

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149580.1 + phase: 0 /pseudo
(260 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
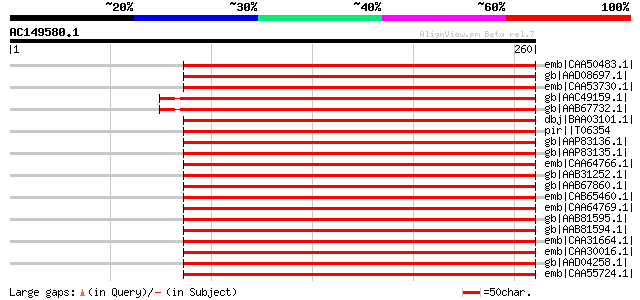
Sequences producing significant alignments: (bits) Value
emb|CAA50483.1| lipoxygenase [Lens culinaris] gi|585416|sp|P3841... 293 2e-78
gb|AAD08697.1| lipoxygenase LoxN3 [Pisum sativum] 289 5e-77
emb|CAA53730.1| lipoxygenase [Pisum sativum] gi|1362063|pir||S56... 285 7e-76
gb|AAC49159.1| lipoxygenase gi|7433156|pir||T06596 lipoxygenase ... 277 2e-73
gb|AAB67732.1| lipoxygenase L-5 [Glycine max] gi|7433153|pir||T0... 276 5e-73
dbj|BAA03101.1| lipxygenase L-4 [Glycine max] gi|585418|sp|P3841... 273 5e-72
pir||T06354 lipoxygenase (EC 1.13.11.12) - soybean gi|439857|gb|... 273 5e-72
gb|AAP83136.1| lipoxygenase [Nicotiana attenuata] gi|32454706|gb... 268 9e-71
gb|AAP83135.1| lipoxygenase [Nicotiana attenuata] 268 9e-71
emb|CAA64766.1| lipoxygenase [Solanum tuberosum] 268 1e-70
gb|AAB31252.1| linoleate:oxygen oxidoreductase; lipoxygenase; LO... 267 2e-70
gb|AAB67860.1| lipoxygenase [Solanum tuberosum] 267 2e-70
emb|CAB65460.1| lipoxygenase [Solanum tuberosum] 266 6e-70
emb|CAA64769.1| lipoxygenase [Solanum tuberosum] 266 6e-70
gb|AAB81595.1| lipoxygenase [Solanum tuberosum] 266 6e-70
gb|AAB81594.1| lipoxygenase [Solanum tuberosum] 266 6e-70
emb|CAA31664.1| unnamed protein product [Glycine max] gi|81793|p... 265 7e-70
emb|CAA30016.1| lipoxygenase [Glycine max] gi|126406|sp|P09186|L... 265 7e-70
gb|AAD04258.1| 5-lipoxygenase [Solanum tuberosum] 265 9e-70
emb|CAA55724.1| lipoxygenase [Solanum tuberosum] gi|585417|sp|P3... 264 2e-69
>emb|CAA50483.1| lipoxygenase [Lens culinaris] gi|585416|sp|P38414|LOX1_LENCU
Lipoxygenase
Length = 866
Score = 293 bits (751), Expect = 2e-78
Identities = 136/174 (78%), Positives = 153/174 (87%)
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
WLLAKA+ VVNDSC HQLVSHWL THAVVEPF+IATNRHLSV+HPIHKLL+PHYR TM I
Sbjct: 506 WLLAKAFVVVNDSCYHQLVSHWLNTHAVVEPFIIATNRHLSVVHPIHKLLLPHYRDTMNI 565
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
NA +RN+L+NA GI+ESTFL+ Y MEMSAVVYKDWVF ++GLP DL RG+AV D SSP
Sbjct: 566 NALARNVLVNAEGIIESTFLWGNYAMEMSAVVYKDWVFPDQGLPNDLIKRGVAVKDPSSP 625
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
HG+RLLIEDYPYASDGLEIWAAIKSWV+EYVNFYY+SD + D EL+AFWKEL
Sbjct: 626 HGVRLLIEDYPYASDGLEIWAAIKSWVEEYVNFYYKSDAAIAQDAELQAFWKEL 679
>gb|AAD08697.1| lipoxygenase LoxN3 [Pisum sativum]
Length = 492
Score = 289 bits (740), Expect = 5e-77
Identities = 134/174 (77%), Positives = 153/174 (87%)
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
WLLAKA+ VVNDSC HQLVSHWL THAVVEPF+IATNRHLSV+HPIHKLL+PHYR TM I
Sbjct: 132 WLLAKAFVVVNDSCYHQLVSHWLNTHAVVEPFIIATNRHLSVVHPIHKLLLPHYRDTMNI 191
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
NA +R++L+NA GI+ESTFL+ Y MEMSAVVYKDWVF ++GLP DL RG+AV D S+P
Sbjct: 192 NALARDVLVNAEGIIESTFLWGNYAMEMSAVVYKDWVFTDQGLPNDLIKRGVAVKDPSAP 251
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
+GLRLLIEDYPYASDGLEIWAAIKSWV+EYVNFYY+SD + D EL+AFWKEL
Sbjct: 252 YGLRLLIEDYPYASDGLEIWAAIKSWVEEYVNFYYKSDGAIAQDAELQAFWKEL 305
>emb|CAA53730.1| lipoxygenase [Pisum sativum] gi|1362063|pir||S56655 lipoxygenase
(EC 1.13.11.12) loxG - garden pea
Length = 868
Score = 285 bits (730), Expect = 7e-76
Identities = 130/174 (74%), Positives = 152/174 (86%)
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
WLLAKAY VVNDSC HQLVSHWL THAVVEPF+IATNRHLSV+HP+HKLL+PHYR TM I
Sbjct: 508 WLLAKAYVVVNDSCYHQLVSHWLNTHAVVEPFIIATNRHLSVVHPVHKLLLPHYRDTMNI 567
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
N+ +RN+L+NA GI+ESTFL+ Y +E+SAVVY+DWVF +GLP DL RG+AV D +SP
Sbjct: 568 NSLARNVLVNAEGIIESTFLWGGYSLELSAVVYRDWVFTNQGLPNDLLKRGVAVVDPASP 627
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
HG+RLLIEDYPYASDGLEIWAAIKSWV+EYVNFYY+SD+ + D ELKA WK+L
Sbjct: 628 HGIRLLIEDYPYASDGLEIWAAIKSWVEEYVNFYYKSDEAIAQDAELKASWKDL 681
>gb|AAC49159.1| lipoxygenase gi|7433156|pir||T06596 lipoxygenase (EC 1.13.11.12) 7
- soybean gi|1588566|prf||2208476A lipoxygenase
Length = 856
Score = 277 bits (708), Expect = 2e-73
Identities = 131/186 (70%), Positives = 153/186 (81%), Gaps = 2/186 (1%)
Query: 75 PNSTAYHAGTFNWLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHK 134
P+S A + WLLAKAY VVNDSC HQLVSHWL THAVVEPFVIATNRHLSV+HPI+K
Sbjct: 486 PSSEGVEA--YIWLLAKAYVVVNDSCYHQLVSHWLNTHAVVEPFVIATNRHLSVVHPIYK 543
Query: 135 LLVPHYRGTMTINARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLK 194
LL PHYR TM IN+ +R L+NA GI+E TFL+ +Y +EMSAVVYKDWVF ++ LP DL
Sbjct: 544 LLFPHYRDTMNINSLARKSLVNADGIIEKTFLWGRYALEMSAVVYKDWVFTDQALPNDLV 603
Query: 195 NRGMAVDDDSSPHGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELK 254
RG+AV D S+PHG+RLLIEDYPYASDGLEIW AIKSWV EYV+FYY+SD ++ D EL+
Sbjct: 604 KRGVAVKDPSAPHGVRLLIEDYPYASDGLEIWDAIKSWVQEYVSFYYKSDAAIQQDPELQ 663
Query: 255 AFWKEL 260
A+WKEL
Sbjct: 664 AWWKEL 669
>gb|AAB67732.1| lipoxygenase L-5 [Glycine max] gi|7433153|pir||T07036 lipoxygenase
(EC 1.13.11.12) L-5 - soybean
Length = 853
Score = 276 bits (705), Expect = 5e-73
Identities = 127/186 (68%), Positives = 156/186 (83%), Gaps = 2/186 (1%)
Query: 75 PNSTAYHAGTFNWLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHK 134
P+S A + WLLAKAY VVND+C HQ++SHWL THAVVEPFVIATNRHLSV+HPI+K
Sbjct: 483 PSSEGVEA--YIWLLAKAYVVVNDACYHQIISHWLNTHAVVEPFVIATNRHLSVVHPIYK 540
Query: 135 LLVPHYRGTMTINARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLK 194
LL PHYR TM IN+ +R L+NA GI+E TFL+ +Y +EMSAV+YKDWVF ++ LP DL
Sbjct: 541 LLFPHYRDTMNINSLARKSLVNADGIIEKTFLWGRYSLEMSAVIYKDWVFTDQALPNDLV 600
Query: 195 NRGMAVDDDSSPHGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELK 254
RG+AV D S+PHG+RLLIEDYPYASDGLEIW AIKSWV+EYV+FYY+SD++++ D EL+
Sbjct: 601 KRGVAVKDPSAPHGVRLLIEDYPYASDGLEIWDAIKSWVEEYVSFYYKSDEELQKDPELQ 660
Query: 255 AFWKEL 260
A+WKEL
Sbjct: 661 AWWKEL 666
>dbj|BAA03101.1| lipxygenase L-4 [Glycine max] gi|585418|sp|P38417|LOX4_SOYBN
Lipoxygenase-4 (L-4) (VSP94)
Length = 853
Score = 273 bits (697), Expect = 5e-72
Identities = 123/174 (70%), Positives = 149/174 (84%)
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
WLLAKAY VVND+C HQ++SHWL THA+VEPFVIATNR LSV+HPI+KLL PHYR TM I
Sbjct: 493 WLLAKAYVVVNDACYHQIISHWLSTHAIVEPFVIATNRQLSVVHPIYKLLFPHYRDTMNI 552
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
N+ +R L+NA GI+E TFL+ +Y MEMSAV+YKDWVF ++ LP DL RG+AV D S+P
Sbjct: 553 NSLARKALVNADGIIEKTFLWGRYSMEMSAVIYKDWVFTDQALPNDLVKRGVAVKDPSAP 612
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
HG+RLLIEDYPYASDGLEIW AIKSWV EYV+FYY+SD++++ D EL+A+WKEL
Sbjct: 613 HGVRLLIEDYPYASDGLEIWDAIKSWVQEYVSFYYKSDEELQKDPELQAWWKEL 666
>pir||T06354 lipoxygenase (EC 1.13.11.12) - soybean gi|439857|gb|AAA03726.1|
lipoxygenase
Length = 839
Score = 273 bits (697), Expect = 5e-72
Identities = 123/174 (70%), Positives = 149/174 (84%)
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
WLLAKAY VVND+C HQ++SHWL THA+VEPFVIATNR LSV+HPI+KLL PHYR TM I
Sbjct: 479 WLLAKAYVVVNDACYHQIISHWLSTHAIVEPFVIATNRQLSVVHPIYKLLFPHYRDTMNI 538
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
N+ +R L+NA GI+E TFL+ +Y MEMSAV+YKDWVF ++ LP DL RG+AV D S+P
Sbjct: 539 NSLARKALVNADGIIEKTFLWGRYSMEMSAVIYKDWVFTDQALPNDLVKRGVAVKDPSAP 598
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
HG+RLLIEDYPYASDGLEIW AIKSWV EYV+FYY+SD++++ D EL+A+WKEL
Sbjct: 599 HGVRLLIEDYPYASDGLEIWDAIKSWVQEYVSFYYKSDEELQKDPELQAWWKEL 652
>gb|AAP83136.1| lipoxygenase [Nicotiana attenuata] gi|32454706|gb|AAP83134.1|
lipoxygenase [Nicotiana attenuata]
Length = 861
Score = 268 bits (686), Expect = 9e-71
Identities = 125/174 (71%), Positives = 144/174 (81%)
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
W LAKAY VNDS HQL+SHWL THAV+EPFVIATNR LSVLHPIHKLL PH+R TM I
Sbjct: 502 WELAKAYVAVNDSGVHQLISHWLNTHAVIEPFVIATNRQLSVLHPIHKLLHPHFRDTMNI 561
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
NA +R ILINAGG++EST KY MEMSAVVYK+W+F ++ LPTDL RGMAV+D SSP
Sbjct: 562 NAMARQILINAGGVLESTVFPSKYAMEMSAVVYKNWIFPDQALPTDLVKRGMAVEDSSSP 621
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
HG+RLLI+DYPYA DGLEIW+AIKSWV EY +FYY+SD + D EL+A+WKEL
Sbjct: 622 HGIRLLIQDYPYAVDGLEIWSAIKSWVTEYCSFYYKSDDSILKDNELQAWWKEL 675
>gb|AAP83135.1| lipoxygenase [Nicotiana attenuata]
Length = 861
Score = 268 bits (686), Expect = 9e-71
Identities = 125/174 (71%), Positives = 144/174 (81%)
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
W LAKAY VNDS HQL+SHWL THAV+EPFVIATNR LSVLHPIHKLL PH+R TM I
Sbjct: 502 WELAKAYVAVNDSGVHQLISHWLNTHAVIEPFVIATNRQLSVLHPIHKLLHPHFRDTMNI 561
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
NA +R ILINAGG++EST KY MEMSAVVYK+W+F ++ LPTDL RGMAV+D SSP
Sbjct: 562 NAMARQILINAGGVLESTVFPSKYAMEMSAVVYKNWIFPDQALPTDLVKRGMAVEDSSSP 621
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
HG+RLLI+DYPYA DGLEIW+AIKSWV EY +FYY+SD + D EL+A+WKEL
Sbjct: 622 HGIRLLIQDYPYAVDGLEIWSAIKSWVTEYCSFYYKSDDSILKDNELQAWWKEL 675
>emb|CAA64766.1| lipoxygenase [Solanum tuberosum]
Length = 861
Score = 268 bits (684), Expect = 1e-70
Identities = 124/174 (71%), Positives = 144/174 (82%)
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
W LAKAY VNDS HQL+SHWL THAV+EPFVIATNR LSVLHPIHKLL PH+R TM I
Sbjct: 502 WQLAKAYVAVNDSGVHQLISHWLNTHAVIEPFVIATNRQLSVLHPIHKLLYPHFRDTMNI 561
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
NA +R ILINAGG++EST K+ MEMSAVVYKDWVF ++ LP DL RG+AV+D SSP
Sbjct: 562 NAMARQILINAGGVLESTVFQSKFAMEMSAVVYKDWVFPDQALPADLVKRGVAVEDSSSP 621
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
HG+RLLIEDYPYA DGLEIW+AIKSWV +Y +FYY SD+++ D EL+A+WKEL
Sbjct: 622 HGVRLLIEDYPYAVDGLEIWSAIKSWVSDYCSFYYGSDEEILKDNELQAWWKEL 675
>gb|AAB31252.1| linoleate:oxygen oxidoreductase; lipoxygenase; LOX [Solanum
tuberosum]
Length = 857
Score = 267 bits (682), Expect = 2e-70
Identities = 124/174 (71%), Positives = 144/174 (82%)
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
W LAKAY VNDS HQL+SHWL THAV+EPFVIATNR LSVLHPIHKLL PH+R TM I
Sbjct: 498 WQLAKAYVAVNDSGVHQLISHWLNTHAVIEPFVIATNRQLSVLHPIHKLLYPHFRDTMNI 557
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
NA +R ILINAGG++EST K+ MEMSAVVYKDWVF ++ LP DL RG+AV+D SSP
Sbjct: 558 NAMARQILINAGGVLESTVFPSKFAMEMSAVVYKDWVFPDQALPADLVKRGVAVEDSSSP 617
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
HG+RLLIEDYPYA DGLEIW+AIKSWV +Y +FYY SD+++ D EL+A+WKEL
Sbjct: 618 HGVRLLIEDYPYAVDGLEIWSAIKSWVTDYCSFYYGSDEEILKDNELQAWWKEL 671
>gb|AAB67860.1| lipoxygenase [Solanum tuberosum]
Length = 860
Score = 267 bits (682), Expect = 2e-70
Identities = 124/174 (71%), Positives = 144/174 (82%)
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
W LAKAY VNDS HQL+SHWL THAV+EPFVIATNR LSVLHPIHKLL PH+R TM I
Sbjct: 501 WQLAKAYVAVNDSGVHQLISHWLNTHAVIEPFVIATNRQLSVLHPIHKLLYPHFRDTMNI 560
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
NA +R ILINAGG++EST K+ MEMSAVVYKDWVF ++ LP DL RG+AV+D SSP
Sbjct: 561 NAMARQILINAGGVLESTVFPSKFAMEMSAVVYKDWVFPDQALPADLVKRGVAVEDSSSP 620
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
HG+RLLIEDYPYA DGLEIW+AIKSWV +Y +FYY SD+++ D EL+A+WKEL
Sbjct: 621 HGVRLLIEDYPYAVDGLEIWSAIKSWVTDYCSFYYGSDEEILKDNELQAWWKEL 674
>emb|CAB65460.1| lipoxygenase [Solanum tuberosum]
Length = 861
Score = 266 bits (679), Expect = 6e-70
Identities = 122/174 (70%), Positives = 144/174 (82%)
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
W LAKAY VND+ HQL+SHWL THAV+EPFVIATNR LSVLHPIHKLL PH+R TM I
Sbjct: 502 WQLAKAYVAVNDTGVHQLISHWLNTHAVIEPFVIATNRQLSVLHPIHKLLYPHFRDTMNI 561
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
NA +R IL+NAGG++EST K+ MEMSAVVYKDWVF ++ LP DL RG+AV+D SSP
Sbjct: 562 NASARQILVNAGGVLESTVFQSKFAMEMSAVVYKDWVFPDQALPADLVKRGVAVEDSSSP 621
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
HG+RLLIEDYPYA DGLEIW+AIKSWV +Y +FYY SD+++ D EL+A+WKEL
Sbjct: 622 HGVRLLIEDYPYAVDGLEIWSAIKSWVTDYCSFYYGSDEEILKDNELQAWWKEL 675
>emb|CAA64769.1| lipoxygenase [Solanum tuberosum]
Length = 697
Score = 266 bits (679), Expect = 6e-70
Identities = 122/174 (70%), Positives = 144/174 (82%)
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
W LAKAY VND+ HQL+SHWL THAV+EPFVIATNR LSVLHPIHKLL PH+R TM I
Sbjct: 338 WQLAKAYVAVNDTGVHQLISHWLNTHAVIEPFVIATNRQLSVLHPIHKLLYPHFRDTMNI 397
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
NA +R IL+NAGG++EST K+ MEMSAVVYKDWVF ++ LP DL RG+AV+D SSP
Sbjct: 398 NASARQILVNAGGVLESTVFQSKFAMEMSAVVYKDWVFPDQALPADLVKRGVAVEDSSSP 457
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
HG+RLLIEDYPYA DGLEIW+AIKSWV +Y +FYY SD+++ D EL+A+WKEL
Sbjct: 458 HGVRLLIEDYPYAVDGLEIWSAIKSWVTDYCSFYYGSDEEILKDNELQAWWKEL 511
>gb|AAB81595.1| lipoxygenase [Solanum tuberosum]
Length = 861
Score = 266 bits (679), Expect = 6e-70
Identities = 122/174 (70%), Positives = 144/174 (82%)
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
W LAKAY VND+ HQL+SHWL THAV+EPFVIATNR LSVLHPIHKLL PH+R TM I
Sbjct: 502 WQLAKAYVAVNDTGVHQLISHWLNTHAVIEPFVIATNRQLSVLHPIHKLLYPHFRDTMNI 561
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
NA +R IL+NAGG++EST K+ MEMSAVVYKDWVF ++ LP DL RG+AV+D SSP
Sbjct: 562 NASARQILVNAGGVLESTVFQSKFAMEMSAVVYKDWVFPDQALPADLVKRGVAVEDSSSP 621
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
HG+RLLIEDYPYA DGLEIW+AIKSWV +Y +FYY SD+++ D EL+A+WKEL
Sbjct: 622 HGVRLLIEDYPYAVDGLEIWSAIKSWVTDYCSFYYGSDEEILKDNELQAWWKEL 675
>gb|AAB81594.1| lipoxygenase [Solanum tuberosum]
Length = 861
Score = 266 bits (679), Expect = 6e-70
Identities = 122/174 (70%), Positives = 144/174 (82%)
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
W LAKAY VND+ HQL+SHWL THAV+EPFVIATNR LSVLHPIHKLL PH+R TM I
Sbjct: 502 WQLAKAYVAVNDTGVHQLISHWLNTHAVIEPFVIATNRQLSVLHPIHKLLYPHFRDTMNI 561
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
NA +R IL+NAGG++EST K+ MEMSAVVYKDWVF ++ LP DL RG+AV+D SSP
Sbjct: 562 NASARQILVNAGGVLESTVFQSKFAMEMSAVVYKDWVFPDQALPADLVKRGVAVEDSSSP 621
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
HG+RLLIEDYPYA DGLEIW+AIKSWV +Y +FYY SD+++ D EL+A+WKEL
Sbjct: 622 HGVRLLIEDYPYAVDGLEIWSAIKSWVTDYCSFYYGSDEEILKDNELQAWWKEL 675
>emb|CAA31664.1| unnamed protein product [Glycine max] gi|81793|pir||S01864
lipoxygenase (EC 1.13.11.12) 3 - soybean
Length = 857
Score = 265 bits (678), Expect = 7e-70
Identities = 122/174 (70%), Positives = 144/174 (82%)
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
WLLAKAY VVNDSC HQLVSHWL THAVVEPF+IATNRHLSV+HPI+KLL PHYR TM I
Sbjct: 498 WLLAKAYVVVNDSCYHQLVSHWLNTHAVVEPFIIATNRHLSVVHPIYKLLHPHYRDTMNI 557
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
N +R L+N GG++E TFL+ +Y +EMSAVVYKDWVF ++ LP DL RGMA++D S P
Sbjct: 558 NGLARLSLVNDGGVIEQTFLWGRYSVEMSAVVYKDWVFTDQALPADLIKRGMAIEDPSCP 617
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
HG+RL+IEDYPYA DGLEIW AIK+WV EYV YY+SD +++D EL+A WKEL
Sbjct: 618 HGIRLVIEDYPYAVDGLEIWDAIKTWVHEYVFLYYKSDDTLREDPELQACWKEL 671
>emb|CAA30016.1| lipoxygenase [Glycine max] gi|126406|sp|P09186|LOX3_SOYBN Seed
lipoxygenase-3 (L-3)
Length = 857
Score = 265 bits (678), Expect = 7e-70
Identities = 122/174 (70%), Positives = 144/174 (82%)
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
WLLAKAY VVNDSC HQLVSHWL THAVVEPF+IATNRHLSV+HPI+KLL PHYR TM I
Sbjct: 498 WLLAKAYVVVNDSCYHQLVSHWLNTHAVVEPFIIATNRHLSVVHPIYKLLHPHYRDTMNI 557
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
N +R L+N GG++E TFL+ +Y +EMSAVVYKDWVF ++ LP DL RGMA++D S P
Sbjct: 558 NGLARLSLVNDGGVIEQTFLWGRYSVEMSAVVYKDWVFTDQALPADLIKRGMAIEDPSCP 617
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
HG+RL+IEDYPYA DGLEIW AIK+WV EYV YY+SD +++D EL+A WKEL
Sbjct: 618 HGIRLVIEDYPYAVDGLEIWDAIKTWVHEYVFLYYKSDDTLREDPELQACWKEL 671
>gb|AAD04258.1| 5-lipoxygenase [Solanum tuberosum]
Length = 864
Score = 265 bits (677), Expect = 9e-70
Identities = 122/174 (70%), Positives = 144/174 (82%)
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
W LAKAY VND+ HQL+SHWL THAV+EPFVIATNR LSVLHPIHKLL PH+R TM I
Sbjct: 505 WQLAKAYVAVNDAGVHQLISHWLNTHAVIEPFVIATNRQLSVLHPIHKLLYPHFRDTMNI 564
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
NA +R ILINAGG++EST K+ +EMSAVVYKDWVF ++ LP DL RG+AV+D SSP
Sbjct: 565 NASARQILINAGGVLESTVFQSKFALEMSAVVYKDWVFPDQALPADLVKRGVAVEDSSSP 624
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
HG+RLLIEDYPYA DGLEIW+AIKSWV +Y +FYY SD+++ D EL+A+WKEL
Sbjct: 625 HGVRLLIEDYPYAVDGLEIWSAIKSWVTDYCSFYYGSDEEILKDNELQAWWKEL 678
>emb|CAA55724.1| lipoxygenase [Solanum tuberosum] gi|585417|sp|P37831|LOX1_SOLTU
Lipoxygenase 1
Length = 861
Score = 264 bits (674), Expect = 2e-69
Identities = 121/174 (69%), Positives = 143/174 (81%)
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
W LAKAY VND HQL+SHWL THAV+EPFVIATNR LSVLHPIHKLL PH+R TM I
Sbjct: 502 WQLAKAYVAVNDVGVHQLISHWLNTHAVIEPFVIATNRQLSVLHPIHKLLYPHFRDTMNI 561
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
NA +R +L+NAGG++EST K+ MEMSAVVYKDWVF ++ LP DL RG+AV+D SSP
Sbjct: 562 NASARQLLVNAGGVLESTVFQSKFAMEMSAVVYKDWVFPDQALPADLVKRGVAVEDSSSP 621
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
HG+RLLIEDYPYA DGLEIW+AIKSWV +Y +FYY SD+++ D EL+A+WKEL
Sbjct: 622 HGVRLLIEDYPYAVDGLEIWSAIKSWVTDYCSFYYGSDEEILKDNELQAWWKEL 675
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.316 0.137 0.435
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 571,715,345
Number of Sequences: 2540612
Number of extensions: 29771673
Number of successful extensions: 195177
Number of sequences better than 10.0: 1540
Number of HSP's better than 10.0 without gapping: 442
Number of HSP's successfully gapped in prelim test: 1177
Number of HSP's that attempted gapping in prelim test: 183900
Number of HSP's gapped (non-prelim): 8579
length of query: 260
length of database: 863,360,394
effective HSP length: 125
effective length of query: 135
effective length of database: 545,783,894
effective search space: 73680825690
effective search space used: 73680825690
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 74 (33.1 bits)
Medicago: description of AC149580.1


