

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149577.7 - phase: 0
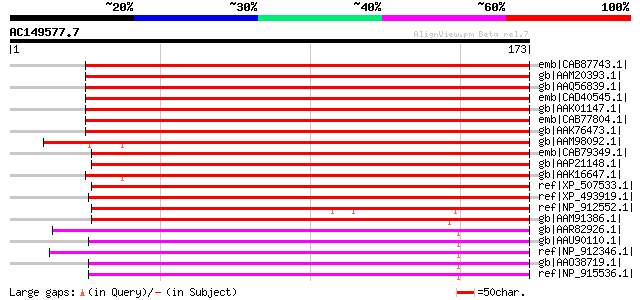
(173 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAB87743.1| transport inhibitor response 1 (TIR1) [Arabidops... 197 8e-50
gb|AAM20393.1| transport inhibitor response 1, putative [Arabido... 196 3e-49
gb|AAQ56839.1| At3g26830 [Arabidopsis thaliana] gi|9279671|dbj|B... 193 2e-48
emb|CAD40545.1| OSJNBa0072K14.18 [Oryza sativa (japonica cultiva... 186 2e-46
gb|AAK01147.1| GRR1-like protein 1 [Arabidopsis thaliana] 183 1e-45
emb|CAB77804.1| putative homolog of transport inhibitor response... 183 2e-45
gb|AAK76473.1| putative F-box protein GRR1 protein 1, AtFBL18 [A... 181 6e-45
gb|AAM98092.1| AT5g49980/K9P8_12 [Arabidopsis thaliana] gi|87774... 171 6e-42
emb|CAB79349.1| transport inhibitor response-like protein [Arabi... 170 2e-41
gb|AAP21148.1| At4g24390/T22A6_220 [Arabidopsis thaliana] gi|201... 170 2e-41
gb|AAK16647.1| F-box containing protein TIR1 [Populus tremula x ... 167 8e-41
ref|XP_507533.1| PREDICTED OJ1175_B01.8-1 gene product [Oryza sa... 167 1e-40
ref|XP_493919.1| similar to Arabidopsis thaliana transport inhib... 157 9e-38
ref|NP_912552.1| Putative F-box containing protein TIR1 [Oryza s... 134 8e-31
gb|AAM91386.1| At2g39940/T28M21.10 [Arabidopsis thaliana] gi|208... 127 2e-28
gb|AAR82926.1| coronatine-insensitive 1 [Lycopersicon esculentum... 124 8e-28
gb|AAU90110.1| putative LRR-containing F-box protein [Oryza sati... 117 2e-25
ref|NP_912346.1| unknown protein [Oryza sativa (japonica cultiva... 116 2e-25
gb|AAO38719.1| COI1 [Oryza sativa (japonica cultivar-group)] gi|... 109 4e-23
ref|NP_915536.1| P0529E05.15 [Oryza sativa (japonica cultivar-gr... 109 4e-23
>emb|CAB87743.1| transport inhibitor response 1 (TIR1) [Arabidopsis thaliana]
gi|25054937|gb|AAN71945.1| putative transport inhibitor
response TIR1, AtFBL1 protein [Arabidopsis thaliana]
gi|18412567|ref|NP_567135.1| transport inhibitor
response 1 (TIR1) (FBL1) [Arabidopsis thaliana]
gi|2352494|gb|AAB69176.1| transport inhibitor response 1
[Arabidopsis thaliana] gi|2352492|gb|AAB69175.1|
transport inhibitor response 1 [Arabidopsis thaliana]
Length = 594
Score = 197 bits (502), Expect = 8e-50
Identities = 90/148 (60%), Positives = 110/148 (73%)
Query: 26 FPDEVLERVLGMMKSRKDRSSVSLVCKEWYNAERWSRRNVFIGNCYAVSPEILTRRFPNI 85
FP+EVLE V ++ KDR+SVSLVCK WY ERW RR VFIGNCYAVSP + RRFP +
Sbjct: 9 FPEEVLEHVFSFIQLDKDRNSVSLVCKSWYEIERWCRRKVFIGNCYAVSPATVIRRFPKV 68
Query: 86 RSVTMKGKPRFSDFNLVPANWGADIHSWLVVFADKYPFLEELRLKRMAVSDESLEFLAFS 145
RSV +KGKP F+DFNLVP WG ++ W+ + Y +LEE+RLKRM V+D+ LE +A S
Sbjct: 69 RSVELKGKPHFADFNLVPDGWGGYVYPWIEAMSSSYTWLEEIRLKRMVVTDDCLELIAKS 128
Query: 146 FPNFKALSLLSCDGFSTDGLAAVATNCK 173
F NFK L L SC+GFSTDGLAA+A C+
Sbjct: 129 FKNFKVLVLSSCEGFSTDGLAAIAATCR 156
>gb|AAM20393.1| transport inhibitor response 1, putative [Arabidopsis thaliana]
gi|8698729|gb|AAF78487.1| Strong similarity to transport
inhibitor response 1 (TIR1) from Arabidopsis thaliana
gb|AF005047 gi|18391439|ref|NP_563915.1| transport
inhibitor response protein, putative [Arabidopsis
thaliana] gi|25083863|gb|AAN72129.1| transport inhibitor
response 1, putative [Arabidopsis thaliana]
gi|25352269|pir||F86261 F13K23.7 protein - Arabidopsis
thaliana
Length = 577
Score = 196 bits (497), Expect = 3e-49
Identities = 89/148 (60%), Positives = 115/148 (77%)
Query: 26 FPDEVLERVLGMMKSRKDRSSVSLVCKEWYNAERWSRRNVFIGNCYAVSPEILTRRFPNI 85
FPDEV+E V + S KDR+S+SLVCK W+ ER+SR+ VFIGNCYA++PE L RRFP +
Sbjct: 4 FPDEVIEHVFDFVASHKDRNSISLVCKSWHKIERFSRKEVFIGNCYAINPERLIRRFPCL 63
Query: 86 RSVTMKGKPRFSDFNLVPANWGADIHSWLVVFADKYPFLEELRLKRMAVSDESLEFLAFS 145
+S+T+KGKP F+DFNLVP WG +H W+ A LEELRLKRM V+DESL+ L+ S
Sbjct: 64 KSLTLKGKPHFADFNLVPHEWGGFVHPWIEALARSRVGLEELRLKRMVVTDESLDLLSRS 123
Query: 146 FPNFKALSLLSCDGFSTDGLAAVATNCK 173
F NFK+L L+SC+GF+TDGLA++A NC+
Sbjct: 124 FANFKSLVLVSCEGFTTDGLASIAANCR 151
>gb|AAQ56839.1| At3g26830 [Arabidopsis thaliana] gi|9279671|dbj|BAB01228.1|
transport inhibitor response-like protein [Arabidopsis
thaliana] gi|17064984|gb|AAL32646.1| transport inhibitor
response-like protein [Arabidopsis thaliana]
gi|18405102|ref|NP_566800.1| transport inhibitor
response protein, putative [Arabidopsis thaliana]
Length = 575
Score = 193 bits (490), Expect = 2e-48
Identities = 89/148 (60%), Positives = 114/148 (76%)
Query: 26 FPDEVLERVLGMMKSRKDRSSVSLVCKEWYNAERWSRRNVFIGNCYAVSPEILTRRFPNI 85
FPDEV+E V + S KDR+++SLVCK WY ER+SR+ VFIGNCYA++PE L RRFP +
Sbjct: 4 FPDEVIEHVFDFVTSHKDRNAISLVCKSWYKIERYSRQKVFIGNCYAINPERLLRRFPCL 63
Query: 86 RSVTMKGKPRFSDFNLVPANWGADIHSWLVVFADKYPFLEELRLKRMAVSDESLEFLAFS 145
+S+T+KGKP F+DFNLVP WG + W+ A LEELRLKRM V+DESLE L+ S
Sbjct: 64 KSLTLKGKPHFADFNLVPHEWGGFVLPWIEALARSRVGLEELRLKRMVVTDESLELLSRS 123
Query: 146 FPNFKALSLLSCDGFSTDGLAAVATNCK 173
F NFK+L L+SC+GF+TDGLA++A NC+
Sbjct: 124 FVNFKSLVLVSCEGFTTDGLASIAANCR 151
>emb|CAD40545.1| OSJNBa0072K14.18 [Oryza sativa (japonica cultivar-group)]
gi|50923929|ref|XP_472325.1| OSJNBa0072K14.18 [Oryza
sativa (japonica cultivar-group)]
Length = 575
Score = 186 bits (473), Expect = 2e-46
Identities = 88/148 (59%), Positives = 112/148 (75%)
Query: 26 FPDEVLERVLGMMKSRKDRSSVSLVCKEWYNAERWSRRNVFIGNCYAVSPEILTRRFPNI 85
FP+EV+E + + +++DR++VSLVCK WY ER SRR VF+GNCYAV + RFPN+
Sbjct: 4 FPEEVVEHIFSFLPAQRDRNTVSLVCKVWYEIERLSRRGVFVGNCYAVRAGRVAARFPNV 63
Query: 86 RSVTMKGKPRFSDFNLVPANWGADIHSWLVVFADKYPFLEELRLKRMAVSDESLEFLAFS 145
R++T+KGKP F+DFNLVP +WG W+ A LEELR+KRM VSDESLE LA S
Sbjct: 64 RALTVKGKPHFADFNLVPPDWGGYAGPWIEAAARGCHGLEELRMKRMVVSDESLELLARS 123
Query: 146 FPNFKALSLLSCDGFSTDGLAAVATNCK 173
FP F+AL L+SC+GFSTDGLAAVA++CK
Sbjct: 124 FPRFRALVLISCEGFSTDGLAAVASHCK 151
>gb|AAK01147.1| GRR1-like protein 1 [Arabidopsis thaliana]
Length = 585
Score = 183 bits (465), Expect = 1e-45
Identities = 84/148 (56%), Positives = 110/148 (73%)
Query: 26 FPDEVLERVLGMMKSRKDRSSVSLVCKEWYNAERWSRRNVFIGNCYAVSPEILTRRFPNI 85
FP +VLE +L + S +DR+SVSLVCK W+ ER +R+ VF+GNCYAVSP +TRRFP +
Sbjct: 5 FPPKVLEHILSFIDSNEDRNSVSLVCKSWFETERKTRKRVFVGNCYAVSPAAVTRRFPEM 64
Query: 86 RSVTMKGKPRFSDFNLVPANWGADIHSWLVVFADKYPFLEELRLKRMAVSDESLEFLAFS 145
RS+T+KGKP F+D+NLVP WG W+ A K LEE+R+KRM V+DE LE +A S
Sbjct: 65 RSLTLKGKPHFADYNLVPDGWGGYAWPWIEAMAAKSSSLEEIRMKRMVVTDECLEKIAAS 124
Query: 146 FPNFKALSLLSCDGFSTDGLAAVATNCK 173
F +FK L L SC+GFSTDG+AA+A+ C+
Sbjct: 125 FKDFKVLVLTSCEGFSTDGIAAIASTCR 152
>emb|CAB77804.1| putative homolog of transport inhibitor response 1 [Arabidopsis
thaliana] gi|23297438|gb|AAN12969.1| putative F-box
protein AtFBL18 [Arabidopsis thaliana]
gi|18412177|ref|NP_567255.1| F-box family protein
(FBL18) [Arabidopsis thaliana] gi|4262147|gb|AAD14447.1|
putative homolog of transport inhibitor response 1
[Arabidopsis thaliana] gi|25352265|pir||E85040
hypothetical protein AT4g03190 [imported] - Arabidopsis
thaliana
Length = 585
Score = 183 bits (464), Expect = 2e-45
Identities = 84/148 (56%), Positives = 109/148 (72%)
Query: 26 FPDEVLERVLGMMKSRKDRSSVSLVCKEWYNAERWSRRNVFIGNCYAVSPEILTRRFPNI 85
FP +VLE +L + S +DR+SVSLVCK W+ ER +R+ VF+GNCYAVSP +TRRFP +
Sbjct: 5 FPPKVLEHILSFIDSNEDRNSVSLVCKSWFETERKTRKRVFVGNCYAVSPAAVTRRFPEM 64
Query: 86 RSVTMKGKPRFSDFNLVPANWGADIHSWLVVFADKYPFLEELRLKRMAVSDESLEFLAFS 145
RS+T+KGKP F+D+NLVP WG W+ A K LEE+R+KRM V+DE LE +A S
Sbjct: 65 RSLTLKGKPHFADYNLVPDGWGGYAWPWIEAMAAKSSSLEEIRMKRMVVTDECLEKIAAS 124
Query: 146 FPNFKALSLLSCDGFSTDGLAAVATNCK 173
F +FK L L SC+GFSTDG+AA+A C+
Sbjct: 125 FKDFKVLVLTSCEGFSTDGIAAIAATCR 152
>gb|AAK76473.1| putative F-box protein GRR1 protein 1, AtFBL18 [Arabidopsis
thaliana]
Length = 585
Score = 181 bits (460), Expect = 6e-45
Identities = 83/148 (56%), Positives = 109/148 (73%)
Query: 26 FPDEVLERVLGMMKSRKDRSSVSLVCKEWYNAERWSRRNVFIGNCYAVSPEILTRRFPNI 85
FP +VLE +L + S +DR+SVSLVCK W+ ER +R+ VF+GNCYAVSP +TRRFP +
Sbjct: 5 FPPKVLEHILSFIDSNEDRNSVSLVCKSWFETERKTRKRVFVGNCYAVSPAAVTRRFPEM 64
Query: 86 RSVTMKGKPRFSDFNLVPANWGADIHSWLVVFADKYPFLEELRLKRMAVSDESLEFLAFS 145
RS+T+KGKP F+D+NLVP WG W+ A K LEE+R+KR+ V+DE LE +A S
Sbjct: 65 RSLTLKGKPHFADYNLVPDGWGGYAWPWIEAMAAKSSSLEEIRMKRIVVTDECLEKIAAS 124
Query: 146 FPNFKALSLLSCDGFSTDGLAAVATNCK 173
F +FK L L SC+GFSTDG+AA+A C+
Sbjct: 125 FKDFKVLVLTSCEGFSTDGIAAIAATCR 152
>gb|AAM98092.1| AT5g49980/K9P8_12 [Arabidopsis thaliana] gi|8777429|dbj|BAA97019.1|
transport inhibitor response 1 protein [Arabidopsis
thaliana] gi|28416503|gb|AAO42782.1| AT5g49980/K9P8_12
[Arabidopsis thaliana] gi|18423092|ref|NP_568718.1|
transport inhibitor response protein, putative
[Arabidopsis thaliana] gi|15912307|gb|AAL08287.1|
AT5g49980/K9P8_12 [Arabidopsis thaliana]
Length = 619
Score = 171 bits (434), Expect = 6e-42
Identities = 86/167 (51%), Positives = 111/167 (65%), Gaps = 5/167 (2%)
Query: 12 SQGEKNNNMDSNSD-FPDEVLERVLG----MMKSRKDRSSVSLVCKEWYNAERWSRRNVF 66
S K+ N SNS FPD VLE VL + SR DR++ SLVCK W+ E +R VF
Sbjct: 35 SSPNKSRNCISNSQTFPDHVLENVLENVLQFLDSRCDRNAASLVCKSWWRVEALTRSEVF 94
Query: 67 IGNCYAVSPEILTRRFPNIRSVTMKGKPRFSDFNLVPANWGADIHSWLVVFADKYPFLEE 126
IGNCYA+SP LT+RF +RS+ +KGKPRF+DFNL+P +WGA+ W+ A YP LE+
Sbjct: 95 IGNCYALSPARLTQRFKRVRSLVLKGKPRFADFNLMPPDWGANFAPWVSTMAQAYPCLEK 154
Query: 127 LRLKRMAVSDESLEFLAFSFPNFKALSLLSCDGFSTDGLAAVATNCK 173
+ LKRM V+D+ L LA SFP FK L L+ C+GF T G++ VA C+
Sbjct: 155 VDLKRMFVTDDDLALLADSFPGFKELILVCCEGFGTSGISIVANKCR 201
>emb|CAB79349.1| transport inhibitor response-like protein [Arabidopsis thaliana]
gi|5051781|emb|CAB45074.1| transport inhibitor
response-like protein [Arabidopsis thaliana]
gi|7487322|pir||T09902 hypothetical protein T22A6.220 -
Arabidopsis thaliana
Length = 614
Score = 170 bits (430), Expect = 2e-41
Identities = 78/146 (53%), Positives = 102/146 (69%)
Query: 28 DEVLERVLGMMKSRKDRSSVSLVCKEWYNAERWSRRNVFIGNCYAVSPEILTRRFPNIRS 87
+ VLE VL + SR DR++VSLVC+ WY E +R VFIGNCY++SP L RF +RS
Sbjct: 47 ENVLENVLQFLTSRCDRNAVSLVCRSWYRVEAQTRLEVFIGNCYSLSPARLIHRFKRVRS 106
Query: 88 VTMKGKPRFSDFNLVPANWGADIHSWLVVFADKYPFLEELRLKRMAVSDESLEFLAFSFP 147
+ +KGKPRF+DFNL+P NWGA W+ A YP+LE++ LKRM V+D+ L LA SFP
Sbjct: 107 LVLKGKPRFADFNLMPPNWGAQFSPWVAATAKAYPWLEKVHLKRMFVTDDDLALLAESFP 166
Query: 148 NFKALSLLSCDGFSTDGLAAVATNCK 173
FK L+L+ C+GF T G+A VA C+
Sbjct: 167 GFKELTLVCCEGFGTSGIAIVANKCR 192
>gb|AAP21148.1| At4g24390/T22A6_220 [Arabidopsis thaliana]
gi|20147209|gb|AAM10320.1| AT4g24390/T22A6_220
[Arabidopsis thaliana] gi|42573021|ref|NP_974607.1|
F-box family protein (FBX14) [Arabidopsis thaliana]
gi|30686516|ref|NP_567702.2| F-box family protein
(FBX14) [Arabidopsis thaliana]
Length = 623
Score = 170 bits (430), Expect = 2e-41
Identities = 78/146 (53%), Positives = 102/146 (69%)
Query: 28 DEVLERVLGMMKSRKDRSSVSLVCKEWYNAERWSRRNVFIGNCYAVSPEILTRRFPNIRS 87
+ VLE VL + SR DR++VSLVC+ WY E +R VFIGNCY++SP L RF +RS
Sbjct: 56 ENVLENVLQFLTSRCDRNAVSLVCRSWYRVEAQTRLEVFIGNCYSLSPARLIHRFKRVRS 115
Query: 88 VTMKGKPRFSDFNLVPANWGADIHSWLVVFADKYPFLEELRLKRMAVSDESLEFLAFSFP 147
+ +KGKPRF+DFNL+P NWGA W+ A YP+LE++ LKRM V+D+ L LA SFP
Sbjct: 116 LVLKGKPRFADFNLMPPNWGAQFSPWVAATAKAYPWLEKVHLKRMFVTDDDLALLAESFP 175
Query: 148 NFKALSLLSCDGFSTDGLAAVATNCK 173
FK L+L+ C+GF T G+A VA C+
Sbjct: 176 GFKELTLVCCEGFGTSGIAIVANKCR 201
>gb|AAK16647.1| F-box containing protein TIR1 [Populus tremula x Populus
tremuloides]
Length = 635
Score = 167 bits (424), Expect = 8e-41
Identities = 81/152 (53%), Positives = 105/152 (68%), Gaps = 4/152 (2%)
Query: 26 FPDEVLERVLG----MMKSRKDRSSVSLVCKEWYNAERWSRRNVFIGNCYAVSPEILTRR 81
+PD+VLE VL + SRKDR++ SLVC+ WY E +R ++FIGNCYAVSP+ R
Sbjct: 65 YPDQVLENVLENVLWFLTSRKDRNAASLVCRSWYRVEALTRSDLFIGNCYAVSPKRAMSR 124
Query: 82 FPNIRSVTMKGKPRFSDFNLVPANWGADIHSWLVVFADKYPFLEELRLKRMAVSDESLEF 141
F IRSVT+KGKPRF+DFNL+P WGA W+ A YP+LE++ LKRM+V+D+ L
Sbjct: 125 FTRIRSVTLKGKPRFADFNLMPPYWGAHFAPWVSAMAMTYPWLEKVHLKRMSVTDDDLAL 184
Query: 142 LAFSFPNFKALSLLSCDGFSTDGLAAVATNCK 173
LA SF FK L L+ C+GF T GLA V + C+
Sbjct: 185 LAESFSGFKELVLVCCEGFGTSGLAIVVSRCR 216
>ref|XP_507533.1| PREDICTED OJ1175_B01.8-1 gene product [Oryza sativa (japonica
cultivar-group)] gi|51964402|ref|XP_506986.1| PREDICTED
OJ1175_B01.8-1 gene product [Oryza sativa (japonica
cultivar-group)] gi|50912987|ref|XP_467901.1| putative
F-box containing protein TIR1 [Oryza sativa (japonica
cultivar-group)] gi|47497357|dbj|BAD19396.1| putative
F-box containing protein TIR1 [Oryza sativa (japonica
cultivar-group)]
Length = 637
Score = 167 bits (422), Expect = 1e-40
Identities = 78/146 (53%), Positives = 101/146 (68%)
Query: 28 DEVLERVLGMMKSRKDRSSVSLVCKEWYNAERWSRRNVFIGNCYAVSPEILTRRFPNIRS 87
+ VLE VL + + +DR++ SLVC+ WY AE +RR +FIGNCYAVSP RF +R+
Sbjct: 69 ENVLESVLEFLTAARDRNAASLVCRSWYRAEAQTRRELFIGNCYAVSPRRAVERFGGVRA 128
Query: 88 VTMKGKPRFSDFNLVPANWGADIHSWLVVFADKYPFLEELRLKRMAVSDESLEFLAFSFP 147
V +KGKPRF+DF+LVP WGA + W+ YP LE + LKRM VS++ L +A SFP
Sbjct: 129 VVLKGKPRFADFSLVPYGWGAYVSPWVAALGPAYPHLERICLKRMTVSNDDLALIAKSFP 188
Query: 148 NFKALSLLSCDGFSTDGLAAVATNCK 173
FK LSL+ CDGFST GLAA+A C+
Sbjct: 189 LFKELSLVCCDGFSTLGLAAIAERCR 214
>ref|XP_493919.1| similar to Arabidopsis thaliana transport inhibitor response 1
(TIR1) (T48087) [Oryza sativa]
Length = 587
Score = 157 bits (398), Expect = 9e-38
Identities = 73/147 (49%), Positives = 93/147 (62%)
Query: 27 PDEVLERVLGMMKSRKDRSSVSLVCKEWYNAERWSRRNVFIGNCYAVSPEILTRRFPNIR 86
PDEV E + + DR + + C W AER SRR + + NCYA +P RFP++R
Sbjct: 21 PDEVWEHAFSFLPAAADRGAAAGACSSWLRAERRSRRRLAVANCYAAAPRDAVERFPSVR 80
Query: 87 SVTMKGKPRFSDFNLVPANWGADIHSWLVVFADKYPFLEELRLKRMAVSDESLEFLAFSF 146
+ +KGKP F+DF LVP WGA W+ AD +P LEEL KRM V+DE LE +A SF
Sbjct: 81 AAEVKGKPHFADFGLVPPAWGAAAAPWIAAAADGWPLLEELSFKRMVVTDECLEMIAASF 140
Query: 147 PNFKALSLLSCDGFSTDGLAAVATNCK 173
NF+ L L+SCDGFST GLAA+A C+
Sbjct: 141 RNFQVLRLVSCDGFSTAGLAAIAAGCR 167
>ref|NP_912552.1| Putative F-box containing protein TIR1 [Oryza sativa (japonica
cultivar-group)] gi|24756871|gb|AAN64135.1| Putative
F-box containing protein TIR1 [Oryza sativa (japonica
cultivar-group)]
Length = 603
Score = 134 bits (338), Expect = 8e-31
Identities = 71/149 (47%), Positives = 98/149 (65%), Gaps = 3/149 (2%)
Query: 28 DEVLERVLGMMKSRKDRSSVSLVCKEWYNAERWSRRNVFIGNCYAVSPEILTRRFPNIRS 87
D VLE VL + S +DR + SLVC+ W AE +R +V + N A SP + RRFP R
Sbjct: 29 DNVLETVLQFLDSARDRCAASLVCRSWSRAESATRASVAVRNLLAASPARVARRFPAARR 88
Query: 88 VTMKGKPRFSDFNLVPANW-GADIHSW-LVVFADKYPFLEELRLKRMAVSDESLEFLAFS 145
V +KG+PRF+DFNL+P W GAD W V A +P L L LKR+ V+D+ L+ ++ S
Sbjct: 89 VLLKGRPRFADFNLLPPGWAGADFRPWAAAVAAAAFPALASLFLKRITVTDDDLDLVSRS 148
Query: 146 FP-NFKALSLLSCDGFSTDGLAAVATNCK 173
P +F+ LSLL CDGFS+ GLA++A++C+
Sbjct: 149 LPASFRDLSLLLCDGFSSAGLASIASHCR 177
>gb|AAM91386.1| At2g39940/T28M21.10 [Arabidopsis thaliana]
gi|2088647|gb|AAB95279.1| coronatine-insensitive 1
(COI1), AtFBL2 [Arabidopsis thaliana]
gi|15010648|gb|AAK73983.1| At2g39940/T28M21.10
[Arabidopsis thaliana] gi|59797640|sp|O04197|COI1_ARATH
Coronatine-insensitive protein 1 (F-box/LRR-repeat
protein 2) (AtFBL2) (COI-1) (AtCOI1)
gi|3158394|gb|AAC17498.1| LRR-containing F-box protein
[Arabidopsis thaliana] gi|18405209|ref|NP_565919.1|
coronatine-insensitive 1 / COI1 (FBL2) [Arabidopsis
thaliana]
Length = 592
Score = 127 bits (318), Expect = 2e-28
Identities = 59/147 (40%), Positives = 94/147 (63%), Gaps = 1/147 (0%)
Query: 28 DEVLERVLGMMKSRKDRSSVSLVCKEWYNAERWSRRNVFIGNCYAVSPEILTRRFPNIRS 87
D+V+E+V+ + KDR S SLVC+ W+ + +R +V + CY +P+ L+RRFPN+RS
Sbjct: 18 DDVIEQVMTYITDPKDRDSASLVCRRWFKIDSETREHVTMALCYTATPDRLSRRFPNLRS 77
Query: 88 VTMKGKPRFSDFNLVPANWGADIHSWLVVFADKYPFLEELRLKRMAVSDESLEFLAFS-F 146
+ +KGKPR + FNL+P NWG + W+ ++ L+ + +RM VSD L+ LA +
Sbjct: 78 LKLKGKPRAAMFNLIPENWGGYVTPWVTEISNNLRQLKSVHFRRMIVSDLDLDRLAKARA 137
Query: 147 PNFKALSLLSCDGFSTDGLAAVATNCK 173
+ + L L C GF+TDGL ++ T+C+
Sbjct: 138 DDLETLKLDKCSGFTTDGLLSIVTHCR 164
>gb|AAR82926.1| coronatine-insensitive 1 [Lycopersicon esculentum]
gi|40218003|gb|AAR82925.1| coronatine-insensitive 1
[Lycopersicon esculentum]
Length = 603
Score = 124 bits (312), Expect = 8e-28
Identities = 62/160 (38%), Positives = 94/160 (58%), Gaps = 1/160 (0%)
Query: 15 EKNNNMDSNSDFPDEVLERVLGMMKSRKDRSSVSLVCKEWYNAERWSRRNVFIGNCYAVS 74
E+ N+ +S D V E V+ ++ +DR +VSLVCK W+ + +R+++ + CY
Sbjct: 2 EERNSTRLSSSTNDTVWECVIPYIQESRDRDAVSLVCKRWWQIDAITRKHITMALCYTAK 61
Query: 75 PEILTRRFPNIRSVTMKGKPRFSDFNLVPANWGADIHSWLVVFADKYPFLEELRLKRMAV 134
PE L+RRFP++ SV +KGKPR + FNL+P +WG + W++ + L+ L +RM V
Sbjct: 62 PEQLSRRFPHLESVKLKGKPRAAMFNLIPEDWGGYVTPWVMEITKSFSKLKALHFRRMIV 121
Query: 135 SDESLEFLAFSFPN-FKALSLLSCDGFSTDGLAAVATNCK 173
D LE LA + L L C GFSTDGL ++ +CK
Sbjct: 122 RDSDLELLANRRGRVLQVLKLDKCSGFSTDGLLHISRSCK 161
>gb|AAU90110.1| putative LRR-containing F-box protein [Oryza sativa (japonica
cultivar-group)]
Length = 597
Score = 117 bits (292), Expect = 2e-25
Identities = 59/148 (39%), Positives = 87/148 (57%), Gaps = 1/148 (0%)
Query: 27 PDEVLERVLGMMKSRKDRSSVSLVCKEWYNAERWSRRNVFIGNCYAVSPEILTRRFPNIR 86
P+E L VLG + +DR +VSLVC+ W+ + +R++V + CYA SP L RFP +
Sbjct: 24 PEEALHLVLGYVDDPRDREAVSLVCRRWHRIDALTRKHVTVPFCYAASPAHLLARFPRLE 83
Query: 87 SVTMKGKPRFSDFNLVPANWGADIHSWLVVFADKYPFLEELRLKRMAVSDESLEFLAFSF 146
S+ +KGKPR + + L+P +WGA W+ A L+ L L+RM V+D+ L L +
Sbjct: 84 SLAVKGKPRAAMYGLIPEDWGAYARPWVAELAAPLECLKALHLRRMVVTDDDLAALVRAR 143
Query: 147 PN-FKALSLLSCDGFSTDGLAAVATNCK 173
+ + L L C GFSTD L VA +C+
Sbjct: 144 GHMLQELKLDKCSGFSTDALRLVARSCR 171
>ref|NP_912346.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|29893584|gb|AAP06838.1| unknown protein [Oryza sativa
(japonica cultivar-group)]
Length = 589
Score = 116 bits (291), Expect = 2e-25
Identities = 59/161 (36%), Positives = 90/161 (55%), Gaps = 1/161 (0%)
Query: 14 GEKNNNMDSNSDFPDEVLERVLGMMKSRKDRSSVSLVCKEWYNAERWSRRNVFIGNCYAV 73
GE+ + PD L V+G ++ DR ++SLVC+ W + SR++V + Y+
Sbjct: 6 GERRLGRAMSFGIPDVALGLVMGFVEDPWDRDAISLVCRHWCRVDALSRKHVTVAMAYST 65
Query: 74 SPEILTRRFPNIRSVTMKGKPRFSDFNLVPANWGADIHSWLVVFADKYPFLEELRLKRMA 133
+P+ L RRFP + S+ +K KPR + FNL+P +WG W+ + + FL+ L L+RM
Sbjct: 66 TPDRLFRRFPCLESLKLKAKPRAAMFNLIPEDWGGSASPWIRQLSASFHFLKALHLRRMI 125
Query: 134 VSDESLEFLAFSFPN-FKALSLLSCDGFSTDGLAAVATNCK 173
VSD+ L+ L + + + L C GFST LA VA CK
Sbjct: 126 VSDDDLDVLVRAKAHMLSSFKLDRCSGFSTSSLALVARTCK 166
>gb|AAO38719.1| COI1 [Oryza sativa (japonica cultivar-group)]
gi|56784261|dbj|BAD81943.1| COI1 [Oryza sativa (japonica
cultivar-group)] gi|64175189|gb|AAY41186.1|
coronatine-insensitive 1 [Oryza sativa (japonica
cultivar-group)]
Length = 595
Score = 109 bits (272), Expect = 4e-23
Identities = 55/148 (37%), Positives = 85/148 (57%), Gaps = 1/148 (0%)
Query: 27 PDEVLERVLGMMKSRKDRSSVSLVCKEWYNAERWSRRNVFIGNCYAVSPEILTRRFPNIR 86
PDE L V+G ++ +DR + S VC+ W+ + +R++V + CYA P L RFP +
Sbjct: 22 PDEALHLVMGHVEDPRDREAASRVCRRWHRIDALTRKHVTVAFCYAARPARLRERFPRLE 81
Query: 87 SVTMKGKPRFSDFNLVPANWGADIHSWLVVFADKYPFLEELRLKRMAVSDESLEFLAFSF 146
S+++KGKPR + + L+P +WGA W+ A L+ L L+RM V+D + L +
Sbjct: 82 SLSLKGKPRAAMYGLIPDDWGAYAAPWIDELAAPLECLKALHLRRMTVTDADIAALVRAR 141
Query: 147 PN-FKALSLLSCDGFSTDGLAAVATNCK 173
+ + L L C GFSTD L VA +C+
Sbjct: 142 GHMLQELKLDKCIGFSTDALRLVARSCR 169
>ref|NP_915536.1| P0529E05.15 [Oryza sativa (japonica cultivar-group)]
Length = 630
Score = 109 bits (272), Expect = 4e-23
Identities = 55/148 (37%), Positives = 85/148 (57%), Gaps = 1/148 (0%)
Query: 27 PDEVLERVLGMMKSRKDRSSVSLVCKEWYNAERWSRRNVFIGNCYAVSPEILTRRFPNIR 86
PDE L V+G ++ +DR + S VC+ W+ + +R++V + CYA P L RFP +
Sbjct: 57 PDEALHLVMGHVEDPRDREAASRVCRRWHRIDALTRKHVTVAFCYAARPARLRERFPRLE 116
Query: 87 SVTMKGKPRFSDFNLVPANWGADIHSWLVVFADKYPFLEELRLKRMAVSDESLEFLAFSF 146
S+++KGKPR + + L+P +WGA W+ A L+ L L+RM V+D + L +
Sbjct: 117 SLSLKGKPRAAMYGLIPDDWGAYAAPWIDELAAPLECLKALHLRRMTVTDADIAALVRAR 176
Query: 147 PN-FKALSLLSCDGFSTDGLAAVATNCK 173
+ + L L C GFSTD L VA +C+
Sbjct: 177 GHMLQELKLDKCIGFSTDALRLVARSCR 204
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.320 0.134 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 296,287,137
Number of Sequences: 2540612
Number of extensions: 11629534
Number of successful extensions: 28174
Number of sequences better than 10.0: 143
Number of HSP's better than 10.0 without gapping: 87
Number of HSP's successfully gapped in prelim test: 56
Number of HSP's that attempted gapping in prelim test: 27959
Number of HSP's gapped (non-prelim): 247
length of query: 173
length of database: 863,360,394
effective HSP length: 119
effective length of query: 54
effective length of database: 561,027,566
effective search space: 30295488564
effective search space used: 30295488564
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 70 (31.6 bits)
Medicago: description of AC149577.7


