

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121239.8 + phase: 0
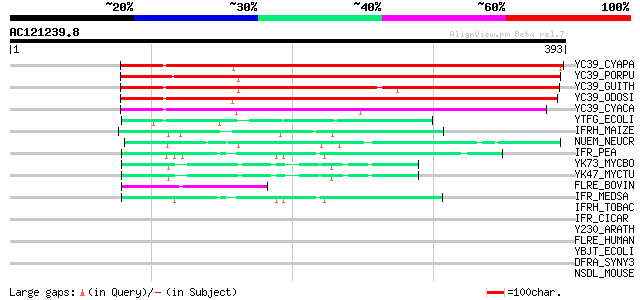
(393 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
YC39_CYAPA (P48279) Hypothetical 36.4 kDa protein ycf39 323 5e-88
YC39_PORPU (P51238) Hypothetical 35.7 kDa protein ycf39 (ORF319) 319 7e-87
YC39_GUITH (O78472) Hypothetical 35.9 kDa protein ycf39 294 3e-79
YC39_ODOSI (P49534) Hypothetical 36.3 kDa protein ycf39 (ORF319) 272 9e-73
YC39_CYACA (O19883) Hypothetical 35.4 kDa protein ycf39 178 2e-44
YTFG_ECOLI (P39315) Hypothetical protein ytfG 52 2e-06
IFRH_MAIZE (P52580) Isoflavone reductase homolog IRL (EC 1.3.1.-) 51 6e-06
NUEM_NEUCR (P25284) NADH-ubiquinone oxidoreductase 40 kDa subuni... 49 3e-05
IFR_PEA (P52576) Isoflavone reductase (EC 1.3.1.45) (IFR) (2'-hy... 47 1e-04
YK73_MYCBO (P65685) Hypothetical protein Mb2073c 46 1e-04
YK47_MYCTU (P65684) Hypothetical protein Rv2047c/MT2107 46 1e-04
FLRE_BOVIN (P52556) Flavin reductase (EC 1.5.1.30) (FR) (NADPH-d... 44 5e-04
IFR_MEDSA (P52575) Isoflavone reductase (EC 1.3.1.45) (IFR) (2'-... 44 7e-04
IFRH_TOBAC (P52579) Isoflavone reductase homolog A622 (EC 1.3.1.-) 44 0.001
IFR_CICAR (Q00016) Isoflavone reductase (EC 1.3.1.45) (IFR) (2'-... 42 0.004
Y230_ARATH (O80934) Protein At2g37660, chloroplast precursor 40 0.008
FLRE_HUMAN (P30043) Flavin reductase (EC 1.5.1.30) (FR) (NADPH-d... 40 0.010
YBJT_ECOLI (P75822) Hypothetical protein ybjT 39 0.018
DFRA_SYNY3 (P73212) Putative dihydroflavonol-4-reductase (EC 1.1... 38 0.039
NSDL_MOUSE (Q9R1J0) NAD(P)-dependent steroid dehydrogenase (EC 1... 38 0.051
>YC39_CYAPA (P48279) Hypothetical 36.4 kDa protein ycf39
Length = 321
Score = 323 bits (828), Expect = 5e-88
Identities = 164/320 (51%), Positives = 224/320 (69%), Gaps = 7/320 (2%)
Query: 79 SILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRDWGATVVNADLSKPETIPA 138
SILV+GATGTLGRQIVR ALDEGY VRCLVR A FL++WGA ++ DLS+PE++
Sbjct: 2 SILVIGATGTLGRQIVRSALDEGYQVRCLVR-NLRKAAFLKEWGAKLIWGDLSQPESLLP 60
Query: 139 TLVGVHTVIDCATGRPEEP--IKTVDWEGKVALIQCAKAMGIQKYVFYSIHNCDKHPEVP 196
L G+ +ID +T RP +P + VD +GK ALI AKAM I+K++F+SI N +K+ +VP
Sbjct: 61 ALTGIRVIIDTSTSRPTDPAGVYQVDLKGKKALIDAAKAMKIEKFIFFSILNSEKYSQVP 120
Query: 197 LMEIKYCTENFLRDSGLNHIVIRLCGFMQGLIGQYAVPILEEKSVWGTDAPTRIAYMDTQ 256
LM IK TE L++SGLN+ + +LCGF QGLIGQYAVPIL++++VW T T IAYMDT
Sbjct: 121 LMRIKTVTEELLKESGLNYTIFKLCGFFQGLIGQYAVPILDQQTVWITTESTSIAYMDTI 180
Query: 257 DIARLTFIALRNEKINGKLLTFAGPRAWTTQEVITLCERLAGQDANVTTVPVSVLRLTRQ 316
DIAR T +L ++ N ++ G R+W + ++I LCERL+GQ+A VT VP++ L L R
Sbjct: 181 DIARFTLRSLVLKETNNRVFPLVGTRSWNSADIIQLCERLSGQNAKVTRVPIAFLELARN 240
Query: 317 LTRFFEWTNDVADRLAFSEVLTSDTVFSVPMAETYNLLGVDTKDIITLEKYLQDYFTNIL 376
+ FFEW ++ADRLAF+EVL+ F+ M E Y + +++ LE YLQ+YF+ IL
Sbjct: 241 TSCFFEWGWNIADRLAFTEVLSKSQFFNSSMDEVYKIFKIESTSTTILESYLQEYFSRIL 300
Query: 377 KKLKDLKAQSKQ----SDIF 392
K+LK++ +Q Q SD+F
Sbjct: 301 KRLKEINSQQSQKKKTSDLF 320
>YC39_PORPU (P51238) Hypothetical 35.7 kDa protein ycf39 (ORF319)
Length = 319
Score = 319 bits (818), Expect = 7e-87
Identities = 155/314 (49%), Positives = 222/314 (70%), Gaps = 3/314 (0%)
Query: 79 SILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRDWGATVVNADLSKPETIPA 138
++LV+GATGTLGRQIVRRALDEGY+V+C+VR A FL++WGA +V DL PE+I
Sbjct: 2 TLLVIGATGTLGRQIVRRALDEGYNVKCMVRNLRKSA-FLKEWGAELVYGDLKLPESILQ 60
Query: 139 TLVGVHTVIDCATGRPEEPIKT--VDWEGKVALIQCAKAMGIQKYVFYSIHNCDKHPEVP 196
+ GV VID +T RP +P T +D +GK+ALI+ AKA +Q+++F+SI N D++P+VP
Sbjct: 61 SFCGVTAVIDASTSRPSDPYNTEQIDLDGKIALIEAAKAAKVQRFIFFSILNADQYPKVP 120
Query: 197 LMEIKYCTENFLRDSGLNHIVIRLCGFMQGLIGQYAVPILEEKSVWGTDAPTRIAYMDTQ 256
LM +K N+L+ S +++ V L GF QGLI QYA+PIL++KSVW T T IAY+DTQ
Sbjct: 121 LMNLKSQVVNYLQKSSISYTVFSLGGFFQGLISQYAIPILDKKSVWVTGESTPIAYIDTQ 180
Query: 257 DIARLTFIALRNEKINGKLLTFAGPRAWTTQEVITLCERLAGQDANVTTVPVSVLRLTRQ 316
D A+L +L ++L G +AWT+ E+ITLCE+L+GQ ++ +P+S+L+ R+
Sbjct: 181 DAAKLVIKSLGVPSTENRILPLVGNKAWTSAEIITLCEKLSGQKTQISQIPLSLLKALRK 240
Query: 317 LTRFFEWTNDVADRLAFSEVLTSDTVFSVPMAETYNLLGVDTKDIITLEKYLQDYFTNIL 376
+T+ +WT +++DRLAF+EVLTS +F PM E Y +L +D ++I+LEKY Q+YF IL
Sbjct: 241 ITKTLQWTWNISDRLAFAEVLTSGEIFMAPMNEVYEILSIDKSEVISLEKYFQEYFGKIL 300
Query: 377 KKLKDLKAQSKQSD 390
K LKD+ + Q D
Sbjct: 301 KVLKDISYEQSQQD 314
>YC39_GUITH (O78472) Hypothetical 35.9 kDa protein ycf39
Length = 314
Score = 294 bits (752), Expect = 3e-79
Identities = 146/316 (46%), Positives = 217/316 (68%), Gaps = 9/316 (2%)
Query: 79 SILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRDWGATVVNADLSKPETIPA 138
S+LV+GATGTLGRQIVRRALDEGY+V CLVR A FL++WGA ++ DLS PET+P
Sbjct: 2 SLLVIGATGTLGRQIVRRALDEGYEVSCLVR-NLRKAYFLKEWGAELLYGDLSLPETLPT 60
Query: 139 TLVGVHTVIDCATGRPEEPIKT--VDWEGKVALIQCAKAMGIQKYVFYSIHNCDKHPEVP 196
L + +ID +T RP +P K +D EGK+AL++ AK GI+++VF+S+ N + +P
Sbjct: 61 NLTKITAIIDASTARPSDPYKAEKIDLEGKIALVEAAKVAGIKRFVFFSVLNAQNYRHLP 120
Query: 197 LMEIKYCTENFLRDSGLNHIVIRLCGFMQGLIGQYAVPILEEKSVWGTDAPTRIAYMDTQ 256
L+ +K E +L+ S L + +L GF QGLI QYA+PILE++++W T T+I Y+DT
Sbjct: 121 LVNLKCRMEEYLQTSELEYTTFQLSGFFQGLISQYAIPILEKQTIWITGEYTKINYIDTN 180
Query: 257 DIARLTFIALRNEKING---KLLTFAGPRAWTTQEVITLCERLAGQDANVTTVPVSVLRL 313
DIA+ A+R+ +NG + + G ++W ++E+I LCERL+GQ AN+T +P+ +L
Sbjct: 181 DIAK---FAVRSLSLNGTIKRTIPLVGLKSWNSEEIIQLCERLSGQKANITKIPLQLLVF 237
Query: 314 TRQLTRFFEWTNDVADRLAFSEVLTSDTVFSVPMAETYNLLGVDTKDIITLEKYLQDYFT 373
+ LT FFEWT ++++RLAF+EVL + + MA TY+ G + DI +LE Y+QDYF
Sbjct: 238 FKYLTGFFEWTTNISERLAFAEVLNARQDITSEMASTYSQFGFEPNDITSLESYMQDYFG 297
Query: 374 NILKKLKDLKAQSKQS 389
IL++LK+L + +++
Sbjct: 298 RILRRLKELSDERERT 313
>YC39_ODOSI (P49534) Hypothetical 36.3 kDa protein ycf39 (ORF319)
Length = 319
Score = 272 bits (696), Expect = 9e-73
Identities = 135/312 (43%), Positives = 200/312 (63%), Gaps = 3/312 (0%)
Query: 79 SILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRDWGATVVNADLSKPETIPA 138
S+L++G TGTLGRQ+V +AL +GY VRCLVR A+FL++WGA ++ DLS+PETIP
Sbjct: 2 SLLIIGGTGTLGRQVVLQALTKGYQVRCLVR-NFRKANFLKEWGAELIYGDLSRPETIPP 60
Query: 139 TLVGVHTVIDCATGRPEE--PIKTVDWEGKVALIQCAKAMGIQKYVFYSIHNCDKHPEVP 196
L G+ VID +T RP + +K VDW+GK ALI+ A+A ++ +VF S N ++ +P
Sbjct: 61 CLQGITAVIDTSTSRPSDLDTLKQVDWDGKCALIEAAQAANVKHFVFCSSQNVEQFLNIP 120
Query: 197 LMEIKYCTENFLRDSGLNHIVIRLCGFMQGLIGQYAVPILEEKSVWGTDAPTRIAYMDTQ 256
LME+K+ E L+ S + + V RL GF QGLI QYA+P+LE + T+ T ++YMDTQ
Sbjct: 121 LMEMKFGIETKLQQSNIPYTVFRLAGFYQGLIEQYAIPVLENLPILVTNENTCVSYMDTQ 180
Query: 257 DIARLTFIALRNEKINGKLLTFAGPRAWTTQEVITLCERLAGQDANVTTVPVSVLRLTRQ 316
DIA+ +L+ + + G + W + E+I LCE+LAGQ A V +P+ +L+L Q
Sbjct: 181 DIAKFCLRSLQLPETKNRTFVLGGQKGWVSSEIINLCEQLAGQSAKVNKIPLFLLKLVSQ 240
Query: 317 LTRFFEWTNDVADRLAFSEVLTSDTVFSVPMAETYNLLGVDTKDIITLEKYLQDYFTNIL 376
+ FFEW +++DRLAF+E+L + FS + Y VD +I L+ Y +YF +L
Sbjct: 241 IFGFFEWGQNISDRLAFAEILNVENDFSKSTFDLYKTFKVDDSEITQLDDYFLEYFIRLL 300
Query: 377 KKLKDLKAQSKQ 388
K+L+D+ + Q
Sbjct: 301 KRLRDINFEDIQ 312
>YC39_CYACA (O19883) Hypothetical 35.4 kDa protein ycf39
Length = 312
Score = 178 bits (452), Expect = 2e-44
Identities = 109/306 (35%), Positives = 167/306 (53%), Gaps = 5/306 (1%)
Query: 79 SILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRDWGATVVNADLSKPETIPA 138
S+LV+GAT TLGRQIV++AL +GY+V+CLVR A FL+ WGA +V DL PET+P
Sbjct: 2 SLLVIGATSTLGRQIVKKALIQGYEVKCLVR-NSKKAAFLKAWGAILVYGDLMVPETLPQ 60
Query: 139 TLVGVHTVIDCATGRPEEPIK--TVDWEGKVALIQCAKAMGIQKYVFYSIHNCDKHPEVP 196
VG +ID +T + ++ TVD K A+++ A ++K+V +S+ N ++ +VP
Sbjct: 61 CFVGASVIIDVSTVKVKDLNNDYTVDIYCKRAVLEAAIQAKVKKFVSFSMFNSSQYLDVP 120
Query: 197 LMEIKYCTENFLRDSGLNHIVIRLCGFMQGLIGQYAVPILEEKSVWGTDAP--TRIAYMD 254
+IK + L SG+N+++ GF Q L QYA PIL + V+ + +I+Y+D
Sbjct: 121 STKIKSDFDRALIKSGINYLIFTPLGFFQDLTSQYAAPILSSQPVFILEQSESVQISYID 180
Query: 255 TQDIARLTFIALRNEKINGKLLTFAGPRAWTTQEVITLCERLAGQDANVTTVPVSVLRLT 314
+D A + A + G R+W +I+LC+ AGQ + V TVP+ ++
Sbjct: 181 ARDAANIVLAATSLAFLKNIDFPLIGNRSWNNVSIISLCQLYAGQASPVVTVPLLLVYFI 240
Query: 315 RQLTRFFEWTNDVADRLAFSEVLTSDTVFSVPMAETYNLLGVDTKDIITLEKYLQDYFTN 374
R++ FF +T L E L+ + S L + K + +LEKY Q YFT+
Sbjct: 241 RRILGFFAFTRQFYFYLTLVENLSRSSYLSFNHENENKFLKIYQKKLFSLEKYFQYYFTH 300
Query: 375 ILKKLK 380
L LK
Sbjct: 301 KLGNLK 306
>YTFG_ECOLI (P39315) Hypothetical protein ytfG
Length = 286
Score = 52.4 bits (124), Expect = 2e-06
Identities = 55/225 (24%), Positives = 89/225 (39%), Gaps = 16/225 (7%)
Query: 80 ILVVGATGTLGRQIVRRALDE--GYDVRCLVRPRPAPADFLRDWGATVVNADLSKPETIP 137
I + GATG LG ++ + + +VR PA A L G TV AD +
Sbjct: 2 IAITGATGQLGHYVIESLMKTVPASQIVAIVR-NPAKAQALAAQGITVRQADYGDEAALT 60
Query: 138 ATLVGVHTVI---DCATGRPEEPIKTVDWEGKVALIQCAKAMGIQKYVFYSIHNCDKHPE 194
+ L GV ++ G+ + V I AKA G++ + S+ + D P
Sbjct: 61 SALQGVEKLLLISSSEVGQRAPQHRNV--------INAAKAAGVKFIAYTSLLHADTSP- 111
Query: 195 VPLMEIKYCTENFLRDSGLNHIVIRLCGFMQGLIGQYAVPILEEKSVWGTDAPTRIAYMD 254
+ L + TE L DSG+ + ++R + + + A LE G +IA
Sbjct: 112 LGLADEHIETEKMLADSGIVYTLLRNGWYSENYLAS-APAALEHGVFIGAAGDGKIASAT 170
Query: 255 TQDIARLTFIALRNEKINGKLLTFAGPRAWTTQEVITLCERLAGQ 299
D A + GK+ AG AWT ++ + +G+
Sbjct: 171 RADYAAAAARVISEAGHEGKVYELAGDSAWTLTQLAAELTKQSGK 215
>IFRH_MAIZE (P52580) Isoflavone reductase homolog IRL (EC 1.3.1.-)
Length = 309
Score = 50.8 bits (120), Expect = 6e-06
Identities = 61/249 (24%), Positives = 100/249 (39%), Gaps = 28/249 (11%)
Query: 78 TSILVVGATGTLGRQIVRRALDEGYDVRCLVRPR----PAPADFLR---DWGATVVNADL 130
+ ILVVG TG LGR +V + G+ LVR PA A L+ D G T++ DL
Sbjct: 6 SKILVVGGTGYLGRHVVAASARLGHPTSALVRDTAPSDPAKAALLKSFQDAGVTLLKGDL 65
Query: 131 SKPETIPATLVGVHTVIDCATGRPEEPIKTVDWEGKVALIQCAKAMGIQKYVFYSIHNCD 190
++ + + G VI + ++ + L+ K G K F S D
Sbjct: 66 YDQASLVSAVKGADVVISV--------LGSMQIADQSRLVDAIKEAGNVKRFFPSEFGLD 117
Query: 191 ------KHPEVPLMEIKYCTENFLRDSGLNHIVIRLCGFMQGL----IGQYAVP-ILEEK 239
P ++ K +G+ + + GF G +GQ P +K
Sbjct: 118 VDRTGIVEPAKSILGAKVGIRRATEAAGIPY-TYAVAGFFAGFGLPKVGQVLAPGPPADK 176
Query: 240 SVWGTDAPTRIAYMDTQDIARLTFIALRNEKINGKLLTFAGP-RAWTTQEVITLCERLAG 298
+V D T+ +++ DIA T +A + + K+L P + E+++L E+ G
Sbjct: 177 AVVLGDGDTKAVFVEEGDIATYTVLAADDPRAENKVLYIKPPANTLSHNELLSLWEKKTG 236
Query: 299 QDANVTTVP 307
+ VP
Sbjct: 237 KTFRREYVP 245
>NUEM_NEUCR (P25284) NADH-ubiquinone oxidoreductase 40 kDa subunit,
mitochondrial precursor (EC 1.6.5.3) (EC 1.6.99.3)
(Complex I-40KD) (CI-40KD)
Length = 375
Score = 48.5 bits (114), Expect = 3e-05
Identities = 76/324 (23%), Positives = 123/324 (37%), Gaps = 26/324 (8%)
Query: 82 VVGATGTLGRQIVRRALDEGYDVRCLVRP--RPAPADFLRDWGATV-VNADLSKPETIPA 138
V GATG LGR IV R +G V R D G V + DL ++I
Sbjct: 56 VFGATGQLGRYIVNRLARQGCTVVIPFRDEYNKRHLKVTGDLGKVVMIEFDLRNTQSIEE 115
Query: 139 TLVGVHTVIDCATGRPEEPIKT-----VDWEGKVALIQCAKAMGIQKYVFYSIHNCDKHP 193
+ V V+ GR + P K V EG + + + +++ S +N D +
Sbjct: 116 S-VRHSDVVYNLIGR-DYPTKNFSFEDVHIEGAERIAEAVAKYDVDRFIHVSSYNADPNS 173
Query: 194 EVPLMEIKYCTENFLRDSGLNHIVIR---LCGFMQGLIGQYA--VPILEEKSVWGTDAPT 248
E K E +R ++R + GF L+ + A IL + P
Sbjct: 174 ECEFFATKARGEQVVRSIFPETTIVRPAPMFGFEDRLLHKLASVKNILTSNGMQEKYNPV 233
Query: 249 RIAYMDTQDIARLTFIALRNEKINGKLLTFAGPRAWTTQEVITLCERLAGQDANVTTVPV 308
+ D+ + L ++ + GP+ +TT E+ + +R + VP
Sbjct: 234 HVI-----DVGQALEQMLWDDNTASETFELYGPKTYTTAEISEMVDREIYKRRRHVNVPK 288
Query: 309 SVLR-LTRQLTRFFEWTNDVADRLAFSEVLTSDTVFSVPMAETYNLLGVDTKDIITLE-K 366
+L+ + L + W AD + E D V P A+T+ LG++ DI
Sbjct: 289 KILKPIAGVLNKALWWPIMSADEI---EREFHDQVID-PEAKTFKDLGIEPADIANFTYH 344
Query: 367 YLQDYFTNILKKLKDLKAQSKQSD 390
YLQ Y +N L + ++ D
Sbjct: 345 YLQSYRSNAYYDLPPATEKERRED 368
>IFR_PEA (P52576) Isoflavone reductase (EC 1.3.1.45) (IFR)
(2'-hydroxyisoflavone reductase) (NADPH:isoflavone
oxidoreductase)
Length = 318
Score = 46.6 bits (109), Expect = 1e-04
Identities = 75/301 (24%), Positives = 120/301 (38%), Gaps = 44/301 (14%)
Query: 80 ILVVGATGTLGRQIVRRALDEGYDVRCLVR--------PRPAPA-------DFLRDW--- 121
IL++GATG +GR IV ++ G LVR P+ A + L+++
Sbjct: 7 ILILGATGAIGRHIVWASIKAGNPTYALVRKTSDNVNKPKLTEAANPETKEELLKNYQAS 66
Query: 122 GATVVNADLSKPETIPATLVGVHTVIDCATGRPEEPIKTVDWEGKVALIQCAKAMGIQKY 181
G ++ D++ ET+ + V TVI CA GR + E +V +I+ K G K
Sbjct: 67 GVILLEGDINDHETLVNAIKQVDTVI-CAAGR-------LLIEDQVKVIKAIKEAGNVKR 118
Query: 182 VFYSIH--NCDKH----PEVPLMEIKYCTENFLRDSGLNHIVIRLC------GFMQGLIG 229
F S + D+H P + E K + G+ + LC F++ L
Sbjct: 119 FFPSEFGLDVDRHDAVEPVRQVFEEKASIRRVVESEGVPYTY--LCCHAFTGYFLRNLAQ 176
Query: 230 QYAVPILEEKSVWGTDAPTRIAYMDTQDIARLTFIALRNEKINGKLLTFAGPRAW-TTQE 288
A +K V D R AY+ D+ T A + K + P + T E
Sbjct: 177 IDATDPPRDKVVILGDGNVRGAYVTEADVGTYTIRAANDPNTLNKAVHIRLPNNYLTANE 236
Query: 289 VITLCERLAGQDANVTTVPVSVLRLTRQLTRFFEWTNDVADRLAFSEVLTSDTVFSVPMA 348
VI L E+ G+ T V + Q + F ++ L S+ + D V+ + A
Sbjct: 237 VIALWEKKIGKTLEKTYVSEEQVLKDIQTSSF---PHNYLLALYHSQQIKGDAVYEIDPA 293
Query: 349 E 349
+
Sbjct: 294 K 294
>YK73_MYCBO (P65685) Hypothetical protein Mb2073c
Length = 854
Score = 46.2 bits (108), Expect = 1e-04
Identities = 55/220 (25%), Positives = 87/220 (39%), Gaps = 38/220 (17%)
Query: 80 ILVVGATGTLGRQIVRRALDEGYDVRCLVRPR----PAPADFLRDWGATVVNADLSKPET 135
I V GA+G LGR + R L +G++V + R R P+ ADF + AD+
Sbjct: 3 IAVTGASGVLGRGLTARLLSQGHEVVGIARHRPDSWPSSADF--------IAADIRDATA 54
Query: 136 IPATLVGVHTVIDCATGRPEEPIKTVDWEGKVALIQCAKAMGIQKYVFYSIHNCDKHPEV 195
+ + + G V CA R +D G +++ G + VF S P V
Sbjct: 55 VESAMTGADVVAHCAWVRGRNDHINID--GTANVLKAMAETGTGRIVFTS---SGHQPRV 109
Query: 196 PLMEIKYCTENFLRDSGLNHIVIRLCGFMQG------LIGQYAVPILEEKSVWGTDAPTR 249
E L D GL + +R C + G + +A+P+L D +
Sbjct: 110 ---------EQMLADCGLEWVAVR-CALIFGRNVDNWVQRLFALPVL---PAGYADRVVQ 156
Query: 250 IAYMDTQDIARLTFIALRNEKINGKLLTFAGPRAWTTQEV 289
+ + D D RL AL + I+ + A P T + +
Sbjct: 157 VVHSD--DAQRLLVRALLDTVIDSGPVNLAAPGELTFRRI 194
>YK47_MYCTU (P65684) Hypothetical protein Rv2047c/MT2107
Length = 854
Score = 46.2 bits (108), Expect = 1e-04
Identities = 55/220 (25%), Positives = 87/220 (39%), Gaps = 38/220 (17%)
Query: 80 ILVVGATGTLGRQIVRRALDEGYDVRCLVRPR----PAPADFLRDWGATVVNADLSKPET 135
I V GA+G LGR + R L +G++V + R R P+ ADF + AD+
Sbjct: 3 IAVTGASGVLGRGLTARLLSQGHEVVGIARHRPDSWPSSADF--------IAADIRDATA 54
Query: 136 IPATLVGVHTVIDCATGRPEEPIKTVDWEGKVALIQCAKAMGIQKYVFYSIHNCDKHPEV 195
+ + + G V CA R +D G +++ G + VF S P V
Sbjct: 55 VESAMTGADVVAHCAWVRGRNDHINID--GTANVLKAMAETGTGRIVFTS---SGHQPRV 109
Query: 196 PLMEIKYCTENFLRDSGLNHIVIRLCGFMQG------LIGQYAVPILEEKSVWGTDAPTR 249
E L D GL + +R C + G + +A+P+L D +
Sbjct: 110 ---------EQMLADCGLEWVAVR-CALIFGRNVDNWVQRLFALPVL---PAGYADRVVQ 156
Query: 250 IAYMDTQDIARLTFIALRNEKINGKLLTFAGPRAWTTQEV 289
+ + D D RL AL + I+ + A P T + +
Sbjct: 157 VVHSD--DAQRLLVRALLDTVIDSGPVNLAAPGELTFRRI 194
>FLRE_BOVIN (P52556) Flavin reductase (EC 1.5.1.30) (FR)
(NADPH-dependent diaphorase) (NADPH-flavin reductase)
(FLR) (Biliverdin reductase B) (EC 1.3.1.24) (BVR-B)
(Biliverdin-IX beta-reductase) (Green heme binding
protein) (GHBP)
Length = 205
Score = 44.3 bits (103), Expect = 5e-04
Identities = 34/104 (32%), Positives = 50/104 (47%), Gaps = 3/104 (2%)
Query: 80 ILVVGATGTLGRQIVRRALDEGYDVRCLVR-PRPAPADFLRDWGATVVNADLSKPETIPA 138
I + GATG G + +A+ GY+V LVR P P++ + A VV D+ +P +
Sbjct: 5 IALFGATGNTGLTTLAQAVQAGYEVTVLVRDPSRLPSEGPQP--AHVVVGDVRQPADVDK 62
Query: 139 TLVGVHTVIDCATGRPEEPIKTVDWEGKVALIQCAKAMGIQKYV 182
T+ G VI R + TV EG ++ KA G+ K V
Sbjct: 63 TVAGQDAVIVLLGTRNDLSPTTVMSEGAQNIVAAMKAHGVDKVV 106
>IFR_MEDSA (P52575) Isoflavone reductase (EC 1.3.1.45) (IFR)
(2'-hydroxyisoflavone reductase) (NADPH:isoflavone
oxidoreductase)
Length = 318
Score = 43.9 bits (102), Expect = 7e-04
Identities = 64/258 (24%), Positives = 97/258 (36%), Gaps = 41/258 (15%)
Query: 80 ILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPA------------------DFLRDW 121
IL++G TG +GR IV ++ G LVR P D +
Sbjct: 7 ILILGPTGAIGRHIVWASIKAGNPTYALVRKTPGNVNKPKLITAANPETKEELIDNYQSL 66
Query: 122 GATVVNADLSKPETIPATLVGVHTVIDCATGRPEEPIKTVDWEGKVALIQCAKAMGIQKY 181
G ++ D++ ET+ + V VI CA GR + E +V +I+ K G K
Sbjct: 67 GVILLEGDINDHETLVKAIKQVDIVI-CAAGR-------LLIEDQVKIIKAIKEAGNVKK 118
Query: 182 VFYSIH--NCDKH----PEVPLMEIKYCTENFLRDSGLNHIVIRLC------GFMQGLIG 229
F S + D+H P + E K + G+ + LC F++ L
Sbjct: 119 FFPSEFGLDVDRHEAVEPVRQVFEEKASIRRVIEAEGVPYTY--LCCHAFTGYFLRNLAQ 176
Query: 230 QYAVPILEEKSVWGTDAPTRIAYMDTQDIARLTFIALRNEKINGKLLTFAGPRAWTTQ-E 288
+K V D + AY+ D+ T A + K + P + TQ E
Sbjct: 177 LDTTDPPRDKVVILGDGNVKGAYVTEADVGTFTIRAANDPNTLNKAVHIRLPENYLTQNE 236
Query: 289 VITLCERLAGQDANVTTV 306
VI L E+ G+ T V
Sbjct: 237 VIALWEKKIGKTLEKTYV 254
>IFRH_TOBAC (P52579) Isoflavone reductase homolog A622 (EC 1.3.1.-)
Length = 310
Score = 43.5 bits (101), Expect = 0.001
Identities = 52/242 (21%), Positives = 100/242 (40%), Gaps = 30/242 (12%)
Query: 78 TSILVVGATGTLGRQIVRRALDEGYDVRCLVR-------PRPAPADFLRDWGATVVNADL 130
+ IL++G TG +G+ +V + G+ L+R + D + +G T++ D+
Sbjct: 7 SKILIIGGTGYIGKYLVETSAKSGHPTFALIRESTLKNPEKSKLIDTFKSYGVTLLFGDI 66
Query: 131 SKPETIPATLVGVHTVIDCATGRPEEPIKTVDWEGKVALIQCAKAMG-IQKYV-----FY 184
S E++ + V VI G+ + +V +I+ K G I++++ F
Sbjct: 67 SNQESLLKAIKQVDVVISTVGGQ--------QFTDQVNIIKAIKEAGNIKRFLPSEFGFD 118
Query: 185 SIHNCDKHPEVPLMEIKYCTENFLRDSGLNHIVIRLCG----FMQGLIGQY--AVPILEE 238
H P L +K + G+ + + +C F +GQ P ++
Sbjct: 119 VDHARAIEPAASLFALKVRIRRMIEAEGIPYTYV-ICNWFADFFLPNLGQLEAKTPPRDK 177
Query: 239 KSVWGTDAPTRIAYMDTQDIARLTFIALRNEKINGKLLTFAGP-RAWTTQEVITLCERLA 297
++G P I Y+ +DIA T A+ + + K L P + E+++L E
Sbjct: 178 VVIFGDGNPKAI-YVKEEDIATYTIEAVDDPRTLNKTLHMRPPANILSFNEIVSLWEDKI 236
Query: 298 GQ 299
G+
Sbjct: 237 GK 238
>IFR_CICAR (Q00016) Isoflavone reductase (EC 1.3.1.45) (IFR)
(2'-hydroxyisoflavone reductase) (NADPH:isoflavone
oxidoreductase)
Length = 318
Score = 41.6 bits (96), Expect = 0.004
Identities = 62/275 (22%), Positives = 110/275 (39%), Gaps = 51/275 (18%)
Query: 80 ILVVGATGTLGRQIVRRALDEGYDVRCLVRPRP---------------APADFLRDW--- 121
ILV+G TG +GR +V ++ G L+R P + + L+ +
Sbjct: 7 ILVLGPTGAIGRHVVWASIKAGNPTYALIRKTPGDINKPSLVAAANPESKEELLQSFKAA 66
Query: 122 GATVVNADLSKPETIPATLVGVHTVIDCATGRPEEPIKTVDWEGKVALIQCAKAMGIQKY 181
G ++ D++ E + + V TVI C GR + +D +V +I+ K G K
Sbjct: 67 GVILLEGDMNDHEALVKAIKQVDTVI-CTFGR----LLILD---QVKIIKAIKEAGNVKR 118
Query: 182 VFYSIH--NCDKH----PEVPLMEIKYCTENFLRDSGLNHIVI---RLCGFMQGLIGQY- 231
F S + D+H P P+ + K + G+ + + G+ + Q+
Sbjct: 119 FFPSEFGLDVDRHDAVDPVRPVFDEKASIRRVVEAEGVPYTYLCCHAFTGYFLRNLAQFD 178
Query: 232 AVPILEEKSVWGTDAPTRIAYMDTQDIARLTFIALRNEKINGKLLTFAGPRAW-TTQEVI 290
A +K + D + AY+ D+ T A + + K + P + T+ EV+
Sbjct: 179 ATEPPRDKVIILGDGNVKGAYVTEADVGTYTIRAANDPRTLNKAVHIRLPHNYLTSNEVV 238
Query: 291 TLCERLAG--------------QDANVTTVPVSVL 311
+L E+ G +D NV+T P + L
Sbjct: 239 SLWEKKIGKTLEKSYISEEKVLKDINVSTFPHNYL 273
>Y230_ARATH (O80934) Protein At2g37660, chloroplast precursor
Length = 325
Score = 40.4 bits (93), Expect = 0.008
Identities = 52/247 (21%), Positives = 96/247 (38%), Gaps = 35/247 (14%)
Query: 62 NSERAVNLGPGTPVRPTSILVVGATGTLGRQIVRRALD--EGYDVRCLVRPRPAPADFLR 119
N +V + P ++LV GA G G+ + ++ + E + R LVR + +
Sbjct: 61 NRRVSVTVSAAATTEPLTVLVTGAGGRTGQIVYKKLKERSEQFVARGLVRTKESKEKING 120
Query: 120 DWGATVVNADLSKPETIPATLVGVHTVIDCATGRPE-----EPIK--------------- 159
+ V D+ +I + G+ ++ + P+ +P K
Sbjct: 121 E--DEVFIGDIRDTASIAPAVEGIDALVILTSAVPQMKPGFDPSKGGRPEFFFDDGAYPE 178
Query: 160 TVDWEGKVALIQCAKAMGIQKYVFYSI-------HNCDKHPEVPLMEIKYCTENFLRDSG 212
VDW G+ I AKA G+++ V H + ++ K E +L DSG
Sbjct: 179 QVDWIGQKNQIDAAKAAGVKQIVLVGSMGGTNINHPLNSIGNANILVWKRKAEQYLADSG 238
Query: 213 LNHIVIRLCGFMQGLIGQYAVPILEEKSVWGTDAPTRIAYMDTQDIARLTFIALRNEKIN 272
+ + +IR G G + + ++ + T+ T + D+A + AL+ E+
Sbjct: 239 IPYTIIRAGGLQDKDGGIRELLVGKDDELLETETRT----IARADVAEVCVQALQLEEAK 294
Query: 273 GKLLTFA 279
K L A
Sbjct: 295 FKALDLA 301
>FLRE_HUMAN (P30043) Flavin reductase (EC 1.5.1.30) (FR)
(NADPH-dependent diaphorase) (NADPH-flavin reductase)
(FLR) (Biliverdin reductase B) (EC 1.3.1.24) (BVR-B)
(Biliverdin-IX beta-reductase) (Green heme binding
protein) (GHBP)
Length = 205
Score = 40.0 bits (92), Expect = 0.010
Identities = 40/146 (27%), Positives = 64/146 (43%), Gaps = 9/146 (6%)
Query: 80 ILVVGATGTLGRQIVRRALDEGYDVRCLVRPRP-APADFLRDWGATVVNADLSKPETIPA 138
I + GATG G + +A+ GY+V LVR P++ R A VV D+ + +
Sbjct: 5 IAIFGATGQTGLTTLAQAVQAGYEVTVLVRDSSRLPSEGPRP--AHVVVGDVLQAADVDK 62
Query: 139 TLVGVHTVIDCATGRPEEPIKTVDWEGKVALIQCAKAMGIQKYVFYSIHNCDKHPEVPLM 198
T+ G VI R + TV EG ++ KA G+ K V + P
Sbjct: 63 TVAGQDAVIVLLGTRNDLSPTTVMSEGARNIVAAMKAHGVDKVVACTSAFLLWDPTKVPP 122
Query: 199 EIKYCTEN------FLRDSGLNHIVI 218
++ T++ LR+SGL ++ +
Sbjct: 123 RLQAVTDDHIRMHKVLRESGLKYVAV 148
>YBJT_ECOLI (P75822) Hypothetical protein ybjT
Length = 476
Score = 39.3 bits (90), Expect = 0.018
Identities = 34/139 (24%), Positives = 62/139 (44%), Gaps = 4/139 (2%)
Query: 77 PTSILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRDWGATVVNADLSKPETI 136
P ILV+GA+G +G+ +VR +G+ + R A L+ + DLS P+ +
Sbjct: 2 PQRILVLGASGYIGQHLVRTLSQQGHQILAAARHVDRLAK-LQLANVSCHKVDLSWPDNL 60
Query: 137 PATLVGVHTVIDCATGRPEEPIKTVDWEGKVAL--IQCAKAMGIQKYVFYSIHNCDKHPE 194
PA L + TV E + E +VAL + + +++ +F S H +
Sbjct: 61 PALLQDIDTVYFLVHSMGEGG-DFIAQERQVALNVRDALREVPVKQLIFLSSLQAPPHEQ 119
Query: 195 VPLMEIKYCTENFLRDSGL 213
+ + T + LR++ +
Sbjct: 120 SDHLRARQATADILREANV 138
>DFRA_SYNY3 (P73212) Putative dihydroflavonol-4-reductase (EC
1.1.1.219) (DFR) (Dihydrokaempferol 4-reductase)
Length = 343
Score = 38.1 bits (87), Expect = 0.039
Identities = 31/109 (28%), Positives = 51/109 (46%), Gaps = 7/109 (6%)
Query: 82 VVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRDWGATVVNADLSKPETIPATLV 141
V G TG +G +VR L++GY VR LVR P D L++ V DL+ + + +
Sbjct: 15 VTGGTGFVGANLVRHLLEQGYQVRALVRASSRP-DNLQNLPIDWVVGDLNDGD-LHQQMQ 72
Query: 142 GVHTVIDCAT-----GRPEEPIKTVDWEGKVALIQCAKAMGIQKYVFYS 185
G + A + E + + G ++ CA+ GI++ V+ S
Sbjct: 73 GCQGLFHVAAHYSLWQKDREALYRSNVLGTRNILACAQKAGIERTVYTS 121
>NSDL_MOUSE (Q9R1J0) NAD(P)-dependent steroid dehydrogenase (EC
1.1.1.-)
Length = 362
Score = 37.7 bits (86), Expect = 0.051
Identities = 30/111 (27%), Positives = 49/111 (44%), Gaps = 15/111 (13%)
Query: 82 VVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRDWGATVVN---ADLSKPETIPA 138
V+G +G LG+ +V + L+ GY V D + + V DL + +
Sbjct: 31 VIGGSGFLGQHMVEQLLERGYTVNVF--------DIHQGFDNPRVQFFIGDLCNQQDLYP 82
Query: 139 TLVGVHTVIDCATGRP----EEPIKTVDWEGKVALIQCAKAMGIQKYVFYS 185
L GV TV CA+ P +E V++ G +I+ + G+QK + S
Sbjct: 83 ALKGVSTVFHCASPPPYSNNKELFYRVNFIGTKTVIETCREAGVQKLILTS 133
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.322 0.138 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 47,184,723
Number of Sequences: 164201
Number of extensions: 2107405
Number of successful extensions: 5637
Number of sequences better than 10.0: 46
Number of HSP's better than 10.0 without gapping: 18
Number of HSP's successfully gapped in prelim test: 28
Number of HSP's that attempted gapping in prelim test: 5598
Number of HSP's gapped (non-prelim): 48
length of query: 393
length of database: 59,974,054
effective HSP length: 112
effective length of query: 281
effective length of database: 41,583,542
effective search space: 11684975302
effective search space used: 11684975302
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 67 (30.4 bits)
Medicago: description of AC121239.8


