFSKIAFYSFIIYCATSMGTHV----LAAHQVMAQTYRMCNVWGEPLSQTAQSFM- 383
Query: 387 CSFAEKDY--NKVTTAATRTLQMSFVLGVGLSLVVG------GGLYFGAGVFSKNVAVIH 438
E Y N+ A L+ ++G L LV+G GL+ G K V +
Sbjct: 384 ---PEMLYGANRNLPKARTLLKSLMIIGATLGLVLGVIGTAVPGLFPGVYTHDK-VIISE 439
Query: 439 LIRLGLPFVAATQPINSLAFVFDGVNYGASDFAYSAYSLVMVS---IASVTSLFFLYKSK 495
+ RL +PF A + + +G D + S VM S I +T +F
Sbjct: 440 MHRLLIPFFMALSAL-PMTVSLEGTLLAGRDLKF--VSSVMSSSFIIGCLTLMFVTRSGY 496
Query: 496 GFIGIWIAL 504
G +G W L
Sbjct: 497 GLLGCWFVL 505
Score = 34.3 bits (77), Expect = 0.80
Identities = 22/63 (34%), Positives = 32/63 (49%), Gaps = 2/63 (3%)
Query: 32 KEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAAAGVSIAVFNQASRITIFPLVSI 91
KEI+ P+ P+ SLIDT IG +ELAA G + + S + +F +S+
Sbjct: 100 KEIVKFTGPAMGMWICGPLMSLIDTVVIGQGSSIELAALGPGTVLCDHMSYVFMF--LSV 157
Query: 92 TTS 94
TS
Sbjct: 158 ATS 160
>YAO6_SCHPO (Q10085) Hypothetical protein C11D3.06 in chromosome I
Length = 455
Score = 60.8 bits (146), Expect = 8e-09
Identities = 77/320 (24%), Positives = 144/320 (44%), Gaps = 24/320 (7%)
Query: 205 AKPLLGAMGLKYDSPMLVPAV-KYLRLRALGAPAVLLSLAMQGIFRGFKDTTTPLYVIVS 263
+KP+L + + +P L A K+LR G + ++ + + T Y+++
Sbjct: 110 SKPILIFL---HQTPELAEASQKFLRYLIPGGLGYVCFELLKKFLQTQEITRAGSYILLV 166
Query: 264 GYALNVAMDPLLIFYFKLGIRGAAISHVLSQYIMATLLLFILMKKVDLLPP----SMKDL 319
LNVA++ LL+ Y+ LG++GA ++ LS Y ++ +LL K V + + L
Sbjct: 167 TSPLNVALNFLLVHYYGLGLKGAPLATGLS-YWLSFILLTQYAKYVKGAEAWNGWNKRCL 225
Query: 320 QIF-RFLKNGGLLLARVIAVTFCVTLSASLAARLGPIPMAAFQTCLQVWMTSSLLADGLA 378
+ F F+K L + V + + A +A +LG +P+AA V MT+ L + +
Sbjct: 226 ENFGPFVKLSLLGIVMVGTEWWAFEIVALVAGKLGAVPLAA----QSVIMTTDQLLNTIP 281
Query: 379 VAIQAILACSFAEKDYNKVTTAATRTLQMSFVLGVG------LSLVVGGGLYFGAGVFSK 432
+ I + A + A+ T +++ ++GV ++++ +Y +F+
Sbjct: 282 FGLGIITSNRVAYYLGAGLPDNASLTAKVAAIVGVAVGSVIMITMIAVRNIY--GRIFTN 339
Query: 433 NVAVIHLIRLGLPFVAATQPINSLAFVFDGVNYGASDFAYSAYSLVMV--SIASVTSLFF 490
+ VI L+ L +P VAA Q +SL G G A + A ++
Sbjct: 340 DPDVIQLVALVMPLVAAFQISDSLNGTMGGALRGTGRQKVGAIVNITAYYLFALPLGIYL 399
Query: 491 LYKSKGFIGIWIALTIYMSL 510
+ KG +G+WI I +S+
Sbjct: 400 AFHGKGLVGLWIGQVIALSI 419
>YPNP_BACSU (P54181) Hypothetical protein ypnP
Length = 445
Score = 55.1 bits (131), Expect = 4e-07
Identities = 84/355 (23%), Positives = 151/355 (41%), Gaps = 29/355 (8%)
Query: 159 SSIVTNSSNKSESKPIRKKRHIASASTALLFGTVLGLIQAATLIFAAKPLLGAMGLKYDS 218
++ +T S + I+ +A LL G +G A F ++ LL LK
Sbjct: 71 NATLTILSQQKGKDDIKGMASYINAFVVLLTGLSIGF--GAAGFFLSELLLRL--LKTPE 126
Query: 219 PMLVPAVKYLRLRALGAPAVLLSLAMQGIFRGFKDTTTPLYVIVSGYALNVAMDPLLIFY 278
M+ A YL+++ +G + + + R D+ TPL I LN + PL I
Sbjct: 127 SMIPLAETYLQIQFIGILFLFGYNFISTVLRALGDSKTPLRFIAFAVVLNTVLAPLFISV 186
Query: 279 FKLGIRGAAISHVLSQYIMATLLLFILMKKVDLLPPSMKDLQIFR----FLKNGGLLLAR 334
F++GI GAA S +LSQ I LF ++K L+P S+ + + LK G +
Sbjct: 187 FRMGIAGAAYSTILSQGIAFLYGLFYVIKH-KLVPFSIPRMPKWEESALILKLGIPAGLQ 245
Query: 335 VIAVTFCVTLSASLAARLGPIPMAAFQTCLQVWMTSSLLADGLAVAIQAILACSFAEKDY 394
++ +T + S+ G ++ F ++ +L A A+ ++ + +
Sbjct: 246 MMVITGGMMAIMSVVNSYGDHVVSGFGAVQRLDSIITLPAMAAGTAVNSMAGQNIGIGNE 305
Query: 395 NKVTTAATRTLQMSFVLGVGLSLVVGGGLYFGAGVFSKNVAVIHLIRLGL--PFVAA--T 450
+V T A ++ + + LV+ ++ VF K +LIRL + P A
Sbjct: 306 KRVGTIA----RLGVIAVISCMLVIAVMIW----VFGK-----YLIRLFISEPDAVAFGE 352
Query: 451 QPINSLAFV--FDGVNYGASDFAYSAYSLVMVSIASVTSLFFL-YKSKGFIGIWI 502
Q + +AF F GVN+ + +A +++ V + ++ S + L Y W+
Sbjct: 353 QYLKWIAFFYPFIGVNFVLNGIVRAAGAMLQVLVLNLISFWVLRYPFTALFSAWL 407
>NORM_THEMA (Q9WZS2) Probable multidrug resistance protein norM
(Na(+)/drug antiporter) (Multidrug-efflux transporter)
Length = 464
Score = 54.7 bits (130), Expect = 6e-07
Identities = 59/295 (20%), Positives = 124/295 (42%), Gaps = 17/295 (5%)
Query: 224 AVKYLRLRALGAPAVLLSLAMQGIFRGFKDTTTPLYVIVSGYALNVAMDPLLIF----YF 279
A +YL++ G+ + + RG DT TP+ V LN+ +D +IF +
Sbjct: 138 AKEYLKVILSGSMGFSIMAVFSAMLRGAGDTRTPMIVTGLTNFLNIFLDYAMIFGKFGFP 197
Query: 280 KLGIRGAAISHVLSQYIMATLLLFILMKKVD------LLPPSMKDLQIFRFLKNGGLLLA 333
++G+RGAA++ +LS+++ A +L +++ K+ + L+PP + L+ G
Sbjct: 198 EMGVRGAAVATILSRFVGAGILTYVIFKREEFQLRKGLVPPKWSSQK--EILRVGFPTAI 255
Query: 334 RVIAVTFCVTLSASLAARLGPIPMAAFQTCLQVWMTSSLLADGLAVAIQAILACSFAEKD 393
+ V + A++ G A + + V S + A G++VAI ++ +
Sbjct: 256 ENFVFSTGVLMFANILLIAGAEAYAGHRIGINVESLSFMPAFGISVAITTLVGRYNGMGN 315
Query: 394 YNKVTTAATRTLQMSFVLGVGLSLVVGGGLYFGAGVFSKNVAVIHLIRLGLPFVAATQPI 453
V + +S + V + +++ +F+ + +I + +L + + Q
Sbjct: 316 KEHVLGVIRQGWILSLLFQVTVGIIIFLFPEPLIRIFTSDPQIIEISKLPVKIIGLFQFF 375
Query: 454 NSLAFVFDGVNYGASDFAYSAYSLVMVSI----ASVTSLFFLYKSKGFIGIWIAL 504
++ +G G + + +SI V + Y G +G WI +
Sbjct: 376 LAIDSTMNGALRGTGN-TLPPMIITFISIWTARLPVAFVMVKYFQLGLLGAWIGM 429
Score = 33.9 bits (76), Expect = 1.0
Identities = 32/127 (25%), Positives = 60/127 (47%), Gaps = 12/127 (9%)
Query: 186 ALLFGTVLGLIQAATLIFAAKPLLGAMGLKYDSPMLVPAVKYLRLRALGAPAVLLSL--A 243
+LLF +G+I + +PL+ + S + + L ++ +G L++
Sbjct: 330 SLLFQVTVGII----IFLFPEPLIRI----FTSDPQIIEISKLPVKIIGLFQFFLAIDST 381
Query: 244 MQGIFRGFKDTTTPLYV-IVSGYALNVAMDPLLIFYFKLGIRGAAISHVLSQYIMATL-L 301
M G RG +T P+ + +S + + + +++ YF+LG+ GA I + +TL L
Sbjct: 382 MNGALRGTGNTLPPMIITFISIWTARLPVAFVMVKYFQLGLLGAWIGMIADIIFRSTLKL 441
Query: 302 LFILMKK 308
LF L K
Sbjct: 442 LFFLSGK 448
>YD38_YEAST (Q05497) Hypothetical 77.8 kDa protein in MRPS28-HXT7
intergenic region
Length = 695
Score = 51.2 bits (121), Expect = 6e-06
Identities = 78/303 (25%), Positives = 121/303 (39%), Gaps = 27/303 (8%)
Query: 226 KYLRLRALGAPAVLLS------LAMQGIFRGFKDTTTPLYVIVSGYALNVAMDPLLIF-- 277
++LR+ LGAPA + L QGIF +YV+ LNV + L++
Sbjct: 349 RFLRVLILGAPAYIFFENLKRFLQAQGIF------DAGIYVLTICAPLNVLVSYTLVWNK 402
Query: 278 YFKLGIRGAAISHVLSQYIMATLLLFILMKKVDLLPPSMKDLQIFRFLKNGGLLLARVIA 337
Y +G GAAI+ VL+ ++M LLLF + + F + G L I
Sbjct: 403 YIGVGFIGAAIAVVLNFWLMFFLLLFYALYIDGRKCWGGFSRKAFTHWNDLGHLAFSGII 462
Query: 338 VTFCVTLSASL----AARLGPIPMAAFQTCLQVWMTSSLLADGLAVAIQAILACSFAEKD 393
+ LS L +A G +AA + ++ + ++ +A K
Sbjct: 463 MLEAEELSYELLTLFSAYYGVSYLAAQSAVSTMAALLYMIPFAIGISTSTRIANFIGAKR 522
Query: 394 YNKVTTAATRTLQMSFVLGVGLSLVVGGGLYFGAGVFSKNVAVIHLIRLGLPFVAATQPI 453
+ ++ L SF+ G ++ G A ++SK+ VI LI LP V Q
Sbjct: 523 TDFAHISSQVGLSFSFIAGFINCCILVFGRNLIANIYSKDPEVIKLIAQVLPLVGIVQNF 582
Query: 454 NSLAFVF------DGVNYGASDFAYSAYSLVMVSIASVTSLFFLYKSKGFIGIWIALTIY 507
+SL V G+ S AY L + +A + S FF K G+WI +
Sbjct: 583 DSLNAVAGSCLRGQGMQSLGSIVNLMAYYLFGIPLALILSWFFDMK---LYGLWIGIGSA 639
Query: 508 MSL 510
M L
Sbjct: 640 MLL 642
>NORM_VIBVU (Q8D9N8) Probable multidrug resistance protein norM
(Na(+)/drug antiporter) (Multidrug-efflux transporter)
Length = 483
Score = 47.0 bits (110), Expect = 1e-04
Identities = 81/349 (23%), Positives = 137/349 (39%), Gaps = 39/349 (11%)
Query: 190 GTVLGLIQAATLI---FAAKPLLGAMGLKYDSPMLVPAVKYLRLRALGAPAVLLSLAMQG 246
G V+ L+ + +I F + +LG M + D+ M + Y+ PA LL ++
Sbjct: 118 GAVMALLISIPIIGVLFQTQWILGYMNV--DAVMATKTIGYIHAVMFAVPAFLLFQTLRS 175
Query: 247 IFRGFKDTTTPLYVIVSGYALNVAMDPLLIFYFKLGIR-----GAAISHVLSQYIMATLL 301
+ G T + + G LN+ ++ + + Y KLG G ++ + +IM LL
Sbjct: 176 LTDGLSLTKPAMVIGFIGLLLNIPLNWMFV-YGKLGAPALGGVGCGVATAIVYWIMFLLL 234
Query: 302 LFIL-----MKKVDLLP---PSMKDLQIFRFLKNGGLLLARVIAVTFCVTLSASLAARLG 353
LF + +++V L P + Q+ + K G + A + + A L A LG
Sbjct: 235 LFYVTTSHRLRQVQLFTTFHPPQLNAQV-KLFKLGFPVAAALFFEVTLFAVVALLVAPLG 293
Query: 354 PIPMAAFQTCLQVWMTSSLLADGLAVAIQAILACSFAEKDYNKVTTAATRTLQMSFVLGV 413
+AA Q + +L + A + EK +T R +L V
Sbjct: 294 STVVAAHQVAINFSSLVFMLPMSIGAATSIRVGHMLGEK-----STEGARIASHVGIL-V 347
Query: 414 GLSLVVGGGLYFG------AGVFSKNVAVIHLIRLGLPFVAATQPINSLAFVFDGVNYGA 467
GLS V L A +++ N VI L L F A Q +++ + G G
Sbjct: 348 GLSTAVFTALLTVILREQIALLYTDNRVVITLAMQLLIFTAIYQCTDAIQVIAAGALRGY 407
Query: 468 SDF------AYSAYSLVMVSIASVTSLF-FLYKSKGFIGIWIALTIYMS 509
D + AY L+ + V L ++ + G G WI + +S
Sbjct: 408 KDMRAIFNRTFIAYWLLGLPTGYVLGLTDWIVEPMGAQGFWIGFIVGLS 456
>YEEO_ECOLI (P76352) Hypothetical protein yeeO
Length = 547
Score = 46.6 bits (109), Expect = 2e-04
Identities = 35/141 (24%), Positives = 68/141 (47%), Gaps = 5/141 (3%)
Query: 169 SESKPIRKKRHIASASTALLFGTVLGLIQAATLIFAAKPLLGAMGLKYDSPMLVPAVKYL 228
S K R++ +A+ + ++ T+ ++ A + + ++ + + + A+ YL
Sbjct: 158 SLGKRDRRRARVATRQSLVIM-TLFAVLLATLIHHFGEQIIDFVAGDATTEVKALALTYL 216
Query: 229 RLRALGAPAVLLSLAMQGIFRGFKDTTTPLYVIVSGYALNVAMDPLLIF----YFKLGIR 284
L L PA ++L G RG +T PL + S LN+ + +LI+ + LG
Sbjct: 217 ELTVLSYPAAAITLIGSGALRGAGNTKIPLLINGSLNILNIIISGILIYGLFSWPGLGFV 276
Query: 285 GAAISHVLSQYIMATLLLFIL 305
GA + +S+YI A +L++L
Sbjct: 277 GAGLGLTISRYIGAVAILWVL 297
>NORM_VIBPA (O82855) Multidrug resistance protein norM (Na(+)/drug
antiporter) (Multidrug-efflux transporter)
Length = 456
Score = 41.6 bits (96), Expect = 0.005
Identities = 52/269 (19%), Positives = 101/269 (37%), Gaps = 21/269 (7%)
Query: 217 DSPMLVPAVKYLRLRALGAPAVLLSLAMQGIFRGFKDTTTPLYVIVSGYALNVAMDPLLI 276
++ M V Y+ PA LL ++ G T + + G LN+ ++ + +
Sbjct: 119 EAVMAGKTVGYIHAVIFAVPAFLLFQTLRSFTDGMSLTKPAMVIGFIGLLLNIPLNWIFV 178
Query: 277 F----YFKLGIRGAAISHVLSQYIMATLLLFILMKKVDLLPPSMKDLQIF---------- 322
+ +LG G ++ + ++M LLL +M +K + +F
Sbjct: 179 YGKFGAPELGGVGCGVATTIVYWVMFALLLAYVMTS-----SRLKSINVFGEYHKPQWKA 233
Query: 323 --RFLKNGGLLLARVIAVTFCVTLSASLAARLGPIPMAAFQTCLQVWMTSSLLADGLAVA 380
R K G + A + + A L + LGPI +AA Q + +L + A
Sbjct: 234 QVRLFKLGFPVAAALFFEVTLFAVVALLVSPLGPIIVAAHQVAINFSSLVFMLPMSVGAA 293
Query: 381 IQAILACSFAEKDYNKVTTAATRTLQMSFVLGVGLSLVVGGGLYFGAGVFSKNVAVIHLI 440
+ + E++ + A+ + + L +++ A +++ N VI L
Sbjct: 294 VSIRVGHRLGEENVDGARVASRVGIMVGLALATITAIITVLSRELIAELYTNNPEVISLA 353
Query: 441 RLGLPFVAATQPINSLAFVFDGVNYGASD 469
L F A Q +++ + G G D
Sbjct: 354 MQLLLFAAVYQCTDAVQVIAAGALRGYKD 382
>NORM_VIBCH (Q9KRU4) Probable multidrug resistance protein norM
(Na(+)/drug antiporter) (Multidrug-efflux transporter)
Length = 461
Score = 41.6 bits (96), Expect = 0.005
Identities = 63/296 (21%), Positives = 112/296 (37%), Gaps = 20/296 (6%)
Query: 190 GTVLGLIQAATLI---FAAKPLLGAMGLKYDSPMLVPAVKYLRLRALGAPAVLLSLAMQG 246
G +L L+ + +I F + ++ M + + M V Y+ PA LL A++
Sbjct: 95 GLILALLVSVPIIAVLFQTQFIIRFMDV--EEAMATKTVGYMHAVIFAVPAYLLFQALRS 152
Query: 247 IFRGFKDTTTPLYVIVSGYALNVAMDPLLIF----YFKLGIRGAAISHVLSQYIMATLLL 302
G T + + G LN+ ++ + ++ +LG G ++ + +IM LLL
Sbjct: 153 FTDGMSLTKPAMVIGFIGLLLNIPLNWIFVYGKFGAPELGGVGCGVATAIVYWIMLLLLL 212
Query: 303 FILMKKVDLL---------PPSMKDLQIFRFLKNGGLLLARVIAVTFCVTLSASLAARLG 353
F ++ L P K+L R + G + A + + A L A LG
Sbjct: 213 FYIVTSKRLAHVKVFETFHKPQPKEL--IRLFRLGFPVAAALFFEVTLFAVVALLVAPLG 270
Query: 354 PIPMAAFQTCLQVWMTSSLLADGLAVAIQAILACSFAEKDYNKVTTAATRTLQMSFVLGV 413
+AA Q L + + A+ + E+D AA L
Sbjct: 271 STVVAAHQVALNFSSLVFMFPMSIGAAVSIRVGHKLGEQDTKGAAIAANVGLMTGLATAC 330
Query: 414 GLSLVVGGGLYFGAGVFSKNVAVIHLIRLGLPFVAATQPINSLAFVFDGVNYGASD 469
+L+ A ++++N V+ L L F A Q ++++ V G G D
Sbjct: 331 ITALLTVLFREQIALLYTENQVVVALAMQLLLFAAIYQCMDAVQVVAAGSLRGYKD 386
>NORM_DEIRA (Q9RY44) Probable multidrug resistance protein norM
(Na(+)/drug antiporter) (Multidrug-efflux transporter)
Length = 451
Score = 41.6 bits (96), Expect = 0.005
Identities = 58/210 (27%), Positives = 90/210 (42%), Gaps = 24/210 (11%)
Query: 186 ALLFGTVLGLIQAATLIFAAKPLLGAMGLKYDSPMLVPA-------VKYLRLRALGAPAV 238
AL G L L+ +A ++ PL+ A L + P PA YLR+ +LG
Sbjct: 94 ALGGGLRLALLLSAVML----PLMWA--LSFVLPNFAPAGVSRDLVAAYLRVYSLGMLPN 147
Query: 239 LLSLAMQGIFRGFKDTTTPLYVIVSGYALNVAMDPLLIFYF----KLGIRGAAISHVLSQ 294
L +A++G G V ++G + + P L F + +LG+ GAA + +
Sbjct: 148 LAFIALRGTLEGTGKPGAVTGVALTGVVWALLVAPALAFGWGPLPRLGLAGAAGASASAA 207
Query: 295 YIMATLLLFILMKKV----DLLPPSMKDLQIFRFLKNGGLLLARVIAVTFCVTLSASLAA 350
+IMA LL + ++V L P + +FR GL L + TL L A
Sbjct: 208 WIMAALLWPLARRRVAYAGPLGPLGDEVRALFRLGWPIGLTLGAEGGMFSVTTL---LMA 264
Query: 351 RLGPIPMAAFQTCLQVWMTSSLLADGLAVA 380
R GP +AA +Q ++ G+A A
Sbjct: 265 RFGPEVLAAHNVTMQTITAFFMVPLGIASA 294
Score = 31.2 bits (69), Expect = 6.8
Identities = 33/116 (28%), Positives = 51/116 (43%), Gaps = 12/116 (10%)
Query: 180 IASASTALLFGTVLGLIQAATLIFAAKPLLG-----AMGLKYDSP----MLVPAVKYLRL 230
+A A A L G LGL A L FA L ++ + + P ++ A +L +
Sbjct: 308 LAQARRAGLVG--LGLSSAVMLTFAVIELAAPRTVFSVFVNVNDPANAGLIAAATGFLSI 365
Query: 231 RALGAPAVLLSLAMQGIFRGFKDTTTPLYV-IVSGYALNVAMDPLLIFYFKLGIRG 285
AL L + G RG +DT PL V +V+ + + + + +L LG RG
Sbjct: 366 AALFQLMDGLQVTANGALRGLQDTRVPLLVSLVAYWVVGLGLGSVLSSVAGLGARG 421
>NORM_SYNY3 (Q55364) Probable multidrug resistance protein norM
(Na(+)/drug antiporter) (Multidrug-efflux transporter)
Length = 461
Score = 41.2 bits (95), Expect = 0.007
Identities = 28/79 (35%), Positives = 41/79 (51%), Gaps = 1/79 (1%)
Query: 20 ECSLVFKMDSLAKEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAAAGVSIAVFNQ 79
+ S+ F M + A+E L +A P A A A +DT +G LG LAA G++ +F
Sbjct: 3 DISMRFGMQAEAREFLKLAVPLAGAQMAQAATGFVDTVMMGWLGQDVLAAGGLATMIF-M 61
Query: 80 ASRITIFPLVSITTSFVAE 98
A +T L+S + VAE
Sbjct: 62 AFMMTGVGLISGVSPLVAE 80
Score = 34.3 bits (77), Expect = 0.80
Identities = 63/315 (20%), Positives = 128/315 (40%), Gaps = 29/315 (9%)
Query: 227 YLRLRALGAPAVLLSLAMQGIFRGFKDTTTPLYVIVSGYALNVAMDPLLIF----YFKLG 282
YL + A G + ++G + ++++ N+ + L F + LG
Sbjct: 135 YLNVMAWGLFPAIAFATLRGCIIALSQARPIMLIVIAATLFNILGNYGLGFGKWGFPALG 194
Query: 283 IRGAAISHVLSQYIMATLLLFILMKKVDLLPPSMKD---------LQIFRFLKNGGLLLA 333
I G AI+ + + +IM LL ++ L S+ D LQ ++ G + +A
Sbjct: 195 ITGLAIASISAHWIMFLSLLVYMLWHKPLHQYSLFDSLHHLKPRILQQLLWV-GGPIGIA 253
Query: 334 RVIAVTFCVTLSASLAARLGPIPMAAFQTCLQVWMTSSLLADGLAVAIQAILACSFAEKD 393
V+ +T S + A LG +AA Q Q + ++ ++ A + F ++
Sbjct: 254 AVLEYGLYLTASFFMGA-LGTPILAAHQVVSQTVLVLFMVPLAMSYAATVRVGQWFGQQH 312
Query: 394 YNKVTTAATRTLQMS--FVLGVGLSLVVGGGLYFGAGVFSKNVAVIHLIRLGLPFV---A 448
+ ++ AA ++ ++ F+L G++L+ G + + A + +G+ + A
Sbjct: 313 WPQIRQAALVSIGLAVLFMLTAGIALLAYPQQIIGLYLDLNDPANGEALNVGISIMKIAA 372
Query: 449 ATQPINSLAFVFDGVNYGASDFAYSAYSLVMVSIA------SVTSLFFLYKSKGFIGIWI 502
++ L +GV G D + +V+ +IA + + L Y G+WI
Sbjct: 373 FGLVLDGLQRTANGVLQGLQD---TRIPMVLGTIAYWGIGLTASYLLGFYTPLSGAGVWI 429
Query: 503 ALTIYMSLRMFAGVW 517
I ++ A +W
Sbjct: 430 GTYIGLAAASIAYLW 444
>MVIN_BDEBA (Q8VNZ2) Virulence factor mviN homolog
Length = 523
Score = 41.2 bits (95), Expect = 0.007
Identities = 48/200 (24%), Positives = 85/200 (42%), Gaps = 24/200 (12%)
Query: 176 KKRHIASASTALLFGTVLGLIQAATLIFAAKPLLGAMGL--KYDSPMLVPAVKYLRLRAL 233
K++ +A + L L A L A+P++ + L K+ + LR+ A+
Sbjct: 313 KEKFQETAEESFLMNLFLAWPAALGLYILAEPIIEVLFLRGKFTVQDVQMTAAILRIYAV 372
Query: 234 GAPAVLLSLAMQGIFRGFKDTTTPLYVIVSGYALNVAMDPLLIFYFKLGIRGAAISHVLS 293
V S + ++ K+T P+ + + A++V++ P+L+ + G+ G IS V++
Sbjct: 373 SLLLVSCSRVLMPLYYSVKNTKVPMVLALVSLAVHVSLAPVLMRQW--GLEGLMISGVVA 430
Query: 294 QYIMATLLLFILMKKVD------LLPPSMK----------DLQIFRFLK----NGGLLLA 333
I A LL+ +L K LL P++K LQ + L G +LA
Sbjct: 431 ALINAVLLMGLLKKYSPGIRMSVLLRPALKFVLAGAGMVISLQAYELLMAQTGRGLQMLA 490
Query: 334 RVIAVTFCVTLSASLAARLG 353
+ + V LA LG
Sbjct: 491 LFVTILLAVVAYFGLAYVLG 510
>MVIN_BUCAP (Q8K9L3) Virulence factor mviN homolog
Length = 514
Score = 40.4 bits (93), Expect = 0.011
Identities = 51/189 (26%), Positives = 87/189 (45%), Gaps = 16/189 (8%)
Query: 156 ISKSSIVTNSSNKSESKPIRKKRHIASASTALLFGTVLGLIQAATLIFAAKPL---LGAM 212
+S S+I+ S + S S + + I + + FG +L L + L +KPL L
Sbjct: 281 VSLSTILFTSFSCSYSNNTQSEYKIL-LNWGIRFGLILSLPISVILFMFSKPLVIILFQY 339
Query: 213 GLKYDSPMLVPAVKYLRLRALGAPAVLLSLAMQGIFRGFKDTTTPLYVIVSGYALNVAMD 272
G D +L+ K L L + G + +L + F ++ P+ + + L M+
Sbjct: 340 GKFTDFDVLMTQ-KALELYSFGLVSFILVKILVSAFYSCQEVNIPMRISILTLFLTQLMN 398
Query: 273 PLLIFYFKLGIRGAAISHVLSQYIMATLLLFILMKKVDLLPPSMKD-LQIFRFLKNGGLL 331
P LIFYF+ G A+S ++ +I LL + L +K ++ + D + IFR L
Sbjct: 399 PFLIFYFQHS--GLALSCSIASWINFFLLYWKLYQK-GIINFKLNDFIFIFR-------L 448
Query: 332 LARVIAVTF 340
L V+ +TF
Sbjct: 449 LIAVLVMTF 457
>NOM1_PSEAE (Q9I3Y3) Probable multidrug resistance protein norM 1
(Na(+)/drug antiporter) (Multidrug-efflux transporter)
Length = 477
Score = 40.0 bits (92), Expect = 0.015
Identities = 25/92 (27%), Positives = 49/92 (53%), Gaps = 2/92 (2%)
Query: 186 ALLFGTVLGLIQAATLIFAAKPLLGAMGLKYDSPMLVPAVKYLRLRALGAPAVLLSLAMQ 245
AL ++G + A L + ++P+LG M ++ + ++ P++ YL+ ALG PA L ++
Sbjct: 101 ALWLALLIGPLSGAVLWWLSEPILGLMKVRPE--LIGPSLLYLKGIALGFPAAALYHVLR 158
Query: 246 GIFRGFKDTTTPLYVIVSGYALNVAMDPLLIF 277
G T + + + G LN+ ++ LI+
Sbjct: 159 CYTNGLGRTRPSMVLGIGGLLLNIPINYALIY 190
>Y447_MYCPN (P75130) Hypothetical protein MG447 homolog (K05_orf499)
Length = 549
Score = 38.9 bits (89), Expect = 0.032
Identities = 37/175 (21%), Positives = 71/175 (40%), Gaps = 18/175 (10%)
Query: 267 LNVAMDPLLIFYFKLGIRGAAISHVLSQYI--MATLLLFILMKKVDLLPPSMKDLQIFR- 323
LNV LL+ + LG+ G+A++ +L +I MA ++ I + K L S++D R
Sbjct: 244 LNVLFVFLLVRFSTLGVVGSAVAAILVYFITFMAYVVYLISLNKRGLTYLSLRDFSFKRV 303
Query: 324 ---------------FLKNGGLLLARVIAVTFCVTLSASLAARLGPIPMAAFQTCLQVWM 368
F +NG L + +F V L+ +L + + + +
Sbjct: 304 SFNLFLVISMVGLASFFRNGSLSILNTFYESFLVNLTKTLTDQSDTFYLVLLTGPIAIAN 363
Query: 369 TSSLLADGLAVAIQAILACSFAEKDYNKVTTAATRTLQMSFVLGVGLSLVVGGGL 423
+S G+ ++ +++ F + + TL + V L L++ GL
Sbjct: 364 LTSAAIFGVLQGVRTVVSYKFGQGQLADIKRINVYTLLVCLVFAALLYLILAVGL 418
>NORM_SALTY (Q8ZPP2) Multidrug resistance protein norM (Na(+)/drug
antiporter) (Multidrug-efflux transporter)
Length = 457
Score = 38.1 bits (87), Expect = 0.055
Identities = 69/313 (22%), Positives = 124/313 (39%), Gaps = 28/313 (8%)
Query: 217 DSPMLVPAVKYLRLRALGAPAVLLSLAMQGIFRGFKDTTTPLYVIVSGYALNVAMDPLLI 276
D + AV YLR GAP L + G T + + G +N+ ++ + I
Sbjct: 120 DPALADKAVGYLRALLWGAPGYLFFQVARNQCEGLAKTKPGMVMGFLGLLVNIPVNYIFI 179
Query: 277 F-YF---KLGIRGAAISHVLSQYIMATLLLFILMKKVDLLPPSMKDLQIFR-FLKNGGLL 331
+ +F +LG G ++ ++M FI M SM+D++ + F K ++
Sbjct: 180 YGHFGMPELGGIGCGVATAAVYWVM-----FIAMLSYIKHARSMRDIRNEKGFGKPDSVV 234
Query: 332 LARVI--------AVTFCVTLSASLA---ARLGPIPMAAFQTCLQVWMTSSLLADGLAVA 380
+ R+I A+ F VTL A +A + LG + +A Q L +L LA A
Sbjct: 235 MKRLIQLGLPIALALFFEVTLFAVVALLVSPLGIVDVAGHQIALNFSSLMFVLPMSLAAA 294
Query: 381 IQAILACSFAEKDYNKVTTAATRTLQMSFVLGVGLSLVVGGGLYFGAGVFSKNVAVIHLI 440
+ + + TAA L + + V ++ A +++ N V+ L
Sbjct: 295 VTIRVGYRLGQGSTLDAQTAARTGLGVGICMAVVTAIFTVTLRKHIALLYNDNPEVVALA 354
Query: 441 RLGLPFVAATQPINSLAFVFDGVNYGASD------FAYSAYSLVMVSIASVTSLFFLYKS 494
+ A Q +S+ + G+ G D ++AY ++ + + +L L
Sbjct: 355 AQLMLLAAVYQISDSIQVIGSGILRGYKDTRSIFFITFTAYWVLGLPSGYILALTDLVVD 414
Query: 495 K-GFIGIWIALTI 506
+ G G W+ I
Sbjct: 415 RMGPAGFWMGFII 427
>NORM_SALTI (Q8Z6N7) Multidrug resistance protein norM (Na(+)/drug
antiporter) (Multidrug-efflux transporter)
Length = 457
Score = 38.1 bits (87), Expect = 0.055
Identities = 69/313 (22%), Positives = 124/313 (39%), Gaps = 28/313 (8%)
Query: 217 DSPMLVPAVKYLRLRALGAPAVLLSLAMQGIFRGFKDTTTPLYVIVSGYALNVAMDPLLI 276
D + AV YLR GAP L + G T + + G +N+ ++ + I
Sbjct: 120 DPALADKAVGYLRALLWGAPGYLFFQVARNQCEGLAKTKPGMVMGFLGLLVNIPVNYIFI 179
Query: 277 F-YF---KLGIRGAAISHVLSQYIMATLLLFILMKKVDLLPPSMKDLQIFR-FLKNGGLL 331
+ +F +LG G ++ ++M FI M SM+D++ + F K ++
Sbjct: 180 YGHFGMPELGGIGCGVATAAVYWVM-----FIAMLSYIKHARSMRDIRNEKGFGKPDSVV 234
Query: 332 LARVI--------AVTFCVTLSASLA---ARLGPIPMAAFQTCLQVWMTSSLLADGLAVA 380
+ R+I A+ F VTL A +A + LG + +A Q L +L LA A
Sbjct: 235 MKRLIQLGLPIALALFFEVTLFAVVALLVSPLGIVDVAGHQIALNFSSLMFVLPMSLAAA 294
Query: 381 IQAILACSFAEKDYNKVTTAATRTLQMSFVLGVGLSLVVGGGLYFGAGVFSKNVAVIHLI 440
+ + + TAA L + + V ++ A +++ N V+ L
Sbjct: 295 VTIRVGYRLGQGSTLDAQTAARTGLGVGICMAVVTAIFTVTLRKHIALLYNDNPEVVALA 354
Query: 441 RLGLPFVAATQPINSLAFVFDGVNYGASD------FAYSAYSLVMVSIASVTSLFFLYKS 494
+ A Q +S+ + G+ G D ++AY ++ + + +L L
Sbjct: 355 AQLMLLAAVYQISDSIQVIGSGILRGYKDTRSIFFITFTAYWVLGLPSGYILALTDLVVD 414
Query: 495 K-GFIGIWIALTI 506
+ G G W+ I
Sbjct: 415 RMGPAGFWMGFII 427
Score = 31.2 bits (69), Expect = 6.8
Identities = 14/52 (26%), Positives = 25/52 (47%)
Query: 26 KMDSLAKEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAAAGVSIAVF 77
K S A+++L +A P LA A +DT G ++AA + +++
Sbjct: 3 KYTSEARQLLALAIPVILAQVAQTAMGFVDTVMAGGYSATDMAAVAIGTSIW 54
>NORM_PASMU (Q9CMZ9) Probable multidrug resistance protein norM
(Na(+)/drug antiporter) (Multidrug-efflux transporter)
Length = 464
Score = 38.1 bits (87), Expect = 0.055
Identities = 39/138 (28%), Positives = 66/138 (47%), Gaps = 10/138 (7%)
Query: 185 TALLFGTVLGLIQAA-TLIFAAKPLLGAMGLKYDSPMLVPAVKYLRLRALGAPAVLLSLA 243
+AL+ G ++ +I A T+IF + + + +K D ++ A L + AL + + +
Sbjct: 322 SALIVGLLIAVITATLTVIFRVE--IAEIFVK-DRDVIAMAGTLLLIAALYQFSDTVQVV 378
Query: 244 MQGIFRGFKDTTTPLYVI-----VSGYALNVAMDPLLIFYFKLGIRGAAISHVLSQYIMA 298
G RG+KDT LY+ V G + + + LG G I V+S I A
Sbjct: 379 AGGALRGYKDTKAILYITLFCYWVVGMPMGYTLARTDLLMPALGAEGFWIGFVVSLTIAA 438
Query: 299 TLLLFILMKKVDLLPPSM 316
TLL+ I M+K+ P ++
Sbjct: 439 TLLM-IRMRKIQAQPDAI 455
Score = 37.4 bits (85), Expect = 0.094
Identities = 68/317 (21%), Positives = 119/317 (37%), Gaps = 35/317 (11%)
Query: 225 VKYLRLRALGAPAVLLSLAMQGIFRGFKDTTTPLYVIVSGYALNVAMDPLLIFYFKLGIR 284
+ YL G PA LL + + + G T + + G LN+ ++ + I Y KLGI
Sbjct: 132 IGYLHAMIWGLPAYLLMINFRCLNDGIAKTKPAMVITFLGLGLNIPLNYIFI-YGKLGIP 190
Query: 285 -----GAAISHVLSQYIMATLLLFILMKKVDLLPPSMKDLQIF-------------RFLK 326
G I+ + + M L++ + +DL++F + L
Sbjct: 191 AFGAVGCGIATAIVNWFMCILMIAYCKN-----ARNQRDLKVFAKILEKPNFTTLKKLLN 245
Query: 327 NGGLLLARVIAVTFCVTLSASLAARLGPIPMAAFQTCLQVWMTSSLLADGLAVAIQAILA 386
G + + L+A L + LG +A+ Q L +L L +A ++
Sbjct: 246 LGFPIAVALFCEVALFALTALLLSPLGTDIVASHQIALNTSSFLFMLPMSLGMAATILVG 305
Query: 387 CSFAEKDYNKVTTAATRTLQMSFVLGVGLSLVVGGGLYFGAGVFSKNVAVIHLIRLGLPF 446
E +K + L + ++ V + + A +F K+ VI + L
Sbjct: 306 QRLGEGAADKAKQVSYSALIVGLLIAVITATLTVIFRVEIAEIFVKDRDVIAMAGTLLLI 365
Query: 447 VAATQPINSLAFVFDGVNYGASD---------FAYSAYSLVM-VSIASVTSLFFLYKSKG 496
A Q +++ V G G D F Y + M ++A L ++G
Sbjct: 366 AALYQFSDTVQVVAGGALRGYKDTKAILYITLFCYWVVGMPMGYTLARTDLLMPALGAEG 425
Query: 497 F-IGIWIALTIYMSLRM 512
F IG ++LTI +L M
Sbjct: 426 FWIGFVVSLTIAATLLM 442
Score = 31.6 bits (70), Expect = 5.2
Identities = 20/59 (33%), Positives = 29/59 (48%), Gaps = 8/59 (13%)
Query: 31 AKEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAAAGVSIAVFNQASRITIFPLV 89
AK++L IA P LA A L+DT G + ++AA V +++ FPLV
Sbjct: 13 AKKLLLIALPILLAQIAQNSMGLVDTIMSGRVSSADMAAISVGASIW--------FPLV 63
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.327 0.139 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 50,311,105
Number of Sequences: 164201
Number of extensions: 1936164
Number of successful extensions: 7699
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 40
Number of HSP's that attempted gapping in prelim test: 7622
Number of HSP's gapped (non-prelim): 113
length of query: 517
length of database: 59,974,054
effective HSP length: 115
effective length of query: 402
effective length of database: 41,090,939
effective search space: 16518557478
effective search space used: 16518557478
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 68 (30.8 bits)
Medicago: description of AC122162.1


