

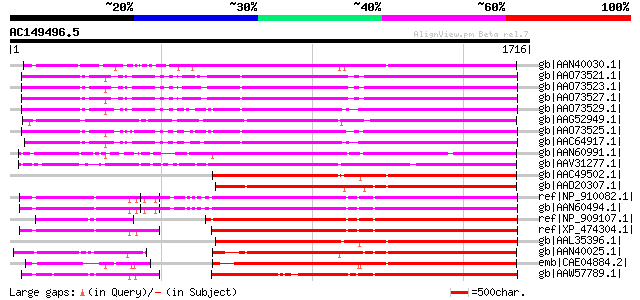
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149496.5 + phase: 0
(1716 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAN40030.1| putative gag-pol polyprotein [Zea mays] 1104 0.0
gb|AAO73521.1| gag-pol polyprotein [Glycine max] 1083 0.0
gb|AAO73523.1| gag-pol polyprotein [Glycine max] 1082 0.0
gb|AAO73527.1| gag-pol polyprotein [Glycine max] 1081 0.0
gb|AAO73529.1| gag-pol polyprotein [Glycine max] 1065 0.0
gb|AAG52949.1| gag/pol polyprotein [Arabidopsis thaliana] 1057 0.0
gb|AAO73525.1| gag-pol polyprotein [Glycine max] 1050 0.0
gb|AAC64917.1| gag-pol polyprotein [Glycine max] 1048 0.0
gb|AAN60991.1| Putative Zea mays retrotransposon Opie-2 [Oryza s... 1041 0.0
gb|AAV31277.1| putative polyprotein [Oryza sativa (japonica cult... 1025 0.0
gb|AAC49502.1| Pol [Zea mays] gi|7489803|pir||T04112 pol protein... 1000 0.0
gb|AAD20307.1| copia-type pol polyprotein [Zea mays] 999 0.0
ref|NP_910082.1| putative gag-pol polyprotein [Oryza sativa (jap... 984 0.0
gb|AAN60494.1| Putative Zea mays retrotransposon Opie-2 [Oryza s... 980 0.0
ref|NP_909107.1| unnamed protein product [Oryza sativa (japonica... 978 0.0
ref|XP_474304.1| OSJNBb0004A17.2 [Oryza sativa (japonica cultiva... 978 0.0
gb|AAL35396.1| Opie2a pol [Zea mays] 967 0.0
gb|AAN40025.1| putative gag-pol polyprotein [Zea mays] 964 0.0
emb|CAE04884.2| OSJNBa0042I15.6 [Oryza sativa (japonica cultivar... 918 0.0
gb|AAW57789.1| putative polyprotein [Oryza sativa (japonica cult... 905 0.0
>gb|AAN40030.1| putative gag-pol polyprotein [Zea mays]
Length = 2319
Score = 1104 bits (2855), Expect = 0.0
Identities = 665/1712 (38%), Positives = 951/1712 (54%), Gaps = 149/1712 (8%)
Query: 45 WKTNMYSFIMGLDEELWDILEDGVDDLDLDEEGAAIDRRIHTPAQKKLYKKHHKIRGIIV 104
W M S ++G+ LW+I+ G+ EE TP + ++ + II
Sbjct: 138 WADKMKSHLIGVHPSLWEIVNVGMYKPAQGEE--------MTPEMMQEVHRNAQAVSIIK 189
Query: 105 ASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYELFRMKDDESIEEM 164
S+ EY K+ + A+ ++ L + EG K K + L + + ES++ +
Sbjct: 190 GSLCPEEYRKVQGREDARDIWNILRMSHEGDPKAKRHRVEALESELARYDWTKGESLQSL 249
Query: 165 YSRFQTLVSGLQILKKSYVASDHVSKI-LRSLPSRWRPKVTAIEEAKDLNTLSVEDLVSS 223
+ R LV+ +++L + V+++ +R+ + + I + D ++ L +
Sbjct: 250 FDRLMVLVNKIRVLGSEDWSDSKVTRLFMRAYKEKDKSLARMIRDRDDYEDMTPHQLFAK 309
Query: 224 LKVHEMSLNEHESSKKSKSIALPSKGKTS-KSSKAYKASESEEESPDRDSDEDQSVKMAM 282
++ HE +E K S AL + + + K SK +KA + E S D DS D+ AM
Sbjct: 310 IQQHE---SEEAPIKTRDSHALITNEQDNLKKSKDHKAKKVVETSSDEDSSSDEDT--AM 364
Query: 283 LSNKLEYLARKQKKFLSKRGGYKNSKKEDQKGCFNCKKPGHFIADCPDLQKEKSKSKSKK 342
+ RK KF +K ++ C+ C + GHFIADCP+ +++++K + KK
Sbjct: 365 FIKTFKKFVRKNDKF---------QRKGKKRACYECGQTGHFIADCPNKKEQEAKKEYKK 415
Query: 343 SSFSSS-------KFRKQIKKSLMATWEDLDSESGSDKEEAEDDTKAAMGLVATVSSEAV 395
F K +K + + W D S S++EE +VA V+ ++
Sbjct: 416 DKFKKGGKTKGYFKKKKYGQAHIGEEWNSDDESSSSEEEE----------VVANVAIQST 465
Query: 396 SEAESDSEDENEVYSKIPRQELVDSLKELLSLFEHRTNELTDLKEKYVDLMKQQKSTLLE 455
S A+ + +++ Y IP + K ++LF + D D K+ +++
Sbjct: 466 SSAQLFTNLQDDSY--IPTCLMAKGDK--VTLFSN------DFSNDDDDDQIAMKNKMIK 515
Query: 456 LKASEEELKGFNLISTTYEDRLKILCQKLQEKCDKGSGNKHEIALDDFIMAGIDRS-KVA 514
E L G+N+I+ KL EK DK A +D ++ +R+ ++
Sbjct: 516 ----EFGLNGYNVIT------------KLMEKLDKRKATLD--AQEDLLILEKERNLELQ 557
Query: 515 SMIYSTYKNNGKGIGYSEEKSKEYSLKSYCDCIKDGLKSTFVP-----------EGTNAV 563
+I++ K I K+ +L + + ++ + S V + NA
Sbjct: 558 ELIHN------KDIEVINLKTSIDNLANEKNALESSMLSLNVQNQELQVQLENCKNINAP 611
Query: 564 TAV-QSKPETSGSQAKITSKPENLKIKVMTKSDPKSQKIKI------------LKRSEPV 610
T V +SK ++ + K +K + + + S ++K+ LK+ EP
Sbjct: 612 TLVFESKSSSNDNSCKHCAK-YHASCCLTNHARKHSPQVKVEEILKRCSSNDGLKKVEPK 670
Query: 611 HQNLIRPESK--IPKQKDQKNKAATASDKTIPKGVKPEVLNDQ--KPLSIHPKVCIRARE 666
+++L + + ++N + PK ++ L D + S + K
Sbjct: 671 YKSLKPNNGRRGLGFNSSKENPSTVHKGWRSPKFIEGTTLYDALGRIHSSNDKSSQVYSS 730
Query: 667 KQRSWYLDSGCSRHMTGEKALFLTLTM-KDGGEVKFGGNQTGKIIGTGTIGNSSI-SINN 724
SW LDSGC+ HMTGEK +F TL + ++ E+ FG + K+IG G I S S++N
Sbjct: 731 GGSSWVLDSGCTNHMTGEKDMFHTLQLTQEAQEIVFGDSGKSKVIGIGKIPISDQQSLSN 790
Query: 725 VWLVDGLKHNLLSISQFCDNGYDVMFSKTNCTLVNKNDKSITFKGKRVENVYKINFSDLA 784
V LVD L +NLLS+SQ C GY+ +FS + ++ + D S+ F G+ +Y ++F+
Sbjct: 791 VLLVDSLSYNLLSVSQLCGMGYNCLFSDVDVKILRREDSSVAFTGRLKGKLYLVDFTTSK 850
Query: 785 DQKVVCLLSMNDKKWVWHKRLGHANWRLISKISKLQLVKGLPNIDYHSDALCGACQKGKI 844
CL++ +DK W+WH+RL H R ++K+ K + GL N+ + D +CGACQ GK
Sbjct: 851 VTPETCLVAKSDKGWLWHRRLAHVGMRNLAKLQKDNHIIGLTNVVFEKDRVCGACQAGKQ 910
Query: 845 VKSTFKSKDIVSTSRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKFIKSKDY 904
+SK++V+T RPLELLH+DLFGPV S+ GSKYGLVIVDD+SR+TWV F+ K
Sbjct: 911 HGVPHQSKNVVTTKRPLELLHMDLFGPVAYISIGGSKYGLVIVDDFSRFTWVFFLSDKGE 970
Query: 905 ACEVFSSFCTQIQSEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNGV 964
E+ F + Q+E ELKI KVRSD+G EF+N E F + GI HEFS P TPQQNGV
Sbjct: 971 TQEILKKFMRRAQNEFELKIKKVRSDNGTEFKNTGVEEFLGEEGIKHEFSVPYTPQQNGV 1030
Query: 965 VERKNRTLQEMARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPN 1024
VERKNRTL E ARTM+ E +FWAEAVNT+C+ NR+Y+ + +KTAYEL G +P
Sbjct: 1031 VERKNRTLIEAARTMLDEYKTPDNFWAEAVNTACHAINRLYLHKIYKKTAYELLTGNKPK 1090
Query: 1025 ISYFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETHCVEESIHVKF 1084
+ YF FGC C+ILN K KF + G LGY+ + YRV+N+ T VE +I V F
Sbjct: 1091 VDYFRVFGCKCFILNKKVKSSKFAPRVDEGFLLGYASNAHGYRVFNNSTGLVEIAIDVTF 1150
Query: 1085 DDREP-------------------------GNKTPEQSESNAGT-------------TDS 1106
D+ G P++ + GT S
Sbjct: 1151 DESNGSQGHVSNDTVGNEKLPCESIKKLAIGEVRPQERDDEEGTLWMTNEVVDVGARVVS 1210
Query: 1107 EDASESDQPSDSEKHSEVESRPEAEITPEAESNSEAETSPVEQNENASEDVQDNTQQVIQ 1166
++ S PS S + E+ E E + P++Q N E +Q + V
Sbjct: 1211 DNVSTQANPSTSSHPNHEENHQRMPTVVEDEQENIDGEVPLDQVTNEEEQIQRHPS-VPH 1269
Query: 1167 PKFKH--KSSHPEELIIGSKDSPRRTRSHFRQEESLIGLLSIIEPKTVEEALSDDGWILA 1224
P+ H + HP + I+GS TRS +S +EP VEEAL D WI A
Sbjct: 1270 PRVHHTIQRDHPVDNILGSIRRGVTTRSRLANFCEFYSFVSSLEPLKVEEALDDPDWITA 1329
Query: 1225 MQEELNQFQRNDVWDLIPKPFQKNIIGTKWVFRNKLNEKGEVTRNKARLVAQGYSQQEGI 1284
MQEELN F RN+VW L+ +P Q N+IGTKWVFRNK +E G VT+NKARLVAQGY+Q EG+
Sbjct: 1330 MQEELNNFTRNEVWSLVQRPKQ-NVIGTKWVFRNKQDENGVVTKNKARLVAQGYTQVEGL 1388
Query: 1285 DYTETFAPVARLEAIRLLLSYAINHGIILYQMDVKSAFLNGVIEEEVYVKQPPGFEDIKH 1344
D+ ET+APVARLE+IR+L++YA NH LYQMDVKSAFLNG ++E VYV+QPPGFED K
Sbjct: 1389 DFGETYAPVARLESIRILIAYATNHDFKLYQMDVKSAFLNGPLQERVYVEQPPGFEDPKK 1448
Query: 1345 PDHVYKLKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQVDTTLFGKTLKKDILIVQIYVD 1404
P+HVY L K+LYGLKQAPRAWYD L +FLIKN F G+ D+TLF + + ++ + QIYVD
Sbjct: 1449 PNHVYLLHKALYGLKQAPRAWYDCLKDFLIKNGFTIGKADSTLFTRKVDNELFVCQIYVD 1508
Query: 1405 DIIFGSTNASLCKEFSKLMQDEFEMSMMGELKFFLGIQINQSKEGVYVHQTKYTKELLKK 1464
DIIFGSTN C+EFSK+M + FEMSMMGELK+FLG Q+ Q KEG ++ QTKYT+++LKK
Sbjct: 1509 DIIFGSTNEKFCEEFSKVMTNRFEMSMMGELKYFLGFQVKQLKEGTFLCQTKYTQDMLKK 1568
Query: 1465 FKLEDCKVMNTPMHPTCTLSKEDTGTVVDQKLYRGMIGSLLYLTASRPDILFSVCLCARF 1524
F +E K TPM L + G VDQKLYR MIGSLLYL ASRPDI+ SVC+CARF
Sbjct: 1569 FGMEKAKHAKTPMPSNGHLDLNEEGKPVDQKLYRSMIGSLLYLCASRPDIMLSVCMCARF 1628
Query: 1525 QSDPRESHLTAVKRIFRYLKGTTNLGLLYRKSLDYKLIGFCDADYAGDRIERKSTSGNCQ 1584
Q++P++ HL AVKRI RYL T NLGL Y K + L+G+ D+DYAG +++RKST+G CQ
Sbjct: 1629 QANPKDCHLVAVKRILRYLVHTQNLGLWYPKGSFFDLLGYSDSDYAGCKVDRKSTTGTCQ 1688
Query: 1585 FLGENLISWASKRQATIAMSTAEAEYISAASCCTQLLWMKHQLEDYQINANSIPIYCDNT 1644
FLG +L+SW+SK+Q +A+STAEAEYI+A +CC QLLWMK L D+ N IP+ CDN
Sbjct: 1689 FLGRSLVSWSSKKQNCVALSTAEAEYIAAGACCAQLLWMKQTLRDFGCEFNKIPLLCDNE 1748
Query: 1645 AAICLSKNPILHSRAKHIEIKHHFIRDYVQKG 1676
+AI L+ NP+ HSR KHI+I+ HF+RD+ KG
Sbjct: 1749 SAIKLANNPVQHSRTKHIDIRRHFLRDHEAKG 1780
>gb|AAO73521.1| gag-pol polyprotein [Glycine max]
Length = 1574
Score = 1083 bits (2802), Expect = 0.0
Identities = 633/1663 (38%), Positives = 918/1663 (55%), Gaps = 166/1663 (9%)
Query: 38 DPEEFSWWKTNMYSFIMGLDEELWDILEDGVDDLD-LDEEGAAIDR----RIHTPAQKKL 92
D + +WK M +F+ LD W + G + LD EG D T + +L
Sbjct: 17 DGSNYEYWKARMVAFLKSLDSRTWKAVIKGWEHPKMLDTEGKPTDELKPEEDWTKEEDEL 76
Query: 93 YKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYEL 152
+ K + + + + ++ + AK + L EG+ KVK ++ +L ++E
Sbjct: 77 ALGNSKALNALFNGVDKNIFRLINTCTVAKDAWEILKITHEGTSKVKISRLQLLATKFEN 136
Query: 153 FRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAKDL 212
+MK++E I + + + + L + V KILRSLP R+ KVTAIEEA+D+
Sbjct: 137 LKMKEEECIHDFHMNILEIANACTALGERITDEKLVRKILRSLPKRFDMKVTAIEEAQDI 196
Query: 213 NTLSVEDLVSSLKVHEMSLNEHESSKKSKSIALPSKGKTSKSSKAYKASESEEESPDRDS 272
+ V++L+ SL+ E+ L++ + KKSK++A S E EE+ D ++
Sbjct: 197 CNMRVDELIGSLQTFELGLSDR-AEKKSKNLAFVSN------------DEGEEDEYDLNT 243
Query: 273 DEDQSVKMAMLSNK----LEYLARKQKKFLSK-----RGGYKNSKKEDQKG-------CF 316
DE + + +L + L + ++QK + R G K KK D K C
Sbjct: 244 DEGLTNAVVLLGKQFNKVLNRMDKRQKPHVQNIPFDIRKGSKYQKKSDVKPSHSKGIQCH 303
Query: 317 NCKKPGHFIADCPDLQKEKSKSKSKKSSFSSSKFRKQIKKSLMATWEDLDSESGSDKEEA 376
C+ GH IA+CP K+ +K L D +SE SD +
Sbjct: 304 GCEGYGHIIAECPTHLKKH-------------------RKGLSVCQSDTESEQESDSDR- 343
Query: 377 EDDTKAAMGLVATVSSEAVSEAESDSEDENEVYSKIPRQELVDSLKELLSLFEHRTNELT 436
D A G+ T +ED ++ S+I EL S ++L E +
Sbjct: 344 --DVNALTGIFET------------AEDSSDTDSEITFDELATSYRKLCIKSEKILQQEA 389
Query: 437 DLKEKYVDLMKQQKSTLLELKASEEELKGFNLISTTYEDRLKILCQKLQEKCDKGSGNKH 496
LK+ DL ++++ E+ + E+ N +K+L NK
Sbjct: 390 QLKKVIADLEAEKEAHKEEISELKGEVGFLNSKLENMTKSIKML-------------NKG 436
Query: 497 EIALDDFIMAGIDRSKVASMIYSTYKNNGKGIGYSEEKSKEYSLKSYCDCIKDGLKSTFV 556
LD+ ++ G + N +G+G++ + + ++ + FV
Sbjct: 437 SDTLDEVLLLGKNAG------------NQRGLGFNPKSAGRTTM------------TEFV 472
Query: 557 PEGTNAVTAVQSKPETSGSQAKITSKPENLKIKVMTKSDPKSQKIKILKRSEPVHQNLIR 616
P +K T + ++ S+ ++ K + + +P +L
Sbjct: 473 P----------AKNRTGATMSQHRSRHHGMQQKKSKRKKWRCHYCGKYGHIKPFCYHL-H 521
Query: 617 PESKIPKQKDQKNKAATASDKTIPKGVKPEVLNDQKPLSIHPKVCIRAREKQRSWYLDSG 676
P +K +PK K +S+ +RA K+ WYLDSG
Sbjct: 522 PHHGTQSSNSRKKMM------WVPK---------HKAVSLVVHTSLRASAKE-DWYLDSG 565
Query: 677 CSRHMTGEKALFLTLTMKDGGEVKFGGNQTGKIIGTGTIGNSSI-SINNVWLVDGLKHNL 735
CSRHMTG K L + V FG GKIIG G + + + S+N V LV GL NL
Sbjct: 566 CSRHMTGVKEFLLNIEPCSTSYVTFGDGSKGKIIGMGKLVHDGLPSLNKVLLVKGLTANL 625
Query: 736 LSISQFCDNGYDVMFSKTNCTLVNKNDKSITFKGKRVENVYKINFSDLADQKVVCLLSMN 795
+SISQ CD G++V F+K+ C + N+ + + KG R ++ + CL S
Sbjct: 626 ISISQLCDEGFNVNFTKSECLVTNEKSE-VLMKGSRSKDNCYLWTPQETSYSSTCLSSKE 684
Query: 796 DKKWVWHKRLGHANWRLISKISKLQLVKGLPNIDYHSDALCGACQKGKIVKSTFKSKDIV 855
D+ +WH+R GH + R + KI V+G+PN+ +CG CQ GK VK + +
Sbjct: 685 DEVRIWHQRFGHLHLRGMKKIIDKGAVRGIPNLKIEEGRICGECQIGKQVKMSHQKLQHQ 744
Query: 856 STSRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKFIKSKDYACEVFSSFCTQ 915
+TSR LELLH+DL GP+ SL G +Y V+VDD+SR+TWVKFI+ K EVF +
Sbjct: 745 TTSRVLELLHMDLMGPMQVESLGGKRYAYVVVDDFSRFTWVKFIREKSETFEVFKELSLR 804
Query: 916 IQSEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEM 975
+Q EK+ I ++RSDHG EFEN FC GI HEFS+ TPQQNG+VERKNRTLQE
Sbjct: 805 LQREKDCVIKRIRSDHGREFENSRLTEFCTSEGITHEFSAAITPQQNGIVERKNRTLQEA 864
Query: 976 ARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNISYFHQFGCTC 1035
AR M+H L + WAEA+NT+CYI NR+ +R T YE++KGR+P++ +FH FG C
Sbjct: 865 ARVMLHAKELPYNLWAEAMNTACYIHNRVTLRRGTPTTLYEIWKGRKPSVKHFHIFGSPC 924
Query: 1036 YILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETHCVEESIHVKFDDREPGNKTPE 1095
YIL ++ +K D K+ GIFLGYS S+AYRV+NS T V ESI+V DD P K
Sbjct: 925 YILADREQRRKMDPKSDAGIFLGYSTNSRAYRVFNSRTRTVMESINVVVDDLSPARKKDV 984
Query: 1096 QSESNAGTTDSEDASESDQPSDSEKHSEVESRPEAEITPEAESNSEAETSPVEQNENASE 1155
+ + + DA++S + +++ S+S + S + Q + S
Sbjct: 985 EEDVRTSGDNVADAAKSGENAEN-------------------SDSATDESNINQPDKRSS 1025
Query: 1156 DVQDNTQQVIQPKFKHKSSHPEELIIGSKDSPRRTRSHFRQEESLIGLLSIIEPKTVEEA 1215
+ + HP+ELIIG + TRS + S +S IEPK V+EA
Sbjct: 1026 T-------------RIQKMHPKELIIGDPNRGVTTRSREVEIVSNSCFVSKIEPKNVKEA 1072
Query: 1216 LSDDGWILAMQEELNQFQRNDVWDLIPKPFQKNIIGTKWVFRNKLNEKGEVTRNKARLVA 1275
L+D+ WI AMQEEL QF+RN+VW+L+P+P N+IGTKW+F+NK NE+G +TRNKARLVA
Sbjct: 1073 LTDEFWINAMQEELEQFKRNEVWELVPRPEGTNVIGTKWIFKNKTNEEGVITRNKARLVA 1132
Query: 1276 QGYSQQEGIDYTETFAPVARLEAIRLLLSYAINHGIILYQMDVKSAFLNGVIEEEVYVKQ 1335
QGY+Q EG+D+ ETFAPVARLE+IRLLL A LYQMDVKSAFLNG + EEVYV+Q
Sbjct: 1133 QGYTQIEGVDFDETFAPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEVYVEQ 1192
Query: 1336 PPGFEDIKHPDHVYKLKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQVDTTLFGKTLKKD 1395
P GF D HPDHVY+LKK+LYGLKQAPRAWY+RL+ FL + + +G +D TLF K ++
Sbjct: 1193 PKGFADPTHPDHVYRLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQDAEN 1252
Query: 1396 ILIVQIYVDDIIFGSTNASLCKEFSKLMQDEFEMSMMGELKFFLGIQINQSKEGVYVHQT 1455
++I QIYVDDI+FG + + + F + MQ EFEMS++GEL +FLG+Q+ Q ++ +++ Q+
Sbjct: 1253 LMIAQIYVDDIVFGGMSNEMLRHFVQQMQSEFEMSLVGELTYFLGLQVKQMEDSIFLSQS 1312
Query: 1456 KYTKELLKKFKLEDCKVMNTPMHPTCTLSKEDTGTVVDQKLYRGMIGSLLYLTASRPDIL 1515
+Y K ++KKF +E+ TP LSK++ GT VDQ LYR MIGSLLYLTASRPDI
Sbjct: 1313 RYAKNIVKKFGMENASHKRTPAPTHLKLSKDEAGTSVDQSLYRSMIGSLLYLTASRPDIT 1372
Query: 1516 FSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLYRKSLDYKLIGFCDADYAGDRIE 1575
++V +CAR+Q++P+ SHLT VKRI +Y+ GT++ G++Y + L+G+CDAD+AG +
Sbjct: 1373 YAVGVCARYQANPKISHLTQVKRILKYVNGTSDYGIMYCHCSNPMLVGYCDADWAGSADD 1432
Query: 1576 RKSTSGNCQFLGENLISWASKRQATIAMSTAEAEYISAASCCTQLLWMKHQLEDYQINAN 1635
RKSTSG C +LG NLISW SK+Q +++STAEAEYI+A S C+QL+WMK L++Y + +
Sbjct: 1433 RKSTSGGCFYLGNNLISWFSKKQNCVSLSTAEAEYIAAGSSCSQLVWMKQMLKEYNVEQD 1492
Query: 1636 SIPIYCDNTAAICLSKNPILHSRAKHIEIKHHFIRDYVQKGIL 1678
+ +YCDN +AI +SKNP+ HSR KHI+I+HH+IRD V ++
Sbjct: 1493 VMTLYCDNMSAINISKNPVQHSRTKHIDIRHHYIRDLVDDKVI 1535
>gb|AAO73523.1| gag-pol polyprotein [Glycine max]
Length = 1576
Score = 1082 bits (2799), Expect = 0.0
Identities = 631/1663 (37%), Positives = 918/1663 (54%), Gaps = 164/1663 (9%)
Query: 38 DPEEFSWWKTNMYSFIMGLDEELWDILEDGVDDLD-LDEEGAAIDR----RIHTPAQKKL 92
D + +WK M +F+ LD W + G + LD EG D T + +L
Sbjct: 17 DGSNYEYWKARMVAFLKSLDSRTWKAVIKGWEHPKMLDTEGKPTDELKPEEDWTKEEDEL 76
Query: 93 YKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYEL 152
+ K + + + + ++ + AK L + EG+ KVK ++ +L ++E
Sbjct: 77 ALGNSKALNALFNGVDKNIFRLINTCTVAKDACEILKSTHEGTSKVKMSRLQLLATKFEN 136
Query: 153 FRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAKDL 212
+MK++E I + + + + L + V KILRSLP R+ KVTAIEEA+D+
Sbjct: 137 LKMKEEECIHDFHMNILEIANACTALGERITDEKLVRKILRSLPKRFDMKVTAIEEAQDI 196
Query: 213 NTLSVEDLVSSLKVHEMSLNEHESSKKSKSIALPSKGKTSKSSKAYKASESEEESPDRDS 272
+ V++L+ SL+ E+ L++ + KKSK++A S E EE+ D D+
Sbjct: 197 CNMRVDELIGSLQTFELGLSDR-AEKKSKNLAFVSN------------DEGEEDEYDLDT 243
Query: 273 DEDQSVKMAMLSNK----LEYLARKQKKFLSK-----RGGYKNSKKEDQKG-------CF 316
DE + + +L + L + ++QK + R G K K+ D K C
Sbjct: 244 DEGLTNAVVLLGKQFNKVLNRMDKRQKPHVQNIPFDIRKGSKYQKRSDVKPSHSKGIQCH 303
Query: 317 NCKKPGHFIADCPDLQKEKSKSKSKKSSFSSSKFRKQIKKSLMATWEDLDSESGSDKEEA 376
C+ GH IA+CP K+ +K L D +SE SD +
Sbjct: 304 GCEGYGHIIAECPTHLKKH-------------------RKGLSVCQSDTESEQESDSDR- 343
Query: 377 EDDTKAAMGLVATVSSEAVSEAESDSEDENEVYSKIPRQELVDSLKELLSLFEHRTNELT 436
D A +G+ T +ED ++ S+I EL S ++L E +
Sbjct: 344 --DVNALIGIFET------------AEDSSDTDSEITFDELAASYRKLCIKSEKILQQEA 389
Query: 437 DLKEKYVDLMKQQKSTLLELKASEEELKGFNLISTTYEDRLKILCQKLQEKCDKGSGNKH 496
LK+ DL ++++ E+ + E+ N +K+L NK
Sbjct: 390 QLKKVIADLEAEKEAHKEEISELKGEVGFLNSKLENMTKSIKML-------------NKG 436
Query: 497 EIALDDFIMAGIDRSKVASMIYSTYKNNGKGIGYSEEKSKEYSLKSYCDCIKDGLKSTFV 556
LD+ ++ G + N +G+G++ + + ++ + FV
Sbjct: 437 SDTLDEVLLLGKNAG------------NQRGLGFNPKSAGRTTM------------TEFV 472
Query: 557 PEGTNAVTAVQSKPETSGSQAKITSKPENLKIKVMTKSDPKSQKIKILKRSEPVHQNLIR 616
P +K T + ++ S+ ++ K + + +P +L
Sbjct: 473 P----------AKNRTGATMSQHRSRHHGMQQKKSKRKKWRCHYCGKYGHIKPFCYHLHG 522
Query: 617 PESKIPKQKDQKNKAATASDKTIPKGVKPEVLNDQKPLSIHPKVCIRAREKQRSWYLDSG 676
+ + + K +PK K +S+ +RA K+ WYLDSG
Sbjct: 523 HPHHGTQSSNSRKKMMW-----VPK---------HKAVSLVVHTSLRASAKE-DWYLDSG 567
Query: 677 CSRHMTGEKALFLTLTMKDGGEVKFGGNQTGKIIGTGTIGNSSI-SINNVWLVDGLKHNL 735
CSRHMTG K L + V FG GKIIG G + + + S+N V LV GL NL
Sbjct: 568 CSRHMTGVKEFLLNIEPCSTSYVTFGDGSKGKIIGMGKLVHDGLPSLNKVLLVKGLTANL 627
Query: 736 LSISQFCDNGYDVMFSKTNCTLVNKNDKSITFKGKRVENVYKINFSDLADQKVVCLLSMN 795
+SISQ CD G++V F+K+ C + N+ + + KG R ++ + CL S
Sbjct: 628 ISISQLCDEGFNVNFTKSECLVTNEKSE-VLMKGSRSKDNCYLWTPQETSYSSTCLSSKE 686
Query: 796 DKKWVWHKRLGHANWRLISKISKLQLVKGLPNIDYHSDALCGACQKGKIVKSTFKSKDIV 855
D+ +WH+R GH + R + KI V+G+PN+ +CG CQ GK VK + +
Sbjct: 687 DEVRIWHQRFGHLHLRGMKKILDKSAVRGIPNLKIEEGRICGECQIGKQVKMSHQKLQHQ 746
Query: 856 STSRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKFIKSKDYACEVFSSFCTQ 915
+TSR LELLH+DL GP+ SL G +Y V+VDD+SR+TWV FI+ K EVF +
Sbjct: 747 TTSRVLELLHMDLMGPMQVESLGGKRYAYVVVDDFSRFTWVNFIREKSGTFEVFKKLSLR 806
Query: 916 IQSEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEM 975
+Q EK+ I ++RSDHG EFEN F FC GI HEFS+ TPQQNG+VERKNRTLQE
Sbjct: 807 LQREKDCVIKRIRSDHGREFENSRFTEFCTSEGITHEFSAAITPQQNGIVERKNRTLQEA 866
Query: 976 ARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNISYFHQFGCTC 1035
AR M+H L + WAEA+NT+CYI NR+ +R T YE++KGR+P++ +FH FG C
Sbjct: 867 ARVMLHAKELPYNLWAEAMNTACYIHNRVTLRRGTPTTLYEIWKGRKPSVKHFHIFGSPC 926
Query: 1036 YILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETHCVEESIHVKFDDREPGNKTPE 1095
YIL ++ +K D K+ GIFLGYS S+AYRV+NS T V ESI+V DD P K
Sbjct: 927 YILADREQRRKMDPKSDAGIFLGYSTNSRAYRVFNSRTRTVMESINVVVDDLSPARKKDV 986
Query: 1096 QSESNAGTTDSEDASESDQPSDSEKHSEVESRPEAEITPEAESNSEAETSPVEQNENASE 1155
+ + + DA++S + +++ S+S + S + Q + S
Sbjct: 987 EEDVRTSGDNVADAAKSGENAEN-------------------SDSATDESNINQPDKRSS 1027
Query: 1156 DVQDNTQQVIQPKFKHKSSHPEELIIGSKDSPRRTRSHFRQEESLIGLLSIIEPKTVEEA 1215
+ + HP+ELIIG + TRS + S +S IEPK V+EA
Sbjct: 1028 T-------------RIQKMHPKELIIGDPNRGVTTRSREVEIVSNSCFVSKIEPKNVKEA 1074
Query: 1216 LSDDGWILAMQEELNQFQRNDVWDLIPKPFQKNIIGTKWVFRNKLNEKGEVTRNKARLVA 1275
L+D+ WI AMQEEL QF+RN+VW+L+P+P N+IGTKW+F+NK NE+G +TRNKARLVA
Sbjct: 1075 LTDEFWINAMQEELEQFKRNEVWELVPRPEGTNVIGTKWIFKNKTNEEGVITRNKARLVA 1134
Query: 1276 QGYSQQEGIDYTETFAPVARLEAIRLLLSYAINHGIILYQMDVKSAFLNGVIEEEVYVKQ 1335
QGY+Q EG+D+ ETFAPVARLE+IRLLL A LYQMDVKSAFLNG + EEVYV+Q
Sbjct: 1135 QGYTQIEGVDFDETFAPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEVYVEQ 1194
Query: 1336 PPGFEDIKHPDHVYKLKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQVDTTLFGKTLKKD 1395
P GF D HPDHVY+LKK+LYGLKQAPRAWY+RL+ FL + + +G +D TLF K ++
Sbjct: 1195 PKGFADPTHPDHVYRLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQDAEN 1254
Query: 1396 ILIVQIYVDDIIFGSTNASLCKEFSKLMQDEFEMSMMGELKFFLGIQINQSKEGVYVHQT 1455
++I QIYVDDI+FG + + + F + MQ EFEMS++GEL +FLG+Q+ Q ++ +++ Q+
Sbjct: 1255 LMIAQIYVDDIVFGGMSNEMLRHFVQQMQSEFEMSLVGELTYFLGLQVKQMEDSIFLSQS 1314
Query: 1456 KYTKELLKKFKLEDCKVMNTPMHPTCTLSKEDTGTVVDQKLYRGMIGSLLYLTASRPDIL 1515
+Y K ++KKF +E+ TP LSK++ GT VDQK YR MIGSLLYLTASRPDI
Sbjct: 1315 RYAKNIVKKFGMENASHKRTPAPTHLKLSKDEAGTSVDQKPYRSMIGSLLYLTASRPDIT 1374
Query: 1516 FSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLYRKSLDYKLIGFCDADYAGDRIE 1575
++V +CAR+Q++P+ SHL VKRI +Y+ GT++ G++Y L+G+CDAD+AG +
Sbjct: 1375 YAVGVCARYQANPKISHLNQVKRILKYVNGTSDYGIMYCHCSSSMLVGYCDADWAGSADD 1434
Query: 1576 RKSTSGNCQFLGENLISWASKRQATIAMSTAEAEYISAASCCTQLLWMKHQLEDYQINAN 1635
RKSTSG C +LG NLISW SK+Q +++STAEAEYI+A S C+QL+WMK L++Y + +
Sbjct: 1435 RKSTSGGCFYLGNNLISWFSKKQNCVSLSTAEAEYIAAGSSCSQLVWMKQMLKEYNVEQD 1494
Query: 1636 SIPIYCDNTAAICLSKNPILHSRAKHIEIKHHFIRDYVQKGIL 1678
+ +YCDN +AI +SKNP+ HSR KHI+I+HH+IRD V ++
Sbjct: 1495 VMTLYCDNMSAINISKNPVQHSRTKHIDIRHHYIRDLVDDKVI 1537
>gb|AAO73527.1| gag-pol polyprotein [Glycine max]
Length = 1576
Score = 1081 bits (2796), Expect = 0.0
Identities = 633/1663 (38%), Positives = 920/1663 (55%), Gaps = 164/1663 (9%)
Query: 38 DPEEFSWWKTNMYSFIMGLDEELWDILEDGVDDLD-LDEEGAAIDR----RIHTPAQKKL 92
D + +WK M +F+ LD W + G + LD EG D T + +L
Sbjct: 17 DGSNYEYWKARMVAFLKSLDSRTWKAVIKGWEHPKMLDTEGKPTDELKPEEDWTKEEDEL 76
Query: 93 YKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYEL 152
+ K + + + + ++ + AK + L EG+ KVK ++ +L ++E
Sbjct: 77 ALGNSKALNALFNGVDKNIFRLINTCTVAKDAWEILKITHEGTSKVKMSRLQLLATKFEN 136
Query: 153 FRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAKDL 212
+MK++E I + + + + L + V KILRSLP R+ KVTAIEEA+D+
Sbjct: 137 LKMKEEECIHDFHMNILEIANACTALGERITDEKLVRKILRSLPKRFDMKVTAIEEAQDI 196
Query: 213 NTLSVEDLVSSLKVHEMSLNEHESSKKSKSIALPSKGKTSKSSKAYKASESEEESPDRDS 272
+ V++L+ SL+ E+ L++ + KKSK++A S E EE+ D D+
Sbjct: 197 CNMRVDELIGSLQTFELGLSDR-AEKKSKNLAFVSN------------DEGEEDEYDLDT 243
Query: 273 DEDQSVKMAMLSNK----LEYLARKQKKFLSK-----RGGYKNSKKEDQKG-------CF 316
DE + + +L + L + ++QK + R G K K+ D K C
Sbjct: 244 DEGLTNAVVLLGKQFNKVLNRMDKRQKPHVQNIPFDIRKGSKYQKRSDVKPSHSKGIQCH 303
Query: 317 NCKKPGHFIADCPDLQKEKSKSKSKKSSFSSSKFRKQIKKSLMATWEDLDSESGSDKEEA 376
C+ GH IA+CP K+ +K L D +SE SD +
Sbjct: 304 GCEGYGHIIAECPTHLKKH-------------------RKGLSVCQSDTESEQESDSDR- 343
Query: 377 EDDTKAAMGLVATVSSEAVSEAESDSEDENEVYSKIPRQELVDSLKELLSLFEHRTNELT 436
D A G+ T +ED ++ S+I EL S ++L E +
Sbjct: 344 --DVNALTGIFET------------AEDSSDTDSEITFDELAASYRKLCIKSEKILQQEA 389
Query: 437 DLKEKYVDLMKQQKSTLLELKASEEELKGFNLISTTYEDRLKILCQKLQEKCDKGSGNKH 496
LK+ DL ++++ E+ + E+ N T + +K+L NK
Sbjct: 390 QLKKVIADLEAEKEAHEEEISELKGEVGFLNSKLETMKKSIKML-------------NKG 436
Query: 497 EIALDDFIMAGIDRSKVASMIYSTYKNNGKGIGYSEEKSKEYSLKSYCDCIKDGLKSTFV 556
LD+ ++ G + N +G+G++ + + ++ + FV
Sbjct: 437 SDTLDEVLLLGKNAG------------NQRGLGFNPKFAGRTTM------------TEFV 472
Query: 557 PEGTNAVTAVQSKPETSGSQAKITSKPENLKIKVMTKSDPKSQKIKILKRSEPVHQNLIR 616
P +K T + ++ S+ + K + + +P +L
Sbjct: 473 P----------AKNRTGTTMSQHLSRHHGTQQKKSKRKKWRCHYCGKYGHIKPFCYHLHG 522
Query: 617 PESKIPKQKDQKNKAATASDKTIPKGVKPEVLNDQKPLSIHPKVCIRAREKQRSWYLDSG 676
+ + + K +PK K +S+ +RA K+ WYLDSG
Sbjct: 523 HPHHGTQSSNSRKKMMW-----VPK---------HKAVSLVVHTSLRASAKE-DWYLDSG 567
Query: 677 CSRHMTGEKALFLTLTMKDGGEVKFGGNQTGKIIGTGTIGNSSI-SINNVWLVDGLKHNL 735
CSRHMTG K L + V FG GKIIG G + + + S+N V LV GL NL
Sbjct: 568 CSRHMTGVKEFLLNIEPCSTSYVTFGDGSKGKIIGMGKLVHDGLPSLNKVLLVKGLTANL 627
Query: 736 LSISQFCDNGYDVMFSKTNCTLVNKNDKSITFKGKRVENVYKINFSDLADQKVVCLLSMN 795
+SISQ CD G++V F+K+ C + N+ + + KG R ++ + CL S
Sbjct: 628 ISISQLCDEGFNVNFTKSECLVTNEKSE-VLMKGSRSKDNCYLWTPQETSYSSTCLSSKE 686
Query: 796 DKKWVWHKRLGHANWRLISKISKLQLVKGLPNIDYHSDALCGACQKGKIVKSTFKSKDIV 855
D+ +WH+R GH + R + KI V+G+PN+ +CG CQ GK VK + +
Sbjct: 687 DEVRIWHQRFGHLHLRGMKKIIDKGAVRGIPNLKIEEGRICGECQIGKQVKMSHQKLRHQ 746
Query: 856 STSRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKFIKSKDYACEVFSSFCTQ 915
+TSR LELLH+DL GP+ SL G +Y V+VDD+SR+TWV FI+ K EVF +
Sbjct: 747 TTSRVLELLHMDLMGPMQVESLGGKRYAYVVVDDFSRFTWVNFIREKSETFEVFKELSLR 806
Query: 916 IQSEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEM 975
+Q EK+ I ++RSDHG EFEN F FC GI HEFS+ TPQQNG+VERKNRTLQE
Sbjct: 807 LQREKDCVIKRIRSDHGREFENSRFTEFCTSEGITHEFSAAITPQQNGIVERKNRTLQEA 866
Query: 976 ARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNISYFHQFGCTC 1035
AR M+H L + WAEA+NT+CYI NR+ +R T YE++KGR+P++ +FH FG C
Sbjct: 867 ARVMLHAKELPYNLWAEAMNTACYIHNRVTLRRGTPTTLYEIWKGRKPSVKHFHIFGSPC 926
Query: 1036 YILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETHCVEESIHVKFDDREPGNKTPE 1095
YIL ++ +K D K+ GIFLGYS S+AYRV+NS T V ESI+V DD P K
Sbjct: 927 YILADREQRRKMDPKSDAGIFLGYSTNSRAYRVFNSRTRTVMESINVVVDDLSPARKKDV 986
Query: 1096 QSESNAGTTDSEDASESDQPSDSEKHSEVESRPEAEITPEAESNSEAETSPVEQNENASE 1155
+ + + DA++S + +++ S+S + S + Q + S
Sbjct: 987 EEDVRTLGDNVADAAKSGENAEN-------------------SDSATDESNINQPDKRSS 1027
Query: 1156 DVQDNTQQVIQPKFKHKSSHPEELIIGSKDSPRRTRSHFRQEESLIGLLSIIEPKTVEEA 1215
+ + HP+ELIIG + TRS + S +S IEPK V+EA
Sbjct: 1028 T-------------RIQKMHPKELIIGDPNRGVTTRSREVEIVSNSCFVSKIEPKNVKEA 1074
Query: 1216 LSDDGWILAMQEELNQFQRNDVWDLIPKPFQKNIIGTKWVFRNKLNEKGEVTRNKARLVA 1275
L+D+ WI AMQEEL QF+RN+VW+L+P+P N+IGTKW+F+NK NE+G +TRNKARLVA
Sbjct: 1075 LTDEFWINAMQEELEQFKRNEVWELVPRPEGTNVIGTKWIFKNKTNEEGVITRNKARLVA 1134
Query: 1276 QGYSQQEGIDYTETFAPVARLEAIRLLLSYAINHGIILYQMDVKSAFLNGVIEEEVYVKQ 1335
QGY+Q EG+D+ ETFAPVARLE+IRLLL A LYQMDVKSAFLNG + EEVYV+Q
Sbjct: 1135 QGYTQIEGVDFDETFAPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEVYVEQ 1194
Query: 1336 PPGFEDIKHPDHVYKLKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQVDTTLFGKTLKKD 1395
P GF D HPDHVY+LKK+LYGLKQAPRAWY+RL+ FL + + +G +D TLF K ++
Sbjct: 1195 PKGFADPTHPDHVYRLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQDAEN 1254
Query: 1396 ILIVQIYVDDIIFGSTNASLCKEFSKLMQDEFEMSMMGELKFFLGIQINQSKEGVYVHQT 1455
++I QIYVDDI+FG + + + F + MQ EFEMS++GEL +FLG+Q+ Q ++ +++ Q+
Sbjct: 1255 LMIAQIYVDDIVFGGMSNEMLRHFVQQMQSEFEMSLVGELTYFLGLQVKQMEDSIFLSQS 1314
Query: 1456 KYTKELLKKFKLEDCKVMNTPMHPTCTLSKEDTGTVVDQKLYRGMIGSLLYLTASRPDIL 1515
+Y K ++KKF +E+ TP LSK++ GT VDQ LYR MIGSLLYLTASRPDI
Sbjct: 1315 RYAKNIVKKFGMENASHKRTPAPTHLKLSKDEAGTSVDQSLYRSMIGSLLYLTASRPDIT 1374
Query: 1516 FSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLYRKSLDYKLIGFCDADYAGDRIE 1575
++V +CAR+Q++P+ SHLT VKRI +Y+ GT++ G++Y + L+G+CDAD+AG +
Sbjct: 1375 YAVGVCARYQANPKISHLTQVKRILKYVNGTSDYGIMYCHCSNPMLVGYCDADWAGSADD 1434
Query: 1576 RKSTSGNCQFLGENLISWASKRQATIAMSTAEAEYISAASCCTQLLWMKHQLEDYQINAN 1635
RKSTSG C +LG NLISW SK+Q +++STAEAEYI+A S C+QL+WMK L++Y + +
Sbjct: 1435 RKSTSGGCFYLGNNLISWFSKKQNCVSLSTAEAEYIAAGSSCSQLVWMKQMLKEYNVEQD 1494
Query: 1636 SIPIYCDNTAAICLSKNPILHSRAKHIEIKHHFIRDYVQKGIL 1678
+ +YCDN +AI +SKNP+ HSR KHI+I+HH+IRD V ++
Sbjct: 1495 VMTLYCDNMSAINISKNPVQHSRTKHIDIRHHYIRDLVDDKVI 1537
>gb|AAO73529.1| gag-pol polyprotein [Glycine max]
Length = 1577
Score = 1065 bits (2755), Expect = 0.0
Identities = 632/1664 (37%), Positives = 916/1664 (54%), Gaps = 165/1664 (9%)
Query: 38 DPEEFSWWKTNMYSFIMGLDEELWDILEDGVDDLD-LDEEGAAIDR----RIHTPAQKKL 92
D + +WK M +F+ LD W + G + LD EG + T + +L
Sbjct: 17 DGTNYEYWKARMVAFLKSLDSRTWKAVIKGWEHPKMLDTEGKPTNELKPEEDWTKEEDEL 76
Query: 93 YKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYEL 152
+ K + + + + ++ + AK + L EG+ KVK ++ +L ++E
Sbjct: 77 ALGNSKALNALFNGVDKNIFRLINTCTVAKDAWEILKTTHEGTSKVKMSRLQLLATKFEN 136
Query: 153 FRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAKDL 212
+MK++E I + + + + L + V KILRSLP R+ KVTAIEEA+D+
Sbjct: 137 LKMKEEECIHDFHMTILEIANACTALGERMTDEKLVRKILRSLPKRFDMKVTAIEEAQDI 196
Query: 213 NTLSVEDLVSSLKVHEMSLNEHESSKKSKSIALPSKGKTSKSSKAYKASESEEESPDRDS 272
+ V++L+ SL+ E+ L++ + KKSK++A S E EE+ D D+
Sbjct: 197 CNMRVDELIGSLQTFELGLSDR-TEKKSKNLAFVSN------------DEGEEDEYDLDT 243
Query: 273 DEDQSVKMAMLSNK----LEYLARKQKKF-----LSKRGGYKNSKKEDQKG-------CF 316
DE + + L + L + R+QK L R G + +K D+K C
Sbjct: 244 DEGLTNAVVFLGKQFNKVLNRMDRRQKPHVRNISLDIRKGSEYQRKSDEKPSHSKGIQCR 303
Query: 317 NCKKPGHFIADCPDLQKEKSKSKSKKSSFSSSKFRKQIKKSLMATWEDLDSESGSDKEEA 376
C+ GH A+CP K++ K S S +D +SE SD +
Sbjct: 304 GCEGYGHIKAECPTHLKKQRKGLSVCRS------------------DDTESEQESDSDR- 344
Query: 377 EDDTKAAMGLVATVSSEAVSEAESDSEDENEVYSKIPRQELVDSLKELLSLFEHRTNELT 436
D A G + +ED ++ S+I EL +EL E +
Sbjct: 345 --DVNALTGRFES------------AEDSSDTDSEITFDELAIFYRELCIKSEKILQQEA 390
Query: 437 DLKEKYVDLMKQQKSTLLELKASEEELKGFNLISTTYEDRLKILCQKLQEKCDKGSGNKH 496
LK+ +L ++++ E+ + E+ N +K+L NK
Sbjct: 391 QLKKVIANLEAEKEAHEEEISKLKGEVGFLNSKLENMTKSIKML-------------NKG 437
Query: 497 EIALDDFIMAGIDRSKVASMIYSTYKNNGKGIGYSEEKSKEYSLKSYCDCIKDGLKSTFV 556
LD+ + G KV N +G+G++ + + ++ + FV
Sbjct: 438 SDMLDZVLQLG---KKVG---------NQRGLGFNHKSAGRTTM------------TEFV 473
Query: 557 P-EGTNAVTAVQSKPETSGSQAKITSKPENLKIKVMTKSDPKSQKIKILKRSEPVHQNLI 615
P + + T Q + G+Q K SK + + K I
Sbjct: 474 PAKNSTGATMSQHRSRHHGTQQK-RSKRKKWRCHYCGK------------------YGHI 514
Query: 616 RPESKIPKQKDQKNKAATASDKTIPKGVKPEVLNDQKPLSIHPKVCIRAREKQRSWYLDS 675
+P ++S + + K ++++ L +H + A+E WYLDS
Sbjct: 515 KPFCYHLHGHPHHGTQGSSSGRKMMWVPKHKIVS----LVVHTSLRASAKE---DWYLDS 567
Query: 676 GCSRHMTGEKALFLTLTMKDGGEVKFGGNQTGKIIGTGTIGNSSI-SINNVWLVDGLKHN 734
GCSRHMTG K + + V FG GKI G G + + + S+N V LV GL N
Sbjct: 568 GCSRHMTGVKEFLVNIEPCSTSYVTFGDGSKGKITGMGKLVHEGLPSLNKVLLVKGLTVN 627
Query: 735 LLSISQFCDNGYDVMFSKTNCTLVNKNDKSITFKGKRVENVYKINFSDLADQKVVCLLSM 794
L+SISQ CD G++V F+K+ C + N+ + + KG R ++ + + CL S
Sbjct: 628 LISISQLCDEGFNVNFTKSECLVTNEKSE-VLMKGSRSKDNCYLWTPQESSHSSTCLFSK 686
Query: 795 NDKKWVWHKRLGHANWRLISKISKLQLVKGLPNIDYHSDALCGACQKGKIVKSTFKSKDI 854
D+ +WH+R GH + R + KI V+G+PN+ +CG CQ GK VK + +
Sbjct: 687 EDEVKIWHQRFGHLHLRGMKKIIDKGAVRGIPNLKIEEGRICGECQIGKQVKMSHQKLQH 746
Query: 855 VSTSRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKFIKSKDYACEVFSSFCT 914
+TSR LELLH+DL GP+ SL G +Y V+VDD+SR+TWV FI+ K EVF
Sbjct: 747 QTTSRVLELLHMDLMGPMQVESLGGKRYAYVVVDDFSRFTWVNFIREKSDTFEVFKELSL 806
Query: 915 QIQSEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNGVVERKNRTLQE 974
++Q EK+ I ++RSDHG EFEN F FC GI HEFS+ TPQQNG+VERKNRTLQE
Sbjct: 807 RLQREKDCVIKRIRSDHGREFENSKFTEFCTSEGITHEFSAAITPQQNGIVERKNRTLQE 866
Query: 975 MARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNISYFHQFGCT 1034
AR M+H L + WAEA+NT+CYI NR+ +R T YE++KGR+P + +FH FG
Sbjct: 867 AARVMLHAKELPYNLWAEAMNTACYIHNRVTLRRGTPTTLYEIWKGRKPTVKHFHIFGSP 926
Query: 1035 CYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETHCVEESIHVKFDDREPGNKTP 1094
CYIL ++ +K D K+ GIFLGYS S+AYRV+NS T V ESI+V DD P K
Sbjct: 927 CYILADREQRRKMDPKSDAGIFLGYSTNSRAYRVFNSRTRTVMESINVVVDDLTPARKKD 986
Query: 1095 EQSESNAGTTDSEDASESDQPSDSEKHSEVESRPEAEITPEAESNSEAETSPVEQNENAS 1154
+ + + D +D+ K +E N+E N +++
Sbjct: 987 VEEDVR---------TSGDNVADTAKSAE---------------NAE--------NSDSA 1014
Query: 1155 EDVQDNTQQVIQPKFKHKSSHPEELIIGSKDSPRRTRSHFRQEESLIGLLSIIEPKTVEE 1214
D + Q +P + + HP+ELIIG + TRS + S +S IEPK V+E
Sbjct: 1015 TDEPNINQPDKRPSIRIQKMHPKELIIGDPNRGVTTRSREIEIVSNSCFVSKIEPKNVKE 1074
Query: 1215 ALSDDGWILAMQEELNQFQRNDVWDLIPKPFQKNIIGTKWVFRNKLNEKGEVTRNKARLV 1274
AL+D+ WI AMQEEL QF+RN+VW+L+P+P N+IGTKW+F+NK NE+G +TRNKARLV
Sbjct: 1075 ALTDEFWINAMQEELEQFKRNEVWELVPRPEGTNVIGTKWIFKNKTNEEGVITRNKARLV 1134
Query: 1275 AQGYSQQEGIDYTETFAPVARLEAIRLLLSYAINHGIILYQMDVKSAFLNGVIEEEVYVK 1334
AQGY+Q EG+D+ ETFAPVARLE+IRLLL A LYQMDVKSAFLNG + EE YV+
Sbjct: 1135 AQGYTQIEGVDFDETFAPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEAYVE 1194
Query: 1335 QPPGFEDIKHPDHVYKLKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQVDTTLFGKTLKK 1394
QP GF D HPDHVY+LKK+LYGLKQAPRAWY+RL+ FL + + +G +D TLF K +
Sbjct: 1195 QPKGFVDPTHPDHVYRLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQDAE 1254
Query: 1395 DILIVQIYVDDIIFGSTNASLCKEFSKLMQDEFEMSMMGELKFFLGIQINQSKEGVYVHQ 1454
+++I QIYVDDI+FG + + + F + MQ EFEMS++GEL +FLG+Q+ Q ++ +++ Q
Sbjct: 1255 NLMIAQIYVDDIVFGGMSNEMLRHFVQQMQSEFEMSLVGELTYFLGLQVKQMEDSIFLSQ 1314
Query: 1455 TKYTKELLKKFKLEDCKVMNTPMHPTCTLSKEDTGTVVDQKLYRGMIGSLLYLTASRPDI 1514
+KY K ++KKF +E+ TP LSK++ GT VDQ LYR MIGSLLYLTASRPDI
Sbjct: 1315 SKYAKNIVKKFGMENASHKRTPAPTHLKLSKDEAGTSVDQSLYRSMIGSLLYLTASRPDI 1374
Query: 1515 LFSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLYRKSLDYKLIGFCDADYAGDRI 1574
++V +CAR+Q++P+ SHL VKRI +Y+ GT++ G++Y L+G+CDAD+AG
Sbjct: 1375 TYAVGVCARYQANPKISHLNQVKRILKYVNGTSDYGIMYCHCSGSMLVGYCDADWAGSAD 1434
Query: 1575 ERKSTSGNCQFLGENLISWASKRQATIAMSTAEAEYISAASCCTQLLWMKHQLEDYQINA 1634
+RKSTSG C +LG NLISW SK+Q +++STAEAEYI+A S C+QL+WMK L++Y +
Sbjct: 1435 DRKSTSGGCFYLGNNLISWFSKKQNCVSLSTAEAEYIAAGSSCSQLVWMKQMLKEYNVEQ 1494
Query: 1635 NSIPIYCDNTAAICLSKNPILHSRAKHIEIKHHFIRDYVQKGIL 1678
+ + +YCDN +AI +SKNP+ HSR KHI+I+HH+IR+ V ++
Sbjct: 1495 DVMTLYCDNMSAINISKNPVQHSRTKHIDIRHHYIRELVDDKVI 1538
>gb|AAG52949.1| gag/pol polyprotein [Arabidopsis thaliana]
Length = 1643
Score = 1057 bits (2733), Expect = 0.0
Identities = 616/1664 (37%), Positives = 917/1664 (55%), Gaps = 110/1664 (6%)
Query: 42 FSWWKTNMYSFIMGLDEELWDIL-----------EDGVDDLDLDEEGAAIDRRIHTPAQK 90
+ WK M + I GL +E W E+G D L +++ T A++
Sbjct: 21 YGHWKVKMRALIRGLGKEAWIATSVGWKAPVVKGENGEDVLKTEDQW--------TDAEE 72
Query: 91 KLYKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQY 150
+ + +I S+ + ++ ++ + +AK + L +EG+ VK ++ ML Q+
Sbjct: 73 AKATANSRALSLIFNSVNQNQFKRIQNCESAKEAWDKLAKAYEGTSSVKRSRIDMLASQF 132
Query: 151 ELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAK 210
E M + E+IEE + + S L K Y V K+LR LPSR+ K TA+ +
Sbjct: 133 ENLTMDESENIEEFSGKISAIASEAHNLGKKYKDKKLVKKLLRCLPSRFESKRTAMGTSL 192
Query: 211 DLNTLSVEDLVSSLKVHEMSLNEHESSKKSKSIALPSKGKTSKSSKAYKASESEEESPDR 270
D +T+ E++V L+ +E+ + KG SK +SE E +
Sbjct: 193 DTDTIDFEEVVGMLQAYELEITS-------------GKGGYSKGVALAVSSEKNEIQELK 239
Query: 271 DSDEDQSVKMAMLSNKLEY--LARKQKKFLSKRGGYKNSKKEDQKGCFNCKKPGHFIADC 328
DS + + ++E AR Q + + K + C C+ GH A+C
Sbjct: 240 DSMSMMAKNFSRAMKRVEKRGFARNQGSDRDRDRDRDRNSKRSEIQCHECQGYGHIKAEC 299
Query: 329 PDLQKEKSKSKSKKSSFSSSKFRKQIKKSLMATWEDLDSESGSDKEEAEDDTKAAMGLVA 388
P L+++ K S+ +KF KS +S+S SD E++E+D K + V
Sbjct: 300 PSLKRKDLKC-SECRGIGHTKFDCIGSKSKPDRSYIAESDSDSDDEDSEEDVKGFVSFVG 358
Query: 389 TVSSEAVSEAESDSE---DENEVYSKIPRQELVDSLKELLSLFEHRTNELTDLKEKYVDL 445
+ + VS SDSE ++ E+ + +D E L+E N L KEK + L
Sbjct: 359 IIEDDNVSSDSSDSEVGCEKEEISADDESDVEMDVDGEFRKLYE---NWLVLSKEKVIWL 415
Query: 446 MKQQKSTLLELKASEEELKGFNLISTTYEDRLKILCQKLQEKCDKGSGNKHEIALDDFIM 505
++ K ++ E+LKG ++ + + + +EK + S + + ++
Sbjct: 416 EEKVK-----VQEQIEQLKGELAVANQIKSEMILKYSAKEEKNRELSQDLSDTRKKIHML 470
Query: 506 AGIDRSKVASMIYSTYKNNGKGIGYSEEKSKEYSLKSYCDCIKDGLKSTFVPEGTNAVTA 565
+ + + + G+GY S K+ FV A T
Sbjct: 471 NKGTKDLDSILAAGRVGKSNFGLGYHGGGSST--------------KTNFVRSKAAAPTQ 516
Query: 566 VQSKPETSGSQAKITSKPENLKIKVMTKSDPKSQKIKILKRSEPVHQNLIRPESKIPKQK 625
QS + + K +N ++ + R + + R +++ K K
Sbjct: 517 SQSVFRSKSNSVPARRKYQNQN-HYHSQRTVTGYECYYCGRHGHIQRYCYRYAARLSKLK 575
Query: 626 DQKNKAATASDKTIP-KGVKPEVLNDQKPLSIHPKVCIRAREKQRSWYLDSGCSRHMTGE 684
Q K P +G ++ ++ L H A ++ WY DSG SRHMTG
Sbjct: 576 RQ--------GKLYPHQGRNSKMYVRREDLYCHVAYTSIAEGVKKPWYFDSGASRHMTGS 627
Query: 685 KALFLTLTMKDGGEVKFGGNQTGKIIGTGTIGNSSIS-INNVWLVDGLKHNLLSISQFCD 743
+A + V FGG G+I G G + + + NV+ V+GL NL+S+SQ CD
Sbjct: 628 QANLNNYSSVKESNVMFGGGAKGRIKGKGDLTETEKPHLTNVYFVEGLTANLISVSQLCD 687
Query: 744 NGYDVMFSKTNCTLVNKNDKSITFKGKRVENVYKINFSDLADQKVVCLLSMNDKKWVWHK 803
G V F+K C N+ +++ + N Y + ++ +CL + + +WH+
Sbjct: 688 EGLTVSFNKVKCWATNERNQNTLTGVRTGNNCY------MWEEPKICLRAEKEDPVLWHQ 741
Query: 804 RLGHANWRLISKISKLQLVKGLPNIDYHSDALCGACQKGKIVKSTFKSKDIVSTSRPLEL 863
RLGH N R +SK+ ++V+G+P + + +CGAC +GK ++ K + + T++ L+L
Sbjct: 742 RLGHMNARSMSKLVNKEMVRGVPELKHIEKIVCGACNQGKQIRVQHKRVEGIQTTQVLDL 801
Query: 864 LHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKFIKSKDYACEVFSSFCTQIQSEKELK 923
+H+DL GP+ T S+ G +Y V+VDD+SR+ WV+FI+ K F Q+++EK++
Sbjct: 802 IHMDLMGPMQTESIAGKRYVFVLVDDFSRYAWVRFIREKSETANSFKILALQLKNEKKMG 861
Query: 924 ILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEMARTMIHEN 983
I ++RSD GGEF NE F FCE GI H++S+PRTPQ NGVVERKNRTLQEMAR MIH N
Sbjct: 862 IKQIRSDRGGEFMNEAFNSFCESQGIFHQYSAPRTPQSNGVVERKNRTLQEMARAMIHGN 921
Query: 984 NLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNISYFHQFGCTCYILNTKDY 1043
+ + FWAEA++T+CY+ NR+Y+R +KT YE++KG++PN+SYF FGC CYI+N KD
Sbjct: 922 GVPEKFWAEAISTACYVINRVYVRLGSDKTPYEIWKGKKPNLSYFRVFGCVCYIMNDKDQ 981
Query: 1044 LKKFDAKAQRGIFLGYSERSKAYRVYNSETHCVEESIHVKFDDREPGNKTPE-------Q 1096
L KFD++++ G FLGY+ S AYRV+N + +EES++V FDD PE +
Sbjct: 982 LGKFDSRSEEGFFLGYATNSLAYRVWNKQRGKIEESMNVVFDD----GSMPELQIIVRNR 1037
Query: 1097 SESNAGTTDSEDASESDQPSDSEKHSEVESRPEAEITPEAESNSEAETSPV--EQNENAS 1154
+E +++ +D D+ ++ + E+ P A + E +
Sbjct: 1038 NEPQTSISNNHGEERNDNQFDNGDINKSGEESDEEVPPAQVHRDHASKDIIGDPSGERVT 1097
Query: 1155 EDVQDNTQQVIQPKFKHKSSHPEELIIGSKDSPRRTRSHFRQEESLIG--LLSIIEPKTV 1212
V+ + +Q+ K KH R + F E ++ +SI+EPK V
Sbjct: 1098 RGVKQDYRQLAGIKQKH-----------------RVMASFACFEEIMFSCFVSIVEPKNV 1140
Query: 1213 EEALSDDGWILAMQEELNQFQRNDVWDLIPKPFQKNIIGTKWVFRNKLNEKGEVTRNKAR 1272
+EAL D WILAM+EEL +F R+ VWDL+P+P Q N+IGTKW+F+NK +E G +TRNKAR
Sbjct: 1141 KEALEDHFWILAMEEELEEFSRHQVWDLVPRPPQVNVIGTKWIFKNKFDEVGNITRNKAR 1200
Query: 1273 LVAQGYSQQEGIDYTETFAPVARLEAIRLLLSYAINHGIILYQMDVKSAFLNGVIEEEVY 1332
LVAQGY+Q EG+D+ ETFAPVARLE IR LL A G L+QMDVK AFLNG+IEEEVY
Sbjct: 1201 LVAQGYTQVEGLDFDETFAPVARLECIRFLLGTACGMGFKLHQMDVKCAFLNGIIEEEVY 1260
Query: 1333 VKQPPGFEDIKHPDHVYKLKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQVDTTLFGKTL 1392
V+QP GFE+++ P++VYKLKK+LYGLKQAPRAWY+RL+ FLI + RG VD TLF K
Sbjct: 1261 VEQPKGFENLEFPEYVYKLKKALYGLKQAPRAWYERLTTFLIVQGYTRGSVDKTLFVKND 1320
Query: 1393 KKDILIVQIYVDDIIFGSTNASLCKEFSKLMQDEFEMSMMGELKFFLGIQINQSKEGVYV 1452
I+I+QIYVDDI+FG T+ L K F K M EF MSM+GELK+FLG+QINQ+ EG+ +
Sbjct: 1321 VHGIIIIQIYVDDIVFGGTSDKLVKTFVKTMTTEFRMSMVGELKYFLGLQINQTDEGITI 1380
Query: 1453 HQTKYTKELLKKFKLEDCKVMNTPMHPTCTLSKEDTGTVVDQKLYRGMIGSLLYLTASRP 1512
Q+ Y + L+K+F + K TPM T L K++ G VD+KLYRGMIGSLLYLTA+RP
Sbjct: 1381 SQSTYAQNLVKRFGMCSSKPAPTPMSTTTKLFKDEKGVKVDEKLYRGMIGSLLYLTATRP 1440
Query: 1513 DILFSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLYRKSLDYKLIGFCDADYAGD 1572
D+ SV LCAR+QS+P+ SHL AVKRI +Y+ GT N GL Y + L+G+CDAD+ G+
Sbjct: 1441 DLCLSVGLCARYQSNPKASHLLAVKRIIKYVSGTINYGLNYTRDTSLVLVGYCDADWGGN 1500
Query: 1573 RIERKSTSGNCQFLGENLISWASKRQATIAMSTAEAEYISAASCCTQLLWMKHQLEDYQI 1632
+R+ST+G FLG NLISW SK+Q +++S+ ++EYI+ SCCTQLLWM+ DY +
Sbjct: 1501 LDDRRSTTGGVFFLGSNLISWHSKKQNCVSLSSTQSEYIALGSCCTQLLWMRQMGLDYGM 1560
Query: 1633 N-ANSIPIYCDNTAAICLSKNPILHSRAKHIEIKHHFIRDYVQK 1675
+ + + CDN +AI +SKNP+ HS KHI I+HHF+R+ V++
Sbjct: 1561 TFPDPLLVKCDNESAIAISKNPVQHSVTKHIAIRHHFVRELVEE 1604
>gb|AAO73525.1| gag-pol polyprotein [Glycine max]
Length = 1576
Score = 1050 bits (2716), Expect = 0.0
Identities = 628/1665 (37%), Positives = 909/1665 (53%), Gaps = 168/1665 (10%)
Query: 38 DPEEFSWWKTNMYSFIMGLDEELWDILEDGVDDLD-LDEEGAAIDR----RIHTPAQKKL 92
D + +WK M +F+ LD W + G + LD EG + T + +L
Sbjct: 17 DGTNYEYWKARMVAFLKSLDSRTWKAVIKGWEHPKMLDTEGKPTNELKPEEDWTKEEDEL 76
Query: 93 YKKHHKIRGIIVASIPRTEYMKMSDKSTAK-AMFASLCANFEGSKKVKEAKALMLVHQYE 151
+ K + + + + ++ + AK A L EG+ KVK ++ +L ++E
Sbjct: 77 ALGNSKALNALFNGVDKNIFRLINTCTVAKDACGEILKTTHEGTSKVKMSRLQLLATKFE 136
Query: 152 LFRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAKD 211
+MK++E I + + + + L + V KILRSLP R+ KVTAIEEA+D
Sbjct: 137 NLKMKEEECIHDFHMNILEIANACTALGERMTDEKLVRKILRSLPKRFDMKVTAIEEAQD 196
Query: 212 LNTLSVEDLVSSLKVHEMSLNEHESSKKSKSIALPSKGKTSKSSKAYKASESEEESPDRD 271
+ + V++L+ SL+ E+ L++ + KKSK++A S E EE+ D D
Sbjct: 197 ICNMRVDELIGSLQTFELGLSDR-NEKKSKNLAFVSN------------DEGEEDEYDLD 243
Query: 272 SDEDQSVKMAMLSNK----LEYLARKQKKFLSK-----RGGYKNSKKEDQKG-------C 315
+DE + + +L + L + R+QK + R G + KK D+K C
Sbjct: 244 TDEGLTNAVGLLGKQFNKVLNRMDRRQKPHVRNIPFDIRKGSEYHKKSDEKPSHSKGIQC 303
Query: 316 FNCKKPGHFIADCPDLQKEKSKSKSKKSSFSSSKFRKQIKKSLMATWEDLDSESGSDKEE 375
C+ GH A+CP K++ K S S +D +SE SD +
Sbjct: 304 HGCEGYGHIKAECPTHLKKQRKGLSVCRS------------------DDTESEQESDSDR 345
Query: 376 AEDDTKAAMGLVATVSSEAVSEAESDSEDENEVYSKIPRQELVDSLKELLSLFEHRTNEL 435
D A G E++ DS D +I EL S ++L E +
Sbjct: 346 ---DVNALTGRF---------ESDEDSSD-----IEITFDELAISYRKLCIKSEKILQQE 388
Query: 436 TDLKEKYVDLMKQQKSTLLELKASEEELKGFNLISTTYEDRLKILCQKLQEKCDKGSGNK 495
LK+ +L ++++ E+ + E+ N +K+L NK
Sbjct: 389 AQLKKVIANLEAEKEAHEEEISELKGEVGFLNSKLENMTKSIKML-------------NK 435
Query: 496 HEIALDDFIMAGIDRSKVASMIYSTYKNNGKGIGYSEEKSKEYSLKSYCDCIKDGLKSTF 555
LD+ + G + N +G+G++ + + ++ + F
Sbjct: 436 GSDMLDEVLQLGKN------------VGNQRGLGFNHKSACRITM------------TEF 471
Query: 556 VP-EGTNAVTAVQSKPETSGSQAKITSKPENLKIKVMTKSDPKSQKIKILKRSEPVHQNL 614
VP + + T Q + G+Q K KS K + +
Sbjct: 472 VPAKNSTGATMSQHRSRHHGTQQK--------------KSKRKKWRCHYCGK-----YGH 512
Query: 615 IRPESKIPKQKDQKNKAATASDKTIPKGVKPEVLNDQKPLSIHPKVCIRAREKQRSWYLD 674
I+P +++S + + K ++++ L +H + A+E WYLD
Sbjct: 513 IKPFCYHLHGHPHHGTQSSSSGRKMMWVPKHKIVS----LVVHTSLRASAKE---DWYLD 565
Query: 675 SGCSRHMTGEKALFLTLTMKDGGEVKFGGNQTGKIIGTGTIGNSSI-SINNVWLVDGLKH 733
SGCSRHMTG K + + V FG GKI G G + + + S+N V LV GL
Sbjct: 566 SGCSRHMTGVKEFLVNIEPCSTSYVTFGDGSKGKITGMGKLVHDGLPSLNKVLLVKGLTA 625
Query: 734 NLLSISQFCDNGYDVMFSKTNCTLVNKNDKSITFKGKRVENVYKINFSDLADQKVVCLLS 793
NL+SISQ CD G++V F+K+ C + N+ + + KG R ++ + CL S
Sbjct: 626 NLISISQLCDEGFNVNFTKSECLVTNEKSE-VLMKGSRSKDNCYLWTPQETSYSSTCLSS 684
Query: 794 MNDKKWVWHKRLGHANWRLISKISKLQLVKGLPNIDYHSDALCGACQKGKIVKSTFKSKD 853
D+ +WH+R GH + R + KI V+G+PN+ +CG CQ GK VK + +
Sbjct: 685 KEDEVKIWHQRFGHLHLRGMKKIIDKGAVRGIPNLKIEEGRICGECQIGKQVKMSHQKLQ 744
Query: 854 IVSTSRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKFIKSKDYACEVFSSFC 913
+TS LELLH+DL GP+ SL G +Y V+VDD+SR+TWV FI+ K EVF
Sbjct: 745 HQTTSMVLELLHMDLMGPMQVESLGGKRYAYVVVDDFSRFTWVNFIREKSDTFEVFKELS 804
Query: 914 TQIQSEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNGVVERKNRTLQ 973
++Q EK+ I ++RSDHG EFEN F FC GI HEFS+ TPQQNG+VERKNRTLQ
Sbjct: 805 LRLQREKDCVIKRIRSDHGREFENSKFTEFCTSEGITHEFSAAITPQQNGIVERKNRTLQ 864
Query: 974 EMARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNISYFHQFGC 1033
E R M+H L + WAEA+NT+CYI NR+ +R T YE++KGR+P + +FH FG
Sbjct: 865 EATRVMLHAKELPYNLWAEAMNTACYIHNRVTLRRGTPTTLYEIWKGRKPTVKHFHIFGS 924
Query: 1034 TCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETHCVEESIHVKFDDREPGNKT 1093
CYIL ++ +K D K+ GIFLGYS S+AYRV+NS T V ESI+V DD P K
Sbjct: 925 PCYILADREQRRKMDPKSDAGIFLGYSTNSRAYRVFNSRTRTVMESINVVVDDLTPARKK 984
Query: 1094 PEQSESNAGTTDSEDASESDQPSDSEKHSEVESRPEAEITPEAESNSEAETSPVEQNENA 1153
+ + + D ++S AE+ +++++ E N N
Sbjct: 985 DVEEDVRTSEDNVADTAKS-----------------------AENAEKSDSTTDEPNINQ 1021
Query: 1154 SEDVQDNTQQVIQPKFKHKSSHPEELIIGSKDSPRRTRSHFRQEESLIGLLSIIEPKTVE 1213
+ P + + P+ELIIG + TRS + S +S IEPK V+
Sbjct: 1022 PDK---------SPFIRIQKMQPKELIIGDPNRGVTTRSREIEIVSNSCFVSKIEPKNVK 1072
Query: 1214 EALSDDGWILAMQEELNQFQRNDVWDLIPKPFQKNIIGTKWVFRNKLNEKGEVTRNKARL 1273
EAL+D+ WI AMQEEL QF+RN+VW+L+P+P N+IGTKW+F+NK NE+G +TRNKARL
Sbjct: 1073 EALTDEFWINAMQEELEQFKRNEVWELVPRPEGTNVIGTKWIFKNKTNEEGVITRNKARL 1132
Query: 1274 VAQGYSQQEGIDYTETFAPVARLEAIRLLLSYAINHGIILYQMDVKSAFLNGVIEEEVYV 1333
VAQGY+Q EG+D+ ETFAPVARLE+IRLLL A LYQMDVKSAFLNG + EE YV
Sbjct: 1133 VAQGYTQIEGVDFDETFAPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEAYV 1192
Query: 1334 KQPPGFEDIKHPDHVYKLKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQVDTTLFGKTLK 1393
+QP GF D H DHVY+LKK+LYGLKQAPRAWY+RL+ FL + + +G +D TLF K
Sbjct: 1193 EQPKGFVDPTHLDHVYRLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQDA 1252
Query: 1394 KDILIVQIYVDDIIFGSTNASLCKEFSKLMQDEFEMSMMGELKFFLGIQINQSKEGVYVH 1453
++++I QIYVDDI+FG + + + F MQ EFEMS++GEL +FLG+Q+ Q ++ +++
Sbjct: 1253 ENLMIAQIYVDDIVFGGMSNEMLRHFVPQMQSEFEMSLVGELHYFLGLQVKQMEDSIFLS 1312
Query: 1454 QTKYTKELLKKFKLEDCKVMNTPMHPTCTLSKEDTGTVVDQKLYRGMIGSLLYLTASRPD 1513
Q+KY K ++KKF +E+ TP LSK++ GT VDQ LYR MIGSLLYLTASRPD
Sbjct: 1313 QSKYAKNIVKKFGMENASHKRTPAPTHLKLSKDEAGTSVDQNLYRSMIGSLLYLTASRPD 1372
Query: 1514 ILFSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLYRKSLDYKLIGFCDADYAGDR 1573
I F+V +CAR+Q++P+ SHL VKRI +Y+ GT++ G++Y D L+G+CDAD+AG
Sbjct: 1373 ITFAVGVCARYQANPKISHLNQVKRILKYVNGTSDYGIMYCHCSDSMLVGYCDADWAGSA 1432
Query: 1574 IERKSTSGNCQFLGENLISWASKRQATIAMSTAEAEYISAASCCTQLLWMKHQLEDYQIN 1633
+RK TSG C +LG NLISW SK+Q +++STAEAEYI+A S C+QL+WMK L++Y +
Sbjct: 1433 DDRKCTSGGCFYLGTNLISWFSKKQNCVSLSTAEAEYIAAGSSCSQLVWMKQMLKEYNVE 1492
Query: 1634 ANSIPIYCDNTAAICLSKNPILHSRAKHIEIKHHFIRDYVQKGIL 1678
+ + +YCDN +AI +SKNP+ H+R KHI+I+HH+IRD V I+
Sbjct: 1493 QDVMTLYCDNMSAINISKNPVQHNRTKHIDIRHHYIRDLVDDKII 1537
>gb|AAC64917.1| gag-pol polyprotein [Glycine max]
Length = 1550
Score = 1048 bits (2711), Expect = 0.0
Identities = 626/1653 (37%), Positives = 903/1653 (53%), Gaps = 165/1653 (9%)
Query: 49 MYSFIMGLDEELWDILEDGVDDLD-LDEEGAAIDR----RIHTPAQKKLYKKHHKIRGII 103
M +F+ LD W + G + LD EG + T + +L + K +
Sbjct: 1 MVAFLKSLDSRTWKAVIKGWEHPKMLDTEGKPTNELKPEEDWTKEEDELALGNSKALNAL 60
Query: 104 VASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYELFRMKDDESIEE 163
+ + + ++ + AK + L EG+ KVK ++ +L ++E +MK++E I E
Sbjct: 61 FNGVDKNIFRLINTCTVAKDAWEILKTTHEGTSKVKMSRLQLLATKFENLKMKEEECIHE 120
Query: 164 MYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAKDLNTLSVEDLVSS 223
+ + + L + V KILRSLP R+ KVTAIEEA+D+ + V++L+ S
Sbjct: 121 FHMNILEIANACTALGERMTDEKLVRKILRSLPKRFDMKVTAIEEAQDICNMRVDELIGS 180
Query: 224 LKVHEMSLNEHESSKKSKSIALPSKGKTSKSSKAYKASESEEESPDRDSDEDQSVKMAML 283
L+ E+ L++ + KKSK++A S E EE+ D D+DE + + +L
Sbjct: 181 LQTFELGLSDR-TEKKSKNLAFVSN------------DEGEEDEYDLDTDEGLTNAVVLL 227
Query: 284 SNK----LEYLARKQKKFLSK-----RGGYKNSKKEDQKG-------CFNCKKPGHFIAD 327
+ L + R+QK + R G + K+ D+K C C+ GH A+
Sbjct: 228 GKQFNKVLNRMDRRQKPHVRNIPFDIRKGSEYQKRSDEKPSHSKGIQCHGCEGYGHIKAE 287
Query: 328 CPDLQKEKSKSKSKKSSFSSSKFRKQIKKSLMATWEDLDSESGSDKEEAEDDTKAAMGLV 387
CP K++ K S S +D +SE SD + D A G
Sbjct: 288 CPTHLKKQRKGLSVCRS------------------DDTESEQESDSDR---DVNALTGRF 326
Query: 388 ATVSSEAVSEAESDSEDENEVYSKIPRQELVDSLKELLSLFEHRTNELTDLKEKYVDLMK 447
+ +ED ++ S+I EL S +EL E + LK+ +L
Sbjct: 327 ES------------AEDSSDTDSEITFDELAISYRELCIKSEKILQQEAQLKKVIANLEA 374
Query: 448 QQKSTLLELKASEEELKGFNLISTTYEDRLKILCQKLQEKCDKGSGNKHEIALDDFIMAG 507
++++ E+ + E+ N +K+L NK LD+ + G
Sbjct: 375 EKEAHEDEISELKGEIGFLNSKLENMTKSIKML-------------NKGSDLLDEVLQLG 421
Query: 508 IDRSKVASMIYSTYKNNGKGIGYSEEKSKEYSLKSYCDCIKDGLKSTFVP-EGTNAVTAV 566
+ N +G+G++ + + ++ + FVP + + T
Sbjct: 422 KN------------VGNQRGLGFNHKSAGRTTM------------TEFVPAKNSTGATMS 457
Query: 567 QSKPETSGSQAKITSKPENLKIKVMTKSDPKSQKIKILKRSEPVHQNLIRPESKIPKQKD 626
Q + G+Q K KS K + + + P
Sbjct: 458 QHRSRHHGTQQK--------------KSKRKKWRCHYCGKYGHIKPFCYHLHGH-PHHGT 502
Query: 627 QKNKAATASDKTIPKGVKPEVLNDQKPLSIHPKVCIRAREKQRSWYLDSGCSRHMTGEKA 686
Q + + G K + K +S+ +RA K+ WYLDSGCSRHMTG K
Sbjct: 503 QSSSS----------GRKMMWVPKHKTVSLVVHTSLRASAKE-DWYLDSGCSRHMTGVKE 551
Query: 687 LFLTLTMKDGGEVKFGGNQTGKIIGTGTIGNSSI-SINNVWLVDGLKHNLLSISQFCDNG 745
+ + V FG GKI G G + + + S+N V LV GL NL+SISQ CD G
Sbjct: 552 FLVNIEPCSTSYVTFGDGSKGKITGMGKLVHDGLPSLNKVLLVKGLTANLISISQLCDEG 611
Query: 746 YDVMFSKTNCTLVNKNDKSITFKGKRVENVYKINFSDLADQKVVCLLSMNDKKWVWHKRL 805
++V F+K+ C + N+ + + KG R ++ + CL S D+ +WH+R
Sbjct: 612 FNVNFTKSECLVTNEKSE-VLMKGSRSKDNCYLWTPQETSYSSTCLFSKEDEVKIWHQRF 670
Query: 806 GHANWRLISKISKLQLVKGLPNIDYHSDALCGACQKGKIVKSTFKSKDIVSTSRPLELLH 865
GH + R + KI V+G+PN+ +CG CQ GK VK + + +TSR LELLH
Sbjct: 671 GHLHLRGMKKIIDKGAVRGIPNLKIEEGRICGECQIGKQVKMSNQKLQHQTTSRVLELLH 730
Query: 866 IDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKFIKSKDYACEVFSSFCTQIQSEKELKIL 925
+DL GP+ SL +Y V+VDD+SR+TWV FI+ K EVF ++Q EK+ I
Sbjct: 731 MDLMGPMQVESLGRKRYAYVVVDDFSRFTWVNFIREKSDTFEVFKELSLRLQREKDCVIK 790
Query: 926 KVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEMARTMIHENNL 985
++RSDHG EFEN F FC GI HEFS+ TPQQNG+VERKNRTLQE AR M+H L
Sbjct: 791 RIRSDHGREFENSKFTEFCTSEGITHEFSAAITPQQNGIVERKNRTLQEAARVMLHAKEL 850
Query: 986 AKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNISYFHQFGCTCYILNTKDYLK 1045
+ WAEA+NT+CYI NR+ +R T YE++KGR+P + +FH G CYIL ++ +
Sbjct: 851 PYNLWAEAMNTACYIHNRVTLRRGTPTTLYEIWKGRKPTVKHFHICGSPCYILADREQRR 910
Query: 1046 KFDAKAQRGIFLGYSERSKAYRVYNSETHCVEESIHVKFDDREPGNKTPEQSESNAGTTD 1105
K D K+ GIFLGYS S+AYRV+NS T V ESI+V DD P K + +
Sbjct: 911 KMDPKSDAGIFLGYSTNSRAYRVFNSRTRTVMESINVVVDDLTPARKKDVEEDVR----- 965
Query: 1106 SEDASESDQPSDSEKHSEVESRPEAEITPEAESNSEAETSPVEQNENASEDVQDNTQQVI 1165
+ D +D+ K +E N+E N +++ D + Q
Sbjct: 966 ----TSGDNVADTAKSAE---------------NAE--------NSDSATDEPNINQPDK 998
Query: 1166 QPKFKHKSSHPEELIIGSKDSPRRTRSHFRQEESLIGLLSIIEPKTVEEALSDDGWILAM 1225
+P + + HP+ELIIG + TRS + S +S IEPK V+EAL+D+ WI AM
Sbjct: 999 RPSIRIQKMHPKELIIGDPNRGVTTRSREIEIISNSCFVSKIEPKNVKEALTDEFWINAM 1058
Query: 1226 QEELNQFQRNDVWDLIPKPFQKNIIGTKWVFRNKLNEKGEVTRNKARLVAQGYSQQEGID 1285
QEEL QF+RN+VW+L+P+P N+IGTKW+F+NK NE+G +TRNKARLVAQGY+Q EG+D
Sbjct: 1059 QEELEQFKRNEVWELVPRPEGTNVIGTKWIFKNKTNEEGVITRNKARLVAQGYTQIEGVD 1118
Query: 1286 YTETFAPVARLEAIRLLLSYAINHGIILYQMDVKSAFLNGVIEEEVYVKQPPGFEDIKHP 1345
+ ETFAP ARLE+IRLLL A LYQMDVKSAFLNG + EE YV+QP GF D HP
Sbjct: 1119 FDETFAPGARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEAYVEQPKGFVDPTHP 1178
Query: 1346 DHVYKLKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQVDTTLFGKTLKKDILIVQIYVDD 1405
DHVY+LKK+LYGLKQAPRAWY+RL+ FL + + +G +D TLF K ++++I QIYVDD
Sbjct: 1179 DHVYRLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQDAENLMIAQIYVDD 1238
Query: 1406 IIFGSTNASLCKEFSKLMQDEFEMSMMGELKFFLGIQINQSKEGVYVHQTKYTKELLKKF 1465
I+FG + + + F + MQ EFEMS++GEL +FLG+Q+ Q ++ +++ Q+KY K ++KKF
Sbjct: 1239 IVFGGMSNEMLRHFVQQMQSEFEMSLVGELTYFLGLQVKQMEDSIFLSQSKYAKNIVKKF 1298
Query: 1466 KLEDCKVMNTPMHPTCTLSKEDTGTVVDQKLYRGMIGSLLYLTASRPDILFSVCLCARFQ 1525
+E+ TP LSK++ GT VDQ LYR MIGSLLYLTASRPDI ++V CAR+Q
Sbjct: 1299 GMENASHKRTPAPTHLKLSKDEAGTSVDQSLYRSMIGSLLYLTASRPDITYAVGGCARYQ 1358
Query: 1526 SDPRESHLTAVKRIFRYLKGTTNLGLLYRKSLDYKLIGFCDADYAGDRIERKSTSGNCQF 1585
++P+ SHL VKRI +Y+ GT++ G++Y D L+G+CDAD+AG +RKST G C +
Sbjct: 1359 ANPKISHLNQVKRILKYVNGTSDYGIMYCHCSDSMLVGYCDADWAGSVDDRKSTFGGCFY 1418
Query: 1586 LGENLISWASKRQATIAMSTAEAEYISAASCCTQLLWMKHQLEDYQINANSIPIYCDNTA 1645
LG N ISW SK+Q +++STAEAEYI+A S C+QL+WMK L++Y + + + +YCDN +
Sbjct: 1419 LGTNFISWFSKKQNCVSLSTAEAEYIAAGSSCSQLVWMKQMLKEYNVEQDVMTLYCDNLS 1478
Query: 1646 AICLSKNPILHSRAKHIEIKHHFIRDYVQKGIL 1678
AI +SKNP+ HSR KHI+I+HH+IRD V ++
Sbjct: 1479 AINISKNPVQHSRTKHIDIRHHYIRDLVDDKVI 1511
>gb|AAN60991.1| Putative Zea mays retrotransposon Opie-2 [Oryza sativa (japonica
cultivar-group)] gi|34902324|ref|NP_912508.1| Putative
Zea mays retrotransposon Opie-2 [Oryza sativa (japonica
cultivar-group)]
Length = 2011
Score = 1041 bits (2692), Expect = 0.0
Identities = 636/1671 (38%), Positives = 911/1671 (54%), Gaps = 159/1671 (9%)
Query: 30 GKAPKFNGDPEEFSWWKTNMYSFIMGLDEELWDILEDGVDDLDLDEEGAAIDRRIHTPAQ 89
GKAP FNG +S WK M + + + +W I++ G AI T
Sbjct: 9 GKAPMFNGT--NYSTWKIKMSTHLKAMSFHIWSIVDVGF----------AITGTPLTEID 56
Query: 90 KKLYKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQ 149
+ K + + ++ S+ + E+ ++S+ TA ++ L E + + K+AK L Q
Sbjct: 57 HRNLKLNAQAMNVLFNSLSQEEFDRVSNLETAYEIWNKLAEIHESTSEYKDAKLHFLKIQ 116
Query: 150 YELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEA 209
YE F M ES+ +MY R +V+ L+ L +Y + K+LR+LP ++ VT + +
Sbjct: 117 YETFSMLPHESVNDMYGRLNVIVNDLKGLGANYTNLEVAQKMLRALPEKYETLVTMLINS 176
Query: 210 KDLNTLSVEDLVSSLKVHEMSLNEHESSKKSKSIALPSKGKTSKSSKAYKASESEEESPD 269
D++ ++ L+ + ++M ++ KK A PSK + ++ S+S+ +
Sbjct: 177 -DMSRMTPASLLGKINTNDM----YKLKKKEMEEASPSKKCIALQAEVEDKSKSKVNEVN 231
Query: 270 RDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGGYKNSKKEDQKG-------CFNCKKPG 322
+D +E+ + +L+ + L ++K+ RG N ++ + CF C
Sbjct: 232 KDLEEE----IVLLARRFNDLLGRRKE--RGRGSNSNRRRNRRPNKTLSNLRCFEC---- 281
Query: 323 HFIADCPDLQKEKSKSKSKKSSFSSSKFRKQIKKSLMATWEDLDSESGSDKEEAEDDTKA 382
D +E S S + K+ K +++A E A
Sbjct: 282 -------DSNEESSASSGSEEEGGDDASSKKKKMAVVAIKE-------------APSLFA 321
Query: 383 AMGLVATVSSEAVSEAESDSEDENEVYSKIPRQELVDSLKELLSLFEHRTNELTDLKEKY 442
+ L+A S+ S ++S+S+D+ + S EL+S+FE EL EK
Sbjct: 322 PLCLMAKSPSKVTSLSDSESDDDCDDVS----------YDELVSMFE----ELHAYSEKE 367
Query: 443 VDLMKQQKSTLLELKASEEELKGFNLISTTYEDRLKILCQKLQEKCDKGSGNKHEIALDD 502
+ K K L+ EELK T +RL I +KL+E D AL D
Sbjct: 368 IIKFKTLKKDHASLEVLYEELK-------TSHERLTISHEKLKEAHDNLLSTTQHGALID 420
Query: 503 FIMAGIDRSKVASMIYSTYKNNGKGIGYSEEKSKEYSLKSYCDCIKDGLKSTFVPEGTNA 562
+G S CD + D +++
Sbjct: 421 -------------------------VGIS------------CDLLDDSATCHIAHVASSS 443
Query: 563 VTA----VQSKPETSGSQAKITSKPENLKIKVMTKSDPKSQKIKILKRSEPVHQNLIRPE 618
++ + P +S S ++ +L V+ ++ K Q K+ K E + +
Sbjct: 444 ISTSCDDLMDMPNSSSSSC-VSICDASL---VVENNELKEQVAKLNKSLERCFKGKNTLD 499
Query: 619 SKIPKQKDQKNKAATASDKTIPKGVKPEVLNDQKPLSIHPKVCIRAREKQR--------- 669
+ +Q+ NK + KG KP + + + K C + RE
Sbjct: 500 KILSEQQCILNKEGLGF--ILKKGKKPSH-RATRFVKSNGKYCSKCREVGHLVNYRTGGS 556
Query: 670 SWYLDSGCSRHMTGEKALFLTLTM--KDGGEVKFGGNQTGKIIGTGTIGNSS-ISINNVW 726
W LDSGC++HMTG++A+F T + + +V FG N GK+IG G I S+ +SI+NV
Sbjct: 557 HWVLDSGCTQHMTGDRAMFTTFEVGGNEQEKVTFGDNSKGKVIGLGKIAISNDLSIDNVS 616
Query: 727 LVDGLKHNLLSISQFCDNGYDVMFSKTNCTLVNKNDKSITFKGKRVENVYKINFSDLADQ 786
LV L NLLS++Q CD F + + DKS FKG R N+Y +F+
Sbjct: 617 LVKSLNFNLLSVAQICDLVLSCAFFPQEVIVSSLLDKSCVFKGFRYGNLYLGDFNSSEAN 676
Query: 787 KVVCLLSMNDKKWVWHKRLGHANWRLISKISKLQLVKGLPNIDYHSDALCGACQKGKIVK 846
CL++ W+WH+RL H +SK+SK LV GL ++ + D LC ACQ GK V
Sbjct: 677 LKTCLVAKTSLGWLWHRRLAHVGMNQLSKLSKRDLVVGLKDVKFEKDKLCSACQAGKQVA 736
Query: 847 STFKSKDIVSTSRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKFIKSKDYAC 906
+ +K I+STSRPLELLH++ FGP S+ G+ + LVIVDDYSR+TW+ F+ K
Sbjct: 737 CSHPTKSIMSTSRPLELLHMEFFGPTTYKSIGGNSHCLVIVDDYSRYTWMFFLHDKSIVA 796
Query: 907 EVFSSFCTQIQSEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNGVVE 966
E+F F + Q+E ++K+RSD+G +F+N E +C+ GI HE S+ +PQQNGVVE
Sbjct: 797 ELFKKFAKRGQNEFNCTLVKIRSDNGSKFKNTNIEDYCDDLGIKHELSATYSPQQNGVVE 856
Query: 967 RKNRTLQEMARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNIS 1026
KNRTL EMARTM+ E ++ FWAEA+NT+C+ NR+Y+ +L+KT+YEL GR+PN++
Sbjct: 857 MKNRTLIEMARTMLDEYGVSDSFWAEAINTACHATNRLYLHRLLKKTSYELIVGRKPNVA 916
Query: 1027 YFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETHCVEESIHVKFDD 1086
YF FGC CYI L KF+++ G LGY+ SKAYRVYN VEE+ V+FD+
Sbjct: 917 YFRVFGCKCYIYRKGVRLTKFESRCDEGFLLGYASNSKAYRVYNKNKGIVEETADVQFDE 976
Query: 1087 REPGNKTPEQSESNAGTTDSEDASESDQPSDSEKHSEVESRPEAEITPEAESNSEAETSP 1146
G++ ++ + G A ++ D K EVE +P E +++ +
Sbjct: 977 TN-GSQEGHENLDDVGDEGLMRAMKNMSIGD-VKPIEVEDKPSTSTQDEPSTSATPSQAQ 1034
Query: 1147 VEQNENASEDVQDNTQQVIQPKFKHKSS--HPEELIIGSKDSPRRTRSHFRQEESLIGLL 1204
VE E ++D+ + P+ S HP + ++G +TRS +
Sbjct: 1035 VEVEEEKAQDLP------MPPRIHTALSKDHPIDQVLGDISKGVQTRSRVASICEHYSFV 1088
Query: 1205 SIIEPKTVEEALSDDGWILAMQEELNQFQRNDVWDLIPKPFQKNIIGTKWVFRNKLNEKG 1264
S +EPK V+EAL D WI AM +ELN F RN VW L+ + N+IGTKWVFRNK +E G
Sbjct: 1089 SCLEPKHVDEALCDPDWINAMHKELNNFARNKVWTLVERLRDHNVIGTKWVFRNKQDENG 1148
Query: 1265 EVTRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLLSYAINHGIILYQMDVKSAFLN 1324
V RNKAR VAQG++Q EG+D+ ETFAPV RLEAI +LL++A I L+QMDVKSAFLN
Sbjct: 1149 LVVRNKARFVAQGFTQVEGLDFGETFAPVTRLEAICILLAFASCFNIKLFQMDVKSAFLN 1208
Query: 1325 GVIEEEVYVKQPPGFEDIKHPDHVYKLKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQVD 1384
G I E V+V+QPPGFED K+P+HVYKL K+LYGLKQAPRAWY+RL +FL+ DF+ G+VD
Sbjct: 1209 GEIAELVFVEQPPGFEDPKYPNHVYKLSKALYGLKQAPRAWYERLRDFLLSKDFKIGKVD 1268
Query: 1385 TTLFGKTLKKDILIVQIYVDDIIFGSTNASLCKEFSKLMQDEFEMSMMGELKFFLGIQIN 1444
TTLF K + D + QIYVDDIIFG TN CKEF +M EFEMSM+GEL FF G+QI
Sbjct: 1269 TTLFTKIIGDDFFVCQIYVDDIIFGCTNEVFCKEFGDMMSREFEMSMIGELSFFHGLQIK 1328
Query: 1445 QSKEGVYVHQTKYTKELLKKFKLEDCKVMNTPMHPTCTLSKEDTGTVVDQKLYRGMIGSL 1504
Q K+G F LED K + TPM L ++ G VD KLYR MIGSL
Sbjct: 1329 QLKDGT--------------FGLEDAKPIKTPMATNGHLDLDEGGKPVDLKLYRSMIGSL 1374
Query: 1505 LYLTASRPDILFSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLYRKSLDYKLIGF 1564
LYLTASRPDI+FSVC+CARFQ+ P+E HL AVKRI RYLK ++ +GL Y K +KL+G+
Sbjct: 1375 LYLTASRPDIMFSVCMCARFQAAPKECHLVAVKRILRYLKHSSTIGLWYPKGAKFKLVGY 1434
Query: 1565 CDADYAGDRIERKSTSGNCQFLGENLISWASKRQATIAMSTAEAEYISAASCCTQLLWMK 1624
D+DYAG +++RKSTSG+CQ LG +L+SW+SK+Q +A+ AEAEY+SA SCC QLLWMK
Sbjct: 1435 SDSDYAGCKVDRKSTSGSCQMLGRSLVSWSSKKQNFVALFIAEAEYVSAGSCCAQLLWMK 1494
Query: 1625 HQLEDYQINANSIPIYCDNTAAICLSKNPILHSRAKHIEIKHHFIRDYVQK 1675
L DY I+ P+ C+N +AI ++ NP+ HSR KHI+I+HHF+RD+V K
Sbjct: 1495 QILLDYGISFTKTPLLCENDSAIKIANNPVQHSRTKHIDIRHHFLRDHVAK 1545
>gb|AAV31277.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
Length = 1577
Score = 1025 bits (2650), Expect = 0.0
Identities = 633/1664 (38%), Positives = 884/1664 (53%), Gaps = 156/1664 (9%)
Query: 30 GKAPKFNGDPEEFSWWKTNMYSFIMGLDEELWDILEDG--VDDLDLDEEGAAIDRR---I 84
GKAP FNG +S WK M + + + +W I++ G + L E ID R +
Sbjct: 9 GKAPMFNGT--NYSTWKIKMSTHLKAMSFHIWSIVDVGFAITGTPLME----IDHRNLQL 62
Query: 85 HTPAQKKLYKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKAL 144
+ A L+ S+ + E+ ++S+ TA ++ L EG+ + K+AK
Sbjct: 63 NAQAMNALFN-----------SLSQEEFDRVSNLETAYEIWNKLAEIHEGTSEYKDAKLH 111
Query: 145 MLVHQYELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVT 204
L QYE F M ES+ +MY R +V+ L+ L +Y + K+LR+LP ++ VT
Sbjct: 112 FLKIQYETFSMLPHESVNDMYGRLNVIVNDLKGLGANYTDLEVAQKMLRALPEKYETLVT 171
Query: 205 AIEEAKDLNTLSVEDLVSSLKVHEMSLNEHESSKKSKSIALPSKGKTSKSSKAYKASESE 264
+ + D++ ++ L+ + ++M +GK S S++
Sbjct: 172 MLINS-DMSRMTPASLLGKINTNDMR---------------KERGKGSNSNRRRN----- 210
Query: 265 EESPDRDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGGYKNSKKEDQKGCFNCKKPGHF 324
R+ K LS CF C + GHF
Sbjct: 211 ---------------------------RRPNKTLSNLR------------CFECGEKGHF 231
Query: 325 IADCPDLQKEKSKSKSKKSS-FSSSKFRKQIKKSLMATWEDLDSESGSDKEEAEDDTKAA 383
+ CP + KS KKS + K K+ K + A E+ DS S
Sbjct: 232 ASKCPSKDDDGDKSSKKKSGGYKLMKKLKKEGKKIEAFIEEWDSNEESSPHPGPRKKMVM 291
Query: 384 MGLVATVSSEAVSEAESDSEDENEV-YSKIPRQELVD-----SLKELLSLFEHRTNELTD 437
M + + V + K P E D S EL+S+FE EL
Sbjct: 292 MQAPGRRRWPLLPSRRLHHSSLHFVSWQKAPLSESDDDCDDVSYDELVSMFE----ELHA 347
Query: 438 LKEKYVDLMKQQKSTLLELKASEEELKGFNLISTTYEDRLKILCQKLQEKCDKGSGNKHE 497
EK + K K L+ EELK T +RL I +KL+E D
Sbjct: 348 YSEKEIVKFKALKKDHASLEVLYEELK-------TSHERLTISHEKLKEAHDNLLSTTQH 400
Query: 498 IALDDFIMAGIDRSKVASMIYSTYKNNGKGIGYSEEKSKEYSLKSYCDCIKDGLKSTFVP 557
AL D + D ++ + Y + S+ + CD + D S+
Sbjct: 401 GALID-VGISCDLLDDSATFHIAYVASS-------------SISTSCDDLVDMSSSS--- 443
Query: 558 EGTNAVTAVQSKPETSGSQAKITSKPENLKIKVMTKSDPKSQKIKILKRSEPVHQNLIRP 617
++ V+ + ++ K N +++ K KI +R + L
Sbjct: 444 -SSSCVSICDASLVVENNELKEQVAKLNKRLERCFKGKNTLDKILSEQRCILNKEGL--- 499
Query: 618 ESKIPKQKDQKNKAATASDKTIPKGVKPEVLNDQKPLSIHPKVCIRAREKQRSWYLDSGC 677
IPK+ + + AT K+ + + + D + + + W LDSGC
Sbjct: 500 -GFIPKKGKKPSHRATRFVKSNGRLMMVALFLD---MWVPLYSVVNYHTGGSHWVLDSGC 555
Query: 678 SRHMTGEKALFLTLTM--KDGGEVKFGGNQTGKIIGTGTIGNSS-ISINNVWLVDGLKHN 734
++HMTG++A+F T + + +V FG N GK+IG G I S+ +SI+NV LV L N
Sbjct: 556 TQHMTGDRAMFTTFEVGRNEQEKVTFGDNSKGKVIGLGKIAISNDLSIDNVSLVKSLNFN 615
Query: 735 LLSISQFCDNGYDVMFSKTNCTLVNKNDKSITFKGKRVENVYKINFSDLADQKVVCLLSM 794
LLS++Q CD F + + DKS FKG R N+Y ++F+ CL++
Sbjct: 616 LLSVAQICDLSLSCAFFPQEVIVSSLLDKSCVFKGFRYGNLYLVDFNSSEANLKTCLVAK 675
Query: 795 NDKKWVWHKRLGHANWRLISKISKLQLVKGLPNIDYHSDALCGACQKGKIVKSTFKSKDI 854
W+WH+RL H +SK SK LV GL ++ + D LC ACQ GK V + +K I
Sbjct: 676 TSLGWLWHRRLAHVGMNQLSKFSKRDLVMGLKDVKFEKDKLCSACQAGKQVACSHPTKSI 735
Query: 855 VSTSRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKFIKSKDYACEVFSSFCT 914
+STS+PLELLH+DLF P S+ G+ + LVIVDDYSR+TWV F+ K ++F F
Sbjct: 736 MSTSKPLELLHMDLFDPTTYKSIGGNSHCLVIVDDYSRYTWVFFLHDKSIVADLFKKFAK 795
Query: 915 QIQSEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNGVVERKNRTLQE 974
+ Q+E ++K+RS+ G EF+N E +C+ GI HE + +PQQNGVVERKNRTL E
Sbjct: 796 RAQNEFSCTLVKIRSNIGSEFKNTNIEDYCDDLGIKHELFATYSPQQNGVVERKNRTLIE 855
Query: 975 MARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNISYFHQFGCT 1034
MARTM+ E ++ FWAEA+NT+C+ NR+Y+ +L+KT+YE+ GR+PNI+YF FGC
Sbjct: 856 MARTMLDEYGVSDSFWAEAINTACHATNRLYLHRVLKKTSYEVIVGRKPNIAYFRVFGCK 915
Query: 1035 CYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETHCVEESIHVKFDDREPGNKTP 1094
CYI L KF+++ G LGY+ +SKAYRVYN VEE+ V+FD
Sbjct: 916 CYIHRKGVRLTKFESRCDEGFLLGYASKSKAYRVYNKNKGIVEETADVQFD--------- 966
Query: 1095 EQSESNAGTTDSEDASESDQPSDSEKHSEVESRP-EAEITPEAESNSEAETSPVEQNENA 1153
E + S G + +D + + S + +P E E P + E TS +
Sbjct: 967 ETNGSQEGHENLDDVGDEGLMRVMKNMSIGDVKPIEVEDKPSTSTQDEPSTSAMPSQAQV 1026
Query: 1154 SEDVQDNTQQVIQPKFKHKSS--HPEELIIGSKDSPRRTRSHFRQEESLIGLLSIIEPKT 1211
+ + + + P+ S HP + ++G +TRS +S +E K
Sbjct: 1027 EVEEEKAQEPPMPPRIHTALSKDHPIDQVLGDISKGVQTRSRVTSICEHYSFVSCLERKH 1086
Query: 1212 VEEALSDDGWILAMQEELNQFQRNDVWDLIPKPFQKNIIGTKWVFRNKLNEKGEVTRNKA 1271
V+EAL D W+ AM EEL F RN VW L+ +P N+IGTKWVFRNK +E G V RNKA
Sbjct: 1087 VDEALCDPDWMNAMHEELKNFARNKVWTLVERPRDHNVIGTKWVFRNKQDENGLVVRNKA 1146
Query: 1272 RLVAQGYSQQEGIDYTETFAPVARLEAIRLLLSYAINHGIILYQMDVKSAFLNGVIEEEV 1331
RLVAQG++Q EG+D+ ETFAPVARLEAI +LL++A I L+QMDVKSAFLN
Sbjct: 1147 RLVAQGFTQVEGLDFGETFAPVARLEAICILLAFASCFDIKLFQMDVKSAFLN------- 1199
Query: 1332 YVKQPPGFEDIKHPDHVYKLKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQVDTTLFGKT 1391
D K+P+HVYKL K+LYGL+QAPRAWY+RL +FL+ DF+ G+VD TLF K
Sbjct: 1200 ---------DTKYPNHVYKLSKALYGLRQAPRAWYERLRDFLLSKDFKIGKVDITLFTKI 1250
Query: 1392 LKKDILIVQIYVDDIIFGSTNASLCKEFSKLMQDEFEMSMMGELKFFLGIQINQSKEGVY 1451
+ D + QIYVDDIIFGSTN CKEF +M EFEMSM+GEL FFLG+QI Q K G +
Sbjct: 1251 IGDDFFVYQIYVDDIIFGSTNEVFCKEFGDMMSREFEMSMIGELSFFLGLQIKQLKNGTF 1310
Query: 1452 VHQTKYTKELLKKFKLEDCKVMNTPMHPTCTLSKEDTGTVVDQKLYRGMIGSLLYLTASR 1511
V QTKY K+LLK+F LED K + TPM L ++ G VD KLYR MIGSLLYLT SR
Sbjct: 1311 VSQTKYIKDLLKRFGLEDAKPIKTPMATNGHLDLDEGGKPVDLKLYRSMIGSLLYLTVSR 1370
Query: 1512 PDILFSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLYRKSLDYKLIGFCDADYAG 1571
PDI+FSVC+CARFQ+ P+E HL AVKRI RYLK ++ +GL Y K +KL+G+ D DYAG
Sbjct: 1371 PDIMFSVCMCARFQAAPKECHLVAVKRILRYLKHSSTIGLWYPKGAKFKLVGYSDPDYAG 1430
Query: 1572 DRIERKSTSGNCQFLGENLISWASKRQATIAMSTAEAEYISAASCCTQLLWMKHQLEDYQ 1631
+++RKSTS +CQ LG +L+SW+SK+Q ++A+STAE EY+SA SCC QLLWMK L DY
Sbjct: 1431 CKVDRKSTSSSCQMLGRSLVSWSSKKQNSVALSTAETEYVSAGSCCAQLLWMKQTLLDYG 1490
Query: 1632 INANSIPIYCDNTAAICLSKNPILHSRAKHIEIKHHFIRDYVQK 1675
I+ P+ CDN AI ++ NP+ HSR KHI+I+HHF+RD+V K
Sbjct: 1491 ISFTKTPLLCDNDGAIKIANNPVQHSRTKHIDIRHHFLRDHVAK 1534
>gb|AAC49502.1| Pol [Zea mays] gi|7489803|pir||T04112 pol protein homolog - maize
retrotransposon Opie-2
Length = 1068
Score = 1000 bits (2586), Expect = 0.0
Identities = 514/1026 (50%), Positives = 681/1026 (66%), Gaps = 28/1026 (2%)
Query: 670 SWYLDSGCSRHMTGEKALFLTLTMKDGGE--VKFGGNQTGKIIGTGTIGNSSI-SINNVW 726
SW +DSGC+ HMTGEK +F + + + FG GK+ G G I S+ SI+NV+
Sbjct: 10 SWIIDSGCTNHMTGEKKMFTSYVKNKDSQDSIIFGDGNQGKVKGLGKIAISNEHSISNVF 69
Query: 727 LVDGLKHNLLSISQFCDNGYDVMFSKTNCTLVNKNDKSITFKGKRVENVYKINFSDLADQ 786
LV+ L +NLLS+SQ C+ GY+ +F+ + ++ + D S+ FKG +Y ++F+
Sbjct: 70 LVESLGYNLLSVSQLCNMGYNCLFTNIDVSVFRRCDGSLAFKGVLDGKLYLVDFAKEEAG 129
Query: 787 KVVCLLSMNDKKWVWHKRLGHANWRLISKISKLQLVKGLPNIDYHSDALCGACQKGKIVK 846
CL++ W+WH+RL H + + K+ K + V GL N+ + D C ACQ GK V
Sbjct: 130 LDACLIAKTSMGWLWHRRLAHVGMKNLHKLLKGEHVIGLTNVQFEKDRPCAACQAGKQVG 189
Query: 847 STFKSKDIVSTSRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKFIKSKDYAC 906
+ +K++++TSRPLE+LH+DLFGPV S+ GSKYGLVIVDD+SR+TWV F++ K
Sbjct: 190 GSHHTKNVMTTSRPLEMLHMDLFGPVAYLSIGGSKYGLVIVDDFSRFTWVFFLQEKSETQ 249
Query: 907 EVFSSFCTQIQSEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNGVVE 966
F + Q+E ELK+ K+RSD+G EF+N E F E+ GI HEFS+P TPQQNGVVE
Sbjct: 250 GTLKRFLRRAQNEFELKVKKIRSDNGSEFKNLQVEEFLEEEGIKHEFSAPYTPQQNGVVE 309
Query: 967 RKNRTLQEMARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNIS 1026
RKNRTL +MARTM+ E + FW EAVNT+C+ NR+Y+ +L+ T+YEL G +PN+S
Sbjct: 310 RKNRTLIDMARTMLGEFKTPECFWTEAVNTACHAINRVYLHRILKNTSYELLTGNKPNVS 369
Query: 1027 YFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETHCVEESIHVKFDD 1086
YF FG CYIL K KF KA G LGY +KAYRV+N + VE S V FD+
Sbjct: 370 YFRVFGSKCYILVKKGRNSKFAPKAVEGFLLGYDSNTKAYRVFNKSSGLVEVSGDVVFDE 429
Query: 1087 REPGNKTPEQSESNAGTTDSEDASESDQPSDSEKHSEV-ESRPEAEITPEAESNSEAETS 1145
N +P + D +D E D P+ + + + E RP+ + E S S
Sbjct: 430 T---NGSPREQ-----VVDCDDVDEEDIPTAAIRTMAIGEVRPQEQDEREQPSPSTMVHP 481
Query: 1146 PVEQNENASED-------VQDN--TQQVIQP------KFKHKSSHPEELIIGSKDSPRRT 1190
P + +E + QD+ ++ QP + + HP + I+G T
Sbjct: 482 PTQDDEQVHQQEVCDQGGAQDDHVLEEEAQPAPPTQVRAMIQRDHPVDQILGDISKGVTT 541
Query: 1191 RSHFRQEESLIGLLSIIEPKTVEEALSDDGWILAMQEELNQFQRNDVWDLIPKPFQKNII 1250
RS +S IEP VEEAL D W+LAMQEELN F+RN+VW L+P+P Q N++
Sbjct: 542 RSRLVNFCEHNSFVSSIEPFRVEEALLDPDWVLAMQEELNNFKRNEVWTLVPRPKQ-NVV 600
Query: 1251 GTKWVFRNKLNEKGEVTRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLLSYAINHG 1310
GTKWVFRNK +E+G VTRNKARLVA+GY+Q G+D+ ETFAPVARLE+IR+LL+YA +H
Sbjct: 601 GTKWVFRNKQDERGVVTRNKARLVAKGYAQVAGLDFEETFAPVARLESIRILLAYAAHHS 660
Query: 1311 IILYQMDVKSAFLNGVIEEEVYVKQPPGFEDIKHPDHVYKLKKSLYGLKQAPRAWYDRLS 1370
LYQMDVKSAFLNG I+EEVYV+QPPGFED ++PDHV KL K+LYGLKQAPRAWY+ L
Sbjct: 661 FRLYQMDVKSAFLNGPIKEEVYVEQPPGFEDERYPDHVCKLSKALYGLKQAPRAWYECLR 720
Query: 1371 NFLIKNDFERGQVDTTLFGKTLKKDILIVQIYVDDIIFGSTNASLCKEFSKLMQDEFEMS 1430
+FLI N F+ G+ D TLF KT D+ + QIYVDDIIFGSTN C+EFS++M +FEMS
Sbjct: 721 DFLIANAFKVGKADPTLFTKTCDGDLFVCQIYVDDIIFGSTNQKSCEEFSRVMTQKFEMS 780
Query: 1431 MMGELKFFLGIQINQSKEGVYVHQTKYTKELLKKFKLEDCKVMNTPMHPTCTLSKEDTGT 1490
MMGEL +FLG Q+ Q K+G ++ QTKYT++LLK+F ++D K TPM G
Sbjct: 781 MMGELNYFLGFQVKQLKDGTFISQTKYTQDLLKRFGMKDAKPAKTPMGTDGHTDLNKGGK 840
Query: 1491 VVDQKLYRGMIGSLLYLTASRPDILFSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLG 1550
VDQK YR MIGSLLYL ASRPDI+ SVC+CARFQSDP+E HL AVKRI RYL T G
Sbjct: 841 SVDQKAYRSMIGSLLYLCASRPDIMLSVCMCARFQSDPKECHLVAVKRILRYLVATPCFG 900
Query: 1551 LLYRKSLDYKLIGFCDADYAGDRIERKSTSGNCQFLGENLISWASKRQATIAMSTAEAEY 1610
L Y K + L+G+ D+DYAG +++RKSTSG CQFLG +L+SW SK+Q ++A+STAEAEY
Sbjct: 901 LWYPKGSTFDLVGYSDSDYAGCKVDRKSTSGTCQFLGRSLVSWNSKKQTSVALSTAEAEY 960
Query: 1611 ISAASCCTQLLWMKHQLEDYQINANSIPIYCDNTAAICLSKNPILHSRAKHIEIKHHFIR 1670
++A CC QLLWM+ L D+ N + +P+ CDN +AI +++NP+ HSR KHI+I+HHF+R
Sbjct: 961 VAAGQCCAQLLWMRQTLRDFGYNLSKVPLLCDNESAIRMAENPVEHSRTKHIDIRHHFLR 1020
Query: 1671 DYVQKG 1676
D+ QKG
Sbjct: 1021 DHQQKG 1026
>gb|AAD20307.1| copia-type pol polyprotein [Zea mays]
Length = 1063
Score = 999 bits (2583), Expect = 0.0
Identities = 522/1028 (50%), Positives = 678/1028 (65%), Gaps = 40/1028 (3%)
Query: 681 MTGEKALFLTLTMKDGGE--VKFGGNQTGKIIGTGTIGNS-SISINNVWLVDGLKHNLLS 737
MTGEK +F + + + FG G + G G I S SI+NV+LVD L +NLLS
Sbjct: 1 MTGEKRMFSSYEKNQDPQRAITFGDGNQGLVKGLGKIAISPDHSISNVFLVDSLDYNLLS 60
Query: 738 ISQFCDNGYDVMFSKTNCTLVNKNDKSITFKGKRVENVYKINFSDLADQKVVCLLSMNDK 797
+SQ C GY+ +F+ T+ ++D SI FKG +Y ++F D A+ CL++ +
Sbjct: 61 VSQLCQMGYNCLFTDIGVTVFRRSDDSIAFKGVLEGQLYLVDF-DRAELDT-CLIAKTNM 118
Query: 798 KWVWHKRLGHANWRLISKISKLQLVKGLPNIDYHSDALCGACQKGKIVKSTFKSKDIVST 857
W+WH+RL H + + K+ K + + GL N+ + D +C ACQ GK V + K+I++T
Sbjct: 119 GWLWHRRLAHVGMKNLHKLLKGEHILGLTNVHFEKDRICSACQAGKQVGTHHPHKNIMTT 178
Query: 858 SRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKFIKSKDYACEVFSSFCTQIQ 917
RPLELLH+DLFGP+ S+ GSKY LVIVDDYSR+TWV F++ K E F + Q
Sbjct: 179 DRPLELLHMDLFGPIAYISIGGSKYCLVIVDDYSRFTWVFFLQEKSQTQETLKGFLRRAQ 238
Query: 918 SEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEMAR 977
+E L+I K+RSD+G EF+N E F E+ GI HEFSSP TPQQNGVVERKNRTL +MAR
Sbjct: 239 NEFGLRIKKIRSDNGTEFKNSQIESFLEEEGIKHEFSSPYTPQQNGVVERKNRTLLDMAR 298
Query: 978 TMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNISYFHQFGCTCYI 1037
TM+ E FWAEAVNT+CY NR+Y+ +L+KT+YEL G++PNISYF FG C+I
Sbjct: 299 TMLDEYKTPDRFWAEAVNTACYAINRLYLHRILKKTSYELLTGKKPNISYFRVFGSKCFI 358
Query: 1038 LNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETHCVEESIHVKFDDREPGNKTPEQS 1097
L + KF K G LGY ++AYRV+N T VE S V FD+ G++ +
Sbjct: 359 LIKRGRKSKFAPKTVEGFLLGYDSNTRAYRVFNKSTGLVEVSCDVVFDETN-GSQVEQVD 417
Query: 1098 ESNAGTTDS-------------------EDASESDQPSDSEKHSEVESRPEAEITPEAES 1138
G + E S DQPS S + S ++ E E + E
Sbjct: 418 LDEIGEEQAPCIALRNMSIGDVCPKESEEPPSTQDQPSSSMQASP-PTQNEDEAQNDEEQ 476
Query: 1139 NSEAETSPVEQNENASEDVQDNTQQVIQPKFKH-------KSSHPEELIIGSKDSPRRTR 1191
N E E + N+ + + +P+ H + HP + I+G TR
Sbjct: 477 NQEDEPPQDDSNDQGGDTNDQEKEDEEEPRPPHPRVHQAIQRDHPVDTILGDIHKGVTTR 536
Query: 1192 S---HFRQEESLIGLLSIIEPKTVEEALSDDGWILAMQEELNQFQRNDVWDLIPKPFQKN 1248
S HF + S + S IEP VEEAL D W++AMQEELN F RN+VW L+P+P Q N
Sbjct: 537 SRVAHFCEHYSFV---SSIEPHRVEEALQDSDWVVAMQEELNNFTRNEVWHLVPRPNQ-N 592
Query: 1249 IIGTKWVFRNKLNEKGEVTRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLLSYAIN 1308
++GTKWVFRNK +E G VTRNKARLVA+GYSQ EG+D+ ET+APVARLE+IR+LL+YA
Sbjct: 593 VVGTKWVFRNKQDEHGVVTRNKARLVAKGYSQVEGLDFGETYAPVARLESIRILLAYATY 652
Query: 1309 HGIILYQMDVKSAFLNGVIEEEVYVKQPPGFEDIKHPDHVYKLKKSLYGLKQAPRAWYDR 1368
HG LYQMDVKSAFLNG I+EEVYV+QPPGFED ++P+HVY+L K+LYGLKQAPRAWY+
Sbjct: 653 HGFKLYQMDVKSAFLNGPIKEEVYVEQPPGFEDSEYPNHVYRLSKALYGLKQAPRAWYEC 712
Query: 1369 LSNFLIKNDFERGQVDTTLFGKTLKKDILIVQIYVDDIIFGSTNASLCKEFSKLMQDEFE 1428
L +FLI N F+ G+ D TLF KTL+ D+ + QIYVDDIIFGSTN S C+EFS++M +FE
Sbjct: 713 LRDFLIANGFKVGKADPTLFTKTLENDLFVCQIYVDDIIFGSTNKSTCEEFSRIMTQKFE 772
Query: 1429 MSMMGELKFFLGIQINQSKEGVYVHQTKYTKELLKKFKLEDCKVMNTPMHPTCTLSKEDT 1488
MSMMGELK+FLG Q+ Q +EG ++ QTKYT+++L KF ++D K + TPM L +
Sbjct: 773 MSMMGELKYFLGFQVKQLQEGTFICQTKYTQDILTKFGMKDAKPIKTPMGTNGHLDLDTG 832
Query: 1489 GTVVDQKLYRGMIGSLLYLTASRPDILFSVCLCARFQSDPRESHLTAVKRIFRYLKGTTN 1548
G VDQK+YR MIGSLLYL ASRPDI+ SVC+CARFQSDP+ESHLTAVKRI RYL T
Sbjct: 833 GKSVDQKVYRSMIGSLLYLCASRPDIMLSVCMCARFQSDPKESHLTAVKRILRYLAYTPK 892
Query: 1549 LGLLYRKSLDYKLIGFCDADYAGDRIERKSTSGNCQFLGENLISWASKRQATIAMSTAEA 1608
GL Y + + LIG+ DAD+AG +I RKSTSG CQFLG +L+SWASK+Q ++A+STAEA
Sbjct: 893 FGLWYPRGSTFDLIGYSDADWAGCKINRKSTSGTCQFLGRSLVSWASKKQNSVALSTAEA 952
Query: 1609 EYISAASCCTQLLWMKHQLEDYQINANSIPIYCDNTAAICLSKNPILHSRAKHIEIKHHF 1668
EYI+A CC QLLWM+ L DY +P+ CDN +AI ++ NP+ HSR KHI I++HF
Sbjct: 953 EYIAAGHCCAQLLWMRQTLLDYGYKLTKVPLLCDNESAIKMADNPVEHSRTKHIAIRYHF 1012
Query: 1669 IRDYVQKG 1676
+RD+ QKG
Sbjct: 1013 LRDHQQKG 1020
>ref|NP_910082.1| putative gag-pol polyprotein [Oryza sativa (japonica cultivar-group)]
gi|28269414|gb|AAO37957.1| putative gag-pol polyprotein
[Oryza sativa (japonica cultivar-group)]
Length = 1969
Score = 984 bits (2544), Expect = 0.0
Identities = 558/1286 (43%), Positives = 768/1286 (59%), Gaps = 81/1286 (6%)
Query: 434 ELTDLKEKYV-----------------DLMKQQ------KSTLLELKASEEELKGF---- 466
EL +LKEK+V D ++QQ K+ LLE A++E +
Sbjct: 510 ELKELKEKFVHDRIGRCENCPILTSDNDELRQQVAMLKTKNDLLESFATKEPIHSSCANC 569
Query: 467 NLISTTYEDRLKIL--------CQK-LQEKCDKGSGNKHEIALDDFIMAGIDRSKVASMI 517
++ T +D ++ C + K D S K L + K +MI
Sbjct: 570 AILETELKDAKTVIDSIKSIDSCSSCISLKVDLESAKKENSYLQQSLERFAQGKKKLNMI 629
Query: 518 YSTYKN--NGKGIGYSEEKSKEYSLKSYCDCIKDGLKSTFVPEGTNAVTAVQSKPETSGS 575
K N +GIGY + + ++ G + T+ + + KP T S
Sbjct: 630 LDQSKVSINNQGIGYD-----------FAESLRIGTHE--ILGVTDGMIELAQKPITFKS 676
Query: 576 QAKITSKPENLKIKVMTKSDPKSQKIKILKRSEPVHQNLIRPESKIPKQKDQKNKAATAS 635
I N S+PK + + +S+PV P K PKQ + K
Sbjct: 677 AGFIG----NTSSSTPKTSEPKM--VPMTSKSKPVEL----PRPKNPKQVEHKQNQRQT- 725
Query: 636 DKTIPKGVKPEVLNDQKPLSIHPKVCIRAREKQRSWYLDSGCSRHMTGEKALFLTLTMKD 695
K + K K E K + V + SW +DSGCSRHMTGE F +LT
Sbjct: 726 -KPVEK-TKYECTYCGKAGHLDFGV-----GRSNSWLVDSGCSRHMTGEAKWFTSLTRAS 778
Query: 696 GGE-VKFGGNQTGKIIGTGTIG-NSSISINNVWLVDGLKHNLLSISQFCDNGYDVMFSKT 753
G E + FG +G+++ GTI N + +V LV LK+NLLS+SQ CD +V F K
Sbjct: 779 GDETITFGDASSGRVMAKGTIKVNDKFMLKDVALVSKLKYNLLSVSQLCDENLEVRFKKD 838
Query: 754 NCTLVNKNDKSITFKGKRVENVYKINFSDLADQKVVCLL-SMNDKKWVWHKRLGHANWRL 812
+++ ++ + F RV V+ NF A CL+ S N + WH+RLGH +
Sbjct: 839 RSRVLDASESPV-FDISRVGRVFFANFDSSAPGPSRCLIASENRDLFFWHRRLGHIGFDH 897
Query: 813 ISKISKLQLVKGLPNIDYHSDALCGACQKGKIVKSTFKSKDIVSTSRPLELLHIDLFGPV 872
+S+IS + L++GLP + D +C C+ GK+ S+ K +V T P +LLH+D GP
Sbjct: 898 LSRISGMDLIRGLPKLKVQKDLVCAPCRHGKMTSSSHKPVTMVMTDGPGQLLHMDTVGPA 957
Query: 873 NTASLYGSKYGLVIVDDYSRWTWVKFIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHG 932
S+ G Y LV+VDD+SR++WV F++SK+ F S + E + +RSD+G
Sbjct: 958 RVQSVGGKWYVLVVVDDFSRYSWVYFLESKEETFGFFQSLARSLALEFPGALRAIRSDNG 1017
Query: 933 GEFENEPFELFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAE 992
EF+N FE FC+ G+ H+FSSP PQQNGVVERKNRTL EMARTM+ E + FW E
Sbjct: 1018 SEFKNSAFESFCDSSGVEHQFSSPYVPQQNGVVERKNRTLVEMARTMLDEFTTPRKFWTE 1077
Query: 993 AVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNISYFHQFGCTCYILNTKDYLKKFDAKAQ 1052
A++ +C+I NR+++R +L KT YEL GRRP +S+ FGC C++L + + L KF++++
Sbjct: 1078 AISAACFISNRVFLRTILHKTPYELRFGRRPKVSHLRVFGCKCFVLKSGN-LDKFESRSL 1136
Query: 1053 RGIFLGYSERSKAYRVYNSETHCVEESIHVKFDDREPGNKTPEQSESNAGTTDSEDASES 1112
GIFLGY+ S+AYRVY T+ + E+ V FD+ PG + + ED+ +
Sbjct: 1137 DGIFLGYATHSRAYRVYVLSTNKIVETCEVTFDEASPGARPEISGVPDESIFVDEDSDDD 1196
Query: 1113 DQPSDSEKHSEVESRPEAEITPEAESNSEAETSPVEQNENASEDVQDNTQQVIQPKFKHK 1172
D D ++S P + T + S + +P + +A+E++ T P+
Sbjct: 1197 D---DDSIPPPLDSTPPVQETGSPSTTSPSGDAPTTSS-SAAEEIDGGTSGPTAPRHIQN 1252
Query: 1173 SSHPEELIIGSKDSPRRTRSHFRQEESLIGLLSIIEPKTVEEALSDDGWILAMQEELNQF 1232
P+ +I G + R RS+ + + + EPK V ALSD+ W+ AM EEL F
Sbjct: 1253 RHPPDSMIGGLGERVTRNRSYELVNSAFV---ASFEPKNVCHALSDENWVNAMHEELENF 1309
Query: 1233 QRNDVWDLIPKPFQKNIIGTKWVFRNKLNEKGEVTRNKARLVAQGYSQQEGIDYTETFAP 1292
+RN VW L+ P N+IGTKWVF+NKL E G + RNKARLVAQG++Q EG+D+ ETFAP
Sbjct: 1310 ERNKVWSLVEPPLGFNVIGTKWVFKNKLGEDGSIVRNKARLVAQGFTQVEGLDFEETFAP 1369
Query: 1293 VARLEAIRLLLSYAINHGIILYQMDVKSAFLNGVIEEEVYVKQPPGFEDIKHPDHVYKLK 1352
VARLEAIR+LL++A + G L+QMDVKSAFLNGVIEEEVYVKQPPGFE+ K P+HV+KL+
Sbjct: 1370 VARLEAIRILLAFAASKGFKLFQMDVKSAFLNGVIEEEVYVKQPPGFENPKFPNHVFKLE 1429
Query: 1353 KSLYGLKQAPRAWYDRLSNFLIKNDFERGQVDTTLFGKTLKKDILIVQIYVDDIIFGSTN 1412
K+LYGLKQAPRAWY+RL FL++N FE G VD TLF D L+VQIYVDDIIFG ++
Sbjct: 1430 KALYGLKQAPRAWYERLKTFLLQNGFEMGAVDKTLFTLHSGIDFLLVQIYVDDIIFGGSS 1489
Query: 1413 ASLCKEFSKLMQDEFEMSMMGELKFFLGIQINQSKEGVYVHQTKYTKELLKKFKLEDCKV 1472
+L +FS +M EFEMSMMGEL FFLG+QI Q+KEG++VHQTKY+KELLKKF + DCK
Sbjct: 1490 HALVAQFSDVMSREFEMSMMGELTFFLGLQIKQTKEGIFVHQTKYSKELLKKFDMADCKP 1549
Query: 1473 MNTPMHPTCTLSKEDTGTVVDQKLYRGMIGSLLYLTASRPDILFSVCLCARFQSDPRESH 1532
+ TPM T +L ++ G VDQ+ YR MIGSLLYLTASRPDI FSVCLCARFQ+ PR SH
Sbjct: 1550 IATPMATTSSLGPDEDGEEVDQREYRSMIGSLLYLTASRPDIHFSVCLCARFQASPRTSH 1609
Query: 1533 LTAVKRIFRYLKGTTNLGLLYRKSLDYKLIGFCDADYAGDRIERKSTSGNCQFLGENLIS 1592
AVKRIFRY+K T G+ Y S + F DAD+AG +I+RKSTSG C FLG +L+S
Sbjct: 1610 RQAVKRIFRYIKSTLEYGIWYSCSSALSVRAFSDADFAGCKIDRKSTSGTCHFLGTSLVS 1669
Query: 1593 WASKRQATIAMSTAEAEYISAASCCTQLLWMKHQLEDYQINANSIPIYCDNTAAICLSKN 1652
W+S++Q+++A STAEAEY++AAS C+Q+LWM L+DY ++ + +P+ CDNT+AI ++KN
Sbjct: 1670 WSSRKQSSVAQSTAEAEYVAAASACSQVLWMISTLKDYGLSFSGVPLLCDNTSAINIAKN 1729
Query: 1653 PILHSRAKHIEIKHHFIRDYVQKGIL 1678
P+ HSR KHIEI++HF+RD V+KG +
Sbjct: 1730 PVQHSRTKHIEIRYHFLRDNVEKGTI 1755
Score = 122 bits (307), Expect = 8e-26
Identities = 118/498 (23%), Positives = 210/498 (41%), Gaps = 53/498 (10%)
Query: 32 APKFNGDPEEFSWWKTNMYSFIMGLDEELWDILEDGV---DDLDLDEEGAAIDRRIHTPA 88
A F D F WK M +++ +W+ ++ DD D+ TPA
Sbjct: 7 AKPFVFDGHNFVIWKARMEAYLQSQGHNVWNKVKSPYTVPDDADI------------TPA 54
Query: 89 QKKLYKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVH 148
+++ R I+ I E+ ++ +A M+ +LC EG+ ++ +
Sbjct: 55 NMAQVDFNYRARNAIIGGISSGEFNRVQHHKSAHDMWTALCNFHEGNNDIQLVRQNQFHK 114
Query: 149 QYELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLP-SRWRPKVTAIE 207
+Y+ F M ESI+ + RF ++S L+ + K + +D+ +L L W KVT+I
Sbjct: 115 EYQRFEMHPGESIDSYFKRFGEIISKLRSVGKEFSDNDNARHLLNCLDYGVWEMKVTSIT 174
Query: 208 EAKDLNTLSVEDLVSSLKVHEMSLNEHESSKKSKSIALPSKGKTSKSSKAYKASESEEES 267
E+ L+ L+++ L S LK HEM + + K S ++ G TS + A+ +
Sbjct: 175 ESAPLSDLTMDKLYSKLKTHEMDVFHRKGLKHSMALVADPSGSTSSNDSAF-VYGGFSLA 233
Query: 268 PDRDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGGYKNSKKEDQKGCFNCKKPGHFIAD 327
E+Q K+ + L ARK + K K + CF C +P H +
Sbjct: 234 ALHSVTEEQLEKIP--EDDLALFARKFSRAYKNVRDRKRGKTNEPFVCFECGEPNHKRVN 291
Query: 328 CPDLQKEKSKSKSKKSSFSSSKFRKQIKK----SLMATWEDLD-SESGSDKEEAEDDTKA 382
CP L K+KS +KK K +K + K ++A E++ S+ SD ++ E K
Sbjct: 292 CPKL-KKKSDKTTKKPEGQGRKGKKDLMKKAIHKVLAALEEVQLSDIDSDDDDQEKGDKD 350
Query: 383 AMGLVATVSSE--------AVSEAESDSEDENEVYSKIPRQELV---------DSLKELL 425
G+ ++E A+ + + SE IP + DS+ + L
Sbjct: 351 FSGMCCLANNEDFINLCLMALEDKDDSSEHPEVCLDDIPSLDGSLCDDSCSDNDSVDDEL 410
Query: 426 SLFEHRTNELTDLKEKY------VDLMKQQKSTL-LELKASEEELKGFNLIST--TYEDR 476
S E + + ++ +KY ++ +K + + LE+ + + ST +Y
Sbjct: 411 SK-ERMAHLMIEISDKYRSSKYKIEKLKSENDGMALEIARLRSMIPEEDTCSTCASYLSE 469
Query: 477 LKILCQKLQEKCDKGSGN 494
+ +L KL + C G+GN
Sbjct: 470 INLLKDKL-KSCALGAGN 486
>gb|AAN60494.1| Putative Zea mays retrotransposon Opie-2 [Oryza sativa (japonica
cultivar-group)] gi|34902378|ref|NP_912535.1| Putative
Zea mays retrotransposon Opie-2 [Oryza sativa (japonica
cultivar-group)]
Length = 2145
Score = 980 bits (2533), Expect = 0.0
Identities = 556/1286 (43%), Positives = 767/1286 (59%), Gaps = 81/1286 (6%)
Query: 434 ELTDLKEKYV-----------------DLMKQQ------KSTLLELKASEEELKGF---- 466
EL +LKEK+V D ++QQ K+ LLE A++E +
Sbjct: 510 ELKELKEKFVHDRIGRCENCPILTSDNDELRQQVAMLRTKNDLLESFATKEPIHSSCANC 569
Query: 467 NLISTTYEDRLKIL--------CQK-LQEKCDKGSGNKHEIALDDFIMAGIDRSKVASMI 517
++ T +D ++ C + K D S K L + K +MI
Sbjct: 570 AILETELKDAKTVIDSIKSIDSCSSCISLKVDLESAKKENSYLQQSLERFAQGKKKLNMI 629
Query: 518 YSTYKN--NGKGIGYSEEKSKEYSLKSYCDCIKDGLKSTFVPEGTNAVTAVQSKPETSGS 575
K N +GIGY + + ++ G + T+ + + KP T S
Sbjct: 630 LDQSKVSINNQGIGYD-----------FAESLRIGTHE--ILGVTDGMIELAQKPITFKS 676
Query: 576 QAKITSKPENLKIKVMTKSDPKSQKIKILKRSEPVHQNLIRPESKIPKQKDQKNKAATAS 635
I N S+PK + + +S+PV P K PKQ + K
Sbjct: 677 AGFIG----NTSSSTPKTSEPKM--VPMTSKSKPVEL----PRPKNPKQVEHKQNQRQT- 725
Query: 636 DKTIPKGVKPEVLNDQKPLSIHPKVCIRAREKQRSWYLDSGCSRHMTGEKALFLTLTMKD 695
K + K K E K + V + SW +DSGCSRHMTGE F +LT
Sbjct: 726 -KPVEK-TKYECTYCGKAGHLDFGV-----GRSNSWLVDSGCSRHMTGEAKWFTSLTRAS 778
Query: 696 GGE-VKFGGNQTGKIIGTGTIG-NSSISINNVWLVDGLKHNLLSISQFCDNGYDVMFSKT 753
E + FG +G+++ GTI N + +V LV LK+NLLS+SQ CD +V F K
Sbjct: 779 SDETITFGDASSGRVMAKGTIKVNDKFMLKDVALVSKLKYNLLSVSQLCDENLEVRFKKD 838
Query: 754 NCTLVNKNDKSITFKGKRVENVYKINFSDLADQKVVCLL-SMNDKKWVWHKRLGHANWRL 812
+++ ++ + F RV V+ NF A CL+ S N + WH+RLGH +
Sbjct: 839 RSRVLDASESPV-FDISRVGRVFFANFDSSAPGPSRCLIASENRDLFFWHRRLGHIGFDH 897
Query: 813 ISKISKLQLVKGLPNIDYHSDALCGACQKGKIVKSTFKSKDIVSTSRPLELLHIDLFGPV 872
+S+IS + L++GLP + D +C C+ GK+ S+ K +V T P +LLH+D GP
Sbjct: 898 LSRISGMDLIRGLPKLKVPKDLVCAPCRHGKMTSSSHKPVTMVMTDGPGQLLHMDTVGPA 957
Query: 873 NTASLYGSKYGLVIVDDYSRWTWVKFIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHG 932
S+ G Y LV+VDD+SR++WV F++SK+ F S + E + +RSD+G
Sbjct: 958 RVQSVGGKWYVLVVVDDFSRYSWVYFLESKEETFGFFQSLARSLALEFPGALRAIRSDNG 1017
Query: 933 GEFENEPFELFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAE 992
EF+N FE FC+ G+ H+FSSP PQQNGVVERKNRTL EMARTM+ E + FW E
Sbjct: 1018 SEFKNSAFESFCDSSGVEHQFSSPYVPQQNGVVERKNRTLVEMARTMLDEFTTPRKFWTE 1077
Query: 993 AVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNISYFHQFGCTCYILNTKDYLKKFDAKAQ 1052
A++ +C+I NR+++R +L KT YEL GRRP +S+ FGC C++L + + L KF++++
Sbjct: 1078 AISAACFISNRVFLRTILHKTPYELRFGRRPKVSHLRVFGCKCFVLKSGN-LDKFESRSL 1136
Query: 1053 RGIFLGYSERSKAYRVYNSETHCVEESIHVKFDDREPGNKTPEQSESNAGTTDSEDASES 1112
GIFLGY+ S+AYRVY T+ + E+ V FD+ PG + + ED+ +
Sbjct: 1137 DGIFLGYATHSRAYRVYVLSTNKIVETCEVTFDEASPGARPEISGVPDESIFVDEDSDDD 1196
Query: 1113 DQPSDSEKHSEVESRPEAEITPEAESNSEAETSPVEQNENASEDVQDNTQQVIQPKFKHK 1172
D D ++S P + T + S + +P + +A+E++ T P+
Sbjct: 1197 D---DDSIPPPLDSTPPVQETGSPSTTSPSGDAPTTSS-SAAEEIDGGTSGPTAPRHIQN 1252
Query: 1173 SSHPEELIIGSKDSPRRTRSHFRQEESLIGLLSIIEPKTVEEALSDDGWILAMQEELNQF 1232
P+ +I G + R RS+ + + + EPK V ALSD+ W+ AM EEL F
Sbjct: 1253 RHPPDSMIGGLGERVTRNRSYELVNSAFV---ASFEPKNVCHALSDENWVNAMHEELENF 1309
Query: 1233 QRNDVWDLIPKPFQKNIIGTKWVFRNKLNEKGEVTRNKARLVAQGYSQQEGIDYTETFAP 1292
+RN VW L+ P N+IGTKWVF+NKL E G + RNKARLVAQG++Q EG+D+ ETFAP
Sbjct: 1310 ERNKVWSLVEPPLGFNVIGTKWVFKNKLGEDGSIVRNKARLVAQGFTQVEGLDFEETFAP 1369
Query: 1293 VARLEAIRLLLSYAINHGIILYQMDVKSAFLNGVIEEEVYVKQPPGFEDIKHPDHVYKLK 1352
VARLEAIR+LL++A + G L+QMDVKSAFLNGVIEEEVYVKQPPGFE+ K P+HV+KL+
Sbjct: 1370 VARLEAIRILLAFAASKGFKLFQMDVKSAFLNGVIEEEVYVKQPPGFENPKFPNHVFKLE 1429
Query: 1353 KSLYGLKQAPRAWYDRLSNFLIKNDFERGQVDTTLFGKTLKKDILIVQIYVDDIIFGSTN 1412
K+LYGLKQAPRAWY+RL FL++N FE G VD TLF D L+VQIYVDDIIFG ++
Sbjct: 1430 KALYGLKQAPRAWYERLKTFLLQNGFEMGAVDKTLFTLHSGIDFLLVQIYVDDIIFGGSS 1489
Query: 1413 ASLCKEFSKLMQDEFEMSMMGELKFFLGIQINQSKEGVYVHQTKYTKELLKKFKLEDCKV 1472
+L +FS +M EFEMSMMGEL FFLG+QI Q+KEG++VHQTKY+KELLKKF + DCK
Sbjct: 1490 HALVAQFSDVMSREFEMSMMGELTFFLGLQIKQTKEGIFVHQTKYSKELLKKFDMADCKP 1549
Query: 1473 MNTPMHPTCTLSKEDTGTVVDQKLYRGMIGSLLYLTASRPDILFSVCLCARFQSDPRESH 1532
+ TPM T +L ++ G VDQ+ YR MIGSLLYLTASRPDI FSVCLCARFQ+ PR SH
Sbjct: 1550 IATPMATTSSLGPDEDGEEVDQREYRSMIGSLLYLTASRPDIHFSVCLCARFQASPRTSH 1609
Query: 1533 LTAVKRIFRYLKGTTNLGLLYRKSLDYKLIGFCDADYAGDRIERKSTSGNCQFLGENLIS 1592
AVKR+FRY+K T G+ Y S + F DAD+AG +I+RKSTSG C FLG +L+S
Sbjct: 1610 RQAVKRMFRYIKSTLEYGIWYSCSSALSVRAFSDADFAGCKIDRKSTSGTCHFLGTSLVS 1669
Query: 1593 WASKRQATIAMSTAEAEYISAASCCTQLLWMKHQLEDYQINANSIPIYCDNTAAICLSKN 1652
W+S++Q+++A STAEAEY++AAS C+Q+LWM L+DY ++ + +P+ CDNT+AI ++KN
Sbjct: 1670 WSSRKQSSVAQSTAEAEYVAAASACSQVLWMISTLKDYGLSFSGVPLLCDNTSAINIAKN 1729
Query: 1653 PILHSRAKHIEIKHHFIRDYVQKGIL 1678
P+ HSR KHIEI++HF+RD V+KG +
Sbjct: 1730 PVQHSRTKHIEIRYHFLRDNVEKGTI 1755
Score = 122 bits (307), Expect = 8e-26
Identities = 118/498 (23%), Positives = 210/498 (41%), Gaps = 53/498 (10%)
Query: 32 APKFNGDPEEFSWWKTNMYSFIMGLDEELWDILEDGV---DDLDLDEEGAAIDRRIHTPA 88
A F D F WK M +++ +W+ ++ DD D+ TPA
Sbjct: 7 AKPFVFDGHNFVIWKARMEAYLQSQGHNVWNKVKSPYTVPDDADI------------TPA 54
Query: 89 QKKLYKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVH 148
+++ R I+ I E+ ++ +A M+ +LC EG+ ++ +
Sbjct: 55 NMAQVDFNYRARNAIIGGISSGEFNRVQHHKSAHDMWTALCNFHEGNNDIQLVRQNQFHK 114
Query: 149 QYELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLP-SRWRPKVTAIE 207
+Y+ F M ESI+ + RF ++S L+ + K + +D+ +L L W KVT+I
Sbjct: 115 EYQRFEMHPGESIDSYFKRFGEIISKLRSVGKEFSDNDNARHLLNCLDYGVWEMKVTSIT 174
Query: 208 EAKDLNTLSVEDLVSSLKVHEMSLNEHESSKKSKSIALPSKGKTSKSSKAYKASESEEES 267
E+ L+ L+++ L S LK HEM + + K S ++ G TS + A+ +
Sbjct: 175 ESAPLSDLTMDKLYSKLKTHEMDVFHRKGLKHSMALVADPSGSTSSNDSAF-VYGGFSLA 233
Query: 268 PDRDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGGYKNSKKEDQKGCFNCKKPGHFIAD 327
E+Q K+ + L ARK + K K + CF C +P H +
Sbjct: 234 ALHSVTEEQLEKIP--EDDLALFARKFSRAYKNVRDRKRGKTNEPFVCFECGEPNHKRVN 291
Query: 328 CPDLQKEKSKSKSKKSSFSSSKFRKQIKK----SLMATWEDLD-SESGSDKEEAEDDTKA 382
CP L K+KS +KK K +K + K ++A E++ S+ SD ++ E K
Sbjct: 292 CPKL-KKKSDKTTKKPEGQGRKGKKDLMKKAIHKVLAALEEVQLSDIDSDDDDQEKGDKD 350
Query: 383 AMGLVATVSSE--------AVSEAESDSEDENEVYSKIPRQELV---------DSLKELL 425
G+ ++E A+ + + SE IP + DS+ + L
Sbjct: 351 FSGMCCLANNEDFINLCLMALEDKDDSSEHPEVCLDDIPSLDGSLCDDSCSDNDSVDDEL 410
Query: 426 SLFEHRTNELTDLKEKY------VDLMKQQKSTL-LELKASEEELKGFNLIST--TYEDR 476
S E + + ++ +KY ++ +K + + LE+ + + ST +Y
Sbjct: 411 SK-ERMAHLMIEISDKYRSSKYKIEKLKSENDGMALEIARLRSMIPEEDTCSTCASYLSE 469
Query: 477 LKILCQKLQEKCDKGSGN 494
+ +L KL + C G+GN
Sbjct: 470 INLLKDKL-KSCALGAGN 486
>ref|NP_909107.1| unnamed protein product [Oryza sativa (japonica cultivar-group)]
Length = 1410
Score = 978 bits (2528), Expect = 0.0
Identities = 504/1034 (48%), Positives = 682/1034 (65%), Gaps = 14/1034 (1%)
Query: 648 LNDQKPLSIHPKVCIRAREKQRSWYLDSGCSRHMTGEKALFLTLTMKDGGE-VKFGGNQT 706
L D+ S HP+ R SW +DSGCSRHMTGE F +LT G E + FG +
Sbjct: 348 LEDKDDSSEHPEDFGVGRSN--SWLVDSGCSRHMTGEAKWFTSLTRASGDETITFGDASS 405
Query: 707 GKIIGTGTIG-NSSISINNVWLVDGLKHNLLSISQFCDNGYDVMFSKTNCTLVNKNDKSI 765
G+++ GTI N + +V LV LK+NLLS+SQ CD +V F K +++ ++ +
Sbjct: 406 GRVMAKGTIKVNDKFMLKDVALVSKLKYNLLSVSQLCDENLEVRFKKDRSRVLDASESPV 465
Query: 766 TFKGKRVENVYKINFSDLADQKVVCLL-SMNDKKWVWHKRLGHANWRLISKISKLQLVKG 824
F RV V+ NF A CL+ S N + WH+RLGH + +S+IS + L++G
Sbjct: 466 -FDISRVGRVFFANFDSSAPGPSRCLVASENRDLFFWHRRLGHIGFDHLSRISGMDLIRG 524
Query: 825 LPNIDYHSDALCGACQKGKIVKSTFKSKDIVSTSRPLELLHIDLFGPVNTASLYGSKYGL 884
LP + D +C C+ GK+ S+ K +V T P +LLH++ GP S+ G Y L
Sbjct: 525 LPKLKAPKDLVCAPCRHGKMTSSSHKPVTMVMTDGPGQLLHMNTVGPARVQSVGGKWYVL 584
Query: 885 VIVDDYSRWTWVKFIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFENEPFELFC 944
V+VDD+SR++WV F++SK+ F S + E + +RSD+G EF+N FE FC
Sbjct: 585 VVVDDFSRYSWVYFLESKEETFGFFQSLARSLALEFPGALRAIRSDNGSEFKNSAFESFC 644
Query: 945 EKHGILHEFSSPRTPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAVNTSCYIQNRI 1004
+ G+ H+FSSP PQQNGVVERKNRTL EMARTM+ E + FW EA++ +C+I NR+
Sbjct: 645 DSSGVEHQFSSPYVPQQNGVVERKNRTLVEMARTMLDEFTTPRKFWTEAISAACFISNRV 704
Query: 1005 YIRPMLEKTAYELFKGRRPNISYFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSK 1064
++R +L KT YEL GRRP +S+ FGC C++L + + L KF++++ GIFLGY+ S+
Sbjct: 705 FLRTILHKTPYELRFGRRPKVSHLRVFGCKCFVLKSGN-LDKFESRSLDGIFLGYATHSR 763
Query: 1065 AYRVYNSETHCVEESIHVKFDDREPGNKTPEQSESNAGTTDSEDASESDQPSDSEKHSEV 1124
AYRVY T+ + E+ V FD+ PG + + ED+ + D D +
Sbjct: 764 AYRVYVLSTNKIVETCEVTFDEASPGARPEISGVLDESIFVDEDSDDDD---DDSIPPPL 820
Query: 1125 ESRPEAEITPEAESNSEAETSPVEQNENASEDVQDNTQQVIQPKFKHKSSHPEELIIGSK 1184
+S P + T + S + +P + +A+E++ T P+ P+ +I G
Sbjct: 821 DSTPPVQETGSPSTTSPSGDAPTTSS-SAAEEIDGGTSGPTAPRHIQNRHPPDSMIGGLG 879
Query: 1185 DSPRRTRSHFRQEESLIGLLSIIEPKTVEEALSDDGWILAMQEELNQFQRNDVWDLIPKP 1244
+ R RS+ + + + EPK V ALSD+ W+ AM EEL F+RN VW L+ P
Sbjct: 880 ERVTRNRSYDLVNSAFV---ASFEPKNVCHALSDENWVNAMHEELENFERNKVWSLVEPP 936
Query: 1245 FQKNIIGTKWVFRNKLNEKGEVTRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLLS 1304
N+IGTKWVF+NKL E G + RNKARLVAQG++Q EG+D+ ETFAPVARLEAIR+LL+
Sbjct: 937 LGFNVIGTKWVFKNKLGEDGSIVRNKARLVAQGFTQVEGLDFEETFAPVARLEAIRILLA 996
Query: 1305 YAINHGIILYQMDVKSAFLNGVIEEEVYVKQPPGFEDIKHPDHVYKLKKSLYGLKQAPRA 1364
+A + G L+QMDVKSAFLNGVIEEEVYVKQPPGFE+ K P+HV+KL K+LYGLKQAPRA
Sbjct: 997 FAASKGFKLFQMDVKSAFLNGVIEEEVYVKQPPGFENPKFPNHVFKLDKALYGLKQAPRA 1056
Query: 1365 WYDRLSNFLIKNDFERGQVDTTLFGKTLKKDILIVQIYVDDIIFGSTNASLCKEFSKLMQ 1424
WY+RL FL++N FE G VD TLF D L+VQIYVDDIIFG ++ +L +FS +M
Sbjct: 1057 WYERLKTFLLQNGFEMGAVDKTLFTLHSGIDFLLVQIYVDDIIFGGSSHALVAQFSDVMS 1116
Query: 1425 DEFEMSMMGELKFFLGIQINQSKEGVYVHQTKYTKELLKKFKLEDCKVMNTPMHPTCTLS 1484
EFEMSMMGEL FFLG+QI Q+KEG++VHQTKY+KELLKKF + DCK + TPM T +L
Sbjct: 1117 REFEMSMMGELTFFLGLQIKQTKEGIFVHQTKYSKELLKKFDMADCKPIATPMATTSSLG 1176
Query: 1485 KEDTGTVVDQKLYRGMIGSLLYLTASRPDILFSVCLCARFQSDPRESHLTAVKRIFRYLK 1544
++ G VDQ+ YR MIGSLLYLTASRPDI FSVCLCARFQ+ PR SH AVKRIFRY+K
Sbjct: 1177 PDEDGEEVDQREYRSMIGSLLYLTASRPDIHFSVCLCARFQASPRTSHRQAVKRIFRYIK 1236
Query: 1545 GTTNLGLLYRKSLDYKLIGFCDADYAGDRIERKSTSGNCQFLGENLISWASKRQATIAMS 1604
T G+ Y S + F DAD+AG +I+RKSTSG C FLG +L+SW+S++Q+++A S
Sbjct: 1237 STLEYGIWYSCSSALSVRAFSDADFAGCKIDRKSTSGTCHFLGTSLVSWSSRKQSSVAQS 1296
Query: 1605 TAEAEYISAASCCTQLLWMKHQLEDYQINANSIPIYCDNTAAICLSKNPILHSRAKHIEI 1664
TAEAEY++AAS C+Q+LWM L+DY ++ + +P+ CDNT+AI ++KNP+ HSR KHIEI
Sbjct: 1297 TAEAEYVAAASACSQVLWMISTLKDYGLSFSGVPLLCDNTSAINIAKNPVQHSRTKHIEI 1356
Query: 1665 KHHFIRDYVQKGIL 1678
++HF+RD V+KG +
Sbjct: 1357 RYHFLRDNVEKGTI 1370
Score = 114 bits (285), Expect = 3e-23
Identities = 86/331 (25%), Positives = 144/331 (42%), Gaps = 12/331 (3%)
Query: 86 TPAQKKLYKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALM 145
TPA +++ R I+ I E+ ++ +A M+ +LC EG+ ++ +
Sbjct: 29 TPANMAQVDFNYRARNAIIGGISSGEFNRVQHHKSAHDMWTALCNFHEGNNDIQLVRQNQ 88
Query: 146 LVHQYELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLP-SRWRPKVT 204
+Y+ F M ESI+ + RF +VS L+ + K + +D+ +L L W KVT
Sbjct: 89 FHKEYQRFEMHPGESIDSYFKRFGEIVSKLRSVGKEFSDNDNARHLLNCLDYGVWEMKVT 148
Query: 205 AIEEAKDLNTLSVEDLVSSLKVHEMSLNEHESSKKSKSIALPSKGKTSKSSKAYKASESE 264
+I E+ L+ L+++ L S LK HEM + + K S ++ G TS + A+
Sbjct: 149 SITESAPLSDLTMDKLYSKLKTHEMDVFHRKGLKHSMALVADPSGSTSSNDSAFVCG-GF 207
Query: 265 EESPDRDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGGYKNSKKEDQKGCFNCKKPGHF 324
+ E+Q K+ + L ARK + K K + CF C +P H
Sbjct: 208 SLAALHSVTEEQLEKIP--EDDLALFARKFSRAYKNVRNKKRGKTNEPFVCFECGEPNHI 265
Query: 325 IADCPDLQKEKSKSKSKKS----SFSSSKFRKQIKKSLMATWEDLDSESGSDKEEAEDDT 380
+CP L+K+ K+ K + +K I K L A E S+ SD ++ E
Sbjct: 266 RVNCPKLKKKSDKTTKKPEGQGRKGKNDLMKKAIHKVLAALEEVQLSDIDSDDDDQEKGD 325
Query: 381 KAAMGLVATVSSEAVSE----AESDSEDENE 407
K G+ ++E A D +D +E
Sbjct: 326 KDFSGMCCLANNEDFINLCLMALEDKDDSSE 356
>ref|XP_474304.1| OSJNBb0004A17.2 [Oryza sativa (japonica cultivar-group)]
gi|32488723|emb|CAE03600.1| OSJNBb0004A17.2 [Oryza sativa
(japonica cultivar-group)]
Length = 1877
Score = 978 bits (2527), Expect = 0.0
Identities = 497/1015 (48%), Positives = 675/1015 (65%), Gaps = 12/1015 (1%)
Query: 667 KQRSWYLDSGCSRHMTGEKALFLTLTMKDGGE-VKFGGNQTGKIIGTGTIG-NSSISINN 724
+ SW +DSGCSRHMTGE F +LT E + FG +G+++ GTI N + +
Sbjct: 832 RSNSWLVDSGCSRHMTGEAKWFTSLTRASSDETITFGDASSGRVMAKGTIKVNDKFMLKD 891
Query: 725 VWLVDGLKHNLLSISQFCDNGYDVMFSKTNCTLVNKNDKSITFKGKRVENVYKINFSDLA 784
V LV LK+NLLS+SQ CD +V F K +++ ++ + F RV V+ NF A
Sbjct: 892 VALVSKLKYNLLSVSQLCDENLEVRFKKDRSRVLDASESPV-FDISRVGRVFFANFDSSA 950
Query: 785 DQKVVCLL-SMNDKKWVWHKRLGHANWRLISKISKLQLVKGLPNIDYHSDALCGACQKGK 843
CL+ S N + WH+RLGH + +S+IS + L++GLP + D +C C+ GK
Sbjct: 951 PGPSRCLIASENRDLFFWHRRLGHIGFDHLSRISGMDLIRGLPKLKVPKDLVCAPCRHGK 1010
Query: 844 IVKSTFKSKDIVSTSRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKFIKSKD 903
+ S+ K +V T P +LLH+D GP S+ G Y LV+VDD+SR++WV F++SK+
Sbjct: 1011 MTSSSHKPVTMVMTDGPGQLLHMDTVGPARVQSVGGKWYVLVVVDDFSRYSWVYFLESKE 1070
Query: 904 YACEVFSSFCTQIQSEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNG 963
F S + E + +RSD+G EF+N FE FC+ G+ H+FSSP PQQNG
Sbjct: 1071 ETFGFFQSLARSLALEFPGALRAIRSDNGSEFKNSAFESFCDSSGVEHQFSSPYVPQQNG 1130
Query: 964 VVERKNRTLQEMARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRP 1023
VVERKNRTL EMARTM+ E + FW EA++ +C+I NR+++R +L KT YEL GRRP
Sbjct: 1131 VVERKNRTLVEMARTMLDEFTTPRKFWTEAISAACFISNRVFLRTILHKTPYELRFGRRP 1190
Query: 1024 NISYFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETHCVEESIHVK 1083
+S+ FGC C++L + + L KF++++ GIFLGY+ S+AYRVY T+ + E+ V
Sbjct: 1191 KVSHLRVFGCKCFVLKSGN-LDKFESRSLDGIFLGYATHSRAYRVYVLSTNKIVETCEVT 1249
Query: 1084 FDDREPGNKTPEQSESNAGTTDSEDASESDQPSDSEKHSEVESRPEAEITPEAESNSEAE 1143
FD+ PG + + ED+ + D D ++S P + T + S +
Sbjct: 1250 FDEASPGARPEISGVPDESIFVDEDSDDDD---DDSIPPPLDSTPPVQETGSPSTTSPSG 1306
Query: 1144 TSPVEQNENASEDVQDNTQQVIQPKFKHKSSHPEELIIGSKDSPRRTRSHFRQEESLIGL 1203
+P + +A+E++ T P+ P+ +I G + R RS+ + +
Sbjct: 1307 DAPTTSS-SAAEEIDGGTSGPTAPRHIQNRHPPDSMIGGLGERVTRNRSYELVNSAFV-- 1363
Query: 1204 LSIIEPKTVEEALSDDGWILAMQEELNQFQRNDVWDLIPKPFQKNIIGTKWVFRNKLNEK 1263
+ EPK V ALSD+ W+ AM EEL F+RN VW L+ P N+IGTKWVF+NKL E
Sbjct: 1364 -ASFEPKNVCHALSDENWVNAMHEELENFERNKVWSLVEPPLGFNVIGTKWVFKNKLGED 1422
Query: 1264 GEVTRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLLSYAINHGIILYQMDVKSAFL 1323
G + RNKARLVAQG++Q EG+D+ ETFAPVARLEAIR+LL++A + G L+QMDVKSAFL
Sbjct: 1423 GSIVRNKARLVAQGFTQVEGLDFEETFAPVARLEAIRILLAFAASKGFKLFQMDVKSAFL 1482
Query: 1324 NGVIEEEVYVKQPPGFEDIKHPDHVYKLKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQV 1383
NGVIEEEVYVKQPPGFE+ K P+HV+KL+K+LYGLKQAPRAWY+RL FL++N FE G V
Sbjct: 1483 NGVIEEEVYVKQPPGFENPKFPNHVFKLEKALYGLKQAPRAWYERLKTFLLQNGFEMGAV 1542
Query: 1384 DTTLFGKTLKKDILIVQIYVDDIIFGSTNASLCKEFSKLMQDEFEMSMMGELKFFLGIQI 1443
D TLF D L+VQIYVDDIIFG ++ +L +FS +M EFEMSMMGEL FFLG+QI
Sbjct: 1543 DKTLFTLHSGIDFLLVQIYVDDIIFGGSSHALVAQFSDVMSREFEMSMMGELTFFLGLQI 1602
Query: 1444 NQSKEGVYVHQTKYTKELLKKFKLEDCKVMNTPMHPTCTLSKEDTGTVVDQKLYRGMIGS 1503
Q+KEG++VHQTKY+KELLKKF + DCK + TPM T +L ++ G VDQ+ YR MIGS
Sbjct: 1603 KQTKEGIFVHQTKYSKELLKKFDMADCKPIATPMATTSSLGPDEDGEEVDQREYRSMIGS 1662
Query: 1504 LLYLTASRPDILFSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLYRKSLDYKLIG 1563
LLYLTASRPDI FSVCLCARFQ+ PR SH AVKR+FRY+K T G+ Y S +
Sbjct: 1663 LLYLTASRPDIHFSVCLCARFQASPRTSHRQAVKRMFRYIKSTLEYGIWYSCSSALSVRA 1722
Query: 1564 FCDADYAGDRIERKSTSGNCQFLGENLISWASKRQATIAMSTAEAEYISAASCCTQLLWM 1623
F DAD+AG +I+RKSTSG C FLG +L+SW+S++Q+++A STAEAEY++AAS C+Q+LWM
Sbjct: 1723 FSDADFAGCKIDRKSTSGTCHFLGTSLVSWSSRKQSSVAQSTAEAEYVAAASACSQVLWM 1782
Query: 1624 KHQLEDYQINANSIPIYCDNTAAICLSKNPILHSRAKHIEIKHHFIRDYVQKGIL 1678
L+DY ++ + +P+ CDNT+AI ++KNP+ HSR KHIEI++HF+RD V+KG +
Sbjct: 1783 ISTLKDYGLSFSGVPLLCDNTSAINIAKNPVQHSRTKHIEIRYHFLRDNVEKGTI 1837
Score = 122 bits (307), Expect = 8e-26
Identities = 118/498 (23%), Positives = 210/498 (41%), Gaps = 53/498 (10%)
Query: 32 APKFNGDPEEFSWWKTNMYSFIMGLDEELWDILEDGV---DDLDLDEEGAAIDRRIHTPA 88
A F D F WK M +++ +W+ ++ DD D+ TPA
Sbjct: 7 AKPFVFDGHNFVIWKARMEAYLQSQGHNVWNKVKSPYTVPDDADI------------TPA 54
Query: 89 QKKLYKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVH 148
+++ R I+ I E+ ++ +A M+ +LC EG+ ++ +
Sbjct: 55 NMAQVDFNYRARNAIIGGISSGEFNRVQHHKSAHDMWTALCNFHEGNNDIQLVRQNQFHK 114
Query: 149 QYELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLP-SRWRPKVTAIE 207
+Y+ F M ESI+ + RF ++S L+ + K + +D+ +L L W KVT+I
Sbjct: 115 EYQRFEMHPGESIDSYFKRFGEIISKLRSVGKEFSDNDNARHLLNCLDYGVWEMKVTSIT 174
Query: 208 EAKDLNTLSVEDLVSSLKVHEMSLNEHESSKKSKSIALPSKGKTSKSSKAYKASESEEES 267
E+ L+ L+++ L S LK HEM + + K S ++ G TS + A+ +
Sbjct: 175 ESAPLSDLTMDKLYSKLKTHEMDVFHRKGLKHSMALVADPSGSTSSNDSAF-VYGGFSLA 233
Query: 268 PDRDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGGYKNSKKEDQKGCFNCKKPGHFIAD 327
E+Q K+ + L ARK + K K + CF C +P H +
Sbjct: 234 ALHSVTEEQLEKIP--EDDLALFARKFSRAYKNVRDRKRGKTNEPFVCFECGEPNHKRVN 291
Query: 328 CPDLQKEKSKSKSKKSSFSSSKFRKQIKK----SLMATWEDLD-SESGSDKEEAEDDTKA 382
CP L K+KS +KK K +K + K ++A E++ S+ SD ++ E K
Sbjct: 292 CPKL-KKKSDKTTKKPEGQGRKGKKDLMKKAIHKVLAALEEVQLSDIDSDDDDQEKGDKD 350
Query: 383 AMGLVATVSSE--------AVSEAESDSEDENEVYSKIPRQELV---------DSLKELL 425
G+ ++E A+ + + SE IP + DS+ + L
Sbjct: 351 FSGMCCLANNEDFINLCLMALEDKDDSSEHPEVCLDDIPSLDGSLCDDSCSDNDSVDDEL 410
Query: 426 SLFEHRTNELTDLKEKY------VDLMKQQKSTL-LELKASEEELKGFNLIST--TYEDR 476
S E + + ++ +KY ++ +K + + LE+ + + ST +Y
Sbjct: 411 SK-ERMAHLMIEISDKYRSSKYKIEKLKSENDGMALEIARLRSMIPEEDTCSTCASYLSE 469
Query: 477 LKILCQKLQEKCDKGSGN 494
+ +L KL + C G+GN
Sbjct: 470 INLLKDKL-KSCALGAGN 486
>gb|AAL35396.1| Opie2a pol [Zea mays]
Length = 1048
Score = 967 bits (2500), Expect = 0.0
Identities = 496/1017 (48%), Positives = 668/1017 (64%), Gaps = 28/1017 (2%)
Query: 681 MTGEKALFLTLTMKDGGE--VKFGGNQTGKIIGTGTIGNSSI-SINNVWLVDGLKHNLLS 737
MTGEK +F + + + FG GK+ G G I SS SI+NV+LV+ L +N LS
Sbjct: 1 MTGEKKMFTSYVKNKDSQDSIIFGDGNQGKVKGLGKIAISSEHSISNVFLVESLGYNFLS 60
Query: 738 ISQFCDNGYDVMFSKTNCTLVNKNDKSITFKGKRVENVYKINFSDLADQKVVCLLSMNDK 797
+SQ C+ GY+ +F+ + ++ ++D S+ FKG +Y ++F+ CL++
Sbjct: 61 VSQLCNMGYNCLFTNVDVSVFRRSDGSLAFKGVLDGKLYLVDFAKEEAGLDACLIAKTSM 120
Query: 798 KWVWHKRLGHANWRLISKISKLQLVKGLPNIDYHSDALCGACQKGKIVKSTFKSKDIVST 857
W+WH+RL H + + K+ K + V GL N+ + D C ACQ GK V+ + +K++++T
Sbjct: 121 GWLWHRRLAHVGMKNLHKLLKGEHVIGLTNVQFKKDRPCVACQAGKQVRGSHHTKNVMTT 180
Query: 858 SRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKFIKSKDYACEVFSSFCTQIQ 917
SRPLE+LH+DLFGPV S+ GSKYGLVIVDD+SR+TWV F++ K + + Q
Sbjct: 181 SRPLEMLHMDLFGPVAYLSIGGSKYGLVIVDDFSRFTWVFFLQDKSETQGTLKRYLRRAQ 240
Query: 918 SEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEMAR 977
+E ELK+ K+RSD+G EF+N E F + GI HEFS+P TPQQNGVVERKNRTL +MAR
Sbjct: 241 NEFELKVKKIRSDNGSEFKNLQVEEFLVEEGIKHEFSAPYTPQQNGVVERKNRTLMDMAR 300
Query: 978 TMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNISYFHQFGCTCYI 1037
TM+ E + FW+EAVNT+C+ NR+Y+ +L+ T+YEL G +PN+SYF FG CYI
Sbjct: 301 TMLGEFKTPERFWSEAVNTACHSINRVYLHRLLKNTSYELLTGNKPNVSYFRVFGSKCYI 360
Query: 1038 LNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETHCVEESIHVKFDDREPGNKTPEQS 1097
L K KF KA G LGY +KAYRV+N + VE S V FD E EQ
Sbjct: 361 LVKKGRTSKFAPKAVEGFLLGYDSNTKAYRVFNKSSGLVEVSSDVVFD--ETNGSLREQ- 417
Query: 1098 ESNAGTTDSEDASESDQPSDSEKHSEV-ESRPEAEITPEAESNSEAETSPVEQNENA--- 1153
+ +D E D P+ + + + + RP+ + + S++ P + +E A
Sbjct: 418 -----VVNLDDVDEEDVPTAAMRTMAIGDVRPQEHLEQDQPSSTTMVHPPTQDDEQAPQV 472
Query: 1154 -------SEDVQDNTQQV-----IQPKFKHKSSHPEELIIGSKDSPRRTRSHFRQEESLI 1201
++DVQ ++ Q + + +HP I+G TRS
Sbjct: 473 GAHDQGGAQDVQVEEEEAPQAPPTQVRATIQRNHPVNQILGDISKGVTTRSRLVNFCEHY 532
Query: 1202 GLLSIIEPKTVEEALSDDGWILAMQEELNQFQRNDVWDLIPKPFQKNIIGTKWVFRNKLN 1261
+S IEP VEE L D W+LAMQEELN F+RN+VW L+P+P Q N++GTKWVFRNK +
Sbjct: 533 SFVSSIEPFRVEEVLLDPDWVLAMQEELNNFKRNEVWTLVPRPKQ-NVVGTKWVFRNKQD 591
Query: 1262 EKGEVTRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLLSYAINHGIILYQMDVKSA 1321
E G VTRNKARLVA+GY+Q G+D+ ETFAPVARL++IR+ L+YA +H LYQMDVKSA
Sbjct: 592 EHGVVTRNKARLVAKGYAQVAGLDFEETFAPVARLKSIRIWLAYAAHHSFRLYQMDVKSA 651
Query: 1322 FLNGVIEEEVYVKQPPGFEDIKHPDHVYKLKKSLYGLKQAPRAWYDRLSNFLIKNDFERG 1381
FLNG I+EEVYV+QPPGFED + PDHV KL K+LYGLKQAPRAWY+ L +FLI N F+ G
Sbjct: 652 FLNGPIKEEVYVEQPPGFEDERFPDHVCKLSKALYGLKQAPRAWYECLRDFLIANAFKVG 711
Query: 1382 QVDTTLFGKTLKKDILIVQIYVDDIIFGSTNASLCKEFSKLMQDEFEMSMMGELKFFLGI 1441
+ D TLF KT D+ + QIYVDDIIFGSTN + C+EFS++M +FEMSMMGEL +FLG
Sbjct: 712 KADPTLFTKTCDGDLFVCQIYVDDIIFGSTNQNSCEEFSRVMTQKFEMSMMGELSYFLGF 771
Query: 1442 QINQSKEGVYVHQTKYTKELLKKFKLEDCKVMNTPMHPTCTLSKEDTGTVVDQKLYRGMI 1501
Q+ Q K+G ++ QTKYT++L+K+F ++D K TPM G VDQK YR I
Sbjct: 772 QVRQLKDGTFISQTKYTQDLIKRFGMKDAKPAKTPMGTDGHTDLNKGGKSVDQKAYRSTI 831
Query: 1502 GSLLYLTASRPDILFSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLYRKSLDYKL 1561
GSLLYL ASRPDI+ SVC+CARFQSDPRE HL AVKRI RYL T G+ Y K + L
Sbjct: 832 GSLLYLCASRPDIMLSVCMCARFQSDPRECHLVAVKRILRYLVATPCFGIWYPKGSTFDL 891
Query: 1562 IGFCDADYAGDRIERKSTSGNCQFLGENLISWASKRQATIAMSTAEAEYISAASCCTQLL 1621
IG+ D+DYA +++RKSTS CQFLG +L+SW SK+Q ++A+STAEAEY++ CC QLL
Sbjct: 892 IGYSDSDYARCKVDRKSTSRMCQFLGRSLVSWNSKKQTSVALSTAEAEYVAVGQCCAQLL 951
Query: 1622 WMKHQLEDYQINANSIPIYCDNTAAICLSKNPILHSRAKHIEIKHHFIRDYVQKGIL 1678
WM+ L D+ N + +P+ CDN +AI +++NP+ HSR KHI+I+HHF+RD+ QKG++
Sbjct: 952 WMRQTLRDFGYNLSEVPLLCDNESAIRIAENPVEHSRTKHIDIRHHFLRDHQQKGVI 1008
>gb|AAN40025.1| putative gag-pol polyprotein [Zea mays]
Length = 1892
Score = 964 bits (2492), Expect = 0.0
Identities = 505/1039 (48%), Positives = 661/1039 (63%), Gaps = 84/1039 (8%)
Query: 670 SWYLDSGCSRHMTGEKALFLTLTMKDGGE--VKFGGNQTGKIIGTGTIGNSSISINNVWL 727
SW LDSGC+ HMTGEK +F + + + FG G + G
Sbjct: 863 SWILDSGCTNHMTGEKKMFSSYEKNKDPQRAITFGDGNQGLVKGV--------------- 907
Query: 728 VDGLKHNLLSISQFCDNGYDVMFSKTNCTLVNKNDKSITFKGKRVENVYKINFSDLADQK 787
T+ ++D SI FKG +Y + F D A+
Sbjct: 908 ----------------------------TVFRRSDDSIAFKGVLEGQLYLVVF-DRAELD 938
Query: 788 VVCLLSMNDKKWVWHKRLGHANWRLISKISKLQLVKGLPNIDYHSDALCGACQKGKIVKS 847
CL++ + W+WH+RL H + + K+ K + + GL N+ + D +C ACQ GK V +
Sbjct: 939 T-CLIAKTNMGWLWHRRLAHVGMKNLHKLLKGEHILGLTNVHFEKDRICSACQAGKQVGT 997
Query: 848 TFKSKDIVSTSRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKFIKSKDYACE 907
K+I++T RPLELLH+DLFGP+ S+ GSKY LVIVDDYSR+TWV F++ K E
Sbjct: 998 HHPHKNIMTTDRPLELLHMDLFGPIAYISIGGSKYCLVIVDDYSRFTWVFFLQEKSQTQE 1057
Query: 908 VFSSFCTQIQSEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNGVVER 967
F + Q+E L+I K+RSD+G EF+N E F E+ GI HEFSSP TPQQNGVVER
Sbjct: 1058 TLKGFLRRAQNEFGLRIKKIRSDNGTEFKNSQIESFLEEEGIKHEFSSPYTPQQNGVVER 1117
Query: 968 KNRTLQEMARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNISY 1027
KNRTL +MARTM+ E FWAEAVNT+CY NR+Y+ +L+KT+YEL G++PNISY
Sbjct: 1118 KNRTLLDMARTMLDEYKTPDRFWAEAVNTACYAINRLYLHRILKKTSYELLTGKKPNISY 1177
Query: 1028 FHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETHCVEESIHVKFDDR 1087
F FG C+IL + KF K G LGY ++AYRV+N T VE S V FD+
Sbjct: 1178 FRVFGSKCFILIKRGRKSKFAPKTVEGFLLGYDSNTRAYRVFNKSTGLVEVSCDVVFDET 1237
Query: 1088 EP--------------------------GNKTPEQSESNAGTTDSEDAS-ESDQPSDSEK 1120
G+ P++SE T D +S ++ P+ +E
Sbjct: 1238 NGSQVEQVDLDEIGEEQAPCIALRNMSIGDVCPKESEEPPSTQDQPPSSMQASPPTQNED 1297
Query: 1121 HSEVESRPEAEITPEAESNSEAETSPVEQNENASEDVQDNTQQVIQPKFKHKSSHPEELI 1180
++ + E+ P + +++ +Q + E+ + +V Q + HP + I
Sbjct: 1298 EAQNDEEQNQEVKPPQDKSNDQGGDTNDQEKEDEEEPRPPHPRVHQAI---QRDHPVDTI 1354
Query: 1181 IGSKDSPRRTRS---HFRQEESLIGLLSIIEPKTVEEALSDDGWILAMQEELNQFQRNDV 1237
+G TRS HF + S + S IEP VEEAL D W++AMQEELN F RN+V
Sbjct: 1355 LGDIHKGVTTRSRVAHFCEHYSFV---SSIEPHRVEEALQDSDWVVAMQEELNNFTRNEV 1411
Query: 1238 WDLIPKPFQKNIIGTKWVFRNKLNEKGEVTRNKARLVAQGYSQQEGIDYTETFAPVARLE 1297
W L+P+P Q N++GTKWVFRNK +E G VTRNKARLVA+GYSQ EG+D+ ET+APVARLE
Sbjct: 1412 WHLVPRPNQ-NVVGTKWVFRNKQDEHGVVTRNKARLVAKGYSQVEGLDFDETYAPVARLE 1470
Query: 1298 AIRLLLSYAINHGIILYQMDVKSAFLNGVIEEEVYVKQPPGFEDIKHPDHVYKLKKSLYG 1357
+IR+LL+YA HG LYQMDVKSAFLNG I+EEVYV+QPPGFED ++P+HVY+L K+LYG
Sbjct: 1471 SIRILLAYATYHGFKLYQMDVKSAFLNGPIKEEVYVEQPPGFEDSEYPNHVYRLSKALYG 1530
Query: 1358 LKQAPRAWYDRLSNFLIKNDFERGQVDTTLFGKTLKKDILIVQIYVDDIIFGSTNASLCK 1417
LKQAPRAWY+ L +FLI N F+ G+ D TLF KTL+ D+ + QIYVDDIIFGSTN S C+
Sbjct: 1531 LKQAPRAWYECLRDFLIANGFKVGKADPTLFTKTLENDLFVCQIYVDDIIFGSTNESTCE 1590
Query: 1418 EFSKLMQDEFEMSMMGELKFFLGIQINQSKEGVYVHQTKYTKELLKKFKLEDCKVMNTPM 1477
EFS++M +FEMSMMGELK+FLG Q+ Q +EG ++ QTKYT+++L KF ++D K + TPM
Sbjct: 1591 EFSRIMTQKFEMSMMGELKYFLGFQVKQLREGTFISQTKYTQDILAKFGMKDAKPIKTPM 1650
Query: 1478 HPTCTLSKEDTGTVVDQKLYRGMIGSLLYLTASRPDILFSVCLCARFQSDPRESHLTAVK 1537
L + G VDQK+YR MIGSLLYL ASRPDI+ SVC+CARFQSDP+ESHLTAVK
Sbjct: 1651 GTNGHLDLDTGGKSVDQKVYRSMIGSLLYLCASRPDIMLSVCMCARFQSDPKESHLTAVK 1710
Query: 1538 RIFRYLKGTTNLGLLYRKSLDYKLIGFCDADYAGDRIERKSTSGNCQFLGENLISWASKR 1597
RI RYL T GL Y + + LIG+ DAD+AG +I RKSTSG CQFLG +L+SWASK+
Sbjct: 1711 RILRYLAYTPKFGLWYPRGSTFDLIGYSDADWAGCKINRKSTSGTCQFLGRSLVSWASKK 1770
Query: 1598 QATIAMSTAEAEYISAASCCTQLLWMKHQLEDYQINANSIPIYCDNTAAICLSKNPILHS 1657
Q ++A+STAEAEYI+A CC QLLWM+ L DY +P+ CDN +AI ++ NP+ HS
Sbjct: 1771 QNSVALSTAEAEYIAAGHCCAQLLWMRQTLRDYGYKLTKVPLLCDNESAIKMADNPVEHS 1830
Query: 1658 RAKHIEIKHHFIRDYVQKG 1676
R KHI I++HF+RD+ QKG
Sbjct: 1831 RTKHIAIRYHFLRDHQQKG 1849
Score = 114 bits (286), Expect = 2e-23
Identities = 121/473 (25%), Positives = 203/473 (42%), Gaps = 57/473 (12%)
Query: 12 KYTSVKHDYDTADKKTDS-----GKAPKFNGDPEEFSWWKTNMYSFIMGLDEELWDILED 66
KY+ + Y K T GK P FNG E+++ W M + L + +WD++E
Sbjct: 108 KYSKIPLRYPRVPKHTPLLSVPLGKPPTFNG--EDYAMWSNLMRFHLTSLHKRIWDVVEY 165
Query: 67 GVDDLDLDEEGAAIDRRIHTPAQKKLYKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFA 126
GV + +E D ++ + + I++AS+ + EY K+ AK ++
Sbjct: 166 GVQVPSIGDEDYDTDE------VAQIEHFNSQAATILLASLSKEEYNKVQGLKNAKEIWD 219
Query: 127 SLCANFEGSKKVKEAKALMLVHQYELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVASD 186
L EG + K K + + F ++ E ++MY+R +TLV+ ++ L +
Sbjct: 220 LLKTAHEGDELTKITKRETIEGELGRFHLRQGEEPQDMYNRLKTLVNQVRNLGSTKWDDH 279
Query: 187 HVSK-ILRSLPSRWRPKVTAIEEAKDLNTLSVEDLVSSLKVHEMSLNEHESSKKSKSIAL 245
V K ILR+L +V I ++ E+++ + E + + SKK +
Sbjct: 280 EVVKVILRALIFLNPTQVQLIRGNPRYPLMTPEEVIGNFVSFECMI---KGSKKINELDE 336
Query: 246 PSKGKTSKSSKAYKASES--EEESPDRDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGG 303
PS + A+KA+E EE +P R Q + + L N+ L K + + K+
Sbjct: 337 PSTSEAQPV--AFKATEEKKEESTPSR-----QPIDASKLDNEEMALIIKSFRQILKQRK 389
Query: 304 YKNSKKEDQKGCFNCKKPGHFIADCPDLQKEKSKSKSKKSSFSSSKFRKQIKKSLMATWE 363
K+ K +K C+ C KPGHFIA CP + + + KK K + K
Sbjct: 390 GKDYKPRSKKVCYKCGKPGHFIAKCP-MSSDSDRGDDKKGRRKEKKRYYKKKGGDAHVCR 448
Query: 364 DLDS-ESGSDKEEAEDDTKAAM--GLV-----------------ATVSSEAVSEAESDSE 403
+ DS ES SD + ED A+ GL+ S E SD +
Sbjct: 449 EWDSDESSSDSSDDEDAANIAVTKGLLFPNVGHKCLMAKDGKKKVKSKSSTKYETSSDED 508
Query: 404 DENE------VYSKIPRQELVDSLKELLSLFEHRTNELTDLKEKYVDLMKQQK 450
D+NE +++ + E + L EL+S H ++L D +E + L+K+ K
Sbjct: 509 DKNEEDNLRILFANL-NMEQKEKLNELISAI-HEKDDLLDSQEDF--LIKENK 557
>emb|CAE04884.2| OSJNBa0042I15.6 [Oryza sativa (japonica cultivar-group)]
Length = 1510
Score = 918 bits (2373), Expect = 0.0
Identities = 487/1036 (47%), Positives = 647/1036 (62%), Gaps = 71/1036 (6%)
Query: 670 SWYLDSGCSRHMTGEKALFLTLTMKDGGEVKFGGNQTGKIIGTGTIGNSSISINNVWLVD 729
SW +DSGC+ HMTGE+++F +L K G
Sbjct: 470 SWVVDSGCTNHMTGERSMFTSLDEKGGS-------------------------------- 497
Query: 730 GLKHNLLSISQFCDNGYDVMFSKTNCTLVNKNDKSITFKGKRVENVYKINFSDLADQKVV 789
+ N++ F D+G + + ++V ++D SI FKG ++Y ++F
Sbjct: 498 --RENIV----FGDDGKEKLQFIIRVSIV-RDDSSIAFKGVLKGDLYLVDFDVDRVNPEA 550
Query: 790 CLLSMNDKKWVWHKRLGHANWRLISKISKLQLVKGLPNIDYHSDALCGACQKGKIVKSTF 849
CL++ + W+WH+RL H R ++ + K + + GL N+ + D +C ACQ GK V S
Sbjct: 551 CLIAKSSMGWLWHRRLAHVGMRNLASLLKGEHILGLSNVSFEMDRVCSACQAGKQVGSPH 610
Query: 850 KSKDIVSTSRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKFIKSKDYACEVF 909
K+I++T+RPLELLH+DLFGPV S+ G+KYG VIVDD+S +TWV F+ K A +VF
Sbjct: 611 PIKNIMTTTRPLELLHMDLFGPVAYISIGGNKYGFVIVDDFSCFTWVYFLHDKSEAQDVF 670
Query: 910 SSFCTQIQSEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNGVVERKN 969
F Q Q+ +L I +VRSD+GGEF+N E F ++ GI HEFS+P P QNG+VERKN
Sbjct: 671 KRFTKQAQNLYDLTIKRVRSDNGGEFKNTQVEEFLDEEGIKHEFSAPYDPPQNGIVERKN 730
Query: 970 RTLQEMARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNISYFH 1029
RTL E AR M+ E + FWAEAV+T+C+ NR+Y+ +L+KT+YEL G++PN+SYF
Sbjct: 731 RTLIEAARAMLDEYKTSDVFWAEAVSTACHAINRLYLHKILKKTSYELLSGKKPNVSYFR 790
Query: 1030 QFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETHCVEESIHVKFDDREP 1089
FG +IL+ KF K G LGY + AYRV+N +T + IHV D +P
Sbjct: 791 VFGSKFFILSKMPRSSKFSPKVDEGFLLGYESNAHAYRVFN-KTSGEQVVIHV-VRDVDP 848
Query: 1090 GNKTPEQSESNAGTTDSEDASES-DQPSDSEKHSEVES-RPEAEITPEAESNSEAETSPV 1147
++ + +++D E DQP S +S E E+ + N P
Sbjct: 849 SQAIGTKAIGDIRPVETQDDQEDRDQPPSSTSNSPTSVVSAEPEVPGPIDRNLRTSPGPE 908
Query: 1148 EQNE-------NASEDV------------------QDNTQQVIQPKFKH--KSSHPEELI 1180
+ SEDV Q V P+ H + HP + I
Sbjct: 909 VPGSTVRNLRTSGSEDVPTAQVDGIDAAGTLGHTDQAQVPLVHHPRIHHTVQRDHPVDNI 968
Query: 1181 IGSKDSPRRTRSHFRQEESLIGLLSIIEPKTVEEALSDDGWILAMQEELNQFQRNDVWDL 1240
+G TRS +S +EP VE+AL D W++AMQEELN F RN VW+L
Sbjct: 969 LGDIRKGVTTRSRVASFCQHYSFVSSLEPTRVEDALGDSDWVMAMQEELNNFARNQVWNL 1028
Query: 1241 IPKPFQKNIIGTKWVFRNKLNEKGEVTRNKARLVAQGYSQQEGIDYTETFAPVARLEAIR 1300
+ +P Q N+IGTKW+FRNK +E V RNKARLV QG++Q EG+D+ ETFAPVARLE+IR
Sbjct: 1029 VERPKQ-NVIGTKWIFRNKQDEHVVVVRNKARLVTQGFTQVEGLDFGETFAPVARLESIR 1087
Query: 1301 LLLSYAINHGIILYQMDVKSAFLNGVIEEEVYVKQPPGFEDIKHPDHVYKLKKSLYGLKQ 1360
+LL+YA +H L+QMDVKSAFLNG I E VYV+QPPGFED K P+HVYKL K+LYGLKQ
Sbjct: 1088 ILLAYAAHHDFRLFQMDVKSAFLNGPISELVYVEQPPGFEDPKLPNHVYKLHKALYGLKQ 1147
Query: 1361 APRAWYDRLSNFLIKNDFERGQVDTTLFGKTLKKDILIVQIYVDDIIFGSTNASLCKEFS 1420
APRAWY+ L +FL+KN FE G DTTLF K K D+ I QIYVDDIIFGSTNAS C+EFS
Sbjct: 1148 APRAWYECLRDFLLKNGFEIGNADTTLFTKKFKSDLFICQIYVDDIIFGSTNASFCEEFS 1207
Query: 1421 KLMQDEFEMSMMGELKFFLGIQINQSKEGVYVHQTKYTKELLKKFKLEDCKVMNTPMHPT 1480
+M FEMSMMGEL FFL +Q+ Q++EG ++ QTKY K++LKKF +ED K + TPM
Sbjct: 1208 SIMTKRFEMSMMGELTFFLWLQVKQAQEGTFISQTKYVKDILKKFGMEDAKPIKTPMPTN 1267
Query: 1481 CTLSKEDTGTVVDQKLYRGMIGSLLYLTASRPDILFSVCLCARFQSDPRESHLTAVKRIF 1540
L +D G VDQK+YR MIGSLLYL ASRPDI+ SVC+CARFQ++P+E HL AVKRI
Sbjct: 1268 GHLDLDDNGKCVDQKVYRSMIGSLLYLCASRPDIMLSVCMCARFQAEPKECHLIAVKRIQ 1327
Query: 1541 RYLKGTTNLGLLYRKSLDYKLIGFCDADYAGDRIERKSTSGNCQFLGENLISWASKRQAT 1600
RYL T NLGL Y K D++L+G+ D+DYAG +++RKS +G CQFLG +L+SW K+Q +
Sbjct: 1328 RYLVHTPNLGLWYPKGCDFELLGYSDSDYAGCKVDRKSITGTCQFLGPSLVSWFPKKQNS 1387
Query: 1601 IAMSTAEAEYISAASCCTQLLWMKHQLEDYQINANSIPIYCDNTAAICLSKNPILHSRAK 1660
I +ST EAEY++A SCC QLLWMK L+D+ N P+ CDN +AI ++ NP+ HS+ K
Sbjct: 1388 IVLSTTEAEYVAAGSCCAQLLWMKQTLKDFGYNFTKTPLLCDNESAIKIANNPVQHSKTK 1447
Query: 1661 HIEIKHHFIRDYVQKG 1676
HI+I HHF+RD+ KG
Sbjct: 1448 HIDIHHHFLRDHETKG 1463
Score = 95.9 bits (237), Expect = 1e-17
Identities = 108/443 (24%), Positives = 179/443 (40%), Gaps = 101/443 (22%)
Query: 53 IMGLDEELWDILEDGVD----DLDLDEEGAAIDRRIHTPAQKKLYKKHHKIRGIIVASIP 108
++ L +W ++ GVD D++L E Q++L ++ + I++++
Sbjct: 5 LISLHPSIWKVVCTGVDVPHDDMELTSE------------QEQLIHRNAQASNAILSTLS 52
Query: 109 RTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYELFRMKDDESIEEMYSRF 168
E+ K+ AK + +L EGS V+EAK +L + F M D E+ +EMY R
Sbjct: 53 LEEFNKVDGLEEAKEICDTLQLAHEGSPAVREAKIELLEGRLGRFVMDDKETPQEMYDRM 112
Query: 169 QTLVSGLQILKKSYVASDHVSK-ILRSLPSRWRPKVTAIEEAKDLNTLSVEDLVSSLKVH 227
LV+ ++ L + + V K +LR R V+ I E KD L++ D++ + H
Sbjct: 113 MILVNKIKGLGSEDMTNHFVVKRLLREFGPRNPTLVSMIREKKDFKRLTLSDILGRIVSH 172
Query: 228 EMSLNEHESSKKSKSIALPSKGKTSKSSKAYKASESEEESPDRDSDEDQSVKMAMLSNKL 287
EM E E+ K + +
Sbjct: 173 EM--QEEEARKTRRRV-------------------------------------------- 186
Query: 288 EYLARKQKKFLSKRGGYKNSKKEDQKG-------CFNCKKPGHFIADCPDLQKEKSKSKS 340
K+ K ++ GY +K+D KG CFNC + GHFIAD P + K+K
Sbjct: 187 -----KRFKHFLRKSGYGKGRKDDDKGKKQSKRACFNCGEYGHFIADFPKSNEAKAKGGK 241
Query: 341 KKSSFSSSKFRKQIKKSLM-ATWEDLDSESGSDKEEAEDDTKA-AMGLVATVSSEAVSEA 398
KK R + ++ M W D E K + + K G VATV+ ++ S +
Sbjct: 242 KKPE------RAHVAEAHMPEVWYSGDEEDPEVKPKPKPKDKVDGEGGVATVTFKSSSSS 295
Query: 399 E-------SDSEDENEVYS-------KIPRQELVDSLKELLSLFEHRTNELTDLKEKYVD 444
+ SD +D++ YS K+ Q+ + ++ S E NEL D+ + +
Sbjct: 296 KERLFNNLSDDDDDSYHYSCFMAQGRKVMTQKPSHTSLDVDSSDEESDNELDDVLKSFSK 355
Query: 445 LMKQQKSTLLE----LKASEEEL 463
Q + L+ LK E L
Sbjct: 356 PAMQHLAKLMRALDTLKKENERL 378
>gb|AAW57789.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
Length = 1799
Score = 905 bits (2340), Expect = 0.0
Identities = 479/1021 (46%), Positives = 656/1021 (63%), Gaps = 45/1021 (4%)
Query: 667 KQRSWYLDSGCSRHMTGEKALFLTLTMKDGGE-VKFGGNQTGKIIGTGTIG-NSSISINN 724
+ SW +DSGCSRHMTGE F +LT G E + FG +G+++ GTI N + +
Sbjct: 775 RSNSWLVDSGCSRHMTGEAKWFTSLTRASGDETITFGDASSGRVMAKGTIKVNDKFMLKD 834
Query: 725 VWLVDGLKHNLLSISQFCDNGYDVMFSKTNCTLVNKNDKSITFKGKRVENVYKINFSDLA 784
V LV LK+NLLS+SQ CD +V F K +++ ++ + F RV V+ NF A
Sbjct: 835 VALVSKLKYNLLSVSQLCDENLEVRFKKDRSRVLDASESPV-FDISRVGRVFFANFDSSA 893
Query: 785 DQKVVCLL-SMNDKKWVWHKRLGHANWRLISKISKLQLVKGLPNIDYHSDALCGACQKGK 843
CL+ S N + WH+RLGH + +S+IS + L++GLP + D +C C+ GK
Sbjct: 894 PGPSRCLIASENRDLFFWHRRLGHIGFDHLSRISGMDLIRGLPKLKVPKDLVCAPCRHGK 953
Query: 844 IVKSTFKSKDIVSTSRPLELLHIDLFGPVNTASLYG------SKYGLVIVDDYSRWTWVK 897
+ S+ K +V T P +LLH+D GP S+ G + + L++V W W
Sbjct: 954 MTSSSHKPVTMVMTDGPGQLLHMDTVGPARVQSVGGKWRKHLASFSLLLV----VWLWSF 1009
Query: 898 FIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPR 957
+ + +A V + ++I L ++ + S+ H+FSSP
Sbjct: 1010 LVLFEPFA--VIMALNSKI-LHLNLSVILLESN--------------------HQFSSPY 1046
Query: 958 TPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYEL 1017
PQQNGVVERKNRTL EMARTM+ E + FW EA++ +C+I NR+++R +L KT YEL
Sbjct: 1047 VPQQNGVVERKNRTLVEMARTMLDEFTTPRKFWTEAISAACFISNRVFLRTILHKTPYEL 1106
Query: 1018 FKGRRPNISYFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETHCVE 1077
GRRP +S+ FGC C++L + + L KF++++ GIFLGY+ S+AYRVY T+ +
Sbjct: 1107 RFGRRPKVSHLRVFGCKCFVLKSGN-LDKFESRSLDGIFLGYATHSRAYRVYVLSTNKIV 1165
Query: 1078 ESIHVKFDDREPGNKTPEQSESNAGTTDSEDASESDQPSDSEKHSEVESRPEAEITPEAE 1137
E+ V FD+ PG + + ED+ + D S S ++S P + T
Sbjct: 1166 ETCEVTFDEASPGARPEISGVPDESIFVDEDSDDDDDDSIS---PPLDSTPPVQETGSPL 1222
Query: 1138 SNSEAETSPVEQNENASEDVQDNTQQVIQPKFKHKSSHPEELIIGSKDSPRRTRSHFRQE 1197
+ S + +P + +A+E++ T P+ P+ +I G + R RS+
Sbjct: 1223 TTSPSGDAPT-TSSSAAEEIDGGTSGPTAPRHIQNRHPPDSMIGGLGERVTRNRSYELVN 1281
Query: 1198 ESLIGLLSIIEPKTVEEALSDDGWILAMQEELNQFQRNDVWDLIPKPFQKNIIGTKWVFR 1257
+ + + EPK V ALSD+ W+ AM EEL F+RN VW L+ P ++IGTKWVF+
Sbjct: 1282 SAFV---ASFEPKNVCHALSDENWVNAMHEELENFERNKVWSLVEPPLGFSVIGTKWVFK 1338
Query: 1258 NKLNEKGEVTRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLLSYAINHGIILYQMD 1317
NKL E G + RNKARLVAQG++Q EG+D+ ETFAPVARLEAIR+LL++A + G L+QMD
Sbjct: 1339 NKLGEDGSIVRNKARLVAQGFTQVEGLDFEETFAPVARLEAIRILLAFAASKGFKLFQMD 1398
Query: 1318 VKSAFLNGVIEEEVYVKQPPGFEDIKHPDHVYKLKKSLYGLKQAPRAWYDRLSNFLIKND 1377
VKSAFLNGVIEEEVYVKQPPGFE+ K P+HV+KL K+LYGLKQAPRAWY+RL FL++N
Sbjct: 1399 VKSAFLNGVIEEEVYVKQPPGFENPKFPNHVFKLDKALYGLKQAPRAWYERLKTFLLQNG 1458
Query: 1378 FERGQVDTTLFGKTLKKDILIVQIYVDDIIFGSTNASLCKEFSKLMQDEFEMSMMGELKF 1437
FE G VD TLF D L+VQIYVDDIIFG ++ +L +F +M EFEMSMMGEL F
Sbjct: 1459 FEMGAVDKTLFTLHSGIDFLLVQIYVDDIIFGGSSHALVAQFFDVMSREFEMSMMGELTF 1518
Query: 1438 FLGIQINQSKEGVYVHQTKYTKELLKKFKLEDCKVMNTPMHPTCTLSKEDTGTVVDQKLY 1497
FLG+QI Q+KEG++VHQTKY+KELLKKF + DCK + TPM T +L ++ G VDQ+ Y
Sbjct: 1519 FLGLQIKQTKEGIFVHQTKYSKELLKKFDMADCKPIATPMATTSSLGPDEDGEEVDQREY 1578
Query: 1498 RGMIGSLLYLTASRPDILFSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLYRKSL 1557
R MIGSLLYLTASRPDI FSVCLCARFQ+ PR SH AVKRIFRY+K T G+ Y S
Sbjct: 1579 RSMIGSLLYLTASRPDIHFSVCLCARFQASPRTSHRQAVKRIFRYIKSTLEYGIWYSCSS 1638
Query: 1558 DYKLIGFCDADYAGDRIERKSTSGNCQFLGENLISWASKRQATIAMSTAEAEYISAASCC 1617
+ F DAD+AG +I+RKSTSG C FLG +L+SW+S++Q+++A STAEAEY++AAS C
Sbjct: 1639 ALSVRAFSDADFAGCKIDRKSTSGTCHFLGTSLVSWSSRKQSSVAQSTAEAEYVAAASAC 1698
Query: 1618 TQLLWMKHQLEDYQINANSIPIYCDNTAAICLSKNPILHSRAKHIEIKHHFIRDYVQKGI 1677
+Q+LWM L+DY ++ + +P+ CDNT+AI ++KNP+ HSR KHIEI++HF+RD V+KG
Sbjct: 1699 SQVLWMISTLKDYGLSFSGVPLLCDNTSAINIAKNPVQHSRTKHIEIRYHFLRDNVEKGT 1758
Query: 1678 L 1678
+
Sbjct: 1759 I 1759
Score = 120 bits (300), Expect = 5e-25
Identities = 118/492 (23%), Positives = 203/492 (40%), Gaps = 56/492 (11%)
Query: 38 DPEEFSWWKTNMYSFIMGLDEELWDILEDGV---DDLDLDEEGAAIDRRIHTPAQKKLYK 94
D F WK M +++ +W+ ++ DD+D+ TPA
Sbjct: 13 DGHNFVIWKARMEAYLPSQGHNVWNKVKSPYTVPDDVDI------------TPANMAQVD 60
Query: 95 KHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVH-QYELF 153
+++ R I+ I E+ ++ +A M+ +LC EG+ ++ L H +Y+ F
Sbjct: 61 FNYRARNAIIGGISSGEFNRVQHHKSAHDMWTALCNFHEGNNDIQ----LNQFHKEYQRF 116
Query: 154 RMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLP-SRWRPKVTAIEEAKDL 212
M ESI+ + RF +VS L+ + K + +D+ +L L W KVT+I E+ L
Sbjct: 117 EMHPGESIDSYFKRFGEIVSKLRSVGKEFSDNDNARHLLNCLDYGVWEMKVTSITESAPL 176
Query: 213 NTLSVEDLVSSLKVHEMSLNEHESSKKSKSIALPSKGKTSKSSKAYKASESEEESPDRDS 272
+ L+++ L S LK HEM + + K S ++ G TS + A+ +
Sbjct: 177 SELTMDKLYSKLKTHEMDVFHRKGLKHSMALVADPSGSTSSNDSAFVCG-GFSLAALHSV 235
Query: 273 DEDQSVKMAMLSNKLEYLARKQKKFLSKRGGYKNSKKEDQKGCFNCKKPGHFIADCPDLQ 332
E+Q K+ + L ARK + K K + CF C +P H +CP L
Sbjct: 236 TEEQLEKIP--EDDLALFARKFSRAYKNVRDRKRGKTNEPFVCFECGEPNHIRVNCPKL- 292
Query: 333 KEKSKSKSKKSSFSSSKFRKQIKK----SLMATWEDLD-SESGSDKEEAEDDTKAAMGLV 387
K+KS +KK K +K + K ++A E++ S+ SD ++ E K G+
Sbjct: 293 KKKSDKTTKKPEGQGRKGKKDLMKKAIHKVLAALEEVQLSDIDSDDDDQEKGDKDFSGMC 352
Query: 388 ATVSSE--------AVSEAESDSEDENEVYSKIPR------------QELVD---SLKEL 424
++E A+ + + SE IP + VD S + +
Sbjct: 353 CLANNEDFINLCLMALEDKDDSSEHPEVCLDDIPSLDGSLCDDSCSDNDTVDDELSKERM 412
Query: 425 LSLFEHRTNELTDLKEKYVDLMKQQKSTLLELKASEEELKGFNLIST--TYEDRLKILCQ 482
L +++ K K L + LE+ + + ST +Y + +L
Sbjct: 413 AHLMIEISDKYRSSKYKIEKLKSENDGMALEIARLRSMIPEEDTCSTCASYLSEINLLKD 472
Query: 483 KLQEKCDKGSGN 494
KL + C G+GN
Sbjct: 473 KL-KSCTLGAGN 483
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.314 0.131 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,806,516,505
Number of Sequences: 2540612
Number of extensions: 121985107
Number of successful extensions: 483728
Number of sequences better than 10.0: 7883
Number of HSP's better than 10.0 without gapping: 3526
Number of HSP's successfully gapped in prelim test: 4545
Number of HSP's that attempted gapping in prelim test: 431717
Number of HSP's gapped (non-prelim): 35724
length of query: 1716
length of database: 863,360,394
effective HSP length: 142
effective length of query: 1574
effective length of database: 502,593,490
effective search space: 791082153260
effective search space used: 791082153260
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 83 (36.6 bits)
Medicago: description of AC149496.5


