

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
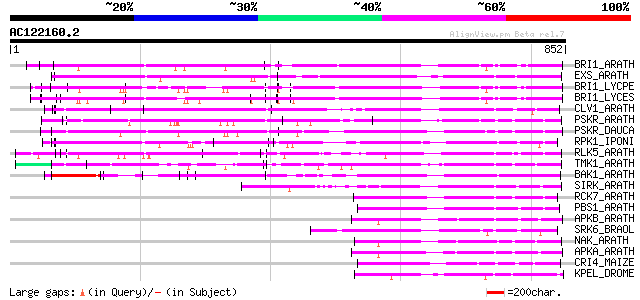
Query= AC122160.2 - phase: 0
(852 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC ... 369 e-101
EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase E... 361 4e-99
BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.... 345 4e-94
BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precurso... 344 5e-94
CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (... 301 6e-81
PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (... 298 3e-80
PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.... 286 1e-76
RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC 2... 285 3e-76
RLK5_ARATH (P47735) Receptor-like protein kinase 5 precursor (EC... 278 4e-74
TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precur... 213 2e-54
BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated rec... 191 5e-48
SIRK_ARATH (O64483) Senescence-induced receptor-like serine/thre... 167 1e-40
RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase RLC... 166 3e-40
PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC 2.7... 166 3e-40
APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor ... 154 1e-36
SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase rec... 147 1e-34
NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK ... 147 1e-34
APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor ... 147 1e-34
CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4 pr... 135 3e-31
KPEL_DROME (Q05652) Probable serine/threonine-protein kinase pel... 134 8e-31
>BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC
2.7.1.37) (AtBRI1) (Brassinosteroid LRR receptor kinase)
Length = 1196
Score = 369 bits (946), Expect = e-101
Identities = 270/822 (32%), Positives = 412/822 (49%), Gaps = 95/822 (11%)
Query: 68 VVELNLSGIGLTGPI-PDTTIGKLNKLHSLDLSNNKIT-TLPSDFWSLTSLKSLNLSSNH 125
++ L+LS +GPI P+ N L L L NN T +P + + L SL+LS N+
Sbjct: 392 LLTLDLSSNNFSGPILPNLCQNPKNTLQELYLQNNGFTGKIPPTLSNCSELVSLHLSFNY 451
Query: 126 ISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGILKC 185
+SG++ +++G+ L + L N EIP+ L + +L+ L LD N IPSG+ C
Sbjct: 452 LSGTIPSSLGSLSKLRDLKLWLNMLEGEIPQELMYVKTLETLILDFNDLTGEIPSGLSNC 511
Query: 186 QSLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGV-SNFSRLKSIVSLNISGN 244
+L I LS+N+L+G +P G L L L+ N+ G + + +S++ L+++ N
Sbjct: 512 TNLNWISLSNNRLTGEIPKWIG-RLENLAILKLSNNSFSGNIPAELGDCRSLIWLDLNTN 570
Query: 245 SFQGSIIEV-----------FVLKLEALDLSRN-------------QFQGHISQVKYNWS 280
F G+I F+ + + + +FQG S+ S
Sbjct: 571 LFNGTIPAAMFKQSGKIAANFIAGKRYVYIKNDGMKKECHGAGNLLEFQGIRSEQLNRLS 630
Query: 281 HLVYLDLSENQLSGEIFQNLNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSL 340
+++ G +N+ ++ L ++ N S +I + L LNL +
Sbjct: 631 TRNPCNITSRVYGGHTSPTFDNNGSMMFLDMSYNMLSGYIPKEIGSMPYLFILNLGHNDI 690
Query: 341 VGHIPDEISHLGNLNALDLSMNHLDGKIPLLKNK--HLQVIDFSHNNLSGPVPSF-ILKS 397
G IPDE+ L LN LDLS N LDG+IP + L ID S+NNLSGP+P ++
Sbjct: 691 SGSIPDEVGDLRGLNILDLSSNKLDGRIPQAMSALTMLTEIDLSNNNLSGPIPEMGQFET 750
Query: 398 LPKMKKYNFSYNNLTLCASEIKPDIMKTSFFGSVNSCPIAANPSFFKKRRDVGHRGMKLA 457
P K NN LC + C + + +R G R LA
Sbjct: 751 FPPAK----FLNNPGLCGYPLP-------------RCDPSNADGYAHHQRSHGRRPASLA 793
Query: 458 LVLTLSLIFALA---GILFLAFGCRRKNKMWEVKQGSYREEQNISGPFSFQTDSTTW-VA 513
+ + L+F+ G++ + R++ + E + Y E SG + ++T W +
Sbjct: 794 GSVAMGLLFSFVCIFGLILVGREMRKRRRKKEAELEMYAEGHGNSGDRT--ANNTNWKLT 851
Query: 514 DVKQATSVPVVIFEKPLLNITFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVK 573
VK+A S+ + FEKPL +TFADLL AT+ F +L+ G FG VY+ L VA+K
Sbjct: 852 GVKEALSINLAAFEKPLRKLTFADLLQATNGFHNDSLIGSGGFGDVYKAILKDGSAVAIK 911
Query: 574 VLVVGSTLTDEEAARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDL 633
L+ S D E E+E +G+IKH NLVPL GYC GD+R+ +Y++M+ G+L+++L+D
Sbjct: 912 KLIHVSGQGDREFMAEMETIGKIKHRNLVPLLGYCKVGDERLLVYEFMKYGSLEDVLHD- 970
Query: 634 PLGVQSTDDWSTDTWEEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPII 693
G+ W R KIA+G+AR LAFLHH CSP II
Sbjct: 971 ------------------------PKKAGVKLNWSTRRKIAIGSARGLAFLHHNCSPHII 1006
Query: 694 HRAVKASSVYLDYDLEPRLSDFGLAKIFGSGLDEEIA----RGSPGYVPPEFSQPEFESP 749
HR +K+S+V LD +LE R+SDFG+A++ S +D ++ G+PGYVPPE+ Q F
Sbjct: 1007 HRDMKSSNVLLDENLEARVSDFGMARLM-SAMDTHLSVSTLAGTPGYVPPEYYQ-SFRCS 1064
Query: 750 TPKSDVYCFGVVLFELLTGKKPV-GDDYTDDKEATTLVSWVRGLVRKNQTSRAIDPKIC- 807
T K DVY +GVVL ELLTGK+P D+ D+ LV WV+ K + S DP++
Sbjct: 1065 T-KGDVYSYGVVLLELLTGKRPTDSPDFGDN----NLVGWVKQHA-KLRISDVFDPELMK 1118
Query: 808 -DTGSDEQIEEALKEVGYLCTADLPFKRPTMQQIVGLLKDIE 848
D + ++ + LK V C D ++RPTM Q++ + K+I+
Sbjct: 1119 EDPALEIELLQHLK-VAVACLDDRAWRRPTMVQVMAMFKEIQ 1159
Score = 115 bits (287), Expect = 6e-25
Identities = 124/406 (30%), Positives = 191/406 (46%), Gaps = 33/406 (8%)
Query: 26 TDEFYVSEFLKKMGLTSSSKVYNFSSSVCSWKGVYCDSNKEHVVELNLSGIGLTGPIPDT 85
T F+ S F + S +Y + S+K V D N N + G
Sbjct: 12 TTLFFFSFFSLSFQASPSQSLYREIHQLISFKDVLPDKNLLPDWSSNKNPCTFDG----- 66
Query: 86 TIGKLNKLHSLDLSNNKI----TTLPSDFWSLTSLKSLNLSSNHISGSLTNNIGNFGLLE 141
+ +K+ S+DLS+ + + + S SLT L+SL LS++HI+GS++ L
Sbjct: 67 VTCRDDKVTSIDLSSKPLNVGFSAVSSSLLSLTGLESLFLSNSHINGSVSG-FKCSASLT 125
Query: 142 NFDLSKNSFSDEIPE--ALSSLVSLKVLKLDHNMFVRSIP---SGILKCQSLVSIDLSSN 196
+ DLS+NS S + +L S LK L + N P SG LK SL +DLS+N
Sbjct: 126 SLDLSRNSLSGPVTTLTSLGSCSGLKFLNVSSNTL--DFPGKVSGGLKLNSLEVLDLSAN 183
Query: 197 QLSGTLPHGF--GDAFPKLRTLNLAENNIYGGVSNFSRLKSIVSLNISGNSFQGSIIEVF 254
+SG G+ D +L+ L ++ N I G V + SR ++ L++S N+F I +
Sbjct: 184 SISGANVVGWVLSDGCGELKHLAISGNKISGDV-DVSRCVNLEFLDVSSNNFSTGIPFLG 242
Query: 255 VLK-LEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQNLNNSMNLKHLSLAC 313
L+ LD+S N+ G S+ + L L++S NQ G I S L++LSLA
Sbjct: 243 DCSALQHLDISGNKLSGDFSRAISTCTELKLLNISSNQFVGPIPPLPLKS--LQYLSLAE 300
Query: 314 NRFSRQKFPKIEMLLG----LEYLNLSKTSLVGHIPDEISHLGNLNALDLSMNHLDGKIP 369
N+F+ + P + L G L L+LS G +P L +L LS N+ G++P
Sbjct: 301 NKFTGE-IP--DFLSGACDTLTGLDLSGNHFYGAVPPFFGSCSLLESLALSSNNFSGELP 357
Query: 370 ---LLKNKHLQVIDFSHNNLSGPVPSFILKSLPKMKKYNFSYNNLT 412
LLK + L+V+D S N SG +P + + + S NN +
Sbjct: 358 MDTLLKMRGLKVLDLSFNEFSGELPESLTNLSASLLTLDLSSNNFS 403
Score = 112 bits (281), Expect = 3e-24
Identities = 107/358 (29%), Positives = 171/358 (46%), Gaps = 20/358 (5%)
Query: 47 YNFSSSVCSWKGVYCDSNKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKITTL 106
++ + + C++ GV C +K ++L+ + + +++ L L SL LSN+ I
Sbjct: 55 WSSNKNPCTFDGVTCRDDKVTSIDLSSKPLNVGFSAVSSSLLSLTGLESLFLSNSHINGS 114
Query: 107 PSDFWSLTSLKSLNLSSNHISGSLT--NNIGNFGLLENFDLSKNS--FSDEIPEALSSLV 162
S F SL SL+LS N +SG +T ++G+ L+ ++S N+ F ++ L L
Sbjct: 115 VSGFKCSASLTSLDLSRNSLSGPVTTLTSLGSCSGLKFLNVSSNTLDFPGKVSGGL-KLN 173
Query: 163 SLKVLKLDHNMFV-RSIPSGILK--CQSLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLA 219
SL+VL L N ++ +L C L + +S N++SG + L L+++
Sbjct: 174 SLEVLDLSANSISGANVVGWVLSDGCGELKHLAISGNKISGDVD---VSRCVNLEFLDVS 230
Query: 220 ENNIYGGVSNFSRLKSIVSLNISGNSFQGSIIEVF--VLKLEALDLSRNQFQGHISQVKY 277
NN G+ ++ L+ISGN G +L+ L++S NQF G I +
Sbjct: 231 SNNFSTGIPFLGDCSALQHLDISGNKLSGDFSRAISTCTELKLLNISSNQFVGPIPPLPL 290
Query: 278 NWSHLVYLDLSENQLSGEIFQNLNNSMN-LKHLSLACNRFSRQKFPKIEMLLGLEYLNLS 336
L YL L+EN+ +GEI L+ + + L L L+ N F P LE L LS
Sbjct: 291 --KSLQYLSLAENKFTGEIPDFLSGACDTLTGLDLSGNHFYGAVPPFFGSCSLLESLALS 348
Query: 337 KTSLVGHIP-DEISHLGNLNALDLSMNHLDGKIP-LLKN--KHLQVIDFSHNNLSGPV 390
+ G +P D + + L LDLS N G++P L N L +D S NN SGP+
Sbjct: 349 SNNFSGELPMDTLLKMRGLKVLDLSFNEFSGELPESLTNLSASLLTLDLSSNNFSGPI 406
>EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase EXS
precursor (EC 2.7.1.37) (Extra sporogenous cells protein)
(EXCESS MICROSPOROCYTES1 protein)
Length = 1192
Score = 361 bits (927), Expect = 4e-99
Identities = 270/829 (32%), Positives = 395/829 (47%), Gaps = 92/829 (11%)
Query: 70 ELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT-TLPSDFWSLTSLKSLNLSSNHISG 128
EL L+ + G IP+ + KL L +LDL +N T +P W T+L S N + G
Sbjct: 405 ELLLTNNQINGSIPED-LWKL-PLMALDLDSNNFTGEIPKSLWKSTNLMEFTASYNRLEG 462
Query: 129 SLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGILKCQSL 188
L IGN L+ LS N + EIP + L SL VL L+ NMF IP + C SL
Sbjct: 463 YLPAEIGNAASLKRLVLSDNQLTGEIPREIGKLTSLSVLNLNANMFQGKIPVELGDCTSL 522
Query: 189 VSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGVSN-------------FSRLKS 235
++DL SN L G +P A +L+ L L+ NN+ G + + S L+
Sbjct: 523 TTLDLGSNNLQGQIPDKI-TALAQLQCLVLSYNNLSGSIPSKPSAYFHQIEMPDLSFLQH 581
Query: 236 IVSLNISGNSFQGSIIEVF--VLKLEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLS 293
++S N G I E L L + LS N G I ++L LDLS N L+
Sbjct: 582 HGIFDLSYNRLSGPIPEELGECLVLVEISLSNNHLSGEIPASLSRLTNLTILDLSGNALT 641
Query: 294 GEIFQNLNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHLGN 353
G I + + NS+ L+ L+LA N+ + +L L LNL+K L G +P + +L
Sbjct: 642 GSIPKEMGNSLKLQGLNLANNQLNGHIPESFGLLGSLVKLNLTKNKLDGPVPASLGNLKE 701
Query: 354 LNALDLSMNHLDGKIP-------------LLKNK-------------HLQVIDFSHNNLS 387
L +DLS N+L G++ + +NK L+ +D S N LS
Sbjct: 702 LTHMDLSFNNLSGELSSELSTMEKLVGLYIEQNKFTGEIPSELGNLTQLEYLDVSENLLS 761
Query: 388 GPVPSFILKSLPKMKKYNFSYNNLT--LCASEIKPDIMKTSFFGSVNSCPIAANPSFFKK 445
G +P+ I LP ++ N + NNL + + + D K G+ C
Sbjct: 762 GEIPTKIC-GLPNLEFLNLAKNNLRGEVPSDGVCQDPSKALLSGNKELCGRVVGS----- 815
Query: 446 RRDVGHRGMKLAL---VLTLSLIFALAGILFLAFGCRRKNKMWEVKQGSYREEQNISGPF 502
D G KL + L L F + +F+ F RR VKQ E S
Sbjct: 816 --DCKIEGTKLRSAWGIAGLMLGFTIIVFVFV-FSLRRWAMTKRVKQRDDPERMEESRLK 872
Query: 503 SFQTDSTTWVADVK--QATSVPVVIFEKPLLNITFADLLSATSNFDRGTLLAEGKFGPVY 560
F + +++ + + S+ + +FE+PLL + D++ AT +F + ++ +G FG VY
Sbjct: 873 GFVDQNLYFLSGSRSREPLSINIAMFEQPLLKVRLGDIVEATDHFSKKNIIGDGGFGTVY 932
Query: 561 RGFLPGNIHVAVKVLVVGSTLTDEEAARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDY 620
+ LPG VAVK L T + E E+E LG++KHPNLV L GYC ++++ +Y+Y
Sbjct: 933 KACLPGEKTVAVKKLSEAKTQGNREFMAEMETLGKVKHPNLVSLLGYCSFSEEKLLVYEY 992
Query: 621 MENGNLQNLLYDLPLGVQSTDDWSTDTWEEADNGIQNVGSEGLLTTWRFRHKIALGTARA 680
M NG+L D W G+ V W R KIA+G AR
Sbjct: 993 MVNGSL-------------------DHWLRNQTGMLEV------LDWSKRLKIAVGAARG 1027
Query: 681 LAFLHHGCSPPIIHRAVKASSVYLDYDLEPRLSDFGLAKIFGS--GLDEEIARGSPGYVP 738
LAFLHHG P IIHR +KAS++ LD D EP+++DFGLA++ + + G+ GY+P
Sbjct: 1028 LAFLHHGFIPHIIHRDIKASNILLDGDFEPKVADFGLARLISACESHVSTVIAGTFGYIP 1087
Query: 739 PEFSQPEFESPTPKSDVYCFGVVLFELLTGKKPVGDDYTDDKEATTLVSWVRGLVRKNQT 798
PE+ Q T K DVY FGV+L EL+TGK+P G D+ + E LV W + + +
Sbjct: 1088 PEYGQS--ARATTKGDVYSFGVILLELVTGKEPTGPDF-KESEGGNLVGWAIQKINQGKA 1144
Query: 799 SRAIDPKICDTGSDEQIEEALKEVGYLCTADLPFKRPTMQQIVGLLKDI 847
IDP + + + L ++ LC A+ P KRP M ++ LK+I
Sbjct: 1145 VDVIDPLLVSVAL-KNSQLRLLQIAMLCLAETPAKRPNMLDVLKALKEI 1192
Score = 152 bits (384), Expect = 4e-36
Identities = 118/353 (33%), Positives = 173/353 (48%), Gaps = 8/353 (2%)
Query: 64 NKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKITTL--PSDFWSLTSLKSLNL 121
N +H+ L+LSG LTG +P + +L +L LDLS+N + PS F SL +L SL++
Sbjct: 111 NLKHLQTLDLSGNSLTGLLP-RLLSELPQLLYLDLSDNHFSGSLPPSFFISLPALSSLDV 169
Query: 122 SSNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSG 181
S+N +SG + IG L N + NSFS +IP + ++ LK F +P
Sbjct: 170 SNNSLSGEIPPEIGKLSNLSNLYMGLNSFSGQIPSEIGNISLLKNFAAPSCFFNGPLPKE 229
Query: 182 ILKCQSLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGV-SNFSRLKSIVSLN 240
I K + L +DLS N L ++P FG+ L LNL + G + KS+ SL
Sbjct: 230 ISKLKHLAKLDLSYNPLKCSIPKSFGE-LHNLSILNLVSAELIGLIPPELGNCKSLKSLM 288
Query: 241 ISGNSFQGSI-IEVFVLKLEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQN 299
+S NS G + +E+ + L RNQ G + W L L L+ N+ SGEI
Sbjct: 289 LSFNSLSGPLPLELSEIPLLTFSAERNQLSGSLPSWMGKWKVLDSLLLANNRFSGEIPHE 348
Query: 300 LNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHLGNLNALDL 359
+ + LKHLSLA N S ++ LE ++LS L G I + +L L L
Sbjct: 349 IEDCPMLKHLSLASNLLSGSIPRELCGSGSLEAIDLSGNLLSGTIEEVFDGCSSLGELLL 408
Query: 360 SMNHLDGKIPL-LKNKHLQVIDFSHNNLSGPVPSFILKSLPKMKKYNFSYNNL 411
+ N ++G IP L L +D NN +G +P + KS + ++ SYN L
Sbjct: 409 TNNQINGSIPEDLWKLPLMALDLDSNNFTGEIPKSLWKS-TNLMEFTASYNRL 460
>BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.37)
(Brassinosteroid LRR receptor kinase)
Length = 1207
Score = 345 bits (884), Expect = 4e-94
Identities = 268/812 (33%), Positives = 414/812 (50%), Gaps = 121/812 (14%)
Query: 63 SNKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT-TLPSDFWSLTSLKSLNL 121
SN +V L+LS LTG IP +++G L+KL L L N+++ +P + L +L++L L
Sbjct: 448 SNCSQLVSLDLSFNYLTGSIP-SSLGSLSKLKDLILWLNQLSGEIPQELMYLQALENLIL 506
Query: 122 SSNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSG 181
N ++G + ++ N L LS N S EIP +L L +L +LKL +N +IP+
Sbjct: 507 DFNDLTGPIPASLSNCTKLNWISLSNNQLSGEIPASLGRLSNLAILKLGNNSISGNIPAE 566
Query: 182 ILKCQSLVSIDLSSNQLSGTLPHGFGDAFPKL--RTLNLAENNIYGGVSNFSRLKSIVSL 239
+ CQSL+ +DL++N L+G++P P L ++ N+A + G + +K+ S
Sbjct: 567 LGNCQSLIWLDLNTNFLNGSIP-------PPLFKQSGNIAVALLTG--KRYVYIKNDGSK 617
Query: 240 NISGNSFQGSIIEVFVLKLEALDLSRNQ--------FQGHISQVKYNWS-HLVYLDLSEN 290
G G+++E ++ E LD + ++G I+Q +N + +++LDLS N
Sbjct: 618 ECHG---AGNLLEFGGIRQEQLDRISTRHPCNFTRVYRG-ITQPTFNHNGSMIFLDLSYN 673
Query: 291 QLSGEIFQNLNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISH 350
+L G I + L + L LNL L G IP ++
Sbjct: 674 KLEGSIPKELG------------------------AMYYLSILNLGHNDLSGMIPQQLGG 709
Query: 351 LGNLNALDLSMNHLDGKIP--LLKNKHLQVIDFSHNNLSGPVPSFILKSLPKMKKYNFSY 408
L N+ LDLS N +G IP L L ID S+NNLSG +P +S P ++ +
Sbjct: 710 LKNVAILDLSYNRFNGTIPNSLTSLTLLGEIDLSNNNLSGMIP----ESAPFDTFPDYRF 765
Query: 409 NNLTLCASEIKPDIMKTSFFGSVNSCPIAANPSFFKKRRDVGHRGM-KLALVLTLSLIFA 467
N +LC + P ++ P + HR LA + + L+F+
Sbjct: 766 ANNSLCGYPLP--------------LPCSSGPKSDANQHQKSHRRQASLAGSVAMGLLFS 811
Query: 468 LA---GILFLAFGCRRKNKMWEVKQGSYREEQNISGPFSFQTDSTTW-VADVKQATSVPV 523
L G++ +A +++ + E +Y + + S T ++ W ++A S+ +
Sbjct: 812 LFCIFGLIIVAIETKKRRRKKEAALEAYMDGHSHSA-----TANSAWKFTSAREALSINL 866
Query: 524 VIFEKPLLNITFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVVGSTLTD 583
FEKPL +TFADLL AT+ F +L+ G FG VY+ L VA+K L+ S D
Sbjct: 867 AAFEKPLRKLTFADLLEATNGFHNDSLVGSGGFGDVYKAQLKDGSVVAIKKLIHVSGQGD 926
Query: 584 EEAARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDW 643
E E+E +G+IKH NLVPL GYC G++R+ +Y+YM+ G+L+++L+D
Sbjct: 927 REFTAEMETIGKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDRK--------- 977
Query: 644 STDTWEEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVY 703
G+ W R KIA+G AR LAFLHH C P IIHR +K+S+V
Sbjct: 978 ----------------KTGIKLNWPARRKIAIGAARGLAFLHHNCIPHIIHRDMKSSNVL 1021
Query: 704 LDYDLEPRLSDFGLAKIFGSGLDEEIA----RGSPGYVPPEFSQPEFESPTPKSDVYCFG 759
LD +LE R+SDFG+A++ S +D ++ G+PGYVPPE+ Q F T K DVY +G
Sbjct: 1022 LDENLEARVSDFGMARLM-SAMDTHLSVSTLAGTPGYVPPEYYQ-SFRCST-KGDVYSYG 1078
Query: 760 VVLFELLTGKKPVGD-DYTDDKEATTLVSWVRGLVRKNQTSRAIDPKICDTGSDEQIE-- 816
VVL ELLTGK+P D+ D+ LV WV+ L K + + D ++ + +IE
Sbjct: 1079 VVLLELLTGKQPTDSADFGDN----NLVGWVK-LHAKGKITDVFDRELLKEDASIEIELL 1133
Query: 817 EALKEVGYLCTADLPFKRPTMQQIVGLLKDIE 848
+ LK V C D +KRPTM Q++ + K+I+
Sbjct: 1134 QHLK-VACACLDDRHWKRPTMIQVMAMFKEIQ 1164
Score = 137 bits (345), Expect = 1e-31
Identities = 120/394 (30%), Positives = 188/394 (47%), Gaps = 40/394 (10%)
Query: 33 EFLKKMGLTSSSKVYNFSSSVCSWKGVYCDSNKEHVVEL---NLSGIGLTGPIPDTTIGK 89
E LK G T S +V + S + S ++ + VEL ++ G L G IP+
Sbjct: 178 EMLK--GATFSLQVLDLSYNNISGFNLFPWVSSMGFVELEFFSIKGNKLAGSIPELDFKN 235
Query: 90 LNKLHSLDLSNNKITTLPSDFWSLTSLKSLNLSSNHISGSLTNNIGNFGLLENFDLSKNS 149
L+ L DLS N +T+ F ++L+ L+LSSN G + +++ + G L +L+ N
Sbjct: 236 LSYL---DLSANNFSTVFPSFKDCSNLQHLDLSSNKFYGDIGSSLSSCGKLSFLNLTNNQ 292
Query: 150 FSDEIPEALSSLVSLKVLK-----------------------LDHNMFVRSIPSGILKCQ 186
F +P+ S + L+ L +N F +P + +C
Sbjct: 293 FVGLVPKLPSESLQYLYLRGNDFQGVYPNQLADLCKTVVELDLSYNNFSGMVPESLGECS 352
Query: 187 SLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGV-SNFSRLKSIVSLNISGNS 245
SL +D+S+N SG LP ++T+ L+ N GG+ +FS L + +L++S N+
Sbjct: 353 SLELVDISNNNFSGKLPVDTLLKLSNIKTMVLSFNKFVGGLPDSFSNLPKLETLDMSSNN 412
Query: 246 FQGSIIEVFVLK-----LEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQNL 300
G II + K L+ L L N F+G I N S LV LDLS N L+G I +L
Sbjct: 413 LTG-IIPSGICKDPMNNLKVLYLQNNLFKGPIPDSLSNCSQLVSLDLSFNYLTGSIPSSL 471
Query: 301 NNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHLGNLNALDLS 360
+ LK L L N+ S + ++ L LE L L L G IP +S+ LN + LS
Sbjct: 472 GSLSKLKDLILWLNQLSGEIPQELMYLQALENLILDFNDLTGPIPASLSNCTKLNWISLS 531
Query: 361 MNHLDGKIP--LLKNKHLQVIDFSHNNLSGPVPS 392
N L G+IP L + +L ++ +N++SG +P+
Sbjct: 532 NNQLSGEIPASLGRLSNLAILKLGNNSISGNIPA 565
Score = 101 bits (251), Expect = 1e-20
Identities = 115/401 (28%), Positives = 184/401 (45%), Gaps = 68/401 (16%)
Query: 50 SSSVCSWKGVYCDSNKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT---TL 106
S+ CS+ GV C +++ ++L+ + + + + + + L+ L SL L N ++ T
Sbjct: 67 STDPCSFTGVSCKNSRVSSIDLSNTFLSVDFSLVTSYLLPLSNLESLVLKNANLSGSLTS 126
Query: 107 PSDFWSLTSLKSLNLSSNHISGSLTNNIGNFGL---LENFDLSKNSFSDEIPEALSSLV- 162
+ +L S++L+ N ISG +++ I +FG+ L++ +LSKN E L
Sbjct: 127 AAKSQCGVTLDSIDLAENTISGPISD-ISSFGVCSNLKSLNLSKNFLDPPGKEMLKGATF 185
Query: 163 SLKVLKLDHNM--------------FVR-------------SIPSGILKCQSLVSIDLSS 195
SL+VL L +N FV SIP L ++L +DLS+
Sbjct: 186 SLQVLDLSYNNISGFNLFPWVSSMGFVELEFFSIKGNKLAGSIPE--LDFKNLSYLDLSA 243
Query: 196 NQLSGTLPHGFGDAFPKLRTLNLAENNIYGGV-SNFSRLKSIVSLNISGNSFQGSIIEVF 254
N S P F D L+ L+L+ N YG + S+ S + LN++ N F G + ++
Sbjct: 244 NNFSTVFP-SFKDC-SNLQHLDLSSNKFYGDIGSSLSSCGKLSFLNLTNNQFVGLVPKLP 301
Query: 255 VLKLEALDLSRNQFQG-HISQVKYNWSHLVYLDLSENQLSGEIFQNLNNSMNLKHLSLAC 313
L+ L L N FQG + +Q+ +V LDLS N SG + ++L +L+ + ++
Sbjct: 302 SESLQYLYLRGNDFQGVYPNQLADLCKTVVELDLSYNNFSGMVPESLGECSSLELVDISN 361
Query: 314 NRFSRQKFPKIEMLLGLEYLN---LSKTSLVGHIPDEISHLGNLNALDLSMNHLDGKIPL 370
N FS K P ++ LL L + LS VG +PD S+L L LD+S N
Sbjct: 362 NNFSG-KLP-VDTLLKLSNIKTMVLSFNKFVGGLPDSFSNLPKLETLDMSSN-------- 411
Query: 371 LKNKHLQVIDFSHNNLSGPVPSFILKSLPKMKKYNFSYNNL 411
NL+G +PS I K K + NNL
Sbjct: 412 --------------NLTGIIPSGICKDPMNNLKVLYLQNNL 438
Score = 100 bits (248), Expect = 2e-20
Identities = 106/360 (29%), Positives = 171/360 (47%), Gaps = 60/360 (16%)
Query: 89 KLNKLHSLDLSNNKITT----LPSDFWSLTSLKSLNLSSNHISGSLTNNI-GNFGL-LEN 142
K +++ S+DLSN ++ + S L++L+SL L + ++SGSLT+ G+ L++
Sbjct: 79 KNSRVSSIDLSNTFLSVDFSLVTSYLLPLSNLESLVLKNANLSGSLTSAAKSQCGVTLDS 138
Query: 143 FDLSKNSFSDEIPEALSSLV--SLKVLKLDHNMFVRSIPSGILKCQ--SLVSIDLSSNQL 198
DL++N+ S I + S V +LK L L N F+ +LK SL +DLS N +
Sbjct: 139 IDLAENTISGPISDISSFGVCSNLKSLNLSKN-FLDPPGKEMLKGATFSLQVLDLSYNNI 197
Query: 199 SGTLPHGFGDAFPKLRTLNLAENNIYGGVSNFSRLKSIVSLNISGNSFQGSIIEVFVLKL 258
SG + FP + ++ E + +I GN GSI E+ L
Sbjct: 198 SGF------NLFPWVSSMGFVELEFF---------------SIKGNKLAGSIPELDFKNL 236
Query: 259 EALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQNLNNSMNLKHLSLACNRFSR 318
LDLS N F K + S+L +LDLS N+ G+I +L++ L L+L N+F
Sbjct: 237 SYLDLSANNFSTVFPSFK-DCSNLQHLDLSSNKFYGDIGSSLSSCGKLSFLNLTNNQFVG 295
Query: 319 --QKFPKIEM----LLGLEY-----------------LNLSKTSLVGHIPDEISHLGNLN 355
K P + L G ++ L+LS + G +P+ + +L
Sbjct: 296 LVPKLPSESLQYLYLRGNDFQGVYPNQLADLCKTVVELDLSYNNFSGMVPESLGECSSLE 355
Query: 356 ALDLSMNHLDGKIP---LLKNKHLQVIDFSHNNLSGPVPSFILKSLPKMKKYNFSYNNLT 412
+D+S N+ GK+P LLK +++ + S N G +P +LPK++ + S NNLT
Sbjct: 356 LVDISNNNFSGKLPVDTLLKLSNIKTMVLSFNKFVGGLPDSF-SNLPKLETLDMSSNNLT 414
Score = 83.2 bits (204), Expect = 3e-15
Identities = 76/274 (27%), Positives = 126/274 (45%), Gaps = 25/274 (9%)
Query: 178 IPSGILKCQSLVSIDLSSNQLSGTLPHGF-GDAFPKLRTLNLAENNIYGGVSNFSRL--- 233
+ S +L +L S+ L + LSG+L L +++LAEN I G +S+ S
Sbjct: 100 VTSYLLPLSNLESLVLKNANLSGSLTSAAKSQCGVTLDSIDLAENTISGPISDISSFGVC 159
Query: 234 KSIVSLNISGNSFQG---SIIEVFVLKLEALDLSRNQFQGH-----ISQVKYNWSHLVYL 285
++ SLN+S N +++ L+ LDLS N G +S + + L +
Sbjct: 160 SNLKSLNLSKNFLDPPGKEMLKGATFSLQVLDLSYNNISGFNLFPWVSSMGF--VELEFF 217
Query: 286 DLSENQLSGEIFQNLNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIP 345
+ N+L+G I + + NL +L L+ N FS FP + L++L+LS G I
Sbjct: 218 SIKGNKLAGSIPEL--DFKNLSYLDLSANNFSTV-FPSFKDCSNLQHLDLSSNKFYGDIG 274
Query: 346 DEISHLGNLNALDLSMNHLDGKIPLLKNKHLQVIDFSHNNLSGPVPSFILKSLPKMKKYN 405
+S G L+ L+L+ N G +P L ++ LQ + N+ G P+ + + + +
Sbjct: 275 SSLSSCGKLSFLNLTNNQFVGLVPKLPSESLQYLYLRGNDFQGVYPNQLADLCKTVVELD 334
Query: 406 FSYNN--------LTLCASEIKPDIMKTSFFGSV 431
SYNN L C+S DI +F G +
Sbjct: 335 LSYNNFSGMVPESLGECSSLELVDISNNNFSGKL 368
>BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precursor (EC
2.7.1.37) (tBRI1) (Altered brassinolide sensitivity 1)
(Systemin receptor SR160)
Length = 1207
Score = 344 bits (883), Expect = 5e-94
Identities = 277/855 (32%), Positives = 426/855 (49%), Gaps = 90/855 (10%)
Query: 32 SEFLKKMGLTSSSKVYNFSSSVCSWK---GVYCDS--NKEHVVELNLSGIGLTGPIPDTT 86
+ F K+ + + SK+ N + V S+ G DS N + L++S LTG IP
Sbjct: 362 NNFSGKLPVDTLSKLSNIKTMVLSFNKFVGGLPDSFSNLLKLETLDMSSNNLTGVIPSGI 421
Query: 87 I-GKLNKLHSLDLSNNKIT-TLPSDFWSLTSLKSLNLSSNHISGSLTNNIGNFGLLENFD 144
+N L L L NN +P + + L SL+LS N+++GS+ +++G+ L++
Sbjct: 422 CKDPMNNLKVLYLQNNLFKGPIPDSLSNCSQLVSLDLSFNYLTGSIPSSLGSLSKLKDLI 481
Query: 145 LSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGILKCQSLVSIDLSSNQLSGTLPH 204
L N S EIP+ L L +L+ L LD N IP+ + C L I LS+NQLSG +P
Sbjct: 482 LWLNQLSGEIPQELMYLQALENLILDFNDLTGPIPASLSNCTKLNWISLSNNQLSGEIPA 541
Query: 205 GFGDAFPKLRTLNLAENNIYGGV-SNFSRLKSIVSLNISGNSFQGSIIEVFVLKLEALDL 263
G L L L N+I G + + +S++ L+++ N GSI +
Sbjct: 542 SLG-RLSNLAILKLGNNSISGNIPAELGNCQSLIWLDLNTNFLNGSIPPPLFKQ------ 594
Query: 264 SRNQFQGHISQVKYNWSHLVYL--DLSE------NQLS-GEIFQNLNNSMNLKHLSLACN 314
G+I+ VY+ D S+ N L G I Q + ++ +H
Sbjct: 595 -----SGNIAVALLTGKRYVYIKNDGSKECHGAGNLLEFGGIRQEQLDRISTRHPCNFTR 649
Query: 315 RFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHLGNLNALDLSMNHLDGKIP--LLK 372
+ P + +L+LS L G IP E+ + L+ L+L N L G IP L
Sbjct: 650 VYRGITQPTFNHNGSMIFLDLSYNKLEGSIPKELGAMYYLSILNLGHNDLSGMIPQQLGG 709
Query: 373 NKHLQVIDFSHNNLSGPVPSFILKSLPKMKKYNFSYNNLTLCASEIKP-DIMKTSFFGSV 431
K++ ++D S+N +G +P+ L SL + + + S NNL+ E P D F +
Sbjct: 710 LKNVAILDLSYNRFNGTIPNS-LTSLTLLGEIDLSNNNLSGMIPESAPFDTFPDYRFANN 768
Query: 432 NSC------PIAANPSFFKKRRDVGHRGM-KLALVLTLSLIFALA---GILFLAFGCRRK 481
+ C P ++ P + HR LA + + L+F+L G++ +A +++
Sbjct: 769 SLCGYPLPIPCSSGPKSDANQHQKSHRRQASLAGSVAMGLLFSLFCIFGLIIVAIETKKR 828
Query: 482 NKMWEVKQGSYREEQNISGPFSFQTDSTTW-VADVKQATSVPVVIFEKPLLNITFADLLS 540
+ E +Y + + S T ++ W ++A S+ + FEKPL +TFADLL
Sbjct: 829 RRKKEAALEAYMDGHSHSA-----TANSAWKFTSAREALSINLAAFEKPLRKLTFADLLE 883
Query: 541 ATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVVGSTLTDEEAARELEFLGRIKHPN 600
AT+ F +L+ G FG VY+ L VA+K L+ S D E E+E +G+IKH N
Sbjct: 884 ATNGFHNDSLVGSGGFGDVYKAQLKDGSVVAIKKLIHVSGQGDREFTAEMETIGKIKHRN 943
Query: 601 LVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTDTWEEADNGIQNVGS 660
LVPL GYC G++R+ +Y+YM+ G+L+++L+D
Sbjct: 944 LVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDRK-------------------------K 978
Query: 661 EGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLEPRLSDFGLAKI 720
G+ W R KIA+G AR LAFLHH C P IIHR +K+S+V LD +LE R+SDFG+A++
Sbjct: 979 IGIKLNWPARRKIAIGAARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARL 1038
Query: 721 FGSGLDEEIA----RGSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFELLTGKKPVGD-D 775
S +D ++ G+PGYVPPE+ Q F T K DVY +GVVL ELLTGK+P D
Sbjct: 1039 M-SAMDTHLSVSTLAGTPGYVPPEYYQ-SFRCST-KGDVYSYGVVLLELLTGKQPTDSAD 1095
Query: 776 YTDDKEATTLVSWVRGLVRKNQTSRAIDPKICDTGSDEQIE--EALKEVGYLCTADLPFK 833
+ D+ LV WV+ L K + + D ++ + +IE + LK V C D +K
Sbjct: 1096 FGDN----NLVGWVK-LHAKGKITDVFDRELLKEDASIEIELLQHLK-VACACLDDRHWK 1149
Query: 834 RPTMQQIVGLLKDIE 848
RPTM Q++ + K+I+
Sbjct: 1150 RPTMIQVMAMFKEIQ 1164
Score = 138 bits (348), Expect = 5e-32
Identities = 113/354 (31%), Positives = 174/354 (48%), Gaps = 39/354 (11%)
Query: 72 NLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKITTLPSDFWSLTSLKSLNLSSNHISGSLT 131
+L G L G IP+ L+ L DLS N +T+ F ++L+ L+LSSN G +
Sbjct: 218 SLKGNKLAGSIPELDFKNLSYL---DLSANNFSTVFPSFKDCSNLQHLDLSSNKFYGDIG 274
Query: 132 NNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGILK-CQSLVS 190
+++ + G L +L+ N F +P+ S SL+ L L N F P+ + C+++V
Sbjct: 275 SSLSSCGKLSFLNLTNNQFVGLVPKLPSE--SLQYLYLRGNDFQGVYPNQLADLCKTVVE 332
Query: 191 IDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYG--GVSNFSRLKSIVSLNISGNSFQG 248
+DLS N SG +P G+ L ++++ NN G V S+L +I ++ +S N F G
Sbjct: 333 LDLSYNNFSGMVPESLGEC-SSLELVDISYNNFSGKLPVDTLSKLSNIKTMVLSFNKFVG 391
Query: 249 SIIEVF--VLKLEALDLSRNQ--------------------------FQGHISQVKYNWS 280
+ + F +LKLE LD+S N F+G I N S
Sbjct: 392 GLPDSFSNLLKLETLDMSSNNLTGVIPSGICKDPMNNLKVLYLQNNLFKGPIPDSLSNCS 451
Query: 281 HLVYLDLSENQLSGEIFQNLNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSL 340
LV LDLS N L+G I +L + LK L L N+ S + ++ L LE L L L
Sbjct: 452 QLVSLDLSFNYLTGSIPSSLGSLSKLKDLILWLNQLSGEIPQELMYLQALENLILDFNDL 511
Query: 341 VGHIPDEISHLGNLNALDLSMNHLDGKIP--LLKNKHLQVIDFSHNNLSGPVPS 392
G IP +S+ LN + LS N L G+IP L + +L ++ +N++SG +P+
Sbjct: 512 TGPIPASLSNCTKLNWISLSNNQLSGEIPASLGRLSNLAILKLGNNSISGNIPA 565
Score = 128 bits (321), Expect = 7e-29
Identities = 113/352 (32%), Positives = 170/352 (48%), Gaps = 38/352 (10%)
Query: 48 NFSSSVCSWKGVYCDSNKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNK----I 103
NFS+ S+K SN +H L+LS G I +++ KL L+L+NN+ +
Sbjct: 245 NFSTVFPSFKDC---SNLQH---LDLSSNKFYGDI-GSSLSSCGKLSFLNLTNNQFVGLV 297
Query: 104 TTLPSDFWSLTSLKS--------------------LNLSSNHISGSLTNNIGNFGLLENF 143
LPS+ L+ L+LS N+ SG + ++G LE
Sbjct: 298 PKLPSESLQYLYLRGNDFQGVYPNQLADLCKTVVELDLSYNNFSGMVPESLGECSSLELV 357
Query: 144 DLSKNSFSDEIP-EALSSLVSLKVLKLDHNMFVRSIPSGILKCQSLVSIDLSSNQLSGTL 202
D+S N+FS ++P + LS L ++K + L N FV +P L ++D+SSN L+G +
Sbjct: 358 DISYNNFSGKLPVDTLSKLSNIKTMVLSFNKFVGGLPDSFSNLLKLETLDMSSNNLTGVI 417
Query: 203 PHGF-GDAFPKLRTLNLAENNIYGG--VSNFSRLKSIVSLNISGNSFQGSIIEVF--VLK 257
P G D L+ L L +NN++ G + S +VSL++S N GSI + K
Sbjct: 418 PSGICKDPMNNLKVLYL-QNNLFKGPIPDSLSNCSQLVSLDLSFNYLTGSIPSSLGSLSK 476
Query: 258 LEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQNLNNSMNLKHLSLACNRFS 317
L+ L L NQ G I Q L L L N L+G I +L+N L +SL+ N+ S
Sbjct: 477 LKDLILWLNQLSGEIPQELMYLQALENLILDFNDLTGPIPASLSNCTKLNWISLSNNQLS 536
Query: 318 RQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHLGNLNALDLSMNHLDGKIP 369
+ + L L L L S+ G+IP E+ + +L LDL+ N L+G IP
Sbjct: 537 GEIPASLGRLSNLAILKLGNNSISGNIPAELGNCQSLIWLDLNTNFLNGSIP 588
Score = 106 bits (264), Expect = 3e-22
Identities = 108/345 (31%), Positives = 170/345 (48%), Gaps = 39/345 (11%)
Query: 79 TGPIPDTTIG-KLNKLHSLDLSNNKITT----LPSDFWSLTSLKSLNLSSNHISGSLTNN 133
TGP T + K +++ S+DLSN ++ + S L++L+SL L + ++SGSLT+
Sbjct: 68 TGPCSFTGVSCKNSRVSSIDLSNTFLSVDFSLVTSYLLPLSNLESLVLKNANLSGSLTSA 127
Query: 134 I-GNFGL-LENFDLSKNSFSDEIPEALSSLV--SLKVLKLDHNMFVRSIPSGILKCQ--S 187
G+ L++ DL++N+ S I + S V +LK L L N F+ +LK S
Sbjct: 128 AKSQCGVTLDSIDLAENTISGPISDISSFGVCSNLKSLNLSKN-FLDPPGKEMLKAATFS 186
Query: 188 LVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGVSNFSRLKSIVSLNISGNSFQ 247
L +DLS N +SG + FP + ++ E + ++ GN
Sbjct: 187 LQVLDLSYNNISGF------NLFPWVSSMGFVELEFF---------------SLKGNKLA 225
Query: 248 GSIIEVFVLKLEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQNLNNSMNLK 307
GSI E+ L LDLS N F K + S+L +LDLS N+ G+I +L++ L
Sbjct: 226 GSIPELDFKNLSYLDLSANNFSTVFPSFK-DCSNLQHLDLSSNKFYGDIGSSLSSCGKLS 284
Query: 308 HLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHL-GNLNALDLSMNHLDG 366
L+L N+F PK+ L+YL L G P++++ L + LDLS N+ G
Sbjct: 285 FLNLTNNQFVGL-VPKLPSE-SLQYLYLRGNDFQGVYPNQLADLCKTVVELDLSYNNFSG 342
Query: 367 KIP--LLKNKHLQVIDFSHNNLSGPVPSFILKSLPKMKKYNFSYN 409
+P L + L+++D S+NN SG +P L L +K S+N
Sbjct: 343 MVPESLGECSSLELVDISYNNFSGKLPVDTLSKLSNIKTMVLSFN 387
Score = 105 bits (262), Expect = 5e-22
Identities = 110/384 (28%), Positives = 180/384 (46%), Gaps = 34/384 (8%)
Query: 50 SSSVCSWKGVYCDSNKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT---TL 106
S+ CS+ GV C +++ ++L+ + + + + + + L+ L SL L N ++ T
Sbjct: 67 STGPCSFTGVSCKNSRVSSIDLSNTFLSVDFSLVTSYLLPLSNLESLVLKNANLSGSLTS 126
Query: 107 PSDFWSLTSLKSLNLSSNHISGSLTNNIGNFGL---LENFDLSKNSFSDEIPEALSSLV- 162
+ +L S++L+ N ISG +++ I +FG+ L++ +LSKN E L +
Sbjct: 127 AAKSQCGVTLDSIDLAENTISGPISD-ISSFGVCSNLKSLNLSKNFLDPPGKEMLKAATF 185
Query: 163 SLKVLKLDHN------MFVRSIPSGILKCQSLVSIDLSSNQLSGTLPHGFGDAFPKLRTL 216
SL+VL L +N +F G ++ L L N+L+G++P F L L
Sbjct: 186 SLQVLDLSYNNISGFNLFPWVSSMGFVE---LEFFSLKGNKLAGSIPEL---DFKNLSYL 239
Query: 217 NLAENNIYGGVSNFSRLKSIVSLNISGNSFQGSIIEVFVL--KLEALDLSRNQFQGHISQ 274
+L+ NN +F ++ L++S N F G I KL L+L+ NQF G + +
Sbjct: 240 DLSANNFSTVFPSFKDCSNLQHLDLSSNKFYGDIGSSLSSCGKLSFLNLTNNQFVGLVPK 299
Query: 275 VKYNWSHLVYLDLSENQLSGEIFQNLNN-SMNLKHLSLACNRFSRQKFPKIEMLLGLEYL 333
+ L YL L N G L + + L L+ N FS + LE +
Sbjct: 300 LPSE--SLQYLYLRGNDFQGVYPNQLADLCKTVVELDLSYNNFSGMVPESLGECSSLELV 357
Query: 334 NLSKTSLVGHIP-DEISHLGNLNALDLSMNHLDGKIP-----LLKNKHLQVIDFSHNNLS 387
++S + G +P D +S L N+ + LS N G +P LLK L+ +D S NNL+
Sbjct: 358 DISYNNFSGKLPVDTLSKLSNIKTMVLSFNKFVGGLPDSFSNLLK---LETLDMSSNNLT 414
Query: 388 GPVPSFILKSLPKMKKYNFSYNNL 411
G +PS I K K + NNL
Sbjct: 415 GVIPSGICKDPMNNLKVLYLQNNL 438
Score = 84.3 bits (207), Expect = 1e-15
Identities = 77/274 (28%), Positives = 126/274 (45%), Gaps = 25/274 (9%)
Query: 178 IPSGILKCQSLVSIDLSSNQLSGTLPHGF-GDAFPKLRTLNLAENNIYGGVSNFSRL--- 233
+ S +L +L S+ L + LSG+L L +++LAEN I G +S+ S
Sbjct: 100 VTSYLLPLSNLESLVLKNANLSGSLTSAAKSQCGVTLDSIDLAENTISGPISDISSFGVC 159
Query: 234 KSIVSLNISGNSFQG---SIIEVFVLKLEALDLSRNQFQGH-----ISQVKYNWSHLVYL 285
++ SLN+S N +++ L+ LDLS N G +S + + L +
Sbjct: 160 SNLKSLNLSKNFLDPPGKEMLKAATFSLQVLDLSYNNISGFNLFPWVSSMGF--VELEFF 217
Query: 286 DLSENQLSGEIFQNLNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIP 345
L N+L+G I + + NL +L L+ N FS FP + L++L+LS G I
Sbjct: 218 SLKGNKLAGSIPEL--DFKNLSYLDLSANNFSTV-FPSFKDCSNLQHLDLSSNKFYGDIG 274
Query: 346 DEISHLGNLNALDLSMNHLDGKIPLLKNKHLQVIDFSHNNLSGPVPSFILKSLPKMKKYN 405
+S G L+ L+L+ N G +P L ++ LQ + N+ G P+ + + + +
Sbjct: 275 SSLSSCGKLSFLNLTNNQFVGLVPKLPSESLQYLYLRGNDFQGVYPNQLADLCKTVVELD 334
Query: 406 FSYNN--------LTLCASEIKPDIMKTSFFGSV 431
SYNN L C+S DI +F G +
Sbjct: 335 LSYNNFSGMVPESLGECSSLELVDISYNNFSGKL 368
>CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (EC
2.7.1.-)
Length = 980
Score = 301 bits (770), Expect = 6e-81
Identities = 243/795 (30%), Positives = 387/795 (48%), Gaps = 94/795 (11%)
Query: 71 LNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKITT-LPSDFWSLTSLKSLNLSSNHISGS 129
L+++ LTG IP T++ L LH+L L N +T +P + L SLKSL+LS N ++G
Sbjct: 246 LDMASCTLTGEIP-TSLSNLKHLHTLFLHINNLTGHIPPELSGLVSLKSLDLSINQLTGE 304
Query: 130 LTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGILKCQSLV 189
+ + N G + +L +N+ +IPEA+ L L+V ++ N F +P+ + + +L+
Sbjct: 305 IPQSFINLGNITLINLFRNNLYGQIPEAIGELPKLEVFEVWENNFTLQLPANLGRNGNLI 364
Query: 190 SIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGV-SNFSRLKSIVSLNISGNSFQG 248
+D+S N L+G +P KL L L+ N +G + + KS+ + I N G
Sbjct: 365 KLDVSDNHLTGLIPKDLCRG-EKLEMLILSNNFFFGPIPEELGKCKSLTKIRIVKNLLNG 423
Query: 249 SI-IEVFVLKL-EALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQNLNNSMNL 306
++ +F L L ++L+ N F G + V + L + LS N SGEI + N NL
Sbjct: 424 TVPAGLFNLPLVTIIELTDNFFSGEL-PVTMSGDVLDQIYLSNNWFSGEIPPAIGNFPNL 482
Query: 307 KHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHLGNLNALDLSMNHLDG 366
+ L L NRF +I L L +N S ++ G IPD IS L ++DLS N ++G
Sbjct: 483 QTLFLDRNRFRGNIPREIFELKHLSRINTSANNITGGIPDSISRCSTLISVDLSRNRING 542
Query: 367 KIPLLKN--KHLQVIDFSHNNLSGPVPSFILKSLPKMKKYNFSYNNLTLCASEIKPDIM- 423
+IP N K+L ++ S N L+G +P+ I ++ + + S+N+L+ ++
Sbjct: 543 EIPKGINNVKNLGTLNISGNQLTGSIPTGI-GNMTSLTTLDLSFNDLSGRVPLGGQFLVF 601
Query: 424 -KTSFFGSVNSC--PIAANPSFFKKRRDVGHRGMKLALVLTLSLIFALAGILFLAFGCRR 480
+TSF G+ C + P+ + D H + + +++I A+ G++ ++ R+
Sbjct: 602 NETSFAGNTYLCLPHRVSCPTRPGQTSDHNHTALFSPSRIVITVIAAITGLILISVAIRQ 661
Query: 481 KNKMWEVKQGSYREEQNISGPFSFQTDSTTWVADVKQATSVPVVIFEKPLLNITFADLLS 540
NK K S W + F+K L+ D+L
Sbjct: 662 MNKKKNQK-------------------SLAW----------KLTAFQK--LDFKSEDVLE 690
Query: 541 ATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLV-VGSTLTDEEAARELEFLGRIKHP 599
++ +G G VYRG +P N+ VA+K LV G+ +D E++ LGRI+H
Sbjct: 691 C---LKEENIIGKGGAGIVYRGSMPNNVDVAIKRLVGRGTGRSDHGFTAEIQTLGRIRHR 747
Query: 600 NLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTDTWEEADNGIQNVG 659
++V L GY D + +Y+YM NG+L LL+ G
Sbjct: 748 HIVRLLGYVANKDTNLLLYEYMPNGSLGELLH---------------------------G 780
Query: 660 SEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLEPRLSDFGLAK 719
S+G W RH++A+ A+ L +LHH CSP I+HR VK++++ LD D E ++DFGLAK
Sbjct: 781 SKGGHLQWETRHRVAVEAAKGLCYLHHDCSPLILHRDVKSNNILLDSDFEAHVADFGLAK 840
Query: 720 IFGSGLDEEI---ARGSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFELLTGKKPVGDDY 776
G E GS GY+ PE++ KSDVY FGVVL EL+ GKKPVG
Sbjct: 841 FLVDGAASECMSSIAGSYGYIAPEYAYT--LKVDEKSDVYSFGVVLLELIAGKKPVG--- 895
Query: 777 TDDKEATTLVSWVRGLVRK-NQTSRA------IDPKICDTGSDEQIEEALKEVGYLCTAD 829
+ E +V WVR + Q S A +DP++ TG + ++ +C +
Sbjct: 896 -EFGEGVDIVRWVRNTEEEITQPSDAAIVVAIVDPRL--TGYPLTSVIHVFKIAMMCVEE 952
Query: 830 LPFKRPTMQQIVGLL 844
RPTM+++V +L
Sbjct: 953 EAAARPTMREVVHML 967
Score = 134 bits (338), Expect = 8e-31
Identities = 125/360 (34%), Positives = 175/360 (47%), Gaps = 14/360 (3%)
Query: 54 CSWKGVYCDSNKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT-TLPSDFWS 112
CS+ GV CD + V+ LN+S L G I IG L L +L L+ N T LP + S
Sbjct: 59 CSFSGVSCDDDAR-VISLNVSFTPLFGTI-SPEIGMLTHLVNLTLAANNFTGELPLEMKS 116
Query: 113 LTSLKSLNLSSN-HISGSLTNNIGNFGL-LENFDLSKNSFSDEIPEALSSLVSLKVLKLD 170
LTSLK LN+S+N +++G+ I + LE D N+F+ ++P +S L LK L
Sbjct: 117 LTSLKVLNISNNGNLTGTFPGEILKAMVDLEVLDTYNNNFNGKLPPEMSELKKLKYLSFG 176
Query: 171 HNMFVRSIPSGILKCQSLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIY-GGV-S 228
N F IP QSL + L+ LSG P F LR + + N Y GGV
Sbjct: 177 GNFFSGEIPESYGDIQSLEYLGLNGAGLSGKSP-AFLSRLKNLREMYIGYYNSYTGGVPP 235
Query: 229 NFSRLKSIVSLNISGNSFQGSI-IEVFVLK-LEALDLSRNQFQGHISQVKYNWSHLVYLD 286
F L + L+++ + G I + LK L L L N GHI L LD
Sbjct: 236 EFGGLTKLEILDMASCTLTGEIPTSLSNLKHLHTLFLHINNLTGHIPPELSGLVSLKSLD 295
Query: 287 LSENQLSGEIFQNLNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPD 346
LS NQL+GEI Q+ N N+ ++L N Q I L LE + + + +P
Sbjct: 296 LSINQLTGEIPQSFINLGNITLINLFRNNLYGQIPEAIGELPKLEVFEVWENNFTLQLPA 355
Query: 347 EISHLGNLNALDLSMNHLDGKIP--LLKNKHLQVIDFSHNNLSGPVPSFI--LKSLPKMK 402
+ GNL LD+S NHL G IP L + + L+++ S+N GP+P + KSL K++
Sbjct: 356 NLGRNGNLIKLDVSDNHLTGLIPKDLCRGEKLEMLILSNNFFFGPIPEELGKCKSLTKIR 415
Score = 80.9 bits (198), Expect = 1e-14
Identities = 47/137 (34%), Positives = 76/137 (55%), Gaps = 2/137 (1%)
Query: 70 ELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT-TLPSDFWSLTSLKSLNLSSNHISG 128
++ LS +G IP IG L +L L N+ +P + + L L +N S+N+I+G
Sbjct: 460 QIYLSNNWFSGEIPPA-IGNFPNLQTLFLDRNRFRGNIPREIFELKHLSRINTSANNITG 518
Query: 129 SLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGILKCQSL 188
+ ++I L + DLS+N + EIP+ ++++ +L L + N SIP+GI SL
Sbjct: 519 GIPDSISRCSTLISVDLSRNRINGEIPKGINNVKNLGTLNISGNQLTGSIPTGIGNMTSL 578
Query: 189 VSIDLSSNQLSGTLPHG 205
++DLS N LSG +P G
Sbjct: 579 TTLDLSFNDLSGRVPLG 595
Score = 61.2 bits (147), Expect = 1e-08
Identities = 34/91 (37%), Positives = 53/91 (57%), Gaps = 2/91 (2%)
Query: 66 EHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT-TLPSDFWSLTSLKSLNLSSN 124
+H+ +N S +TG IPD +I + + L S+DLS N+I +P ++ +L +LN+S N
Sbjct: 504 KHLSRINTSANNITGGIPD-SISRCSTLISVDLSRNRINGEIPKGINNVKNLGTLNISGN 562
Query: 125 HISGSLTNNIGNFGLLENFDLSKNSFSDEIP 155
++GS+ IGN L DLS N S +P
Sbjct: 563 QLTGSIPTGIGNMTSLTTLDLSFNDLSGRVP 593
Score = 43.5 bits (101), Expect = 0.002
Identities = 31/81 (38%), Positives = 48/81 (58%), Gaps = 4/81 (4%)
Query: 333 LNLSKTSLVGHIPDEISHLGNLNALDLSMNHLDGKIPLLKNK--HLQVIDFSHN-NLSGP 389
LN+S T L G I EI L +L L L+ N+ G++PL L+V++ S+N NL+G
Sbjct: 75 LNVSFTPLFGTISPEIGMLTHLVNLTLAANNFTGELPLEMKSLTSLKVLNISNNGNLTGT 134
Query: 390 VPSFILKSLPKMKKYNFSYNN 410
P ILK++ ++ + +YNN
Sbjct: 135 FPGEILKAMVDLEVLD-TYNN 154
Score = 33.5 bits (75), Expect = 2.4
Identities = 19/42 (45%), Positives = 28/42 (66%), Gaps = 1/42 (2%)
Query: 63 SNKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT 104
+N +++ LN+SG LTG IP T IG + L +LDLS N ++
Sbjct: 549 NNVKNLGTLNISGNQLTGSIP-TGIGNMTSLTTLDLSFNDLS 589
>PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (EC
2.7.1.37) (Phytosulfokine LRR receptor kinase)
Length = 1008
Score = 298 bits (764), Expect = 3e-80
Identities = 254/865 (29%), Positives = 390/865 (44%), Gaps = 154/865 (17%)
Query: 88 GKLNKLHSLDLSNNKIT-TLPSDFWSLTSLKSLNLSSNHISGSLTNNIGNFGLLENFDLS 146
GK L L L N +T +P D + L L L + N +SGSL+ I N L D+S
Sbjct: 193 GKCVLLEHLCLGMNDLTGNIPEDLFHLKRLNLLGIQENRLSGSLSREIRNLSSLVRLDVS 252
Query: 147 KNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGI---------------------LKC 185
N FS EIP+ L LK N F+ IP + L C
Sbjct: 253 WNLFSGEIPDVFDELPQLKFFLGQTNGFIGGIPKSLANSPSLNLLNLRNNSLSGRLMLNC 312
Query: 186 QSLV---SIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGV-SNFSRLKSIVSLNI 241
+++ S+DL +N+ +G LP D +L+ +NLA N +G V +F +S+ ++
Sbjct: 313 TAMIALNSLDLGTNRFNGRLPENLPDC-KRLKNVNLARNTFHGQVPESFKNFESLSYFSL 371
Query: 242 SGNS-------------------------FQGSII-----------EVFVL--------- 256
S +S F G + +V V+
Sbjct: 372 SNSSLANISSALGILQHCKNLTTLVLTLNFHGEALPDDSSLHFEKLKVLVVANCRLTGSM 431
Query: 257 --------KLEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQNLNNSMNLKH 308
+L+ LDLS N+ G I ++ L YLDLS N +GEI ++L +L
Sbjct: 432 PRWLSSSNELQLLDLSWNRLTGAIPSWIGDFKALFYLDLSNNSFTGEIPKSLTKLESLTS 491
Query: 309 LSLACNRFSRQKFP----KIEMLLGLEY---------LNLSKTSLVGHIPDEISHLGNLN 355
+++ N S FP + E L+Y + L +L G I +E +L L+
Sbjct: 492 RNISVNEPSPD-FPFFMKRNESARALQYNQIFGFPPTIELGHNNLSGPIWEEFGNLKKLH 550
Query: 356 ALDLSMNHLDGKIP--LLKNKHLQVIDFSHNNLSGPVPSFILKSLPKMKKYNFSYNNLT- 412
DL N L G IP L L+ +D S+N LSG +P L+ L + K++ +YNNL+
Sbjct: 551 VFDLKWNALSGSIPSSLSGMTSLEALDLSNNRLSGSIP-VSLQQLSFLSKFSVAYNNLSG 609
Query: 413 LCASEIKPDIMKTSFFGSVNSCPIAANP-------SFFKKRRDVGHRGMKLALVLTLSLI 465
+ S + S F S + C P + K+ R + +A+ + +
Sbjct: 610 VIPSGGQFQTFPNSSFESNHLCGEHRFPCSEGTESALIKRSRRSRGGDIGMAIGIAFGSV 669
Query: 466 FALAGILFLAFGCRRKNKMWEVKQGSYREEQNISGPFSFQTDSTTWVADVKQATSVPVVI 525
F L + + RR++ G E S + + ++ + S VV+
Sbjct: 670 FLLTLLSLIVLRARRRS-------GEVDPEIEESESMNRK--------ELGEIGSKLVVL 714
Query: 526 FEKPLLNITFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVVGSTLTDEE 585
F+ +++ DLL +T++FD+ ++ G FG VY+ LP VA+K L + E
Sbjct: 715 FQSNDKELSYDDLLDSTNSFDQANIIGCGGFGMVYKATLPDGKKVAIKKLSGDCGQIERE 774
Query: 586 AARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWST 645
E+E L R +HPNLV L G+C + R+ IY YMENG+L L++
Sbjct: 775 FEAEVETLSRAQHPNLVLLRGFCFYKNDRLLIYSYMENGSLDYWLHE------------- 821
Query: 646 DTWEEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLD 705
+N G L W+ R +IA G A+ L +LH GC P I+HR +K+S++ LD
Sbjct: 822 ----------RNDGP--ALLKWKTRLRIAQGAAKGLLYLHEGCDPHILHRDIKSSNILLD 869
Query: 706 YDLEPRLSDFGLAKIFGSGLDEEIAR---GSPGYVPPEFSQPEFESPTPKSDVYCFGVVL 762
+ L+DFGLA++ S + ++ G+ GY+PPE+ Q T K DVY FGVVL
Sbjct: 870 ENFNSHLADFGLARLM-SPYETHVSTDLVGTLGYIPPEYGQASV--ATYKGDVYSFGVVL 926
Query: 763 FELLTGKKPVGDDYTDDKEATTLVSWVRGLVRKNQTSRAIDPKICDTGSDEQIEEALKEV 822
ELLT K+PV D K L+SWV + +++ S DP I +D+++ L E+
Sbjct: 927 LELLTDKRPV--DMCKPKGCRDLISWVVKMKHESRASEVFDPLIYSKENDKEMFRVL-EI 983
Query: 823 GYLCTADLPFKRPTMQQIVGLLKDI 847
LC ++ P +RPT QQ+V L D+
Sbjct: 984 ACLCLSENPKQRPTTQQLVSWLDDV 1008
Score = 149 bits (376), Expect = 3e-35
Identities = 158/578 (27%), Positives = 262/578 (44%), Gaps = 90/578 (15%)
Query: 50 SSSVCSWKGVYCDSNKE-HVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKI-TTLP 107
S+ C+W G+ C+SN V+ L L L+G + + ++GKL+++ L+LS N I ++P
Sbjct: 59 STDCCNWTGITCNSNNTGRVIRLELGNKKLSGKLSE-SLGKLDEIRVLNLSRNFIKDSIP 117
Query: 108 SDFWSLTSLKSLNLSSNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEAL-SSLVSLKV 166
++L +L++L+LSSN +SG + +I N L++FDLS N F+ +P + + ++V
Sbjct: 118 LSIFNLKNLQTLDLSSNDLSGGIPTSI-NLPALQSFDLSSNKFNGSLPSHICHNSTQIRV 176
Query: 167 LKLDHNMFVRSIPSGILKCQSLVSIDLSSNQLSGTLPHGFGDAF--PKLRTLNLAENNIY 224
+KL N F + SG KC L + L N L+G +P D F +L L + EN +
Sbjct: 177 VKLAVNYFAGNFTSGFGKCVLLEHLCLGMNDLTGNIPE---DLFHLKRLNLLGIQENRLS 233
Query: 225 GGVSNFSR-LKSIVSLNISGNSFQGSIIEVF----VLK---------------------- 257
G +S R L S+V L++S N F G I +VF LK
Sbjct: 234 GSLSREIRNLSSLVRLDVSWNLFSGEIPDVFDELPQLKFFLGQTNGFIGGIPKSLANSPS 293
Query: 258 LEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQNLNNSMNLKHLSLACNRFS 317
L L+L N G + L LDL N+ +G + +NL + LK+++LA N F
Sbjct: 294 LNLLNLRNNSLSGRLMLNCTAMIALNSLDLGTNRFNGRLPENLPDCKRLKNVNLARNTFH 353
Query: 318 RQKFPKIEMLLGLEYLNLSKTSLVG--------------------------HIPDEIS-H 350
Q + L Y +LS +SL +PD+ S H
Sbjct: 354 GQVPESFKNFESLSYFSLSNSSLANISSALGILQHCKNLTTLVLTLNFHGEALPDDSSLH 413
Query: 351 LGNLNALDLSMNHLDGKIP--LLKNKHLQVIDFSHNNLSGPVPSFI--LKSLPKMKKYNF 406
L L ++ L G +P L + LQ++D S N L+G +PS+I K+L + N
Sbjct: 414 FEKLKVLVVANCRLTGSMPRWLSSSNELQLLDLSWNRLTGAIPSWIGDFKALFYLDLSNN 473
Query: 407 SYNNLTLCASEIKPDIMKTSFFGSVNSCPIAANPSF-FKKRRDVGHRGMKLALVL----T 461
S+ EI + K S N +P F F +R+ R ++ + T
Sbjct: 474 SF------TGEIPKSLTKLESLTSRNISVNEPSPDFPFFMKRNESARALQYNQIFGFPPT 527
Query: 462 LSLIF-ALAGILFLAFGCRRKNKMWEVKQGSYREE--QNISGPFSFQTDSTTWVADVKQA 518
+ L L+G ++ FG +K ++++K + ++SG S + +++ + +
Sbjct: 528 IELGHNNLSGPIWEEFGNLKKLHVFDLKWNALSGSIPSSLSGMTSLEALD---LSNNRLS 584
Query: 519 TSVPVVIFEKPLLNITFADLLSATSNFDRGTLLAEGKF 556
S+PV L ++F S N G + + G+F
Sbjct: 585 GSIPV-----SLQQLSFLSKFSVAYNNLSGVIPSGGQF 617
Score = 110 bits (276), Expect = 1e-23
Identities = 102/343 (29%), Positives = 158/343 (45%), Gaps = 12/343 (3%)
Query: 81 PIPDTTIGKLNKLHSLDLSNNKITTLPSDFWSLTSLKSLNLSSNHISGSLTNNIGNFGLL 140
P PD G +N S D N T S+ + + L L + +SG L+ ++G +
Sbjct: 48 PKPD---GWINSSSSTDCCNWTGITCNSN--NTGRVIRLELGNKKLSGKLSESLGKLDEI 102
Query: 141 ENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGILKCQSLVSIDLSSNQLSG 200
+LS+N D IP ++ +L +L+ L L N IP+ I +L S DLSSN+ +G
Sbjct: 103 RVLNLSRNFIKDSIPLSIFNLKNLQTLDLSSNDLSGGIPTSI-NLPALQSFDLSSNKFNG 161
Query: 201 TLPHGFGDAFPKLRTLNLAENNIYGG-VSNFSRLKSIVSLNISGNSFQGSIIE-VFVLK- 257
+LP ++R + LA N G S F + + L + N G+I E +F LK
Sbjct: 162 SLPSHICHNSTQIRVVKLAVNYFAGNFTSGFGKCVLLEHLCLGMNDLTGNIPEDLFHLKR 221
Query: 258 LEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQNLNNSMNLKHLSLACNRFS 317
L L + N+ G +S+ N S LV LD+S N SGEI + LK N F
Sbjct: 222 LNLLGIQENRLSGSLSREIRNLSSLVRLDVSWNLFSGEIPDVFDELPQLKFFLGQTNGFI 281
Query: 318 RQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHLGNLNALDLSMNHLDGKIP--LLKNKH 375
+ L LNL SL G + + + LN+LDL N +G++P L K
Sbjct: 282 GGIPKSLANSPSLNLLNLRNNSLSGRLMLNCTAMIALNSLDLGTNRFNGRLPENLPDCKR 341
Query: 376 LQVIDFSHNNLSGPVPSFILKSLPKMKKYNFSYNNLTLCASEI 418
L+ ++ + N G VP K+ + ++ S ++L +S +
Sbjct: 342 LKNVNLARNTFHGQVPE-SFKNFESLSYFSLSNSSLANISSAL 383
>PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.37)
(Phytosulfokine LRR receptor kinase)
Length = 1021
Score = 286 bits (733), Expect = 1e-76
Identities = 251/871 (28%), Positives = 396/871 (44%), Gaps = 142/871 (16%)
Query: 64 NKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT-TLPSDFWSLTSLKSLNLS 122
N V L L+ L+G IP +L+ L L L NN+++ L S L++L L++S
Sbjct: 204 NCSSVEYLGLASNNLSGSIPQELF-QLSNLSVLALQNNRLSGALSSKLGKLSNLGRLDIS 262
Query: 123 SNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGI 182
SN SG + + L F N F+ E+P +LS+ S+ +L L +N I
Sbjct: 263 SNKFSGKIPDVFLELNKLWYFSAQSNLFNGEMPRSLSNSRSISLLSLRNNTLSGQIYLNC 322
Query: 183 LKCQSLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGV-SNFSRLKSIVSLNI 241
+L S+DL+SN SG++P + +L+T+N A+ + +F +S+ SL+
Sbjct: 323 SAMTNLTSLDLASNSFSGSIPSNLPNCL-RLKTINFAKIKFIAQIPESFKNFQSLTSLSF 381
Query: 242 SGNSFQGSIIEVFVLK-----------------------------LEALDLSRNQFQGHI 272
S +S Q + +L+ L+ L ++ Q +G +
Sbjct: 382 SNSSIQNISSALEILQHCQNLKTLVLTLNFQKEELPSVPSLQFKNLKVLIIASCQLRGTV 441
Query: 273 SQVKYNWSHLVYLDLSENQLSGEIFQNLNNSMNLKHLSLACNRFSRQ---KFPKIEMLL- 328
Q N L LDLS NQLSG I L + +L +L L+ N F + ++ L+
Sbjct: 442 PQWLSNSPSLQLLDLSWNQLSGTIPPWLGSLNSLFYLDLSNNTFIGEIPHSLTSLQSLVS 501
Query: 329 -----------------------GLEY---------LNLSKTSLVGHIPDEISHLGNLNA 356
GL+Y ++LS SL G I E L L+
Sbjct: 502 KENAVEEPSPDFPFFKKKNTNAGGLQYNQPSSFPPMIDLSYNSLNGSIWPEFGDLRQLHV 561
Query: 357 LDLSMNHLDGKIP--LLKNKHLQVIDFSHNNLSGPVPSFILKSLPKMKKYNFSYNNLT-- 412
L+L N+L G IP L L+V+D SHNNLSG +P ++K L + ++ +YN L+
Sbjct: 562 LNLKNNNLSGNIPANLSGMTSLEVLDLSHNNLSGNIPPSLVK-LSFLSTFSVAYNKLSGP 620
Query: 413 LCASEIKPDIMKTSFFGSVNSCPIAANP----------SFFKKRRDVGHRGMKLALVLTL 462
+ +SF G+ C A+P S K ++++ + + +A+ L
Sbjct: 621 IPTGVQFQTFPNSSFEGNQGLCGEHASPCHITDQSPHGSAVKSKKNI-RKIVAVAVGTGL 679
Query: 463 SLIFALAGILFLAFGCRRKNKMWEVKQGSYREEQNISGPFSFQTDSTTWVADVKQATSVP 522
+F L L + + ++ K+ AD + S
Sbjct: 680 GTVFLLTVTLLIILRTTSRGEVDPEKKAD---------------------ADEIELGSRS 718
Query: 523 VVIFEKPLLN--ITFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVVGST 580
VV+F N ++ D+L +TS+F++ ++ G FG VY+ LP VA+K L +
Sbjct: 719 VVLFHNKDSNNELSLDDILKSTSSFNQANIIGCGGFGLVYKATLPDGTKVAIKRLSGDTG 778
Query: 581 LTDEEAARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQST 640
D E E+E L R +HPNLV L GYC + ++ IY YM+NG+L L++ G S
Sbjct: 779 QMDREFQAEVETLSRAQHPNLVHLLGYCNYKNDKLLIYSYMDNGSLDYWLHEKVDGPPSL 838
Query: 641 DDWSTDTWEEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKAS 700
D W+ R +IA G A LA+LH C P I+HR +K+S
Sbjct: 839 D-------------------------WKTRLRIARGAAEGLAYLHQSCEPHILHRDIKSS 873
Query: 701 SVYLDYDLEPRLSDFGLAKI---FGSGLDEEIARGSPGYVPPEFSQPEFESPTPKSDVYC 757
++ L L+DFGLA++ + + + ++ G+ GY+PPE+ Q T K DVY
Sbjct: 874 NILLSDTFVAHLADFGLARLILPYDTHVTTDLV-GTLGYIPPEYGQASV--ATYKGDVYS 930
Query: 758 FGVVLFELLTGKKPVGDDYTDDKEATTLVSWVRGLVRKNQTSRAIDPKICDTGSDEQIEE 817
FGVVL ELLTG++P+ D + + L+SWV + + + S DP I D E++
Sbjct: 931 FGVVLLELLTGRRPM--DVCKPRGSRDLISWVLQMKTEKRESEIFDPFIYDKDHAEEMLL 988
Query: 818 ALKEVGYLCTADLPFKRPTMQQIVGLLKDIE 848
L E+ C + P RPT QQ+V L++I+
Sbjct: 989 VL-EIACRCLGENPKTRPTTQQLVSWLENID 1018
Score = 137 bits (345), Expect = 1e-31
Identities = 123/422 (29%), Positives = 184/422 (43%), Gaps = 74/422 (17%)
Query: 48 NFSSSVCSWKGVYCDSNKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKITTLP 107
+FSS+ C W G+ C S+ V L L + +G + + +G+ L S K
Sbjct: 58 SFSSNCCDWVGISCKSS----VSLGLDDVNESGRVVELELGRRKLSGKLSESVAK----- 108
Query: 108 SDFWSLTSLKSLNLSSNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVL 167
L LK LNL+ N +SGS+ ++ N LE DLS N FS P +L +L SL+VL
Sbjct: 109 -----LDQLKVLNLTHNSLSGSIAASLLNLSNLEVLDLSSNDFSGLFP-SLINLPSLRVL 162
Query: 168 K-------------------------LDHNMFVRSIPSGILKCQSLVSIDLSSNQLSGTL 202
L N F SIP GI C S+ + L+SN LSG++
Sbjct: 163 NVYENSFHGLIPASLCNNLPRIREIDLAMNYFDGSIPVGIGNCSSVEYLGLASNNLSGSI 222
Query: 203 PHGFGDAFPKLRTLNLAENNIYGGVSN-FSRLKSIVSLNISGNSFQGSIIEVF--VLKLE 259
P L L L N + G +S+ +L ++ L+IS N F G I +VF + KL
Sbjct: 223 PQELFQ-LSNLSVLALQNNRLSGALSSKLGKLSNLGRLDISSNKFSGKIPDVFLELNKLW 281
Query: 260 ALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQNLNNSMNLKHLSLACNRFSRQ 319
N F G + + N + L L N LSG+I+ N + NL L LA N FS
Sbjct: 282 YFSAQSNLFNGEMPRSLSNSRSISLLSLRNNTLSGQIYLNCSAMTNLTSLDLASNSFSGS 341
Query: 320 KFPKIEMLLGLEYLNLSKTSLVGHIPDE--------------------------ISHLGN 353
+ L L+ +N +K + IP+ + H N
Sbjct: 342 IPSNLPNCLRLKTINFAKIKFIAQIPESFKNFQSLTSLSFSNSSIQNISSALEILQHCQN 401
Query: 354 LNALDLSMNHLDGK---IPLLKNKHLQVIDFSHNNLSGPVPSFILKSLPKMKKYNFSYNN 410
L L L++N + +P L+ K+L+V+ + L G VP ++ S P ++ + S+N
Sbjct: 402 LKTLVLTLNFQKEELPSVPSLQFKNLKVLIIASCQLRGTVPQWLSNS-PSLQLLDLSWNQ 460
Query: 411 LT 412
L+
Sbjct: 461 LS 462
>RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC
2.7.1.37)
Length = 1109
Score = 285 bits (730), Expect = 3e-76
Identities = 244/810 (30%), Positives = 377/810 (46%), Gaps = 120/810 (14%)
Query: 71 LNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT-TLPSDFWSLTSLKSLNLSSNHISGS 129
L+L L+G +P +I K+ L SL L N ++ LP D L L SL L NH +G
Sbjct: 361 LHLYTNNLSGEVP-LSIWKIQSLQSLQLYQNNLSGELPVDMTELKQLVSLALYENHFTGV 419
Query: 130 LTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGILKCQSLV 189
+ ++G LE DL++N F+ IP L S LK L L +N S+PS + C +L
Sbjct: 420 IPQDLGANSSLEVLDLTRNMFTGHIPPNLCSQKKLKRLLLGYNYLEGSVPSDLGGCSTLE 479
Query: 190 SIDLSSNQLSGTLPHGFGDAFPKLRTL--NLAENNIYGGVS-NFSRLKSIVSLNISGNSF 246
+ L N L G LP D K L +L+ NN G + + LK++ ++ +S N
Sbjct: 480 RLILEENNLRGGLP----DFVEKQNLLFFDLSGNNFTGPIPPSLGNLKNVTAIYLSSNQL 535
Query: 247 QGSIIEVF--VLKLEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQNLNNSM 304
GSI ++KLE L+LS N +G + N L LD S N L+G I L +
Sbjct: 536 SGSIPPELGSLVKLEHLNLSHNILKGILPSELSNCHKLSELDASHNLLNGSIPSTLGSLT 595
Query: 305 NLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHLGNLNALDLSMNHL 364
L LSL N FS + L L L L G IP + L L +L+LS N L
Sbjct: 596 ELTKLSLGENSFSGGIPTSLFQSNKLLNLQLGGNLLAGDIPP-VGALQALRSLNLSSNKL 654
Query: 365 DGKIP--LLKNKHLQVIDFSHNNLSGPVPSFILKSLPKMKKYNFSYNNLTLCASEIKPDI 422
+G++P L K K L+ +D SHNNLSG + +L ++ + N S+N L + + P +
Sbjct: 655 NGQLPIDLGKLKMLEELDVSHNNLSGTLR--VLSTIQSLTFINISHN---LFSGPVPPSL 709
Query: 423 MK------TSFFGSVN----------SCPIAANPSFFKKRRDVGHRGMK---LALVLTLS 463
K TSF G+ + +CP ++ + + G G+ +A+++ +
Sbjct: 710 TKFLNSSPTSFSGNSDLCINCPADGLACPESSILRPCNMQSNTGKGGLSTLGIAMIVLGA 769
Query: 464 LIFALAGILFLAF---GCRRKNKMWEVKQGSYREEQNISGPFSFQTDSTTWVADVKQATS 520
L+F + LF AF C++ Q +
Sbjct: 770 LLFIICLFLFSAFLFLHCKKS----------------------------------VQEIA 795
Query: 521 VPVVIFEKPLLNITFADLLSATSNFDRGTLLAEGKFGPVYRGFL-PGNIHVAVKVLVVGS 579
+ + LLN +L AT N + ++ +G G +Y+ L P ++ K++ G
Sbjct: 796 ISAQEGDGSLLN----KVLEATENLNDKYVIGKGAHGTIYKATLSPDKVYAVKKLVFTGI 851
Query: 580 TLTDEEAARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQS 639
RE+E +G+++H NL+ L + + + + +Y YMENG+L ++L++
Sbjct: 852 KNGSVSMVREIETIGKVRHRNLIKLEEFWLRKEYGLILYTYMENGSLHDILHE--TNPPK 909
Query: 640 TDDWSTDTWEEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKA 699
DWST RH IA+GTA LA+LH C P I+HR +K
Sbjct: 910 PLDWST------------------------RHNIAVGTAHGLAYLHFDCDPAIVHRDIKP 945
Query: 700 SSVYLDYDLEPRLSDFGLAKIF---GSGLDEEIARGSPGYVPPEFSQPEFESPTPKSDVY 756
++ LD DLEP +SDFG+AK+ + + +G+ GY+ PE + +S +SDVY
Sbjct: 946 MNILLDSDLEPHISDFGIAKLLDQSATSIPSNTVQGTIGYMAPENAFTTVKS--RESDVY 1003
Query: 757 CFGVVLFELLTGKKPVGDDYTDDKEATTLVSWVRGL-VRKNQTSRAIDPKICDTGSD--- 812
+GVVL EL+T KK + + + T +V WVR + + + + +DP + D D
Sbjct: 1004 SYGVVLLELITRKKALDPSFNGE---TDIVGWVRSVWTQTGEIQKIVDPSLLDELIDSSV 1060
Query: 813 -EQIEEALKEVGYLCTADLPFKRPTMQQIV 841
EQ+ EAL + C KRPTM+ +V
Sbjct: 1061 MEQVTEAL-SLALRCAEKEVDKRPTMRDVV 1089
Score = 146 bits (368), Expect = 3e-34
Identities = 119/362 (32%), Positives = 174/362 (47%), Gaps = 11/362 (3%)
Query: 51 SSVCSWKGVYCDSNKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT-TLPSD 109
S+ CSW GV CD ++ V LNLS G++G I L L + LS N ++PS
Sbjct: 54 STPCSWLGVECD-RRQFVDTLNLSSYGISGEF-GPEISHLKHLKKVVLSGNGFFGSIPSQ 111
Query: 110 FWSLTSLKSLNLSSNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKL 169
+ + L+ ++LSSN +G++ + +G L N L NS PE+L S+ L+ +
Sbjct: 112 LGNCSLLEHIDLSSNSFTGNIPDTLGALQNLRNLSLFFNSLIGPFPESLLSIPHLETVYF 171
Query: 170 DHNMFVRSIPSGILKCQSLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGVS- 228
N SIPS I L ++ L NQ SG +P G+ L+ L L +NN+ G +
Sbjct: 172 TGNGLNGSIPSNIGNMSELTTLWLDDNQFSGPVPSSLGN-ITTLQELYLNDNNLVGTLPV 230
Query: 229 NFSRLKSIVSLNISGNSFQGSIIEVFV--LKLEALDLSRNQFQGHISQVKYNWSHLVYLD 286
+ L+++V L++ NS G+I FV +++ + LS NQF G + N + L
Sbjct: 231 TLNNLENLVYLDVRNNSLVGAIPLDFVSCKQIDTISLSNNQFTGGLPPGLGNCTSLREFG 290
Query: 287 LSENQLSGEIFQNLNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPD 346
LSG I L L LA N FS + P++ + L L + L G IP
Sbjct: 291 AFSCALSGPIPSCFGQLTKLDTLYLAGNHFSGRIPPELGKCKSMIDLQLQQNQLEGEIPG 350
Query: 347 EISHLGNLNALDLSMNHLDGKIPL--LKNKHLQVIDFSHNNLSG--PVPSFILKSLPKMK 402
E+ L L L L N+L G++PL K + LQ + NNLSG PV LK L +
Sbjct: 351 ELGMLSQLQYLHLYTNNLSGEVPLSIWKIQSLQSLQLYQNNLSGELPVDMTELKQLVSLA 410
Query: 403 KY 404
Y
Sbjct: 411 LY 412
Score = 141 bits (356), Expect = 6e-33
Identities = 121/405 (29%), Positives = 176/405 (42%), Gaps = 79/405 (19%)
Query: 70 ELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT-TLPSDFWSLTSLKSLNLSSNHISG 128
EL L+ L G +P T+ L L LD+ NN + +P DF S + +++LS+N +G
Sbjct: 216 ELYLNDNNLVGTLP-VTLNNLENLVYLDVRNNSLVGAIPLDFVSCKQIDTISLSNNQFTG 274
Query: 129 SLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGILKCQSL 188
L +GN L F + S IP L L L L N F IP + KC+S+
Sbjct: 275 GLPPGLGNCTSLREFGAFSCALSGPIPSCFGQLTKLDTLYLAGNHFSGRIPPELGKCKSM 334
Query: 189 VSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGVS-------------------- 228
+ + L NQL G +P G +L+ L+L NN+ G V
Sbjct: 335 IDLQLQQNQLEGEIPGELG-MLSQLQYLHLYTNNLSGEVPLSIWKIQSLQSLQLYQNNLS 393
Query: 229 -----NFSRLKSIVSLNISGNSFQGSIIEVFVL--KLEALDLSRNQFQGHI-----SQVK 276
+ + LK +VSL + N F G I + LE LDL+RN F GHI SQ K
Sbjct: 394 GELPVDMTELKQLVSLALYENHFTGVIPQDLGANSSLEVLDLTRNMFTGHIPPNLCSQKK 453
Query: 277 -------YNW-----------------------------------SHLVYLDLSENQLSG 294
YN+ +L++ DLS N +G
Sbjct: 454 LKRLLLGYNYLEGSVPSDLGGCSTLERLILEENNLRGGLPDFVEKQNLLFFDLSGNNFTG 513
Query: 295 EIFQNLNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHLGNL 354
I +L N N+ + L+ N+ S P++ L+ LE+LNLS L G +P E+S+ L
Sbjct: 514 PIPPSLGNLKNVTAIYLSSNQLSGSIPPELGSLVKLEHLNLSHNILKGILPSELSNCHKL 573
Query: 355 NALDLSMNHLDGKIP--LLKNKHLQVIDFSHNNLSGPVPSFILKS 397
+ LD S N L+G IP L L + N+ SG +P+ + +S
Sbjct: 574 SELDASHNLLNGSIPSTLGSLTELTKLSLGENSFSGGIPTSLFQS 618
Score = 107 bits (266), Expect = 2e-22
Identities = 77/252 (30%), Positives = 129/252 (50%), Gaps = 15/252 (5%)
Query: 65 KEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT-TLPSDFWSLTSLKSLNLSS 123
K++++ +LSG TGPIP ++G L + ++ LS+N+++ ++P + SL L+ LNLS
Sbjct: 498 KQNLLFFDLSGNNFTGPIP-PSLGNLKNVTAIYLSSNQLSGSIPPELGSLVKLEHLNLSH 556
Query: 124 NHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGIL 183
N + G L + + N L D S N + IP L SL L L L N F IP+ +
Sbjct: 557 NILKGILPSELSNCHKLSELDASHNLLNGSIPSTLGSLTELTKLSLGENSFSGGIPTSLF 616
Query: 184 KCQSLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGVS-NFSRLKSIVSLNIS 242
+ L+++ L N L+G +P A LR+LNL+ N + G + + +LK + L++S
Sbjct: 617 QSNKLLNLQLGGNLLAGDIPP--VGALQALRSLNLSSNKLNGQLPIDLGKLKMLEELDVS 674
Query: 243 GNSFQGSI-IEVFVLKLEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQNLN 301
N+ G++ + + L +++S N F G + S +L+ S SG N +
Sbjct: 675 HNNLSGTLRVLSTIQSLTFINISHNLFSGPVPP-----SLTKFLNSSPTSFSG----NSD 725
Query: 302 NSMNLKHLSLAC 313
+N LAC
Sbjct: 726 LCINCPADGLAC 737
>RLK5_ARATH (P47735) Receptor-like protein kinase 5 precursor (EC
2.7.1.37)
Length = 999
Score = 278 bits (711), Expect = 4e-74
Identities = 247/833 (29%), Positives = 386/833 (45%), Gaps = 145/833 (17%)
Query: 87 IGKLNKLHSLDLSN-NKITTLPSDFWSLTSLKSLNLSSNHISGSLTNNIGNFGLLENFDL 145
+G L +L L L+ N + +P LTSL +L+L+ N ++GS+ + I +E +L
Sbjct: 208 LGNLTELQVLWLAGCNLVGPIPPSLSRLTSLVNLDLTFNQLTGSIPSWITQLKTVEQIEL 267
Query: 146 SKNSFSDEIPEALSSLVSLK-----------------------VLKLDHNMFVRSIPSGI 182
NSFS E+PE++ ++ +LK L L NM +P I
Sbjct: 268 FNNSFSGELPESMGNMTTLKRFDASMNKLTGKIPDNLNLLNLESLNLFENMLEGPLPESI 327
Query: 183 LKCQSLVSIDLSSNQLSGTLPHGFGDAFP-----------------------KLRTLNLA 219
+ ++L + L +N+L+G LP G P KL L L
Sbjct: 328 TRSKTLSELKLFNNRLTGVLPSQLGANSPLQYVDLSYNRFSGEIPANVCGEGKLEYLILI 387
Query: 220 ENNIYGGVS-NFSRLKSIVSLNISGNSFQGSIIEVF--VLKLEALDLSRNQFQGHISQVK 276
+N+ G +S N + KS+ + +S N G I F + +L L+LS N F G I +
Sbjct: 388 DNSFSGEISNNLGKCKSLTRVRLSNNKLSGQIPHGFWGLPRLSLLELSDNSFTGSIPKTI 447
Query: 277 YNWSHLVYLDLSENQLSGEIFQNLNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLS 336
+L L +S+N+ SG I + + + +S A N FS + + L L L+LS
Sbjct: 448 IGAKNLSNLRISKNRFSGSIPNEIGSLNGIIEISGAENDFSGEIPESLVKLKQLSRLDLS 507
Query: 337 KTSLVGHIPDEISHLGNLNALDLSMNHLDGKIPLLKN--KHLQVIDFSHNNLSGPVPSFI 394
K L G IP E+ NLN L+L+ NHL G+IP L +D S N SG +P
Sbjct: 508 KNQLSGEIPRELRGWKNLNELNLANNHLSGEIPKEVGILPVLNYLDLSSNQFSGEIP-LE 566
Query: 395 LKSLPKMKKYNFSYNNLTLCASEIKP----DIMKTSFFGSVNSC-PIAANPSFFKKRRDV 449
L++L K+ N SYN+L + +I P I F G+ C + + +++
Sbjct: 567 LQNL-KLNVLNLSYNHL---SGKIPPLYANKIYAHDFIGNPGLCVDLDGLCRKITRSKNI 622
Query: 450 GHRGMKLALVLTLSLIFALAGILFLAFGCRRKNKMWEVKQGSYREEQNISGPFSFQTDST 509
G+ + L + L L+F + ++F+A CR K+ +K + ++
Sbjct: 623 GYVWILLTIFLLAGLVFVVGIVMFIA-KCR---KLRALKSSTLA--------------AS 664
Query: 510 TWVADVKQATSVPVVIFEKPLLNITFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIH 569
W + K S + AD L D ++ G G VY+ L G
Sbjct: 665 KWRSFHKLHFSEHEI-----------ADCL------DEKNVIGFGSSGKVYKVELRGGEV 707
Query: 570 VAVKVL----------VVGSTLTDEEAARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYD 619
VAVK L +L + A E+E LG I+H ++V L C +GD ++ +Y+
Sbjct: 708 VAVKKLNKSVKGGDDEYSSDSLNRDVFAAEVETLGTIRHKSIVRLWCCCSSGDCKLLVYE 767
Query: 620 YMENGNLQNLLYDLPLGVQSTDDWSTDTWEEADNGIQNVGSEGLLTTWRFRHKIALGTAR 679
YM NG+L ++L+ G G++ W R +IAL A
Sbjct: 768 YMPNGSLADVLHGDRKG-------------------------GVVLGWPERLRIALDAAE 802
Query: 680 ALAFLHHGCSPPIIHRAVKASSVYLDYDLEPRLSDFGLAKI---FGSGLDEEIA--RGSP 734
L++LHH C PPI+HR VK+S++ LD D +++DFG+AK+ GS E ++ GS
Sbjct: 803 GLSYLHHDCVPPIVHRDVKSSNILLDSDYGAKVADFGIAKVGQMSGSKTPEAMSGIAGSC 862
Query: 735 GYVPPEFSQPEFESPTPKSDVYCFGVVLFELLTGKKPVGDDYTDDKEATTLVSWVRGLVR 794
GY+ PE+ KSD+Y FGVVL EL+TGK+P D DK+ + WV +
Sbjct: 863 GYIAPEYVYT--LRVNEKSDIYSFGVVLLELVTGKQPT-DSELGDKD---MAKWVCTALD 916
Query: 795 KNQTSRAIDPKICDTGSDEQIEEALKEVGYLCTADLPFKRPTMQQIVGLLKDI 847
K IDPK+ D E+I + + +G LCT+ LP RP+M+++V +L+++
Sbjct: 917 KCGLEPVIDPKL-DLKFKEEISKVI-HIGLLCTSPLPLNRPSMRKVVIMLQEV 967
Score = 162 bits (411), Expect = 3e-39
Identities = 118/311 (37%), Positives = 160/311 (50%), Gaps = 8/311 (2%)
Query: 78 LTGPIPDTTIGKLNKLHSLDLSNNKIT-TLPSDFWSLTSLKSLNLSSNHISGSLTNNIGN 136
LTG IPD L L SL+L N + LP +L L L +N ++G L + +G
Sbjct: 296 LTGKIPDNL--NLLNLESLNLFENMLEGPLPESITRSKTLSELKLFNNRLTGVLPSQLGA 353
Query: 137 FGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGILKCQSLVSIDLSSN 196
L+ DLS N FS EIP + L+ L L N F I + + KC+SL + LS+N
Sbjct: 354 NSPLQYVDLSYNRFSGEIPANVCGEGKLEYLILIDNSFSGEISNNLGKCKSLTRVRLSNN 413
Query: 197 QLSGTLPHGFGDAFPKLRTLNLAENNIYGGV-SNFSRLKSIVSLNISGNSFQGSIIEVFV 255
+LSG +PHGF P+L L L++N+ G + K++ +L IS N F GSI
Sbjct: 414 KLSGQIPHGFW-GLPRLSLLELSDNSFTGSIPKTIIGAKNLSNLRISKNRFSGSIPNEIG 472
Query: 256 LKLEALDLS--RNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQNLNNSMNLKHLSLAC 313
+++S N F G I + L LDLS+NQLSGEI + L NL L+LA
Sbjct: 473 SLNGIIEISGAENDFSGEIPESLVKLKQLSRLDLSKNQLSGEIPRELRGWKNLNELNLAN 532
Query: 314 NRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHLGNLNALDLSMNHLDGKIPLLKN 373
N S + ++ +L L YL+LS G IP E+ +L LN L+LS NHL GKIP L
Sbjct: 533 NHLSGEIPKEVGILPVLNYLDLSSNQFSGEIPLELQNL-KLNVLNLSYNHLSGKIPPLYA 591
Query: 374 KHLQVIDFSHN 384
+ DF N
Sbjct: 592 NKIYAHDFIGN 602
Score = 142 bits (357), Expect = 5e-33
Identities = 135/471 (28%), Positives = 210/471 (43%), Gaps = 90/471 (19%)
Query: 9 VLVLTFLFKHLASQQPNTDEFYVSEFLKKMGLTS-----SSKVYNFSSSVCSWKGVYCDS 63
+L+L +L S N D + + K+GL+ SS N + C W GV CD+
Sbjct: 6 ILLLCLSSTYLPSLSLNQDATILRQ--AKLGLSDPAQSLSSWSDNNDVTPCKWLGVSCDA 63
Query: 64 NKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT------------------- 104
+VV ++LS L GP P + + L LHSL L NN I
Sbjct: 64 TS-NVVSVDLSSFMLVGPFP-SILCHLPSLHSLSLYNNSINGSLSADDFDTCHNLISLDL 121
Query: 105 -------TLPSDF-WSLTSLKSLNLSSNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPE 156
++P ++L +LK L +S N++S ++ ++ G F LE+ +L+ N S IP
Sbjct: 122 SENLLVGSIPKSLPFNLPNLKFLEISGNNLSDTIPSSFGEFRKLESLNLAGNFLSGTIPA 181
Query: 157 ALSSLVSLKVLKLDHNMF-------------------------VRSIPSGILKCQSLVSI 191
+L ++ +LK LKL +N+F V IP + + SLV++
Sbjct: 182 SLGNVTTLKELKLAYNLFSPSQIPSQLGNLTELQVLWLAGCNLVGPIPPSLSRLTSLVNL 241
Query: 192 DLSSNQLSGTLP----------------HGFGDAFPK-------LRTLNLAENNIYGGVS 228
DL+ NQL+G++P + F P+ L+ + + N + G +
Sbjct: 242 DLTFNQLTGSIPSWITQLKTVEQIELFNNSFSGELPESMGNMTTLKRFDASMNKLTGKIP 301
Query: 229 NFSRLKSIVSLNISGNSFQGSIIEVFVLK--LEALDLSRNQFQGHI-SQVKYNWSHLVYL 285
+ L ++ SLN+ N +G + E L L L N+ G + SQ+ N S L Y+
Sbjct: 302 DNLNLLNLESLNLFENMLEGPLPESITRSKTLSELKLFNNRLTGVLPSQLGAN-SPLQYV 360
Query: 286 DLSENQLSGEIFQNLNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIP 345
DLS N+ SGEI N+ L++L L N FS + + L + LS L G IP
Sbjct: 361 DLSYNRFSGEIPANVCGEGKLEYLILIDNSFSGEISNNLGKCKSLTRVRLSNNKLSGQIP 420
Query: 346 DEISHLGNLNALDLSMNHLDGKIP--LLKNKHLQVIDFSHNNLSGPVPSFI 394
L L+ L+LS N G IP ++ K+L + S N SG +P+ I
Sbjct: 421 HGFWGLPRLSLLELSDNSFTGSIPKTIIGAKNLSNLRISKNRFSGSIPNEI 471
Score = 88.2 bits (217), Expect = 8e-17
Identities = 74/230 (32%), Positives = 113/230 (48%), Gaps = 7/230 (3%)
Query: 71 LNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT-TLPSDFWSLTSLKSLNLSSNHISGS 129
++LS +G IP G+ KL L L +N + + ++ SL + LS+N +SG
Sbjct: 360 VDLSYNRFSGEIPANVCGE-GKLEYLILIDNSFSGEISNNLGKCKSLTRVRLSNNKLSGQ 418
Query: 130 LTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGILKCQSLV 189
+ + L +LS NSF+ IP+ + +L L++ N F SIP+ I ++
Sbjct: 419 IPHGFWGLPRLSLLELSDNSFTGSIPKTIIGAKNLSNLRISKNRFSGSIPNEIGSLNGII 478
Query: 190 SIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGVSNFSR-LKSIVSLNISGNSFQG 248
I + N SG +P +L L+L++N + G + R K++ LN++ N G
Sbjct: 479 EISGAENDFSGEIPESL-VKLKQLSRLDLSKNQLSGEIPRELRGWKNLNELNLANNHLSG 537
Query: 249 SI-IEVFVLK-LEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEI 296
I EV +L L LDLS NQF G I N L L+LS N LSG+I
Sbjct: 538 EIPKEVGILPVLNYLDLSSNQFSGEIPLELQNLK-LNVLNLSYNHLSGKI 586
Score = 42.7 bits (99), Expect = 0.004
Identities = 44/160 (27%), Positives = 71/160 (43%), Gaps = 24/160 (15%)
Query: 285 LDLSENQLSGEIFQNLNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHI 344
L LS+ S + + N+ K L ++C+ S + ++LS LVG
Sbjct: 33 LGLSDPAQSLSSWSDNNDVTPCKWLGVSCDATSN-----------VVSVDLSSFMLVGPF 81
Query: 345 PDEISHLGNLNALDLSMNHLDGKIPL--LKNKH-LQVIDFSHNNLSGPVPSFILKSLPKM 401
P + HL +L++L L N ++G + H L +D S N L G +P + +LP +
Sbjct: 82 PSILCHLPSLHSLSLYNNSINGSLSADDFDTCHNLISLDLSENLLVGSIPKSLPFNLPNL 141
Query: 402 KKYNFSYNNLTLCASEIKPDIMKTSF--FGSVNSCPIAAN 439
K S NNL+ D + +SF F + S +A N
Sbjct: 142 KFLEISGNNLS--------DTIPSSFGEFRKLESLNLAGN 173
>TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precursor
(EC 2.7.1.-)
Length = 942
Score = 213 bits (541), Expect = 2e-54
Identities = 233/847 (27%), Positives = 374/847 (43%), Gaps = 127/847 (14%)
Query: 64 NKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKITTLPSD-FWSLTSLKSLNLS 122
N + L L ++GP+P ++ L L L LSNN ++PSD F LTSL+S+ +
Sbjct: 86 NLSELERLELQWNNISGPVP--SLSGLASLQVLMLSNNNFDSIPSDVFQGLTSLQSVEID 143
Query: 123 SNHI-SGSLTNNIGNFGLLENFDLSKNSFSDEIPEALS--SLVSLKVLKLDHNMFVRSIP 179
+N S + ++ N L+NF + + S +P L L +L L N +P
Sbjct: 144 NNPFKSWEIPESLRNASALQNFSANSANVSGSLPGFLGPDEFPGLSILHLAFNNLEGELP 203
Query: 180 SGILKCQSLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGVSNFSRLKSIVSL 239
+ Q + S+ L+ +L+G + L+ + L N G + +FS LK + SL
Sbjct: 204 MSLAGSQ-VQSLWLNGQKLTGDIT--VLQNMTGLKEVWLHSNKFSGPLPDFSGLKELESL 260
Query: 240 NISGNSFQGSIIEVFVLKLEAL---DLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEI 296
++ NSF G + +L LE+L +L+ N QG + K + V +DL ++ S +
Sbjct: 261 SLRDNSFTGPV-PASLLSLESLKVVNLTNNHLQGPVPVFKSS----VSVDLDKDSNSFCL 315
Query: 297 FQNLNNSMNLKHLSLACNRFSRQKFPKI-EMLLG----------------LEYLNLSKTS 339
+K L L + F P++ E G + ++L K
Sbjct: 316 SSPGECDPRVKSLLLIASSFDYP--PRLAESWKGNDPCTNWIGIACSNGNITVISLEKME 373
Query: 340 LVGHIPDEISHLGNLNALDLSMNHLDGKIP--LLKNKHLQVIDFSHNNLSGPVPSFILKS 397
L G I E + +L + L +N+L G IP L +L+ +D S N L G VP F
Sbjct: 374 LTGTISPEFGAIKSLQRIILGINNLTGMIPQELTTLPNLKTLDVSSNKLFGKVPGF---- 429
Query: 398 LPKMKKYNFSYNNLTLCASEIKPDIMK--TSFFGSVNSCPIAANPSFFKKRRDVGHRGMK 455
+ N N + PDI K +S +S P + S +D RGMK
Sbjct: 430 -----RSNVVVN------TNGNPDIGKDKSSLSSPGSSSPSGGSGSGINGDKD--RRGMK 476
Query: 456 LALVLTLSLIFALAGIL------FLAFGCRRKNKMWEVKQGSYREEQN--ISGPFSFQTD 507
+ + + + L G+L L F +K + K+ S E N + P +D
Sbjct: 477 SSTFIGIIVGSVLGGLLSIFLIGLLVFCWYKKRQ----KRFSGSESSNAVVVHPRHSGSD 532
Query: 508 ----------STTWVADVKQATSVP--------VVIFEKPLLNITFADLLSATSNFDRGT 549
S+ V + ++P + + E + I+ L S T+NF
Sbjct: 533 NESVKITVAGSSVSVGGISDTYTLPGTSEVGDNIQMVEAGNMLISIQVLRSVTNNFSSDN 592
Query: 550 LLAEGKFGPVYRGFLPGNIHVAVKVLVVGSTLTD--EEAARELEFLGRIKHPNLVPLTGY 607
+L G FG VY+G L +AVK + G E E+ L +++H +LV L GY
Sbjct: 593 ILGSGGFGVVYKGELHDGTKIAVKRMENGVIAGKGFAEFKSEIAVLTKVRHRHLVTLLGY 652
Query: 608 CVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTDTWEEADNGIQNVGSEGLLTTW 667
C+ G++++ +Y+YM G L L+ +WS + G + LL W
Sbjct: 653 CLDGNEKLLVYEYMPQGTLSRHLF----------EWSEE------------GLKPLL--W 688
Query: 668 RFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLEPRLSDFGLAKIF--GSGL 725
+ R +AL AR + +LH IHR +K S++ L D+ +++DFGL ++ G G
Sbjct: 689 KQRLTLALDVARGVEYLHGLAHQSFIHRDLKPSNILLGDDMRAKVADFGLVRLAPEGKGS 748
Query: 726 DEEIARGSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFELLTGKKPVGDDYTDDKEATTL 785
E G+ GY+ PE++ T K DVY FGV+L EL+TG+K + D + +E+ L
Sbjct: 749 IETRIAGTFGYLAPEYAVT--GRVTTKVDVYSFGVILMELITGRKSL--DESQPEESIHL 804
Query: 786 VSWVRGLVRKNQTS--RAIDPKICDTGSDEQIEEALKEVGYL---CTADLPFKRPTMQQI 840
VSW + + + S +AID I DE+ ++ V L C A P++RP M
Sbjct: 805 VSWFKRMYINKEASFKKAIDTTI---DLDEETLASVHTVAELAGHCCAREPYQRPDMGHA 861
Query: 841 VGLLKDI 847
V +L +
Sbjct: 862 VNILSSL 868
Score = 85.5 bits (210), Expect = 5e-16
Identities = 99/385 (25%), Positives = 153/385 (39%), Gaps = 79/385 (20%)
Query: 10 LVLTFLFKHLASQQPNTDEFYVSEFLK-KMGLTSSSKVYNFSSSVCSWKGVYCDSNKEHV 68
L+ +F F L S + +S L K L S C W + C K V
Sbjct: 8 LLFSFTFLLLLSLSKADSDGDLSAMLSLKKSLNPPSSFGWSDPDPCKWTHIVCTGTKR-V 66
Query: 69 VELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKITTLPSDFWSLTSLKSLNLSSNHISG 128
+ + GL G TL D +L+ L+ L L N+ISG
Sbjct: 67 TRIQIGHSGLQG------------------------TLSPDLRNLSELERLELQWNNISG 102
Query: 129 SLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVR-SIPSGILKCQS 187
+ + + L+ LS N+F + L SL+ +++D+N F IP + +
Sbjct: 103 PVPS-LSGLASLQVLMLSNNNFDSIPSDVFQGLTSLQSVEIDNNPFKSWEIPESLRNASA 161
Query: 188 LVSIDLSSNQLSGTLPHGFG-DAFPKLRTLNLAENNIYGGVSNFSRLKSIVSLNISGNSF 246
L + +S +SG+LP G D FP L L+LA NN+ G + ++++G
Sbjct: 162 LQNFSANSANVSGSLPGFLGPDEFPGLSILHLAFNNLEGELP----------MSLAG--- 208
Query: 247 QGSIIEVFVLKLEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQNLNNSMNL 306
SQV+ W L+ +L+G+I L N L
Sbjct: 209 --------------------------SQVQSLW-------LNGQKLTGDITV-LQNMTGL 234
Query: 307 KHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHLGNLNALDLSMNHLDG 366
K + L N+FS P L LE L+L S G +P + L +L ++L+ NHL G
Sbjct: 235 KEVWLHSNKFSGP-LPDFSGLKELESLSLRDNSFTGPVPASLLSLESLKVVNLTNNHLQG 293
Query: 367 KIPLLKNKHLQVIDFSHNN--LSGP 389
+P+ K+ +D N+ LS P
Sbjct: 294 PVPVFKSSVSVDLDKDSNSFCLSSP 318
Score = 71.6 bits (174), Expect = 8e-12
Identities = 79/285 (27%), Positives = 127/285 (43%), Gaps = 65/285 (22%)
Query: 119 LNLSSNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSI 178
+ + + + G+L+ ++ N LE +L N+ S +P +LS L SL+VL L +N F SI
Sbjct: 69 IQIGHSGLQGTLSPDLRNLSELERLELQWNNISGPVP-SLSGLASLQVLMLSNNNF-DSI 126
Query: 179 PSGILK-CQSLVSIDLSSNQL-SGTLPHGFGDAFPKLRTLNLAENNIYGGVSNFSRLKSI 236
PS + + SL S+++ +N S +P +A L+ + N+ G + F
Sbjct: 127 PSDVFQGLTSLQSVEIDNNPFKSWEIPESLRNA-SALQNFSANSANVSGSLPGF------ 179
Query: 237 VSLNISGNSFQGSIIEVFVLKLEALDLSRNQFQGHI------SQVKYNWSHLVYLDLSEN 290
+ + F G L L L+ N +G + SQV+ W L+
Sbjct: 180 ----LGPDEFPG---------LSILHLAFNNLEGELPMSLAGSQVQSLW-------LNGQ 219
Query: 291 QLSGEIFQNLNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISH 350
+L+G+I L N LK + L N+FS G +PD S
Sbjct: 220 KLTGDITV-LQNMTGLKEVWLHSNKFS------------------------GPLPD-FSG 253
Query: 351 LGNLNALDLSMNHLDGKIP--LLKNKHLQVIDFSHNNLSGPVPSF 393
L L +L L N G +P LL + L+V++ ++N+L GPVP F
Sbjct: 254 LKELESLSLRDNSFTGPVPASLLSLESLKVVNLTNNHLQGPVPVF 298
Score = 36.6 bits (83), Expect = 0.29
Identities = 20/78 (25%), Positives = 38/78 (48%), Gaps = 2/78 (2%)
Query: 333 LNLSKTSLVGHIPDEISHLGNLNALDLSMNHLDGKIPLLKN-KHLQVIDFSHNNLSGPVP 391
+ + + L G + ++ +L L L+L N++ G +P L LQV+ S+NN +P
Sbjct: 69 IQIGHSGLQGTLSPDLRNLSELERLELQWNNISGPVPSLSGLASLQVLMLSNNNFDS-IP 127
Query: 392 SFILKSLPKMKKYNFSYN 409
S + + L ++ N
Sbjct: 128 SDVFQGLTSLQSVEIDNN 145
>BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated
receptor kinase 1 precursor (EC 2.7.1.37)
(BRI1-associated receptor kinase 1) (Somatic
embryogenesis receptor-like kinase 3)
Length = 615
Score = 191 bits (486), Expect = 5e-48
Identities = 162/566 (28%), Positives = 253/566 (44%), Gaps = 79/566 (13%)
Query: 285 LDLSENQLSGEIFQNLNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHI 344
+DL LSG++ L NL++L L N + ++ L L L+L +L G I
Sbjct: 73 VDLGNANLSGQLVMQLGQLPNLQYLELYSNNITGTIPEQLGNLTELVSLDLYLNNLSGPI 132
Query: 345 PDEISHLGNLNALDLSMNHLDGKIP--LLKNKHLQVIDFSHNNLSGPVPSFILKSLPKMK 402
P + L L L L+ N L G+IP L LQV+D S+N L+G +P + S
Sbjct: 133 PSTLGRLKKLRFLRLNNNSLSGEIPRSLTAVLTLQVLDLSNNPLTGDIP--VNGSFSLFT 190
Query: 403 KYNFSYNNLTLCASEIKPDIMKTSFFGSVNSCPIAANPSFFKKRRDVGHRGMKLALVLTL 462
+F+ LT + P I T PS R G + +
Sbjct: 191 PISFANTKLTPLPASPPPPISPTP-------------PSPAGSNRITG--AIAGGVAAGA 235
Query: 463 SLIFALAGILFLAFGCRRKNKMWEVKQGSYREEQNISGPFSFQTDSTTWVADVKQATSVP 522
+L+FA+ I LA+ R+K Q+ + D + +K+
Sbjct: 236 ALLFAVPAIA-LAWWRRKK-------------PQDHFFDVPAEEDPEVHLGQLKR----- 276
Query: 523 VVIFEKPLLNITFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVVGSTLT 582
+ +L A+ NF +L G FG VY+G L VAVK L T
Sbjct: 277 ----------FSLRELQVASDNFSNKNILGRGGFGKVYKGRLADGTLVAVKRLKEERTQG 326
Query: 583 DE-EAARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTD 641
E + E+E + H NL+ L G+C+ +R+ +Y YM NG++ + L + P D
Sbjct: 327 GELQFQTEVEMISMAVHRNLLRLRGFCMTPTERLLVYPYMANGSVASCLRERPESQPPLD 386
Query: 642 DWSTDTWEEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASS 701
W R +IALG+AR LA+LH C P IIHR VKA++
Sbjct: 387 -------------------------WPKRQRIALGSARGLAYLHDHCDPKIIHRDVKAAN 421
Query: 702 VYLDYDLEPRLSDFGLAKI--FGSGLDEEIARGSPGYVPPEFSQPEFESPTPKSDVYCFG 759
+ LD + E + DFGLAK+ + RG+ G++ PE+ S K+DV+ +G
Sbjct: 422 ILLDEEFEAVVGDFGLAKLMDYKDTHVTTAVRGTIGHIAPEYLSTGKSS--EKTDVFGYG 479
Query: 760 VVLFELLTGKKPVGDDYTDDKEATTLVSWVRGLVRKNQTSRAIDPKICDTGSDEQIEEAL 819
V+L EL+TG++ + + L+ WV+GL+++ + +D + DE++E+ L
Sbjct: 480 VMLLELITGQRAFDLARLANDDDVMLLDWVKGLLKEKKLEALVDVDLQGNYKDEEVEQ-L 538
Query: 820 KEVGYLCTADLPFKRPTMQQIVGLLK 845
+V LCT P +RP M ++V +L+
Sbjct: 539 IQVALLCTQSSPMERPKMSEVVRMLE 564
Score = 69.3 bits (168), Expect = 4e-11
Identities = 48/151 (31%), Positives = 78/151 (50%), Gaps = 27/151 (17%)
Query: 54 CSWKGVYCDSNKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT-TLPSDFWS 112
C+W V C+S+ V ++L L+G + +G+L L L+L +N IT T+P +
Sbjct: 57 CTWFHVTCNSDNS-VTRVDLGNANLSGQLV-MQLGQLPNLQYLELYSNNITGTIPEQLGN 114
Query: 113 LTSLKSLNLSSNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHN 172
LT L SL+L N++SG + + +G L L+ NS S EIP +L+++++L+VL
Sbjct: 115 LTELVSLDLYLNNLSGPIPSTLGRLKKLRFLRLNNNSLSGEIPRSLTAVLTLQVL----- 169
Query: 173 MFVRSIPSGILKCQSLVSIDLSSNQLSGTLP 203
DLS+N L+G +P
Sbjct: 170 -------------------DLSNNPLTGDIP 181
Score = 54.7 bits (130), Expect = 1e-06
Identities = 42/133 (31%), Positives = 62/133 (46%), Gaps = 2/133 (1%)
Query: 261 LDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQNLNNSMNLKHLSLACNRFSRQK 320
+DL G + +L YL+L N ++G I + L N L L L N S
Sbjct: 73 VDLGNANLSGQLVMQLGQLPNLQYLELYSNNITGTIPEQLGNLTELVSLDLYLNNLSGPI 132
Query: 321 FPKIEMLLGLEYLNLSKTSLVGHIPDEISHLGNLNALDLSMNHLDGKIPLLKNKHL-QVI 379
+ L L +L L+ SL G IP ++ + L LDLS N L G IP+ + L I
Sbjct: 133 PSTLGRLKKLRFLRLNNNSLSGEIPRSLTAVLTLQVLDLSNNPLTGDIPVNGSFSLFTPI 192
Query: 380 DFSHNNLSGPVPS 392
F++ L+ P+P+
Sbjct: 193 SFANTKLT-PLPA 204
Score = 49.3 bits (116), Expect = 4e-05
Identities = 40/131 (30%), Positives = 54/131 (40%), Gaps = 26/131 (19%)
Query: 144 DLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGILKCQSLVSIDLSSNQLSGTLP 203
DL + S ++ L L +L+ L+L N +IP + LVS+DL N LSG +P
Sbjct: 74 DLGNANLSGQLVMQLGQLPNLQYLELYSNNITGTIPEQLGNLTELVSLDLYLNNLSGPIP 133
Query: 204 HGFGDAFPKLRTLNLAENNIYGGVSNFSRLKSIVSLNISGNSFQGSIIE--VFVLKLEAL 261
S RLK + L ++ NS G I VL L+ L
Sbjct: 134 ------------------------STLGRLKKLRFLRLNNNSLSGEIPRSLTAVLTLQVL 169
Query: 262 DLSRNQFQGHI 272
DLS N G I
Sbjct: 170 DLSNNPLTGDI 180
Score = 46.2 bits (108), Expect = 4e-04
Identities = 29/77 (37%), Positives = 49/77 (62%), Gaps = 3/77 (3%)
Query: 64 NKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT-TLPSDFWSLTSLKSLNLS 122
N +V L+L L+GPIP +T+G+L KL L L+NN ++ +P ++ +L+ L+LS
Sbjct: 114 NLTELVSLDLYLNNLSGPIP-STLGRLKKLRFLRLNNNSLSGEIPRSLTAVLTLQVLDLS 172
Query: 123 SNHISGSLTNNIGNFGL 139
+N ++G + N G+F L
Sbjct: 173 NNPLTGDIPVN-GSFSL 188
>SIRK_ARATH (O64483) Senescence-induced receptor-like
serine/threonine kinase precursor (FLG22-induced
receptor-like kinase 1)
Length = 876
Score = 167 bits (423), Expect = 1e-40
Identities = 147/504 (29%), Positives = 233/504 (46%), Gaps = 88/504 (17%)
Query: 356 ALDLSMNHLDGKI-PLLKN-KHLQVIDFSHNNLSGPVPSFILKSLPKMKKYNFSYNNLTL 413
+L++S + L G+I P N ++ +D S N L+G +P+F L +LP + + N N LT
Sbjct: 418 SLNISFSELRGQIDPAFSNLTSIRKLDLSGNTLTGEIPAF-LANLPNLTELNVEGNKLTG 476
Query: 414 CASEIKPDIMKTSF----FGSVNSCPIAANPSFFKKRRDVGHRGMKLALVLTLSLIFALA 469
+ + K FG ++ + S KK+ G+ + + + + L+ ALA
Sbjct: 477 IVPQRLHERSKNGSLSLRFGRNPDLCLSDSCSNTKKKNKNGYIIPLVVVGIIVVLLTALA 536
Query: 470 GILFLAFGCRRKNKMWEVKQGSYREEQNISGPFSFQTDSTTWVADVKQATSVPVVIFEKP 529
LF F K K ++G+ E +GP +K A
Sbjct: 537 --LFRRF----KKKQ---QRGTLGER---NGP-------------LKTAKRY-------- 563
Query: 530 LLNITFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVVGSTLTDEEAARE 589
++++++ T+NF+R ++ +G FG VY G + G VAVKVL S +E E
Sbjct: 564 ---FKYSEVVNITNNFER--VIGKGGFGKVYHGVINGE-QVAVKVLSEESAQGYKEFRAE 617
Query: 590 LEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTDTWE 649
++ L R+ H NL L GYC + + IY+YM N NL + L
Sbjct: 618 VDLLMRVHHTNLTSLVGYCNEINHMVLIYEYMANENLGDYL------------------- 658
Query: 650 EADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLE 709
G + +W R KI+L A+ L +LH+GC PPI+HR VK +++ L+ L+
Sbjct: 659 --------AGKRSFILSWEERLKISLDAAQGLEYLHNGCKPPIVHRDVKPTNILLNEKLQ 710
Query: 710 PRLSDFGLAKIF---GSGLDEEIARGSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFELL 766
+++DFGL++ F GSG + GS GY+ PE+ KSDVY GVVL E++
Sbjct: 711 AKMADFGLSRSFSVEGSGQISTVVAGSIGYLDPEYYST--RQMNEKSDVYSLGVVLLEVI 768
Query: 767 TGKKPVGDDYTDDKEATTLVSWVRGLVRKNQTSRAIDPKI---CDTGSDEQIEEALKEVG 823
TG+ + T E + VR ++ +D ++ D GS + + E+
Sbjct: 769 TGQPAIASSKT---EKVHISDHVRSILANGDIRGIVDQRLRERYDVGSAWK----MSEIA 821
Query: 824 YLCTADLPFKRPTMQQIVGLLKDI 847
CT +RPTM Q+V LK I
Sbjct: 822 LACTEHTSAQRPTMSQVVMELKQI 845
Score = 40.0 bits (92), Expect = 0.026
Identities = 24/84 (28%), Positives = 41/84 (48%)
Query: 96 LDLSNNKITTLPSDFWSLTSLKSLNLSSNHISGSLTNNIGNFGLLENFDLSKNSFSDEIP 155
+D S I + SD + + SLN+S + + G + N + DLS N+ + EIP
Sbjct: 396 VDYSWEGIDCIQSDNTTNPRVVSLNISFSELRGQIDPAFSNLTSIRKLDLSGNTLTGEIP 455
Query: 156 EALSSLVSLKVLKLDHNMFVRSIP 179
L++L +L L ++ N +P
Sbjct: 456 AFLANLPNLTELNVEGNKLTGIVP 479
Score = 39.7 bits (91), Expect = 0.034
Identities = 28/82 (34%), Positives = 40/82 (48%), Gaps = 2/82 (2%)
Query: 236 IVSLNISGNSFQGSIIEVF--VLKLEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLS 293
+VSLNIS + +G I F + + LDLS N G I N +L L++ N+L+
Sbjct: 416 VVSLNISFSELRGQIDPAFSNLTSIRKLDLSGNTLTGEIPAFLANLPNLTELNVEGNKLT 475
Query: 294 GEIFQNLNNSMNLKHLSLACNR 315
G + Q L+ LSL R
Sbjct: 476 GIVPQRLHERSKNGSLSLRFGR 497
Score = 35.0 bits (79), Expect = 0.84
Identities = 47/233 (20%), Positives = 104/233 (44%), Gaps = 31/233 (13%)
Query: 142 NFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGILKCQSLVSIDLSSNQLSGT 201
+ ++S + +I A S+L S++ L L N IP+ + +L +++ N+L+G
Sbjct: 418 SLNISFSELRGQIDPAFSNLTSIRKLDLSGNTLTGEIPAFLANLPNLTELNVEGNKLTGI 477
Query: 202 LPHGFGDAFPKLRTLNLAENNIYGGVSNFSRLKSIVSL---NISGNSFQGSIIEVFVLKL 258
+P + R+ N + + +G + S + N +G ++ + V+ L
Sbjct: 478 VPQRLHE-----RSKNGSLSLRFGRNPDLCLSDSCSNTKKKNKNGYIIPLVVVGIIVVLL 532
Query: 259 EALDLSR-----------NQFQGHISQVKYNWSHLVYLDLSENQLS-------GEIFQNL 300
AL L R + G + K + + ++++ N G+++ +
Sbjct: 533 TALALFRRFKKKQQRGTLGERNGPLKTAKRYFKYSEVVNITNNFERVIGKGGFGKVYHGV 592
Query: 301 NN--SMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHL 351
N + +K LS + ++ ++++L+ + + NL TSLVG+ +EI+H+
Sbjct: 593 INGEQVAVKVLSEESAQGYKEFRAEVDLLMRVHHTNL--TSLVGYC-NEINHM 642
Score = 33.9 bits (76), Expect = 1.9
Identities = 20/61 (32%), Positives = 35/61 (56%)
Query: 257 KLEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQNLNNSMNLKHLSLACNRF 316
++ +L++S ++ +G I N + + LDLS N L+GEI L N NL L++ N+
Sbjct: 415 RVVSLNISFSELRGQIDPAFSNLTSIRKLDLSGNTLTGEIPAFLANLPNLTELNVEGNKL 474
Query: 317 S 317
+
Sbjct: 475 T 475
>RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase
RLCKVII (EC 2.7.1.37)
Length = 423
Score = 166 bits (419), Expect = 3e-40
Identities = 109/319 (34%), Positives = 159/319 (49%), Gaps = 35/319 (10%)
Query: 528 KPLLNITFADLLSATSNFDRGTLLAEGKFGPVYRGFLPG-NIHVAVKVLVVGSTLTDEEA 586
K TF +L AT NF L EG FG V++G + + VA+K L E
Sbjct: 86 KKAQTFTFQELAEATGNFRSDCFLGEGGFGKVFKGTIEKLDQVVAIKQLDRNGVQGIREF 145
Query: 587 ARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTD 646
E+ L HPNLV L G+C GDQR+ +Y+YM G+L++ L+ LP G + D
Sbjct: 146 VVEVLTLSLADHPNLVKLIGFCAEGDQRLLVYEYMPQGSLEDHLHVLPSGKKPLD----- 200
Query: 647 TWEEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDY 706
W R KIA G AR L +LH +PP+I+R +K S++ L
Sbjct: 201 --------------------WNTRMKIAAGAARGLEYLHDRMTPPVIYRDLKCSNILLGE 240
Query: 707 DLEPRLSDFGLAKIFGSGLDEEIA---RGSPGYVPPEFSQPEFESPTPKSDVYCFGVVLF 763
D +P+LSDFGLAK+ SG ++ G+ GY P+++ T KSD+Y FGVVL
Sbjct: 241 DYQPKLSDFGLAKVGPSGDKTHVSTRVMGTYGYCAPDYAMT--GQLTFKSDIYSFGVVLL 298
Query: 764 ELLTGKKPVGDDYTDDKEATTLVSWVRGLVR-KNQTSRAIDPKICDTGSDEQIEEALKEV 822
EL+TG+K + D T ++ LV W R L + + + +DP + + +AL +
Sbjct: 299 ELITGRKAI--DNTKTRKDQNLVGWARPLFKDRRNFPKMVDPLLQGQYPVRGLYQAL-AI 355
Query: 823 GYLCTADLPFKRPTMQQIV 841
+C + P RP + +V
Sbjct: 356 SAMCVQEQPTMRPVVSDVV 374
>PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC
2.7.1.37) (AvrPphB susceptible protein 1)
Length = 456
Score = 166 bits (419), Expect = 3e-40
Identities = 110/315 (34%), Positives = 159/315 (49%), Gaps = 35/315 (11%)
Query: 535 FADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHV-AVKVLVVGSTLTDEEAARELEFL 593
F +L +AT NF T L EG FG VY+G L V AVK L + E E+ L
Sbjct: 76 FRELAAATMNFHPDTFLGEGGFGRVYKGRLDSTGQVVAVKQLDRNGLQGNREFLVEVLML 135
Query: 594 GRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTDTWEEADN 653
+ HPNLV L GYC GDQR+ +Y++M G+L++ L+DLP ++ D
Sbjct: 136 SLLHHPNLVNLIGYCADGDQRLLVYEFMPLGSLEDHLHDLPPDKEALD------------ 183
Query: 654 GIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLEPRLS 713
W R KIA G A+ L FLH +PP+I+R K+S++ LD P+LS
Sbjct: 184 -------------WNMRMKIAAGAAKGLEFLHDKANPPVIYRDFKSSNILLDEGFHPKLS 230
Query: 714 DFGLAKIFGSGLDEEIA---RGSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFELLTGKK 770
DFGLAK+ +G ++ G+ GY PE++ T KSDVY FGVV EL+TG+K
Sbjct: 231 DFGLAKLGPTGDKSHVSTRVMGTYGYCAPEYAMT--GQLTVKSDVYSFGVVFLELITGRK 288
Query: 771 PVGDDYTDDKEATTLVSWVRGLVR-KNQTSRAIDPKICDTGSDEQIEEALKEVGYLCTAD 829
+ + ++ LV+W R L + + + DP++ + +AL V +C +
Sbjct: 289 AIDSEMPHGEQ--NLVAWARPLFNDRRKFIKLADPRLKGRFPTRALYQAL-AVASMCIQE 345
Query: 830 LPFKRPTMQQIVGLL 844
RP + +V L
Sbjct: 346 QAATRPLIADVVTAL 360
>APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor (EC
2.7.1.-)
Length = 412
Score = 154 bits (388), Expect = 1e-36
Identities = 117/342 (34%), Positives = 175/342 (50%), Gaps = 52/342 (15%)
Query: 525 IFEKP-LLNITFADLLSATSNFDRGTLLAEGKFGPVYRGFL---------PGN-IHVAVK 573
I + P L + TFA+L +AT NF ++L EG FG V++G++ PG + +AVK
Sbjct: 48 ILQSPNLKSFTFAELKAATRNFRPDSVLGEGGFGSVFKGWIDEQTLTASKPGTGVVIAVK 107
Query: 574 VLVVGSTLTDEEAARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDL 633
L +E E+ +LG+ HPNLV L GYC+ + R+ +Y++M G+L+N L+
Sbjct: 108 KLNQDGWQGHQEWLAEVNYLGQFSHPNLVKLIGYCLEDEHRLLVYEFMPRGSLENHLF-- 165
Query: 634 PLGVQSTDDWSTDTWEEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPII 693
GS +W R K+ALG A+ LAFLH+ +I
Sbjct: 166 -----------------------RRGSYFQPLSWTLRLKVALGAAKGLAFLHNA-ETSVI 201
Query: 694 HRAVKASSVYLDYDLEPRLSDFGLAKIFGSGLDEEIA---RGSPGYVPPEFSQPEFESPT 750
+R K S++ LD + +LSDFGLAK +G ++ G+ GY PE+ T
Sbjct: 202 YRDFKTSNILLDSEYNAKLSDFGLAKDGPTGDKSHVSTRIMGTYGYAAPEYLAT--GHLT 259
Query: 751 PKSDVYCFGVVLFELLTGKKPVGDDYTDDKEATTLVSWVRGLV-RKNQTSRAIDPKICDT 809
KSDVY +GVVL E+L+G++ V + ++ LV W R L+ K + R ID ++ D
Sbjct: 260 TKSDVYSYGVVLLEVLSGRRAVDKNRPPGEQ--KLVEWARPLLANKRKLFRVIDNRLQDQ 317
Query: 810 GSDEQIEEALKEVGYLCTADLPFK---RPTMQQIVGLLKDIE 848
S +EEA K V L L F+ RP M ++V L+ I+
Sbjct: 318 YS---MEEACK-VATLALRCLTFEIKLRPNMNEVVSHLEHIQ 355
>SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase
receptor precursor (EC 2.7.1.37) (S-receptor kinase)
(SRK)
Length = 849
Score = 147 bits (371), Expect = 1e-34
Identities = 119/397 (29%), Positives = 186/397 (45%), Gaps = 58/397 (14%)
Query: 462 LSLIFALAGILFLAFGCRRKNKMWEVKQGSY---REEQNISGPFSFQTDSTTWVADVKQA 518
+SL ++ +L L C K K K + ++N + P + V K+
Sbjct: 448 ISLTVGVSVLLLLIMFCLWKRKQKRAKASAISIANTQRNQNLPMN------EMVLSSKRE 501
Query: 519 TSVPVVIFEKPLLNITFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVVG 578
S E L I ++ AT NF L +G FG VY+G L +AVK L
Sbjct: 502 FSGEYKFEELELPLIEMETVVKATENFSSCNKLGQGGFGIVYKGRLLDGKEIAVKRLSKT 561
Query: 579 STLTDEEAARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQ 638
S +E E+ + R++H NLV + G C+ GD+++ IY+Y+EN +L + L+
Sbjct: 562 SVQGTDEFMNEVTLIARLQHINLVQVLGCCIEGDEKMLIYEYLENLSLDSYLFG------ 615
Query: 639 STDDWSTDTWEEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVK 698
+ W R I G AR L +LH IIHR +K
Sbjct: 616 --------------------KTRRSKLNWNERFDITNGVARGLLYLHQDSRFRIIHRDLK 655
Query: 699 ASSVYLDYDLEPRLSDFGLAKIFGSGLDEEIAR-----GSPGYVPPEFSQPEFESPTPKS 753
S++ LD ++ P++SDFG+A+IF DE A G+ GY+ PE++ + + KS
Sbjct: 656 VSNILLDKNMIPKISDFGMARIFER--DETEANTMKVVGTYGYMSPEYAM--YGIFSEKS 711
Query: 754 DVYCFGVVLFELLTGKKPVGD---DYTDDKEATTLVSWVRGLVRKNQTSRAIDPKICDTG 810
DV+ FGV++ E+++GKK G DY +D L+S+V ++ + +DP I D+
Sbjct: 712 DVFSFGVIVLEIVSGKKNRGFYNLDYEND-----LLSYVWSRWKEGRALEIVDPVIVDSL 766
Query: 811 SDE----QIEEALK--EVGYLCTADLPFKRPTMQQIV 841
S + Q +E LK ++G LC +L RP M +V
Sbjct: 767 SSQPSIFQPQEVLKCIQIGLLCVQELAEHRPAMSSVV 803
>NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK (EC
2.7.1.37)
Length = 389
Score = 147 bits (371), Expect = 1e-34
Identities = 110/334 (32%), Positives = 167/334 (49%), Gaps = 49/334 (14%)
Query: 530 LLNITFADLLSATSNFDRGTLLAEGKFGPVYRGFL---------PGN-IHVAVKVLVVGS 579
L N + ++L SAT NF +++ EG FG V++G++ PG I +AVK L
Sbjct: 53 LKNFSLSELKSATRNFRPDSVVGEGGFGCVFKGWIDESSLAPSKPGTGIVIAVKRLNQEG 112
Query: 580 TLTDEEAARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQS 639
E E+ +LG++ HPNLV L GYC+ + R+ +Y++M G+L+N L+
Sbjct: 113 FQGHREWLAEINYLGQLDHPNLVKLIGYCLEEEHRLLVYEFMTRGSLENHLF-------- 164
Query: 640 TDDWSTDTWEEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKA 699
G+ +W R ++ALG AR LAFLH+ P +I+R KA
Sbjct: 165 -----------------RRGTFYQPLSWNTRVRMALGAARGLAFLHN-AQPQVIYRDFKA 206
Query: 700 SSVYLDYDLEPRLSDFGLAKIFGSGLDEEIA---RGSPGYVPPEFSQPEFESPTPKSDVY 756
S++ LD + +LSDFGLA+ G + ++ G+ GY PE+ S KSDVY
Sbjct: 207 SNILLDSNYNAKLSDFGLARDGPMGDNSHVSTRVMGTQGYAAPEYLATGHLS--VKSDVY 264
Query: 757 CFGVVLFELLTGKKPVGDDYTDDKEATTLVSWVRG-LVRKNQTSRAIDPKICDTGSDEQI 815
FGVVL ELL+G++ + D LV W R L K + R +DP++ +
Sbjct: 265 SFGVVLLELLSGRRAI--DKNQPVGEHNLVDWARPYLTNKRRLLRVMDPRL---QGQYSL 319
Query: 816 EEALK--EVGYLCTADLPFKRPTMQQIVGLLKDI 847
ALK + C + RPTM +IV ++++
Sbjct: 320 TRALKIAVLALDCISIDAKSRPTMNEIVKTMEEL 353
>APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor (EC
2.7.1.-)
Length = 410
Score = 147 bits (371), Expect = 1e-34
Identities = 115/341 (33%), Positives = 168/341 (48%), Gaps = 50/341 (14%)
Query: 525 IFEKP-LLNITFADLLSATSNFDRGTLLAEGKFGPVYRGFL---------PGN-IHVAVK 573
I + P L + +FA+L SAT NF ++L EG FG V++G++ PG + +AVK
Sbjct: 47 ILQSPNLKSFSFAELKSATRNFRPDSVLGEGGFGCVFKGWIDEKSLTASRPGTGLVIAVK 106
Query: 574 VLVVGSTLTDEEAARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDL 633
L +E E+ +LG+ H +LV L GYC+ + R+ +Y++M G+L+N L+
Sbjct: 107 KLNQDGWQGHQEWLAEVNYLGQFSHRHLVKLIGYCLEDEHRLLVYEFMPRGSLENHLFRR 166
Query: 634 PLGVQSTDDWSTDTWEEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPII 693
L Q +W+ R K+ALG A+ LAFL H +I
Sbjct: 167 GLYFQP-------------------------LSWKLRLKVALGAAKGLAFL-HSSETRVI 200
Query: 694 HRAVKASSVYLDYDLEPRLSDFGLAKIFGSGLDEEIA---RGSPGYVPPEFSQPEFESPT 750
+R K S++ LD + +LSDFGLAK G ++ G+ GY PE+ T
Sbjct: 201 YRDFKTSNILLDSEYNAKLSDFGLAKDGPIGDKSHVSTRVMGTHGYAAPEYLAT--GHLT 258
Query: 751 PKSDVYCFGVVLFELLTGKKPVGDDYTDDKEATTLVSWVRG-LVRKNQTSRAIDPKICDT 809
KSDVY FGVVL ELL+G++ V D LV W + LV K + R ID ++ D
Sbjct: 259 TKSDVYSFGVVLLELLSGRRAV--DKNRPSGERNLVEWAKPYLVNKRKIFRVIDNRLQDQ 316
Query: 810 GSDEQIEEALK--EVGYLCTADLPFKRPTMQQIVGLLKDIE 848
S +EEA K + C RP M ++V L+ I+
Sbjct: 317 YS---MEEACKVATLSLRCLTTEIKLRPNMSEVVSHLEHIQ 354
>CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4
precursor (EC 2.7.1.-)
Length = 901
Score = 135 bits (341), Expect = 3e-31
Identities = 93/318 (29%), Positives = 156/318 (48%), Gaps = 38/318 (11%)
Query: 534 TFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVVGSTL--TDEEAARELE 591
++ +L AT F + + +G F V++G L VAVK + S + + +E EL+
Sbjct: 494 SYEELEQATGGFSEDSQVGKGSFSCVFKGILRDGTVVAVKRAIKASDVKKSSKEFHNELD 553
Query: 592 FLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTDTWEEA 651
L R+ H +L+ L GYC G +R+ +Y++M +G+L L+ ++ +W+
Sbjct: 554 LLSRLNHAHLLNLLGYCEDGSERLLVYEFMAHGSLYQHLHGKDPNLKKRLNWAR------ 607
Query: 652 DNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLEPR 711
R IA+ AR + +LH PP+IHR +K+S++ +D D R
Sbjct: 608 ------------------RVTIAVQAARGIEYLHGYACPPVIHRDIKSSNILIDEDHNAR 649
Query: 712 LSDFGLAKIFG---SGLD-EEIARGSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFELLT 767
++DFGL+ I G SG E+ G+ GY+ PE+ + + T KSDVY FGVVL E+L+
Sbjct: 650 VADFGLS-ILGPADSGTPLSELPAGTLGYLDPEYYRLHY--LTTKSDVYSFGVVLLEILS 706
Query: 768 GKKPVGDDYTDDKEATTLVSWVRGLVRKNQTSRAIDPKICDTGSDEQIEEALKEVGYLCT 827
G+K + + E +V W L++ +DP + SD + + + V C
Sbjct: 707 GRKAIDMQF----EEGNIVEWAVPLIKAGDIFAILDP-VLSPPSDLEALKKIASVACKCV 761
Query: 828 ADLPFKRPTMQQIVGLLK 845
RP+M ++ L+
Sbjct: 762 RMRGKDRPSMDKVTTALE 779
>KPEL_DROME (Q05652) Probable serine/threonine-protein kinase pelle
(EC 2.7.1.37)
Length = 501
Score = 134 bits (338), Expect = 8e-31
Identities = 101/330 (30%), Positives = 158/330 (47%), Gaps = 40/330 (12%)
Query: 530 LLNITFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVVGSTLTDE----- 584
LL I +A+L +AT + L +G FG VYRG + VA+KV+ S D+
Sbjct: 198 LLEIDYAELENATDGWSPDNRLGQGGFGDVYRGKWK-QLDVAIKVMNYRSPNIDQKMVEL 256
Query: 585 -EAARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDW 643
++ EL++L I+H N++ L GY + G + +Y M+ G+L+ L
Sbjct: 257 QQSYNELKYLNSIRHDNILALYGYSIKGGKPCLVYQLMKGGSLEARLRA----------- 305
Query: 644 STDTWEEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVY 703
+A N + + TW+ R I+LGTAR + FLH P+IH +K +++
Sbjct: 306 -----HKAQNPLPAL-------TWQQRFSISLGTARGIYFLHTARGTPLIHGDIKPANIL 353
Query: 704 LDYDLEPRLSDFGLAKIFGSGLDEEI----ARGSPGYVPPEFSQPEFESPTPKSDVYCFG 759
LD L+P++ DFGL + LD + G+ Y+PPEF F + DVY FG
Sbjct: 354 LDQCLQPKIGDFGLVREGPKSLDAVVEVNKVFGTKIYLPPEFR--NFRQLSTGVDVYSFG 411
Query: 760 VVLFELLTGKKPVGDDYTDDKEATTLVSWVRGLVRKNQTSRAIDPKICDTGSDEQIEEAL 819
+VL E+ TG++ V D +++ L+ +V+ R+N+ G + +
Sbjct: 412 IVLLEVFTGRQ-VTDRVPENETKKNLLDYVKQQWRQNRMELLEKHLAAPMGKELDMCMCA 470
Query: 820 KEVGYLCTADLPFKRPTMQQIVGLLKDIEP 849
E G CTA P RP+M + LK EP
Sbjct: 471 IEAGLHCTALDPQDRPSMNAV---LKRFEP 497
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.137 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 101,783,962
Number of Sequences: 164201
Number of extensions: 4493177
Number of successful extensions: 16918
Number of sequences better than 10.0: 1608
Number of HSP's better than 10.0 without gapping: 432
Number of HSP's successfully gapped in prelim test: 1176
Number of HSP's that attempted gapping in prelim test: 11826
Number of HSP's gapped (non-prelim): 3429
length of query: 852
length of database: 59,974,054
effective HSP length: 119
effective length of query: 733
effective length of database: 40,434,135
effective search space: 29638220955
effective search space used: 29638220955
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 70 (31.6 bits)
Medicago: description of AC122160.2


