

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121237.9 + phase: 0 /pseudo
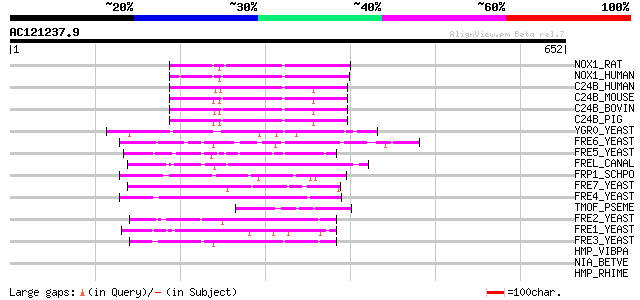
(652 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
NOX1_RAT (Q9WV87) NADPH oxidase homolog 1 (NOX-1) (NOH-1) (NADH/... 115 4e-25
NOX1_HUMAN (Q9Y5S8) NADPH oxidase homolog 1 (NOX-1) (NOH-1) (NAD... 115 5e-25
C24B_HUMAN (P04839) Cytochrome B-245 heavy chain (P22 phagocyte ... 107 7e-23
C24B_MOUSE (Q61093) Cytochrome B-245 heavy chain (P22 phagocyte ... 107 1e-22
C24B_BOVIN (O46522) Cytochrome B-245 heavy chain (P22 phagocyte ... 104 6e-22
C24B_PIG (P52649) Cytochrome B-245 heavy chain (P22 phagocyte B-... 103 1e-21
YGR0_YEAST (P53109) Hypothetical 65.8 kDa protein in SUT1-RCK1 i... 70 1e-11
FRE6_YEAST (Q12473) Ferric reductase transmembrane component 6 p... 67 1e-10
FRE5_YEAST (Q08908) Ferric reductase transmembrane component 5 p... 66 3e-10
FREL_CANAL (P78588) Probable ferric reductase transmembrane comp... 65 4e-10
FRP1_SCHPO (Q04800) Ferric reductase transmembrane component (EC... 64 1e-09
FRE7_YEAST (Q12333) Ferric reductase transmembrane component 7 (... 60 2e-08
FRE4_YEAST (P53746) Ferric reductase transmembrane component 4 p... 54 1e-06
TMOF_PSEME (Q03304) Toluene-4-monooxygenase electron transfer co... 51 1e-05
FRE2_YEAST (P36033) Ferric reductase transmembrane component 2 p... 50 2e-05
FRE1_YEAST (P32791) Ferric reductase transmembrane component 1 p... 49 5e-05
FRE3_YEAST (Q08905) Ferric reductase transmembrane component 3 p... 45 5e-04
HMP_VIBPA (P40609) Flavohemoprotein (Hemoglobin-like protein) (F... 40 0.025
NIA_BETVE (P27783) Nitrate reductase [NAD(P)H] (EC 1.7.1.2) (NR) 38 0.073
HMP_RHIME (Q7WUM8) Flavohemoprotein (Hemoglobin-like protein) (F... 38 0.073
>NOX1_RAT (Q9WV87) NADPH oxidase homolog 1 (NOX-1) (NOH-1)
(NADH/NADPH mitogenic oxidase subunit P65-MOX)
(Mitogenic oxidase 1) (MOX1)
Length = 563
Score = 115 bits (288), Expect = 4e-25
Identities = 71/258 (27%), Positives = 120/258 (45%), Gaps = 50/258 (19%)
Query: 188 TYVSNVAGEIASLIALAMWITSIPQIRRKMYEVFFYTHHLYILYILFFAIHAGVGSMC-- 245
T ++ + G +A++ + M +++ IRR +E+F+YTHHL+I+YI+ IH G+G +
Sbjct: 170 TSIAGLTGVVATVALVLMVTSAMEFIRRNYFELFWYTHHLFIIYIICLGIH-GLGGIVRG 228
Query: 246 -------------------------------------------VIAPGVFLFLIDRHLRF 262
++AP F ++ +R LRF
Sbjct: 229 QTEESMSESHPRNCSYSFHEWDKYERSCRSPHFVGQPPESWKWILAPIAF-YIFERILRF 287
Query: 263 LQSRQHARLLSARLLPCDALELNFSKNPSLYYNPTSLVFINVPKVSKLQWHPFTVTSSCN 322
+SRQ + + PC LEL K +F+N P +S L+WHPFT+TS+
Sbjct: 288 YRSRQKVVITKVVMHPCKVLELQMRKR-GFTMGIGQYIFVNCPSISFLEWHPFTLTSA-- 344
Query: 323 METNYLSVAIKNVGSWSNKLYQELSSSSLDHLNISVEGPYGPHTAQFLRHEQIVMVSGGS 382
E + S+ I+ G W+ L + I V+GP+G + ++E V+V G
Sbjct: 345 PEEEFFSIHIRAAGDWTENLIRTFEQQHSPMPRIEVDGPFGTVSEDVFQYEVAVLVGAGI 404
Query: 383 GVTPFISIIRDLIFQSQQ 400
GVTPF S ++ + ++ Q+
Sbjct: 405 GVTPFASFLKSIWYKFQR 422
>NOX1_HUMAN (Q9Y5S8) NADPH oxidase homolog 1 (NOX-1) (NOH-1)
(NADH/NADPH mitogenic oxidase subunit P65-MOX)
(Mitogenic oxidase 1) (MOX1)
Length = 564
Score = 115 bits (287), Expect = 5e-25
Identities = 77/258 (29%), Positives = 122/258 (46%), Gaps = 52/258 (20%)
Query: 188 TYVSNVAGEIASLIALAMWITSIPQ-IRRKMYEVFFYTHHLYILYILFFAIHAGVGSMC- 245
T ++ + G I + IAL + +TS + IRR +EVF+YTHHL+I YIL IH G+G +
Sbjct: 171 TSIAGLTGVIMT-IALILMVTSATEFIRRSYFEVFWYTHHLFIFYILGLGIH-GIGGIVR 228
Query: 246 --------------------------------------------VIAPGVFLFLIDRHLR 261
++AP V L++ +R LR
Sbjct: 229 GQTEESMNESHPRKCAESFEMWDDRDSHCRRPKFEGHPPESWKWILAP-VILYICERILR 287
Query: 262 FLQSRQHARLLSARLLPCDALELNFSKNPSLYYNPTSLVFINVPKVSKLQWHPFTVTSSC 321
F +S+Q + + P LEL +K +F+N P +S L+WHPFT+TS+
Sbjct: 288 FYRSQQKVVITKVVMHPSKVLELQMNKR-GFSMEVGQYIFVNCPSISLLEWHPFTLTSA- 345
Query: 322 NMETNYLSVAIKNVGSWSNKLYQELSSSSLDHLNISVEGPYGPHTAQFLRHEQIVMVSGG 381
E ++ S+ I+ G W+ L + I V+GP+G + ++E V+V G
Sbjct: 346 -PEEDFFSIHIRAAGDWTENLIRAFEQQYSPIPRIEVDGPFGTASEDVFQYEVAVLVGAG 404
Query: 382 SGVTPFISIIRDLIFQSQ 399
GVTPF SI++ + ++ Q
Sbjct: 405 IGVTPFASILKSIWYKFQ 422
>C24B_HUMAN (P04839) Cytochrome B-245 heavy chain (P22 phagocyte
B-cytochrome) (Neutrophil cytochrome B, 91 kDa
polypeptide) (CGD91-PHOX) (GP91-PHOX) (GP91-1) (Heme
binding membrane glycoprotein GP91PHOX) (Cytochrome
B(558) beta chain) (Superoxide-generat
Length = 569
Score = 107 bits (268), Expect = 7e-23
Identities = 69/258 (26%), Positives = 119/258 (45%), Gaps = 51/258 (19%)
Query: 188 TYVSNVAGEIASLIALAMWITSIPQIRRKMYEVFFYTHHLYILYILFFAIHAG------- 240
T ++ + G + +L + + +S IRR +EVF+YTHHL++++ + AIH
Sbjct: 171 TLLAGITGVVITLCLILIITSSTKTIRRSYFEVFWYTHHLFVIFFIGLAIHGAERIVRGQ 230
Query: 241 -VGSMCV----------------------------------IAPGVFLFLIDRHLRFLQS 265
S+ V I +FL+L +R +RF +S
Sbjct: 231 TAESLAVHNITVCEQKISEWGKIKECPIPQFAGNPPMTWKWIVGPMFLYLCERLVRFWRS 290
Query: 266 RQHARLLSARLLPCDALELNFSKNPSLYYNPTSLVFINVPKVSKLQWHPFTVTSSCNMET 325
+Q + P +EL K +F+ PKVSKL+WHPFT+TS+ E
Sbjct: 291 QQKVVITKVVTHPFKTIELQMKKK-GFKMEVGQYIFVKCPKVSKLEWHPFTLTSA--PEE 347
Query: 326 NYLSVAIKNVGSWSNKLYQELSSSSLDHLN------ISVEGPYGPHTAQFLRHEQIVMVS 379
++ S+ I+ VG W+ L+ + + I+V+GP+G + +E +++V
Sbjct: 348 DFFSIHIRIVGDWTEGLFNACGCDKQEFQDAWKLPKIAVDGPFGTASEDVFSYEVVMLVG 407
Query: 380 GGSGVTPFISIIRDLIFQ 397
G GVTPF SI++ + ++
Sbjct: 408 AGIGVTPFASILKSVWYK 425
>C24B_MOUSE (Q61093) Cytochrome B-245 heavy chain (P22 phagocyte
B-cytochrome) (Neutrophil cytochrome B, 91 kDa
polypeptide) (CGD91-PHOX) (GP91-PHOX) (GP91-1) (Heme
binding membrane glycoprotein GP91PHOX) (Cytochrome
B(558) beta chain) (Superoxide-generat
Length = 570
Score = 107 bits (266), Expect = 1e-22
Identities = 70/259 (27%), Positives = 121/259 (46%), Gaps = 53/259 (20%)
Query: 188 TYVSNVAGEIASLIALAMWITSIPQIRRKMYEVFFYTHHLYILYILFFAIH--------- 238
T ++ + G + +L + + +S IRR +EVF+YTHHL++++ + AIH
Sbjct: 172 TRLAGITGIVITLCLILIITSSTKTIRRSYFEVFWYTHHLFVIFFIGLAIHGAERIVRGQ 231
Query: 239 -------------------------------AGVGSMC---VIAPGVFLFLIDRHLRFLQ 264
AG M ++ P +FL+L +R +RF +
Sbjct: 232 TAESLEEHNLDICADKIEEWGKIKECPVPKFAGNPPMTWKWIVGP-MFLYLCERLVRFWR 290
Query: 265 SRQHARLLSARLLPCDALELNFSKNPSLYYNPTSLVFINVPKVSKLQWHPFTVTSSCNME 324
S+Q + P +EL K +F+ PKVSKL+WHPFT+TS+ E
Sbjct: 291 SQQKVVITKVVTHPFKTIELQMKKK-GFKMEVGQYIFVKCPKVSKLEWHPFTLTSA--PE 347
Query: 325 TNYLSVAIKNVGSWSNKLYQELSSSSLDHLN------ISVEGPYGPHTAQFLRHEQIVMV 378
++ S+ I+ VG W+ L+ + + I+V+GP+G + +E +++V
Sbjct: 348 EDFFSIHIRIVGDWTEGLFNACGCDKQEFQDAWKLPKIAVDGPFGTASEDVFSYEVVMLV 407
Query: 379 SGGSGVTPFISIIRDLIFQ 397
G GVTPF SI++ + ++
Sbjct: 408 GAGIGVTPFASILKSVWYK 426
>C24B_BOVIN (O46522) Cytochrome B-245 heavy chain (P22 phagocyte
B-cytochrome) (Neutrophil cytochrome B, 91 kDa
polypeptide) (CGD91-PHOX) (GP91-PHOX) (GP91-1) (Heme
binding membrane glycoprotein GP91PHOX) (Cytochrome
B(558) beta chain) (Superoxide-generat
Length = 570
Score = 104 bits (260), Expect = 6e-22
Identities = 68/259 (26%), Positives = 120/259 (46%), Gaps = 53/259 (20%)
Query: 188 TYVSNVAGEIASLIALAMWITSIPQIRRKMYEVFFYTHHLYILYILFFAIHA-------- 239
T ++ + G + +L + + +S IRR +EVF+YTHHL++++ + AIH
Sbjct: 172 TRLAGITGVVITLCLILIITSSTKTIRRSYFEVFWYTHHLFVIFFIGLAIHGAQRIVRGQ 231
Query: 240 --------------------GVGSMC---------------VIAPGVFLFLIDRHLRFLQ 264
G C ++ P +FL+L +R +RF +
Sbjct: 232 TAESLLKHQPRNCYQNISQWGKIENCPIPEFSGNPPMTWKWIVGP-MFLYLCERLVRFWR 290
Query: 265 SRQHARLLSARLLPCDALELNFSKNPSLYYNPTSLVFINVPKVSKLQWHPFTVTSSCNME 324
S+Q + P +EL K +F+ P VSKL+WHPFT+TS+ E
Sbjct: 291 SQQKVVITKVVTHPFKTIELQMKKK-GFKMEVGQYIFVKCPVVSKLEWHPFTLTSA--PE 347
Query: 325 TNYLSVAIKNVGSWSNKLYQELSSSSLDHLN------ISVEGPYGPHTAQFLRHEQIVMV 378
++ S+ I+ VG W+ L++ + + I+V+GP+G + +E +++V
Sbjct: 348 EDFFSIHIRIVGDWTEGLFKACGCDKQEFQDAWKLPKIAVDGPFGTASEDVFSYEVVMLV 407
Query: 379 SGGSGVTPFISIIRDLIFQ 397
G GVTPF SI++ + ++
Sbjct: 408 GAGIGVTPFASILKSVWYK 426
>C24B_PIG (P52649) Cytochrome B-245 heavy chain (P22 phagocyte
B-cytochrome) (Neutrophil cytochrome B, 91 kDa
polypeptide) (CGD91-PHOX) (GP91-PHOX) (GP91-1) (Heme
binding membrane glycoprotein GP91PHOX) (Cytochrome
B(558) beta chain) (Superoxide-generatin
Length = 484
Score = 103 bits (257), Expect = 1e-21
Identities = 68/259 (26%), Positives = 121/259 (46%), Gaps = 53/259 (20%)
Query: 188 TYVSNVAGEIASLIALAMWITSIPQIRRKMYEVFFYTHHLYILYILFFAIH--------- 238
T ++ + G + +L + + +S IRR +EVF+YTHHL++++ + AIH
Sbjct: 86 TRLAGITGVVITLCLILIITSSTKTIRRSYFEVFWYTHHLFVIFFIGLAIHGAERIVRRQ 145
Query: 239 -------------------------------AGVGSMC---VIAPGVFLFLIDRHLRFLQ 264
AG M ++ P +FL+L +R +RF +
Sbjct: 146 TPKSLLVHDPKACAQNISQWGKIKDCPIPEFAGNPPMTWKWIVGP-MFLYLCERLVRFWR 204
Query: 265 SRQHARLLSARLLPCDALELNFSKNPSLYYNPTSLVFINVPKVSKLQWHPFTVTSSCNME 324
S+Q + P +EL K +F+ P VSKL+WHPFT+TS+ E
Sbjct: 205 SQQKVVITKVVTHPFKTIELQMKKK-GFRMEVGQYIFVKRPAVSKLEWHPFTLTSA--PE 261
Query: 325 TNYLSVAIKNVGSWSNKLYQELSSSSLDHLN------ISVEGPYGPHTAQFLRHEQIVMV 378
++ S+ I+ VG W+ L++ + + I+V+GP+G + ++ +++V
Sbjct: 262 EDFFSIHIRIVGDWTEGLFKACGCDKQEFQDAWKLPKIAVDGPFGTASEDVFSYQVVMLV 321
Query: 379 SGGSGVTPFISIIRDLIFQ 397
G GVTPF SI++ + ++
Sbjct: 322 GAGIGVTPFASILKSVWYK 340
>YGR0_YEAST (P53109) Hypothetical 65.8 kDa protein in SUT1-RCK1
intergenic region
Length = 570
Score = 70.5 bits (171), Expect = 1e-11
Identities = 88/359 (24%), Positives = 154/359 (42%), Gaps = 57/359 (15%)
Query: 114 YIPVSITSACTNKMRKFTRMS-SILPL-------------VGLTSESSIKYHIWLGQLSM 159
Y S+T+ T +++ R+S +++PL T I +H W ++
Sbjct: 85 YYHYSLTTQSTVYLKRLGRLSYALIPLNLFLTLRPNWFLRKNCTYTDFIPFHKWFSRIIT 144
Query: 160 VLFAAHSIGFIIYWAIT-NQMIEMLEWSKTYVSNVAGEIASLIALAMWITSIPQIRRKMY 218
V+ H I FII WAI N ++ KT+ N AG I S++ L + I SI +RR Y
Sbjct: 145 VIGLLHGIFFIIKWAIDDNVSLKQKLILKTF--NFAGFIISILVLFLLICSIGPMRRYNY 202
Query: 219 EVFFYTHHLY-ILYILFFAIHAGVGSMCVIAPGVFLFLIDRHLRFLQSRQHARLLSARLL 277
+F+ H+L + +IL IH+ PGV + + L R++ A+ L
Sbjct: 203 RLFYIVHNLVNVAFILLTPIHS--------RPGVKFPFLLLNCTLLFIHIINRIVFAKSL 254
Query: 278 PCDALELNFSKNPSL-----------YYNPTSLVFIN-VPKVSKLQW----HPFTVTSSC 321
N+SK + Y+ P S + I+ +++ L W HP+T+ S
Sbjct: 255 MILNKNANYSKTNLVHVRLPRAILPDYFEPGSHIRISPYRRINPLYWLLPSHPYTIASLA 314
Query: 322 NMETNYLSVAIKNV-----GSWSNKLYQELSSSSLD-HLNISVEGPYGPHTAQ--FLRHE 373
E N + + IK GS L S LD ++ Y P + + +
Sbjct: 315 --EDNSIDLIIKETSTAEPGSQIESLRSNPKSFHLDQEKTYTLINSYPPSVPEECYSQGT 372
Query: 374 QIVMVSGGSGVTPFISIIRDLIFQSQQQEFQPPKLLLVCIFKNYADLTMLDLMLPISGL 432
I ++ GGSG++ + + R F + ++ L ++ + K+Y++ ++ L +GL
Sbjct: 373 NIAIICGGSGISFALPLFRH-FFNKENVKY----LKMIWLIKDYSEYELVLDYLKTNGL 426
>FRE6_YEAST (Q12473) Ferric reductase transmembrane component 6
precursor (EC 1.16.1.7) (Ferric-chelate reductase 6)
Length = 712
Score = 67.4 bits (163), Expect = 1e-10
Identities = 84/385 (21%), Positives = 160/385 (40%), Gaps = 68/385 (17%)
Query: 130 FTRMSSILP-LVGLTSESSIKYHIWLGQLSMVLFAAHSIGFIIYWAITNQMIEMLEWSKT 188
FT +S L L G+ S I +H W+G++ ++ HS+ + ++ AI N ++
Sbjct: 301 FTARNSFLEFLTGVKFNSFISFHKWIGRIMVLNATIHSLSYSLF-AIINHAFKISNKQLY 359
Query: 189 YVSNVAGEIASLIALAMWITSIPQIRRKMYEVFFYTHHLYILYILFFAI---HAGV--GS 243
+ +A + L + S+ +R++ YE F YTH IL +LFF H + G
Sbjct: 360 WKFGIASITVLCVLLVL---SLGIVRKRHYEFFLYTH--IILALLFFYCCWQHVKIFNGW 414
Query: 244 MCVIAPGVFLFLIDRHLRFLQSRQHARLLSARLLPCDALELNFSKNPSLYYNPTSLVFIN 303
I + ++ +++ R +L R + LN S NP + +
Sbjct: 415 KEWIVVSLLIWGLEKLFRIWN------ILQFRFPKATLINLNTSNNPH-----DEMFKVI 463
Query: 304 VPKVSKL--------------------QWHPFTVTSSCNMETNYLSVAIKNVGSWSNKLY 343
+PK ++ Q HPFT+ E + IK + +Y
Sbjct: 464 IPKYNRRWHSKPGQYCFIYFLHPLVFWQCHPFTIID----EGEKCVLVIKPKSGLTRFIY 519
Query: 344 QEL--SSSSLDHLNISVEGPYGPHTAQFLRHEQIVMVSGGSGVTPFISIIRDLIFQSQQQ 401
+ S + L +++EGPYGP + + ++++SGG+G+ + D + +
Sbjct: 520 NHILQSLNGKLQLRVAIEGPYGPSNLHLDKFDHLLLLSGGTGLPGPL----DHAIKLSRN 575
Query: 402 EFQPPKLLLVCIFKNYADLTMLDLMLPISGLKTRISQLQ-----LQIEAYITREKQEPSK 456
+P + L+ KN + L +G K+ I +L+ + ++ Y+T +K +K
Sbjct: 576 PDKPKSIDLIMAIKNPSFL---------NGYKSEILELKNSRSHVNVQVYLT-QKTAVTK 625
Query: 457 DTQKQIQTIWFKTNLSDCPISAVLG 481
+ Q I F +++ A +G
Sbjct: 626 AANARDQLIHFDDIMTELTSFAHIG 650
>FRE5_YEAST (Q08908) Ferric reductase transmembrane component 5
precursor (EC 1.16.1.7) (Ferric-chelate reductase 5)
Length = 694
Score = 65.9 bits (159), Expect = 3e-10
Identities = 77/263 (29%), Positives = 120/263 (45%), Gaps = 24/263 (9%)
Query: 134 SSILPLVGLTSESSIKYHIWLGQLSMVLFAAHSIGFIIYWAITNQMIEMLEWSKTYVSNV 193
S++ L G+ + I YH WLG+ +V H+IG+ Y A + +++S + S
Sbjct: 293 STMTWLTGIRYTAFITYHKWLGRFMLVDCTIHAIGY-TYHAYIENYWKYVKYSDLWTS-- 349
Query: 194 AGEIASLIALAMWITSIPQIRRKMYEVFFYTHHLYILYILFF------AIHAGVGSMCVI 247
G A +I + S RR YE+F TH IL I FF G G ++
Sbjct: 350 -GRHAMIIVGILVFFSFFFFRRHYYELFVITH--IILAIGFFHACWKHCYKLGWGEW-IM 405
Query: 248 APGVFLFLIDRHLRFLQSRQHARLLSARLLPC--DALELNFSKNPSLY-YNPTSLVFINV 304
A +F ++ DR LR ++ + A+L C +E+ SK+ + P +++
Sbjct: 406 ACALF-WIADRILRLIKIAIFG-MPWAKLKLCGESMIEVRISKSSKWWKAEPGQYIYLYF 463
Query: 305 --PKVSKLQWHPFTVTSSCNMETNYLSVAIKNVGSWSNKLYQE-LSSSSLDHLNISVEGP 361
PK+ Q HPFTV S +E L V I + KL + L S + + EGP
Sbjct: 464 LRPKIF-WQSHPFTVMDSL-VEDGELVVVITVKNGLTKKLQEYLLESEGYTEMRVLAEGP 521
Query: 362 YGPHTAQFLRHEQIVMVSGGSGV 384
YG T L E ++ ++GG+GV
Sbjct: 522 YGQSTRTHL-FESLLFIAGGAGV 543
>FREL_CANAL (P78588) Probable ferric reductase transmembrane
component (EC 1.16.1.7) (Ferric-chelate reductase)
Length = 669
Score = 65.5 bits (158), Expect = 4e-10
Identities = 71/292 (24%), Positives = 130/292 (44%), Gaps = 23/292 (7%)
Query: 139 LVGLTSESSIKYHIWLGQLSMVLFAAHSIGFIIYWAITNQMIEMLEWSKTYVSNVAGEIA 198
L G + I YH W+ ++ ++L H+I F + T + ++ + ++ + G ++
Sbjct: 263 LTGWDFATFIMYHRWISRVDVLLIIVHAITFSVSDKATGKYKNRMK--RDFM--IWGTVS 318
Query: 199 SLIALAMWITSIPQIRRKMYEVFFYTHHLYILYILFFAIHA-------GVGSMCVIAPGV 251
++ + ++ RRK YEVFF H I+ ++FF + G G A V
Sbjct: 319 TICGGFILFQAMLFFRRKCYEVFFLIH---IVLVVFFVVGGYYHLESQGYGDFMWAAIAV 375
Query: 252 FLFLIDRHLRFLQSRQH-ARLLSARLLPCDALELNFSKNPSLYYNPTSLVFINVPKVSK- 309
+ F DR +R + AR + + D L++ K FI+ K +
Sbjct: 376 WAF--DRVVRLGRIFFFGARKATVSIKGDDTLKIEVPKPKYWKSVAGGHAFIHFLKPTLF 433
Query: 310 LQWHPFTVTSSCNMETNYLSVAIKNVGSWSNKLYQELSSSSLDHLNISVEGPYGPHTAQF 369
LQ HPFT T++ + + L IKN + + Y + + + VEGPYG ++
Sbjct: 434 LQSHPFTFTTTESNDKIVLYAKIKNGITSNIAKYLSPLPGNTATIRVLVEGPYGEPSSAG 493
Query: 370 LRHEQIVMVSGGSGVTPFISIIRDLIFQSQQQEFQPPKLLLVCIFKNYADLT 421
+ +V V+GG+G+ S DL +S+ Q + L+ I +++ L+
Sbjct: 494 RNCKNVVFVAGGNGIPGIYSECVDLAKKSKNQSIK-----LIWIIRHWKSLS 540
>FRP1_SCHPO (Q04800) Ferric reductase transmembrane component (EC
1.16.1.7) (Ferric-chelate reductase)
Length = 564
Score = 63.9 bits (154), Expect = 1e-09
Identities = 74/317 (23%), Positives = 137/317 (42%), Gaps = 65/317 (20%)
Query: 130 FTRMSSILPLVGLTSESSIKY-HIWLGQLSMVLFAAHSIGFIIYWAITNQMIEMLEWSKT 188
F+ ++ L+ ++S + Y H L Q ++++ A H +I A + + T
Sbjct: 135 FSIKNNPFALLLISSHEKMNYVHRRLSQYAIMIGAIHGFAYIGLAAQGKRAL------LT 188
Query: 189 YVSNVAGEIASLIALAMWITSIPQIRRKMYEVFFYTHHL-YILYILFFAIHAGVGSMCVI 247
+ G + + + M ++S+P RR+ YE FF HH+ I +++ +H CV+
Sbjct: 189 ARVTIIGYVILGLMVIMIVSSLPFFRRRFYEWFFVLHHMCSIGFLITIWLH---HRRCVV 245
Query: 248 --APGVFLFLIDRHLRFLQSRQHARLLSARLLPCDALELNFSKNP-------SLYYNPTS 298
V +++ DR R L+S + L+ D L + K P L + +
Sbjct: 246 YMKVCVAVYVFDRGCRMLRSFLNRSKFDVVLVEDD---LIYMKGPRPKKSFFGLPWGAGN 302
Query: 299 LVFINVPKVSKLQWHPFTVTSSCNMETNYLSVAIKNVGSWSNKLYQELSSSSL---DHLN 355
++IN+P +S Q HPFT+ S + + L VA++ ++ +L +++SS SL +N
Sbjct: 303 HMYINIPSLSYWQIHPFTIASVPSDDFIELFVAVR--AGFTKRLAKKVSSKSLSDVSDIN 360
Query: 356 IS-------------------------------------VEGPYGPHTAQFLRHEQIVMV 378
IS ++GPYGP + + + + +
Sbjct: 361 ISDEKIEKNGDVGIEVMERHSLSQEDLVFESSAAKVSVLMDGPYGPVSNPYKDYSYLFLF 420
Query: 379 SGGSGVTPFISIIRDLI 395
+GG GV+ + II D I
Sbjct: 421 AGGVGVSYILPIILDTI 437
>FRE7_YEAST (Q12333) Ferric reductase transmembrane component 7 (EC
1.16.1.7) (Ferric-chelate reductase 7)
Length = 629
Score = 59.7 bits (143), Expect = 2e-08
Identities = 63/264 (23%), Positives = 113/264 (41%), Gaps = 20/264 (7%)
Query: 139 LVGLTSESSIKYHIWLGQLSMVLFAAHSIGFIIYWAITNQMIEMLEWSKTYVSNVAGEIA 198
LVGL+ E YH W L + H I F+ M + K +G
Sbjct: 185 LVGLSYEKINIYHQWASILCLFFSWVHVIPFLRQARHEGGYERMHQRWKASDMWRSGVPP 244
Query: 199 SLIALAMWITSIPQIRRKMYEVFFYTHHLYIL--YI-LFFAIHAGVGS-MCVIAPGVFLF 254
L +W++S+P RR YE+F H + + YI LF+ ++ + S M ++A V F
Sbjct: 245 ILFLNLLWLSSLPIARRHFYEIFLQLHWILAVGFYISLFYHVYPELNSHMYLVATIVVWF 304
Query: 255 -------LIDRHLRFLQSRQHARLLSARLLPCDALELNFSKNPSLYYNPTSLVFINVPKV 307
+ +LR +S + + + ++ +EL K+ + Y+P +F+
Sbjct: 305 AQLFYRLAVKGYLRPGRSFMASTIANVSIVGEGCVEL-IVKDVEMAYSPGQHIFVRTIDK 363
Query: 308 SKLQWHPFTVTSSCNMETNYLSVAIKNVGSWSNKLYQELSSSSLDHLNISVEGPYGPHTA 367
+ HPF++ S + + I+ +S +LY+ S+ D I ++GPYG
Sbjct: 364 GIISNHPFSIFPSAKYPGG-IKMLIRAQKGFSKRLYE----SNDDMKKILIDGPYGGIER 418
Query: 368 QFLRHEQIVMVSGGSGVT---PFI 388
+ ++ GSG++ PF+
Sbjct: 419 DIRSFTNVYLICSGSGISTCLPFL 442
>FRE4_YEAST (P53746) Ferric reductase transmembrane component 4
precursor (EC 1.16.1.7) (Ferric-chelate reductase 4)
Length = 719
Score = 54.3 bits (129), Expect = 1e-06
Identities = 63/270 (23%), Positives = 114/270 (41%), Gaps = 19/270 (7%)
Query: 130 FTRMSSILPLV-GLTSESSIKYHIWLGQLSMVLFAAHSIGFIIYWAITNQMIEMLEWS-- 186
F ++ L L+ GL S I +H WLG++ + H+ GF TN + +W+
Sbjct: 287 FAGRNNFLQLISGLKHTSFIVFHKWLGRMMFLDAIIHAAGF------TNYYLYYKKWNTV 340
Query: 187 KTYVSNVAGEIASLIALAMWITSIPQIRRKMYEVFFYTHHLYILYILFFAIH--AGVGSM 244
+ V G + +A + SI RR YE F H ++ L+ +
Sbjct: 341 RLRVYWKFGIATTCLAGMLIFFSIAAFRRHYYETFMALHIVFAALFLYTCWEHVTNFSGI 400
Query: 245 CVIAPGVFLFLIDRHLRFLQ-SRQHARLLSARLLPCDALELNFSKNPSLY-YNPTSLVFI 302
I + ++ +DR +R + + +L+ D + + K + P VF+
Sbjct: 401 EWIYAAIAIWGVDRIVRITRIALLGFPKADLQLVGSDLVRVTVKKPKKFWKAKPGQYVFV 460
Query: 303 N-VPKVSKLQWHPFTVTSSCNMETNYLSV--AIKNVGSWSNKLYQELSSSSLDHLNISVE 359
+ + + Q HPFTV SC + + V A K V + + + +++E
Sbjct: 461 SFLRPLCFWQSHPFTVMDSCVNDRELVIVLKAKKGVTKLVRNFVERKGGKA--SMRLAIE 518
Query: 360 GPYGPHTAQFLRHEQIVMVSGGSGVTPFIS 389
GPYG + R + +++++GGSG+ IS
Sbjct: 519 GPYGSKSTAH-RFDNVLLLAGGSGLPGPIS 547
>TMOF_PSEME (Q03304) Toluene-4-monooxygenase electron transfer
component [Includes: Ferredoxin; Ferredoxin--NAD(+)
reductase (EC 1.18.1.3)]
Length = 326
Score = 50.8 bits (120), Expect = 1e-05
Identities = 34/137 (24%), Positives = 69/137 (49%), Gaps = 11/137 (8%)
Query: 266 RQHARLLSARLLPCDALELNFSKNPSLYYNPTSLVFINVPKVSKLQWHPFTVTSSCN-ME 324
R R++S R L + EL + ++P ++VP++ + S+ N ++
Sbjct: 102 RFSTRVVSKRFLSDEMFELRLEAEQKVVFSPGQYFMVDVPELGTRAY------SAANPVD 155
Query: 325 TNYLSVAIKNVGSWSNKLYQELSSSSLDHLNISVEGPYGPHTAQFLRHEQIVMVSGGSGV 384
N L++ +K V + K+ L++ +++ L + +GPYG + Q V ++GGSG+
Sbjct: 156 GNTLTLIVKAVPN--GKVSCALANETIETLQL--DGPYGLSVLKTADETQSVFIAGGSGI 211
Query: 385 TPFISIIRDLIFQSQQQ 401
P +S++ LI Q ++
Sbjct: 212 APMVSMVNTLIAQGYEK 228
>FRE2_YEAST (P36033) Ferric reductase transmembrane component 2
precursor (EC 1.16.1.7) (Ferric-chelate reductase 2)
Length = 711
Score = 50.1 bits (118), Expect = 2e-05
Identities = 55/252 (21%), Positives = 108/252 (42%), Gaps = 16/252 (6%)
Query: 141 GLTSESSIKYHIWLGQLSMVLFAAHSIGFIIYWAITNQMIEMLEW--SKTYVSNVAGEIA 198
G+ S I +H WLG++ + H + Y + N+ W SK + G A
Sbjct: 306 GVKYTSFIMFHKWLGRMMFLDAMIHGSAYTSY-TVANK-----TWATSKNRLYWQFGVAA 359
Query: 199 SLIALAMWITSIPQIRRKMYEVFFYTHHLYILYILFFAIHAGVGSMCVIA---PGVFLFL 255
+A M S R+ YE F + H+ + + F+A V S+ I + +++
Sbjct: 360 LCLAGTMVFFSFAVFRKYFYEAFLFL-HIVLGAMFFYACWEHVVSLSGIEWIYTAIAIWI 418
Query: 256 IDRHLRFLQSRQHA-RLLSARLLPCDALELNFSKNPSLY-YNPTSLVFIN-VPKVSKLQW 312
+DR +R +++ S +L+ D + L K + P VF++ + + Q
Sbjct: 419 VDRIIRIIKASYFGFPKASLQLIGDDLIRLTVKKPARPWRAKPGQYVFVSFLHPLYFWQS 478
Query: 313 HPFTVTSSCNMETNYLSVAIKNVGSWSNKLYQELSSSSLDHLNISVEGPYGPHTAQFLRH 372
HPFTV S + + + + G + + +++EGPYG ++ +
Sbjct: 479 HPFTVLDSVSKNGELVIILKEKKGVTRLVKKYVCRNGGKTSMRLAIEGPYG-SSSPVNNY 537
Query: 373 EQIVMVSGGSGV 384
+++++GG+G+
Sbjct: 538 NNVLLLTGGTGL 549
>FRE1_YEAST (P32791) Ferric reductase transmembrane component 1
precursor (EC 1.16.1.7) (Ferric-chelate reductase 1)
Length = 686
Score = 48.5 bits (114), Expect = 5e-05
Identities = 69/271 (25%), Positives = 118/271 (43%), Gaps = 24/271 (8%)
Query: 132 RMSSILPLVGLTSESSIKYHIWLGQLSMVLFAAHSIGFIIYWAITNQMIEMLEWSKTYVS 191
R + +P+ GL+ + YH W + +L HSI + + + + L K Y
Sbjct: 275 RNNPFIPITGLSFSTFNFYHKWSAYVCFMLAVVHSI-VMTASGVKRGVFQSLV-RKFYFR 332
Query: 192 NVAGEIASLIALAMWITSIPQIRRKMYEVFFYTHH-LYILYILFFAIHA-GVGSMCVIAP 249
G +A+++ + S R + YE+F H + I++I+ H +G M I
Sbjct: 333 --WGIVATILMSIIIFQSEKVFRNRGYEIFLLIHKAMNIMFIIAMYYHCHTLGWMGWIWS 390
Query: 250 GVFLFLIDRHLRFLQSRQHARLLSARLLPCD---ALELNFSKNPSLYYNPTSLVFINV-- 304
+ DR R ++ + L +A L D ++++ K Y + ++
Sbjct: 391 MAGILCFDRFCRIVRIIMNGGLKTATLSTTDDSNVIKISVKKPKFFKYQVGAFAYMYFLS 450
Query: 305 PKVS---KLQWHPFTVTSSCNMETN---YLSVAIK-NVGSWSNKLYQELSS-SSLDHLNI 356
PK + Q HPFTV S + + N L++ +K N G L + LS+ + I
Sbjct: 451 PKSAWFYSFQSHPFTVLSERHRDPNNPDQLTMYVKANKGITRVLLSKVLSAPNHTVDCKI 510
Query: 357 SVEGPYG---PHTAQFLRHEQIVMVSGGSGV 384
+EGPYG PH A+ R+ +V V+ G GV
Sbjct: 511 FLEGPYGVTVPHIAKLKRN--LVGVAAGLGV 539
>FRE3_YEAST (Q08905) Ferric reductase transmembrane component 3
precursor (EC 1.16.1.7) (Ferric-chelate reductase 3)
Length = 711
Score = 45.4 bits (106), Expect = 5e-04
Identities = 57/254 (22%), Positives = 112/254 (43%), Gaps = 20/254 (7%)
Query: 141 GLTSESSIKYHIWLGQLSMVLFAAHSIGFIIYWAITNQMIEMLEW--SKTYVSNVAGEIA 198
G+ S I +H WLG++ + H A T+ + +W SK G A
Sbjct: 306 GVKYTSFIMFHKWLGRMMFLDAVIHGA------AYTSYSVFYKDWAASKEETYWQFGVAA 359
Query: 199 SLIALAMWITSIPQIRRKMYEVFFYTHHLYILYILFFAI----HAGVGSMCVIAPGVFLF 254
I M S+ R+ YE F + H +L LFF + + I + ++
Sbjct: 360 LCIVGVMVFFSLAMFRKFFYEAFLFLH--IVLGALFFYTCWEHVVELSGIEWIYAAIAIW 417
Query: 255 LIDRHLRFLQ-SRQHARLLSARLLPCDALELNFSKNPSLY-YNPTSLVFIN-VPKVSKLQ 311
IDR +R ++ S S +L+ D + + + L+ P VF++ + + Q
Sbjct: 418 TIDRLIRIVRVSYFGFPKASLQLVGDDIIRVTVKRPVRLWKAKPGQYVFVSFLHHLYFWQ 477
Query: 312 WHPFTVTSSCNMETNYLSVAIKNVGSWSNKLYQEL-SSSSLDHLNISVEGPYGPHTAQFL 370
HPFTV S ++ L++ +K + + + + + + +++EGPYG ++
Sbjct: 478 SHPFTVLDSI-IKDGELTIILKEKKGVTKLVKKYVCCNGGKASMRLAIEGPYG-SSSPVN 535
Query: 371 RHEQIVMVSGGSGV 384
++ +++++GG+G+
Sbjct: 536 NYDNVLLLTGGTGL 549
>HMP_VIBPA (P40609) Flavohemoprotein (Hemoglobin-like protein)
(Flavohemoglobin) (Nitric oxide dioxygenase) (EC
1.14.12.17) (NO oxygenase) (NOD)
Length = 394
Score = 39.7 bits (91), Expect = 0.025
Identities = 24/95 (25%), Positives = 45/95 (47%), Gaps = 4/95 (4%)
Query: 300 VFINVPKVSKLQWHPFTVTSSCNMETNYLSVAIKNVGSWSNKLYQELSSSSLDHLNISVE 359
++IN K + ++++SS T +SV + G SN L+ EL+ + +
Sbjct: 191 IYINSDKFENQEIRQYSLSSSVQENTYRISVKREQGGKVSNYLHDELNIGD----KVKLA 246
Query: 360 GPYGPHTAQFLRHEQIVMVSGGSGVTPFISIIRDL 394
P G + +V++S G G+TP +S++ L
Sbjct: 247 APAGDFFMDVDTNTPVVLISAGVGLTPTLSMLESL 281
>NIA_BETVE (P27783) Nitrate reductase [NAD(P)H] (EC 1.7.1.2) (NR)
Length = 898
Score = 38.1 bits (87), Expect = 0.073
Identities = 42/144 (29%), Positives = 65/144 (44%), Gaps = 26/144 (18%)
Query: 300 VFINVPKVSKLQWHPFTVTSSCNMETNYLSVAIKNVGSWSNKLYQE--LSSSSLDHLNIS 357
VF+ KL +T TS+ + E YL + +K SN + L S LD L I
Sbjct: 681 VFLCATIDDKLCMRAYTPTSTID-EVGYLDLVVKIYFKNSNPRFPNGGLMSQHLDSLPIG 739
Query: 358 ----VEGPYG----PHTAQFLRH------EQIVMVSGGSGVTPFISIIRDLIFQSQQQEF 403
V+GP G FL H +++ MV+GG+G+TP +I+ I + + E
Sbjct: 740 SVLHVKGPLGHVEYTGRGNFLVHGEPKFAKRLAMVAGGTGITPIYQVIQ-AILKDPEDET 798
Query: 404 QPPKLLLVCIFKNYADLTMLDLML 427
+ +F YA+ T D++L
Sbjct: 799 E--------MFVVYANRTEDDILL 814
>HMP_RHIME (Q7WUM8) Flavohemoprotein (Hemoglobin-like protein)
(Flavohemoglobin) (Nitric oxide dioxygenase) (EC
1.14.12.17) (NO oxygenase) (NOD)
Length = 403
Score = 38.1 bits (87), Expect = 0.073
Identities = 34/129 (26%), Positives = 60/129 (46%), Gaps = 13/129 (10%)
Query: 300 VFINVPKVSKLQWHPFTVTSSCNMETNYLSVAIKNVGSWSNKLYQELSSSSLDHLNISVE 359
V + VPK+ Q ++++ S N + +SV ++ G + +SS D +N+ E
Sbjct: 193 VAVQVPKLGYQQIRQYSLSDSPNGRSYRISVKREDGGLGTPGY---VSSLLHDEINVGDE 249
Query: 360 ----GPYGPHTAQFLRHEQIVMVSGGSGVTPFISIIRDLIFQSQQQEFQPPKLLLVCIFK 415
PYG IV++SGG G+TP +S+++ + + P K++ V +
Sbjct: 250 LKLAAPYGNFYIDVSATTPIVLISGGVGLTPMVSMLKKAL------QTPPRKVVFVHGAR 303
Query: 416 NYADLTMLD 424
N A M D
Sbjct: 304 NSAVHAMRD 312
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.329 0.140 0.449
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 75,984,036
Number of Sequences: 164201
Number of extensions: 3079009
Number of successful extensions: 9411
Number of sequences better than 10.0: 64
Number of HSP's better than 10.0 without gapping: 14
Number of HSP's successfully gapped in prelim test: 50
Number of HSP's that attempted gapping in prelim test: 9351
Number of HSP's gapped (non-prelim): 81
length of query: 652
length of database: 59,974,054
effective HSP length: 117
effective length of query: 535
effective length of database: 40,762,537
effective search space: 21807957295
effective search space used: 21807957295
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 69 (31.2 bits)
Medicago: description of AC121237.9


