

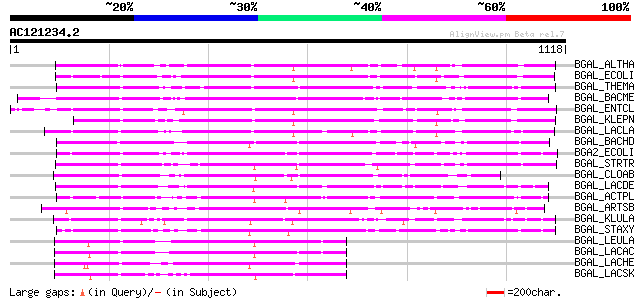
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121234.2 + phase: 0
(1118 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BGAL_ALTHA (P81650) Beta-galactosidase (EC 3.2.1.23) (Lactase) (... 729 0.0
BGAL_ECOLI (P00722) Beta-galactosidase (EC 3.2.1.23) (Lactase) 686 0.0
BGAL_THEMA (Q56307) Beta-galactosidase (EC 3.2.1.23) (Lactase) 672 0.0
BGAL_BACME (O52847) Beta-galactosidase (EC 3.2.1.23) (Lactase) 665 0.0
BGAL_ENTCL (Q47077) Beta-galactosidase (EC 3.2.1.23) (Lactase) 657 0.0
BGAL_KLEPN (P06219) Beta-galactosidase (EC 3.2.1.23) (Lactase) 625 e-178
BGAL_LACLA (Q48727) Beta-galactosidase (EC 3.2.1.23) (Lactase) 580 e-165
BGAL_BACHD (Q9K9C6) Beta-galactosidase (EC 3.2.1.23) (Lactase) 531 e-150
BGA2_ECOLI (P06864) Evolved beta-galactosidase alpha-subunit (EC... 531 e-150
BGAL_STRTR (P23989) Beta-galactosidase (EC 3.2.1.23) (Lactase) 514 e-145
BGAL_CLOAB (P24131) Beta-galactosidase (EC 3.2.1.23) (Lactase) 488 e-137
BGAL_LACDE (P20043) Beta-galactosidase (EC 3.2.1.23) (Lactase) 472 e-132
BGAL_ACTPL (P70753) Beta-galactosidase (EC 3.2.1.23) (Lactase) 467 e-131
BGAL_ARTSB (Q59140) Beta-galactosidase (EC 3.2.1.23) (Lactase) 465 e-130
BGAL_KLULA (P00723) Beta-galactosidase (EC 3.2.1.23) (Lactase) 451 e-126
BGAL_STAXY (O33815) Beta-galactosidase (EC 3.2.1.23) (Lactase) 412 e-114
BGAL_LEULA (Q02603) Beta-galactosidase large subunit (EC 3.2.1.2... 331 8e-90
BGAL_LACAC (O07684) Beta-galactosidase large subunit (EC 3.2.1.2... 328 6e-89
BGAL_LACHE (Q7WTB4) Beta-galactosidase large subunit (EC 3.2.1.2... 320 2e-86
BGAL_LACSK (Q48846) Beta-galactosidase large subunit (EC 3.2.1.2... 315 6e-85
>BGAL_ALTHA (P81650) Beta-galactosidase (EC 3.2.1.23) (Lactase)
(Beta-D-galactoside galactohydrolase)
Length = 1038
Score = 729 bits (1881), Expect = 0.0
Identities = 419/1047 (40%), Positives = 589/1047 (56%), Gaps = 100/1047 (9%)
Query: 93 KSLSGYWKFFLASNPCNVPAKF---HDSEFQDSEWSTLPVPSNWQLHGFDRPIYTNVTYP 149
KSL+G W F L P V S+ +W ++ VPSNWQLHGFD+PIY NV YP
Sbjct: 47 KSLNGQWDFKLFDKPEAVDESLLYEKISKELSGDWQSITVPSNWQLHGFDKPIYCNVKYP 106
Query: 150 FPLDPPFVPTENPTGCYRMDFHLPKEWEGRRILLHFEAVDSAFCAWINGHPIGYSQDSRL 209
F ++PPFVP++NPTGCYR +F + E +R + FE V+SAF W NG +GYSQDSRL
Sbjct: 107 FAVNPPFVPSDNPTGCYRTEFTITPEQLTQRNHIIFEGVNSAFHLWCNGQWVGYSQDSRL 166
Query: 210 PAEFEVTDFCHPCGSDLKNVLAVQVFRWSDGCYLEDQDHWRMSGIHRDVLLLAKPEVFIT 269
P+EF++++ G+ N +AV V RWSDG YLEDQD W +SGI RDV LL KP+ I
Sbjct: 167 PSEFDLSELL-VVGT---NRIAVMVIRWSDGSYLEDQDMWWLSGIFRDVNLLTKPQSQIR 222
Query: 270 DYFFKSNLAEDFSSAEMLVEVKIDRLQDTSIDNVLTNYTIEATLYDSGSWESSDGNPDLL 329
D F +L + A + ++ I N NY + ++D ++S P +
Sbjct: 223 DVFITPDLDACYRDATLHIKTAI---------NAPNNYQVAVQIFDG---KTSLCEPKIQ 270
Query: 330 SSNVADITFQPTTTPLGFYGYTLVGKLQSPKLWSAEQPYLYTLVVVLKDKSGRVLDCESS 389
S+N + + + + F + +SPK W+AE PYLY VV L D+ G +D E+
Sbjct: 271 STNNKRVDEKGGWSDVVFQTIAI----RSPKKWTAETPYLYRCVVSLLDEQGNTVDVEAY 326
Query: 390 QVGFKNVSKAHKQLLVNGHPVVIRGVNRHEHHPEVGKANIESCMVKDLVLMKQNNINAVR 449
+GF+ V + QL VNG P++IRGVNRHEHHPE G A + M++D+ LMKQNN NAVR
Sbjct: 327 NIGFRKVEMLNGQLCVNGKPLLIRGVNRHEHHPENGHAVSTADMIEDIKLMKQNNFNAVR 386
Query: 450 NSHYPQHPRWYELCDLFGMYMIDEANIETHGFDYSKHLKHPTLEPMWATAMLDRVIGMVE 509
+HYP HP +YELCD G+Y++DEANIETHG L +P+WA A + R MVE
Sbjct: 387 TAHYPNHPLFYELCDELGLYVVDEANIETHGMFPMGRL---ASDPLWAGAFMSRYTQMVE 443
Query: 510 RDKNHTCIISWSLGNESGFGTNHFAMAGWIRGRDSSRVIHYEGGGSRTPCTDIVCPMYMR 569
RDKNH II WSLGNE G G NH AM GW + D SR + YEGGG+ T TDI+CPMY R
Sbjct: 444 RDKNHASIIIWSLGNECGHGANHDAMYGWSKSFDPSRPVQYEGGGANTTATDIICPMYSR 503
Query: 570 V-----------WDMLKIANDPTETRPLILCEYSHAMGNSNGNLHIYWEAIDNTFGLQGG 618
V + + K + P ETRPLILCEY+HAMGNS G+ YW+A LQGG
Sbjct: 504 VDTDIKDDAVPKYSIKKWLSLPGETRPLILCEYAHAMGNSLGSFDDYWQAFREYPRLQGG 563
Query: 619 FIWDWVDQALRKVQADGTKQWAYGGEFGDIPNDLNFCLNGLVWPDRTAHPVLHEVKFLYQ 678
FIWDWVDQ L K+ +G WAYGG+FGD ND FC+NGL++PDRT HP L E K+ Q
Sbjct: 564 FIWDWVDQGLSKIDENGKHYWAYGGDFGDELNDRQFCINGLLFPDRTPHPSLFEAKYSQQ 623
Query: 679 PIKVNLSDG---------KLEIKNTHFFQTTEGLEFSWYISADGYKLGSDKLSLPPIKPQ 729
++ L + +++ + + F+ T+ + W + +G + +++L I PQ
Sbjct: 624 HLQFTLREQNQNQNQNQYSIDVFSDYVFRHTDNEKLVWQLIQNGVCVEQGEMAL-NIAPQ 682
Query: 730 SNYVFDWKSGPWYSLWDSSSSEEIFLTITAKLLNSTRWVEAGHVVTTAQVQLPAKRDIVP 789
S + K+ + + +L + L+N + + A HV+ + Q +L
Sbjct: 683 STHTLTIKTKTAF-----EHGAQYYLNLDVALINDSHFANANHVMDSEQFKL-------I 730
Query: 790 HAINIGSGNLVVETLGDTIKVSQQD--------VWDITFNTKTGLIESWKVKGVHVMNKG 841
++ N+ S + T I V++ D + + FN ++GLIE W V++
Sbjct: 731 NSNNLNSKSFASATEKSVISVNETDSHLSIENNTFKLVFNQQSGLIEQWLQDDTQVISSP 790
Query: 842 IHPCFWRASIDNDKGGG------ADSYLSRWKAAGIDSVHFIAESC-SVQSTTGNAVKLL 894
+ F+RA +DND G +++ +RW AGI S +VQS+ V++
Sbjct: 791 LVDNFYRAPLDNDIGVSEVDNLDPNAWEARWSRAGIGQWQRTCSSINAVQSSVD--VRIT 848
Query: 895 VVFHGVTKGEEGSLPNQDKSKVL-FTTEMTYTIYASGDVILECNVKPNADLPPLPRVGIE 953
VF N + + VL T+ YT+ +G + L +V N LPP+PR+G+
Sbjct: 849 CVF------------NYEFNGVLQAQTQWLYTLNNTGTISLNVDVNLNDTLPPMPRIGLS 896
Query: 954 MNLEKSLD-QVSWYGRGPFECYPDRKAAAQVAVYEKSVDELHVPYIVPGESGGRADVRWA 1012
+ K D +V+W G GPFE YPDRK+AA+ Y S++EL+ PYI P ++G R+D +
Sbjct: 897 TTINKQSDTKVNWLGLGPFENYPDRKSAARFGYYSLSLNELYTPYIFPTDNGLRSDCQLL 956
Query: 1013 TFLNKNGFGIYTSKYGSSPPMQMSASYYSTSELDRAGHDYELVKGDNIEVHLDHKHMGLG 1072
+ N G + +AS YS + L +A H EL+ D I VH+DH+HMG+G
Sbjct: 957 SINNLIVTGAFL----------FAASEYSQNMLTQAKHTNELIADDCIHVHIDHQHMGVG 1006
Query: 1073 GDDSWSPCVHDQYLVPPVPYSFSVRLS 1099
GDDSWSP H +YL+ Y++S+ L+
Sbjct: 1007 GDDSWSPSTHKEYLLEQKNYNYSLTLT 1033
>BGAL_ECOLI (P00722) Beta-galactosidase (EC 3.2.1.23) (Lactase)
Length = 1023
Score = 686 bits (1769), Expect = 0.0
Identities = 397/1027 (38%), Positives = 564/1027 (54%), Gaps = 81/1027 (7%)
Query: 92 VKSLSGYWKFFLASNPCNVPAKFHDSEFQDSEWSTLPVPSNWQLHGFDRPIYTNVTYPFP 151
++SL+G W+F P VP + + + +++ T+ VPSNWQ+HG+D PIYTNVTYP
Sbjct: 51 LRSLNGEWRFAWFPAPEAVPESWLECDLPEAD--TVVVPSNWQMHGYDAPIYTNVTYPIT 108
Query: 152 LDPPFVPTENPTGCYRMDFHLPKEW--EGRRILLHFEAVDSAFCAWINGHPIGYSQDSRL 209
++PPFVPTENPTGCY + F++ + W EG+ ++ F+ V+SAF W NG +GY QDSRL
Sbjct: 109 VNPPFVPTENPTGCYSLTFNVDESWLQEGQTRII-FDGVNSAFHLWCNGRWVGYGQDSRL 167
Query: 210 PAEFEVTDFCHPCGSDLKNVLAVQVFRWSDGCYLEDQDHWRMSGIHRDVLLLAKPEVFIT 269
P+EF+++ F +N LAV V RWSDG YLEDQD WRMSGI RDV LL KP I+
Sbjct: 168 PSEFDLSAFLRAG----ENRLAVMVLRWSDGSYLEDQDMWRMSGIFRDVSLLHKPTTQIS 223
Query: 270 DYFFKSNLAEDFSSAEMLVEVKI-DRLQDTSIDNVLTNYTIEATLYDSGSWESSDGNPDL 328
D+ + +DFS A + EV++ L+D + +L+ + +S P
Sbjct: 224 DFHVATRFNDDFSRAVLEAEVQMCGELRDY--------LRVTVSLWQGETQVASGTAP-- 273
Query: 329 LSSNVADITFQPTTTPLGFYG--YTLVGKLQSPKLWSAEQPYLYTLVVVLKDKSGRVLDC 386
+ D G Y TL +++PKLWSAE P LY VV L G +++
Sbjct: 274 FGGEIID--------ERGGYADRVTLRLNVENPKLWSAEIPNLYRAVVELHTADGTLIEA 325
Query: 387 ESSQVGFKNVSKAHKQLLVNGHPVVIRGVNRHEHHPEVGKANIESCMVKDLVLMKQNNIN 446
E+ VGF+ V + LL+NG P++IRGVNRHEHHP G+ E MV+D++LMKQNN N
Sbjct: 326 EACDVGFREVRIENGLLLLNGKPLLIRGVNRHEHHPLHGQVMDEQTMVQDILLMKQNNFN 385
Query: 447 AVRNSHYPQHPRWYELCDLFGMYMIDEANIETHGFDYSKHLKHPTLEPMWATAMLDRVIG 506
AVR SHYP HP WY LCD +G+Y++DEANIETHG L T +P W AM +RV
Sbjct: 386 AVRCSHYPNHPLWYTLCDRYGLYVVDEANIETHGMVPMNRL---TDDPRWLPAMSERVTR 442
Query: 507 MVERDKNHTCIISWSLGNESGFGTNHFAMAGWIRGRDSSRVIHYEGGGSRTPCTDIVCPM 566
MV+RD+NH +I WSLGNESG G NH A+ WI+ D SR + YEGGG+ T TDI+CPM
Sbjct: 443 MVQRDRNHPSVIIWSLGNESGHGANHDALYRWIKSVDPSRPVQYEGGGADTTATDIICPM 502
Query: 567 YMRV-----------WDMLKIANDPTETRPLILCEYSHAMGNSNGNLHIYWEAIDNTFGL 615
Y RV W + K + P ETRPLILCEY+HAMGNS G YW+A L
Sbjct: 503 YARVDEDQPFPAVPKWSIKKWLSLPGETRPLILCEYAHAMGNSLGGFAKYWQAFRQYPRL 562
Query: 616 QGGFIWDWVDQALRKVQADGTKQWAYGGEFGDIPNDLNFCLNGLVWPDRTAHPVLHEVKF 675
QGGF+WDWVDQ+L K +G AYGG+FGD PND FC+NGLV+ DRT HP L E K
Sbjct: 563 QGGFVWDWVDQSLIKYDENGNPWSAYGGDFGDTPNDRQFCMNGLVFADRTPHPALTEAKH 622
Query: 676 LYQPIKVNLSDGKLEIKNTHFFQTTEGLEFSWYISADGYKLGSDKLSLPPIKPQSNYVFD 735
Q + LS +E+ + + F+ ++ W ++ DG L S ++ L + PQ + +
Sbjct: 623 QQQFFQFRLSGQTIEVTSEYLFRHSDNELLHWMVALDGKPLASGEVPL-DVAPQGKQLIE 681
Query: 736 WKSGPWYSLWDSSSSEEIFLTITAKLLNSTRWVEAGHVVTTAQVQLPAKRDIVPHAINIG 795
P S+ +++LT+ N+T W EAGH+ Q +L + A +
Sbjct: 682 LPELP-----QPESAGQLWLTVRVVQPNATAWSEAGHISAWQQWRLAENLSVTLPAASHA 736
Query: 796 SGNLVVETLGDTIKVSQQDVWDITFNTKTGLIESWKVKGVHVMNKGIHPCFWRASIDNDK 855
+L + I++ + W FN ++G + + + + F RA +DND
Sbjct: 737 IPHLTTSEMDFCIELGNKR-WQ--FNRQSGFLSQMWIGDKKQLLTPLRDQFTRAPLDNDI 793
Query: 856 GGG------ADSYLSRWKAAGIDSVHFIAESCSVQSTTGNAVKLLVVFHGVTKGEEGSLP 909
G ++++ RWKAAG H+ AE+ +Q T +++ +G
Sbjct: 794 GVSEATRIDPNAWVERWKAAG----HYQAEAALLQCTADTLADAVLITTAHAWQHQG--- 846
Query: 910 NQDKSKVLFTTEMTYTIYASGDVILECNVKPNADLPPLPRVGIEMNLEKSLDQVSWYGRG 969
K LF + TY I SG + + +V+ +D P R+G+ L + ++V+W G G
Sbjct: 847 -----KTLFISRKTYRIDGSGQMAITVDVEVASDTPHPARIGLNCQLAQVAERVNWLGLG 901
Query: 970 PFECYPDRKAAAQVAVYEKSVDELHVPYIVPGESGGRADVRWATFLNKNGFGIYTSKYGS 1029
P E YPDR AA ++ + +++ PY+ P E+G R R + G +
Sbjct: 902 PQENYPDRLTAACFDRWDLPLSDMYTPYVFPSENGLRCGTRELNYGPHQWRGDF------ 955
Query: 1030 SPPMQMSASYYSTSELDRAGHDYELVKGDNIEVHLDHKHMGLGGDDSWSPCVHDQYLVPP 1089
Q + S YS +L H + L + +++D HMG+GGDDSWSP V ++ +
Sbjct: 956 ----QFNISRYSQQQLMETSHRHLLHAEEGTWLNIDGFHMGIGGDDSWSPSVSAEFQLSA 1011
Query: 1090 VPYSFSV 1096
Y + +
Sbjct: 1012 GRYHYQL 1018
>BGAL_THEMA (Q56307) Beta-galactosidase (EC 3.2.1.23) (Lactase)
Length = 1084
Score = 672 bits (1735), Expect = 0.0
Identities = 396/1019 (38%), Positives = 551/1019 (53%), Gaps = 93/1019 (9%)
Query: 94 SLSGYWKFFLASNPCNVPAKFHDSEFQDSEWSTLPVPSNWQLHGFDRPIYTNVTYPFPLD 153
SL+G W+F A NP VP F +F DS W + VPSNW++ G+ +PIYTNV YPF +
Sbjct: 41 SLNGNWRFLFAKNPFEVPEDFFSEKFDDSNWDEIEVPSNWEMKGYGKPIYTNVVYPFEPN 100
Query: 154 PPFVPTE-NPTGCYRMDFHLPKEWEGRRILLHFEAVDSAFCAWINGHPIGYSQDSRLPAE 212
PPFVP + NPTG YR +P++W + I LHFE V S F W+NG IG+S+DS PAE
Sbjct: 101 PPFVPKDDNPTGVYRRWIEIPEDWFKKEIFLHFEGVRSFFYLWVNGKKIGFSKDSCTPAE 160
Query: 213 FEVTDFCHPCGSDLKNVLAVQVFRWSDGCYLEDQDHWRMSGIHRDVLLLAKPEVFITDYF 272
F +TD P KN++ V+V +WSDG YLEDQD W +GI+RDV L A P+ I D F
Sbjct: 161 FRLTDVLRPG----KNLITVEVLKWSDGSYLEDQDMWWFAGIYRDVYLYALPKFHIRDVF 216
Query: 273 FKSNLAEDFSSAEMLVEVKIDRLQDTSIDNVLTNYTIEATLYDSGSWESSDGNPDLLSSN 332
+++L E++ + ++ ++V++ L + + +E TL PD
Sbjct: 217 VRTDLDENYRNGKIFLDVEMRNLGEEEEKD------LEVTLI----------TPDGDEKT 260
Query: 333 VADITFQPTTTPLGFYGYTLVGKLQSPKLWSAEQPYLYTLVVVLKDKSGRVLDCESSQVG 392
+ T +P L F ++ PK WSAE P+LY L + L + +V G
Sbjct: 261 LVKETVKPEDRVLSF-----AFDVKDPKKWSAETPHLYVLKLKLGEDEKKV------NFG 309
Query: 393 FKNVSKAHKQLLVNGHPVVIRGVNRHEHHPEVGKANIESCMVKDLVLMKQNNINAVRNSH 452
F+ + LL NG P+ I+GVNRHE P+ G A M++D+ LMKQ+NIN VR SH
Sbjct: 310 FRKIEIKDGTLLFNGKPLYIKGVNRHEFDPDRGHAVTVERMIQDIKLMKQHNINTVRTSH 369
Query: 453 YPQHPRWYELCDLFGMYMIDEANIETHGFDYSKHLKHPTLEPMWA--TAMLDRVIGMVER 510
YP +WY+LCD FG+Y+IDEANIE+HG D+ + TL W A DR+ MVER
Sbjct: 370 YPNQTKWYDLCDYFGLYVIDEANIESHGIDWDPEV---TLANRWEWEKAHFDRIKRMVER 426
Query: 511 DKNHTCIISWSLGNESGFGTNHFAMAGWIRGRDSSRVIHYEGGGSRTPC--TDIVCPMYM 568
DKNH II WSLGNE+G G N A WI+ RD++R+IHYEG R D+ MY
Sbjct: 427 DKNHPSIIFWSLGNEAGDGVNFEKAALWIKKRDNTRLIHYEGTTRRGESYYVDVFSLMYP 486
Query: 569 RVWDMLKIANDPTETRPLILCEYSHAMGNSNGNLHIYWEAIDNTFGLQGGFIWDWVDQAL 628
++ +L+ A+ E +P I+CEY+HAMGNS GNL YW+ I+ L GG IWDWVDQ +
Sbjct: 487 KMDILLEYASKKRE-KPFIMCEYAHAMGNSVGNLKDYWDVIEKYPYLHGGCIWDWVDQGI 545
Query: 629 RKVQADGTKQWAYGGEFGDIPNDLNFCLNGLVWPDRTAHPVLHEVKFLYQPIKV-NLSDG 687
RK +G + WAYGG+FGD PND NFC+NG+V PDRT P L+EVK +YQ +K+ +S
Sbjct: 546 RKKDENGREFWAYGGDFGDTPNDGNFCINGVVLPDRTPEPELYEVKKVYQNVKIRQVSKD 605
Query: 688 KLEIKNTHFFQTTEGLEFSWYISADGYKLGSDKLSLPPIKPQSNYVFDWKSGPWYSLWDS 747
E++N + F E + +W I DG + + + + P + DS
Sbjct: 606 TYEVENRYLFTNLEMFDGAWKIRKDGEVIEEKTFKIFAEPGEKRLL----KIPLPEMDDS 661
Query: 748 SSSEEIFLTITAKLLNSTRWVEAGHVVTTAQVQLPA----KRDIVPHAINIGSGNLVVET 803
E FL I+ L T W E GHVV Q L A K+ I G +
Sbjct: 662 ----EYFLEISFSLSEDTPWAEKGHVVAWEQFLLKAPAFEKKSISDGVSLREDGKHLTVE 717
Query: 804 LGDTIKVSQQDVWDITFNTKTGLIESWKVKGVHVMNKGIHPCFWRASIDNDKGGGADSYL 863
DT+ V F+ TGL+E + ++ + P FWR DND G L
Sbjct: 718 AKDTVYV---------FSKLTGLLEQILHRRKKILKSPVVPNFWRVPTDNDIGNRMPQRL 768
Query: 864 SRWKAAG----IDSVHFIAESCSVQSTTGNAVKLLVVFHGVTKGEEGSLPNQDKSKVLFT 919
+ WK A + +H+ E N V + VF LP + ++T
Sbjct: 769 AIWKRASKERKLFKMHWKKEE--------NRVSVHSVF---------QLPG---NSWVYT 808
Query: 920 TEMTYTIYASGDVILECNVKPNADLPPLPRVGIEMNLEKSLDQVSWYGRGPFECYPDRKA 979
TYT++ +GDV+++ ++ P D+P +PR+G + + + V WYGRGP E Y DRK
Sbjct: 809 ---TYTVFGNGDVLVDLSLIPAEDVPEIPRIGFQFTVPEEFGTVEWYGRGPHETYWDRKE 865
Query: 980 AAQVAVYEKSVDELHVPYIVPGESGGRADVRWATFLNKNGFGIYTSKYGSSPPMQMSASY 1039
+ A Y K+V E+ Y+ P E+G R+DVRW L+ ++ S P + S
Sbjct: 866 SGLFARYRKAVGEMMHRYVRPQETGNRSDVRWFA-LSDGETKLFVS---GMPQIDFSVWP 921
Query: 1040 YSTSELDRAGHDYELVKGDNIEVHLDHKHMGLGGDDSWSPCVHDQYLVPPVPYSFSVRL 1098
+S +L+R H EL + D + V++D + MGLGGDDSW H +Y + P PY FS R+
Sbjct: 922 FSMEDLERVQHISELPERDFVTVNVDFRQMGLGGDDSWGAMPHLEYRLLPKPYRFSFRM 980
>BGAL_BACME (O52847) Beta-galactosidase (EC 3.2.1.23) (Lactase)
Length = 1034
Score = 665 bits (1715), Expect = 0.0
Identities = 405/1086 (37%), Positives = 577/1086 (52%), Gaps = 96/1086 (8%)
Query: 16 APNNGYKVWED-PSFIKWRKRDPHVHLHCHESVEGSLKYWYQRSKVDYLVSQSAVWKDDA 74
AP NGY W + P + + H L +++VE +LK ++S V Y
Sbjct: 12 APANGYPEWNNNPEIFQLNRSKAHALLMPYQTVEEALKN-DRKSSVYY------------ 58
Query: 75 VNGALESAAFWVKDLPFVKSLSGYWKFFLASNPCNVPAKFHDSEFQDSEWSTLPVPSNWQ 134
+SL+G W F A N F EF +W ++ VPS+WQ
Sbjct: 59 ------------------QSLNGSWYFHFAENADGRVKNFFAPEFSYEKWDSISVPSHWQ 100
Query: 135 LHGFDRPIYTNVTYPF----PLDPPFVPTE-NPTGCYRMDFHLPKEWEGRRILLHFEAVD 189
L G+D P YTNVTYP+ L+PPF PT+ NP G Y F EW+ + + + F+ V+
Sbjct: 101 LQGYDYPQYTNVTYPWVENEELEPPFAPTKYNPVGQYVRTFTPKSEWKDQPVYISFQGVE 160
Query: 190 SAFCAWINGHPIGYSQDSRLPAEFEVTDFCHPCGSDLKNVLAVQVFRWSDGCYLEDQDHW 249
SAF WING +GYS+DS PAEF++T + + +N +AV+V+RWSD +LEDQD W
Sbjct: 161 SAFYVWINGEFVGYSEDSFTPAEFDITSYLQ----EGENTIAVEVYRWSDASWLEDQDFW 216
Query: 250 RMSGIHRDVLLLAKPEVFITDYFFKSNLAEDFSSAEMLVEVKIDRLQDTSIDNVLTNYTI 309
RMSGI RDV L + P+V I D+ +S+L ++ E+ V I + ++ T
Sbjct: 217 RMSGIFRDVYLYSTPQVHIYDFSVRSSLDNNYEDGELSVSADILNYFEHDTQDL----TF 272
Query: 310 EATLYDSGSWESSDGNPDLLSSNVADITFQPTTTPLGFYGYTLVGKLQSPKLWSAEQPYL 369
E LYD+ + E L +N++ ++ Q T + L ++SP WSAE P L
Sbjct: 273 EVMLYDANAQEVLQAP---LQTNLS-VSDQRTVS--------LRTHIKSPAKWSAESPNL 320
Query: 370 YTLVVVLKDKSGRVLDCESSQVGFKNVSKAHKQLLVNGHPVVIRGVNRHEHHPEVGKANI 429
YTLV+ LK+ +G +++ ES +VGF+ + + +NG +V+RGVNRHE G+A I
Sbjct: 321 YTLVLSLKNAAGSIIETESCKVGFRTFEIKNGLMTINGKRIVLRGVNRHEFDSVKGRAGI 380
Query: 430 -ESCMVKDLVLMKQNNINAVRNSHYPQHPRWYELCDLFGMYMIDEANIETHG----FDYS 484
M+ D++LMKQ+NINAVR SHYP WYELC+ +G+Y+IDE N+ETHG
Sbjct: 381 TREDMIHDILLMKQHNINAVRTSHYPNDSVWYELCNEYGLYVIDETNLETHGTWTYLQEG 440
Query: 485 KHLKHPTLEPMWATAMLDRVIGMVERDKNHTCIISWSLGNESGFGTNHFAMAGWIRGRDS 544
+ P +P W +LDR M ERDKNH II WSLGNES G N M + + +DS
Sbjct: 441 EQKAVPGSKPEWKENVLDRCRSMYERDKNHPSIIIWSLGNESFGGENFQHMYTFFKEKDS 500
Query: 545 SRVIHYEGGGSRTP--CTDIVCPMYMRVWDMLKIANDPTETRPLILCEYSHAMGNSNGNL 602
+R++HYEG +DI MY++ D+ + A +P ILCEYSHAMGNS GNL
Sbjct: 501 TRLVHYEGIFHHRDYDASDIESTMYVKPADVERYAL-MNPKKPYILCEYSHAMGNSCGNL 559
Query: 603 HIYWEAIDNTFGLQGGFIWDWVDQALRKVQADGTKQWAYGGEFGDIPNDLNFCLNGLVWP 662
+ YWE D LQGGFIWDW DQAL+ DGT AYGG+FGD PND NFC NGL++
Sbjct: 560 YKYWELFDQYPILQGGFIWDWKDQALQATAEDGTSYLAYGGDFGDTPNDGNFCGNGLIFA 619
Query: 663 DRTAHPVLHEVKFLYQPIK---VNLSDGKLEIKNTHFFQTTEGLEFSWYISADGYKLGSD 719
D TA P + EVK YQP+K V+ + GK ++N H F +F W + +G +L
Sbjct: 620 DGTASPKIAEVKKCYQPVKWTAVDPAKGKFAVQNKHLFTNLNAYDFVWTVEKNG-ELVEK 678
Query: 720 KLSLPPIKPQSNYVFDWKSGPWYSLWDSSSSEEIFLTITAKLLNSTRWVEAGHVVTTAQV 779
SL + P S P Y + ++E LT++ +L T W AG+ V Q
Sbjct: 679 HASLLNVAPDGTDELT-LSYPLYE--QENETDEFVLTLSLRLSKDTAWASAGYEVAYEQF 735
Query: 780 QLPAKRDIVPHAINIGSGNLVVETLGDTIKVSQQDVWDITFNTKTGLIESWKVKGVHVMN 839
LPAK + ++ L V+ T+ V+ + I F+ + G S+ + ++
Sbjct: 736 VLPAKAAM--PSVKAAHPALTVDQNEQTLTVTGTNFTAI-FDKRKGQFISYNYERTELLA 792
Query: 840 KGIHPCFWRASIDNDKGGGADSYLSRWKAAGIDSVHFIAESCSVQSTTGNAVKLLVVFHG 899
G P FWRA DND G W+ A ++ Q V+ V F
Sbjct: 793 SGFRPNFWRAVTDNDLGNKLHERCQTWRQASLE-----------QHVKKVTVQPQVDF-- 839
Query: 900 VTKGEEGSLPNQDKSKVLFTTEMTYTIYASGDVILECNVKPNADLPPLPRVGIEMNLEKS 959
V E +L N L + +TYT+Y G++ +E ++ P+ +P +P +G+ + +
Sbjct: 840 VIISVELALDNS-----LASCYVTYTLYNDGEMKIEQSLAPSETMPEIPEIGMLFTMNAA 894
Query: 960 LDQVSWYGRGPFECYPDRKAAAQVAVYEKSVDELHVPYIVPGESGGRADVRWATFLNKNG 1019
D ++WYGRGP E Y DRK A++A+++ SV E PY+ P E G + DVRWAT N G
Sbjct: 895 FDSLTWYGRGPHENYWDRKTGAKLALHKGSVKEQVTPYLRPQECGNKTDVRWATITNDQG 954
Query: 1020 FGIYTSKYGSSPPMQMSASYYSTSELDRAGHDYELVKGDNIEVHLDHKHMGLGGDDSWSP 1079
G P ++++A YS EL+ H Y+L D++ V +++K MG+GGDDSW
Sbjct: 955 RGFLIK---GLPTVELNALPYSPFELEAYDHFYKLPASDSVTVRVNYKQMGVGGDDSWQA 1011
Query: 1080 CVHDQY 1085
H Y
Sbjct: 1012 KTHPNY 1017
>BGAL_ENTCL (Q47077) Beta-galactosidase (EC 3.2.1.23) (Lactase)
Length = 1028
Score = 657 bits (1694), Expect = 0.0
Identities = 408/1134 (35%), Positives = 583/1134 (50%), Gaps = 140/1134 (12%)
Query: 1 MASSPSSSLVGPLLLAPNNGYKVWEDPSFIKWRKRDPHVHLHCHESVEGSLKYWYQRSKV 60
M+ +PS +L LLA + WE+P +W + H LH + +
Sbjct: 1 MSDTPSLTLSA--LLARRD----WENPVVTQWNRLAAHAPLHSWRNEPSA---------- 44
Query: 61 DYLVSQSAVWKDDAVNGALESAAFWVKDLPFVKSLSGYWKFFLASNPCNVPAKFHDSEFQ 120
+DDA + ++ L+G W+F + P VP + +
Sbjct: 45 ----------RDDAGSARRQT-------------LNGLWRFSYFTAPEQVPQAWVTEDCA 81
Query: 121 DSEWSTLPVPSNWQLHGFDRPIYTNVTYPFPLDPPFVPTENPTGCYRMDFHLPKEW-EGR 179
D+ +PVPSNWQ+ GFD PIYTNVTYP P++PPFVP ENPTGCY + F + W +
Sbjct: 82 DAV--AMPVPSNWQMQGFDTPIYTNVTYPIPVNPPFVPQENPTGCYSLTFEVDDAWLQSG 139
Query: 180 RILLHFEAVDSAFCAWINGHPIGYSQDSRLPAEFEVTDFCHPCGSDLKNVLAVQVFRWSD 239
+ + F+ V+SAF W NG IGYSQDSRLPAEF+++ P +N LAV V RW D
Sbjct: 140 QTRIIFDGVNSAFHLWCNGQWIGYSQDSRLPAEFDLSAALRPG----QNRLAVMVLRWCD 195
Query: 240 GCYLEDQDHWRMSGIHRDVLLLAKPEVFITDYFFKSNLAEDFSSAEMLVEVKIDRLQDTS 299
G YLEDQD WRMSGI RDV LL KPE I DY ++L + A + V+V T
Sbjct: 196 GSYLEDQDMWRMSGIFRDVTLLHKPETQIADYHVVTDLNAELDRAVLKVDV-------TL 248
Query: 300 IDNVLTNYTIEATLYDSGSWESSDGNPDLLSSNVADITFQPTTTPLGFYG-----YTLVG 354
+ + TL+ G A ++ QP + + G T+
Sbjct: 249 AGAGFADGEVVFTLWRKGE-------------KCASVSRQPGSAIVDERGSWAERLTVTI 295
Query: 355 KLQSPKLWSAEQPYLYTLVVVLKDKSGRVLDCESSQVGFKNVSKAHKQLLVNGHPVVIRG 414
+++P LWSAE P LY L + L + G VL+ E+ VGF+ V ++ L +NG P++IRG
Sbjct: 296 PVETPALWSAETPELYRLTMALLNPQGEVLEIEACDVGFRRVEISNGLLKLNGKPLLIRG 355
Query: 415 VNRHEHHPEVGKANIESCMVKDLVLMKQNNINAVRNSHYPQHPRWYELCDLFGMYMIDEA 474
VNRHEHH E G+ E+ M +D+ MKQ++ NAVR SHYP HP WY+LCD +G+Y++DEA
Sbjct: 356 VNRHEHHSENGQVMDEATMRRDIETMKQHSFNAVRCSHYPNHPLWYQLCDRYGLYVVDEA 415
Query: 475 NIETHGFDYSKHLKHPTLEPMWATAMLDRVIGMVERDKNHTCIISWSLGNESGFGTNHFA 534
NIETHG L +P W AM +RV MV+RD+NH II WSLGNESG G NH A
Sbjct: 416 NIETHGMVPMSRLAD---DPRWLPAMSERVTRMVQRDRNHPSIIIWSLGNESGHGANHDA 472
Query: 535 MAGWIRGRDSSRVIHYEGGGSRTPCTDIVCPMYMRV-----------WDMLKIANDPTET 583
+ W++ D +R + YEGGG+ T TDIVCPMY RV W + K P ET
Sbjct: 473 LYRWLKTTDPTRPVQYEGGGANTAATDIVCPMYARVDRDQPFPAVPKWSIKKWIGMPDET 532
Query: 584 RPLILCEYSHAMGNSNGNLHIYWEAIDNTFGLQGGFIWDWVDQALRKVQADGTKQWAYGG 643
RPLILCEY+HAMGNS G YW+A + LQGGF+WDWVDQAL K DG WAYGG
Sbjct: 533 RPLILCEYAHAMGNSFGGFAKYWQAFRSHPRLQGGFVWDWVDQALTKRDEDGNTFWAYGG 592
Query: 644 EFGDIPNDLNFCLNGLVWPDRTAHPVLHEV---KFLYQPIKVNLSDGKLEIKNTHFFQTT 700
+FGD PND FCLNGLV+PDRT HP L+E + + +V+ S +E+++ + F+ T
Sbjct: 593 DFGDKPNDRQFCLNGLVFPDRTPHPALYEAHGPQQFFTFTRVSTSPLVIEVQSGYLFRHT 652
Query: 701 EGLEFSWYISADGYKLGSDKLSLPPIKPQSNYVF----DWKSGPWYSLWDSSSSEEIFLT 756
+ +W ++ DG L + +++L + + + + +GP E++L
Sbjct: 653 DNEVLNWTVARDGDVLVAGEVTLAMVPEGTQRLEIALPELNAGPG----------EVWLN 702
Query: 757 ITAKLLNSTRWVEAGHVVTTAQVQLPAKRDIVPHAINIGSGNLVVETLGDTIK--VSQQD 814
+ + +T W A + + + ++ V T D I +Q
Sbjct: 703 VEVRQPRATPWSPAAIAAAGSSGRFRLRSLLLRQPRR----RAAVLTQTDRILEIAHRQQ 758
Query: 815 VWDITFNTKTGLIESWKVKGVHVMNKGIHPCFWRASIDNDKGGGA------DSYLSRWKA 868
W F+ +G + W GV + + RA +DND G ++++ RWKA
Sbjct: 759 RWQ--FDRASGNLTQWWRNGVETLLSPLTDNVSRAPLDNDIGVSEATKIDPNAWVERWKA 816
Query: 869 AGIDSVHFIAESCSVQSTTGNAVKLLVVFHGVTKGEEGSLPNQDKSKVLFTTEMTYTIYA 928
AG+ + C + G V + H + + + K LF + + I
Sbjct: 817 AGMYDLTPRVLHCEAEQHAGEVV--VTTQHVL----------EYRGKALFLSRKVWRIDE 864
Query: 929 SGDVILECNVKPNADLPPLPRVGIEMNLEKSLDQVSWYGRGPFECYPDRKAAAQVAVYEK 988
G + + V +D+P R+G+ ++L ++ + V W G GP E YPDRK AAQ +
Sbjct: 865 QGVLHGDIQVDMASDIPEPARIGLSVHLAETPENVRWLGLGPHENYPDRKLAAQQGRWTL 924
Query: 989 SVDELHVPYIVPGESGGRADVRWATFLNKNGFGIYTSKYGSSPPMQMSASYYSTSELDRA 1048
++ +H PYI P E+G R D R + + + S S YS +L
Sbjct: 925 PLEAMHTPYIFPTENGLRCDTR----------ELVVGMHQLNGHFHFSVSRYSQQQLRET 974
Query: 1049 GHDYELVKGDNIEVHLDHKHMGLGGDDSWSPCVHDQYLVP--PVPYSFSVRLSP 1100
H + L + ++LD HMG+GGDDSWSP V ++++ + Y+FS + +P
Sbjct: 975 THHHLLREEPGCWLNLDAFHMGVGGDDSWSPSVSPEFILQTRQLRYTFSWQQNP 1028
>BGAL_KLEPN (P06219) Beta-galactosidase (EC 3.2.1.23) (Lactase)
Length = 1034
Score = 625 bits (1611), Expect = e-178
Identities = 383/996 (38%), Positives = 519/996 (51%), Gaps = 80/996 (8%)
Query: 128 PVPSNWQLHGFDRPIYTNVTYPFPLDPPFVPTENPTGCYRMDFHLPKEW-EGRRILLHFE 186
PVPSNWQ+ G+D PIYTNV YP PP VP +NPTGCY + F + W E + + F+
Sbjct: 91 PVPSNWQMEGYDAPIYTNVRYPIDTTPPRVPEDNPTGCYSLHFTVEDTWRENGQTQIIFD 150
Query: 187 AVDSAFCAWINGHPIGYSQDSRLPAEFEVTDFCHPCGSDLKNVLAVQVFRWSDGCYLEDQ 246
V+SAF W NG +GYSQDSRLPA F+++ F P N L V V RWS G +LEDQ
Sbjct: 151 GVNSAFHLWCNGVWVGYSQDSRLPAAFDLSPFLRPGD----NRLCVMVMRWSAGSWLEDQ 206
Query: 247 DHWRMSGIHRDVLLLAKPEVFITDYFFKSNLAEDFSSAEMLVEVKIDRLQDTSIDNVLTN 306
D WRMSGI R V LL KP+ + D L + + V+ I+ + + L
Sbjct: 207 DMWRMSGIFRSVWLLNKPQQRLCDVQLTPALDALYRDGTLQVQATIE-----ATEAALAG 261
Query: 307 YTIEATLYDSGSWESSDGNPDLLSSNVADITFQPTTTPLGFYGYTLVGKLQ--SPKLWSA 364
++ +L+ G + + G L + PT G Y + L +P WSA
Sbjct: 262 LSVGVSLW-RGEEQFAAGRQPLGT---------PTVDERGHYAERVDFSLAVATPAHWSA 311
Query: 365 EQPYLYTLVVVLKDKSGRVLDCESSQVGFKNVSKAHKQLLVNGHPVVIRGVNRHEHHPEV 424
E P Y VV L + +L+ E+ +GF+ + A L +NG P++IRGVNRHEHH
Sbjct: 312 ETPNCYRAVVTLW-RGDELLEAEAWDIGFRRIEIADGLLRLNGKPLLIRGVNRHEHHHLR 370
Query: 425 GKANIESCMVKDLVLMKQNNINAVRNSHYPQHPRWYELCDLFGMYMIDEANIETHGFDYS 484
G+ E+ MV+D++LMKQNN NAVR SHYP PRWYELC+ +G+Y++DEANIETHG
Sbjct: 371 GQVVTEADMVQDILLMKQNNFNAVRCSHYPNAPRWYELCNRYGLYVVDEANIETHGMVPM 430
Query: 485 KHLKHPTLEPMWATAMLDRVIGMVERDKNHTCIISWSLGNESGFGTNHFAMAGWIRGRDS 544
L +P W A RV MV+ ++NH CII WSLGNESG G NH A+ W++ D
Sbjct: 431 NRLSD---DPAWLPAFSARVTRMVQSNRNHPCIIIWSLGNESGGGGNHEALYHWLKRNDP 487
Query: 545 SRVIHYEGGGSRTPCTDIVCPMYMRV-----------WDMLKIANDPTETRPLILCEYSH 593
SR + YEGGG+ T TDI+CPMY RV W + K + P E RPLILCEY+H
Sbjct: 488 SRPVQYEGGGADTTATDIICPMYARVERDQPIPAVPKWGIKKWISLPGEQRPLILCEYAH 547
Query: 594 AMGNSNGNLHIYWEAIDNTFGLQGGFIWDWVDQALRKVQADGTKQWAYGGEFGDIPNDLN 653
AMGNS GN YW+A LQGGFIWDW DQA+RK ADG+ WAYGG+FGD PND
Sbjct: 548 AMGNSLGNFADYWQAFREYPRLQGGFIWDWADQAIRKTFADGSVGWAYGGDFGDKPNDRQ 607
Query: 654 FCLNGLVWPDRTAHPVLHEVKFLYQPIKVNL---SDGKLEIKNTHFFQTTEGLEFSWYIS 710
FC+NGLV+PDRT HP L E K Q + L S ++ I + + F+ T+ W +
Sbjct: 608 FCMNGLVFPDRTPHPSLVEAKHAQQYFQFTLLSTSPLRVRIISEYLFRPTDNEVVRWQVQ 667
Query: 711 ADGYKL--GSDKLSLPPIKPQSNYVFDWKSGPWYSLWDSSSSEEIFLTITAKLLNSTRWV 768
A G L G L+LPP + D SL + ++LT+ +T W
Sbjct: 668 AAGEPLYHGDLTLALPPEGSDEITLLD-------SLILPEGARAVWLTLEVTQPQATAWS 720
Query: 769 EAGHVVTTAQVQLPAKRDIVPHAINIGSGNLVVETLGDTIKVSQQDVWDITFNTKTGLIE 828
EA H V Q LPA +L+V I+ Q W T + +TGL+
Sbjct: 721 EAEHRVAWQQFPLPAPLGCRRPPCLPALPDLIVSDEVWQIRAGSQ-CW--TIDRRTGLLS 777
Query: 829 SWKVKGVHVMNKGIHPCFWRASIDNDKGGG------ADSYLSRWKAAGIDSVHFIAESCS 882
W V G + + F RA +DND G ++++ RW++AG+ + C
Sbjct: 778 RWSVGGQEQLLTPLRDQFIRAPLDNDIGVSEVERIDPNAWVERWRSAGLYDLEAHCVQCD 837
Query: 883 VQSTTGNAVKLLVVFHGVTKGEEGSLPNQDKSKVLFTTEMTYTIYASGDVILECNVKPNA 942
Q L+ +GEE V+ + A G + L + +
Sbjct: 838 AQRLANET--LVDCRWHYLRGEE----------VVIVSHWRMHFTADGTLRLAVDGERAE 885
Query: 943 DLPPLPRVGIEMNLEKSLDQVSWYGRGPFECYPDRKAAAQVAVYEKSVDELHVPYIVPGE 1002
LPPLPRVG+ + VSW G GP E YPDR+++A A +E+ + + PYI P E
Sbjct: 886 TLPPLPRVGLHFQVADQQAPVSWLGLGPHENYPDRRSSACFARWEQPLAAMTTPYIFPTE 945
Query: 1003 SGGRADVRWATFLNKNGFGIYTSKYGSSPPMQMSASYYSTSELDRAGHDYELVKGDNIEV 1062
+G R D + + ++ S S +ST +L H +++ D + +
Sbjct: 946 NGLRCDTQ----------ALDWGRWHISGHFHFSVQPWSTRQLMETDHWHKMQAEDGVWI 995
Query: 1063 HLDHKHMGLGGDDSWSPCVHDQYLVPPVPYSFSVRL 1098
LD HMG+GGDDSW+P V Q+L+ + + V L
Sbjct: 996 TLDGLHMGVGGDDSWTPSVLPQWLLSQTRWQYEVSL 1031
>BGAL_LACLA (Q48727) Beta-galactosidase (EC 3.2.1.23) (Lactase)
Length = 998
Score = 580 bits (1494), Expect = e-165
Identities = 371/1056 (35%), Positives = 533/1056 (50%), Gaps = 106/1056 (10%)
Query: 70 WKDDAVNGALESAAFWVKDLPFVKSLSGYWKFFLASNPCNVPAKFHDSEFQDSEWSTLPV 129
W++ V+ DL +SL+G W F S +VP + + +E + V
Sbjct: 15 WENPVVSNWNRLPMHTPMDLLEKQSLNGLWNFDHFSRISDVPKNWLELTESKTE---IIV 71
Query: 130 PSNWQLHGFDR---PIYTNVTYPFPLDPPFVPTENPTGCYRMDFHLPKEW-EGRRILLHF 185
PSNWQ+ D+ PIYTNVTYP P+ PP+VP NP G Y F + KEW E + L F
Sbjct: 72 PSNWQIEFKDKSDVPIYTNVTYPIPIQPPYVPEANPVGAYSRYFDITKEWLESGHVHLTF 131
Query: 186 EAVDSAFCAWINGHPIGYSQDSRLPAEFEVTDFCHPCGSDLKNVLAVQVFRWSDGCYLED 245
E V SAF W+NG GYS+DSRLPAEF++++ + +N L V VFRWS Y ED
Sbjct: 132 EGVGSAFHFWLNGEYGGYSEDSRLPAEFDISNLA----KEGQNCLKVLVFRWSKVTYFED 187
Query: 246 QDHWRMSGIHRDVLLLAKPEVFITDYFFKSNLAEDFSSAEMLVEVKIDRLQDTSIDNVLT 305
QD WRMSGI R V L P+ ++ D+ K++L ED A + ++ + D +
Sbjct: 188 QDMWRMSGIFRSVNLQWLPDNYLLDFSIKTDLDEDLDFANVKLQAYAKNIDDACL----- 242
Query: 306 NYTIEATLYDSGSWESSDGNPDLLSSNVADITFQPTTTPLGFYGYTLVGKLQSPKLWSAE 365
E LYD E G +G+ + +PKLWS E
Sbjct: 243 ----EFKLYDD---EQLIGE---------------------CHGFDAEIGVVNPKLWSDE 274
Query: 366 QPYLYTLVVVLKDKSGRVLDCESSQVGFKNVSKAHKQLLVNGHPVVIRGVNRHEHHPEVG 425
PYLY L + L D+SG V E+ ++G + ++ QL +NG +++RGVN+HE PE G
Sbjct: 275 IPYLYRLELTLMDRSGAVFHKETKKIGIRKIAIEKGQLKINGKALLVRGVNKHEFTPEHG 334
Query: 426 KANIESCMVKDLVLMKQNNINAVRNSHYPQHPRWYELCDLFGMYMIDEANIETHGFDYSK 485
E M+KD+ LMK++N NAVR SHYP RWYELCD +G+Y++DEANIETHG
Sbjct: 335 YVVSEEVMIKDIKLMKEHNFNAVRCSHYPNDSRWYELCDEYGLYVMDEANIETHGMTPMN 394
Query: 486 HLKHPTLEPMWATAMLDRVIGMVERDKNHTCIISWSLGNESGFGTNHFAMAGWIRGRDSS 545
L T +P + M +RV MV RD+NH II WSLGNESG+G+NH A+ W + DSS
Sbjct: 395 RL---TNDPTYLPLMSERVTRMVMRDRNHPSIIIWSLGNESGYGSNHQALYDWCKSFDSS 451
Query: 546 RVIHYEGGG----SRTPCTDIVCPMYMRV--------WDMLKIANDPTETRPLILCEYSH 593
R +HYEGG T TDI+CPMY RV + + E RPLILCEY+H
Sbjct: 452 RPVHYEGGDDASRGATDATDIICPMYARVDSPSINAPYSLKTWMGVAGENRPLILCEYAH 511
Query: 594 AMGNSNGNLHIYWEAIDNTFGLQGGFIWDWVDQALRKVQADGTKQWAYGGEFGDIPNDLN 653
MGNS G YW+A LQGGFIWDWVDQ L K DG +AYGG+FGD PND
Sbjct: 512 DMGNSLGGFGKYWQAFREIDRLQGGFIWDWVDQGLLK---DG--NYAYGGDFGDKPNDRQ 566
Query: 654 FCLNGLVWPDRTAHPVLHEVKFLYQPIKVNLSDGKL------EIKNTHFFQTTEGLEFSW 707
F LNGLV+P+R A P L E K+ Q + L L + N + F++T+ + +
Sbjct: 567 FSLNGLVFPNRQAKPALREAKYWQQYYQFELEKTPLGQVFAFTVTNEYLFRSTDNEKLCY 626
Query: 708 YISADGYKLGSDKLSLPPIKPQSNYVFDWKSGPWYSLWDSSSSEEIFLTITAKLLNSTRW 767
+ L ++L L + + D P ++ +FL I K +
Sbjct: 627 QLINGLEVLWENELIL-NMPAGGSMRIDLSELP------IDGTDNLFLNIQVKTIEKCNL 679
Query: 768 VEAGHVVTTAQVQLPAKRDIVPHAINIGSGNLVVETLGDTIKVSQQDVWDITFNTKTGLI 827
+E+ V Q L K + + L + T++ ++Q FN G +
Sbjct: 680 LESDFEVAHQQFVLQEKINFTDRIDSNEEITLFEDEELLTVRSAKQ---KFIFNKSNGNL 736
Query: 828 ESW-KVKGVHVMNKGIHPCFWRASIDNDKGGG------ADSYLSRWKAAGIDSVHFIAES 880
W KG + + F RA +DND G +++L RWK G + + ++
Sbjct: 737 SRWLDEKGNEKLLHELSEQFTRAPLDNDIGVSEVEHIDPNAWLERWKGIGFYELKTLLKT 796
Query: 881 CSVQSTTGNAVKLLVVFHGVTKGEEGSLPNQDKSKVLFTTEMTYTIYASGDVILECNVKP 940
+Q+T + + + + K K+ F+T Y I+ +G+++L+ + K
Sbjct: 797 MIIQATENEVIISVQTDY------------EAKGKIAFSTIREYHIFRNGELLLKVDFKR 844
Query: 941 NADLPPLPRVGIEMNLEKSLDQVSWYGRGPFECYPDRKAAAQVAVYEKSVDELHVPYIVP 1000
N + P R+G+ + L + + V+++G GP E YPDR+ A+ + + ++ PYI P
Sbjct: 845 NIEFPEPARIGLSLQLAEKAENVTYFGLGPDENYPDRRGASLFGQWNLRITDMTTPYIFP 904
Query: 1001 GESGGRADVRWATFLNKNGFGIYTSKYGSSPPMQMSASYYSTSELDRAGHDYELVKGDNI 1060
E+G R + R N + G S + S YS ++L + GH + L +
Sbjct: 905 SENGLRMETR-----ELNYDRLKVRAMGQS--FAFNLSPYSQNQLAKKGHWHLLEEEAGT 957
Query: 1061 EVHLDHKHMGLGGDDSWSPCVHDQYLVPPVPYSFSV 1096
+++D HMG+GGDDSWSP V +YL+ Y + V
Sbjct: 958 WLNIDGFHMGVGGDDSWSPSVAQEYLLTKGNYHYEV 993
>BGAL_BACHD (Q9K9C6) Beta-galactosidase (EC 3.2.1.23) (Lactase)
Length = 1014
Score = 531 bits (1369), Expect = e-150
Identities = 335/1025 (32%), Positives = 501/1025 (48%), Gaps = 103/1025 (10%)
Query: 94 SLSGYWKFFLASNPCNVPAKFHDSEFQDSEWSTLPVPSNWQLHGFDRPIYTNVTYPFP-- 151
SL+G+WKF A NP P +F+ F W + VP + QL G+ +P Y N YP+
Sbjct: 47 SLNGHWKFHYAINPNTRPKEFYQLGFDCKCWDDILVPGHIQLQGYGKPQYVNTMYPWDGH 106
Query: 152 --LDPPFVPTE-NPTGCYRMDFHLPKEWEGRRILLHFEAVDSAFCAWINGHPIGYSQDSR 208
L PP +P + NP G Y F +P + + F+ V++AF W+NG +GYS+DS
Sbjct: 107 HHLRPPEIPEDDNPVGSYVKYFDIPNNMSNHPLFISFQGVETAFYVWLNGEFVGYSEDSF 166
Query: 209 LPAEFEVTDFCHPCGSDLKNVLAVQVFRWSDGCYLEDQDHWRMSGIHRDVLLLAKPEVFI 268
PAEF++T P + +N L V+V++ S G +LEDQD WR SGI RDV L P + +
Sbjct: 167 TPAEFDLT----PYAVEGENKLCVEVYQRSTGSWLEDQDFWRFSGIFRDVYLYTIPNIHV 222
Query: 269 TDYFFKSNLAEDFSSAEMLVEVKIDRLQDTSIDNVLTNYTIEATLYDSGSWESSDGNPDL 328
D +++L + + +++ R Q+ + V Y E + + +++ +
Sbjct: 223 YDMHVRADLDRSLQTGILETTLELKRSQEKEVMIVAELYDAEGAVVATADMKTNQDQATV 282
Query: 329 LSSNVADITFQPTTTPLGFYGYTLVGKLQSPKLWSAEQPYLYTLVVVLKDKSGRVLDCES 388
S + SP LWSAE PYLY L + L D++G +++
Sbjct: 283 SMS------------------------VDSPALWSAEDPYLYKLFLKLFDENGTLVEVVP 318
Query: 389 SQVGFKNVSKAHKQLLVNGHPVVIRGVNRHEHHPEVGKANIESCMVKDLVLMKQNNINAV 448
++GF+ + + +NG +V +GVNRHE + G+ + M++D+ MK++NINAV
Sbjct: 319 QKIGFRRFELVNNIMTLNGKRIVFKGVNRHEFNGRTGRVVTKEDMLEDIKTMKKHNINAV 378
Query: 449 RNSHYPQHPRWYELCDLFGMYMIDEANIETHG-------FDYSKHLKHPTLEPMWATAML 501
R SHYP + WY+LCD +G+Y+IDE N+ETHG + S ++ LE W ++
Sbjct: 379 RTSHYPNNSEWYQLCDEYGLYVIDEMNLETHGSWQKLGKVEPSWNIPGNHLE--WEPIVM 436
Query: 502 DRVIGMVERDKNHTCIISWSLGNESGFGTNHFAMAGWIRGRDSSRVIHYEG---GGSRTP 558
DR + M ERDKNH I+ WS GNES G ++ + + D SR++HYEG +
Sbjct: 437 DRAVSMFERDKNHPSILIWSCGNESYAGEVILNVSRYFKSVDPSRLVHYEGVFHARAYDA 496
Query: 559 CTDIVCPMYMRVWDMLK-IANDPTETRPLILCEYSHAMGNSNGNLHIYWEAIDNTFGLQG 617
+D+ MY + D+ + NDP +P I CEY HAMGNS G +H Y E QG
Sbjct: 497 TSDMESRMYAKPKDIEDYLTNDP--KKPYISCEYMHAMGNSLGGMHKYTELEQKYPMYQG 554
Query: 618 GFIWDWVDQALRKVQADGTKQWAYGGEFGDIPNDLNFCLNGLVWPDRTAHPVLHEVKFLY 677
GFIWD++DQAL K G + +AYGG+FGD P D +FC NG+V+ DR P + EVKFLY
Sbjct: 555 GFIWDYIDQALLKKDRYGKEYFAYGGDFGDRPTDYSFCANGIVYADRKPSPKMQEVKFLY 614
Query: 678 QPIKVNLSDGKLEIKNTHFFQTTEGLEFSWYISADGYKLGSDKLSLPPIKPQSNYV-FDW 736
Q IK+ + +KN + F T + + + +G +L K + + Y+ FDW
Sbjct: 615 QNIKLVPDREGVLVKNENLFTDTSAYQLEYVLYWEGTELYRKKQDVFVAPQEEVYLPFDW 674
Query: 737 KSGPWYSLWDSSSSEEIFLTITAKLLNSTRWVEAGHVVTTAQVQLPAKRDIVPHAINIGS 796
+ S E + L W E GH V Q H +G+
Sbjct: 675 LE------QGMNESGEYCIHTMLTLKQDQLWAEKGHEVAFGQ-----------HVYRMGA 717
Query: 797 GNLVVETLGDTIKVSQQDV--------WDITFNTKTGLIESWKVKGVHVMNKGIHPCFWR 848
G +KV DV + + F+ G + S G ++ + P FWR
Sbjct: 718 IQKERNARG-ALKVVHGDVNIGIHGEDFSVLFSKAVGSLVSLHYAGKEMIEQPPMPLFWR 776
Query: 849 ASIDNDKGGGADSYLSRWKAAGIDSVHFIAESC---SVQSTTGNAVKLLVVFHGVTKGEE 905
A+ DNDKG + W AA +A C V+ G+ V +L +H
Sbjct: 777 ATTDNDKGCSQLYHSGIWYAAS------LARKCVNMEVEEKPGH-VSVLFTYH------- 822
Query: 906 GSLPNQDKSKVLFTTEMTYTIYASGDVILECNV---KPNADLPPLPRVGIEMNLEKSLDQ 962
D+ +V ++ YT++ G + + K LP LP + L +Q
Sbjct: 823 --FAISDRVEV----KVGYTVFPDGSLRVRSTYQASKGGETLPQLPMFALSFKLPADYEQ 876
Query: 963 VSWYGRGPFECYPDRKAAAQVAVYEKSVDELHVPYIVPGESGGRADVRWATFLNKNGFGI 1022
+ WY GP E Y DR A++ ++ V++ Y+ P ESG R VR L+ NG GI
Sbjct: 877 LEWYALGPEENYADRATGARLCTFKNKVEDSLSQYVTPQESGNRTGVRTVKILDANGQGI 936
Query: 1023 YTSKYGSSPPMQMSASYYSTSELDRAGHDYELVKGDNIEVHLDHKHMGLGGDDSWSPCVH 1082
P++ S Y+ EL++A H YEL V++ K MG+GGDDSW VH
Sbjct: 937 EVCSV--EEPIECQISPYTAFELEQASHPYELPNVHYTVVNVAGKQMGVGGDDSWGAPVH 994
Query: 1083 DQYLV 1087
D+Y++
Sbjct: 995 DEYVL 999
>BGA2_ECOLI (P06864) Evolved beta-galactosidase alpha-subunit (EC
3.2.1.23) (Lactase)
Length = 1030
Score = 531 bits (1369), Expect = e-150
Identities = 332/1018 (32%), Positives = 523/1018 (50%), Gaps = 70/1018 (6%)
Query: 95 LSGYWKFFLASNPCNVPAKFHDSEFQDSEWSTLPVPSNWQLHGFDRPIYTNVTYPFPLDP 154
LSG W F +P VP F D W + VP+ WQ+ G + YT+ +PFP+D
Sbjct: 44 LSGQWNFHFFDHPLQVPEAFTSELMAD--WGHITVPAMWQMEGHGKLQYTDEGFPFPIDV 101
Query: 155 PFVPTENPTGCYRMDFHLPKEWEGRRILLHFEAVDSAFCAWINGHPIGYSQDSRLPAEFE 214
PFVP++NPTG Y+ F L W+G++ L+ F+ V++ F ++NG +G+S+ SRL AEF+
Sbjct: 102 PFVPSDNPTGAYQRIFTLSDGWQGKQTLIKFDGVETYFEVYVNGQYVGFSKGSRLTAEFD 161
Query: 215 VTDFCHPCGSDLKNVLAVQVFRWSDGCYLEDQDHWRMSGIHRDVLLLAKPEVFITDYFFK 274
++ N+L V+V +W+D Y+EDQD W +GI RDV L+ K I D+ +
Sbjct: 162 ISAMVKTGD----NLLCVRVMQWADSTYVEDQDMWWSAGIFRDVYLVGKHLTHINDFTVR 217
Query: 275 SNLAEDFSSAEMLVEVKIDRLQDTSIDNVLTNYTIEATLYDSGSWESSDGNPDLLSSNVA 334
++ E + A + EV ++ L + + T+E TL+D G + SS +
Sbjct: 218 TDFDEAYCDATLSCEVVLENLAASPVVT-----TLEYTLFD--------GERVVHSSAID 264
Query: 335 DITFQPTTTPLGFYGYTLVGKLQSPKLWSAEQPYLYTLVVVLKDKSGRVLDCESSQVGFK 394
+ + T+ + +T+ + P+ WSAE PYLY LV+ LKD +G VL+ +VGF+
Sbjct: 265 HLAIEKLTS--ASFAFTV----EQPQQWSAESPYLYHLVMTLKDANGNVLEVVPQRVGFR 318
Query: 395 NVSKAHKQLLVNGHPVVIRGVNRHEHHPEVGKANIESCMVKDLVLMKQNNINAVRNSHYP 454
++ +N V++ GVNRH++ G+A + KDL LMKQ+NIN+VR +HYP
Sbjct: 319 DIKVRDGLFWINNRYVMLHGVNRHDNDHRKGRAVGMDRVEKDLQLMKQHNINSVRTAHYP 378
Query: 455 QHPRWYELCDLFGMYMIDEANIETHGFDYSKHLKHPTLEPMWATAMLDRVIGMVERDKNH 514
PR+YELCD++G++++ E ++E+HGF + T +P W ++R++ + KNH
Sbjct: 379 NDPRFYELCDIYGLFVMAETDVESHGFANVGDISRITDDPQWEKVYVERIVRHIHAQKNH 438
Query: 515 TCIISWSLGNESGFGTNHFAMAGWIRGRDSSRVIHYEGGGSRTPCTDIVCPMYMRVWDML 574
II WSLGNESG+G N AM + D +R++HYE DI+ MY RV M
Sbjct: 439 PSIIIWSLGNESGYGCNIRAMYHAAKALDDTRLVHYEEDRD-AEVVDIISTMYTRVPLMN 497
Query: 575 KIANDPTETRPLILCEYSHAMGNSNGNLHIYWEAIDNTFGLQGGFIWDWVDQALRKVQAD 634
+ P +P I+CEY+HAMGN G L Y +QG ++W+W D ++
Sbjct: 498 EFGEYP-HPKPRIICEYAHAMGNGPGGLTEYQNVFYKHDCIQGHYVWEWCDHGIQAQDDH 556
Query: 635 GTKQWAYGGEFGDIPNDLNFCLNGLVWPDRTAHPVLHEVKFLYQPIKVNLSD---GKLEI 691
G + +GG++GD PN+ NFCL+GL++ D+T P L E K + P+K++ D G+L++
Sbjct: 557 GNVWYKFGGDYGDYPNNYNFCLDGLIYSDQTPGPGLKEYKQVIAPVKIHARDLTRGELKV 616
Query: 692 KNTHFFQTTEGLEFSWYISADGYKLGSDKLSLPPIKPQSNYVFDWKSGPWYSLWDSSSSE 751
+N +F T + + A+G L + ++ L + P S P +
Sbjct: 617 ENKLWFTTLDDYTLHAEVRAEGETLATQQIKLRDVAPNS-------EAPLQITLPQLDAR 669
Query: 752 EIFLTITAKLLNSTRWVEAGHVVTTAQVQLPAKRDI---VPHAINIGSG-NLVVETLGDT 807
E FL IT + TR+ EAGH + T Q P K + VP A N L + L T
Sbjct: 670 EAFLNITVTKDSRTRYSEAGHPIAT--YQFPLKENTAQPVPFAPNNARPLTLEDDRLSCT 727
Query: 808 IKVSQQDVWDITFNTKTGLIESWKVKGVHVMNKGIHPCFWRASIDNDKGGGADSYLSRWK 867
++ + ITF+ +G SW+V G ++ + F++ IDN K Y W+
Sbjct: 728 VRGYN---FAITFSKMSGKPTSWQVNGESLLTREPKINFFKPMIDNHK----QEYEGLWQ 780
Query: 868 AAGIDSVHFIAESCSVQSTTGNAVKLLVVFHGVTKGEEGSLPNQDKSKVLFTTEMTY--T 925
+ + +V+ + G ++L++ V + P D F TY
Sbjct: 781 PNHLQIMQEHLRDFAVEQSDG---EVLIISRTVI-----APPVFD-----FGMRCTYIWR 827
Query: 926 IYASGDVILECNVKPNADLPP-LPRVGIEMNLEKSLDQVSWYGRGPFECYPDRKAAAQVA 984
I A G V + + + D P +P +G M + DQV++YGRGP E Y D + A +
Sbjct: 828 IAADGQVNVALSGERYGDYPHIIPCIGFTMGINGEYDQVAYYGRGPGENYADSQQANIID 887
Query: 985 VYEKSVDELHVPYIVPGESGGRADVRWATFLNKNGFGIYTSKYGSSPPMQMSASYYSTSE 1044
++ +VD + Y P +G R VRW N++G G+ P+ SA +Y+
Sbjct: 888 IWRSTVDAMFENYPFPQNNGNRQHVRWTALTNRHGNGLLVV---PQRPINFSAWHYTQEN 944
Query: 1045 LDRAGHDYELVKGDNIEVHLDHKHMGLGGDDSWSPCVHDQYLVPPVPYSFSVRLSPVT 1102
+ A H EL + D+I ++LDH+ +GL G +SW V D + V +S+ L PV+
Sbjct: 945 IHAAQHCNELQRSDDITLNLDHQLLGL-GSNSWGSEVLDSWRVWFRDFSYGFTLLPVS 1001
>BGAL_STRTR (P23989) Beta-galactosidase (EC 3.2.1.23) (Lactase)
Length = 1026
Score = 514 bits (1325), Expect = e-145
Identities = 341/1053 (32%), Positives = 519/1053 (48%), Gaps = 117/1053 (11%)
Query: 93 KSLSGYWKFFLASNPCNVPAKFHDSEFQDSEWSTLPVPSNWQLHGFDRPIYTNVTYPFP- 151
+SL+G WK A N V F+ +EF +++ + + VP + +L GF P Y N YP+
Sbjct: 46 QSLNGKWKIHYAQNTNQVLKDFYKTEFDETDLNFINVPGHLELQGFGSPQYVNTQYPWDG 105
Query: 152 ---LDPPFVPTE-NPTGCYRMDFHLPKEWEGRRILLHFEAVDSAFCAWINGHPIGYSQDS 207
L PP VP E N Y F L + +++ + F+ V ++ W+NG+ +GYS+DS
Sbjct: 106 KEFLRPPQVPQESNAVASYVKHFTLNDALKDKKVFISFQGVATSIFVWVNGNFVGYSEDS 165
Query: 208 RLPAEFEVTDFCHPCGSDLKNVLAVQVFRWSDGCYLEDQDHWRMSGIHRDVLLLAKPEVF 267
P+EFE++D+ + N LAV V+R+S +LEDQD WR+ GI RDV L A P+V
Sbjct: 166 FTPSEFEISDYL----VEGDNKLAVAVYRYSTASWLEDQDFWRLYGIFRDVYLYAIPKVH 221
Query: 268 ITDYFFKSNLAEDFSSAEMLVEVK-IDRLQDTSIDNVLTNYTIEATLYDSGSWESSDGNP 326
+ D F K + + ++ +++K + +D I VL++Y E + + + + DG
Sbjct: 222 VQDLFVKGDYDYQTKAGQLDIDLKTVGDYEDKKIKYVLSDY--EGIVTEGDASVNGDGE- 278
Query: 327 DLLSSNVADITFQPTTTPLGFYGYTLVGKLQSPKLWSAEQPYLYTLVVVLKDKSGRVLDC 386
LS ++ ++ +P WSAE P LY L++ + D +V++
Sbjct: 279 --LSVSLENLKIKP---------------------WSAESPKLYDLILHVLDDD-QVVEV 314
Query: 387 ESSQVGFKNVSKAHKQLLVNGHPVVIRGVNRHEHHPEVGKANIESCMVKDLVLMKQNNIN 446
+VGF+ K +L+NG +V +GVNRHE + G+ E M+ D+ +MKQ+NIN
Sbjct: 315 VPVKVGFRRFEIKDKLMLLNGKRIVFKGVNRHEFNARTGRCITEEDMLWDIKVMKQHNIN 374
Query: 447 AVRNSHYPQHPRWYELCDLFGMYMIDEANIETHGFDYSKHLKHPT-----LEPMWATAML 501
AVR SHYP RWYELCD +G+Y+IDEAN+ETHG L P+ EP W A L
Sbjct: 375 AVRTSHYPNQTRWYELCDEYGLYVIDEANLETHGTWQKLGLCEPSWNIPASEPEWLPACL 434
Query: 502 DRVIGMVERDKNHTCIISWSLGNESGFGTNHFAMAGWIRGRDSSRVIHYEGGG---SRTP 558
DR M +RDKNH +I WS GNES G + MA + R D++R +HYEG
Sbjct: 435 DRANNMFQRDKNHASVIIWSCGNESYAGKDIADMADYFRSVDNTRPVHYEGVAWCREFDY 494
Query: 559 CTDIVCPMYMRVWDMLK----------------------IANDPTETRPLILCEYSHAMG 596
TDI MY + D+ + + N P +P I CEY H MG
Sbjct: 495 ITDIESRMYAKPADIEEYLTTGKLVDLSSVSDKHFASGNLTNKP--QKPYISCEYMHTMG 552
Query: 597 NSNGNLHIYWEAIDNTFGLQGGFIWDWVDQALRKVQADGTKQWAYGGEFGDIPNDLNFCL 656
NS G L +Y + ++ QGGFIWD++DQA+ K +G++ +YGG++ D P+D FC
Sbjct: 553 NSGGGLQLYTD-LEKYPEYQGGFIWDFIDQAIYKTLPNGSEFLSYGGDWHDRPSDYEFCG 611
Query: 657 NGLVWPDRTAHPVLHEVKFLYQPIKVNLSDGKLEIKNTHFFQTTEGLEFSWYISADGYKL 716
NG+V+ DRT P L VK LY IK+ + + + IKN + F+ F + DG K+
Sbjct: 612 NGIVFADRTLTPKLQTVKHLYSNIKIAVDEKSVTIKNDNLFEDLSAYTFLARVYEDGRKV 671
Query: 717 GSDKLSLPPIKPQSNYVFDWKSG-----PWYSLWDSSSSEEIFLTITAKLLNSTRWVEAG 771
+S Y FD K G P + ++S+SE+I+ + L +T W G
Sbjct: 672 S-----------ESEYHFDVKPGEEATFPVNFVVEASNSEQIY-EVACVLREATEWAPKG 719
Query: 772 HVVTTAQ--VQLPAKRDIVPHAINIGSGNLVVETLGDTIKVSQQDVWDITFNTKTGLIES 829
H + Q V+ + V +N+ G+ + G + I + + S
Sbjct: 720 HEIVRGQYVVEKISTETPVKAPLNVVEGDFNIGIQGQN--------FSILLSRAQNTLVS 771
Query: 830 WKVKGVHVMNKGIHPCFWRASIDNDKGGGADSYLSRWKAAGIDSVHFIAESCSVQSTTGN 889
K GV + KG F RA DND+G G ++ WK AG ++ + + +
Sbjct: 772 AKYNGVEFIEKGPKLSFTRAYTDNDRGAGYPFEMAGWKVAG----NYSKVTDTQIQIEDD 827
Query: 890 AVKLLVVFHGVTKGEEGSLPNQDKSKVLFTTEMTYTIYASGDVILECNVKPNADLPPLPR 949
+VK+ V LP +V ++TY + G + + N A LP P
Sbjct: 828 SVKVTYVH---------ELPGLSDVEV----KVTYQVDYKGRIFVTANYDGKAGLPNFPE 874
Query: 950 VGIEMNLEKSLDQVSWYGRGPFECYPDRKAAAQVAVYEKSVDELHVPYIVPGESGGRADV 1009
G+E + +S+YG G E Y D+ A + YE SV++ PY++P ESG
Sbjct: 875 FGLEFAIGSQFTNLSYYGYGAEESYRDKLPGAYLGRYETSVEKTFAPYLMPQESGNHYGT 934
Query: 1010 RWATFLNKNGFGIYTSKYGSSPPMQMSASYYSTSELDRAGHDYELVKGDNIEVHLDHKHM 1069
R T + N G+ + + + SA ST +++ A H YEL + D + + M
Sbjct: 935 REFTVSDDNHNGLKFTALNKA--FEFSALRNSTEQIENARHQYELQESDATWIKVLAAQM 992
Query: 1070 GLGGDDSWSPCVHDQYLVPPV-PYSFSVRLSPV 1101
G+GGDD+W VHD++L+ Y S + P+
Sbjct: 993 GVGGDDTWGAPVHDEFLLSSADSYQLSFMIEPL 1025
>BGAL_CLOAB (P24131) Beta-galactosidase (EC 3.2.1.23) (Lactase)
Length = 897
Score = 488 bits (1255), Expect = e-137
Identities = 306/921 (33%), Positives = 472/921 (51%), Gaps = 87/921 (9%)
Query: 89 LPFVKSLSGYWKFFLASNPCNVPAKFHDSEFQDSEWSTLPVPSNWQLHGFDRPIYTNVTY 148
+P ++L+G W+F + N +F+ EF S + VP + QL G+D+ Y N Y
Sbjct: 43 MPLKQNLNGKWRFSYSENSSLRIKEFYKDEFDVSWIDYIEVPGHIQLQGYDKCQYINTMY 102
Query: 149 PFP----LDPPFVP-TENPTGCYRMDFHLPKEWEGRRILLHFEAVDSAFCAWINGHPIGY 203
P+ L PP + T NP G Y F + E + ++ + F+ V++AF W+NG +GY
Sbjct: 103 PWEGHDELRPPHISKTYNPVGSYVTFFEVKDELKNKQTFISFQGVETAFYVWVNGEFVGY 162
Query: 204 SQDSRLPAEFEVTDFCHPCGSDLKNVLAVQVFRWSDGCYLEDQDHWRMSGIHRDVLLLAK 263
S+D+ P+EF++TD+ + +N LAV+V++ S ++EDQD WR SGI RDV L A
Sbjct: 163 SEDTFTPSEFDITDYLR----EGENKLAVEVYKRSSASWIEDQDFWRFSGIFRDVYLYAV 218
Query: 264 PEVFITDYFFKSNLAEDFSSAEMLVEVKIDRLQDTSIDNVLTNYTIEATLYDSGSWESSD 323
PE + D F K++L +DF +A++ E+K+ +T+++ L E +
Sbjct: 219 PETHVNDIFIKTDLYDDFKNAKLNAELKMIGNSETTVETYL---------------EDKE 263
Query: 324 GNPDLLSSNVADITFQPTTTPLGFYGYTLVGKLQSPKLWSAEQPYLYTLVVVLKDKSGRV 383
GN +S + P + L TL Q+ LWSAE+P LYTL +++ K G +
Sbjct: 264 GNKIAISEKI------PFSDEL-----TLYLDAQNINLWSAEEPNLYTLYILVNKKDGNL 312
Query: 384 LDCESSQVGFKNVSKAHKQLLVNGHPVVIRGVNRHEHHPEVGKANIESCMVKDLVLMKQN 443
++ + ++GF++ K + + ++ +GVNRHE G++ + M+ D+ +KQ+
Sbjct: 313 IEVVTQKIGFRHFEMKDKIMCLKWKRIIFKGVNRHEFSARRGRSITKEDMLWDIKFLKQH 372
Query: 444 NINAVRNSHYPQHPRWYELCDLFGMYMIDEANIETHGFDYSKHLKHPTLE-----PMWAT 498
NINAVR SHYP WY LCD +G+Y+IDE N+E+HG P+ P W
Sbjct: 373 NINAVRTSHYPNQSLWYRLCDEYGIYLIDETNLESHGSWQKMGQIEPSWNVPGSLPQWQA 432
Query: 499 AMLDRVIGMVERDKNHTCIISWSLGNESGFGTNHFAMAGWIRGRDSSRVIHYEGGGSRTP 558
A+LDR MVERDKNH ++ WS GNES G + + M+ + R +D SR++HYEG T
Sbjct: 433 AVLDRASSMVERDKNHPSVLIWSCGNESYAGEDIYQMSKYFRKKDPSRLVHYEGV---TR 489
Query: 559 CTDIVCP-----MYMRVWDMLKIANDPTETRPLILCEYSHAMGNSNGNLHIYWEAIDNTF 613
C + + MY + ++ + ND + +P I CEY H+MGNS G + Y E D
Sbjct: 490 CREFMTRRHESRMYAKAAEIEEYLNDNPK-KPYISCEYMHSMGNSTGGMMKYTELEDKYL 548
Query: 614 GLQGGFIWDWVDQALRKVQADGTKQWAYGGEFGDIPNDLNFCLNGLVWPDRTAHPVLHEV 673
QGGFIWD+ DQAL + DG + AYGG+F D P D NF NGL++ DRT P EV
Sbjct: 549 MYQGGFIWDYGDQALYRKLPDGKEVLAYGGDFTDRPTDYNFSGNGLIYADRTISPKAQEV 608
Query: 674 KFLYQPIKVNLSDGKLEIKNTHFFQTTEGLEFSWYISADGYKLGSDKLSLPPIKPQSNYV 733
K+LYQ +K+ + + IKN + F T+ + + + DG + L++ + Y+
Sbjct: 609 KYLYQNVKLEPDEKGVTIKNQNLFVNTDKYDLYYIVERDGKLIKDGYLNVSVAPDEEKYI 668
Query: 734 FDWKSGPWYSLWDSSSSEEIFLTITAKLLNSTRWVEAGHVVTTAQVQLPAKRDIVPHAIN 793
+ + + EEI LT + +L +T W E G+ + Q + K D+ H N
Sbjct: 669 -------ELPIGNYNFPEEIVLTTSLRLAQATLWAEKGYEIAFGQKVIKEKSDMNNH--N 719
Query: 794 IGSGNLVVETLGD-TIKVSQQDVWDITFNTKTGLIESWKVKGVHVMNKGIHPCFWRASID 852
S ++ GD I V +D + F+ + G I S + + + +WRA+ D
Sbjct: 720 SESKMKIIH--GDVNIGVHGKD-FKAIFSKQEGGIVSLRYNNKEFITRTPKTFYWRATTD 776
Query: 853 NDKGGGADSYLSRWKAA--GIDSVHFIAESCSVQSTTGNAVKLLVVFHGVTKGEEGSLPN 910
ND+G + S+W AA G V F E + +T LP
Sbjct: 777 NDRGNRHEFRCSQWLAATMGQKYVDFSVEEFDEK---------------ITLYYTYQLPT 821
Query: 911 QDKSKVLFTTEMTYTIYASGDVILECNVKPN--ADLPPLPRVGIEMNLEKSLDQVSWYGR 968
+ V T E+ SG+ I++ NVK + LP LP +G++ L + SWYG
Sbjct: 822 VPSTNVKITYEV------SGEGIIKVNVKYKGVSGLPELPVLGMDFKLLAEFNSFSWYGM 875
Query: 969 GPFECYPDRKAAAQVAVYEKS 989
GP E Y DR A++ +YE +
Sbjct: 876 GPEENYIDRCEGAKLGIYEST 896
>BGAL_LACDE (P20043) Beta-galactosidase (EC 3.2.1.23) (Lactase)
Length = 1006
Score = 472 bits (1215), Expect = e-132
Identities = 333/1010 (32%), Positives = 485/1010 (47%), Gaps = 91/1010 (9%)
Query: 92 VKSLSGYWKFFLASNPCNVPAKFHDSEFQDSEWSTLPVPSNWQLHGFDRPIYTNVTYPFP 151
V+SL G W A N P F+ +F DS + ++ VP N +L GF +P Y NV YP+
Sbjct: 52 VQSLDGDWLIDYAENGQG-PVNFYAEDFDDSNFKSVKVPGNLELQGFGQPQYVNVQYPWD 110
Query: 152 ----LDPPFVPTENPTGCYRMDFHLPKEWEGRRILLHFEAVDSAFCAWINGHPIGYSQDS 207
+ PP +P++NP Y F L + + + + L F+ +A W+NGH +GY +DS
Sbjct: 111 GSEEIFPPQIPSKNPLASYVRYFDLDEAFWDKEVSLKFDGAATAIYVWLNGHFVGYGEDS 170
Query: 208 RLPAEFEVTDFCHPCGSDLKNVLAVQVFRWSDGCYLEDQDHWRMSGIHRDVLLLAKPEVF 267
P+EF VT F N LAV ++++S +LEDQD WRMSG+ R V L AKP +
Sbjct: 171 FTPSEFMVTKFL----KKENNRLAVALYKYSSASWLEDQDFWRMSGLFRSVTLQAKPRLH 226
Query: 268 ITDYFFKSNLAEDFSSAEMLVEVKID-RLQDTSIDNVLTNYTIEATLYDSGSWESSDGNP 326
+ D ++L +++ ++ VE I RL + S E D
Sbjct: 227 LEDLKLTASLTDNYQKGKLEVEANIAYRLPNASF-----------------KLEVRDSEG 269
Query: 327 DLLSSNVADITFQPTTTPLGFYGYTLVGKLQSPKLWSAEQPYLYTLVVVLKDKSGRVLDC 386
DL++ + I + +TL WSAE+P LY + + L ++G +L+
Sbjct: 270 DLVAEKLGPIRSEQLE-------FTLAD--LPVAAWSAEKPNLYQVRLYLY-QAGSLLEV 319
Query: 387 ESSQVGFKNVSKAHKQLLVNGHPVVIRGVNRHEHHPEVGKANIESCMVKDLVLMKQNNIN 446
+VGF+N + +NG +V +G NRHE ++G+A E M+ D+ MK++NIN
Sbjct: 320 SRQEVGFRNFELKDGIMYLNGQRIVFKGANRHEFDSKLGRAITEEDMIWDIKTMKRSNIN 379
Query: 447 AVRNSHYPQHPRWYELCDLFGMYMIDEANIETHGFDYSKHLKH-------PTLEPMWATA 499
AVR SHYP +Y LCD +G+Y+IDEAN+E+HG + K H P + W A
Sbjct: 380 AVRCSHYPNQSLFYRLCDKYGLYVIDEANLESHG-TWEKVGGHEDPSFNVPGDDQHWLGA 438
Query: 500 MLDRVIGMVERDKNHTCIISWSLGNESGFGTNHFAMAGWIRGRDSSRVIHYEG---GGSR 556
L RV M+ RDKNH I+ WSLGNES GT MA ++R D +RV HYEG
Sbjct: 439 SLSRVKNMMARDKNHASILIWSLGNESYAGTVFAQMADYVRKADPTRVQHYEGVTHNRKF 498
Query: 557 TPCTDIVCPMYMRVWDMLK-IANDPTETRPLILCEYSHAMGNSNGNLHIYWEAIDNTFGL 615
T I MY + + + N P +P I EY+HAMGNS G+L Y A++
Sbjct: 499 DDATQIESRMYAPAKVIEEYLTNKP--AKPFISVEYAHAMGNSVGDLAAY-TALEKYPHY 555
Query: 616 QGGFIWDWVDQALRKVQADGTKQWAYGGEFGDIPNDLNFCLNGLVWPDRTAHPVLHEVKF 675
QGGFIWDW+DQ L K DG YGG+F D P D FC NGLV+ DRT P L VK
Sbjct: 556 QGGFIWDWIDQGLEK---DG--HLLYGGDFDDRPTDYEFCGNGLVFADRTESPKLANVKA 610
Query: 676 LYQPIKVNLSDGKLEIKNTHFFQTTEGLEFSWYISADGYKLGSDKLSLPPIKPQSNYVFD 735
LY +K+ + DG+L +KN + F + F + DG + ++P + F
Sbjct: 611 LYANLKLEVKDGQLFLKNDNLFTNSSSYYFLTSLLVDGKLTYQSRPLTFGLEPGESGTF- 669
Query: 736 WKSGPWYSLWDSSSSEEIFLTITAKLLNSTRWVEAGHVVTTAQVQLPAKRDIVPHAINIG 795
+ PW + D E+ +TA L W + G V A+ + P G
Sbjct: 670 --ALPWPEVADEKG--EVVYRVTAHLKEDLPWADEGFTVAEAEEVAQKLPEFKPE----G 721
Query: 796 SGNLVVETLGDTIKVSQQDVWDITFNTKTGLIESWKVKGVHVMNKGIHPCFWRASIDNDK 855
+LV +K + + I F+ G S K G + + FWRA DND+
Sbjct: 722 RPDLVDSDYNLGLKGNN---FQILFSKVKGWPVSLKYAGREYLKRLPEFTFWRALTDNDR 778
Query: 856 GGGADSYLSRWKAAGIDSVHFIAESCSVQSTTGNAVKLLVVFHGVTKGEEGSLPNQDKSK 915
G G L+RW+ AG SC V+ + VK KG+
Sbjct: 779 GAGYGYDLARWENAG-KYARLKDISCEVKEDS-VLVKTAFTLPVALKGD----------- 825
Query: 916 VLFTTEMTYTIYASGDVILECNVKPNADLPPLPRVGIEMNLEKSLDQVSWYGRGPFECYP 975
+TY + G + + + + LP G+ + L K L +YG GP E YP
Sbjct: 826 ----LTVTYEVDGRGKIAVTADFPGAEEAGLLPAFGLNLALPKELTDYRYYGLGPNESYP 881
Query: 976 DRKAAAQVAVYEKSVDELHVPYIVPGESGGRADVRWATFLNKNGFGIYTSKYGSSPPMQM 1035
DR + +Y+ +V + PY P E+G R+ VRW ++ G +T+ + + +
Sbjct: 882 DRLEGNYLGIYQGAVKKNFSPY-RPQETGNRSKVRWYQLFDEKGGLEFTA---NGADLNL 937
Query: 1036 SASYYSTSELDRAGHDYELVKGDNIEVHLDHKHMGLGGDDSWSPCVHDQY 1085
SA YS ++++ A H +EL + V MG+GGDDSW VH ++
Sbjct: 938 SALPYSAAQIEAADHAFELT-NNYTWVRALSAQMGVGGDDSWGQKVHPEF 986
>BGAL_ACTPL (P70753) Beta-galactosidase (EC 3.2.1.23) (Lactase)
Length = 1005
Score = 467 bits (1202), Expect = e-131
Identities = 325/1027 (31%), Positives = 496/1027 (47%), Gaps = 125/1027 (12%)
Query: 95 LSGYWKFFLASNPCNVPAKFHDSEFQDSEWSTLPVPSNWQLHGFDRPIYTNVTYPFPLDP 154
L+G W F + ++P F D F+ +PVP+NWQ HGFD YTN+ YPFP +P
Sbjct: 47 LNGQWDFNYFQSYHDLPDNFLDIAFEHK----IPVPANWQNHGFDHHHYTNINYPFPFEP 102
Query: 155 PFVPTENPTGCYRMDFHL-PKEWEGRRILLHFEAVDSAFCAWINGHPIGYSQDSRLPAEF 213
PFVP +NP G Y L PK +R LL+FE VDS ++N +GYSQ S +EF
Sbjct: 103 PFVPHQNPCGVYHRTLQLTPKH---KRYLLNFEGVDSCLFVYVNKQFVGYSQISHNTSEF 159
Query: 214 EVTDFCHPCGSDLKNVLAVQVFRWSDGCYLEDQDHWRMSGIHRDVLLLAKPEVFITDYFF 273
+V+D+ G+ N L V V +W DG YLEDQD +RMSGI RDV LL + + ++ D+F
Sbjct: 160 DVSDYLQ-AGT---NHLTVVVLKWCDGSYLEDQDKFRMSGIFRDVYLLEREQNYLQDFFI 215
Query: 274 KSNLAEDFSSAEMLVEVKIDRLQDTSIDNVLTNYTIEATLYDSGSWESSDGNPDLLSSNV 333
+ L ++ A + VE R Q +I+ L N P+ +
Sbjct: 216 QYELDDELRHANLKVETLFSR-QPQAIEYQLLN-------------------PNGFT--- 252
Query: 334 ADITFQPTTTPLGFYGYTLVGKLQSPKLWSAEQPYLYTLVVVLKDKSGRVLDCESSQVGF 393
F T T L +++ +LW+AE+P LYTL++ + + ++GF
Sbjct: 253 ---VFNQTDTHLNI-------EVEDIQLWNAEKPQLYTLILHTAE------EVIVQKIGF 296
Query: 394 KNVSKAHKQLLVNGHPVVIRGVNRHEHHPEVGKANIESCMVKDLVLMKQNNINAVRNSHY 453
+ V LL N P+ +GVNRH+ P+ G A + KDL LMKQ+NINA+R +HY
Sbjct: 297 RKVEIKDGILLFNQQPIKFKGVNRHDSDPKTGYAITYAQAHKDLQLMKQHNINAIRTAHY 356
Query: 454 PQHPRWYELCDLFGMYMIDEANIETHGFDY-SKHLKHPTL-------------------- 492
P P + ELCD +G Y+I E+++E+HG + + P++
Sbjct: 357 PNAPWFSELCDQYGFYLIGESDVESHGASMLTVKMPEPSILLNHQNDLQTERIRQDMIDN 416
Query: 493 ------EPMWATAMLDRVIGMVERDKNHTCIISWSLGNESGFGTNHFAMAGWIRGRDSSR 546
+P++ A+LDR VERDKN T II WSLGNE+G+G N A A WI+ RD SR
Sbjct: 417 YCYFARDPLFKKAILDRQQANVERDKNRTSIIIWSLGNEAGYGANFEAAAAWIKQRDKSR 476
Query: 547 VIHYEGG-------GSRTPCTDIVCPMYMRVWDMLKIANDPTETRPLILCEYSHAMGNSN 599
++HYE + D MY D+ + + +P +LCEYSHAMGNSN
Sbjct: 477 LVHYESSIYQHSADNNDLSNLDFYSEMYGSTEDIDRYC-ATAQRKPFVLCEYSHAMGNSN 535
Query: 600 GNLHIYWEAIDNTFGLQGGFIWDWVDQALRKVQADGTKQWAYGGEFGDIPNDLNFCLNGL 659
G+ YW+A GGF+W+W D A +A+G Q+ YGG+FG+ P+D NFC++GL
Sbjct: 536 GDAEDYWQAFHRHPQSCGGFVWEWCDHA--PYRANG--QFGYGGDFGESPHDGNFCMDGL 591
Query: 660 VWPDRTAHPVLHEVKFLYQPIKVNLSDGKLEIKN-THFFQTTEGLEFSWYISADGYKLGS 718
V PDR H L E+K + +P + L D ++ I N F + L + +G
Sbjct: 592 VSPDRIPHSNLLELKNVNRPARAELIDNQIVIHNYLDFTDLADYLTIDYEFVENGVVTSG 651
Query: 719 DKLSLPPIKPQSNYVFDWKSGPWYSLWDSSSSEEIFLTITAKLLNSTRWVEAGHVVTTAQ 778
LS+ KP S+ + + ++ L + +L +T +EA H + Q
Sbjct: 652 GNLSV-SCKPHSSVILPIE-------LPKNNGHLWLLNLDYRLNTATELLEAEHSLGFEQ 703
Query: 779 VQLPAKRDIVPHAINIGSGNLVVETLGDTIKVSQQDVWDITFNTKTGLIESWKVKGVHVM 838
+ L ++ +V I V+ I V + + + G+ + G ++
Sbjct: 704 LNLFSENKLVLPKFTIEKSTFEVQEDHFRINV-HNGQFSYQLDKQKGIFSRIEKAGKAII 762
Query: 839 NKGIHPCFWRASIDNDKGGGADSYLSRWKAAGIDSVHFIAESCSVQSTTGNAVKLLVVFH 898
+ + WRA DND+ W+ AG D + A Q + AV+ V
Sbjct: 763 QQPLDFNIWRAPTDNDR-----LIREAWQNAGYDKAYTRAYEIQWQQSE-QAVEFSV--- 813
Query: 899 GVTKGEEGSLPNQDKSKVLFTTEMTYTIYASGDVILECNVKPNADLPPLPRVGIEMNLEK 958
+ ++ + + ++L T ++ Y I+ G + +E N +LP LPR G+ L K
Sbjct: 814 ------KSAIVSISRGRIL-TLDIRYRIFNDGQISVEINAIRPTELPYLPRFGLRFWLAK 866
Query: 959 SLDQVSWYGRGPFECYPDRKAAAQVAVYEKSVDELHVPYIVPGESGGRADVRWATFLNKN 1018
+ + V ++G G E Y D+ A + +Y + + H Y+ P E+G + N
Sbjct: 867 AENTVEYFGYGEQESYVDKHHLANLGIYHTTAKQNHTDYVKPQENGSHYGCEYLKSEN-- 924
Query: 1019 GFGIYTSKYGSSPPMQMSASYYSTSELDRAGHDYELVKGDNIEVHLDHKHMGLGGDDSWS 1078
++ S +S P + S Y+ EL H YEL + D + +D+K G+ G +S
Sbjct: 925 ---LFVS---ASQPFSFNLSPYTQEELTEKKHYYELQESDYSVLCIDYKMSGI-GSNSCG 977
Query: 1079 PCVHDQY 1085
P + DQY
Sbjct: 978 PNLKDQY 984
>BGAL_ARTSB (Q59140) Beta-galactosidase (EC 3.2.1.23) (Lactase)
Length = 1015
Score = 465 bits (1196), Expect = e-130
Identities = 339/1085 (31%), Positives = 510/1085 (46%), Gaps = 160/1085 (14%)
Query: 64 VSQSAVWKDDAVNGALESAAFWVKDLPFVKSLSGYWKFFLASNPCNVPAK---------- 113
+S S + +G A W+ SL+G W+F L P
Sbjct: 1 MSSSYITDQGPGSGLRVPARSWLNSDAPSLSLNGDWRFRLLPTAPGTPGAGSVLATGETV 60
Query: 114 --FHDSEFQDSEWSTLPVPSNWQLHG---FDRPIYTNVTYPFPLDPPFVPTENPTGCYRM 168
F DS W TL VPS+W L + RPIYTNV YPFP+DPPFVP NPTG YR
Sbjct: 61 EAVASESFDDSSWDTLAVPSHWVLAEDGKYGRPIYTNVQYPFPIDPPFVPDANPTGDYRR 120
Query: 169 DFHLPKEW---EGRRILLHFEAVDSAFCAWINGHPIGYSQDSRLPAEFEVTDFCHPCGSD 225
F +P W + L F+ V+S + W+NG IG SRL EF+V++ P
Sbjct: 121 TFDVPDSWFESTTAALTLRFDGVESRYKVWVNGVEIGVGSGSRLAQEFDVSEALRPG--- 177
Query: 226 LKNVLAVQVFRWSDGCYLEDQDHWRMSGIHRDVLLLAKPEVFITDYFFKSNLAEDFSSAE 285
KN+L V+V +WS YLEDQD W + GI RDV L A+P +TD + +++ + S
Sbjct: 178 -KNLLVVRVHQWSAASYLEDQDQWWLPGIFRDVKLQARPVGGLTDVWLRTDWS---GSGT 233
Query: 286 MLVEVKIDRLQDTSIDNVLTNYTIEATLYDSGS---WESSDGNPDLLSSNVADITFQPTT 342
+ E+ D + + + + G W+S ++VA ++
Sbjct: 234 ITPEITADPAA----------FPVTLRVPELGLEVIWDSP--------ADVAPVS----- 270
Query: 343 TPLGFYGYTLVGKLQSPKLWSAEQPYLYTLVVVLKDKSGRVLDCESSQVGFKNVSKAHKQ 402
+ + + WSAE P LY V + S ++GF+ V Q
Sbjct: 271 -------------IDAVEPWSAEVPRLYDASV------SSAAESISLRLGFRTVKIVGDQ 311
Query: 403 LLVNGHPVVIRGVNRHEHHPEVGKANIESCMVKDLVLMKQNNINAVRNSHYPQHPRWYEL 462
LVNG V+ GVNRHE + + G+ E+ +DL LMK+ N+NA+R SHYP HPR+ +L
Sbjct: 312 FLVNGRKVIFHGVNRHETNADRGRVFDEASAREDLALMKRFNVNAIRTSHYPPHPRFLDL 371
Query: 463 CDLFGMYMIDEANIETHGFDYSKHLKHPTLEPMWATAMLDRVIGMVERDKNHTCIISWSL 522
D G ++I E ++ETHGF K + +P+ +P W A++DR+ VERDKNH I+ WSL
Sbjct: 372 ADELGFWVILECDLETHGFHALKWVGNPSDDPAWRDALVDRMERTVERDKNHASIVMWSL 431
Query: 523 GNESGFGTNHFAMAGWIRGRDSSRVIHYEGGGSRTPCTDIVCPMYMRVWDMLKIANDPT- 581
GNESG G N AMA W RD SR +HYEG + TD+ MY + + I + +
Sbjct: 432 GNESGTGANLAAMAAWTHARDLSRPVHYEGDYTGA-YTDVYSRMYSSIPETDSIGRNDSH 490
Query: 582 --------------ETRPLILCEYSHAMGNSNGNLHIYWEAIDNTFGLQGGFIWDWVDQA 627
TRP ILCEY HAMGN G + Y + +D L GGF+W+W D
Sbjct: 491 ALLLGCNAIESARQRTRPFILCEYVHAMGNGPGAIDQYEDLVDKYPRLHGGFVWEWRDHG 550
Query: 628 LRKVQADGTKQWAYGGEFGDIPNDLNFCLNGLVWPDRTAHPVLHEVKFLYQPIKVNLS-- 685
+R ADGT+ +AYGG+F ++ +D NF ++G++ D T P L E K + PI++ L+
Sbjct: 551 IRTRTADGTEFFAYGGDFDEVIHDGNFVMDGMILSDSTPTPGLFEYKQIVSPIRLALTLN 610
Query: 686 ---DGKLEIKNTHFFQTTEGLEFSWYISADGYKLGSDKLSLPPIKPQSNYVFDWKSGPWY 742
+ L + N + W + +G ++ + +L+ D +GP
Sbjct: 611 AEGNAGLTVANLRHTSDASDVVLRWRVEHNGTRVDAGELTT-----------DGANGP-L 658
Query: 743 SLWDS----------SSSEEIFLTITAKLLNSTRWVEAGHVVTTAQVQL-PAKRDI-VPH 790
DS ++ E +L++ A L +T W AGH ++ Q+ L PA+ + VP
Sbjct: 659 QAGDSLTLTLPTIVAAAEGETWLSVEAVLREATAWAPAGHPLSETQLDLSPAQPPLRVPR 718
Query: 791 AINIGSGNLVVETLGDTIKVSQQDVWDITFNTKTGLIESWKVKGVHVMNKGIHPCFWRAS 850
+ +G VE LG TF+ + + + G+ V + WRA
Sbjct: 719 PASPIAGAAPVE-LGPA-----------TFDAGSLV----TLAGLPVAGPRLE--LWRAP 760
Query: 851 IDNDKG------GGADSYLSRWKAAGIDSVHFIAESCSVQSTTGNAVKLLVVFHGVTKGE 904
DNDKG G D +++ + S + + + T + + G+
Sbjct: 761 TDNDKGQGFGAYGPEDPWINSGRGVPAPSSAVVWQQAGLDRLTRRVEDVAALPQGLRVRS 820
Query: 905 EGSLPNQDKSKVLFTTEMTYTIYASGDVI-LECNVKPNA--DLPPLPRVGIEMNLEKSLD 961
+ N + E + + SGD + L ++ P+A DL PR+G+ ++L +D
Sbjct: 821 RYAAANSEHD---VAVEENWQL--SGDELWLRIDIAPSAGWDL-VFPRIGVRLDLPSEVD 874
Query: 962 QVSWYGRGPFECYPDRKAAAQVAVYEKSVDELHVPYIVPGESGGRADVRWATFLNKNG-- 1019
SW+G GP E YPD +A V + S++EL+V Y P E+G +DVRW L+++G
Sbjct: 875 GASWFGAGPRESYPDSLHSAVVGTHGGSLEELNVNYARPQETGHHSDVRWVE-LSRDGAP 933
Query: 1020 ---FGIYTSKYGSSPPMQMSASYYSTSELDRAGHDYELVKGDNIEVHLDHKHMGLG---- 1072
G P ++ + + E+ A H +EL + + ++LD GLG
Sbjct: 934 WLRIEADPDALGRRPGFSLAKN--TAQEVALAPHPHELPESQHSYLYLDAAQHGLGSRAC 991
Query: 1073 GDDSW 1077
G D W
Sbjct: 992 GPDVW 996
>BGAL_KLULA (P00723) Beta-galactosidase (EC 3.2.1.23) (Lactase)
Length = 1025
Score = 451 bits (1161), Expect = e-126
Identities = 352/1075 (32%), Positives = 509/1075 (46%), Gaps = 142/1075 (13%)
Query: 88 DLPFVKSLSGYWKFFLASNPCNVP-AKFHDSEFQDSEWSTLPVPSNWQLHG---FDRPIY 143
D +SL+G W F L P + P AK D E +WST+ VPS+W+L + +PIY
Sbjct: 28 DQDIFESLNGPWAFALFDAPLDAPDAKNLDWETA-KKWSTISVPSHWELQEDWKYGKPIY 86
Query: 144 TNVTYPFPLDPPFVPTENPTGCYRMDFHLP-KEWEGRRILLHFEAVDSAFCAWINGHPIG 202
TNV YP P+D P PT NPTG Y F L K E L FE VD+ + ++NG +G
Sbjct: 87 TNVQYPIPIDIPNPPTVNPTGVYARTFELDSKSIESFEHRLRFEGVDNCYELYVNGQYVG 146
Query: 203 YSQDSRLPAEFEVTDFCHPCGSDLKNVLAVQVFRWSDGCYLEDQDHWRMSGIHRDVLLLA 262
+++ SR AEF++ + S+ +N++ V+VF+WSD Y+EDQD W +SGI+RDV LL
Sbjct: 147 FNKGSRNGAEFDIQKYV----SEGENLVVVKVFKWSDSTYIEDQDQWWLSGIYRDVSLLK 202
Query: 263 KP------EVFITDYFFKSNLAEDFSSAEMLVEVKIDRLQDTSIDNVLTNYTIE-----A 311
P +V +T F S + AE+ V+V + Q +S D++ N+T+ +
Sbjct: 203 LPKKAHIEDVRVTTTFVDSQ----YQDAELSVKVDV---QGSSYDHI--NFTLYEPEDGS 253
Query: 312 TLYDSGSWESSDGNPDLLSSNVADITFQPTTTPLGFYGYTLVGKLQSPKLWSAEQPYLYT 371
+YD+ S + + N + S I+F + + ++P+ W+AE P LY
Sbjct: 254 KVYDASSLLNEE-NGNTTFSTKEFISFSTKKNEETAFKINV----KAPEHWTAENPTLYK 308
Query: 372 LVVVLKDKSGRVLDCESSQVGFKNVSKAHKQLLVNGHPVVIRGVNRHEHHPEVGKANIES 431
+ L G V+ VGF+ V + VNG ++ RGVNRH+HHP G+A
Sbjct: 309 YQLDLIGSDGSVIQSIKHHVGFRQVELKDGNITVNGKDILFRGVNRHDHHPRFGRAVPLD 368
Query: 432 CMVKDLVLMKQNNINAVRNSHYPQHPRWYELCDLFGMYMIDEANIETHGFD--------- 482
+V+DL+LMK+ NINAVRNSHYP HP+ Y+L D G ++IDEA++ETHG
Sbjct: 369 FVVRDLILMKKFNINAVRNSHYPNHPKVYDLFDKLGFWVIDEADLETHGVQEPFNRHTNL 428
Query: 483 -----------YSKHLKHPTLEPMWATAMLDRVIGMVERDKNHTCIISWSLGNESGFGTN 531
Y + + + P + A LDR +V RD NH II WSLGNE+ +G N
Sbjct: 429 EAEYPDTKNKLYDVNAHYLSDNPEYEVAYLDRASQLVLRDVNHPSIIIWSLGNEACYGRN 488
Query: 532 HFAMAGWIRGRDSSRVIHYEGGGSRTPCTDIVCPMY-----MRVWDMLKIANDPTETRPL 586
H AM I+ D +R++HYE G DI MY M W + +PL
Sbjct: 489 HKAMYKLIKQLDPTRLVHYE-GDLNALSADIFSFMYPTFEIMERWRKNHTDENGKFEKPL 547
Query: 587 ILCEYSHAMGNSNGNLHIYWEAIDNTFGLQGGFIWDWVDQALR---KVQADGT--KQWAY 641
ILCEY HAMGN G+L Y E QGGFIW+W + + ADG K +AY
Sbjct: 548 ILCEYGHAMGNGPGSLKEYQELFYKEKFYQGGFIWEWANHGIEFEDVSTADGKLHKAYAY 607
Query: 642 GGEFGDIPNDLNFCLNGLVWPDRTAHPVLHEVKFLYQPIKVNLSDGKLEIKNTHFFQTTE 701
GG+F + +D F ++GL + P L E K + +P+ + ++ G + I N H F TT+
Sbjct: 608 GGDFKEEVHDGVFIMDGLCNSEHNPTPGLVEYKKVIEPVHIKIAHGSVTITNKHDFITTD 667
Query: 702 GLEFSWYISADGYKLGSDKLSLPPIKPQSNYVFDWKSGPWYSLWDSSSSEEIFLTITAKL 761
L F I D K + +P +KP+ + S Y + A L
Sbjct: 668 HLLF---IDKDTGK----TIDVPSLKPEESVTI--PSDTTY--------------VVAVL 704
Query: 762 LNSTRWVEAGHVVTTAQVQLPAK-RDIVPHA------INIGSGNLVVETLGDTIKVSQQD 814
+ ++AGH + Q +LP K D V IN G + VE+ G
Sbjct: 705 KDDAGVLKAGHEIAWGQAELPLKVPDFVTETAEKAAKINDGKRYVSVESSG--------- 755
Query: 815 VWDITFNTKTGLIESWKVKGVHVMNK--GIHPCFWRASIDNDKGGGADSYLSRWKAAGID 872
+ G IES KVKG + +K G FWR +ND+ WK ID
Sbjct: 756 -LHFILDKLLGKIESLKVKGKEISSKFEGSSITFWRPPTNNDE----PRDFKNWKKYNID 810
Query: 873 SVHFIAESCSVQSTTGNAVKLLVVFHGVTKGEEGSLPNQDKSKVL----FTTEMTYTIYA 928
+ SV+ + ++ ++ V N S V+ F T YTI+A
Sbjct: 811 LMKQNIHGVSVEKGSNGSLAVVTV-------------NSRISPVVFYYGFETVQKYTIFA 857
Query: 929 SGDVILECNVKPNADL--PPLPRVGIEMNLEKSLDQVSWYGRGPFECYPDRKAAAQVAVY 986
+ + L ++K + P PRVG E L S + W GRGP E YPD+K + + +Y
Sbjct: 858 N-KINLNTSMKLTGEYQPPDFPRVGYEFWLGDSYESFEWLGRGPGESYPDKKESQRFGLY 916
Query: 987 E-KSVDELHVPYIVPGESGGRADVRWATFLNKNGFGI-YTSKYGSSPPMQMSASYYSTSE 1044
+ K V+E Y P E+G D FLN G S + P S
Sbjct: 917 DSKDVEEFVYDY--PQENGNHTDTH---FLNIKFEGAGKLSIFQKEKPFNFKIS--DEYG 969
Query: 1045 LDRAGHDYELVKGDNIEVHLDHKHMGLGGDDSWSPCVHDQYLVPPVPYSFSVRLS 1099
+D A H ++ + + LDH G+ G ++ P V DQY + ++F L+
Sbjct: 970 VDEAAHACDVKRYGRHYLRLDHAIHGV-GSEACGPAVLDQYRLKAQDFNFEFDLA 1023
>BGAL_STAXY (O33815) Beta-galactosidase (EC 3.2.1.23) (Lactase)
Length = 994
Score = 412 bits (1060), Expect = e-114
Identities = 304/1023 (29%), Positives = 488/1023 (46%), Gaps = 98/1023 (9%)
Query: 95 LSGYWKFFLASNPCNVPAKFHDSEFQDSEWSTLPVPSNWQLHGFDRPIYTNVTYPFPLDP 154
L+G W F + + A + S Q+S+ TL VPS W L+G+D+ Y N YP P +P
Sbjct: 48 LNGEWYFQYFES---LEAYLNHSNKQESK--TLTVPSVWNLYGYDQIQYLNTQYPIPFNP 102
Query: 155 PFVPTENPTGCYRMDFHLPKEWEGRRILLHFEAVDSAFCAWINGHPIGYSQDSRLPAEFE 214
P+VP +NP G Y F + + + L+FE VDSAF WIN IGYSQ S +EF+
Sbjct: 103 PYVPKDNPCGHYTRKFTIDEYDQQYDYHLNFEGVDSAFYVWINNEFIGYSQISHAISEFD 162
Query: 215 VTDFCHPCGSDLKNVLAVQVFRWSDGCYLEDQDHWRMSGIHRDVLLLAKPEVFITDYFFK 274
+++F +N + V V ++SDG YLEDQD +R SGI RDV +L + + D+ +
Sbjct: 163 ISNFV----KQGENNIEVLVLKYSDGTYLEDQDMFRHSGIFRDVYILKRATERVDDFKVE 218
Query: 275 SNLAEDFSSAEMLVEVKIDRLQDTSIDNVLTNYTIEATLYDSGSWESSDGNPDLLSSNVA 334
+NL++D ++A+ ++VKI+R + ++E LY+ E + S V
Sbjct: 219 TNLSDDLNAAQ--IDVKIERAHNLK--------SVEFQLYNPKGEEVAS------ISGVN 262
Query: 335 DITFQPTTTPLGFYGYTLVGKLQSPKLWSAEQPYLYTLVVVLKDKSGRVLDCESSQVGFK 394
+ F + +P LWS E P LYTL ++ + + +VG +
Sbjct: 263 EHQFD----------------VHNPHLWSTENPVLYTLYILTDQ------EVITQKVGIR 300
Query: 395 NVSKAHKQLLVNGHPVVIRGVNRHEHHPEVGKANIESCMVKDLVLMKQNNINAVRNSHYP 454
V+ + Q +NG + IRG N H+ HPE G ES KDL LMKQ N NA+R +HYP
Sbjct: 301 EVAIQNNQFYINGQSIKIRGTNYHDSHPETGYVMTESHFKKDLELMKQGNFNAIRTAHYP 360
Query: 455 QHPRWYELCDLFGMYMIDEANIETHGF------DYSKHLKHPTLEPMWATAMLDRVIGMV 508
+ P +YE+ D +G Y++ EA+IETHG D ++ + + TA+++R+ +
Sbjct: 361 KSPLFYEMTDQYGFYVMSEADIETHGVVRLYGEDNNEDFNIIADDSKFETAIIERIEASI 420
Query: 509 ERDKNHTCIISWSLGNESGFGTNHFAMAGWIRGRDSSRVIHYEGGGSRTPCT-------D 561
KN++ I+SWSLGNESGFG N A + D++R IHYEG R D
Sbjct: 421 MPLKNYSSIVSWSLGNESGFGKNMVKGAARAKSLDNTRPIHYEGTLYRDKQQHYDLSNID 480
Query: 562 IVCPMYMRVWDMLKI-ANDPTETRPLILCEYSHAMGNSNGNLHIYWEAIDNTFGLQGGFI 620
++ MY ++ + ++P +P ILCEY+HAMGNS G+LH Y ++ GGF+
Sbjct: 481 MISRMYPSPEEIEETYLSNPDLDKPFILCEYAHAMGNSPGDLHAYQTLVEQYDSFIGGFV 540
Query: 621 WDWVDQALRKVQADGTKQWAYGGEFGDIPNDLNFCLNGLVWPDRTAHPVLHEVKFLYQPI 680
W+W D A++ DG + YGG+FG+ +D NFC++G+V+P+R H +E K ++P+
Sbjct: 541 WEWCDHAIQTGMKDGNPIFRYGGDFGEKLHDGNFCVDGIVFPNRVPHEGYYEFKQEHRPL 600
Query: 681 K-VNLSDGKLEIKNTHFFQTTEGLEF--SWYISADGYKLGSDKLSLPPIKPQSNYVFDWK 737
V+ D K+ ++N F E F + + +G K S+ + L P + D
Sbjct: 601 NLVSQEDFKIVLRNQLDFIPAEKYMFVEATVTNLNGGKTISE-IPLSNFLPHTAQTIDLS 659
Query: 738 SGPWYSLWDSSSSEEIFLTITAKLLNSTRW--VEAGHVVTTAQVQLPAKRDIVPHAINIG 795
+ ++ L K + R E GH Q +R +
Sbjct: 660 -----DYINIQHISDVILRYKLKYDDIFRHENFELGHDQIVYQ-----RRTLKEQNEQSD 709
Query: 796 SGNLVVETLGDTIKVSQQDVWDITFNTKTGLIESWKVKGVHVMNKGIHPCFWRASIDNDK 855
++V IKVS FN +ES V+++ WRA DND
Sbjct: 710 ETEILVTQTDKLIKVSVGKSETYVFNKDNASLESVLKHNHIVISQNTTNNIWRAPTDNDT 769
Query: 856 GGGADSYLSRWKAAGIDSVHFIAESCSVQSTTGNAVKLLVVFHGVTKGEEGSLPNQDKSK 915
D W +G + + N ++ ++F+ ++ N
Sbjct: 770 NIKND-----WAYSGYKDITTRVHDYQI---VENETEVSLIFN-------IAMVNDAVPP 814
Query: 916 VLFTTEMTYTIYASGDVILECNVKPNADLPPLPRVGIEMNLEKSLDQVSWYGRGPFECYP 975
VLF T +T+ + +G + + +++ + P LPR G+ + L K+ +QV +YG+GPF Y
Sbjct: 815 VLFGT-VTWHVQRNGTLNVTYDLERDMKAPYLPRFGLGLTLPKAFEQVKYYGKGPFSSYQ 873
Query: 976 DRKAAAQVAVYEKSVDELHVPYIVPGESGGRADVRWATFLNKNGFGIYTSKYGSSPPMQM 1035
D+ A + + +V + +I P E+G + + + I T S
Sbjct: 874 DKGVANYLDDFGTTVTDNGEIHIRPQETGSHNETTFVEISDGCKKVIVT----SDNTFSF 929
Query: 1036 SASYYSTSELDRAGHDYELVKGDNIEVHLDHKHMGLGGDDSWSPCVHDQYLVPPVPYSFS 1095
+ ++YS +L H L D +++D+ G+ G +S P +++ Y + FS
Sbjct: 930 NTTHYSLKQLTETTHKDALEPEDQTYLYIDYAQSGI-GSNSCGPELNEAYRLNNRHIEFS 988
Query: 1096 VRL 1098
L
Sbjct: 989 FNL 991
>BGAL_LEULA (Q02603) Beta-galactosidase large subunit (EC 3.2.1.23)
(Lactase)
Length = 626
Score = 331 bits (848), Expect = 8e-90
Identities = 212/620 (34%), Positives = 306/620 (49%), Gaps = 70/620 (11%)
Query: 91 FVKSLSGYWKFFLASNPCNVPAKFHDSEFQDSEWSTLPVPSNWQLHGFDRPIYTNVTYPF 150
F+KSL+G W+F A P P F+ +F +++ T+ VP + +L G+ + Y N YP+
Sbjct: 40 FIKSLNGAWRFNFAKTPAERPVDFYQPDFDATDFDTIQVPGHIELAGYGQIQYINTLYPW 99
Query: 151 P----LDPPFV-------------PTENPTGCYRMDFHLPKEWEGRRILLHFEAVDSAFC 193
PP+ +N G Y F L ++G+RI++ F+ V+ A
Sbjct: 100 EGKIYRRPPYTLNQDQLTPGLFSDAADNTVGSYLKTFDLDDVFKGQRIIIQFQGVEEALY 159
Query: 194 AWINGHPIGYSQDSRLPAEFEVTDFCHPCGSDLKNVLAVQVFRWSDGCYLEDQDHWRMSG 253
W+NGH IGYS+DS P+EF++T + G NVLAV+V++ S ++EDQD +R SG
Sbjct: 160 VWLNGHFIGYSEDSFTPSEFDLTPYIQDQG----NVLAVRVYKHSTAAFIEDQDMFRFSG 215
Query: 254 IHRDVLLLAKPEVFITDYFFKSNLAEDFSSAEMLVEVKIDRLQDTSIDNVLTNYTIEATL 313
I RDV +LA+P ITD + + S E+ + K+
Sbjct: 216 IFRDVNILAEPASHITDLDIRPVPNANLKSGELNITTKVT-------------------- 255
Query: 314 YDSGSWESSDGNPDLLSSNVAD----ITFQPTTTPLGFYGYTLVGKLQSPKLWSAEQPYL 369
G P L+ V D + T T G + + LWS + PYL
Sbjct: 256 ----------GEPATLALTVKDHDGRVLTSQTQTGSGSVTFDTM-LFDQLHLWSPQTPYL 304
Query: 370 YTLVVVLKDKSGRVLDCESSQVGFKNVS-KAHKQLLVNGHPVVIRGVNRHEHHPEVGKAN 428
Y L + + D ++L+ Q GF+ V + K + VN +VI GVNRHE + G+
Sbjct: 305 YQLTIEVYDADHQLLEVVPYQFGFRTVELRDDKVIYVNNKRLVINGVNRHEWNAHTGRVI 364
Query: 429 IESCMVKDLVLMKQNNINAVRNSHYPQHPRWYELCDLFGMYMIDEANIETHGFDYSKHLK 488
+ M D+ M NNINA R HYP WY+LCD G+Y++ E N+E+HG
Sbjct: 365 SMADMRADIQTMLANNINADRTCHYPDQLPWYQLCDEAGIYLMAETNLESHGSWQKMGAI 424
Query: 489 HPTL-----EPMWATAMLDRVIGMVERDKNHTCIISWSLGNESGFGTNHFAMAGWIRGRD 543
P+ P W A++DR E KNH II WSLGNES G + AM + + D
Sbjct: 425 EPSYNVPGDNPHWPAAVIDRARSNYEWFKNHPSIIFWSLGNESYAGEDIAAMQAFYKEHD 484
Query: 544 SSRVIHYEG----GGSRTPCTDIVCPMYMRVWDMLKIANDPTETRPLILCEYSHAMGNSN 599
SR++HYEG + +D+ MY + +++ D T+P + CEY H MGNS
Sbjct: 485 DSRLVHYEGVFYTPELKDRISDVESRMYEKPQNIVAYLED-NPTKPFLNCEYMHDMGNSL 543
Query: 600 GNLHIYWEAIDNTFGLQGGFIWDWVDQALRKVQADGTKQ--WAYGGEFGDIPNDLNFCLN 657
G + Y + ID QGGFIWD++DQAL V T Q YGG+F + +D F N
Sbjct: 544 GGMQSYNDLIDKYPMYQGGFIWDFIDQAL-FVHDPITDQDVLRYGGDFDERHSDYAFSGN 602
Query: 658 GLVWPDRTAHPVLHEVKFLY 677
GL++ DRT P + EVK+ Y
Sbjct: 603 GLMFADRTPKPAMQEVKYYY 622
>BGAL_LACAC (O07684) Beta-galactosidase large subunit (EC 3.2.1.23)
(Lactase)
Length = 628
Score = 328 bits (840), Expect = 6e-89
Identities = 210/617 (34%), Positives = 311/617 (50%), Gaps = 63/617 (10%)
Query: 91 FVKSLSGYWKFFLASNPCNVPAKFHDSEFQDSEWSTLPVPSNWQLHGFDRPIYTNVTYPF 150
F +SL+G W+F + +P + P F+ +F S ++T+PVPS +L+ F + Y N YP+
Sbjct: 41 FKQSLNGMWQFKFSKDPQSRPVDFYKKDFNTSSFTTIPVPSEIELNNFAQNQYINTLYPW 100
Query: 151 P----LDPPFVPT-------------ENPTGCYRMDFHLPKEWEGRRILLHFEAVDSAFC 193
P + +N G Y F L E I + FE + A
Sbjct: 101 EGKIFRRPAYALNKSDAEEGSFSEGKDNTVGSYIKHFDLNPELRDHDIHIVFEGAERAMY 160
Query: 194 AWINGHPIGYSQDSRLPAEFEVTDFCHPCGSDLKNVLAVQVFRWSDGCYLEDQDHWRMSG 253
W+NGH IGY++DS P+EF++T + + N+LAV+VF+ S +LEDQD +R SG
Sbjct: 161 VWLNGHFIGYAEDSFTPSEFDLTKYI----KEKDNILAVEVFKHSTASWLEDQDMFRFSG 216
Query: 254 IHRDVLLLAKPEVFITDYFFKSNLAEDFSSAEMLVEVKIDRLQDTSIDNVLTNYTIEATL 313
+ R V LLA PE + D K + +++ E+K
Sbjct: 217 LFRSVELLAFPETHLVDLDLKPTVCDNYQDGIFNAELKFTG------------------- 257
Query: 314 YDSGSWESSDGNPDLLSSNVADITFQPTTTPLGFYGYTLVGKLQSPKLWSAEQPYLYTLV 373
S +G+ L +V PL L++ LW PYLY L+
Sbjct: 258 -------SLNGHVHLSVEDVNGSAILEQDVPLDSEVEFTSSTLENIHLWDNNHPYLYQLL 310
Query: 374 VVLKDKSGRVLDCESSQVGFKNVS-KAHKQLLVNGHPVVIRGVNRHEHHPEVGKANIESC 432
+ + D++G +++ Q GF+ + K +L+NG ++I GVNRHE + + G++ +
Sbjct: 311 IEVHDENGHLVELIPYQFGFRRIEINQDKVILLNGKCLIINGVNRHEWNAKTGRSITLND 370
Query: 433 MVKDLVLMKQNNINAVRNSHYPQHPRWYELCDLFGMYMIDEANIETHGFDYSKHLKHPT- 491
M KD+ K+NNINAVR HYP WY LCD G+YM+ E N+E+HG P+
Sbjct: 371 MEKDIDTFKENNINAVRTCHYPNQIPWYYLCDQNGIYMMAENNLESHGTWQKMGQVEPSD 430
Query: 492 ----LEPMWATAMLDRVIGMVERDKNHTCIISWSLGNESGFGTNHFAMAGWIRGRDSSRV 547
P W A++DR E KNHT I+ WSLGNES G+N AM + + DSSR+
Sbjct: 431 NVPGSVPEWREAVIDRARNNYETFKNHTSILFWSLGNESYAGSNIVAMNEFYKEHDSSRL 490
Query: 548 IHYEGGGSR----TPCTDIVCPMYMRVWDMLK-IANDPTETRPLILCEYSHAMGNSNGNL 602
+HYEG R +DI MY+ ++ + + N+P +P + CEY H MGNS+G +
Sbjct: 491 VHYEGVVHRPELKDQISDIESHMYLPPKEVAEYLRNNP--QKPFMECEYMHDMGNSDGGM 548
Query: 603 HIYWEAIDNTFGLQGGFIWDWVDQALRKVQADGTKQ--WAYGGEFGDIPNDLNFCLNGLV 660
Y + ID GGFIWD++DQAL V +KQ YGG+F D +D F +GL+
Sbjct: 549 GSYIKLIDEYPQYVGGFIWDFIDQAL-LVYDPISKQNVLRYGGDFDDRHSDYEFSGDGLM 607
Query: 661 WPDRTAHPVLHEVKFLY 677
+ DRT P + EV++ Y
Sbjct: 608 FADRTPKPAMQEVRYYY 624
>BGAL_LACHE (Q7WTB4) Beta-galactosidase large subunit (EC 3.2.1.23)
(Lactase)
Length = 628
Score = 320 bits (819), Expect = 2e-86
Identities = 205/618 (33%), Positives = 308/618 (49%), Gaps = 65/618 (10%)
Query: 91 FVKSLSGYWKFFLASNPCNVPAKFHDSEFQDSEWSTLPVPSNWQLHGFDRPIYTNVTYP- 149
+ +SL+G WKF ++NP + P F+ +F S + ++PVPS +L + + Y NV +P
Sbjct: 41 YQQSLNGKWKFHFSANPMDRPQDFYQRDFDSSNFDSIPVPSEIELSNYTQNQYINVLFPW 100
Query: 150 ---------FPLDPP-------FVPTENPTGCYRMDFHLPKEWEGRRILLHFEAVDSAFC 193
+ LDP +N G Y F L G+ + + FE V+ A
Sbjct: 101 EGKIFRRPAYALDPNDHEEGSFSKGADNTVGSYLKRFDLSSALIGKDVHIKFEGVEQAMY 160
Query: 194 AWINGHPIGYSQDSRLPAEFEVTDFCHPCGSDLKNVLAVQVFRWSDGCYLEDQDHWRMSG 253
W+NGH +GY++DS P+EF++T + D N+LAV+VF+ S +LEDQD +R SG
Sbjct: 161 VWLNGHFVGYAEDSFTPSEFDLTPYIQ----DKDNLLAVEVFKHSTASWLEDQDMFRFSG 216
Query: 254 IHRDVLLLAKPEVFITDYFFKSNLAEDFSSAEMLVEVKIDRLQDTSIDNVLTNYTIEATL 313
I R V LL P + D K +A+++ + N +
Sbjct: 217 IFRSVELLGIPATHLMDMDLKPRVADNYQDG-------------------IFNLKLHFIG 257
Query: 314 YDSGSWESSDGNPDLLSSNVADITFQPTTTPLGFYGYTLVGKLQSPKLWSAEQPYLYTLV 373
+GS+ LL ++ T + K ++ LW+ PYLY L+
Sbjct: 258 KKAGSFH-------LLVKDIKGHTLLEKNEDIKENVQINNEKFENVHLWNNHDPYLYQLL 310
Query: 374 VVLKDKSGRVLDCESSQVGFKNVS-KAHKQLLVNGHPVVIRGVNRHEHHPEVGKANIESC 432
+ + D+ +L+ Q GF+ + K +L+NG ++I GVNRHE + G++ S
Sbjct: 311 IEVYDEQQNLLELIPFQFGFRRIEISPEKVVLLNGKRLIINGVNRHEWDAKRGRSITMSD 370
Query: 433 MVKDLVLMKQNNINAVRNSHYPQHPRWYELCDLFGMYMIDEANIETHGF-----DYSKHL 487
M D+ K+NNINAVR HYP WY LCD G+Y++ E N+E+HG +
Sbjct: 371 MTTDINTFKENNINAVRTCHYPNQIPWYYLCDQNGIYVMAENNLESHGTWQKMGEIEPSD 430
Query: 488 KHPTLEPMWATAMLDRVIGMVERDKNHTCIISWSLGNESGFGTNHFAMAGWIRGRDSSRV 547
P P W A++DR E KNHT I+ WSLGNES G N AM + + D +R+
Sbjct: 431 NVPGSIPQWKEAVIDRARINYETFKNHTSILFWSLGNESYAGDNIIAMNEFYKSHDDTRL 490
Query: 548 IHYEGGGSR----TPCTDIVCPMYM---RVWDMLKIANDPTETRPLILCEYSHAMGNSNG 600
+HYEG R +D+ MY+ +V + L+ NDP +P + CEY H MGNS+G
Sbjct: 491 VHYEGVVHRPELKDKISDVESCMYLPPKKVEEYLQ--NDP--PKPFMECEYMHDMGNSDG 546
Query: 601 NLHIYWEAIDNTFGLQGGFIWDWVDQALR-KVQADGTKQWAYGGEFGDIPNDLNFCLNGL 659
+ Y + +D GGFIWD++DQAL + G YGG+F D +D F +GL
Sbjct: 547 GMGSYIKLLDKYPQYFGGFIWDFIDQALLVHDEISGHDVLRYGGDFDDRHSDYEFSGDGL 606
Query: 660 VWPDRTAHPVLHEVKFLY 677
++ DRT P + EV++ Y
Sbjct: 607 MFADRTPKPAMQEVRYYY 624
>BGAL_LACSK (Q48846) Beta-galactosidase large subunit (EC 3.2.1.23)
(Lactase)
Length = 625
Score = 315 bits (806), Expect = 6e-85
Identities = 201/614 (32%), Positives = 306/614 (49%), Gaps = 60/614 (9%)
Query: 91 FVKSLSGYWKFFLASNPCNVPAKFHDSEFQDSEWSTLPVPSNWQLHGFDRPIYTNVTYPF 150
F +SL+G W+F + N + P F++ F ++ + VP + +L G+++ Y N YP+
Sbjct: 41 FEQSLNGTWQFHYSVNAASRPKSFYELAFDAQDFEPITVPQHIELAGYEQLHYINTMYPW 100
Query: 151 P----LDPPFVPTE-------------NPTGCYRMDFHLPKEWEGRRILLHFEAVDSAFC 193
P F ++ NP G Y F L +R+++ FE V+ A
Sbjct: 101 EGHYYRRPAFSTSDDKQHLGMFSEADYNPVGSYLHHFDLTPALRNQRVIIRFEGVEQAMY 160
Query: 194 AWINGHPIGYSQDSRLPAEFEVTDFCHPCGSDLKNVLAVQVFRWSDGCYLEDQDHWRMSG 253
W+NG IGY++DS P+EF++T + + N LAV+V + S ++EDQD +R G
Sbjct: 161 VWLNGQFIGYAEDSFTPSEFDLTPYL----KETDNCLAVEVHKRSSAAFIEDQDFFRFFG 216
Query: 254 IHRDVLLLAKPEVFITDYFFKSNLAEDFSSAEMLVEVKIDRLQDTSIDNVLTNYTIEATL 313
I RDV LLAKP + D + + ++ + +VK+ RLQ + +N +
Sbjct: 217 IFRDVKLLAKPRTHLEDLW----VIPEYDVVQQTGQVKL-RLQFSGDENRV--------- 262
Query: 314 YDSGSWESSDGNPDLLSSNVADITFQPTTTPLGFYGYTLVGKLQSPKLWSAEQPYLYTLV 373
D + +L+ AD+T L Y + +Q+ WS + P LYTL
Sbjct: 263 ----HLRIRDQHQIILT---ADLTSGAQVNDL----YKMPELVQA---WSNQTPNLYTLE 308
Query: 374 VVLKDKSGRVLDCESSQVGFKNVSKAHKQLLVNGHPVVIRGVNRHEHHPEVGKANIESCM 433
+ + D++G ++ GF+ + K +L+NG +VI GVNRHE HPE G+
Sbjct: 309 LEVVDQAGETIEISQQPFGFRKIEIKDKVMLLNGKRLVINGVNRHEWHPETGRTITAEDE 368
Query: 434 VKDLVLMKQNNINAVRNSHYPQHPRWYELCDLFGMYMIDEANIETHGFDYSKHLKHPTLE 493
D+ M++N+INAVR SHYP +Y CD G+YM+ E N+E+HG P+
Sbjct: 369 AWDIACMQRNHINAVRTSHYPDRLSFYNGCDQAGIYMMAETNLESHGSWQKMGAVEPSWN 428
Query: 494 -----PMWATAMLDRVIGMVERDKNHTCIISWSLGNESGFGTNHFAMAGWIRGRDSSRVI 548
W A LDR E KNH I+ WSLGNES G+ M + + +D +R++
Sbjct: 429 VPGSYDEWEAATLDRARTNFETFKNHVSILFWSLGNESYAGSVLEKMNAYYKQQDPTRLV 488
Query: 549 HYEG----GGSRTPCTDIVCPMYMRVWDMLKIANDPTETRPLILCEYSHAMGNSNGNLHI 604
HYEG + +D+ MY + +K D +P ILCEY H MGNS G +
Sbjct: 489 HYEGVFRAPEYKATISDVESRMYATPAE-IKAYLDNAPQKPFILCEYMHDMGNSLGGMQS 547
Query: 605 YWEAIDNTFGLQGGFIWDWVDQALRKVQ-ADGTKQWAYGGEFGDIPNDLNFCLNGLVWPD 663
Y + + QGGFIWD++DQAL G ++ YGG+F D P+D F +GLV+
Sbjct: 548 YIDLLSQYDMYQGGFIWDFIDQALLVTDPVTGQRELRYGGDFDDRPSDYEFSGDGLVFAT 607
Query: 664 RTAHPVLHEVKFLY 677
R P + EV++ Y
Sbjct: 608 RDEKPAMQEVRYYY 621
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.136 0.436
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 147,022,712
Number of Sequences: 164201
Number of extensions: 6713688
Number of successful extensions: 13265
Number of sequences better than 10.0: 39
Number of HSP's better than 10.0 without gapping: 33
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 12990
Number of HSP's gapped (non-prelim): 73
length of query: 1118
length of database: 59,974,054
effective HSP length: 121
effective length of query: 997
effective length of database: 40,105,733
effective search space: 39985415801
effective search space used: 39985415801
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 71 (32.0 bits)
Medicago: description of AC121234.2


