

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122160.9 + phase: 0
(435 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
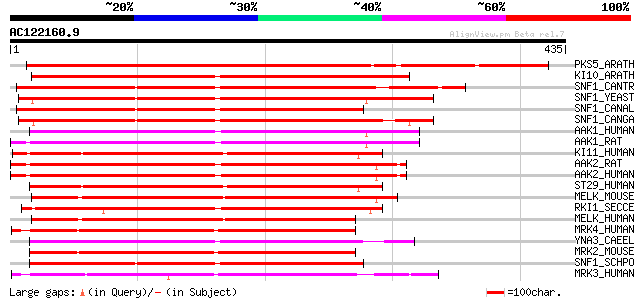
Score E
Sequences producing significant alignments: (bits) Value
PKS5_ARATH (O22932) SOS2-like protein kinase PKS5 (EC 2.7.1.37) 396 e-110
KI10_ARATH (Q38997) SNF1-related protein kinase KIN10 (EC 2.7.1.... 250 4e-66
SNF1_CANTR (O94168) Carbon catabolite derepressing protein kinas... 247 4e-65
SNF1_YEAST (P06782) Carbon catabolite derepressing protein kinas... 241 3e-63
SNF1_CANAL (P52497) Carbon catabolite derepressing protein kinas... 239 8e-63
SNF1_CANGA (Q00372) Carbon catabolite derepressing protein kinas... 236 1e-61
AAK1_HUMAN (Q13131) 5'-AMP-activated protein kinase, catalytic a... 233 7e-61
AAK1_RAT (P54645) 5'-AMP-activated protein kinase, catalytic alp... 233 9e-61
KI11_HUMAN (Q8TDC3) Probable serine/threonine-protein kinase KIA... 232 1e-60
AAK2_RAT (Q09137) 5'-AMP-activated protein kinase, catalytic alp... 232 1e-60
AAK2_HUMAN (P54646) 5'-AMP-activated protein kinase, catalytic a... 232 1e-60
ST29_HUMAN (Q8IWQ3) Serine/threonine-protein kinase 29 (EC 2.7.1... 228 2e-59
MELK_MOUSE (Q61846) Maternal embryonic leucine zipper kinase (EC... 224 3e-58
RKI1_SECCE (Q02723) Carbon catabolite derepressing protein kinas... 224 4e-58
MELK_HUMAN (Q14680) Maternal embryonic leucine zipper kinase (EC... 220 5e-57
MRK4_HUMAN (Q96L34) MAP/microtubule affinity-regulating kinase 4... 218 2e-56
YNA3_CAEEL (P45894) Putative serine/threonine-protein kinase PAR... 217 4e-56
MRK2_MOUSE (Q05512) MAP/microtubule affinity-regulating kinase 2... 214 3e-55
SNF1_SCHPO (O74536) SNF1-like protein kinase ssp2 (EC 2.7.1.-) 211 4e-54
MRK3_HUMAN (P27448) MAP/microtubule affinity-regulating kinase 3... 207 4e-53
>PKS5_ARATH (O22932) SOS2-like protein kinase PKS5 (EC 2.7.1.37)
Length = 435
Score = 396 bits (1018), Expect = e-110
Identities = 207/412 (50%), Positives = 283/412 (68%), Gaps = 10/412 (2%)
Query: 14 LDGKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVI-KVGMIEQIKREISVMKM 72
L GKYELG++LG G FAKV+HA++ TG++VA+K++ K+K++ + IKREIS+M+
Sbjct: 17 LFGKYELGKLLGCGAFAKVFHARDRRTGQSVAVKILNKKKLLTNPALANNIKREISIMRR 76
Query: 73 VKHPNIVQLHEVMASKSKIYIAMELVRGGELFNKIVK-GRLKEDVARVYFQQLISAVDFC 131
+ HPNIV+LHEVMA+KSKI+ AME V+GGELFNKI K GRL ED++R YFQQLISAV +C
Sbjct: 77 LSHPNIVKLHEVMATKSKIFFAMEFVKGGELFNKISKHGRLSEDLSRRYFQQLISAVGYC 136
Query: 132 HSRGVYHRDLKPENLLLDEDGNLKVSDFGLCTFSEHLRQDGLLHTTCGTPAYVSPEVIAK 191
H+RGVYHRDLKPENLL+DE+GNLKVSDFGL ++ +R DGLLHT CGTPAYV+PE+++K
Sbjct: 137 HARGVYHRDLKPENLLIDENGNLKVSDFGLSALTDQIRPDGLLHTLCGTPAYVAPEILSK 196
Query: 192 KGYDGAKADIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKCPPWFSLEARRLITK 251
KGY+GAK D+WSCG++L+VL+AG+LPF D N++ MYKKIY+G+++ P W S + +R +++
Sbjct: 197 KGYEGAKVDVWSCGIVLFVLVAGYLPFNDPNVMNMYKKIYKGEYRFPRWMSPDLKRFVSR 256
Query: 252 LLDPNPNTRISISKIMDSSWFKKPVPKSIAMRKKEKEEEEDKVFEFMECEKSSTTMNAFH 311
LLD NP TRI+I +I+ WF + K I + E E+ KV +E KS +NAF
Sbjct: 257 LLDINPETRITIDEILKDPWFVRGGFKQIKFH--DDEIEDQKVESSLEAVKS---LNAFD 311
Query: 312 IISLSEGFDLSPLFEEKKREEREEMRFATGGTPSRVISKLEQVAKAVKFDVKSSDTKVRL 371
+IS S G DLS LF E RF + +P + ++E A+ +K K
Sbjct: 312 LISYSSGLDLSGLFAGCSNSSGESERFLSEKSPEMLAEEVEGFAREENLRMKKK--KEEE 369
Query: 372 QGQE-RGRKGKLAIAADIYAVTPSFLVVEVKKDKGDTLEYNQFCSKELRPAL 422
G E G+ GK I I + +VVE ++ GD Y + + +LR L
Sbjct: 370 YGFEMEGQNGKFGIGICISRLNDLLVVVEARRRGGDGDCYKEMWNGKLRVQL 421
>KI10_ARATH (Q38997) SNF1-related protein kinase KIN10 (EC 2.7.1.-)
(AKIN10)
Length = 535
Score = 250 bits (639), Expect = 4e-66
Identities = 129/297 (43%), Positives = 195/297 (65%), Gaps = 4/297 (1%)
Query: 18 YELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQIKREISVMKMVKHPN 77
Y+LGR LG G+F +V A++ TG VA+K++ + K+ + M E+++REI ++++ HP+
Sbjct: 42 YKLGRTLGIGSFGRVKIAEHALTGHKVAIKILNRRKIKNMEMEEKVRREIKILRLFMHPH 101
Query: 78 IVQLHEVMASKSKIYIAMELVRGGELFNKIV-KGRLKEDVARVYFQQLISAVDFCHSRGV 136
I++L+EV+ + + IY+ ME V GELF+ IV KGRL+ED AR +FQQ+IS V++CH V
Sbjct: 102 IIRLYEVIETPTDIYLVMEYVNSGELFDYIVEKGRLQEDEARNFFQQIISGVEYCHRNMV 161
Query: 137 YHRDLKPENLLLDEDGNLKVSDFGLCTFSEHLRQDGLLHTTCGTPAYVSPEVIAKKGYDG 196
HRDLKPENLLLD N+K++DFGL S +R L T+CG+P Y +PEVI+ K Y G
Sbjct: 162 VHRDLKPENLLLDSKCNVKIADFGL---SNIMRDGHFLKTSCGSPNYAAPEVISGKLYAG 218
Query: 197 AKADIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKCPPWFSLEARRLITKLLDPN 256
+ D+WSCGVILY LL G LPF D+N+ ++KKI G + P S AR LI ++L +
Sbjct: 219 PEVDVWSCGVILYALLCGTLPFDDENIPNLFKKIKGGIYTLPSHLSPGARDLIPRMLVVD 278
Query: 257 PNTRISISKIMDSSWFKKPVPKSIAMRKKEKEEEEDKVFEFMECEKSSTTMNAFHII 313
P R++I +I WF+ +P+ +A+ + ++ K+ E + E + + H+I
Sbjct: 279 PMKRVTIPEIRQHPWFQAHLPRYLAVPPPDTVQQAKKIDEEILQEVINMGFDRNHLI 335
>SNF1_CANTR (O94168) Carbon catabolite derepressing protein kinase
(EC 2.7.1.-)
Length = 619
Score = 247 bits (631), Expect = 4e-65
Identities = 133/353 (37%), Positives = 217/353 (60%), Gaps = 17/353 (4%)
Query: 6 NPNSHSALLDGKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQIKR 65
+PN + A G+Y++ + LG G+F KV A+++ TG+ VA+K++ ++ + K M +++R
Sbjct: 40 DPNVNPANRIGRYQIIKTLGEGSFGKVKLAQHVGTGQKVALKIINRKTLAKSDMQGRVER 99
Query: 66 EISVMKMVKHPNIVQLHEVMASKSKIYIAMELVRGGELFNKIV-KGRLKEDVARVYFQQL 124
EIS +++++HP+I++L++V+ SK +I + +E G ELF+ IV +G++ ED AR +FQQ+
Sbjct: 100 EISYLRLLRHPHIIKLYDVIKSKDEIIMVIEFA-GKELFDYIVQRGKMPEDEARRFFQQI 158
Query: 125 ISAVDFCHSRGVYHRDLKPENLLLDEDGNLKVSDFGLCTFSEHLRQDGLLHTTCGTPAYV 184
I+AV++CH + HRDLKPENLLLD+ N+K++DFGL S + L T+CG+P Y
Sbjct: 159 IAAVEYCHRHKIVHRDLKPENLLLDDQLNVKIADFGL---SNIMTDGNFLKTSCGSPNYA 215
Query: 185 SPEVIAKKGYDGAKADIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKCPPWFSLE 244
+PEVI+ K Y G + D+WS GVILYV+L G LPF D+ + A++KKI G + P + S
Sbjct: 216 APEVISGKLYAGPEVDVWSSGVILYVMLCGRLPFDDEFIPALFKKISNGVYTLPNYLSPG 275
Query: 245 ARRLITKLLDPNPNTRISISKIMDSSWFKKPVPKSIAMRKKEKEEEEDKVFEFMECEKSS 304
A+ L+T++L NP RI+I +IM+ WFK+ +P + K ++ K
Sbjct: 276 AKHLLTRMLVVNPLNRITIHEIMEDEWFKQDMPDYLLPPDLSK----------IKTSKID 325
Query: 305 TTMNAFHIISLSEGFDLSPLFEEKKREEREEMRFATGGTPSRVISKLEQVAKA 357
+ +S++ G+D + ++ RE A G TP+ +V A
Sbjct: 326 IDEDVISALSVTMGYDRDEIISVIEKANREAA--AGGATPTNQSKSTNEVLDA 376
>SNF1_YEAST (P06782) Carbon catabolite derepressing protein kinase
(EC 2.7.1.-)
Length = 633
Score = 241 bits (614), Expect = 3e-63
Identities = 131/335 (39%), Positives = 204/335 (60%), Gaps = 14/335 (4%)
Query: 8 NSHSALLDG----KYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQI 63
N S+L DG Y++ + LG G+F KV A + TG+ VA+K++ K+ + K M +I
Sbjct: 41 NPKSSLADGAHIGNYQIVKTLGEGSFGKVKLAYHTTTGQKVALKIINKKVLAKSDMQGRI 100
Query: 64 KREISVMKMVKHPNIVQLHEVMASKSKIYIAMELVRGGELFNKIV-KGRLKEDVARVYFQ 122
+REIS +++++HP+I++L++V+ SK +I + +E G ELF+ IV + ++ E AR +FQ
Sbjct: 101 EREISYLRLLRHPHIIKLYDVIKSKDEIIMVIEYA-GNELFDYIVQRDKMSEQEARRFFQ 159
Query: 123 QLISAVDFCHSRGVYHRDLKPENLLLDEDGNLKVSDFGLCTFSEHLRQDGLLHTTCGTPA 182
Q+ISAV++CH + HRDLKPENLLLDE N+K++DFGL S + L T+CG+P
Sbjct: 160 QIISAVEYCHRHKIVHRDLKPENLLLDEHLNVKIADFGL---SNIMTDGNFLKTSCGSPN 216
Query: 183 YVSPEVIAKKGYDGAKADIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKCPPWFS 242
Y +PEVI+ K Y G + D+WSCGVILYV+L LPF D+++ ++K I G + P + S
Sbjct: 217 YAAPEVISGKLYAGPEVDVWSCGVILYVMLCRRLPFDDESIPVLFKNISNGVYTLPKFLS 276
Query: 243 LEARRLITKLLDPNPNTRISISKIMDSSWFKKPVPK-----SIAMRKKEKEEEEDKVFEF 297
A LI ++L NP RISI +IM WFK +P+ + +E+ E D +
Sbjct: 277 PGAAGLIKRMLIVNPLNRISIHEIMQDDWFKVDLPEYLLPPDLKPHPEEENENNDSKKDG 336
Query: 298 MECEKSSTTMNAFHIISLSEGFDLSPLFEEKKREE 332
+ N +I+S + G++ ++E + E
Sbjct: 337 SSPDNDEIDDNLVNILSSTMGYEKDEIYESLESSE 371
>SNF1_CANAL (P52497) Carbon catabolite derepressing protein kinase
(EC 2.7.1.-)
Length = 620
Score = 239 bits (611), Expect = 8e-63
Identities = 120/274 (43%), Positives = 192/274 (69%), Gaps = 6/274 (2%)
Query: 6 NPNSHSALLDGKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQIKR 65
+P ++ A G+Y++ + LG G+F KV A++L TG+ VA+K++ ++ + K M +++R
Sbjct: 41 DPAANPANRIGRYQILKTLGEGSFGKVKLAQHLGTGQKVALKIINRKTLAKSDMQGRVER 100
Query: 66 EISVMKMVKHPNIVQLHEVMASKSKIYIAMELVRGGELFNKIV-KGRLKEDVARVYFQQL 124
EIS +++++HP+I++L++V+ SK +I + +E G ELF+ IV +G++ ED AR +FQQ+
Sbjct: 101 EISYLRLLRHPHIIKLYDVIKSKDEIIMVIEFA-GKELFDYIVQRGKMPEDEARRFFQQI 159
Query: 125 ISAVDFCHSRGVYHRDLKPENLLLDEDGNLKVSDFGLCTFSEHLRQDGLLHTTCGTPAYV 184
I+AV++CH + HRDLKPENLLLD+ N+K++DFGL S + L T+CG+P Y+
Sbjct: 160 IAAVEYCHRHKIVHRDLKPENLLLDDQLNVKIADFGL---SNIMTDGNFLKTSCGSPNYM 216
Query: 185 -SPEVIAKKGYDGAKADIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKCPPWFSL 243
+PEVI+ K Y G + D+WS GVILYV+L G LPF D+ + A++KKI G + P + S
Sbjct: 217 PAPEVISGKLYAGPEVDVWSAGVILYVMLCGRLPFDDEFIPALFKKISNGVYTLPNYLSA 276
Query: 244 EARRLITKLLDPNPNTRISISKIMDSSWFKKPVP 277
A+ L+T++L NP RI+I +IM+ WFK+ +P
Sbjct: 277 GAKHLLTRMLVVNPLNRITIHEIMEDDWFKQDMP 310
>SNF1_CANGA (Q00372) Carbon catabolite derepressing protein kinase
(EC 2.7.1.-)
Length = 612
Score = 236 bits (601), Expect = 1e-61
Identities = 132/337 (39%), Positives = 206/337 (60%), Gaps = 24/337 (7%)
Query: 8 NSHSALLDGK----YELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQI 63
N S+L DG Y++ + LG G+F KV A ++ TG+ VA+K++ K+ + K M +I
Sbjct: 25 NKVSSLADGSRVGNYQIVKTLGEGSFGKVKLAYHVTTGQKVALKIINKKVLAKSDMQGRI 84
Query: 64 KREISVMKMVKHPNIVQLHEVMASKSKIYIAMELVRGGELFNKIV-KGRLKEDVARVYFQ 122
+REIS +++++HP+I++L++V+ SK +I + +E G ELF+ IV + ++ E AR +FQ
Sbjct: 85 EREISYLRLLRHPHIIKLYDVIKSKDEIIMVIEYA-GNELFDYIVQRNKMSEQEARRFFQ 143
Query: 123 QLISAVDFCHSRGVYHRDLKPENLLLDEDGNLKVSDFGLCTFSEHLRQDGLLHTTCGTPA 182
Q+ISAV++CH + HRDLKPENLLLDE N+K++DFGL S + L T+CG+P
Sbjct: 144 QIISAVEYCHRHKIVHRDLKPENLLLDEHLNVKIADFGL---SNIMTDGNFLKTSCGSPN 200
Query: 183 YVSPEVIAKKGYDGAKADIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKCPPWFS 242
Y +PEVI+ K Y G + D+WSCGVILYV+L LPF D+++ ++K I G + P + S
Sbjct: 201 YAAPEVISGKLYAGPEVDVWSCGVILYVMLCRRLPFDDESIPVLFKNISNGVYTLPKFLS 260
Query: 243 LEARRLITKLLDPNPNTRISISKIMDSSWFKKPVPKSIAMRKKEKEEEEDKVFEFMECEK 302
A LI ++L NP RISI +IM WFK + + + + +++E+ +K K
Sbjct: 261 PGASDLIKRMLIVNPLNRISIHEIMQDEWFKVDLAEYLVPQDLKQQEQFNK--------K 312
Query: 303 SSTTMNAFHI-------ISLSEGFDLSPLFEEKKREE 332
S N I +S + G+D ++E + E
Sbjct: 313 SGNEENVEEIDDEMVVTLSKTMGYDKDEIYEALESSE 349
>AAK1_HUMAN (Q13131) 5'-AMP-activated protein kinase, catalytic
alpha-1 chain (EC 2.7.1.-) (AMPK alpha-1 chain)
Length = 550
Score = 233 bits (594), Expect = 7e-61
Identities = 133/317 (41%), Positives = 190/317 (58%), Gaps = 15/317 (4%)
Query: 16 GKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQIKREISVMKMVKH 75
G Y LG LG GTF KV K+ TG VA+K++ ++K+ + ++ +I+REI +K+ +H
Sbjct: 16 GHYILGDTLGVGTFGKVKVGKHELTGHKVAVKILNRQKIRSLDVVGKIRREIQNLKLFRH 75
Query: 76 PNIVQLHEVMASKSKIYIAMELVRGGELFNKIVK-GRLKEDVARVYFQQLISAVDFCHSR 134
P+I++L++V+++ S I++ ME V GGELF+ I K GRL E +R FQQ++S VD+CH
Sbjct: 76 PHIIKLYQVISTPSDIFMVMEYVSGGELFDYICKNGRLDEKESRRLFQQILSGVDYCHRH 135
Query: 135 GVYHRDLKPENLLLDEDGNLKVSDFGLCTFSEHLRQDG-LLHTTCGTPAYVSPEVIAKKG 193
V HRDLKPEN+LLD N K++DFGL ++ DG L T+CG+P Y +PEVI+ +
Sbjct: 136 MVVHRDLKPENVLLDAHMNAKIADFGL----SNMMSDGEFLRTSCGSPNYAAPEVISGRL 191
Query: 194 YDGAKADIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKCPPWFSLEARRLITKLL 253
Y G + DIWS GVILY LL G LPF DD++ ++KKI G F P + + L+ +L
Sbjct: 192 YAGPEVDIWSSGVILYALLCGTLPFDDDHVPTLFKKICDGIFYTPQYLNPSVISLLKHML 251
Query: 254 DPNPNTRISISKIMDSSWFKKPVPK---------SIAMRKKEKEEEEDKVFEFMECEKSS 304
+P R SI I + WFK+ +PK S M E +E + FE E E S
Sbjct: 252 QVDPMKRASIKDIREHEWFKQDLPKYLFPEDPSYSSTMIDDEALKEVCEKFECSEEEVLS 311
Query: 305 TTMNAFHIISLSEGFDL 321
N H L+ + L
Sbjct: 312 CLYNRNHQDPLAVAYHL 328
>AAK1_RAT (P54645) 5'-AMP-activated protein kinase, catalytic
alpha-1 chain (EC 2.7.1.-) (AMPK alpha-1 chain)
Length = 548
Score = 233 bits (593), Expect = 9e-61
Identities = 135/332 (40%), Positives = 196/332 (58%), Gaps = 17/332 (5%)
Query: 1 MCEKENPNSHSALLDGKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMI 60
M EK+ + + G Y LG LG GTF KV K+ TG VA+K++ ++K+ + ++
Sbjct: 1 MAEKQKHDGRVKI--GHYILGDTLGVGTFGKVKVGKHELTGHKVAVKILNRQKIRSLDVV 58
Query: 61 EQIKREISVMKMVKHPNIVQLHEVMASKSKIYIAMELVRGGELFNKIVK-GRLKEDVARV 119
+I+REI +K+ +HP+I++L++V+++ S I++ ME V GGELF+ I K GRL E +R
Sbjct: 59 GKIRREIQNLKLFRHPHIIKLYQVISTPSDIFMVMEYVSGGELFDYICKNGRLDEKESRR 118
Query: 120 YFQQLISAVDFCHSRGVYHRDLKPENLLLDEDGNLKVSDFGLCTFSEHLRQDG-LLHTTC 178
FQQ++S VD+CH V HRDLKPEN+LLD N K++DFGL ++ DG L T+C
Sbjct: 119 LFQQILSGVDYCHRHMVVHRDLKPENVLLDAHMNAKIADFGL----SNMMSDGEFLRTSC 174
Query: 179 GTPAYVSPEVIAKKGYDGAKADIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKCP 238
G+P Y +PEVI+ + Y G + DIWS GVILY LL G LPF DD++ ++KKI G F P
Sbjct: 175 GSPNYAAPEVISGRLYAGPEVDIWSSGVILYALLCGTLPFDDDHVPTLFKKICDGIFYTP 234
Query: 239 PWFSLEARRLITKLLDPNPNTRISISKIMDSSWFKKPVPK---------SIAMRKKEKEE 289
+ + L+ +L +P R +I I + WFK+ +PK S M E +
Sbjct: 235 QYLNPSVISLLKHMLQVDPMKRATIKDIREHEWFKQDLPKYLFPEDPSYSSTMIDDEALK 294
Query: 290 EEDKVFEFMECEKSSTTMNAFHIISLSEGFDL 321
E + FE E E S N H L+ + L
Sbjct: 295 EVCEKFECSEEEVLSCLYNRNHQDPLAVAYHL 326
>KI11_HUMAN (Q8TDC3) Probable serine/threonine-protein kinase
KIAA1811 (EC 2.7.1.37)
Length = 794
Score = 232 bits (592), Expect = 1e-60
Identities = 121/302 (40%), Positives = 188/302 (62%), Gaps = 17/302 (5%)
Query: 3 EKENPNSHSALLDGKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQ 62
E E H+ + G Y L + LG G V + TG+ VA+K+V +EK+ + ++ +
Sbjct: 36 EAEERGRHAQYV-GPYRLEKTLGKGQTGLVKLGVHCITGQKVAIKIVNREKLSE-SVLMK 93
Query: 63 IKREISVMKMVKHPNIVQLHEVMASKSKIYIAMELVRGGELFNKIVK-GRLKEDVARVYF 121
++REI+++K+++HP++++LH+V +K +Y+ +E V GGELF+ +VK GRL AR +F
Sbjct: 94 VEREIAILKLIEHPHVLKLHDVYENKKYLYLVLEHVSGGELFDYLVKKGRLTPKEARKFF 153
Query: 122 QQLISAVDFCHSRGVYHRDLKPENLLLDEDGNLKVSDFGLCTFSEHLRQDGLLHTTCGTP 181
+Q++SA+DFCHS + HRDLKPENLLLDE N++++DFG+ + D LL T+CG+P
Sbjct: 154 RQIVSALDFCHSYSICHRDLKPENLLLDEKNNIRIADFGMASLQVG---DSLLETSCGSP 210
Query: 182 AYVSPEVIAKKGYDGAKADIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKCPPWF 241
Y PEVI + YDG +AD+WSCGVIL+ LL G LPF DDNL + +K+ RG F P +
Sbjct: 211 HYACPEVIKGEKYDGRRADMWSCGVILFALLVGALPFDDDNLRQLLEKVKRGVFHMPHFI 270
Query: 242 SLEARRLITKLLDPNPNTRISISKIMDSSWF----------KKPVP-KSIAMRKKEKEEE 290
+ + L+ +++ P R+S+ +I W+ +P P + +AMR E
Sbjct: 271 PPDCQSLLRGMIEVEPEKRLSLEQIQKHPWYLGGKHEPDPCLEPAPGRRVAMRSLPSNGE 330
Query: 291 ED 292
D
Sbjct: 331 LD 332
>AAK2_RAT (Q09137) 5'-AMP-activated protein kinase, catalytic
alpha-2 chain (EC 2.7.1.-) (AMPK alpha-2 chain)
Length = 552
Score = 232 bits (592), Expect = 1e-60
Identities = 128/319 (40%), Positives = 194/319 (60%), Gaps = 15/319 (4%)
Query: 1 MCEKENPNSHSALLDGKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMI 60
M EK+ + + G Y LG LG GTF KV ++ TG VA+K++ ++K+ + ++
Sbjct: 1 MAEKQKHDGRVKI--GHYVLGDTLGVGTFGKVKIGEHQLTGHKVAVKILNRQKIRSLDVV 58
Query: 61 EQIKREISVMKMVKHPNIVQLHEVMASKSKIYIAMELVRGGELFNKIVK-GRLKEDVARV 119
+IKREI +K+ +HP+I++L++V+++ + ++ ME V GGELF+ I K GR++E AR
Sbjct: 59 GKIKREIQNLKLFRHPHIIKLYQVISTPTDFFMVMEYVSGGELFDYICKHGRVEEVEARR 118
Query: 120 YFQQLISAVDFCHSRGVYHRDLKPENLLLDEDGNLKVSDFGLCTFSEHLRQDG-LLHTTC 178
FQQ++SAVD+CH V HRDLKPEN+LLD N K++DFGL ++ DG L T+C
Sbjct: 119 LFQQILSAVDYCHRHMVVHRDLKPENVLLDAQMNAKIADFGL----SNMMSDGEFLRTSC 174
Query: 179 GTPAYVSPEVIAKKGYDGAKADIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKCP 238
G+P Y +PEVI+ + Y G + DIWSCGVILY LL G LPF D+++ ++KKI G F P
Sbjct: 175 GSPNYAAPEVISGRLYAGPEVDIWSCGVILYALLCGTLPFDDEHVPTLFKKIRGGVFYIP 234
Query: 239 PWFSLEARRLITKLLDPNPNTRISISKIMDSSWFKKPVPKSIAMRKKE------KEEEED 292
+ + L+ +L +P R +I I + WFK+ +P + +E
Sbjct: 235 EYLNRSIATLLMHMLQVDPLKRATIKDIREHEWFKQDLPSYLFPEDPSYDANVIDDEAVK 294
Query: 293 KVFEFMECEKSSTTMNAFH 311
+V E EC +S MN+ +
Sbjct: 295 EVCEKFECTESE-VMNSLY 312
>AAK2_HUMAN (P54646) 5'-AMP-activated protein kinase, catalytic
alpha-2 chain (EC 2.7.1.-) (AMPK alpha-2 chain)
Length = 552
Score = 232 bits (592), Expect = 1e-60
Identities = 128/319 (40%), Positives = 194/319 (60%), Gaps = 15/319 (4%)
Query: 1 MCEKENPNSHSALLDGKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMI 60
M EK+ + + G Y LG LG GTF KV ++ TG VA+K++ ++K+ + ++
Sbjct: 1 MAEKQKHDGRVKI--GHYVLGDTLGVGTFGKVKIGEHQLTGHKVAVKILNRQKIRSLDVV 58
Query: 61 EQIKREISVMKMVKHPNIVQLHEVMASKSKIYIAMELVRGGELFNKIVK-GRLKEDVARV 119
+IKREI +K+ +HP+I++L++V+++ + ++ ME V GGELF+ I K GR++E AR
Sbjct: 59 GKIKREIQNLKLFRHPHIIKLYQVISTPTDFFMVMEYVSGGELFDYICKHGRVEEMEARR 118
Query: 120 YFQQLISAVDFCHSRGVYHRDLKPENLLLDEDGNLKVSDFGLCTFSEHLRQDG-LLHTTC 178
FQQ++SAVD+CH V HRDLKPEN+LLD N K++DFGL ++ DG L T+C
Sbjct: 119 LFQQILSAVDYCHRHMVVHRDLKPENVLLDAHMNAKIADFGL----SNMMSDGEFLRTSC 174
Query: 179 GTPAYVSPEVIAKKGYDGAKADIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKCP 238
G+P Y +PEVI+ + Y G + DIWSCGVILY LL G LPF D+++ ++KKI G F P
Sbjct: 175 GSPNYAAPEVISGRLYAGPEVDIWSCGVILYALLCGTLPFDDEHVPTLFKKIRGGVFYIP 234
Query: 239 PWFSLEARRLITKLLDPNPNTRISISKIMDSSWFKKPVPKSIAMRKKE------KEEEED 292
+ + L+ +L +P R +I I + WFK+ +P + +E
Sbjct: 235 EYLNRSVATLLMHMLQVDPLKRATIKDIREHEWFKQDLPSYLFPEDPSYDANVIDDEAVK 294
Query: 293 KVFEFMECEKSSTTMNAFH 311
+V E EC +S MN+ +
Sbjct: 295 EVCEKFECTESE-VMNSLY 312
>ST29_HUMAN (Q8IWQ3) Serine/threonine-protein kinase 29 (EC
2.7.1.37) (BRSK2) (HUSSY-12)
Length = 736
Score = 228 bits (582), Expect = 2e-59
Identities = 115/287 (40%), Positives = 182/287 (63%), Gaps = 14/287 (4%)
Query: 16 GKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQIKREISVMKMVKH 75
G Y L + LG G V + T + VA+K+V +EK+ + ++ +++REI+++K+++H
Sbjct: 17 GPYRLEKTLGKGQTGLVKLGVHCVTCQKVAIKIVNREKLSE-SVLMKVEREIAILKLIEH 75
Query: 76 PNIVQLHEVMASKSKIYIAMELVRGGELFNKIVK-GRLKEDVARVYFQQLISAVDFCHSR 134
P++++LH+V +K +Y+ +E V GGELF+ +VK GRL AR +F+Q+ISA+DFCHS
Sbjct: 76 PHVLKLHDVYENKKYLYLVLEHVSGGELFDYLVKKGRLTPKEARKFFRQIISALDFCHSH 135
Query: 135 GVYHRDLKPENLLLDEDGNLKVSDFGLCTFSEHLRQDGLLHTTCGTPAYVSPEVIAKKGY 194
+ HRDLKPENLLLDE N++++DFG+ + D LL T+CG+P Y PEVI + Y
Sbjct: 136 SICHRDLKPENLLLDEKNNIRIADFGMASLQVG---DSLLETSCGSPHYACPEVIRGEKY 192
Query: 195 DGAKADIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKCPPWFSLEARRLITKLLD 254
DG KAD+WSCGVIL+ LL G LPF DDNL + +K+ RG F P + + + L+ + +
Sbjct: 193 DGRKADVWSCGVILFALLVGALPFDDDNLRQLLEKVKRGVFHMPHFIPPDCQSLLRGMSE 252
Query: 255 PNPNTRISISKIMDSSWF---------KKPVPKSIAMRKKEKEEEED 292
+ R+++ I W+ ++P+P+ + +R E+ D
Sbjct: 253 VDAARRLTLEHIQKHIWYIGGKNEPEPEQPIPRKVQIRSLPSLEDID 299
>MELK_MOUSE (Q61846) Maternal embryonic leucine zipper kinase (EC
2.7.1.37) (Protein kinase PK38) (mPK38)
Length = 643
Score = 224 bits (571), Expect = 3e-58
Identities = 121/290 (41%), Positives = 184/290 (62%), Gaps = 6/290 (2%)
Query: 18 YELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQIKREISVMKMVKHPN 77
YEL +G G FAKV A ++ TG+ VA+K++ K + + ++K EI +K ++H +
Sbjct: 11 YELYETIGTGGFAKVKLACHVLTGEMVAIKIMDKNAL--GSDLPRVKTEIDALKSLRHQH 68
Query: 78 IVQLHEVMASKSKIYIAMELVRGGELFNKIV-KGRLKEDVARVYFQQLISAVDFCHSRGV 136
I QL+ V+ +K+KI++ +E GGELF+ I+ + RL E+ RV F+Q++SAV + HS+G
Sbjct: 69 ICQLYHVLETKNKIFMVLEYCPGGELFDYIISQDRLSEEETRVVFRQILSAVAYVHSQGY 128
Query: 137 YHRDLKPENLLLDEDGNLKVSDFGLCTFSEHLRQDGLLHTTCGTPAYVSPEVIAKKGYDG 196
HRDLKPENLL DE+ LK+ DFGLC + +D L T CG+ AY +PE+I K Y G
Sbjct: 129 AHRDLKPENLLFDENHKLKLIDFGLCAKPKG-NKDYHLQTCCGSLAYAAPELIQGKSYLG 187
Query: 197 AKADIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKCPPWFSLEARRLITKLLDPN 256
++AD+WS G++LYVL+ GFLPF DDN++A+YKKI RG ++ P W S + L+ ++L +
Sbjct: 188 SEADVWSMGILLYVLMCGFLPFDDDNVMALYKKIMRGKYEVPKWLSPSSILLLQQMLQVD 247
Query: 257 PNTRISISKIMDSSWFKKPVPKSIAMRKKE--KEEEEDKVFEFMECEKSS 304
P RIS+ +++ W + + + K +ED V E +SS
Sbjct: 248 PKKRISMRNLLNHPWVMQDYSCPVEWQSKTPLTHLDEDCVTELSVHHRSS 297
>RKI1_SECCE (Q02723) Carbon catabolite derepressing protein kinase
(EC 2.7.1.-)
Length = 502
Score = 224 bits (570), Expect = 4e-58
Identities = 127/293 (43%), Positives = 190/293 (64%), Gaps = 15/293 (5%)
Query: 10 HSALLDGKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQIKREISV 69
HS L Y LG++LG GTFAKV A++ +T VA+KV+ + ++ M E+ KREI +
Sbjct: 7 HSEALKNYY-LGKILGVGTFAKVIIAEHKHTRHKVAIKVLNRRQMRAPEMEEKAKREIKI 65
Query: 70 MKM---VKHPNIVQLHEVMASKSKIYIAMELVRGGELFNKIV-KGRLKEDVARVYFQQLI 125
+++ + HP+I++++EV+ + I++ ME + G+L + I+ K RL+ED AR FQQ+I
Sbjct: 66 LRLFIDLIHPHIIRVYEVIVTPKDIFVVMEYCQNGDLLDYILEKRRLQEDEARRTFQQII 125
Query: 126 SAVDFCHSRGVYHRDLKPENLLLDEDGNLKVSDFGLCTFSEHLRQDG-LLHTTCGTPAYV 184
SAV++CH V HRDLKPENLLLD N+K++DFGL ++ DG L T+CG+ Y
Sbjct: 126 SAVEYCHRNKVVHRDLKPENLLLDSKYNVKLADFGL----SNVMHDGHFLKTSCGSLNYA 181
Query: 185 SPEVIAKKGYDGAKADIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKCPPWFSLE 244
+PEVI+ K Y G + D+WSCGVILY LL G +PF DDN+ ++KKI G + P + S
Sbjct: 182 APEVISGKLYAGPEIDVWSCGVILYALLCGAVPFDDDNIPNLFKKIKGGTYILPIYLSDL 241
Query: 245 ARRLITKLLDPNPNTRISISKIMDSSWFKKPVPKSIA-----MRKKEKEEEED 292
R LI+++L +P RI+I +I SWF+ +P+ +A M ++ K +ED
Sbjct: 242 VRDLISRMLIVDPMKRITIGEIRKHSWFQNRLPRYLAVPPPDMMQQAKMIDED 294
>MELK_HUMAN (Q14680) Maternal embryonic leucine zipper kinase (EC
2.7.1.37) (hMELK) (Protein kinase PK38) (hPK38)
Length = 651
Score = 220 bits (561), Expect = 5e-57
Identities = 114/255 (44%), Positives = 169/255 (65%), Gaps = 4/255 (1%)
Query: 18 YELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQIKREISVMKMVKHPN 77
YEL +G G FAKV A ++ TG+ VA+K++ K + + +IK EI +K ++H +
Sbjct: 11 YELHETIGTGGFAKVKLACHILTGEMVAIKIMDKNTL--GSDLPRIKTEIEALKNLRHQH 68
Query: 78 IVQLHEVMASKSKIYIAMELVRGGELFNKIV-KGRLKEDVARVYFQQLISAVDFCHSRGV 136
I QL+ V+ + +KI++ +E GGELF+ I+ + RL E+ RV F+Q++SAV + HS+G
Sbjct: 69 ICQLYHVLETANKIFMVLEYCPGGELFDYIISQDRLSEEETRVVFRQIVSAVAYVHSQGY 128
Query: 137 YHRDLKPENLLLDEDGNLKVSDFGLCTFSEHLRQDGLLHTTCGTPAYVSPEVIAKKGYDG 196
HRDLKPENLL DE LK+ DFGLC + +D L T CG+ AY +PE+I K Y G
Sbjct: 129 AHRDLKPENLLFDEYHKLKLIDFGLCAKPKG-NKDYHLQTCCGSLAYAAPELIQGKSYLG 187
Query: 197 AKADIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKCPPWFSLEARRLITKLLDPN 256
++AD+WS G++LYVL+ GFLPF DDN++A+YKKI RG + P W S + L+ ++L +
Sbjct: 188 SEADVWSMGILLYVLMCGFLPFDDDNVMALYKKIMRGKYDVPKWLSPSSILLLQQMLQVD 247
Query: 257 PNTRISISKIMDSSW 271
P RIS+ +++ W
Sbjct: 248 PKKRISMKNLLNHPW 262
>MRK4_HUMAN (Q96L34) MAP/microtubule affinity-regulating kinase 4
(EC 2.7.1.37) (MAP/microtubule affinity-regulating
kinase like 1)
Length = 752
Score = 218 bits (556), Expect = 2e-56
Identities = 109/271 (40%), Positives = 168/271 (61%), Gaps = 11/271 (4%)
Query: 2 CEKENPNSHSALLDGKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIE 61
C +E P+ G Y L R +G G FAKV A+++ TG+ VA+K++ K + + ++
Sbjct: 49 CPEEQPHV------GNYRLLRTIGKGNFAKVKLARHILTGREVAIKIIDKTQ-LNPSSLQ 101
Query: 62 QIKREISVMKMVKHPNIVQLHEVMASKSKIYIAMELVRGGELFNKIVK-GRLKEDVARVY 120
++ RE+ +MK + HPNIV+L EV+ ++ +Y+ ME GE+F+ +V GR+KE AR
Sbjct: 102 KLFREVRIMKGLNHPNIVKLFEVIETEKTLYLVMEYASAGEVFDYLVSHGRMKEKEARAK 161
Query: 121 FQQLISAVDFCHSRGVYHRDLKPENLLLDEDGNLKVSDFGLCTFSEHLRQDGLLHTTCGT 180
F+Q++SAV +CH + + HRDLK ENLLLD + N+K++DFG FS L T CG+
Sbjct: 162 FRQIVSAVHYCHQKNIVHRDLKAENLLLDAEANIKIADFG---FSNEFTLGSKLDTFCGS 218
Query: 181 PAYVSPEVIAKKGYDGAKADIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKCPPW 240
P Y +PE+ K YDG + DIWS GVILY L++G LPF NL + +++ RG ++ P +
Sbjct: 219 PPYAAPELFQGKKYDGPEVDIWSLGVILYTLVSGSLPFDGHNLKELRERVLRGKYRVPFY 278
Query: 241 FSLEARRLITKLLDPNPNTRISISKIMDSSW 271
S + ++ + L NP R ++ +IM W
Sbjct: 279 MSTDCESILRRFLVLNPAKRCTLEQIMKDKW 309
>YNA3_CAEEL (P45894) Putative serine/threonine-protein kinase PAR2.3
(EC 2.7.1.37)
Length = 622
Score = 217 bits (553), Expect = 4e-56
Identities = 113/303 (37%), Positives = 179/303 (58%), Gaps = 20/303 (6%)
Query: 16 GKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQIKREISVMKMVKH 75
G + + +G G F V ++ TG +VA+K++ + ++ +G + + + EI ++ + H
Sbjct: 22 GNFVIKETIGKGAFGAVKRGTHIQTGYDVAIKILNRGRMKGLGTVNKTRNEIDNLQKLTH 81
Query: 76 PNIVQLHEVMASKSKIYIAMELVRGGELFNKIV-KGRLKEDVARVYFQQLISAVDFCHSR 134
P+I +L V+++ S I++ MELV GGELF+ I KG L +R YFQQ+IS V +CH+
Sbjct: 82 PHITRLFRVISTPSDIFLVMELVSGGELFSYITRKGALPIRESRRYFQQIISGVSYCHNH 141
Query: 135 GVYHRDLKPENLLLDEDGNLKVSDFGLCTFSEHLRQDGLLHTTCGTPAYVSPEVIAKKGY 194
+ HRDLKPENLLLD + N+K++DFGL S ++ LL T CG+P Y +PE+I+ K Y
Sbjct: 142 MIVHRDLKPENLLLDANKNIKIADFGL---SNYMTDGDLLSTACGSPNYAAPELISNKLY 198
Query: 195 DGAKADIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKCPPWFSLEARRLITKLLD 254
G + D+WSCGVILY +L G LPF D N+ ++ KI G + P +A LI+ +L
Sbjct: 199 VGPEVDLWSCGVILYAMLCGTLPFDDQNVPTLFAKIKSGRYTVPYSMEKQAADLISTMLQ 258
Query: 255 PNPNTRISISKIMDSSWFKKPVPKSIAMRKKEKEEEEDKVFEFMECEKSSTTMNAFHIIS 314
+P R + +I++ SWF+ +P + F ECE S+ ++ + S
Sbjct: 259 VDPVKRADVKRIVNHSWFRIDLP----------------YYLFPECENESSIVDIDVVQS 302
Query: 315 LSE 317
++E
Sbjct: 303 VAE 305
>MRK2_MOUSE (Q05512) MAP/microtubule affinity-regulating kinase 2
(EC 2.7.1.37) (Serine/threonine-protein kinase Emk)
Length = 774
Score = 214 bits (545), Expect = 3e-55
Identities = 106/257 (41%), Positives = 163/257 (63%), Gaps = 5/257 (1%)
Query: 16 GKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQIKREISVMKMVKH 75
G Y L + +G G FAKV A+++ TGK VA+K++ K + + ++++ RE+ +MK++ H
Sbjct: 51 GNYRLLKTIGKGNFAKVKLARHILTGKEVAVKIIDKTQ-LNSSSLQKLFREVRIMKVLNH 109
Query: 76 PNIVQLHEVMASKSKIYIAMELVRGGELFNKIVK-GRLKEDVARVYFQQLISAVDFCHSR 134
PNIV+L EV+ ++ +Y+ ME GGE+F+ +V GR+KE AR F+Q++ V +CH +
Sbjct: 110 PNIVKLFEVIETEKTLYLVMEYASGGEVFDYLVAHGRMKEKEARAKFRQIVLHVQYCHQK 169
Query: 135 GVYHRDLKPENLLLDEDGNLKVSDFGLCTFSEHLRQDGLLHTTCGTPAYVSPEVIAKKGY 194
+ HRDLK ENLLLD D N+K++DFG FS L T CG+P Y +PE+ K
Sbjct: 170 FIVHRDLKAENLLLDADMNIKIADFG---FSNEFTFGNKLDTFCGSPPYAAPELFQGKKI 226
Query: 195 DGAKADIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKCPPWFSLEARRLITKLLD 254
DG + D+WS GVILY L++G LPF NL + +++ RG ++ P + S + L+ K L
Sbjct: 227 DGPEVDVWSLGVILYTLVSGSLPFDGQNLKELRERVLRGKYRIPFYMSTDCENLLKKFLI 286
Query: 255 PNPNTRISISKIMDSSW 271
NP+ R ++ +IM W
Sbjct: 287 LNPSKRGTLEQIMKDRW 303
>SNF1_SCHPO (O74536) SNF1-like protein kinase ssp2 (EC 2.7.1.-)
Length = 576
Score = 211 bits (536), Expect = 4e-54
Identities = 107/263 (40%), Positives = 170/263 (63%), Gaps = 5/263 (1%)
Query: 16 GKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIEQIKREISVMKMVKH 75
G Y + LG G+F KV A + T + VA+K + ++ + K M +++REIS +K+++H
Sbjct: 32 GPYIIRETLGEGSFGKVKLATHYKTQQKVALKFISRQLLKKSDMHMRVEREISYLKLLRH 91
Query: 76 PNIVQLHEVMASKSKIYIAMELVRGGELFNKIV-KGRLKEDVARVYFQQLISAVDFCHSR 134
P+I++L++V+ + + I + +E GGELF+ IV K R+ ED R +FQQ+I A+++CH
Sbjct: 92 PHIIKLYDVITTPTDIVMVIEYA-GGELFDYIVEKKRMTEDEGRRFFQQIICAIEYCHRH 150
Query: 135 GVYHRDLKPENLLLDEDGNLKVSDFGLCTFSEHLRQDGLLHTTCGTPAYVSPEVIAKKGY 194
+ HRDLKPENLLLD++ N+K++DFGL S + L T+CG+P Y +PEVI K Y
Sbjct: 151 KIVHRDLKPENLLLDDNLNVKIADFGL---SNIMTDGNFLKTSCGSPNYAAPEVINGKLY 207
Query: 195 DGAKADIWSCGVILYVLLAGFLPFQDDNLVAMYKKIYRGDFKCPPWFSLEARRLITKLLD 254
G + D+WSCG++LYV+L G LPF D+ + ++KK+ + P + S A+ LI +++
Sbjct: 208 AGPEVDVWSCGIVLYVMLVGRLPFDDEFIPNLFKKVNSCVYVMPDFLSPGAQSLIRRMIV 267
Query: 255 PNPNTRISISKIMDSSWFKKPVP 277
+P RI+I +I WF +P
Sbjct: 268 ADPMQRITIQEIRRDPWFNVNLP 290
>MRK3_HUMAN (P27448) MAP/microtubule affinity-regulating kinase 3
(EC 2.7.1.37) (Cdc25C-associated protein kinase 1)
(cTAK1) (C-TAK1) (Serine/threonine protein kinase p78)
(Ser/Thr protein kinase PAR-1) (Protein kinase STK10)
Length = 776
Score = 207 bits (527), Expect = 4e-53
Identities = 121/359 (33%), Positives = 192/359 (52%), Gaps = 48/359 (13%)
Query: 2 CEKENPNSHSALLDGKYELGRVLGHGTFAKVYHAKNLNTGKNVAMKVVGKEKVIKVGMIE 61
C E P+ G Y L + +G G FAKV A+++ TG+ VA+K++ K ++ + +
Sbjct: 46 CADEQPHI------GNYRLLKTIGKGNFAKVKLARHILTGREVAIKIIDKTQLNPTSL-Q 98
Query: 62 QIKREISVMKMVKHPNIVQLHEVMASKSKIYIAMELVRGGELFNKIVK-GRLKEDVARVY 120
++ RE+ +MK++ HPNIV+L EV+ ++ +Y+ ME GGE+F+ +V GR+KE AR
Sbjct: 99 KLFREVRIMKILNHPNIVKLFEVIETEKTLYLIMEYASGGEVFDYLVAHGRMKEKEARSK 158
Query: 121 FQQ-----------------------LISAVDFCHSRGVYHRDLKPENLLLDEDGNLKVS 157
F+Q ++SAV +CH + + HRDLK ENLLLD D N+K++
Sbjct: 159 FRQGCQAGQTIKVQVSFDLLSLMFTFIVSAVQYCHQKRIVHRDLKAENLLLDADMNIKIA 218
Query: 158 DFGLCTFSEHLRQDGLLHTTCGTPAYVSPEVIAKKGYDGAKADIWSCGVILYVLLAGFLP 217
DFG FS G L T CG+P Y +PE+ K YDG + D+WS GVILY L++G LP
Sbjct: 219 DFG---FSNEFTVGGKLDTFCGSPPYAAPELFQGKKYDGPEVDVWSLGVILYTLVSGSLP 275
Query: 218 FQDDNLVAMYKKIYRGDFKCPPWFSLEARRLITKLLDPNPNTRISISKIMDSSWFKKPVP 277
F NL + +++ RG ++ P + S + L+ + L NP R ++ +IM W
Sbjct: 276 FDGQNLKELRERVLRGKYRIPFYMSTDCENLLKRFLVLNPIKRGTLEQIMKDRWI----- 330
Query: 278 KSIAMRKKEKEEEEDKVFEFMECEKSSTTMNAFHIISLSEGFDLSPLFEEKKREEREEM 336
EED++ F+E E + I+ + G+ + E + + +E+
Sbjct: 331 --------NAGHEEDELKPFVEPELDISDQKRIDIM-VGMGYSQEEIQESLSKMKYDEI 380
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.136 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 51,529,529
Number of Sequences: 164201
Number of extensions: 2202698
Number of successful extensions: 11923
Number of sequences better than 10.0: 1710
Number of HSP's better than 10.0 without gapping: 1475
Number of HSP's successfully gapped in prelim test: 235
Number of HSP's that attempted gapping in prelim test: 6444
Number of HSP's gapped (non-prelim): 2052
length of query: 435
length of database: 59,974,054
effective HSP length: 113
effective length of query: 322
effective length of database: 41,419,341
effective search space: 13337027802
effective search space used: 13337027802
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 67 (30.4 bits)
Medicago: description of AC122160.9


