

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149576.2 + phase: 0
(538 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
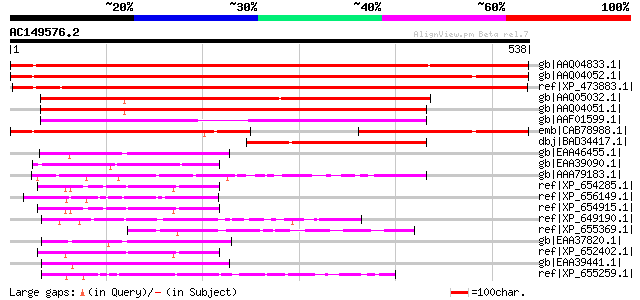
Score E
Sequences producing significant alignments: (bits) Value
gb|AAQ04833.1| lecithine cholesterol acyltransferase-like protei... 770 0.0
gb|AAQ04052.1| lecithine cholesterol acyltransferase-like protei... 738 0.0
ref|XP_473883.1| OSJNBa0008M17.7 [Oryza sativa (japonica cultiva... 686 0.0
gb|AAQ05032.1| phospholipase A1 [Nicotiana tabacum] 451 e-125
gb|AAQ04051.1| phospholipase A1 [Arabidopsis thaliana] gi|213605... 434 e-120
gb|AAF01599.1| unknown protein [Arabidopsis thaliana] 343 1e-92
emb|CAB78988.1| putative protein [Arabidopsis thaliana] gi|32506... 335 2e-90
dbj|BAD34417.1| lecithin cholesterol acyltransferase-like [Oryza... 213 2e-53
gb|EAA46455.1| GLP_90_25807_22631 [Giardia lamblia ATCC 50803] 94 1e-17
gb|EAA39090.1| GLP_305_4486_7113 [Giardia lamblia ATCC 50803] 84 1e-14
gb|AAA79183.1| esterase 80 2e-13
ref|XP_654285.1| lecithin:cholesterol acyltransferase, putative ... 79 5e-13
ref|XP_656149.1| Lecithin:cholesterol acyltransferase, putative ... 78 6e-13
ref|XP_654915.1| lecithin:cholesterol acyltransferase, putative ... 77 1e-12
ref|XP_649190.1| 1-O-acylceramide synthase, putative [Entamoeba ... 76 2e-12
ref|XP_655369.1| 1-O-acylceramide synthase, putative [Entamoeba ... 74 9e-12
gb|EAA37820.1| GLP_661_6782_9100 [Giardia lamblia ATCC 50803] 72 3e-11
ref|XP_652402.1| Lecithin:cholesterol acyltransferase, putative ... 70 1e-10
gb|EAA39441.1| GLP_762_23988_17611 [Giardia lamblia ATCC 50803] 69 4e-10
ref|XP_655259.1| Lecithin:cholesterol acyltransferase, putative ... 64 1e-08
>gb|AAQ04833.1| lecithine cholesterol acyltransferase-like protein [Lycopersicon
esculentum]
Length = 535
Score = 770 bits (1988), Expect = 0.0
Identities = 354/538 (65%), Positives = 443/538 (81%), Gaps = 4/538 (0%)
Query: 1 MTMLLEDVLRSVELWLRLIKKPQPQAYVNPNLDPVLLVPGVGGSILNAVNESDGSQERVW 60
M ML+E++++S+E+WL+LIKKPQ Y++P LDPVLLVPGV GSILNAV++ G ERVW
Sbjct: 1 MAMLIEELIKSIEMWLKLIKKPQE--YIDPTLDPVLLVPGVAGSILNAVDKKTGRTERVW 58
Query: 61 VRFLSAEYKLKTKLWSRYDPSTGKTVTLDQKSRIVVPEDRHGLHAIDVLDPDLVIGSEAV 120
VR L A+++ KLW R+DPSTGKT LD + I VPEDR+GL+AID LDPD++IGS+ V
Sbjct: 59 VRILGADHEFCDKLWCRFDPSTGKTTNLDPDTSIEVPEDRYGLYAIDNLDPDMIIGSDCV 118
Query: 121 YYFHDMIVQMQKWGYQEGKTLFGFGYDFRQSNRLQETMDRFAEKLELIYNAAGGKKIDLI 180
YY+HDMIV+M WGYQEGKTLFGFGYDFRQSNRLQETM+ FA+KLE I+ A+GGKKI++I
Sbjct: 119 YYYHDMIVEMLSWGYQEGKTLFGFGYDFRQSNRLQETMECFAQKLESIHTASGGKKINII 178
Query: 181 SHSMGGLLVKCFMTLHSDIFEKYVKNWIAICAPFQGAPGCTNSTFLNGMSFVEGWEQNFF 240
SHSMGGLLVKCFM LHSDIFEKYVKNWIAI APFQGAPG S+ LNG SFV GWE+ FF
Sbjct: 179 SHSMGGLLVKCFMALHSDIFEKYVKNWIAIAAPFQGAPGYITSSLLNGTSFVHGWEERFF 238
Query: 241 ISKWSMHQLLIECPSIYELMACPNFHWKHVPLLELWRERLHEDGKSHVILESYPPRDSIE 300
ISKWSMHQLLIECPSIYELM CP+FHW+H P+LE+W+E+ + +G+S V+LESY P +++
Sbjct: 239 ISKWSMHQLLIECPSIYELMGCPDFHWEHAPVLEIWKEKSNSNGESSVVLESYSPLEAVS 298
Query: 301 IFKQALVNNKVNHEGEELPLPFNSHIFEWANKTREILSSAKLPSGVKFYNIYGTNLATPH 360
+++ AL NNKV + GE++ LPFN + +WANKTREIL AK+P VKFYNIYGTN TPH
Sbjct: 299 VYELALANNKVTYNGEKISLPFNLELLKWANKTREILCHAKVPDKVKFYNIYGTNYETPH 358
Query: 361 SICYGNADKPVSDLQELRYLQARYVCVDGDGTVPVESAKADGFNAEERVGIPGEHRGILC 420
S+CYG+ + P+SDLQ+L ++Q+ Y+ VDGDGTVP ESAKADG AE RVG+PG+HRGI+C
Sbjct: 359 SVCYGSQNAPISDLQQLPFVQSNYISVDGDGTVPTESAKADGLKAEARVGVPGDHRGIVC 418
Query: 421 EPHLFRILKHWLKAGDPDPFYNPLNDYVILPTAFEMERHKEKGLEVASLKEEWEIISKDQ 480
+ H+FR++KHWL+A D DP+YNP+NDYVILPT+F++ERH EKGL+V SL+EEWEI+S+ Q
Sbjct: 419 DRHVFRVIKHWLRA-DHDPYYNPINDYVILPTSFDIERHHEKGLDVTSLREEWEIVSESQ 477
Query: 481 DGQSNTGDNKMTLSSISVSQEG-ANKSHSEAHATVFVHTDNDGKQHIELNAVAVSVDA 537
DG+ N K + SISVS G N + EAHAT+ VH ++GKQH+ELNA++VS A
Sbjct: 478 DGKENADSGKTKVGSISVSHVGDDNTTWEEAHATLIVHPKSEGKQHVELNAMSVSARA 535
>gb|AAQ04052.1| lecithine cholesterol acyltransferase-like protein [Arabidopsis
thaliana] gi|42566968|ref|NP_193721.2|
lecithin:cholesterol acyltransferase family protein /
LACT family protein [Arabidopsis thaliana]
gi|63003822|gb|AAY25440.1| At4g19860 [Arabidopsis
thaliana]
Length = 535
Score = 738 bits (1905), Expect = 0.0
Identities = 352/540 (65%), Positives = 438/540 (80%), Gaps = 8/540 (1%)
Query: 1 MTMLLEDVLRSVELWLRLIKKPQPQAYVNPNLDPVLLVPGVGGSILNAVNESDGSQERVW 60
M++LLE+++RSVE L+L + Q + YV+PNL+PVLLVPG+ GSILNAV+ +G++ERVW
Sbjct: 1 MSLLLEEIIRSVEALLKLRNRNQ-EPYVDPNLNPVLLVPGIAGSILNAVDHENGNEERVW 59
Query: 61 VRFLSAEYKLKTKLWSRYDPSTGKTVTLDQKSRIVVPEDRHGLHAIDVLDPDLVIGSEAV 120
VR A+++ +TK+WSR+DPSTGKT++LD K+ IVVP+DR GLHAIDVLDPD+++G E+V
Sbjct: 60 VRIFGADHEFRTKMWSRFDPSTGKTISLDPKTSIVVPQDRAGLHAIDVLDPDMIVGRESV 119
Query: 121 YYFHDMIVQMQKWGYQEGKTLFGFGYDFRQSNRLQETMDRFAEKLELIYNAAGGKKIDLI 180
YYFH+MIV+M WG++EGKTLFGFGYDFRQSNRLQET+D+FA+KLE +Y A+G KKI++I
Sbjct: 120 YYFHEMIVEMIGWGFEEGKTLFGFGYDFRQSNRLQETLDQFAKKLETVYKASGEKKINVI 179
Query: 181 SHSMGGLLVKCFMTLHSDIFEKYVKNWIAICAPFQGAPGCTNSTFLNGMSFVEGWEQNFF 240
SHSMGGLLVKCFM LHSDIFEKYV+NWIAI APF+GAPG ST LNGMSFV GWEQNFF
Sbjct: 180 SHSMGGLLVKCFMGLHSDIFEKYVQNWIAIAAPFRGAPGYITSTLLNGMSFVNGWEQNFF 239
Query: 241 ISKWSMHQLLIECPSIYELMACPNFHWKHVPLLELWRERLHED--GKSHVILESYPPRDS 298
+SKWSMHQLLIECPSIYELM CP F W+ P+LELWRE+ D G S+V+LESY +S
Sbjct: 240 VSKWSMHQLLIECPSIYELMCCPYFKWELPPVLELWREKESNDGVGTSYVVLESYCSLES 299
Query: 299 IEIFKQALVNNKVNHEGEELPLPFNSHIFEWANKTREILSSAKLPSGVKFYNIYGTNLAT 358
+E+F ++L NN ++ GE + LPFN I EWA+KT+++L+SAKLP VKFYNIYGTNL T
Sbjct: 300 LEVFTKSLSNNTADYCGESIDLPFNWKIMEWAHKTKQVLASAKLPPKVKFYNIYGTNLET 359
Query: 359 PHSICYGNADKPVSDLQELRYLQARYVCVDGDGTVPVESAKADGFNAEERVGIPGEHRGI 418
PHS+CYGN PV DL LRY Q Y+CVDGDGTVP+ESA ADG A RVG+PGEHRGI
Sbjct: 360 PHSVCYGNEKMPVKDLTNLRYFQPTYICVDGDGTVPMESAMADGLEAVARVGVPGEHRGI 419
Query: 419 LCEPHLFRILKHWLKAGDPDPFYNPLNDYVILPTAFEMERHKEKGLEVASLKEEWEIISK 478
L + +FR+LK WL G+PDPFYNP+NDYVILPT +E E+ E GLEVAS+KE W+IIS
Sbjct: 420 LNDHRVFRMLKKWLNVGEPDPFYNPVNDYVILPTTYEFEKFHENGLEVASVKESWDIISD 479
Query: 479 DQDGQSNTGDNKMTLSSISVSQEGANKS-HSEAHATVFVHTDNDGKQHIELNAVAVSVDA 537
D +N G T++SISVSQ G +++ +EA AT+ V +DG+QH+ELNAV+VSVDA
Sbjct: 480 D----NNIGTTGSTVNSISVSQPGDDQNPQAEARATLTVQPQSDGRQHVELNAVSVSVDA 535
>ref|XP_473883.1| OSJNBa0008M17.7 [Oryza sativa (japonica cultivar-group)]
gi|38344254|emb|CAD41792.2| OSJNBa0008M17.7 [Oryza
sativa (japonica cultivar-group)]
Length = 533
Score = 686 bits (1769), Expect = 0.0
Identities = 314/533 (58%), Positives = 422/533 (78%), Gaps = 3/533 (0%)
Query: 4 LLEDVLRSVELWLRLIKKPQPQAYVNPNLDPVLLVPGVGGSILNAVNESDGSQERVWVRF 63
+LED++R++ELWLR+ K+ P V+P LDPVLLVPG+GGSIL AV+E+ G +ERVWVR
Sbjct: 3 VLEDLIRAIELWLRIAKEQVP--LVDPTLDPVLLVPGIGGSILEAVDEA-GKKERVWVRI 59
Query: 64 LSAEYKLKTKLWSRYDPSTGKTVTLDQKSRIVVPEDRHGLHAIDVLDPDLVIGSEAVYYF 123
L+A+++ + LWS++D STGKTV++D+K+ IVVPEDR+GL+AID LDPD++IG ++V Y+
Sbjct: 60 LAADHEFRAHLWSKFDASTGKTVSVDEKTNIVVPEDRYGLYAIDTLDPDMIIGDDSVCYY 119
Query: 124 HDMIVQMQKWGYQEGKTLFGFGYDFRQSNRLQETMDRFAEKLELIYNAAGGKKIDLISHS 183
HDMIVQM KWGYQEGKTLFGFGYDFRQSNRL ET+DRF+ KLE +Y A+G KKI+LI+HS
Sbjct: 120 HDMIVQMIKWGYQEGKTLFGFGYDFRQSNRLSETLDRFSRKLESVYIASGEKKINLITHS 179
Query: 184 MGGLLVKCFMTLHSDIFEKYVKNWIAICAPFQGAPGCTNSTFLNGMSFVEGWEQNFFISK 243
MGGLLVKCFM+LHSD+FEKY+K+WIAI APFQGAPG ++ LNGMSFVEGWE FFISK
Sbjct: 180 MGGLLVKCFMSLHSDVFEKYIKSWIAIAAPFQGAPGYITTSLLNGMSFVEGWESRFFISK 239
Query: 244 WSMHQLLIECPSIYELMACPNFHWKHVPLLELWRERLHEDGKSHVILESYPPRDSIEIFK 303
WSM QLL+ECPSIYEL+A F W+ P L++WR++L +GK +LESY P ++I++ +
Sbjct: 240 WSMQQLLLECPSIYELLANSTFQWEDTPYLQIWRQKLDTNGKKSAMLESYEPDEAIKMIR 299
Query: 304 QALVNNKVNHEGEELPLPFNSHIFEWANKTREILSSAKLPSGVKFYNIYGTNLATPHSIC 363
+AL +++ +G +PLP + I WA +T+++L +AKLP VKFYNIYGT+ T H++
Sbjct: 300 EALSKHEIISDGMHIPLPLDMDILRWAKETQDVLCNAKLPKSVKFYNIYGTDYDTAHTVR 359
Query: 364 YGNADKPVSDLQELRYLQARYVCVDGDGTVPVESAKADGFNAEERVGIPGEHRGILCEPH 423
YG+ P+S+L +L Y Q Y+CVDGDG+VPVESAKADG +A RVG+ +HRGI+C+ H
Sbjct: 360 YGSEHHPISNLSDLLYTQGNYICVDGDGSVPVESAKADGLDAVARVGVAADHRGIVCDRH 419
Query: 424 LFRILKHWLKAGDPDPFYNPLNDYVILPTAFEMERHKEKGLEVASLKEEWEIISKDQDGQ 483
+FRI++HWL AG+PDPFY+PLNDYVILPTAFE+E++ EK ++ S++E+WEIIS D
Sbjct: 420 VFRIIQHWLHAGEPDPFYDPLNDYVILPTAFEIEKYHEKHGDITSVREDWEIISHRDDES 479
Query: 484 SNTGDNKMTLSSISVSQEGANKSHSEAHATVFVHTDNDGKQHIELNAVAVSVD 536
+ +++S S+EG + S EA AT+FVH ++ G+QH+E+ AV V+ D
Sbjct: 480 KRPAELPPMFNTLSASREGEDGSLEEAQATIFVHPESKGRQHVEVRAVGVTHD 532
>gb|AAQ05032.1| phospholipase A1 [Nicotiana tabacum]
Length = 452
Score = 451 bits (1159), Expect = e-125
Identities = 212/408 (51%), Positives = 293/408 (70%), Gaps = 5/408 (1%)
Query: 33 DPVLLVPGVGGSILNAVNESDGSQE-RVWVRFLSAEYKLKTKLWSRYDPSTGKTVTLDQK 91
DPVLLV G+ GSIL++ ++ G E RVWVR L AE + K KLWS Y+P TG T +LD+
Sbjct: 26 DPVLLVSGLAGSILHSKSKKLGGFETRVWVRLLLAELEFKNKLWSIYNPKTGYTESLDES 85
Query: 92 SRIVVPEDRHGLHAIDVLDPDLVIGS---EAVYYFHDMIVQMQKWGYQEGKTLFGFGYDF 148
+ IVVP+D +GL+AID+LDP +++ VY+FHDMI + K GY++G TLFGFGYDF
Sbjct: 86 TEIVVPQDDYGLYAIDILDPSMMVKCVHLTGVYHFHDMIDMLVKCGYKKGTTLFGFGYDF 145
Query: 149 RQSNRLQETMDRFAEKLELIYNAAGGKKIDLISHSMGGLLVKCFMTLHSDIFEKYVKNWI 208
RQSNR+ + M+ KLE Y A+GG+K+D+ISHSMGGLL+KCF++L+SD+F KYV WI
Sbjct: 146 RQSNRIDKAMNDLKAKLETAYKASGGRKVDIISHSMGGLLIKCFISLYSDVFSKYVNKWI 205
Query: 209 AICAPFQGAPGCTNSTFLNGMSFVEGWEQNFFISKWSMHQLLIECPSIYELMACPNFHWK 268
I PFQGAPGC N + L G+ FV+G+E NFF+S+W+MHQLL+ECPSIYE++ P+F W
Sbjct: 206 TIATPFQGAPGCINDSLLTGVQFVDGFESNFFVSRWTMHQLLVECPSIYEMLPNPDFEWA 265
Query: 269 HVPLLELWRERLHEDGKSHVILESYPPRDSIEIFKQALVNNKVNHEGEELPLPFNSHIFE 328
P + +WR++ ++G+ V LE Y S+ +F++AL +N++N G+ + LPFN I +
Sbjct: 266 KQPEILVWRKK-SKEGEPVVELERYGASTSVTLFQEALKSNELNLNGKTVALPFNLSILD 324
Query: 329 WANKTREILSSAKLPSGVKFYNIYGTNLATPHSICYGNADKPVSDLQELRYLQARYVCVD 388
WA TR+IL++A+LP G+ FY+IYGT+ TP +CYG+ P+ DL + + +Y VD
Sbjct: 325 WAASTRKILNTAQLPQGIPFYSIYGTSFDTPFDVCYGSKASPIEDLTNVCHTMPQYSYVD 384
Query: 389 GDGTVPVESAKADGFNAEERVGIPGEHRGILCEPHLFRILKHWLKAGD 436
GDGTVP ESAKAD F A ERVG+ G HR +L + +F+++K WL D
Sbjct: 385 GDGTVPAESAKADNFEAVERVGVQGGHRELLRDEKVFQLIKKWLGVTD 432
>gb|AAQ04051.1| phospholipase A1 [Arabidopsis thaliana] gi|21360561|gb|AAM47477.1|
AT3g03310/T21P5_27 [Arabidopsis thaliana]
gi|15912273|gb|AAL08270.1| AT3g03310/T21P5_27
[Arabidopsis thaliana] gi|15809942|gb|AAL06898.1|
AT3g03310/T21P5_27 [Arabidopsis thaliana]
gi|18396510|ref|NP_566201.1| lecithin:cholesterol
acyltransferase family protein / LACT family protein
[Arabidopsis thaliana]
Length = 447
Score = 434 bits (1117), Expect = e-120
Identities = 206/403 (51%), Positives = 270/403 (66%), Gaps = 3/403 (0%)
Query: 33 DPVLLVPGVGGSILNAVNESDGSQERVWVRFLSAEYKLKTKLWSRYDPSTGKTVTLDQKS 92
DPVLLV G+GGSIL++ ++ S+ RVWVR A K LWS Y+P TG T LD
Sbjct: 24 DPVLLVSGIGGSILHSKKKNSKSEIRVWVRIFLANLAFKQSLWSLYNPKTGYTEPLDDNI 83
Query: 93 RIVVPEDRHGLHAIDVLDPDLVIGS---EAVYYFHDMIVQMQKWGYQEGKTLFGFGYDFR 149
++VP+D HGL+AID+LDP + VY+FHDMI + GY++G TLFG+GYDFR
Sbjct: 84 EVLVPDDDHGLYAIDILDPSWFVKLCHLTEVYHFHDMIEMLVGCGYKKGTTLFGYGYDFR 143
Query: 150 QSNRLQETMDRFAEKLELIYNAAGGKKIDLISHSMGGLLVKCFMTLHSDIFEKYVKNWIA 209
QSNR+ + +KLE Y +GG+K+ +ISHSMGGL+V CFM LH + F KYV WI
Sbjct: 144 QSNRIDLLILGLKKKLETAYKRSGGRKVTIISHSMGGLMVSCFMYLHPEAFSKYVNKWIT 203
Query: 210 ICAPFQGAPGCTNSTFLNGMSFVEGWEQNFFISKWSMHQLLIECPSIYELMACPNFHWKH 269
I PFQGAPGC N + L G+ FVEG E FF+S+W+MHQLL+ECPSIYE+MA P+F WK
Sbjct: 204 IATPFQGAPGCINDSILTGVQFVEGLESFFFVSRWTMHQLLVECPSIYEMMANPDFKWKK 263
Query: 270 VPLLELWRERLHEDGKSHVILESYPPRDSIEIFKQALVNNKVNHEGEELPLPFNSHIFEW 329
P + +WR++ D + V LES+ +SI++F AL NN++++ G ++ LPFN I +W
Sbjct: 264 QPEIRVWRKKSENDVDTSVELESFGLIESIDLFNDALKNNELSYGGNKIALPFNFAILDW 323
Query: 330 ANKTREILSSAKLPSGVKFYNIYGTNLATPHSICYGNADKPVSDLQELRYLQARYVCVDG 389
A KTREIL+ A+LP GV FYNIYG +L TP +CYG P+ DL E+ Y VDG
Sbjct: 324 AAKTREILNKAQLPDGVSFYNIYGVSLNTPFDVCYGTETSPIDDLSEICQTMPEYTYVDG 383
Query: 390 DGTVPVESAKADGFNAEERVGIPGEHRGILCEPHLFRILKHWL 432
DGTVP ESA A F A VG+ G HRG+L + +F +++ WL
Sbjct: 384 DGTVPAESAAAAQFKAVASVGVSGSHRGLLRDERVFELIQQWL 426
>gb|AAF01599.1| unknown protein [Arabidopsis thaliana]
Length = 393
Score = 343 bits (879), Expect = 1e-92
Identities = 175/400 (43%), Positives = 233/400 (57%), Gaps = 51/400 (12%)
Query: 33 DPVLLVPGVGGSILNAVNESDGSQERVWVRFLSAEYKLKTKLWSRYDPSTGKTVTLDQKS 92
DPVLLV G+GGSIL++ ++ S+ RVWVR A K LWS Y+P TG T LD
Sbjct: 24 DPVLLVSGIGGSILHSKKKNSKSEIRVWVRIFLANLAFKQSLWSLYNPKTGYTEPLDDNI 83
Query: 93 RIVVPEDRHGLHAIDVLDPDLVIGSEAVYYFHDMIVQMQKWGYQEGKTLFGFGYDFRQSN 152
++VP+D HGL+AID+LDP + VY+FHDMI + GY++G TLFG+GYDFRQSN
Sbjct: 84 EVLVPDDDHGLYAIDILDPSWLCHLTEVYHFHDMIEMLVGCGYKKGTTLFGYGYDFRQSN 143
Query: 153 RLQETMDRFAEKLELIYNAAGGKKIDLISHSMGGLLVKCFMTLHSDIFEKYVKNWIAICA 212
R+ + +KLE Y +GG+K+ +ISHSMGGL+V CFM L
Sbjct: 144 RIDLLILGLKKKLETAYKRSGGRKVTIISHSMGGLMVSCFMYL----------------- 186
Query: 213 PFQGAPGCTNSTFLNGMSFVEGWEQNFFISKWSMHQLLIECPSIYELMACPNFHWKHVPL 272
H L+ECPSIYE+MA P+F WK P
Sbjct: 187 ----------------------------------HPELVECPSIYEMMANPDFKWKKQPE 212
Query: 273 LELWRERLHEDGKSHVILESYPPRDSIEIFKQALVNNKVNHEGEELPLPFNSHIFEWANK 332
+ +WR++ D + V LES+ +SI++F AL NN++++ G ++ LPFN I +WA K
Sbjct: 213 IRVWRKKSENDVDTSVELESFGLIESIDLFNDALKNNELSYGGNKIALPFNFAILDWAAK 272
Query: 333 TREILSSAKLPSGVKFYNIYGTNLATPHSICYGNADKPVSDLQELRYLQARYVCVDGDGT 392
TREIL+ A+LP GV FYNIYG +L TP +CYG P+ DL E+ Y VDGDGT
Sbjct: 273 TREILNKAQLPDGVSFYNIYGVSLNTPFDVCYGTETSPIDDLSEICQTMPEYTYVDGDGT 332
Query: 393 VPVESAKADGFNAEERVGIPGEHRGILCEPHLFRILKHWL 432
VP ESA A F A VG+ G HRG+L + +F +++ WL
Sbjct: 333 VPAESAAAAQFKAVASVGVSGSHRGLLRDERVFELIQQWL 372
>emb|CAB78988.1| putative protein [Arabidopsis thaliana] gi|3250695|emb|CAA19703.1|
putative protein [Arabidopsis thaliana]
gi|7452457|pir||T04767 hypothetical protein T16H5.220 -
Arabidopsis thaliana
Length = 493
Score = 335 bits (860), Expect = 2e-90
Identities = 165/286 (57%), Positives = 209/286 (72%), Gaps = 40/286 (13%)
Query: 1 MTMLLEDVLRSVELWLRLIKKPQPQAYVNPNLDPVLLVPGVGGSILNAVNESDGSQERVW 60
M++LLE+++RSVE L+L + Q + YV+PNL+PVLLVPG+ GSILNAV+ +G++ERVW
Sbjct: 1 MSLLLEEIIRSVEALLKLRNRNQ-EPYVDPNLNPVLLVPGIAGSILNAVDHENGNEERVW 59
Query: 61 VRFLSAEYKLKTKLWSRYDPSTGKTVTLDQKSRIVVPEDRHGLHAIDVLDPDLVIGSEAV 120
VR A+++ +TK+WSR+DPSTGKT++LD K+ IVVP+DR GLHAIDVLDPD+++G E+V
Sbjct: 60 VRIFGADHEFRTKMWSRFDPSTGKTISLDPKTSIVVPQDRAGLHAIDVLDPDMIVGRESV 119
Query: 121 YYFHDMIVQMQKWGYQEGKTLFGFGYDFRQSNRLQETMDRFAEKLELIYNAAGGKKIDLI 180
YYFH+MIV+M WG++EGKTLFGFGYDFRQSNRLQET+D+FA+KLE +Y A+G KKI++I
Sbjct: 120 YYFHEMIVEMIGWGFEEGKTLFGFGYDFRQSNRLQETLDQFAKKLETVYKASGEKKINVI 179
Query: 181 SHSMGGLLVKCFMTLHSDIF-------------------------------------EKY 203
SHSMGGLLVKCFM LHSD+ + Y
Sbjct: 180 SHSMGGLLVKCFMGLHSDVCKSLFLYSYSRSMYRIGLLLLLHFEVSSLTCGTSDSTGDNY 239
Query: 204 VKNWIAICAPFQGAPGCTNSTFLNGMSFVEGWEQNFFISKWSMHQL 249
+W I GAPG ST LNGMSFV GWEQNFF+SKWSMHQL
Sbjct: 240 HTDWFRIID--SGAPGYITSTLLNGMSFVNGWEQNFFVSKWSMHQL 283
Score = 214 bits (546), Expect = 4e-54
Identities = 107/177 (60%), Positives = 131/177 (73%), Gaps = 5/177 (2%)
Query: 362 ICYGNADKPVSDLQELRYLQARYVCVDGDGTVPVESAKADGFNAEERVGIPGEHRGILCE 421
I YGN PV DL LRY Q Y+CVDGDGTVP+ESA ADG A RVG+PGEHRGIL +
Sbjct: 321 IHYGNEKMPVKDLTNLRYFQPTYICVDGDGTVPMESAMADGLEAVARVGVPGEHRGILND 380
Query: 422 PHLFRILKHWLKAGDPDPFYNPLNDYVILPTAFEMERHKEKGLEVASLKEEWEIISKDQD 481
+FR+LK WL G+PDPFYNP+NDYVILPT +E E+ E GLEVAS+KE W+IIS D
Sbjct: 381 HRVFRMLKKWLNVGEPDPFYNPVNDYVILPTTYEFEKFHENGLEVASVKESWDIISDD-- 438
Query: 482 GQSNTGDNKMTLSSISVSQEGANKS-HSEAHATVFVHTDNDGKQHIELNAVAVSVDA 537
+N G T++SISVSQ G +++ +EA AT+ V +DG+QH+ELNAV+VSVDA
Sbjct: 439 --NNIGTTGSTVNSISVSQPGDDQNPQAEARATLTVQPQSDGRQHVELNAVSVSVDA 493
>dbj|BAD34417.1| lecithin cholesterol acyltransferase-like [Oryza sativa (japonica
cultivar-group)]
Length = 208
Score = 213 bits (541), Expect = 2e-53
Identities = 92/187 (49%), Positives = 134/187 (71%), Gaps = 2/187 (1%)
Query: 246 MHQLLIECPSIYELMACPNFHWKHVPLLELWRERLHEDGKSHVILESYPPRDSIEIFKQA 305
MHQLL+ECPSIYE++ P+F WK P++++WR++ +DG + ++L Y D + +F++A
Sbjct: 1 MHQLLVECPSIYEMLPNPHFKWKKAPVVQVWRKKPEKDGIAELVL--YEATDCLSLFQEA 58
Query: 306 LVNNKVNHEGEELPLPFNSHIFEWANKTREILSSAKLPSGVKFYNIYGTNLATPHSICYG 365
L NN++ + G+ + LPFN +F+WA +TR IL A+LP V FYNIYGT+ TP+ +CYG
Sbjct: 59 LRNNELKYNGKTIALPFNMSVFKWATETRRILEDAELPDTVSFYNIYGTSYDTPYDVCYG 118
Query: 366 NADKPVSDLQELRYLQARYVCVDGDGTVPVESAKADGFNAEERVGIPGEHRGILCEPHLF 425
+ P+ DL E+ + Y VDGDGTVP+ES ADGF A+ERVGI +HRG+LC+ ++F
Sbjct: 119 SESSPIGDLSEVCHTMPVYTYVDGDGTVPIESTMADGFAAKERVGIEADHRGLLCDENVF 178
Query: 426 RILKHWL 432
+LK WL
Sbjct: 179 ELLKKWL 185
>gb|EAA46455.1| GLP_90_25807_22631 [Giardia lamblia ATCC 50803]
Length = 1058
Score = 93.6 bits (231), Expect = 1e-17
Identities = 62/213 (29%), Positives = 106/213 (49%), Gaps = 21/213 (9%)
Query: 32 LDPVLLVPGVGGSILNAVNESDGSQERVW---------------VRFLSAEYKLKTKLWS 76
L P++L+PGVGGS L+AVN+ + ERVW V +L +T+L++
Sbjct: 5 LPPIILIPGVGGSKLDAVNKKNDKVERVWISKDVLPVPQLGKKFVHYLWGRPDPETQLYT 64
Query: 77 RYDPSTGKTVTLDQKSRIVVPEDRHGLHAIDVLDPDLVIGSEAVYYFHDMIVQ-MQKWGY 135
Y +T +D D ++ ++ L + +G YF +I + MQ +GY
Sbjct: 65 SYTEEYAETRIVDGLEGCWRLLDHWVINTVEQLFKNTTLGK----YFVTIIGRLMQDYGY 120
Query: 136 QEGKTLFGFGYDFRQSNRLQETMDRFAEKLELIYNAAGGKKIDLISHSMGGLLVKCFMTL 195
Q K LFGF YD+RQ + + + + G +++I+HS+GGL+ + + L
Sbjct: 121 QPNKNLFGFSYDWRQPLYAECIKGELHKLIIRVRELNNGMPVNIIAHSLGGLVGRTYCQL 180
Query: 196 HSDIFEKYVKNWIAICAPFQGAPGCTNSTFLNG 228
D + +++ +I I PF G+ T ++F+NG
Sbjct: 181 TPD-WMTHIRRFITIATPFDGSSSMTLNSFING 212
Score = 40.8 bits (94), Expect = 0.11
Identities = 33/99 (33%), Positives = 49/99 (49%), Gaps = 9/99 (9%)
Query: 336 ILSSAK--LPSGVKFYNIYGTNLATPHSICYGNADKPVSDLQELRYLQARYVCVDGDGTV 393
ILSS K +F +I G N+ TP Y KP+S EL+ ++ GDGTV
Sbjct: 929 ILSSMKPCTTDDFRFISINGGNVPTPIHTIY---PKPISTYDELQSQLPVFIMGRGDGTV 985
Query: 394 PVESAKADGFN---AEERVGIP-GEHRGILCEPHLFRIL 428
+ A +D F+ +RV IP H G+L + +F ++
Sbjct: 986 LLSGALSDNFDDALVHDRVVIPDATHGGLLHDEAVFYLI 1024
>gb|EAA39090.1| GLP_305_4486_7113 [Giardia lamblia ATCC 50803]
Length = 875
Score = 83.6 bits (205), Expect = 1e-14
Identities = 61/202 (30%), Positives = 93/202 (45%), Gaps = 14/202 (6%)
Query: 24 PQAYVNPNLDPVLLVPGVGGSILNAVNESDGSQERVWVRFLSAEYKLKTKLWSRYDPSTG 83
P NP P++LVPGV GS+L A N + A KL L+ D T
Sbjct: 36 PDNLFNP---PIILVPGVCGSLLVADNNEVAWLNETLTPYPQASAKLMQYLYGSRDSVTN 92
Query: 84 KTVTLDQKSRIVVPEDRHGL--------HAIDVLDPDLVIGSEAVYYFHDMIVQMQKWGY 135
K V+ ++ + GL H + L P + +YY + +K+GY
Sbjct: 93 KFVSFIERQGFSSVKAVPGLSGCSRLLNHKLTRL-PGIAQKKLGIYYETFAVYLAEKFGY 151
Query: 136 QEGKTLFGFGYDFRQSNRLQETMDRFAEKLELIYNAAGGKKIDLISHSMGGLLVKCFMTL 195
+EG LF F YD+RQ+ + F E L+ G + I ++ HSMGGLLV +M L
Sbjct: 152 KEGLNLFAFTYDWRQALHIASIQSAFDELLKAACQTTGQRCI-VVGHSMGGLLVTTYMRL 210
Query: 196 HSDIFEKYVKNWIAICAPFQGA 217
H D + Y+ ++++ P+ G+
Sbjct: 211 HPD-WNDYIAKFVSLGVPYAGS 231
Score = 43.9 bits (102), Expect = 0.013
Identities = 35/109 (32%), Positives = 58/109 (53%), Gaps = 11/109 (10%)
Query: 328 EWANKTREILSSAKL-PSGV---KFYNIYGTNLATPHSICYGNADKPVSDLQELRYLQAR 383
E+ N+++EILS + P +FY++ G+NL T Y D+ +S L EL Y + +
Sbjct: 753 EYWNQSKEILSREIVYPEDTELFRFYSLNGSNLQTAVHAYY---DEILSSLYELAYRKPK 809
Query: 384 YVCVDGDGTVPVESAKADGF---NAEERVGIPG-EHRGILCEPHLFRIL 428
++ GDGTVP+ S+ +D ++RV P H +L +F +L
Sbjct: 810 FIFTIGDGTVPLISSLSDPIPDRYVDDRVAFPEMGHFEMLQSKEVFELL 858
>gb|AAA79183.1| esterase
Length = 715
Score = 80.1 bits (196), Expect = 2e-13
Identities = 110/427 (25%), Positives = 177/427 (40%), Gaps = 72/427 (16%)
Query: 23 QPQAY---VNPNLDPVLLVPGVGGSILNAVNESDGSQERVWVRFLSAEYKLKTKLWSRY- 78
+P AY ++ PV+L+PG+GGS L E DG +W+ ++ R
Sbjct: 214 KPHAYKLSISNATIPVILIPGIGGSRLEV--EEDGKTSEIWLGLWDMGVGIRDPRHRRIL 271
Query: 79 --DPSTGKTVTLD--QKSRIVVPEDRHG-LHAIDVLDP---DLVIGSEAVYYFHDMIVQM 130
+P +V + Q V PE G AI+ L DL + + V + M +
Sbjct: 272 SLEPVKNGSVNVQPRQPGIKVFPERADGGFRAIEYLSYTKLDLDVIKKQVEQYASMAKHL 331
Query: 131 QKWGYQEGKTLFGFGYDFRQSNRLQETMDRFAEKLELIYNAAGGKKIDLISHSMGGLLVK 190
+K GY++ +TLF YD+R SN + +K++ +G ++ L++HSMGGLLV+
Sbjct: 332 EKMGYRKNRTLFAMPYDWRYSN--ADNAKFLKQKIDEALKESGASQVQLVAHSMGGLLVR 389
Query: 191 CFMTLHSDI-FEKYVKNWIAICAPFQGAPGCTNST---FLNGMSFVEGWEQNFFISKWSM 246
TL S++ ++ VK I + PF G+P + + G+ F+ E IS ++
Sbjct: 390 --ETLLSNVSYQPKVKRIIYMGTPFLGSPRAYQAIKYGYNFGIPFLHE-ETGKVISAYA- 445
Query: 247 HQLLIECPSIYELMACPNFHWKHVPLLELWRERLHEDGKSHVILESYPPRDSIEIFKQAL 306
P++YEL+ + + V L+ +D E + I + L
Sbjct: 446 -------PAVYELLPSRKY-FDTVAFLK-------KDVVHPYSYEEFLQDREIRLDYDPL 490
Query: 307 VNNKVN-HEGEELPLPFNSHIFEWANKTREILSSAKLPSGVKFYNIYGTNLATPHSICYG 365
V N HE +W NKT V Y+I G T G
Sbjct: 491 VKNAGQLHE-------------KWDNKT----------INVPQYSIVGHGQTT----LLG 523
Query: 366 NADKPVSDLQELRYLQARYVCVDGDGTVPVESAKADGFNAEERVGIPGEHRGILCEPHLF 425
PVS R L + GDGTVP SA + +++ + EH + P++
Sbjct: 524 YVMNPVS-----RQLVPYFDPGVGDGTVPYLSANYAQKDIKKQYYVKEEHAKLPVNPYVI 578
Query: 426 RILKHWL 432
+ + H L
Sbjct: 579 QQVGHLL 585
>ref|XP_654285.1| lecithin:cholesterol acyltransferase, putative [Entamoeba
histolytica HM-1:IMSS] gi|56471320|gb|EAL48899.1|
lecithin:cholesterol acyltransferase, putative
[Entamoeba histolytica HM-1:IMSS]
Length = 411
Score = 78.6 bits (192), Expect = 5e-13
Identities = 73/216 (33%), Positives = 106/216 (48%), Gaps = 37/216 (17%)
Query: 30 PNLDPVLLVPGVGGSILNA-VNESDGSQ-------------ERVWV-----RFLSAEYKL 70
P P++ +PG+ S+L VN +D S+ ER+WV R L + L
Sbjct: 21 PAKKPIVFIPGILASMLEGDVNIADISKTPLPEKCDTHVEYERLWVALKNVRPLKNDCSL 80
Query: 71 K--TKLWSRYDPSTGKTVTLDQKSRIVVPEDRHG-LHAIDVLDPDLVIGSEAVYYFHDMI 127
T +W+ ST K + IV P R G +A D +DP+ + A FHD+I
Sbjct: 81 GYLTPMWN----STSKEQIDIEGVNIVSP--RFGSTYACDEIDPNWPVSMFAKC-FHDLI 133
Query: 128 VQMQKWGYQEGKTLFGFGYDFR--QSNRLQETMDRFAEKLELI---YNAAGGKKIDLISH 182
+ +K GY +G + G YD+R + + + FA+ ELI YN G K+ +ISH
Sbjct: 134 KKFKKLGYVDGDNMVGASYDWRYYRYGEYKHKRNWFADTKELIINTYNKYG--KVVVISH 191
Query: 183 SMGGLLVKCFMTLHSDIF-EKYVKNWIAICAPFQGA 217
SMGGL+ F+ F +KY+ NWIAI PF G+
Sbjct: 192 SMGGLMFYKFLDYEGKEFADKYIDNWIAISTPFLGS 227
>ref|XP_656149.1| Lecithin:cholesterol acyltransferase, putative [Entamoeba
histolytica HM-1:IMSS] gi|56473329|gb|EAL50763.1|
Lecithin:cholesterol acyltransferase, putative
[Entamoeba histolytica HM-1:IMSS]
Length = 412
Score = 78.2 bits (191), Expect = 6e-13
Identities = 71/225 (31%), Positives = 109/225 (47%), Gaps = 30/225 (13%)
Query: 15 WLRLIKKPQPQAYVNPNLDPVLLVPGVGGSILNA-VNESDGSQE--------------RV 59
W ++++K + + + PV+L+PG+ SI+ A +N +D Q R
Sbjct: 12 WGKVVQKSELKKDTCDSRSPVVLIPGLMASIIEAKINVADDFQPWPKSGKCEKNKDWFRA 71
Query: 60 WVRFLSAEYKLKTKLWSRY-----DPSTGKTVTLDQKSRIVVPEDRHGLHAIDVLDPDLV 114
WV + K++ + Y + T K T+ + +PE +A D LDP +
Sbjct: 72 WVN-VDIAVPWKSECYINYLSGIWNNQTNKLETIPGID-LRIPEFG-STYACDQLDPVFL 128
Query: 115 IGSEAVYYFHDMIVQMQKWGYQEGKTLFGFGYDFRQSNRLQETMDRFAEKLELIYNAA-- 172
IGS FH +I ++ GY++ +FG YD+R + L +T F LIY
Sbjct: 129 IGS-FTNSFHKIIEHLKSVGYKDQIDMFGASYDWRTVD-LPKTY--FEGVKGLIYEGFKN 184
Query: 173 GGKKIDLISHSMGGLL-VKCFMTLHSDIFEKYVKNWIAICAPFQG 216
GKK+ +ISHSMGG + K F L D +KY++ WIAI APF G
Sbjct: 185 SGKKVVIISHSMGGFVSYKLFDYLGKDFCDKYIQKWIAISAPFIG 229
>ref|XP_654915.1| lecithin:cholesterol acyltransferase, putative [Entamoeba
histolytica HM-1:IMSS] gi|56472007|gb|EAL49529.1|
lecithin:cholesterol acyltransferase, putative
[Entamoeba histolytica HM-1:IMSS]
Length = 411
Score = 77.4 bits (189), Expect = 1e-12
Identities = 72/216 (33%), Positives = 107/216 (49%), Gaps = 37/216 (17%)
Query: 30 PNLDPVLLVPGVGGSILNA-VNESDGSQ-------------ERVWV-----RFLSAEYKL 70
P P++ +PG+ S+L VN +D S+ ER+WV R L + L
Sbjct: 21 PAKKPIVFIPGILASMLEGDVNIADISKTPLPEKCDTHVEYERLWVALKNVRPLKNDCSL 80
Query: 71 K--TKLWSRYDPSTGKTVTLDQKSRIVVPEDRHG-LHAIDVLDPDLVIGSEAVYYFHDMI 127
T +W+ ST K + IV P R G +A D +DP+ + A FHD+I
Sbjct: 81 GYLTPMWN----STSKEQIDIEGVNIVSP--RFGSTYACDEIDPNWPVSIFAKC-FHDLI 133
Query: 128 VQMQKWGYQEGKTLFGFGYDFR--QSNRLQETMDRFAEKLELI---YNAAGGKKIDLISH 182
+ +K GY +G + G YD+R + + + FA+ ELI YN G K+ +ISH
Sbjct: 134 KKFKKLGYVDGDNMVGASYDWRYYRYGEYKHKRNWFADTKELIINTYNKYG--KVVVISH 191
Query: 183 SMGGLLVKCFMT-LHSDIFEKYVKNWIAICAPFQGA 217
SMGGL+ F+ + + +KY+ NWIAI PF G+
Sbjct: 192 SMGGLMFYKFLDYVGKEFADKYIDNWIAISTPFLGS 227
>ref|XP_649190.1| 1-O-acylceramide synthase, putative [Entamoeba histolytica
HM-1:IMSS] gi|67463382|ref|XP_648348.1| 1-O-acylceramide
synthase, putative [Entamoeba histolytica HM-1:IMSS]
gi|56465566|gb|EAL43804.1| 1-O-acylceramide synthase,
putative [Entamoeba histolytica HM-1:IMSS]
gi|56464478|gb|EAL42962.1| 1-O-acylceramide synthase,
putative [Entamoeba histolytica HM-1:IMSS]
Length = 399
Score = 76.3 bits (186), Expect = 2e-12
Identities = 89/360 (24%), Positives = 161/360 (44%), Gaps = 73/360 (20%)
Query: 34 PVLLVPGVGGSILNA-VN------------ESDGSQE--RVWVRFLSAEYKLK----TKL 74
PV+++PG+ S+LNA +N + D S++ +W+ L + L
Sbjct: 22 PVIIIPGIMASMLNAKINIPKNVDFCDRKLDCDRSKDWFTLWLNLLDGIPYVNDCYIAYL 81
Query: 75 WSRYDPSTGKTVTLDQKSRIVVPEDRHG-LHAIDVLDPDLVIGSEAVYYFHDMIVQMQKW 133
Y+ ++G+ ++ + + R G +A+D + P + + + FH++I +QK
Sbjct: 82 TCHYNSTSGR---MENVEGVQIEPPRFGSTYAVDTISPSFPLKTFS-RAFHEVIKGLQKI 137
Query: 134 GYQEGKTLFGFGYDFRQSNRLQETMDRFAEKL-ELIYNA--AGGKKIDLISHSMGGLLVK 190
GY++ LF YD+R + D + EK+ ELI A G K+ L+SHSMGGL
Sbjct: 138 GYKDEFDLFSAPYDWRYYHH-----DEYYEKVKELIIKAYENTGNKVVLVSHSMGGLTTY 192
Query: 191 CFM-TLHSDIFEKYVKNWIAICAPFQGAPGCTNSTFLNGMSFVEGWEQNFFISKWSMHQL 249
+ L + +KY+ W+A+ PF G +T N + + G+ + +SK +L
Sbjct: 193 ILLDKLGKEFCDKYIHRWVAMSTPFIG------TTIANDV-VLAGYNMGYPVSK----EL 241
Query: 250 LIECPSIYELMACPNFHWKHVPLLELWRERLHEDGKSHVILE-----SYPPRDSIEIFKQ 304
+ + +E +A P+ E W ++ V++E Y P+D IE+F Q
Sbjct: 242 IKKAARTFETVAMMG------PIGEYW-------DQNEVLVELANGKKYYPKDQIELFSQ 288
Query: 305 ALVNNKVNHEGEELPLPFNSHIFEWANKTREILSSAKLPSGVKFYNIYGTNLATPHSICY 364
EE+ PF E + ++++P GV+ + + + T H I +
Sbjct: 289 L----------EEMK-PFAKEAVENSFAPYLKKYNSQVPHGVEMHCGITSGIETAHKITF 337
>ref|XP_655369.1| 1-O-acylceramide synthase, putative [Entamoeba histolytica
HM-1:IMSS] gi|56472500|gb|EAL49982.1| 1-O-acylceramide
synthase, putative [Entamoeba histolytica HM-1:IMSS]
Length = 395
Score = 74.3 bits (181), Expect = 9e-12
Identities = 73/305 (23%), Positives = 132/305 (42%), Gaps = 60/305 (19%)
Query: 123 FHDMIVQMQKWGYQEGKTLFGFGYDFRQSNRLQETMDRFAEKLELIYNAA---GGKKIDL 179
++ MI Q+++ GY++ K+LFG GYD+R ++ + +++K++ + +A KK+ +
Sbjct: 126 WYKMIQQLKRIGYKDKKSLFGLGYDWRYAD---VNYNNWSKKVKEVIESAYILNNKKVMI 182
Query: 180 ISHSMGG-LLVKCFMTLHSDIFEKYVKNWIAICAPFQGAPGCTNSTFLNGMSFVEGWEQN 238
++HS+GG + ++ L EKY++ I I APF G T SF+ G +
Sbjct: 183 VTHSLGGPMALQLLFQLGDSFCEKYIEKIITISAPFIG-------TIKALRSFLSGETEG 235
Query: 239 FFISKWSMHQLLIECPSIYELMACPNFHWKHVPLLELWRERLHEDGKSHVILESYPPRDS 298
++ S S+Y+LM PN+ W + +L ++ + + + IL
Sbjct: 236 IPVNPLSFRNFERNIDSVYQLM--PNYQWWNDTIL-IFNGTSYSASQMNQILN------- 285
Query: 299 IEIFKQALVNNKVNHEGEELPLPFNSHIFEWANKTREILSSAKLPSGVKFYNIYGTNLAT 358
L+N ++ N + W K VK Y +Y + + T
Sbjct: 286 -------LINETKDYASFIYTNAMNRYPINWTPK-------------VKLYCLYSSGIET 325
Query: 359 PHSICYGNA--DKPVSDLQELRYLQARYVCVDGDGTVPVESAK-ADGFNAEERVGI-PGE 414
+ Y + ++P+ DGDGTVP+ S N EE + I +
Sbjct: 326 EVLLNYSTSFDNQPIQTFG------------DGDGTVPLNSLSFCKTMNLEESINIGKYD 373
Query: 415 HRGIL 419
H GI+
Sbjct: 374 HFGII 378
>gb|EAA37820.1| GLP_661_6782_9100 [Giardia lamblia ATCC 50803]
Length = 772
Score = 72.4 bits (176), Expect = 3e-11
Identities = 53/214 (24%), Positives = 97/214 (44%), Gaps = 25/214 (11%)
Query: 34 PVLLVPGVGGSILNAVNESDGSQERVWVRFLSAEYKLKTKLWSRYDPSTGKTVTLDQKSR 93
P++L+PG+ + L+ V+ + G +ER WV SA Y K+++ + T D + +
Sbjct: 15 PIILIPGLCSTKLDIVHRTTGMRERAWV---SAHYIPKSRMGEKMINDVWGKPTSDGRYQ 71
Query: 94 IVVPE--DRH---------------GLHAIDVLDPDLVIGSEAVYYFHDMIVQMQKWGYQ 136
+ + D H G+ I L+P ++G YF + ++++ GYQ
Sbjct: 72 SFIEDVGDMHILEGFKGCSHLAQHWGISVIHALNPKFMLGR----YFTTLKNRLKRHGYQ 127
Query: 137 EGKTLFGFGYDFRQSNRLQETMDRFAEKLELIYNAAGGKKIDLISHSMGGLLVKCFMTLH 196
LF YD+RQ + + + N + LI HS G LLV+ +M L+
Sbjct: 128 VDVDLFCHSYDWRQPLSSDAVLGSLRRLILSVLNRTSSLHVILIGHSHGALLVRLYMQLY 187
Query: 197 SDIFEKYVKNWIAICAPFQGAPGCTNSTFLNGMS 230
D + +++ +IAI P+ + + +NG +
Sbjct: 188 DD-WHQHIFRFIAIGPPYDNSSAYMAMSLINGFA 220
Score = 37.7 bits (86), Expect = 0.93
Identities = 23/72 (31%), Positives = 37/72 (50%), Gaps = 3/72 (4%)
Query: 332 KTREILSSAKLPSGVKFYNIYGTNLATPHSICYGNADKPVSDLQELRYLQARYVCVDGDG 391
+T+ I+ + +++ I G+ TP + Y ++PVS EL + DGDG
Sbjct: 670 RTKPIIFPQPNETNFRYFAICGSGCKTPLHVVY---NQPVSSYHELCTQIPTTIDSDGDG 726
Query: 392 TVPVESAKADGF 403
TV + SA +DGF
Sbjct: 727 TVLLHSALSDGF 738
>ref|XP_652402.1| Lecithin:cholesterol acyltransferase, putative [Entamoeba
histolytica HM-1:IMSS] gi|56469251|gb|EAL47014.1|
Lecithin:cholesterol acyltransferase, putative
[Entamoeba histolytica HM-1:IMSS]
Length = 412
Score = 70.5 bits (171), Expect = 1e-10
Identities = 58/210 (27%), Positives = 98/210 (46%), Gaps = 25/210 (11%)
Query: 30 PNLDPVLLVPGVGGSILNA-VNESDGSQ-------------ERVWVRFLSAEYKLKTKLW 75
P P++ +PG+ SIL A V+ +D SQ +R+W+ +
Sbjct: 22 PAKKPIVFIPGILASILEAEVDIADISQTPLPSDCDTHLDYQRLWIALKDLNPFNNDCIL 81
Query: 76 SRYDPSTGKTVT--LDQKSRIVVPEDRHGLHAIDVLDPDLVIGSEAVYYFHDMIVQMQKW 133
P+ +D + ++ +A D +DP+ + A FHD+I + +K
Sbjct: 82 GYLTPTWNSETKEQIDIEGVNILSPKFGSTYACDEIDPNFPLSIFAKC-FHDLIKKFKKL 140
Query: 134 GYQEGKTLFGFGYDFR--QSNRLQETMDRFAEKLELI---YNAAGGKKIDLISHSMGGLL 188
GY +G + G YD+R + + + F + ELI YN G K+ +ISHSMGGL+
Sbjct: 141 GYVDGDNMVGASYDWRYYRYGEYKHKRNWFEDTKELIINTYNKYG--KVVVISHSMGGLM 198
Query: 189 VKCFMT-LHSDIFEKYVKNWIAICAPFQGA 217
F+ + + +KY+ NW+A+ PF G+
Sbjct: 199 FYKFLDYVGKEFADKYIDNWVAMSTPFLGS 228
>gb|EAA39441.1| GLP_762_23988_17611 [Giardia lamblia ATCC 50803]
Length = 2125
Score = 68.9 bits (167), Expect = 4e-10
Identities = 54/204 (26%), Positives = 94/204 (45%), Gaps = 10/204 (4%)
Query: 34 PVLLVPGVGGSILNAVNESDGSQERVWVRF-----LSAEYKLKTKLWSRYDPSTGKTVTL 88
PVLL+ G GS + A + E WV + K+ LW D T +
Sbjct: 7 PVLLIHGTVGSKMRAKSRISAHAEDAWVNSQIIPRMMIATKVANDLWCAPDLETLWVESH 66
Query: 89 DQKSRIVVP-EDRHGLHAIDVLDP-DLVIGSEAVYYFHDMIVQMQK--WGYQEGKTLFGF 144
K V P G + L + ++ + Y+++ ++Q K GY+EG T+ F
Sbjct: 67 VAKYVDVTPYPGLEGARRLLTLKGFERMLRKRRIGYYYETLLQWFKKYCGYEEGVTIDAF 126
Query: 145 GYDFRQSNRLQETMDRFAEKLELIYNAAGGKKIDLISHSMGGLLVKCFMTLHSDIFEKYV 204
YD+RQ + + + ++ + GG+++ +I+HS+GGL+V+ +M + D + +
Sbjct: 127 SYDWRQEIGHPKLQEDLRKCIKAMRCRNGGQRLTVIAHSLGGLVVQAYMQTYPD-WNDDI 185
Query: 205 KNWIAICAPFQGAPGCTNSTFLNG 228
+ AI PF G G + + FL G
Sbjct: 186 SRFAAISVPFDGVGGYSMAGFLTG 209
>ref|XP_655259.1| Lecithin:cholesterol acyltransferase, putative [Entamoeba
histolytica HM-1:IMSS] gi|56472381|gb|EAL49871.1|
Lecithin:cholesterol acyltransferase, putative
[Entamoeba histolytica HM-1:IMSS]
Length = 394
Score = 63.9 bits (154), Expect = 1e-08
Identities = 91/385 (23%), Positives = 157/385 (40%), Gaps = 61/385 (15%)
Query: 34 PVLLVPGVGGSILNAVNESDGSQE-------------RVWVRFLSA-EYKLKTKLW---S 76
P++LVPG+ +IL + + D + + R WV A + LW
Sbjct: 19 PIILVPGLMSTILESKIDVDDNYQPFPQKCSRHKGWFRSWVTVKDAISFTDDCYLWYLHG 78
Query: 77 RYDPSTGKTVTLDQKSRIVVPEDRHGLHAIDVLDPDLVIGSEAVYYFHDMIVQMQKWGYQ 136
++P T K + S I +P+ + +AID L P + I + FH +I ++ GY
Sbjct: 79 VWNPITNKLENIPGIS-IRIPQFGN-TYAIDTLCP-IPIVKRLTHAFHGLIQHLKNQGYV 135
Query: 137 EGKTLFGFGYDFRQSNRLQETMDRFAEKLELIYNAAGGKKIDLISHSMGGLLV-KCFMTL 195
E LFG GYD+R ++ E + + + Y +K+ +ISHSMG + K L
Sbjct: 136 ELFDLFGAGYDWRSNDVSDEYLKSVKDFIVSGYENT-KRKVVIISHSMGAFITYKLLDYL 194
Query: 196 HSDIFEKYVKNWIAICAPFQGAPGCTNSTFLNGMSFVEGWEQNFFISKWSMHQLLIECPS 255
+ + Y+ WI + APF G+ G L G + I++ L S
Sbjct: 195 GKEFCDTYIDKWIPLSAPFLGS-GLAIKELLVGENI------GLPINEKLARDLGRSIQS 247
Query: 256 IYELMACPNFHWKHVPLLELWRERLHEDGKSHVILESYPPRDSIEIFKQALVNNKVNHEG 315
I L P++ W + PL+ + K ++++Y D ++ + ++ N +
Sbjct: 248 IISLSPNPDY-WSNEPLIVFKKSGKQFFAKD--LVDAYNMFDEMKDKAEYILTNSIRAYY 304
Query: 316 EELPLPFNSHIFEWANKTREILSSAKLPSGVKFYNIYGTNLATPHSICYGNADKPVSDLQ 375
E+ + W +P GV+ + Y TP+ I + D
Sbjct: 305 EK---------YNWT-----------IPFGVETHCGYSLGYETPYRIEF------EGDSF 338
Query: 376 ELRYLQARYVCVDGDGTVPVESAKA 400
+ +Y + + +GD V ES KA
Sbjct: 339 DSKY---KVIFSNGDKLVNEESLKA 360
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.318 0.137 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,024,267,514
Number of Sequences: 2540612
Number of extensions: 46679648
Number of successful extensions: 98773
Number of sequences better than 10.0: 176
Number of HSP's better than 10.0 without gapping: 32
Number of HSP's successfully gapped in prelim test: 144
Number of HSP's that attempted gapping in prelim test: 98498
Number of HSP's gapped (non-prelim): 300
length of query: 538
length of database: 863,360,394
effective HSP length: 133
effective length of query: 405
effective length of database: 525,458,998
effective search space: 212810894190
effective search space used: 212810894190
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 78 (34.7 bits)
Medicago: description of AC149576.2


