

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121237.18 - phase: 0
(387 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
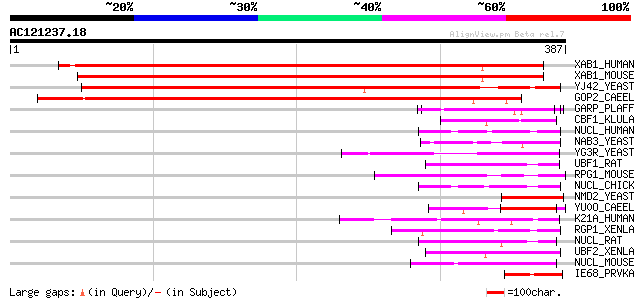
Score E
Sequences producing significant alignments: (bits) Value
XAB1_HUMAN (Q9HCN4) XPA-binding protein 1 (HUSSY-23) 362 e-100
XAB1_MOUSE (Q8VCE2) XPA-binding protein 1 358 2e-98
YJ42_YEAST (P47122) Hypothetical 43.2 kDa protein in HAM1-PEM2 i... 328 1e-89
GOP2_CAEEL (P46577) Gro-1 operon protein 2 320 4e-87
GARP_PLAFF (P13816) Glutamic acid-rich protein precursor 54 9e-07
CBF1_KLULA (P49379) Centromere-binding protein 1 (CBP-1) (Centro... 47 8e-05
NUCL_HUMAN (P19338) Nucleolin (Protein C23) 46 1e-04
NAB3_YEAST (P38996) Nuclear polyadenylated RNA-binding protein 3 46 1e-04
YG3R_YEAST (P53288) Hypothetical 22.2 kDa protein in NSR1-TIF463... 45 2e-04
UBF1_RAT (P25977) Nucleolar transcription factor 1 (Upstream bin... 45 2e-04
RPG1_MOUSE (Q9EPQ2) X-linked retinitis pigmentosa GTPase regulat... 45 2e-04
NUCL_CHICK (P15771) Nucleolin (Protein C23) 45 3e-04
NMD2_YEAST (P38798) Nonsense-mediated mRNA decay protein 2 (Up-f... 45 3e-04
YU0O_CAEEL (Q19753) Hypothetical protein F23B12.7 in chromosome V 45 4e-04
K21A_HUMAN (Q7Z4S6) Kinesin family member 21A (Kinesin-like prot... 45 4e-04
RGP1_XENLA (O13066) Ran GTPase-activating protein 1 44 5e-04
NUCL_RAT (P13383) Nucleolin (Protein C23) 44 5e-04
UBF2_XENLA (P25980) Nucleolar transcription factor 2 (Upstream b... 44 7e-04
NUCL_MOUSE (P09405) Nucleolin (Protein C23) 44 7e-04
IE68_PRVKA (P24827) Immediate-early protein RSP40 44 7e-04
>XAB1_HUMAN (Q9HCN4) XPA-binding protein 1 (HUSSY-23)
Length = 374
Score = 362 bits (929), Expect = e-100
Identities = 186/348 (53%), Positives = 240/348 (68%), Gaps = 13/348 (3%)
Query: 35 EGSSSGSPNFKRKPVIIIVVGMAGSGKTTLMHRLVTHTHLSNIRGYVMNLDPAVMTLPYA 94
E +SG P R PV ++V+GMAGSGKTT + RL H H YV+NLDPAV +P+
Sbjct: 9 ELQASGGP---RHPVCLLVLGMAGSGKTTFVQRLTGHLHAQGTPPYVINLDPAVHEVPFP 65
Query: 95 SNIDIRDTVKYKEVMKQFNLGPNGGILTSLNLFATKFDEVISVIEKRADQLDYVLVDTPG 154
+NIDIRDTVKYKEVMKQ+ LGPNGGI+TSLNLFAT+FD+V+ IEK + YVL+DTPG
Sbjct: 66 ANIDIRDTVKYKEVMKQYGLGPNGGIVTSLNLFATRFDQVMKFIEKAQNMSKYVLIDTPG 125
Query: 155 QIEIFTWSASGAIITEAFASTFPTVVAYVVDTPRAENPTTFMSNMLYACSILYKTRLPLI 214
QIE+FTWSASG IITEA AS+FPTVV YV+DT R+ NP TFMSNMLYACSILYKT+LP I
Sbjct: 126 QIEVFTWSASGTIITEALASSFPTVVIYVMDTSRSTNPVTFMSNMLYACSILYKTKLPFI 185
Query: 215 LAFNKVDVAQHEFALEWMKDFEVFQAAASSDQSYTSNLTQSLSLALDEFYSNLRSAGVSA 274
+ NK D+ H FA+EWM+DFE FQ A + + +Y SNLT+S+SL LDEFYS+LR GVSA
Sbjct: 186 VVMNKTDIIDHSFAVEWMQDFEAFQDALNQETTYVSNLTRSMSLVLDEFYSSLRVVGVSA 245
Query: 275 VTGAGIEGFFKAVEASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREM------ 328
V G G++ F V ++AEEY Y+ + ++ ++ E +++E +++LR++M
Sbjct: 246 VLGTGLDELFVQVTSAAEEYEREYRPEYERLKKSLANAESQQQREQLERLRKDMGSVALD 305
Query: 329 ---EKSGGETVVLSTGLKNKEEDEDEEDEEMDDD-DNVDFGTYTEDED 372
K V+ + L DEEDEE D D D++D E +
Sbjct: 306 AGTAKDSLSPVLHPSDLILTRGTLDEEDEEADSDTDDIDHRVTEESHE 353
>XAB1_MOUSE (Q8VCE2) XPA-binding protein 1
Length = 372
Score = 358 bits (918), Expect = 2e-98
Identities = 181/335 (54%), Positives = 233/335 (69%), Gaps = 10/335 (2%)
Query: 48 PVIIIVVGMAGSGKTTLMHRLVTHTHLSNIRGYVMNLDPAVMTLPYASNIDIRDTVKYKE 107
PV ++V+GMAGSGKTT + RL H H YV+NLDPAV +P+ +NIDIRDTVKYKE
Sbjct: 19 PVCLLVLGMAGSGKTTFVQRLTGHLHNKGCPPYVINLDPAVHEVPFPANIDIRDTVKYKE 78
Query: 108 VMKQFNLGPNGGILTSLNLFATKFDEVISVIEKRADQLDYVLVDTPGQIEIFTWSASGAI 167
VMKQ+ LGPNGGI+TSLNLFAT+FD+V+ IEK + YVL+DTPGQIE+FTWSASG I
Sbjct: 79 VMKQYGLGPNGGIVTSLNLFATRFDQVMKFIEKAQNTFRYVLIDTPGQIEVFTWSASGTI 138
Query: 168 ITEAFASTFPTVVAYVVDTPRAENPTTFMSNMLYACSILYKTRLPLILAFNKVDVAQHEF 227
ITEA AS+FPTVV YV+DT R+ NP TFMSNMLYACSILYKT+LP I+ NK D+ H F
Sbjct: 139 ITEALASSFPTVVIYVMDTSRSTNPVTFMSNMLYACSILYKTKLPFIVVMNKTDIIDHSF 198
Query: 228 ALEWMKDFEVFQAAASSDQSYTSNLTQSLSLALDEFYSNLRSAGVSAVTGAGIEGFFKAV 287
A+EWM+DFE FQ A + + +Y SNLT+S+SL LDEFYS+LR GVSAV G G + V
Sbjct: 199 AVEWMQDFEAFQDALNQETTYVSNLTRSMSLVLDEFYSSLRVVGVSAVVGTGFDELCTQV 258
Query: 288 EASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREM---------EKSGGETVVL 338
++AEEY Y+ + ++ ++ + N++KE +++LR++M K V+
Sbjct: 259 TSAAEEYEREYRPEYERLKKSLANAQSNQQKEQLERLRKDMGSVALDPEAGKGNASPVLD 318
Query: 339 STGLKNKEEDEDEEDEEMDDD-DNVDFGTYTEDED 372
+ L DEEDEE D D D++D E +
Sbjct: 319 PSDLILTRGTLDEEDEEADSDTDDIDHRVTEESRE 353
>YJ42_YEAST (P47122) Hypothetical 43.2 kDa protein in HAM1-PEM2
intergenic region
Length = 385
Score = 328 bits (842), Expect = 1e-89
Identities = 167/344 (48%), Positives = 231/344 (66%), Gaps = 25/344 (7%)
Query: 51 IIVVGMAGSGKTTLMHRLVTHTHLSNIRGYVMNLDPAVMTLPYASNIDIRDTVKYKEVMK 110
II +GMAGSGKTT M RL +H YV+NLDPAV+ +PY +NIDIRD++KYK+VM+
Sbjct: 6 IICIGMAGSGKTTFMQRLNSHLRAEKTPPYVINLDPAVLRVPYGANIDIRDSIKYKKVME 65
Query: 111 QFNLGPNGGILTSLNLFATKFDEVISVIEKRADQLDYVLVDTPGQIEIFTWSASGAIITE 170
+ LGPNG I+TSLNLF+TK D+VI ++E++ D+ ++DTPGQIE F WSASGAIITE
Sbjct: 66 NYQLGPNGAIVTSLNLFSTKIDQVIRLVEQKKDKFQNCIIDTPGQIECFVWSASGAIITE 125
Query: 171 AFASTFPTVVAYVVDTPRAENPTTFMSNMLYACSILYKTRLPLILAFNKVDVAQHEFALE 230
+FAS+FPTV+AY+VDTPR +PTTFMSNMLYACSILYKT+LP+I+ FNK DV + +FA E
Sbjct: 126 SFASSFPTVIAYIVDTPRNSSPTTFMSNMLYACSILYKTKLPMIVVFNKTDVCKADFAKE 185
Query: 231 WMKDFEVFQAAASSDQ----------SYTSNLTQSLSLALDEFYSNLRSAGVSAVTGAGI 280
WM DFE FQAA DQ Y S+L S+SL L+EFYS L GVS+ TG G
Sbjct: 186 WMTDFESFQAAIKEDQDLNGDNGLGSGYMSSLVNSMSLMLEEFYSQLDVVGVSSFTGDGF 245
Query: 281 EGFFKAVEASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLST 340
+ F + V+ +EY + YK + +K K+ EE R++++++ L +++
Sbjct: 246 DEFMQCVDKKVDEYDQYYKQEREKALNLKKKKEEMRKQKSLNGLMKDL------------ 293
Query: 341 GLKNKEEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVDK 384
GL K +++ +D +++ ++ +D DEDE V++
Sbjct: 294 GLNEKSSAAASDNDSIDAISDLE---EDANDGLVDRDEDEGVER 334
>GOP2_CAEEL (P46577) Gro-1 operon protein 2
Length = 355
Score = 320 bits (820), Expect = 4e-87
Identities = 171/352 (48%), Positives = 228/352 (64%), Gaps = 15/352 (4%)
Query: 20 KQKEELSESMKKLDIEGSSSGSPNFKRKPVIIIVVGMAGSGKTTLMHRLVTHTHLSNIRG 79
++ E L S + E S PN +KP I+ V+GMAGSGKTT + RL H
Sbjct: 3 EKAENLPSSSAEASEEPSPQTGPNVNQKPSIL-VLGMAGSGKTTFVQRLTAFLHARKTPP 61
Query: 80 YVMNLDPAVMTLPYASNIDIRDTVKYKEVMKQFNLGPNGGILTSLNLFATKFDEVISVIE 139
YV+NLDPAV +PY N+DIRDTVKYKEVMK+F +GPNG I+T LNL T+FD+VI +I
Sbjct: 62 YVINLDPAVSKVPYPVNVDIRDTVKYKEVMKEFGMGPNGAIMTCLNLMCTRFDKVIELIN 121
Query: 140 KRADQLDYVLVDTPGQIEIFTWSASGAIITEAFASTFPTVVAYVVDTPRAENPTTFMSNM 199
KR+ L+DTPGQIE FTWSASG+IIT++ AS+ PTVV Y+VD+ RA NPTTFMSNM
Sbjct: 122 KRSSDFSVCLLDTPGQIEAFTWSASGSIITDSLASSHPTVVMYIVDSARATNPTTFMSNM 181
Query: 200 LYACSILYKTRLPLILAFNKVDVAQHEFALEWMKDFEVF-QAAASSDQSYTSNLTQSLSL 258
LYACSILY+T+LP I+ FNK D+ + FAL+WM+DFE F +A + SY ++L++SLSL
Sbjct: 182 LYACSILYRTKLPFIVVFNKADIVKPTFALKWMQDFERFDEALEDARSSYMNDLSRSLSL 241
Query: 259 ALDEFYSNLRSAGVSAVTGAGIEGFFKAVEASAEEYMETYKADLDKRREEKQLLEENRRK 318
LDEFY L++ VS+ TG G E A++ S E Y + Y +K EK+LL+E RK
Sbjct: 242 VLDEFYCGLKTVCVSSATGEGFEDVMTAIDESVEAYKKEYVPMYEKVLAEKKLLDEEERK 301
Query: 319 ENMD-----KLRREMEKSGGETVVLSTGLKNK--------EEDEDEEDEEMD 357
+ + K ++ K L + L +K ++E+EED E++
Sbjct: 302 KRDEETLKGKAVHDLNKVANPDEFLESELNSKIDRIHLGGVDEENEEDAELE 353
>GARP_PLAFF (P13816) Glutamic acid-rich protein precursor
Length = 678
Score = 53.5 bits (127), Expect = 9e-07
Identities = 31/97 (31%), Positives = 55/97 (55%), Gaps = 2/97 (2%)
Query: 285 KAVEASAEEYMETYKADLDKRREEKQLLEENRR-KENMDKLRREMEKSGGETVVLSTGLK 343
K EA ++ K + DK+ E K++ EE++ +E+ +++ + E+ E +
Sbjct: 533 KIEEAELQKQKHVDKEE-DKKEESKEVQEESKEVQEDEEEVEEDEEEEEEEEEEEEEEEE 591
Query: 344 NKEEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDE 380
+EE+E+EE+EE +D+D D EDED +EDED+
Sbjct: 592 EEEEEEEEEEEEEEDEDEEDEDDAEEDEDDAEEDEDD 628
Score = 48.9 bits (115), Expect = 2e-05
Identities = 35/104 (33%), Positives = 53/104 (50%), Gaps = 4/104 (3%)
Query: 285 KAVEASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKN 344
K V+ EE E + + ++ EE++ EE +E ++ E E E +
Sbjct: 563 KEVQEDEEEVEEDEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEDEDEEDEDDA-EEDEDD 621
Query: 345 KEEDED--EEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVDKFS 386
EEDED EED++ +DDD D EDED DE+E+EE ++ S
Sbjct: 622 AEEDEDDAEEDDDEEDDDEED-DDEDEDEDEEDEEEEEEEEEES 664
Score = 47.8 bits (112), Expect = 5e-05
Identities = 30/106 (28%), Positives = 53/106 (49%), Gaps = 12/106 (11%)
Query: 288 EASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKEE 347
E EE E + + + +E+++ E+ +E ++ E E+ E + +EE
Sbjct: 549 EDKKEESKEVQEESKEVQEDEEEVEEDEEEEEEEEEEEEEEEEEEEEE---EEEEEEEEE 605
Query: 348 DEDEEDEE---------MDDDDNVDFGTYTEDEDAIDEDEDEEVDK 384
DEDEEDE+ +D+D+ + ED+D D+DEDE+ D+
Sbjct: 606 DEDEEDEDDAEEDEDDAEEDEDDAEEDDDEEDDDEEDDDEDEDEDE 651
Score = 43.9 bits (102), Expect = 7e-04
Identities = 26/97 (26%), Positives = 51/97 (51%), Gaps = 7/97 (7%)
Query: 288 EASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKEE 347
E EE E + + ++ EE++ EE ++ D+ E ++ E + EE
Sbjct: 577 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEDEDEEDEDDAEEDEDDAEEDEDDAEEDDDEE 636
Query: 348 DEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVDK 384
D+DEED++ D+D++ E+++ +E+E+EE +K
Sbjct: 637 DDDEEDDDEDEDED-------EEDEEEEEEEEEESEK 666
Score = 41.6 bits (96), Expect = 0.003
Identities = 24/85 (28%), Positives = 49/85 (57%), Gaps = 6/85 (7%)
Query: 299 KADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKEEDEDEEDEEMDD 358
+A+L K++ + EE++++E+ + E E V + +EE+E+EE+EE ++
Sbjct: 536 EAELQKQKHVDK--EEDKKEESKEVQEESKEVQEDEEEVEEDEEEEEEEEEEEEEEEEEE 593
Query: 359 DDNVDFGTYTEDEDAIDEDEDEEVD 383
++ + E+E+ DEDE++E D
Sbjct: 594 EEEEE----EEEEEEEDEDEEDEDD 614
Score = 37.0 bits (84), Expect = 0.085
Identities = 22/86 (25%), Positives = 50/86 (57%), Gaps = 1/86 (1%)
Query: 299 KADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKEEDEDEEDEEMDD 358
+ D K++ K E ++++++DK + E+S E S ++ EE+ +E++EE ++
Sbjct: 523 RRDNHKKKMAKIEEAELQKQKHVDKEEDKKEESK-EVQEESKEVQEDEEEVEEDEEEEEE 581
Query: 359 DDNVDFGTYTEDEDAIDEDEDEEVDK 384
++ + E+E+ +E+E+EE D+
Sbjct: 582 EEEEEEEEEEEEEEEEEEEEEEEEDE 607
Score = 31.2 bits (69), Expect = 4.7
Identities = 27/92 (29%), Positives = 41/92 (44%), Gaps = 10/92 (10%)
Query: 285 KAVEASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKN 344
K E +E +E K +++ ++KQ E RK+ K R++ EK + + K
Sbjct: 268 KEKEMKEQEKIEKKKKKQEEKEKKKQ---EKERKKQEKKERKQKEKEMKKQKKIEKERKK 324
Query: 345 KEEDE------DEEDEE-MDDDDNVDFGTYTE 369
KEE E D+E+EE M D T E
Sbjct: 325 KEEKEKKKKKHDKENEETMQQPDQTSEETNNE 356
>CBF1_KLULA (P49379) Centromere-binding protein 1 (CBP-1)
(Centromere-binding factor 1) (Centromere promoter
factor 1)
Length = 359
Score = 47.0 bits (110), Expect = 8e-05
Identities = 30/88 (34%), Positives = 48/88 (54%), Gaps = 8/88 (9%)
Query: 301 DLDKRREEKQLLEENRRKENMDKLRREMEKS------GGETVVLSTGLKNKEEDEDEEDE 354
D+ K E Q E++ +N + + +++ S + +++EDEDEE+E
Sbjct: 72 DVVKGESEGQAAEQSSADQNDENINVDVDPSVTAVARAAQHASQRREAGDEDEDEDEEEE 131
Query: 355 EMDDDDNVDFGTYTEDEDA-IDEDEDEE 381
E D+DD+VD +D DA IDED+DEE
Sbjct: 132 E-DEDDHVDIDDVDKDPDAVIDEDDDEE 158
>NUCL_HUMAN (P19338) Nucleolin (Protein C23)
Length = 706
Score = 46.2 bits (108), Expect = 1e-04
Identities = 33/99 (33%), Positives = 54/99 (54%), Gaps = 11/99 (11%)
Query: 286 AVEASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNK 345
A AS +E E + D D +E E++ +E M+ + +K+ V+ KN
Sbjct: 179 AAPASEDEDDEDDEDDEDDDDDE----EDDSEEEAMETTPAKGKKAAK---VVPVKAKNV 231
Query: 346 EEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVDK 384
EDEDEE+++ D+DD+ D +DED DED++EE ++
Sbjct: 232 AEDEDEEEDDEDEDDDDD----EDDEDDDDEDDEEEEEE 266
Score = 41.2 bits (95), Expect = 0.005
Identities = 22/84 (26%), Positives = 45/84 (53%)
Query: 301 DLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKEEDEDEEDEEMDDDD 360
D D +E +++ +++ ++ E + G+ +K K EDE++EE D+D+
Sbjct: 185 DEDDEDDEDDEDDDDDEEDDSEEEAMETTPAKGKKAAKVVPVKAKNVAEDEDEEEDDEDE 244
Query: 361 NVDFGTYTEDEDAIDEDEDEEVDK 384
+ D ED+D D++E+EE ++
Sbjct: 245 DDDDDEDDEDDDDEDDEEEEEEEE 268
Score = 38.9 bits (89), Expect = 0.022
Identities = 30/136 (22%), Positives = 57/136 (41%), Gaps = 25/136 (18%)
Query: 271 GVSAVTGAGIEGFFKAVEASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREMEK 330
G + V G +G A+ A + + K + E+ E+ E+ D+ E+E
Sbjct: 114 GKALVATPGKKG--AAIPAKGAKNGKNAKKEDSDEEEDDDSEEDEEDDEDEDEDEDEIEP 171
Query: 331 SGGETVVLSTGLKNKEEDEDEEDEEMDDDDN-----------------------VDFGTY 367
+ + + +++++++DE+DE+ DDD+ V
Sbjct: 172 AAMKAAAAAPASEDEDDEDDEDDEDDDDDEEDDSEEEAMETTPAKGKKAAKVVPVKAKNV 231
Query: 368 TEDEDAIDEDEDEEVD 383
EDED ++DEDE+ D
Sbjct: 232 AEDEDEEEDDEDEDDD 247
Score = 33.5 bits (75), Expect = 0.94
Identities = 16/44 (36%), Positives = 27/44 (61%), Gaps = 7/44 (15%)
Query: 341 GLKNKEEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVDK 384
G KN + + E+ +E +DDD+ E+++ DEDEDE+ D+
Sbjct: 132 GAKNGKNAKKEDSDEEEDDDS-------EEDEEDDEDEDEDEDE 168
Score = 33.1 bits (74), Expect = 1.2
Identities = 15/37 (40%), Positives = 25/37 (67%), Gaps = 8/37 (21%)
Query: 346 EEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEV 382
E+D+D+ED+E DDD+ +DE+ +E+E+E V
Sbjct: 244 EDDDDDEDDEDDDDE--------DDEEEEEEEEEEPV 272
>NAB3_YEAST (P38996) Nuclear polyadenylated RNA-binding protein 3
Length = 802
Score = 46.2 bits (108), Expect = 1e-04
Identities = 32/103 (31%), Positives = 54/103 (52%), Gaps = 16/103 (15%)
Query: 287 VEASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKE 346
+E A E E +A+ ++ EE+ LE+ +E DK E + GE + N E
Sbjct: 39 IEFDAPE--EEREAEREEENEEQHELEDVNDEEEEDK--EEKGEENGEVI-------NTE 87
Query: 347 EDEDEEDEEM-----DDDDNVDFGTYTEDEDAIDEDEDEEVDK 384
E+E+EE ++ DDDDN + ED+D D+D+D++ ++
Sbjct: 88 EEEEEEHQQKGGNDDDDDDNEEEEEEEEDDDDDDDDDDDDEEE 130
Score = 43.5 bits (101), Expect = 0.001
Identities = 39/144 (27%), Positives = 75/144 (52%), Gaps = 12/144 (8%)
Query: 244 SDQSYTSNLTQ--SLSLALDEFYSNLRSAGVSAVTGAGIEGFFKAVEASAEEYMETYKA- 300
SD+++ S++ S L++D SN ++ GIE E AE E +
Sbjct: 2 SDENHNSDVQDIPSPELSVDS-NSNENELMNNSSADDGIEFDAPEEEREAEREEENEEQH 60
Query: 301 DLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKEEDEDEEDEEMDDDD 360
+L+ +E++ +E + +EN + + E E+ E G + ++D++EE+EE ++DD
Sbjct: 61 ELEDVNDEEEEDKEEKGEENGEVINTEEEE---EEEHQQKGGNDDDDDDNEEEEEEEEDD 117
Query: 361 NVDFGTYTEDEDAIDEDEDEEVDK 384
+ D +D+D DE+E+EE ++
Sbjct: 118 DDD-----DDDDDDDEEEEEEEEE 136
Score = 34.3 bits (77), Expect = 0.55
Identities = 26/100 (26%), Positives = 48/100 (48%), Gaps = 4/100 (4%)
Query: 288 EASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKEE 347
E EE E ++ EE++ ++ ++ D E E+ + + EE
Sbjct: 70 EEDKEEKGEENGEVINTEEEEEEEHQQKGGNDDDDDDNEEEEEEEEDDDDDDDDDDDDEE 129
Query: 348 DEDEEDEEMDDDDNVDFGTYTED----EDAIDEDEDEEVD 383
+E+EE+EE +D+ +V + ED ED D+ +D+EV+
Sbjct: 130 EEEEEEEEGNDNSSVGSDSAAEDGEDEEDKKDKTKDKEVE 169
>YG3R_YEAST (P53288) Hypothetical 22.2 kDa protein in NSR1-TIF4631
intergenic region
Length = 203
Score = 45.4 bits (106), Expect = 2e-04
Identities = 38/152 (25%), Positives = 65/152 (42%), Gaps = 30/152 (19%)
Query: 232 MKDFEVFQAAASSDQSYTSNLTQSLSLALDEFYSNLRSAGVSAVTGAGIEGFFKAVEASA 291
++D ++ASSD + S L S SL+L E S + + + F + +S+
Sbjct: 40 LEDSSSSSSSASSDLRFLS-LDSSFSLSLSEDEDEDESELEDSFDSSFLVSSFSSSSSSS 98
Query: 292 EEYMETYKADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKEEDEDE 351
EE E E E+S + ++S L E+DE+E
Sbjct: 99 EEESE-----------------------------EEEEESLDSSFLVSASLSLSEDDEEE 129
Query: 352 EDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVD 383
+ E D+D++ D + ++ + DEDEDE+ D
Sbjct: 130 DSESEDEDEDEDSDSDSDSDSDSDEDEDEDED 161
>UBF1_RAT (P25977) Nucleolar transcription factor 1 (Upstream
binding factor 1) (UBF-1)
Length = 764
Score = 45.4 bits (106), Expect = 2e-04
Identities = 32/94 (34%), Positives = 45/94 (47%), Gaps = 8/94 (8%)
Query: 291 AEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLS-TGLKNKEEDE 349
AEE YK LD + + KE + R+ M K G S T L++K E E
Sbjct: 619 AEEQQRQYKVHLDLWVKSLSPQDRAAYKEYISNKRKNMTKLRGPNPKSSRTTLQSKSESE 678
Query: 350 DEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVD 383
+++DEE DDDD+ E+E+ DE+ D D
Sbjct: 679 EDDDEEDDDDDD-------EEEEEDDENGDSSED 705
Score = 36.6 bits (83), Expect = 0.11
Identities = 31/134 (23%), Positives = 62/134 (46%), Gaps = 22/134 (16%)
Query: 248 YTSNLTQSLSLALDEFYSNLRSAGVSAVTGAGIEGFFKAVEASAEEYMETYKADLDKRRE 307
+ +L+ A E+ SN R ++ + G + +++ +E + + D D E
Sbjct: 633 WVKSLSPQDRAAYKEYISNKRK-NMTKLRGPNPKSSRTTLQSKSESEEDDDEEDDDDDDE 691
Query: 308 EKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKEEDEDEEDEEMDDDDNVDFGTY 367
E++ +EN + + GG++ ++ EDE E+ +E +DDD+ +
Sbjct: 692 EEEEDDENG----------DSSEDGGDSS------ESSSEDESEDGDENEDDDDDE---- 731
Query: 368 TEDEDAIDEDEDEE 381
+D++ DEDED E
Sbjct: 732 -DDDEDDDEDEDNE 744
>RPG1_MOUSE (Q9EPQ2) X-linked retinitis pigmentosa GTPase
regulator-interacting protein 1 (RPGR-interacting
protein 1)
Length = 1331
Score = 45.4 bits (106), Expect = 2e-04
Identities = 35/133 (26%), Positives = 62/133 (46%), Gaps = 18/133 (13%)
Query: 255 SLSLALDEFYSNLRSAGVSAVTGAGIEGFFKAVEASAEEYMETYKADLDKRREEKQLLEE 314
S+ + LD L G EG K E EE E + ++ EE++ ++E
Sbjct: 882 SIKVQLDWKSHYLAPEGFQMSEAEKPEGEEKEEEGGEEEVKEEEVEEEEEEEEEEEEVKE 941
Query: 315 NRRKENMDKLRREMEKSGGETVVLSTGLKNKEEDEDEEDEEMDDDDNVDFGTYTEDEDAI 374
+ +E ++ E EK K +EE+EDE++EE +++ E+E+
Sbjct: 942 EKEEEEEEEREEEEEKEEE---------KEEEEEEDEKEEEEEEE---------EEEEEE 983
Query: 375 DEDEDEEVDKFSF 387
+EDE+++V + SF
Sbjct: 984 EEDENKDVLEASF 996
Score = 38.9 bits (89), Expect = 0.022
Identities = 29/103 (28%), Positives = 51/103 (49%), Gaps = 4/103 (3%)
Query: 280 IEGFFKAVEASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLR-REMEKSGGETVVL 338
I+G F + S E+ + K LD + L E + +K E E+ GGE V
Sbjct: 866 IKGDFNLTD-SGEKSNGSIKVQLDWKSH--YLAPEGFQMSEAEKPEGEEKEEEGGEEEVK 922
Query: 339 STGLKNKEEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEE 381
++ +EE+E+EE+E ++ + + E+E+ +E E+EE
Sbjct: 923 EEEVEEEEEEEEEEEEVKEEKEEEEEEEREEEEEKEEEKEEEE 965
>NUCL_CHICK (P15771) Nucleolin (Protein C23)
Length = 694
Score = 45.1 bits (105), Expect = 3e-04
Identities = 34/99 (34%), Positives = 49/99 (49%), Gaps = 14/99 (14%)
Query: 286 AVEASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNK 345
AV + +E E + D ++ EE ++ E MD ++K T +T K K
Sbjct: 163 AVVPAKQESEEEEEEDDEEEDEE----DDESEDEAMDTTPAPVKKP---TPAKATPAKAK 215
Query: 346 EEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVDK 384
E EDEEDEE +D+D EDED +EDE+E D+
Sbjct: 216 AESEDEEDEEDEDEDE-------EDEDDEEEDEEESEDE 247
Score = 37.0 bits (84), Expect = 0.085
Identities = 20/57 (35%), Positives = 31/57 (54%), Gaps = 4/57 (7%)
Query: 325 RREMEKSGGETVVLSTGLKNKEEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEE 381
++ + S + V+ G KN + + EE EE D+DD D EDED +E ++EE
Sbjct: 88 KKAVAPSPKKAAVVGKGAKNGKNAKKEESEEEDEDDEDD----EEDEDEEEESDEEE 140
Score = 35.8 bits (81), Expect = 0.19
Identities = 28/111 (25%), Positives = 50/111 (44%), Gaps = 13/111 (11%)
Query: 273 SAVTGAGIEGFFKAVEASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREMEKSG 332
+AV G G + A EE E + D D +E + E + +E ++ +KS
Sbjct: 98 AAVVGKGAKN---GKNAKKEESEEEDEDDEDDEEDEDEEEESDEEEEPAVPVKPAAKKSA 154
Query: 333 GETVVLSTGLKNKEEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVD 383
+ +++ +EE+EE D+++ ++ED DE EDE +D
Sbjct: 155 AAVPAKKPAVVPAKQESEEEEEEDDEEE--------DEED--DESEDEAMD 195
>NMD2_YEAST (P38798) Nonsense-mediated mRNA decay protein 2
(Up-frameshift suppressor 2)
Length = 1089
Score = 45.1 bits (105), Expect = 3e-04
Identities = 18/43 (41%), Positives = 33/43 (75%)
Query: 344 NKEEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVDKFS 386
++++DED++D++ DDDD+ + G +DED DED+D+E ++ S
Sbjct: 891 DEDDDEDDDDDDDDDDDDGEEGDEDDDEDDDDEDDDDEEEEDS 933
Score = 38.9 bits (89), Expect = 0.022
Identities = 17/39 (43%), Positives = 27/39 (68%)
Query: 345 KEEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVD 383
++E EDE+DE+ D+DD+ D +D + DED+DE+ D
Sbjct: 883 QDESEDEDDEDDDEDDDDDDDDDDDDGEEGDEDDDEDDD 921
Score = 35.8 bits (81), Expect = 0.19
Identities = 21/75 (28%), Positives = 38/75 (50%), Gaps = 4/75 (5%)
Query: 313 EENRRKENMDKLRREMEKSGGETVVLST----GLKNKEEDEDEEDEEMDDDDNVDFGTYT 368
+E+ EN D + E E +T G ++DE E++++ DDD++ D
Sbjct: 845 DEDEDDENDDGVDLLGEDEDAEISTPNTESAPGKHQAKQDESEDEDDEDDDEDDDDDDDD 904
Query: 369 EDEDAIDEDEDEEVD 383
+D+D + DED++ D
Sbjct: 905 DDDDGEEGDEDDDED 919
Score = 32.3 bits (72), Expect = 2.1
Identities = 13/38 (34%), Positives = 25/38 (65%)
Query: 346 EEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVD 383
+ED+DE+D++ DDDD + + ++ E D D D +++
Sbjct: 913 DEDDDEDDDDEDDDDEEEEDSDSDLEYGGDLDADRDIE 950
Score = 32.0 bits (71), Expect = 2.7
Identities = 13/41 (31%), Positives = 26/41 (62%)
Query: 344 NKEEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVDK 384
++++DED++DE+ DD++ D + E +D D D E+ +
Sbjct: 913 DEDDDEDDDDEDDDDEEEEDSDSDLEYGGDLDADRDIEMKR 953
Score = 31.6 bits (70), Expect = 3.6
Identities = 27/110 (24%), Positives = 41/110 (36%), Gaps = 30/110 (27%)
Query: 304 KRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKEED--------EDEEDEE 355
K + L+E R E++ K ++ S + N +E ED+EDE+
Sbjct: 790 KFERSENLVESASRLESLLKSLNAIKSKDDRVKGSSASIHNGKESAVPIESITEDDEDED 849
Query: 356 MDDDDNVDF----------------------GTYTEDEDAIDEDEDEEVD 383
++DD VD E ED DED+DE+ D
Sbjct: 850 DENDDGVDLLGEDEDAEISTPNTESAPGKHQAKQDESEDEDDEDDDEDDD 899
>YU0O_CAEEL (Q19753) Hypothetical protein F23B12.7 in chromosome V
Length = 953
Score = 44.7 bits (104), Expect = 4e-04
Identities = 17/39 (43%), Positives = 29/39 (73%)
Query: 343 KNKEEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEE 381
+NK++D +E+D +MD+DD++D E+ED ED+DE+
Sbjct: 810 RNKKKDVEEDDVDMDEDDDIDLNDLNEEEDGGMEDDDED 848
Score = 43.9 bits (102), Expect = 7e-04
Identities = 27/98 (27%), Positives = 51/98 (51%), Gaps = 17/98 (17%)
Query: 293 EYMETYKADLDKRREEKQLLEEN---RRKENMDKLRREMEKSGGETVVLSTGLKNKEEDE 349
+Y + + A+ KR ++K + E++ +++D E+ GG ED+
Sbjct: 798 DYSKEFGAEKSKRNKKKDVEEDDVDMDEDDDIDLNDLNEEEDGGM------------EDD 845
Query: 350 DEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVDKFSF 387
DE+D +MDDDD D ED+D +D+D++++ + F
Sbjct: 846 DEDDGDMDDDDEDDEDA--EDDDEVDDDDEDDDEDGGF 881
Score = 31.6 bits (70), Expect = 3.6
Identities = 25/97 (25%), Positives = 46/97 (46%), Gaps = 17/97 (17%)
Query: 304 KRREEKQLLEE--NRRKENMDKLRREMEKSGGETVVLSTGLKNKEEDE-------DEEDE 354
KRREE E +R ++ K ++ +++G + + + + E D+ E +E
Sbjct: 734 KRREEIPADERFLHRYTSSLQKEKKVKKENGDDDWEYAESVSSAEFDQLLERFEPGELNE 793
Query: 355 EMDDDDNVDFGTYTE--------DEDAIDEDEDEEVD 383
E D D + +FG +ED +D DED+++D
Sbjct: 794 EFDIDYSKEFGAEKSKRNKKKDVEEDDVDMDEDDDID 830
>K21A_HUMAN (Q7Z4S6) Kinesin family member 21A (Kinesin-like protein
KIF2) (NY-REN-62 antigen)
Length = 1674
Score = 44.7 bits (104), Expect = 4e-04
Identities = 45/177 (25%), Positives = 73/177 (40%), Gaps = 40/177 (22%)
Query: 231 WMKDFEVFQAAASSDQSYTSNLTQSLSLALDEFYSNLRSAGVSAVTGAGIEGFFKAVEAS 290
++K+ E +A ++ NL + NL A A +G F + +S
Sbjct: 482 YIKEIEDLRAKLLESEAVNENLRK-----------NLTRATARAPYFSGSSTFSPTILSS 530
Query: 291 AEEYMETYKADLDKRREEKQLLEENRRKENMDKLR---REMEKSGGETVVLSTGLKNKEE 347
+E +E DL K+ EK +E R+K+ + KL RE G+ T + KEE
Sbjct: 531 DKETIEII--DLAKKDLEKLKRKEKRKKKRLQKLEESNREERSVAGKEDNTDTDQEKKEE 588
Query: 348 D---------------------EDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVD 383
EDEE+EE +++D++D G E D D + DE+ +
Sbjct: 589 KGVSERENNELEVEESQEVSDHEDEEEEEEEEEDDIDGG---ESSDESDSESDEKAN 642
>RGP1_XENLA (O13066) Ran GTPase-activating protein 1
Length = 580
Score = 44.3 bits (103), Expect = 5e-04
Identities = 38/125 (30%), Positives = 62/125 (49%), Gaps = 13/125 (10%)
Query: 267 LRSAGVSAVTGAGIEGFFKA-------VEASAEEYMETYKADLDKRREEKQLLEENRRKE 319
+RS G A+ A EG K E A+ + ++ DK EK L N E
Sbjct: 276 VRSKGAQAIASALKEGLHKLKDLNLSYCEIKADAAVSLAESVEDKSDLEKLDLNGNCLGE 335
Query: 320 NMDKLRREMEKSGGETVVLSTGLKNKEEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDED 379
+ +E+ +S +L G + +EDED++D++ DDDD+ D E++D E+E+
Sbjct: 336 EGCEQVQEILESINMANIL--GSLSDDEDEDDDDDDEDDDDDED----DENDDEEVEEEE 389
Query: 380 EEVDK 384
EEV++
Sbjct: 390 EEVEE 394
>NUCL_RAT (P13383) Nucleolin (Protein C23)
Length = 712
Score = 44.3 bits (103), Expect = 5e-04
Identities = 30/98 (30%), Positives = 53/98 (53%), Gaps = 9/98 (9%)
Query: 286 AVEASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTG--LK 343
A AS +E E + D +E++ E++ +E M+ + +K+ + V + +
Sbjct: 182 AAPASEDEDEEDDDDEDDDDDDEEEEEEDDSEEEVMEITPAKGKKTPAKVVPVKAKSVAE 241
Query: 344 NKEEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEE 381
+E+DED+EDEE D+D+ ++ED DEDE+EE
Sbjct: 242 EEEDDEDDEDEEEDEDEE-------DEEDDEDEDEEEE 272
Score = 37.0 bits (84), Expect = 0.085
Identities = 21/83 (25%), Positives = 40/83 (47%), Gaps = 9/83 (10%)
Query: 299 KADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKEEDEDEEDEEMDD 358
K D D+ +E+ + + ++ D+ + K +++EDE+++D+E DD
Sbjct: 141 KEDSDEDEDEEDEDDSDEDEDEEDEFEPPVVKGVKPAKAAPAAPASEDEDEEDDDDEDDD 200
Query: 359 DDNVDFGTYTEDEDAIDEDEDEE 381
DD DE+ +ED+ EE
Sbjct: 201 DD---------DEEEEEEDDSEE 214
Score = 33.5 bits (75), Expect = 0.94
Identities = 19/45 (42%), Positives = 26/45 (57%), Gaps = 14/45 (31%)
Query: 341 GLKNKEEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVDKF 385
G K+ED DE+++E D+DD+ DEDEDEE D+F
Sbjct: 136 GKNAKKEDSDEDEDEEDEDDS-------------DEDEDEE-DEF 166
Score = 31.6 bits (70), Expect = 3.6
Identities = 15/36 (41%), Positives = 24/36 (66%), Gaps = 5/36 (13%)
Query: 343 KNKEEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDE 378
KN ++++ +EDE+ +D+D+ D EDED DE E
Sbjct: 137 KNAKKEDSDEDEDEEDEDDSD-----EDEDEEDEFE 167
>UBF2_XENLA (P25980) Nucleolar transcription factor 2 (Upstream
binding factor-2) (UBF-2) (xUBF-2)
Length = 701
Score = 43.9 bits (102), Expect = 7e-04
Identities = 29/99 (29%), Positives = 46/99 (46%), Gaps = 5/99 (5%)
Query: 291 AEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREMEK----SGGETVVLSTGLKNKE 346
AE+ Y+ D + + KE R+ K S +V+ + + E
Sbjct: 559 AEDQQRLYRTQFDTWMKGLSTQDRAAYKEQNTNKRKSTTKIQAPSSKSKLVIQSKSDDDE 618
Query: 347 EDEDEEDEE-MDDDDNVDFGTYTEDEDAIDEDEDEEVDK 384
+DED+EDEE DDDD+ D +ED D+ D DE+ ++
Sbjct: 619 DDEDDEDEEDDDDDDDEDKEDSSEDGDSSDSSSDEDSEE 657
Score = 32.7 bits (73), Expect = 1.6
Identities = 22/80 (27%), Positives = 36/80 (44%), Gaps = 1/80 (1%)
Query: 301 DLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKEEDEDE-EDEEMDDD 359
D D EE +++ KE+ + + S E +++E++ED+ ED E DDD
Sbjct: 620 DEDDEDEEDDDDDDDEDKEDSSEDGDSSDSSSDEDSEEGEENEDEEDEEDDDEDNEEDDD 679
Query: 360 DNVDFGTYTEDEDAIDEDED 379
DN + + A D D
Sbjct: 680 DNESGSSSSSSSSADSSDSD 699
Score = 31.2 bits (69), Expect = 4.7
Identities = 12/39 (30%), Positives = 25/39 (63%)
Query: 343 KNKEEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEE 381
++ ++D+DE+ E+ +D + + ED + +E+EDEE
Sbjct: 627 EDDDDDDDEDKEDSSEDGDSSDSSSDEDSEEGEENEDEE 665
Score = 31.2 bits (69), Expect = 4.7
Identities = 16/46 (34%), Positives = 25/46 (53%), Gaps = 8/46 (17%)
Query: 346 EEDEDEEDEEMD--------DDDNVDFGTYTEDEDAIDEDEDEEVD 383
++DED+ED D D+D+ + ++ED D+DED E D
Sbjct: 632 DDDEDKEDSSEDGDSSDSSSDEDSEEGEENEDEEDEEDDDEDNEED 677
Score = 30.8 bits (68), Expect = 6.1
Identities = 21/83 (25%), Positives = 45/83 (53%), Gaps = 13/83 (15%)
Query: 299 KADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKEEDEDEEDEEMDD 358
K+D D+ E+ + E++ ++ DK E G++ S +++ +E EE+E+ +D
Sbjct: 613 KSDDDEDDEDDEDEEDDDDDDDEDK---EDSSEDGDS---SDSSSDEDSEEGEENEDEED 666
Query: 359 DDNVDFGTYTEDEDAIDEDEDEE 381
+++ +DED ++D+D E
Sbjct: 667 EED-------DDEDNEEDDDDNE 682
>NUCL_MOUSE (P09405) Nucleolin (Protein C23)
Length = 706
Score = 43.9 bits (102), Expect = 7e-04
Identities = 29/102 (28%), Positives = 49/102 (47%), Gaps = 2/102 (1%)
Query: 280 IEGFFKAVEASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLS 339
++G A A A E + D D+ EE +E ++ +++ G +T
Sbjct: 173 VKGVKPAKAAPAAPASEDEEDDEDEDDEEDD--DEEEEDDSEEEVMEITTAKGKKTPAKV 230
Query: 340 TGLKNKEEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEE 381
+K K E+E+DEE D+DD + +DED +E+E+EE
Sbjct: 231 VPMKAKSVAEEEDDEEEDEDDEDEDDEEEDDEDDDEEEEEEE 272
Score = 39.7 bits (91), Expect = 0.013
Identities = 29/100 (29%), Positives = 53/100 (53%), Gaps = 7/100 (7%)
Query: 285 KAVEASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKN 344
KA A+ E D D ++ + E++ +E M+ + +K+ + V + K+
Sbjct: 180 KAAPAAPASEDEEDDEDEDDEEDDDEEEEDDSEEEVMEITTAKGKKTPAKVVPMKA--KS 237
Query: 345 KEEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVDK 384
E+ED+E EE +DD++ D E+ED D+DE+EE ++
Sbjct: 238 VAEEEDDE-EEDEDDEDED----DEEEDDEDDDEEEEEEE 272
Score = 36.6 bits (83), Expect = 0.11
Identities = 18/40 (45%), Positives = 25/40 (62%), Gaps = 8/40 (20%)
Query: 341 GLKNKEEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDE 380
G K+ED DE+++E D+DD+ DED DE+EDE
Sbjct: 136 GKNAKKEDSDEDEDEEDEDDS--------DEDEDDEEEDE 167
Score = 31.6 bits (70), Expect = 3.6
Identities = 20/83 (24%), Positives = 40/83 (48%), Gaps = 11/83 (13%)
Query: 300 ADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKEEDEDEEDEEMDDD 359
+D D+ E++ +E+ E D+ + K +++E++DE++++ +DD
Sbjct: 144 SDEDEDEEDEDDSDEDEDDEEEDEFEPPIVKGVKPAKAAPAAPASEDEEDDEDEDDEEDD 203
Query: 360 DNVDFGTYTEDEDAIDEDEDEEV 382
D E+ED D +EEV
Sbjct: 204 DE-------EEED----DSEEEV 215
>IE68_PRVKA (P24827) Immediate-early protein RSP40
Length = 364
Score = 43.9 bits (102), Expect = 7e-04
Identities = 20/40 (50%), Positives = 29/40 (72%), Gaps = 2/40 (5%)
Query: 346 EEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVDKF 385
EE+EDEE+EE D+D D Y ED++A DE+++E+ D F
Sbjct: 230 EEEEDEEEEEGDEDGETD--VYEEDDEAEDEEDEEDGDDF 267
Score = 35.4 bits (80), Expect = 0.25
Identities = 17/41 (41%), Positives = 26/41 (62%), Gaps = 3/41 (7%)
Query: 345 KEEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVDKF 385
++EDE+E+ EE D+D+ G EDE+ + DED E D +
Sbjct: 212 EDEDEEEDGEEEDEDEE---GEEEEDEEEEEGDEDGETDVY 249
Score = 35.4 bits (80), Expect = 0.25
Identities = 21/55 (38%), Positives = 30/55 (54%), Gaps = 2/55 (3%)
Query: 331 SGGETVVLSTGLKNKEEDEDEEDEEMDDDDNVDFGTYTED--EDAIDEDEDEEVD 383
S GE G ++EDEDE+++E +DD + ED ED D +EDE+ D
Sbjct: 285 SDGEGSGSDDGGDGEDEDEDEDEDEDEDDGEDEEDEEGEDGGEDGEDGEEDEDED 339
Score = 35.0 bits (79), Expect = 0.32
Identities = 33/106 (31%), Positives = 46/106 (43%), Gaps = 29/106 (27%)
Query: 301 DLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKEEDEDEEDEEMDD-- 358
D D+ + ++ E+ +E D+ E ++ G V + +E EDEEDEE D
Sbjct: 213 DEDEEEDGEEEDEDEEGEEEEDEEEEEGDEDGETDVY-----EEDDEAEDEEDEEDGDDF 267
Query: 359 ------DDNV---------------DFGTYTEDEDAIDEDEDEEVD 383
DD+V D G EDED DEDEDE+ D
Sbjct: 268 DGASVGDDDVFEPPEDGSDGEGSGSDDGGDGEDEDE-DEDEDEDED 312
Score = 34.3 bits (77), Expect = 0.55
Identities = 15/39 (38%), Positives = 26/39 (66%)
Query: 343 KNKEEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEE 381
++++EDEDE+D E ++D+ + G ++ DEDED E
Sbjct: 303 EDEDEDEDEDDGEDEEDEEGEDGGEDGEDGEEDEDEDGE 341
Score = 33.1 bits (74), Expect = 1.2
Identities = 15/38 (39%), Positives = 25/38 (65%)
Query: 347 EDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVDK 384
EDEDE+++E +D+D+ + E ED ++ ED E D+
Sbjct: 299 EDEDEDEDEDEDEDDGEDEEDEEGEDGGEDGEDGEEDE 336
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.316 0.133 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 45,197,006
Number of Sequences: 164201
Number of extensions: 2022480
Number of successful extensions: 30972
Number of sequences better than 10.0: 828
Number of HSP's better than 10.0 without gapping: 355
Number of HSP's successfully gapped in prelim test: 501
Number of HSP's that attempted gapping in prelim test: 20017
Number of HSP's gapped (non-prelim): 4488
length of query: 387
length of database: 59,974,054
effective HSP length: 112
effective length of query: 275
effective length of database: 41,583,542
effective search space: 11435474050
effective search space used: 11435474050
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 67 (30.4 bits)
Medicago: description of AC121237.18


