

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122160.3 - phase: 0 /pseudo
(835 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
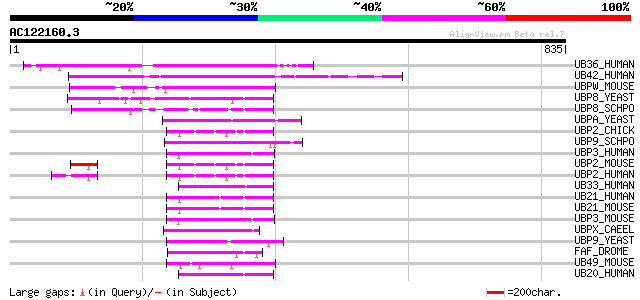
Score E
Sequences producing significant alignments: (bits) Value
UB36_HUMAN (Q9P275) Ubiquitin carboxyl-terminal hydrolase 36 (EC... 236 2e-61
UB42_HUMAN (Q9H9J4) Ubiquitin carboxyl-terminal hydrolase 42 (EC... 225 3e-58
UBPW_MOUSE (Q61068) Ubiquitin carboxyl-terminal hydrolase DUB-1 ... 185 5e-46
UBP8_YEAST (P50102) Ubiquitin carboxyl-terminal hydrolase 8 (EC ... 122 4e-27
UBP8_SCHPO (Q09738) Probable ubiquitin carboxyl-terminal hydrola... 100 2e-20
UBPA_YEAST (P53874) Ubiquitin carboxyl-terminal hydrolase 10 (EC... 97 2e-19
UBP2_CHICK (O57429) Ubiquitin carboxyl-terminal hydrolase 2 (EC ... 97 2e-19
UBP9_SCHPO (Q9P7V9) Probable ubiquitin carboxyl-terminal hydrola... 96 5e-19
UBP3_HUMAN (Q9Y6I4) Ubiquitin carboxyl-terminal hydrolase 3 (EC ... 94 1e-18
UBP2_MOUSE (O88623) Ubiquitin carboxyl-terminal hydrolase 2 (EC ... 94 1e-18
UBP2_HUMAN (O75604) Ubiquitin carboxyl-terminal hydrolase 2 (EC ... 94 1e-18
UB33_HUMAN (Q8TEY7) Ubiquitin carboxyl-terminal hydrolase 33 (EC... 94 2e-18
UB21_HUMAN (Q9UK80) Ubiquitin carboxyl-terminal hydrolase 21 (EC... 92 6e-18
UB21_MOUSE (Q9QZL6) Ubiquitin carboxyl-terminal hydrolase 21 (EC... 92 7e-18
UBP3_MOUSE (Q91W36) Ubiquitin carboxyl-terminal hydrolase 3 (EC ... 90 3e-17
UBPX_CAEEL (P34547) Probable ubiquitin carboxyl-terminal hydrola... 89 4e-17
UBP9_YEAST (P39967) Ubiquitin carboxyl-terminal hydrolase 9 (EC ... 87 2e-16
FAF_DROME (P55824) Probable ubiquitin carboxyl-terminal hydrolas... 87 2e-16
UB49_MOUSE (Q6P9L4) Ubiquitin carboxyl-terminal hydrolase 49 (EC... 86 4e-16
UB20_HUMAN (Q9Y2K6) Ubiquitin carboxyl-terminal hydrolase 20 (EC... 86 4e-16
>UB36_HUMAN (Q9P275) Ubiquitin carboxyl-terminal hydrolase 36 (EC
3.1.2.15) (Ubiquitin thiolesterase 36)
(Ubiquitin-specific processing protease 36)
(Deubiquitinating enzyme 36)
Length = 1121
Score = 236 bits (601), Expect = 2e-61
Identities = 165/465 (35%), Positives = 237/465 (50%), Gaps = 59/465 (12%)
Query: 21 RKIEFIPVKKPFKGFSNDFNIETL-------NPTTSEHRQLVSANIQSSQPQPSKNSEFS 73
+KIEF P K F + +E L NP T + S + ++ Q S+++ S
Sbjct: 38 QKIEFEPASKSFS-----YQLEALKSKYVLLNPKTEGASRHKSGDDPPARRQGSEHTYES 92
Query: 74 --------EFGLDPELSLGITFRRI---GAGLWNLGNTCFLNSVLQCLTYTEPLAAYLQS 122
+ L P L + + R+ GAGL NLGNTCFLN+ +QCLTYT PLA YL S
Sbjct: 93 CGDGVPAPQKVLFPTERLSLRWERVFRVGAGLHNLGNTCFLNATIQCLTYTPPLANYLLS 152
Query: 123 GKHKSSCHIAGFCALCAIQNHVSCALQATGRIVSPQDMVGNLRCILMSCFF-NEQSRH-- 179
+H SCH FC LC +QNH+ A +G + P + +L+ I F N++ H
Sbjct: 153 KEHARSCHQGSFCMLCVMQNHIVQAFANSGNAIKPVSFIRDLKKIARHFRFGNQEDAHEF 212
Query: 180 ----ITQFPKG*AGRCT*VYGELT*VNA*MLPAFWSTE*ITWCFREKFGS*---DLWWSS 232
I K C + + T + FG + S
Sbjct: 213 LRYTIDAMQKACLNGCAKLDRQTQ---------------ATTLVHQIFGGYLRSRVKCSV 257
Query: 233 SKSASNKFDPFLDLSLEILKADSLQKALANFTAAELLDGGEKQYHCQRCKQKVQAVKQLT 292
KS S+ +DP+LD++LEI +A ++ +AL F A++L G E Y C +CK+KV A K+ T
Sbjct: 258 CKSVSDTYDPYLDVALEIRQAANIVRALELFVKADVLSG-ENAYMCAKCKKKVPASKRFT 316
Query: 293 IHKAPYVLAIHLKRFYAHDPNIKIKKKVRFDSALNLKPFVSGSSDGDVKYSLYGVLVHSG 352
IH+ VL + LKRF A+ KI K V + LN++P++S ++ V Y LY VLVHSG
Sbjct: 317 IHRTSNVLTLSLKRF-ANFSGGKITKDVGYPEFLNIRPYMSQNNGDPVMYGLYAVLVHSG 375
Query: 353 FSTHSGHYYCYVRTSNNMWYTLDDTRVSHVGEQEVLNQQAYMLFYVRDKKSITPRKPVNI 412
+S H+GHYYCYV+ SN WY ++D+ V + VLNQQAY+LFY+R S + P +
Sbjct: 376 YSCHAGHYYCYVKASNGQWYQMNDSLVHSSNVKVVLNQQAYVLFYLRIPGS--KKSPEGL 433
Query: 413 AKEEISKTNVIGSRYPSTPTNALKDYPNGHVENKFCGVPLTAETQ 457
IS+T S P P + + D+ ++ N PLT + Q
Sbjct: 434 ----ISRTG--SSSLPGRP-SVIPDHSKKNIGNGIISSPLTGKRQ 471
>UB42_HUMAN (Q9H9J4) Ubiquitin carboxyl-terminal hydrolase 42 (EC
3.1.2.15) (Ubiquitin thiolesterase 42)
(Ubiquitin-specific processing protease 42)
(Deubiquitinating enzyme 42) (Fragment)
Length = 1198
Score = 225 bits (574), Expect = 3e-58
Identities = 164/511 (32%), Positives = 246/511 (48%), Gaps = 51/511 (9%)
Query: 89 RIGAGLWNLGNTCFLNSVLQCLTYTEPLAAYLQSGKHKSSCHIAGFCALCAIQNHVSCAL 148
R+GAGL NLGNTCF N+ LQCLTYT PLA Y+ S +H +CH GFC +C +Q H++ AL
Sbjct: 108 RVGAGLQNLGNTCFANAALQCLTYTPPLANYMLSHEHSKTCHAEGFCMMCTMQAHITQAL 167
Query: 149 QATGRIVSPQDMVGNLRCILMSCFFN--EQSRHITQFPKG*AGR-CT*VYGELT*VNA*M 205
G ++ P ++ +R I F E + Q+ + C +L
Sbjct: 168 SNPGDVIKPMFVINEMRRIARHFRFGNQEDAHEFLQYTVDAMQKACLNGSNKLDRHTQ-- 225
Query: 206 LPAFWSTE*ITWCFREKFGS*DLWWSSSKSASNKFDPFLDLSLEILKADSLQKALANFTA 265
+T + F S + + K S+ FDP+LD++LEI A S+ KAL F
Sbjct: 226 -----ATTLVCQIFGGYLRS-RVKCLNCKGVSDTFDPYLDITLEIKAAQSVNKALEQFVK 279
Query: 266 AELLDGGEKQYHCQRCKQKVQAVKQLTIHKAPYVLAIHLKRFYAHDPNIKIKKKVRFDSA 325
E LDG E Y C +CK+ V A K+ TIH++ VL + LKRF A+ KI K V++
Sbjct: 280 PEQLDG-ENSYKCSKCKKMVPASKRFTIHRSSNVLTLSLKRF-ANFTGGKIAKDVKYPEY 337
Query: 326 LNLKPFVSGSSDGDVKYSLYGVLVHSGFSTHSGHYYCYVRTSNNMWYTLDDTRVSHVGEQ 385
L+++P++S + + Y LY VLVH+GF+ H+GHY+CY++ SN +WY ++D+ VS +
Sbjct: 338 LDIRPYMSQPNGEPIVYVLYAVLVHTGFNCHAGHYFCYIKASNGLWYQMNDSIVSTSDIR 397
Query: 386 EVLNQQAYMLFYVRDKKSITPRKPVNIAKEEISKTNVIGSRYPSTPTNALKDYPNGHVEN 445
VL+QQAY+LFY+R V E T+ G P P + + N
Sbjct: 398 SVLSQQAYVLFYIRS-------HDVKNGGELTHPTHSPGQSSP-RPVISQRVVTNKQAAP 449
Query: 446 KFCGVPLTAETQKN---LLNADPSRISDALIQQKNSGILAKSLMHNETPVSERTSKELTQ 502
F G L + KN L P + + + +G S ++ +PV+ S +
Sbjct: 450 GFIGPQLPSHMIKNPPHLNGTGPLKDTPSSSMSSPNG---NSSVNRASPVNASASVQNWS 506
Query: 503 MNSSDELPVAKSELECLSSLDHSGKD--NVSGNQKCLVAPAGDKPNLFTEDTLLKEGVDS 560
+N S +P +H K +S + K V +PNL + +
Sbjct: 507 VNRSSVIP------------EHPKKQKITISIHNKLPVRQCQSQPNLHS----------N 544
Query: 561 PLVEPIVSNPQTFIGKHASDRTSPSDEVSLS 591
L P P + I A TS + +S+S
Sbjct: 545 SLENPTKPVPSSTITNSAVQSTSNASTMSVS 575
>UBPW_MOUSE (Q61068) Ubiquitin carboxyl-terminal hydrolase DUB-1 (EC
3.1.2.15) (Ubiquitin thiolesterase DUB-1)
(Ubiquitin-specific processing protease DUB-1)
(Deubiquitinating enzyme 1)
Length = 526
Score = 185 bits (469), Expect = 5e-46
Identities = 115/321 (35%), Positives = 172/321 (52%), Gaps = 35/321 (10%)
Query: 91 GAGLWNLGNTCFLNSVLQCLTYTEPLAAYLQSGKHKSSCHIAGFCALCAIQNHVSCAL-- 148
G GL N GN+C+LN+ LQCLT+T PLA Y+ S +H +C C LCA++ V+ +L
Sbjct: 50 GCGLQNTGNSCYLNAALQCLTHTPPLADYMLSQEHSQTCCSPEGCKLCAMEALVTQSLLH 109
Query: 149 QATGRIVSPQDMVGNLRCILMSCFFNEQSRHITQFP----KG*AGRCT*VYGELT*VNA* 204
+G ++ P IL S F Q +F + C V+ + +
Sbjct: 110 SHSGDVMKPSH-------ILTSAFHKHQQEDAHEFLMFTLETMHESCLQVHRQSKPTSED 162
Query: 205 MLPAFWSTE*ITWCFREKFGS*DLWWSSS------KSASNKFDPFLDLSLEILKADSLQK 258
P + FG WW S + S+ +D FLD+ L+I A S+++
Sbjct: 163 SSP-----------IHDIFGG---WWRSQIKCLLCQGTSDTYDRFLDIPLDISSAQSVKQ 208
Query: 259 ALANFTAAELLDGGEKQYHCQRCKQKVQAVKQLTIHKAPYVLAIHLKRFYAHDPNIKIKK 318
AL + +E L G + Y+C +C+QK+ A K L +H AP VL + L RF A N K+ +
Sbjct: 209 ALWDTEKSEELCG-DNAYYCGKCRQKMPASKTLHVHIAPKVLMVVLNRFSAFTGN-KLDR 266
Query: 319 KVRFDSALNLKPFVSGSSDGDVKYSLYGVLVHSGFSTHSGHYYCYVRTSNNMWYTLDDTR 378
KV + L+LKP++S + G + Y+LY VLVH G ++HSGHY+C V+ + WY +DDT+
Sbjct: 267 KVSYPEFLDLKPYLSEPTGGPLPYALYAVLVHDGATSHSGHYFCCVKAGHGKWYKMDDTK 326
Query: 379 VSHVGEQEVLNQQAYMLFYVR 399
V+ VLN+ AY+LFYV+
Sbjct: 327 VTRCDVTSVLNENAYVLFYVQ 347
>UBP8_YEAST (P50102) Ubiquitin carboxyl-terminal hydrolase 8 (EC
3.1.2.15) (Ubiquitin thiolesterase 8)
(Ubiquitin-specific processing protease 8)
(Deubiquitinating enzyme 8)
Length = 471
Score = 122 bits (306), Expect = 4e-27
Identities = 103/346 (29%), Positives = 159/346 (45%), Gaps = 48/346 (13%)
Query: 88 RRIG-AGLWNLGNTCFLNSVLQCLTYTEPLAAYLQSGKHKSSCHIAG-----FCALCAIQ 141
RR G +GL N+G+TCF++S+LQCL + + S H ++C + CAL I
Sbjct: 132 RRDGLSGLINMGSTCFMSSILQCLIHNPYFIRHSMSQIHSNNCKVRSPDKCFSCALDKIV 191
Query: 142 NHVSCAL---QATGRIVSPQDMVGNLRCILMSCF---------FNEQSRHITQFPKG*AG 189
+ + AL QA+ S G + L++C +++Q H +F +
Sbjct: 192 HELYGALNTKQASSSSTSTNRQTGFI--YLLTCAWKINQNLAGYSQQDAH--EFWQFIIN 247
Query: 190 RCT*VY-------GELT*VNA*MLPAFWSTE*ITWCFREKFGS*DLWWSSSKSASNKFDP 242
+ Y E++ N T F S + ++ DP
Sbjct: 248 QIHQSYVLDLPNAKEVSRANNKQCECIVHT-----VFEGSLESSIVCPGCQNNSKTTIDP 302
Query: 243 FLDLSLEILKADSLQKALANFTAAELLDGGEKQYHCQRCKQKVQAVKQLTIHKAPYVLAI 302
FLDLSL+I L + L +F E L + YHC C A+KQL IHK P VL +
Sbjct: 303 FLDLSLDIKDKKKLYECLDSFHKKEQLK--DFNYHCGECNSTQDAIKQLGIHKLPSVLVL 360
Query: 303 HLKRF--YAHDPNIKIKKKVRFDSALNLKPFVSG------SSDG---DVKYSLYGVLVHS 351
LKRF + N K+ + F + LN+K + S S +G D+ Y L G++ H
Sbjct: 361 QLKRFEHLLNGSNRKLDDFIEFPTYLNMKNYCSTKEKDKHSENGKVPDIIYELIGIVSHK 420
Query: 352 GFSTHSGHYYCYVRTSNNMWYTLDDTRVSHVGEQEVLNQQAYMLFY 397
G + + GHY + + S W+ +D+ VS + ++EVL +QAY+LFY
Sbjct: 421 G-TVNEGHYIAFCKISGGQWFKFNDSMVSSISQEEVLKEQAYLLFY 465
>UBP8_SCHPO (Q09738) Probable ubiquitin carboxyl-terminal hydrolase
8 (EC 3.1.2.15) (Ubiquitin thiolesterase 8)
(Ubiquitin-specific processing protease 8)
(Deubiquitinating enzyme 8)
Length = 449
Score = 100 bits (249), Expect = 2e-20
Identities = 91/320 (28%), Positives = 138/320 (42%), Gaps = 48/320 (15%)
Query: 93 GLWNLGNTCFLNSVLQCLTYTEPLAAYLQSGKHKSSCHIAGFCALCAIQNHVSCALQATG 152
G+ NLG TCF++ +LQ + + + SG H S+ C CAI + S +
Sbjct: 146 GIQNLGATCFMSVILQSILHNPLVRNLFFSGFHTSTDCKRPTCMTCAIDDMFSSIYNSKN 205
Query: 153 R--IVSPQDMVGNLRCILMS-CFFNEQSRHI----------TQFPKG*AGRCT*VYGELT 199
+ P ++ + + S C +++Q H T+ G + CT +
Sbjct: 206 KSTFYGPTAVLNLMWKLSKSLCGYSQQDGHEFFVYLLDQMHTESGGGTSMPCTCPIHRIF 265
Query: 200 *VNA*MLPAFWSTE*ITWCFREKFGS*DLWWSSSKSASNKFDPFLDLSLEILKADSLQKA 259
S + + C K DP +D+SL+I +LQ
Sbjct: 266 ---------SGSLKNVVTCL------------DCKKERVAVDPLMDISLDI-NEPTLQGC 303
Query: 260 LANFTAAELLDGGEKQYHCQRCKQKVQAVKQLTIHKAPYVLAIHLKRFYAHD--PNIKIK 317
L F + E + QY C C K A+KQL K P + + LKRF ++ + KI
Sbjct: 304 LERFVSKEKV-----QYSCHSCGSK-NAIKQLVFDKLPPTICMQLKRFEQNNFAMSTKID 357
Query: 318 KKVRFDSALNLKPFVSGSSDGDVKYSLYGVLVHSGFSTHSGHYYCYVRTSNNMWYTLDDT 377
K+V + + L ++ + DV Y LY V+ H G + +GHY Y N W+ LDDT
Sbjct: 358 KQVSYPAFLRMR---YNFNQDDVDYQLYSVVCHKG-TLDTGHYIAYTYYQNQ-WFLLDDT 412
Query: 378 RVSHVGEQEVLNQQAYMLFY 397
+ V E EVLN QAY+LFY
Sbjct: 413 TIVEVKESEVLNSQAYLLFY 432
>UBPA_YEAST (P53874) Ubiquitin carboxyl-terminal hydrolase 10 (EC
3.1.2.15) (Ubiquitin thiolesterase 10)
(Ubiquitin-specific processing protease 10)
(Deubiquitinating enzyme 10) (Disrupter of telomere
silencing protein 4)
Length = 792
Score = 96.7 bits (239), Expect = 2e-19
Identities = 69/217 (31%), Positives = 111/217 (50%), Gaps = 14/217 (6%)
Query: 231 SSSKSASNKFDPFLDLSLEILKAD---SLQKALANFTAAELL--DGGEKQYHCQRCKQKV 285
SSS N PF S ++ A+ S++K++ +F EL+ D +K Y C++C +
Sbjct: 559 SSSSVNLNNGSPFAAAS-DLSSANRRFSIEKSIKDFFNPELIKVDKEQKGYVCEKCHKTT 617
Query: 286 QAVKQLTIHKAPYVLAIHLKRF-YAHDPNIKIKKKVRFDSALNLKPFVSGSSDGDVKYSL 344
AVK +I +AP L +HLK+F + + K+K+ V + L+L + S + VKY L
Sbjct: 618 NAVKHSSILRAPETLLVHLKKFRFNGTSSSKMKQAVSYPMFLDLTEYCE-SKELPVKYQL 676
Query: 345 YGVLVHSGFSTHSGHYYCYVRTSNNMWYTLDDTRVSHVGEQEVLNQ-QAYMLFYVRDKKS 403
V+VH G S SGHY + + + W T DD ++ + E++VL + AY L Y R
Sbjct: 677 LSVVVHEGRSLSSGHYIAHCKQPDGSWATYDDEYINIISERDVLKEPNAYYLLYTR---- 732
Query: 404 ITPRK-PVNIAKEEISKTNVIGSRYPSTPTNALKDYP 439
+TP+ P+ +AK ++ NV N + P
Sbjct: 733 LTPKSVPLPLAKSAMATGNVTSKSKQEQAVNEPNNRP 769
>UBP2_CHICK (O57429) Ubiquitin carboxyl-terminal hydrolase 2 (EC
3.1.2.15) (Ubiquitin thiolesterase 2)
(Ubiquitin-specific processing protease 2)
(Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific
protease)
Length = 357
Score = 96.7 bits (239), Expect = 2e-19
Identities = 65/170 (38%), Positives = 94/170 (55%), Gaps = 14/170 (8%)
Query: 237 SNKFDPFLDLSLEILKAD----SLQKALANFTAAELLDGGEKQYHCQRCKQKVQAVKQLT 292
S FDPF DLSL I K +L L FT ++LDG EK C RCK + + K+ +
Sbjct: 184 STAFDPFWDLSLPIPKKGYGEVTLMDCLRLFTKEDVLDGDEKPTCC-RCKARTRCTKKFS 242
Query: 293 IHKAPYVLAIHLKRFYAHDPNIKIKKKVRFDS----ALNLKPFVSGSSDGDVKYSLYGVL 348
I K P +L +HLKRF + I+ K F + L+L+ F S S + V Y+LY V
Sbjct: 243 IQKFPKILVLHLKRF--SEARIRASKLTTFVNFPLKDLDLREFASQSCNHAV-YNLYAVS 299
Query: 349 VHSGFSTHSGHYYCYVRTS-NNMWYTLDDTRVSHVGEQEVLNQQAYMLFY 397
HSG +T GHY Y ++ ++ W++ +D+RV+ + V + AY+LFY
Sbjct: 300 NHSG-TTMGGHYTAYCKSPISSEWHSFNDSRVTPMSSSHVRSSDAYLLFY 348
Score = 42.4 bits (98), Expect = 0.005
Identities = 18/27 (66%), Positives = 22/27 (80%)
Query: 93 GLWNLGNTCFLNSVLQCLTYTEPLAAY 119
GL NLGNTCF+NS+LQCL+ T+ L Y
Sbjct: 20 GLRNLGNTCFMNSILQCLSNTKELRDY 46
>UBP9_SCHPO (Q9P7V9) Probable ubiquitin carboxyl-terminal hydrolase
9 (EC 3.1.2.15) (Ubiquitin thiolesterase 9)
(Ubiquitin-specific processing protease 9)
(Deubiquitinating enzyme 9)
Length = 585
Score = 95.5 bits (236), Expect = 5e-19
Identities = 71/232 (30%), Positives = 113/232 (48%), Gaps = 31/232 (13%)
Query: 234 KSASNKFDPFLDLSLEILKADSLQKALANFTAAELLDGGEKQYHCQRCKQKVQAVKQLTI 293
++ +++ + FLDLS++I S+ L +F+A+E+L K +HC CK +A K++ I
Sbjct: 248 ENITSRDESFLDLSIDIENHTSVTSCLRSFSASEMLSSKNK-FHCDVCKSLQEAEKRMKI 306
Query: 294 HKAPYVLAIHLKRFYAHDP---NIKIKKKVRFDSALNLKPFVSGSSDGDVKYSLYGVLVH 350
K P +L++HLKRF ++ + K+ + F + + L + + + Y L V+VH
Sbjct: 307 KKLPKILSLHLKRFKYNETQEGHDKLFYTIVFTNEMRLFTTTEDAENAERMYYLSSVIVH 366
Query: 351 SGFSTHSGHYYCYVRTSNNMWYTLDDTRVSHVGE---QEVLNQQ-----AYMLFY----- 397
G H GHY VRT W DD V+ V E Q Q AY+LFY
Sbjct: 367 VGGGPHRGHYVSIVRTKTYGWVLFDDENVTPVNENYLQRFFGDQPGQATAYVLFYTAADE 426
Query: 398 -------VRDKKSITPRK-PVNIAKEEISKTNVIGSRYPSTP-TNALKDYPN 440
V K+SI P P + +E + +N+ STP +N+ YP+
Sbjct: 427 EDDDVSEVDTKESIKPMSIPSQLKQESVEVSNL-----SSTPRSNSTITYPD 473
Score = 32.0 bits (71), Expect = 6.9
Identities = 19/51 (37%), Positives = 27/51 (52%), Gaps = 2/51 (3%)
Query: 93 GLWNLGNTCFLNSVLQCLTYTEPLAAYLQSGKHKSSCHIAGFCALCAIQNH 143
GL N GNTC+++SVL L + +P L S S+ F ++C NH
Sbjct: 42 GLTNYGNTCYVSSVLVSLYHLKPFRDSLNSYPLPSA--PPNFKSVCTKTNH 90
>UBP3_HUMAN (Q9Y6I4) Ubiquitin carboxyl-terminal hydrolase 3 (EC
3.1.2.15) (Ubiquitin thiolesterase 3)
(Ubiquitin-specific processing protease 3)
(Deubiquitinating enzyme 3)
Length = 521
Score = 94.0 bits (232), Expect = 1e-18
Identities = 68/182 (37%), Positives = 93/182 (50%), Gaps = 23/182 (12%)
Query: 237 SNKFDPFLDLSLEILK--------------ADSLQKALANFTAAELLDGGEKQYHCQRCK 282
S KFDPFLDLSL+I SL+ L +FT E LD E Y C +CK
Sbjct: 332 SRKFDPFLDLSLDIPSQFRSKRSKNQENGPVCSLRDCLRSFTDLEELDETEL-YMCHKCK 390
Query: 283 QKVQAVKQLTIHKAPYVLAIHLKRFY--AHDPNIKIKKKVRFD-SALNLKPFV---SGSS 336
+K ++ K+ I K P VL +HLKRF+ A+ N K+ V F L++K ++ S
Sbjct: 391 KKQKSTKKFWIQKLPKVLCLHLKRFHWTAYLRN-KVDTYVEFPLRGLDMKWYLLEPENSG 449
Query: 337 DGDVKYSLYGVLVHSGFSTHSGHYYCYVRTSNNMWYTLDDTRVSHVGEQEVLNQQAYMLF 396
Y L V+VH G SGHY Y T W+ +D+ V+ E+ V+ +AY+LF
Sbjct: 450 PESCLYDLAAVVVHHGSGVGSGHYTAYA-THEGRWFHFNDSTVTLTDEETVVKAKAYILF 508
Query: 397 YV 398
YV
Sbjct: 509 YV 510
Score = 37.4 bits (85), Expect = 0.17
Identities = 17/38 (44%), Positives = 22/38 (57%)
Query: 84 GITFRRIGAGLWNLGNTCFLNSVLQCLTYTEPLAAYLQ 121
G T GL NLGNTCF+N++LQ L+ E Y +
Sbjct: 151 GSTTAICATGLRNLGNTCFMNAILQSLSNIEQFCCYFK 188
>UBP2_MOUSE (O88623) Ubiquitin carboxyl-terminal hydrolase 2 (EC
3.1.2.15) (Ubiquitin thiolesterase 2)
(Ubiquitin-specific processing protease 2)
(Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific
protease)
Length = 353
Score = 94.0 bits (232), Expect = 1e-18
Identities = 61/170 (35%), Positives = 93/170 (53%), Gaps = 14/170 (8%)
Query: 237 SNKFDPFLDLSLEILKAD----SLQKALANFTAAELLDGGEKQYHCQRCKQKVQAVKQLT 292
S FDPF DLSL I K +L + FT ++LDG EK C RC+ + + +K+ +
Sbjct: 180 STVFDPFWDLSLPIAKRGYPEVTLMDCMRLFTKEDILDGDEKPTCC-RCRARKRCIKKFS 238
Query: 293 IHKAPYVLAIHLKRFYAHDPNIKIKKKVRFDS----ALNLKPFVSGSSDGDVKYSLYGVL 348
+ + P +L +HLKRF + I+ K F + L+L+ F S +++ V Y+LY V
Sbjct: 239 VQRFPKILVLHLKRF--SESRIRTSKLTTFVNFPLRDLDLREFASENTNHAV-YNLYAVS 295
Query: 349 VHSGFSTHSGHYYCYVRTS-NNMWYTLDDTRVSHVGEQEVLNQQAYMLFY 397
HSG +T GHY Y R+ W+T +D+ V+ + +V AY+LFY
Sbjct: 296 NHSG-TTMGGHYTAYCRSPVTGEWHTFNDSSVTPMSSSQVRTSDAYLLFY 344
Score = 46.2 bits (108), Expect = 4e-04
Identities = 24/47 (51%), Positives = 29/47 (61%), Gaps = 6/47 (12%)
Query: 92 AGLWNLGNTCFLNSVLQCLTYTEPL------AAYLQSGKHKSSCHIA 132
AGL NLGNTCF+NS+LQCL+ T L Y++ H SS H A
Sbjct: 15 AGLRNLGNTCFMNSILQCLSNTRELRDYCLQRLYMRDLGHTSSAHTA 61
>UBP2_HUMAN (O75604) Ubiquitin carboxyl-terminal hydrolase 2 (EC
3.1.2.15) (Ubiquitin thiolesterase 2)
(Ubiquitin-specific processing protease 2)
(Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific
protease)
Length = 605
Score = 94.0 bits (232), Expect = 1e-18
Identities = 62/170 (36%), Positives = 93/170 (54%), Gaps = 14/170 (8%)
Query: 237 SNKFDPFLDLSLEILKAD----SLQKALANFTAAELLDGGEKQYHCQRCKQKVQAVKQLT 292
S FDPF DLSL I K +L + FT ++LDG EK C RC+ + + +K+ +
Sbjct: 432 STVFDPFWDLSLPIAKRGYPEVTLMDCMRLFTKEDVLDGDEKPTCC-RCRGRKRCIKKFS 490
Query: 293 IHKAPYVLAIHLKRFYAHDPNIKIKKKVRFDS----ALNLKPFVSGSSDGDVKYSLYGVL 348
I + P +L +HLKRF + I+ K F + L+L+ F S +++ V Y+LY V
Sbjct: 491 IQRFPKILVLHLKRF--SESRIRTSKLTTFVNFPLRDLDLREFASENTNHAV-YNLYAVS 547
Query: 349 VHSGFSTHSGHYYCYVRT-SNNMWYTLDDTRVSHVGEQEVLNQQAYMLFY 397
HSG +T GHY Y R+ W+T +D+ V+ + +V AY+LFY
Sbjct: 548 NHSG-TTMGGHYTAYCRSPGTGEWHTFNDSSVTPMSSSQVRTSDAYLLFY 596
Score = 50.1 bits (118), Expect = 2e-05
Identities = 29/75 (38%), Positives = 42/75 (55%), Gaps = 12/75 (16%)
Query: 64 PQPSKNSEFSEFGLDPELSLGITFRRIGAGLWNLGNTCFLNSVLQCLTYTEPL------A 117
P PS++S G++ + + G+ AGL NLGNTCF+NS+LQCL+ T L
Sbjct: 245 PGPSRSSSPGRDGMNSKSAQGL------AGLRNLGNTCFMNSILQCLSNTRELRDYCLQR 298
Query: 118 AYLQSGKHKSSCHIA 132
Y++ H S+ H A
Sbjct: 299 LYMRDLHHGSNAHTA 313
>UB33_HUMAN (Q8TEY7) Ubiquitin carboxyl-terminal hydrolase 33 (EC
3.1.2.15) (Ubiquitin thiolesterase 33)
(Ubiquitin-specific processing protease 33)
(Deubiquitinating enzyme 33) (VHL-interacting
deubiquitinating enzyme 1)
Length = 942
Score = 93.6 bits (231), Expect = 2e-18
Identities = 60/148 (40%), Positives = 82/148 (54%), Gaps = 8/148 (5%)
Query: 255 SLQKALANFTAAELLDGGEKQYHCQRCKQKVQAVKQLTIHKAPYVLAIHLKRFYAHDP-- 312
+LQ LA F A + L G + Y C++CK+ VK + P +L IHLKRF H+
Sbjct: 568 TLQDCLAAFFARDELKG-DNMYSCEKCKKLRNGVKFCKVQNFPEILCIHLKRF-RHELMF 625
Query: 313 NIKIKKKVRFD-SALNLKPFVSGSSDGD-VKYSLYGVLVHSGFSTHSGHYYCYVRTS-NN 369
+ KI V F L+L+PF++ S V Y L V+ H G ++ SGHY Y R + NN
Sbjct: 626 STKISTHVSFPLEGLDLQPFLAKDSPAQIVTYDLLSVICHHGTAS-SGHYIAYCRNNLNN 684
Query: 370 MWYTLDDTRVSHVGEQEVLNQQAYMLFY 397
+WY DD V+ V E V N +AY+LFY
Sbjct: 685 LWYEFDDQSVTEVSESTVQNAEAYVLFY 712
Score = 33.9 bits (76), Expect = 1.8
Identities = 13/27 (48%), Positives = 19/27 (70%)
Query: 93 GLWNLGNTCFLNSVLQCLTYTEPLAAY 119
GL N+GNTC++N+ LQ L+ PL +
Sbjct: 186 GLKNIGNTCYMNAALQALSNCPPLTQF 212
>UB21_HUMAN (Q9UK80) Ubiquitin carboxyl-terminal hydrolase 21 (EC
3.1.2.15) (Ubiquitin thiolesterase 21)
(Ubiquitin-specific processing protease 21)
(Deubiquitinating enzyme 21) (NEDD8-specific protease)
Length = 565
Score = 92.0 bits (227), Expect = 6e-18
Identities = 65/170 (38%), Positives = 87/170 (50%), Gaps = 14/170 (8%)
Query: 237 SNKFDPFLDLSLEILKAD------SLQKALANFTAAELLDGGEKQYHCQRCKQKVQAVKQ 290
S F+ F DLSL I K SL+ FT E L+ E C RC+QK ++ K+
Sbjct: 391 STTFEVFCDLSLPIPKKGFAGGKVSLRDCFNLFTKEEELES-ENAPVCDRCRQKTRSTKK 449
Query: 291 LTIHKAPYVLAIHLKRFYAHDPNIKIKKKVRFD---SALNLKPFVSGSSDGDVKYSLYGV 347
LT+ + P +L +HL RF A +IK K V D L+L F S + G Y LY +
Sbjct: 450 LTVQRFPRILVLHLNRFSASRGSIK-KSSVGVDFPLQRLSLGDFASDKA-GSPVYQLYAL 507
Query: 348 LVHSGFSTHSGHYYCYVRTSNNMWYTLDDTRVSHVGEQEVLNQQAYMLFY 397
HSG S H GHY R W+ +D+RVS V E +V + + Y+LFY
Sbjct: 508 CNHSG-SVHYGHYTALCRCQTG-WHVYNDSRVSPVSENQVASSEGYVLFY 555
Score = 44.3 bits (103), Expect = 0.001
Identities = 33/86 (38%), Positives = 44/86 (50%), Gaps = 13/86 (15%)
Query: 32 FKGFSNDFNIETLNPTTSEHRQLVSANIQSSQPQPSKNSEFSEFGLDPELSLGITFRRIG 91
F G F+I T P + ++SA +SS+P F D +++ G
Sbjct: 163 FPGPPTLFSIRTEPPASHGSFHMISA--RSSEP----------FYSDDKMAHHTLLLGSG 210
Query: 92 -AGLWNLGNTCFLNSVLQCLTYTEPL 116
GL NLGNTCFLN+VLQCL+ T PL
Sbjct: 211 HVGLRNLGNTCFLNAVLQCLSSTRPL 236
>UB21_MOUSE (Q9QZL6) Ubiquitin carboxyl-terminal hydrolase 21 (EC
3.1.2.15) (Ubiquitin thiolesterase 21)
(Ubiquitin-specific processing protease 21)
(Deubiquitinating enzyme 21)
Length = 566
Score = 91.7 bits (226), Expect = 7e-18
Identities = 64/170 (37%), Positives = 87/170 (50%), Gaps = 14/170 (8%)
Query: 237 SNKFDPFLDLSLEILKAD------SLQKALANFTAAELLDGGEKQYHCQRCKQKVQAVKQ 290
S F+ F DLSL I K SL+ + FT E L+ E C RC+QK ++ K+
Sbjct: 392 STTFEVFCDLSLPIPKKGFAGGKVSLRDCFSLFTKEEELES-ENAPVCDRCRQKTRSTKK 450
Query: 291 LTIHKAPYVLAIHLKRFYAHDPNIKIKKKVRFD---SALNLKPFVSGSSDGDVKYSLYGV 347
LT+ + P +L +HL RF +IK K V D L+L F S + G Y LY +
Sbjct: 451 LTVQRFPRILVLHLNRFSTSRGSIK-KSSVGVDFPLQRLSLGDFASDKA-GSPVYQLYAL 508
Query: 348 LVHSGFSTHSGHYYCYVRTSNNMWYTLDDTRVSHVGEQEVLNQQAYMLFY 397
HSG S H GHY R W+ +D+RVS V E +V + + Y+LFY
Sbjct: 509 CNHSG-SVHYGHYTALCRCQTG-WHVYNDSRVSPVSENQVASSEGYVLFY 556
Score = 43.5 bits (101), Expect = 0.002
Identities = 19/24 (79%), Positives = 21/24 (87%)
Query: 93 GLWNLGNTCFLNSVLQCLTYTEPL 116
GL NLGNTCFLN+VLQCL+ T PL
Sbjct: 213 GLRNLGNTCFLNAVLQCLSSTRPL 236
>UBP3_MOUSE (Q91W36) Ubiquitin carboxyl-terminal hydrolase 3 (EC
3.1.2.15) (Ubiquitin thiolesterase 3)
(Ubiquitin-specific processing protease 3)
(Deubiquitinating enzyme 3)
Length = 520
Score = 89.7 bits (221), Expect = 3e-17
Identities = 66/182 (36%), Positives = 91/182 (49%), Gaps = 23/182 (12%)
Query: 237 SNKFDPFLDLSLEILK--------------ADSLQKALANFTAAELLDGGEKQYHCQRCK 282
S KFDPFLDLSL+I SL+ L +FT E LD E Y C +CK
Sbjct: 331 SRKFDPFLDLSLDIPSQFRSKRSKNQENGPVCSLRDCLRSFTDLEELDETEL-YMCHKCK 389
Query: 283 QKVQAVKQLTIHKAPYVLAIHLKRFY--AHDPNIKIKKKVRFD-SALNLKPFV---SGSS 336
+K ++ K+ I K P L +HLKRF+ A+ N K+ V+F L++K ++ S
Sbjct: 390 KKQKSTKKFWIQKLPKALCLHLKRFHWTAYLRN-KVDTYVQFPLRGLDMKCYLLEPENSG 448
Query: 337 DGDVKYSLYGVLVHSGFSTHSGHYYCYVRTSNNMWYTLDDTRVSHVGEQEVLNQQAYMLF 396
Y L V+VH G SGHY Y W+ +D+ V+ E+ V +AY+LF
Sbjct: 449 PDSCLYDLAAVVVHHGSGVGSGHYTAYA-VHEGRWFHFNDSTVTVTDEETVGKAKAYILF 507
Query: 397 YV 398
YV
Sbjct: 508 YV 509
Score = 37.4 bits (85), Expect = 0.17
Identities = 17/38 (44%), Positives = 22/38 (57%)
Query: 84 GITFRRIGAGLWNLGNTCFLNSVLQCLTYTEPLAAYLQ 121
G T GL NLGNTCF+N++LQ L+ E Y +
Sbjct: 151 GSTTAICATGLRNLGNTCFMNAILQSLSNIEQFCCYFK 188
>UBPX_CAEEL (P34547) Probable ubiquitin carboxyl-terminal hydrolase
R10E11.3 (EC 3.1.2.15) (Ubiquitin thiolesterase)
(Ubiquitin-specific processing protease)
(Deubiquitinating enzyme)
Length = 410
Score = 89.4 bits (220), Expect = 4e-17
Identities = 53/148 (35%), Positives = 80/148 (53%), Gaps = 5/148 (3%)
Query: 232 SSKSASNKFDPFLDLSLEILKADSLQKALANFTAAELLDGGEKQYHCQRCKQKVQAVKQL 291
S ++ S+K + FLDLS+++ + S+ L F+ E L G +K Y C+ C K +A K++
Sbjct: 213 SCETVSSKDEDFLDLSIDVEQNTSISHCLRVFSETETLCGDQK-YFCETCSSKQEAQKRM 271
Query: 292 TIHKAPYVLAIHLKRFYAHDP---NIKIKKKVRFDSALNLKPFVSGSSDGDVKYSLYGVL 348
I K P +LA+HLKRF +P + K+ +V F L L + GD Y L +
Sbjct: 272 RIKKPPQLLALHLKRFKFVEPLNRHTKLSYRVVFPLELRLFNVSDDAEYGDRMYDLVATV 331
Query: 349 VHSGFSTHSGHYYCYVRTSNNMWYTLDD 376
VH G + + GHY V+ SN+ W DD
Sbjct: 332 VHCGATPNRGHYITLVK-SNSFWLVFDD 358
Score = 33.5 bits (75), Expect = 2.4
Identities = 22/72 (30%), Positives = 34/72 (46%), Gaps = 6/72 (8%)
Query: 93 GLWNLGNTCFLNSVLQCLTYTEPLAAYLQSGKHKSSCHIAGFCALCAIQNHVSCALQATG 152
GL N GNTC+ NSV+Q L + P + + +K + +G + N V+C
Sbjct: 28 GLVNFGNTCYCNSVIQALFFCRPFREKVLN--YKQTLKKSG----ASKDNLVTCLADLFH 81
Query: 153 RIVSPQDMVGNL 164
I S + VG +
Sbjct: 82 SIASQKRRVGTI 93
>UBP9_YEAST (P39967) Ubiquitin carboxyl-terminal hydrolase 9 (EC
3.1.2.15) (Ubiquitin thiolesterase 9)
(Ubiquitin-specific processing protease 9)
(Deubiquitinating enzyme 9)
Length = 754
Score = 87.0 bits (214), Expect = 2e-16
Identities = 58/191 (30%), Positives = 97/191 (50%), Gaps = 20/191 (10%)
Query: 237 SNKFDPFLDLSLEIL--KADSLQKALANFTAAELLDGGEKQYHCQRCKQKVQAVKQLTIH 294
+++ +PFLD +E+ + +QK L ++ E+L+G K ++C +C +A + + +
Sbjct: 494 TSRDEPFLDFPIEVQGDEETDIQKMLKSYHQREMLNGVNK-FYCNKCYGLQEAERMVGLK 552
Query: 295 KAPYVLAIHLKRF-YAHDP--NIKIKKKVRFDSALNLKPFVSGSSDGDV--KYSLYGVLV 349
+ P++L++HLKRF Y+ + NIK+ K+ + L+ VS + + V KY L GV++
Sbjct: 553 QLPHILSLHLKRFKYSEEQKSNIKLFNKILYPLTLD----VSSTFNTSVYKKYELSGVVI 608
Query: 350 HSGFSTHSGHYYCYVRTSNNMWYTLDDTRVSHVGEQEVL--------NQQAYMLFYVRDK 401
H G GHY C R W DD V + E+ VL AY+LFY +
Sbjct: 609 HMGSGPQHGHYVCICRNEKFGWLLYDDETVESIKEETVLQFTGHPGDQTTAYVLFYKETQ 668
Query: 402 KSITPRKPVNI 412
T + NI
Sbjct: 669 ADKTENQNENI 679
Score = 32.3 bits (72), Expect = 5.3
Identities = 13/18 (72%), Positives = 14/18 (77%)
Query: 93 GLWNLGNTCFLNSVLQCL 110
G N GNTC+ NSVLQCL
Sbjct: 135 GYENFGNTCYCNSVLQCL 152
>FAF_DROME (P55824) Probable ubiquitin carboxyl-terminal hydrolase FAF
(EC 3.1.2.15) (Ubiquitin thiolesterase FAF)
(Ubiquitin-specific processing protease FAF)
(Deubiquitinating enzyme FAF) (Fat facets protein)
Length = 2778
Score = 87.0 bits (214), Expect = 2e-16
Identities = 59/168 (35%), Positives = 86/168 (51%), Gaps = 27/168 (16%)
Query: 238 NKFDPFLDLSLEILKADSLQKALANFTAAELLDGGEKQYHCQRCKQKVQAVKQLTIHKAP 297
+K +PF S++I SL ++L + ELL+G + YHC +C +KV VK++ + K P
Sbjct: 1847 SKEEPFSVFSVDIRNHSSLTESLEQYVKGELLEGADA-YHCDKCDKKVVTVKRVCVKKLP 1905
Query: 298 YVLAIHLKRF---YAHDPNIKIKKKVRFDSALNLKPF-VSGSS--DGDV----------- 340
VLAI LKRF Y IK F L+++P+ VSG + +G+V
Sbjct: 1906 PVLAIQLKRFEYDYERVCAIKFNDYFEFPRILDMEPYTVSGLAKLEGEVVEVGDNCQTNV 1965
Query: 341 ---KYSLYGVLVHSGFSTHSGHYYCYVRTSNN-----MWYTLDDTRVS 380
KY L G++VHSG GHY+ Y+ + N WY DD V+
Sbjct: 1966 ETTKYELTGIVVHSG-QASGGHYFSYILSKNPANGKCQWYKFDDGEVT 2012
>UB49_MOUSE (Q6P9L4) Ubiquitin carboxyl-terminal hydrolase 49 (EC
3.1.2.15) (Ubiquitin thiolesterase 49)
(Ubiquitin-specific processing protease 49)
(Deubiquitinating enzyme 49)
Length = 685
Score = 85.9 bits (211), Expect = 4e-16
Identities = 66/198 (33%), Positives = 89/198 (44%), Gaps = 38/198 (19%)
Query: 237 SNKFDPFLDLSLEILKADS---------------LQKALANFTAAELLDGGEKQYHCQRC 281
SN +PF DLSLE + L + LA FT E L+G + Y C +C
Sbjct: 459 SNTIEPFWDLSLEFPERYHCIEKGFVPLNQTECLLTEMLAKFTETEALEG--RIYACDQC 516
Query: 282 KQK-----------VQAVKQLTIHKAPYVLAIHLKRF--YAHDPNIKIKKKVRFDSALNL 328
K +A KQL I++ P VL +HLKRF + KI V FD L +
Sbjct: 517 NSKRRKSNPKPLVLSEARKQLMIYRLPQVLRLHLKRFRWSGRNHREKIGVHVIFDQVLTM 576
Query: 329 KPF-----VSGSSDGDVKYSLYGVLVHSGFSTHSGHY--YCYVRTSNNMWYTLDDTRVSH 381
+P+ +S Y L V++H G SGHY YCY T W +D+++
Sbjct: 577 EPYCCRDMLSSLDKETFAYDLSAVVMHHGKGFGSGHYTAYCY-NTEGGFWVHCNDSKLDV 635
Query: 382 VGEQEVLNQQAYMLFYVR 399
+EV QAY+LFY R
Sbjct: 636 CSVEEVCKTQAYILFYTR 653
Score = 35.0 bits (79), Expect = 0.82
Identities = 16/36 (44%), Positives = 25/36 (69%), Gaps = 6/36 (16%)
Query: 79 PELSLGITFRRIGAGLWNLGNTCFLNSVLQCLTYTE 114
P ++ G+T GL NLGNTC++NS+LQ L++ +
Sbjct: 243 PAVAPGVT------GLRNLGNTCYMNSILQVLSHLQ 272
>UB20_HUMAN (Q9Y2K6) Ubiquitin carboxyl-terminal hydrolase 20 (EC
3.1.2.15) (Ubiquitin thiolesterase 20)
(Ubiquitin-specific processing protease 20)
(Deubiquitinating enzyme 20)
Length = 913
Score = 85.9 bits (211), Expect = 4e-16
Identities = 56/148 (37%), Positives = 78/148 (51%), Gaps = 8/148 (5%)
Query: 255 SLQKALANFTAAELLDGGEKQYHCQRCKQKVQAVKQLTIHKAPYVLAIHLKRFYAHDP-- 312
+L+ LA F AA+ L G + Y C+RCK+ VK + + P +L IHLKRF H+
Sbjct: 537 TLEDCLAAFFAADELKG-DNMYSCERCKKLRNGVKYCKVLRLPEILCIHLKRF-RHEVMY 594
Query: 313 NIKIKKKVRFD-SALNLKPFVSGSSDGDV-KYSLYGVLVHSGFSTHSGHYYCYVR-TSNN 369
+ KI V F L+L+PF++ + Y L V+ H G + SGHY Y + N
Sbjct: 595 SFKINSHVSFPLEGLDLRPFLAKECTSQITTYDLLSVICHHG-TAGSGHYIAYCQNVING 653
Query: 370 MWYTLDDTRVSHVGEQEVLNQQAYMLFY 397
WY DD V+ V E V N + Y+LFY
Sbjct: 654 QWYEFDDQYVTEVHETVVQNAEGYVLFY 681
Score = 32.7 bits (73), Expect = 4.1
Identities = 23/89 (25%), Positives = 38/89 (41%), Gaps = 15/89 (16%)
Query: 31 PFKGFSNDFNIETLNPTTSEHRQLVSANIQSSQPQPSKNSEFSEFGLDPELSLGITFRRI 90
P G S+ F+ E +P S + V + S++ + GL
Sbjct: 99 PLLGSSSKFS-EQDSPPPSHPLKAVPIAVADEGESESEDDDLKPRGL------------- 144
Query: 91 GAGLWNLGNTCFLNSVLQCLTYTEPLAAY 119
G+ NLGN+C++N+ LQ L+ PL +
Sbjct: 145 -TGMKNLGNSCYMNAALQALSNCPPLTQF 172
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.320 0.134 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 94,260,072
Number of Sequences: 164201
Number of extensions: 3947771
Number of successful extensions: 12286
Number of sequences better than 10.0: 123
Number of HSP's better than 10.0 without gapping: 92
Number of HSP's successfully gapped in prelim test: 31
Number of HSP's that attempted gapping in prelim test: 11843
Number of HSP's gapped (non-prelim): 372
length of query: 835
length of database: 59,974,054
effective HSP length: 119
effective length of query: 716
effective length of database: 40,434,135
effective search space: 28950840660
effective search space used: 28950840660
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 70 (31.6 bits)
Medicago: description of AC122160.3


