

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149495.3 + phase: 0
(412 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
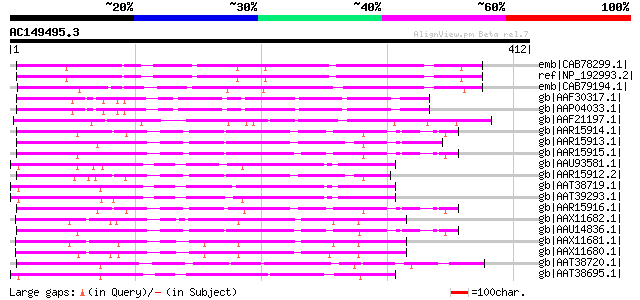
emb|CAB78299.1| putative protein [Arabidopsis thaliana] gi|47259... 191 4e-47
ref|NP_192993.2| F-box family protein [Arabidopsis thaliana] 191 4e-47
emb|CAB79194.1| putative protein [Arabidopsis thaliana] gi|57383... 135 2e-30
gb|AAF30317.1| hypothetical protein [Arabidopsis thaliana] gi|21... 114 7e-24
gb|AAP04033.1| putative F-box protein family [Arabidopsis thalia... 114 7e-24
gb|AAF21197.1| unknown protein [Arabidopsis thaliana] gi|1598347... 110 8e-23
gb|AAR15914.1| S1 self-incompatibility locus-linked putative F-b... 106 1e-21
gb|AAR15913.1| S3 self-incompatibility locus-linked putative F-b... 106 1e-21
gb|AAR15915.1| S2 self-incompatibility locus-linked putative F-b... 106 2e-21
gb|AAU93581.1| putative F-box protein [Solanum demissum] 104 4e-21
gb|AAR15912.2| S2 self-incompatibility locus-linked putative F-b... 103 7e-21
gb|AAT38719.1| putative F-Box protein [Solanum demissum] 102 2e-20
gb|AAT39293.1| putative F-box-like protein [Solanum demissum] 102 3e-20
gb|AAR15916.1| S3 self-incompatibility locus-linked putative F-b... 100 6e-20
gb|AAX11682.1| non-S F-box protein 1 [Petunia axillaris subsp. a... 100 8e-20
gb|AAU14836.1| S3 putative F-box protein SLF-S3B [Petunia x hybr... 100 8e-20
gb|AAX11681.1| S19-locus linked F-box protein [Petunia axillaris... 100 1e-19
gb|AAX11680.1| S17-locus linked F-box protein [Petunia axillaris... 100 1e-19
gb|AAT38720.1| putative F-Box protein [Solanum demissum] 99 2e-19
gb|AAT38695.1| putative F-box protein [Solanum demissum] 98 4e-19
>emb|CAB78299.1| putative protein [Arabidopsis thaliana] gi|4725955|emb|CAB41726.1|
putative protein [Arabidopsis thaliana]
gi|7487230|pir||T07648 hypothetical protein T1P17.150 -
Arabidopsis thaliana
Length = 408
Score = 191 bits (485), Expect = 4e-47
Identities = 119/389 (30%), Positives = 206/389 (52%), Gaps = 43/389 (11%)
Query: 6 LPPDTLAEIFSRLPVKSLLRFRSTSKSLKSIIDSHNFI--NLHRKNSLNRSFILRLRSNI 63
+P D + +IF RLP K+L+R R+ SK +I+ +FI +LHR ++ LR +
Sbjct: 4 IPMDIVNDIFLRLPAKTLVRCRALSKPCYHLINDPDFIESHLHRVLQTGDHLMILLRGAL 63
Query: 64 YQIEDDFSNLTTAVPLNHPFTRNSTNIALIGSCNGLLAVSNGEIALRHPNAANEITIWNP 123
D +L + + HP R + GS NGL+ +SN + ++ ++NP
Sbjct: 64 RLYSVDLDSLDSVSDVEHPMKRGGPT-EVFGSSNGLIGLSN---------SPTDLAVFNP 113
Query: 124 NIRKHHIIPFLPLPITPRSPSDMNCSLCVHGFGFDPLTGDYKILRL-SWLVSLQNPF--- 179
+ R+ H +P + + S + +G G+D ++ DYK++R+ + + ++
Sbjct: 114 STRQIHRLPPSSIDLPDGSSTR---GYVFYGLGYDSVSDDYKVVRMVQFKIDSEDELGCS 170
Query: 180 YDPHVRLFSLKTNSWKIIPTMP----------YALVFAQTMGVLVEDSIHWIMAKKLDGL 229
+ V++FSLK NSWK I ++ Y L++ + GVL +S+HW++ ++ +
Sbjct: 171 FPYEVKVFSLKKNSWKRIESVASSIQLLFYFYYHLLYRRGYGVLAGNSLHWVLPRRPGLI 230
Query: 230 HPSLIVAFNLTLEIFNEVPLPDEIGEEEVNSNDSVEIDVAALGGCLCMTVNYETTKIDVW 289
+LIV F+L LE F V P+ + N N +++D+ L GCLC+ NY+ + +DVW
Sbjct: 231 AFNLIVRFDLALEEFEIVRFPEAVA----NGNVDIQMDIGVLDGCLCLMCNYDQSYVDVW 286
Query: 290 VMKQYGLKDSWCKLFTMMKSCVTSHLKSSSPLCYSSDGSKVLIEGIEVLLEVHHKKLFWY 349
+MK+Y ++DSW K+FT+ K PL YS D K VLLE+++ KL W+
Sbjct: 287 MMKEYNVRDSWTKVFTVQKPKSVKSFSYMRPLVYSKDKKK-------VLLELNNTKLVWF 339
Query: 350 DLKTEQVS---YEEIPDFNGAKICVGSLV 375
DL+++++S ++ P A++ V SLV
Sbjct: 340 DLESKKMSTLRIKDCPSSYSAELVVSSLV 368
>ref|NP_192993.2| F-box family protein [Arabidopsis thaliana]
Length = 413
Score = 191 bits (485), Expect = 4e-47
Identities = 119/389 (30%), Positives = 206/389 (52%), Gaps = 43/389 (11%)
Query: 6 LPPDTLAEIFSRLPVKSLLRFRSTSKSLKSIIDSHNFI--NLHRKNSLNRSFILRLRSNI 63
+P D + +IF RLP K+L+R R+ SK +I+ +FI +LHR ++ LR +
Sbjct: 4 IPMDIVNDIFLRLPAKTLVRCRALSKPCYHLINDPDFIESHLHRVLQTGDHLMILLRGAL 63
Query: 64 YQIEDDFSNLTTAVPLNHPFTRNSTNIALIGSCNGLLAVSNGEIALRHPNAANEITIWNP 123
D +L + + HP R + GS NGL+ +SN + ++ ++NP
Sbjct: 64 RLYSVDLDSLDSVSDVEHPMKRGGPT-EVFGSSNGLIGLSN---------SPTDLAVFNP 113
Query: 124 NIRKHHIIPFLPLPITPRSPSDMNCSLCVHGFGFDPLTGDYKILRL-SWLVSLQNPF--- 179
+ R+ H +P + + S + +G G+D ++ DYK++R+ + + ++
Sbjct: 114 STRQIHRLPPSSIDLPDGSSTR---GYVFYGLGYDSVSDDYKVVRMVQFKIDSEDELGCS 170
Query: 180 YDPHVRLFSLKTNSWKIIPTMP----------YALVFAQTMGVLVEDSIHWIMAKKLDGL 229
+ V++FSLK NSWK I ++ Y L++ + GVL +S+HW++ ++ +
Sbjct: 171 FPYEVKVFSLKKNSWKRIESVASSIQLLFYFYYHLLYRRGYGVLAGNSLHWVLPRRPGLI 230
Query: 230 HPSLIVAFNLTLEIFNEVPLPDEIGEEEVNSNDSVEIDVAALGGCLCMTVNYETTKIDVW 289
+LIV F+L LE F V P+ + N N +++D+ L GCLC+ NY+ + +DVW
Sbjct: 231 AFNLIVRFDLALEEFEIVRFPEAVA----NGNVDIQMDIGVLDGCLCLMCNYDQSYVDVW 286
Query: 290 VMKQYGLKDSWCKLFTMMKSCVTSHLKSSSPLCYSSDGSKVLIEGIEVLLEVHHKKLFWY 349
+MK+Y ++DSW K+FT+ K PL YS D K VLLE+++ KL W+
Sbjct: 287 MMKEYNVRDSWTKVFTVQKPKSVKSFSYMRPLVYSKDKKK-------VLLELNNTKLVWF 339
Query: 350 DLKTEQVS---YEEIPDFNGAKICVGSLV 375
DL+++++S ++ P A++ V SLV
Sbjct: 340 DLESKKMSTLRIKDCPSSYSAELVVSSLV 368
>emb|CAB79194.1| putative protein [Arabidopsis thaliana] gi|5738370|emb|CAB52813.1|
putative protein [Arabidopsis thaliana]
gi|15235636|ref|NP_193970.1| F-box family
protein-related [Arabidopsis thaliana]
gi|25407584|pir||D85256 hypothetical protein AT4g22390
[imported] - Arabidopsis thaliana
Length = 394
Score = 135 bits (341), Expect = 2e-30
Identities = 108/389 (27%), Positives = 184/389 (46%), Gaps = 49/389 (12%)
Query: 7 PPDTLAEIFSRLPVKSLLRFRSTSKSLKSIIDSHNFINLHRKNSLNRS----FILRLRSN 62
P D + E+F RL +L++ R SK S+IDS F++ H + L +LR
Sbjct: 5 PTDLINEMFLRLRATTLVKCRVLSKPCFSLIDSPEFVSSHLRRRLETGEHLMILLRGPRL 64
Query: 63 IYQIEDDFSNLTTAVPLNHPFTRNSTNIALIGSCNGLLAVSNGEIALRHPNAANEITIWN 122
+ +E D + +P HP + GS NG++ + N + ++ I+N
Sbjct: 65 LRTVELDSPENVSDIP--HPLQAGGFT-EVFGSFNGVIGLCNSPV---------DLAIFN 112
Query: 123 PNIRKHHIIPFLPLPITPRSPSDMNCSLCVHGFGFDPLTGDYKILRLSW--LVSLQNPFY 180
P+ RK H +P P+ R D+ +G G+D + D+K++R+ L + F
Sbjct: 113 PSTRKIHRLPIEPIDFPER---DITREYVFYGLGYDSVGDDFKVVRIVQCKLKEGKKKFP 169
Query: 181 DP-HVRLFSLKTNSWKIIPTM----------PYALVFAQTMGVLVEDSIHWIMAKKLDGL 229
P V++FSLK NSWK + M Y L+ + GV+V + +HWI+ ++ +
Sbjct: 170 CPVEVKVFSLKKNSWKRVCLMFEFQILWISYYYHLLPRRGYGVVVNNHLHWILPRRQGVI 229
Query: 230 HPSLIVAFNLTLEIFNEVPLPDEIGEEEVNSNDSVEIDVAALGGCLCMTVNYETTKIDVW 289
+ I+ ++L + + P E+ E+ +D+ L GC+C+ E + +DVW
Sbjct: 230 AFNAIIKYDLASDDIGVLSFPQELYIED-------NMDIGVLDGCVCLMCYDEYSHVDVW 282
Query: 290 VMKQYGLKDSWCKLFTMMKSCVTSHLKSSSPLCYSSDGSKVLIEGIEVLLEVHH-KKLFW 348
V+K+Y SW KL+ + K ++ PL S D SK +LLE+++ L W
Sbjct: 283 VLKEYEDYKSWTKLYRVPKPESVESVEFIRPLICSKDRSK-------ILLEINNAANLMW 335
Query: 349 YDLKTEQVSYE--EIPDFNGAKICVGSLV 375
+DL+++ ++ E A I V SLV
Sbjct: 336 FDLESQSLTTAGIECDSSFTADILVSSLV 364
>gb|AAF30317.1| hypothetical protein [Arabidopsis thaliana]
gi|21536665|gb|AAM60997.1| unknown [Arabidopsis
thaliana]
Length = 416
Score = 114 bits (284), Expect = 7e-24
Identities = 104/375 (27%), Positives = 163/375 (42%), Gaps = 78/375 (20%)
Query: 6 LPPDTLAEIFSRLPVKSLLRFRSTSKSLKSIIDSHNFINLHRK--------NSLNRSFIL 57
LPP+ + EI RLP KS+ RFR SK ++ F +H SL+R I+
Sbjct: 25 LPPEIITEILLRLPAKSIGRFRCVSKLFCTLSSDPGFAKIHLDLILRNESVRSLHRKLIV 84
Query: 58 RLRSNIYQIEDDFSNL------TTAVPLNHPFT----------RNSTN------------ 89
N+Y + DF+++ AV N+P RN
Sbjct: 85 SSH-NLYSL--DFNSIGDGIRDLAAVEHNYPLKDDPSIFSEMIRNYVGDHLYDDRRVMLK 141
Query: 90 ----------IALIGSCNGLLAVSNGEIALRHPNAANEITIWNPNIRKHHIIPFLPLPIT 139
+ ++GS NGL+ +S GE A + ++NP LP
Sbjct: 142 LNAKSYRRNWVEIVGSSNGLVCISPGEGA---------VFLYNPTTGDSKR---LPENFR 189
Query: 140 PRSPSDMNCSLCVHGFGFDPLTGDYKILRLSWLVSLQNPFYDPHVRLFSLKTNSWKIIPT 199
P+S + +GFGFD LT DYK+++L V+ D V +SLK +SW+ I
Sbjct: 190 PKSVEYERDNFQTYGFGFDGLTDDYKLVKL---VATSEDILDASV--YSLKADSWRRICN 244
Query: 200 MPYALVFAQ-TMGVLVEDSIHWIMAKKLDGLHPSLIVAFNLTLEIFNEVPLPDEIGEEEV 258
+ Y T GV +IHW+ + + ++VAF++ E F E+P+PDE +
Sbjct: 245 LNYEHNDGSYTSGVHFNGAIHWVFTESRH--NQRVVVAFDIQTEEFREMPVPDEAEDCSH 302
Query: 259 NSNDSVEIDVAALGGCLCMTVNYETTKIDVWVMKQYGLKDSWCKLFTMMKSCVTSHLKSS 318
++ V V +L G LC+ + D+WVM +YG SW ++ + +S
Sbjct: 303 RFSNFV---VGSLNGRLCVVNSCYDVHDDIWVMSEYGEAKSWSRI------RINLLYRSM 353
Query: 319 SPLCYSSDGSKVLIE 333
PLC + + +VL+E
Sbjct: 354 KPLCSTKNDEEVLLE 368
>gb|AAP04033.1| putative F-box protein family [Arabidopsis thaliana]
gi|26451648|dbj|BAC42921.1| unknown protein [Arabidopsis
thaliana] gi|30679777|ref|NP_566277.2| F-box family
protein [Arabidopsis thaliana]
Length = 427
Score = 114 bits (284), Expect = 7e-24
Identities = 104/375 (27%), Positives = 163/375 (42%), Gaps = 78/375 (20%)
Query: 6 LPPDTLAEIFSRLPVKSLLRFRSTSKSLKSIIDSHNFINLHRK--------NSLNRSFIL 57
LPP+ + EI RLP KS+ RFR SK ++ F +H SL+R I+
Sbjct: 36 LPPEIITEILLRLPAKSIGRFRCVSKLFCTLSSDPGFAKIHLDLILRNESVRSLHRKLIV 95
Query: 58 RLRSNIYQIEDDFSNL------TTAVPLNHPFT----------RNSTN------------ 89
N+Y + DF+++ AV N+P RN
Sbjct: 96 SSH-NLYSL--DFNSIGDGIRDLAAVEHNYPLKDDPSIFSEMIRNYVGDHLYDDRRVMLK 152
Query: 90 ----------IALIGSCNGLLAVSNGEIALRHPNAANEITIWNPNIRKHHIIPFLPLPIT 139
+ ++GS NGL+ +S GE A + ++NP LP
Sbjct: 153 LNAKSYRRNWVEIVGSSNGLVCISPGEGA---------VFLYNPTTGDSKR---LPENFR 200
Query: 140 PRSPSDMNCSLCVHGFGFDPLTGDYKILRLSWLVSLQNPFYDPHVRLFSLKTNSWKIIPT 199
P+S + +GFGFD LT DYK+++L V+ D V +SLK +SW+ I
Sbjct: 201 PKSVEYERDNFQTYGFGFDGLTDDYKLVKL---VATSEDILDASV--YSLKADSWRRICN 255
Query: 200 MPYALVFAQ-TMGVLVEDSIHWIMAKKLDGLHPSLIVAFNLTLEIFNEVPLPDEIGEEEV 258
+ Y T GV +IHW+ + + ++VAF++ E F E+P+PDE +
Sbjct: 256 LNYEHNDGSYTSGVHFNGAIHWVFTESRH--NQRVVVAFDIQTEEFREMPVPDEAEDCSH 313
Query: 259 NSNDSVEIDVAALGGCLCMTVNYETTKIDVWVMKQYGLKDSWCKLFTMMKSCVTSHLKSS 318
++ V V +L G LC+ + D+WVM +YG SW ++ + +S
Sbjct: 314 RFSNFV---VGSLNGRLCVVNSCYDVHDDIWVMSEYGEAKSWSRI------RINLLYRSM 364
Query: 319 SPLCYSSDGSKVLIE 333
PLC + + +VL+E
Sbjct: 365 KPLCSTKNDEEVLLE 379
>gb|AAF21197.1| unknown protein [Arabidopsis thaliana] gi|15983479|gb|AAL11607.1|
AT3g07870/F17A17_21 [Arabidopsis thaliana]
gi|18398079|ref|NP_566322.1| F-box family protein
[Arabidopsis thaliana]
Length = 417
Score = 110 bits (275), Expect = 8e-23
Identities = 105/418 (25%), Positives = 176/418 (41%), Gaps = 67/418 (16%)
Query: 4 DRLPPDTLAEIFSRLPVKSLLRFRSTSKSLKSIIDSHNFINLHRKNSLNRSFILRLRSNI 63
+ LP D +A+IFSRLP+ S+ R +S +S++ H ++ + +L S I
Sbjct: 26 ESLPEDIIADIFSRLPISSIARLMFVCRSWRSVLTQHGRLSSSSSSPTKPCLLLHCDSPI 85
Query: 64 ------YQIEDDFSNLTTAVPLNHPFTRNSTNIALIGSCNGLLAVS----NGEIALRHPN 113
+ ++ + T F + ++GSCNGLL +S N + L +P
Sbjct: 86 RNGLHFLDLSEEEKRIKTK-KFTLRFASSMPEFDVVGSCNGLLCLSDSLYNDSLYLYNPF 144
Query: 114 AANEITIWNPNIRKHHIIPFLPLPITPRSPSDMNCSLCVHGFGFDPLTGDYKILRLSWL- 172
N + + P + + V GFGF +T +YK+L++ +
Sbjct: 145 TTNSLEL-------------------PECSNKYHDQELVFGFGFHEMTKEYKVLKIVYFR 185
Query: 173 --VSLQNPFYDPHVRL-----------FSLKTN----SWKIIPTMPYALVFAQTMGVLVE 215
S N Y R+ S KT SW+ + PY V ++ LV
Sbjct: 186 GSSSNNNGIYRGRGRIQYKQSEVQILTLSSKTTDQSLSWRSLGKAPYKFV-KRSSEALVN 244
Query: 216 DSIHWIMAKKLDGLHPSLIVAFNLTLEIFNEVPLPDEIGEEEVNSNDSVEIDVAALGGCL 275
+H++ + + V+F+L E F E+P PD G N + L GCL
Sbjct: 245 GRLHFVTRPRRH-VPDRKFVSFDLEDEEFKEIPKPDCGGLNRTNHR------LVNLKGCL 297
Query: 276 CMTVNYETTKIDVWVMKQYGLKDSWCKLF---TMMKSCVTSHLKSSSPLCYSSDGSKV-- 330
C V K+D+WVMK YG+K+SW K + T + + +L + +++ KV
Sbjct: 298 CAVVYGNYGKLDIWVMKTYGVKESWGKEYSIGTYLPKGLKQNLDRPMWIWKNAENGKVVR 357
Query: 331 ---LIEGIEVLLEVHHKKLFWYDLKT---EQVSYEEIPDFNGAKICVGSLVPPSFPVD 382
L+E E+LLE + L YD K + + + +P++ + G+L P+D
Sbjct: 358 VLCLLENGEILLEYKSRVLVAYDPKLGKFKDLLFHGLPNWFHTVVHAGTLSWFDTPLD 415
>gb|AAR15914.1| S1 self-incompatibility locus-linked putative F-box protein S1-A134
[Petunia integrifolia subsp. inflata]
Length = 379
Score = 106 bits (265), Expect = 1e-21
Identities = 108/380 (28%), Positives = 167/380 (43%), Gaps = 72/380 (18%)
Query: 6 LPPDTLAEIFSRLPVKSLLRFRSTSKSLKSIIDSHNFINLHRKNSLN------------- 52
LP D + IF LPVKSLLRF+ T K+ II S FINLH ++ N
Sbjct: 4 LPQDVVIYIFVMLPVKSLLRFKCTCKTFYHIIKSSTFINLHLNHTTNFNDELVLLKRSFE 63
Query: 53 ---RSFILRLRSNIYQIED-DFSNLTTAVPLNHPFTRNSTNIA--LIGSCNGLLAVSNGE 106
+F + S ++ ED DF ++ V + H T + I LIG CNGL+ +++
Sbjct: 64 TDEYNFYKSILSFLFAKEDYDFKPISPDVEIPH-LTTTAACICHRLIGPCNGLIVLTDSL 122
Query: 107 IALRHPNAANEITIWNPNIRKHHIIPFLPLPITPRSPSDMNCSLCVHGFGFDPLTGDYKI 166
+ ++NP K+ +IP P I P S+ GFGFD DYK+
Sbjct: 123 TTI----------VFNPATLKYRLIPPCPFGI----PRGFRRSISGIGFGFDSDANDYKV 168
Query: 167 LRLSWLVSLQNPFYDPHVRLFSLKTNSWKIIPTMPYALVF-AQTMGVLVEDSIHWIMAKK 225
+RLS V + + V ++ +SW+ + VF +L + + HW
Sbjct: 169 VRLS-EVYKEPCDKEMKVDIYDFSVDSWRELLGQDVPFVFWFPCAEILYKRNFHWFAF-- 225
Query: 226 LDGLHPSLIVAFNLTLEIFNEVPLPDEIGEEEVNSNDSVEIDVAALGGCLCMTV------ 279
+I+ F++ E F+ + +PD + +D + L C+ +
Sbjct: 226 ---ADDVVILCFDMNTEKFHNMGMPD-----ACHFDDGKSYGLVILFKCMTLICYPDPMP 277
Query: 280 NYETTKI-DVWVMKQYGLKDSWCKLFTMMKSCVTSHLKSSSPLCYSSDGSKVLIEGIEVL 338
+ T K+ D+W+MK+YG K+SW +K C + L SPL D E+L
Sbjct: 278 SSPTEKLTDIWIMKEYGEKESW------IKRC-SIRLLPESPLAVWKD---------EIL 321
Query: 339 LEVHHK--KLFWYDLKTEQV 356
L +H K L YDL + +V
Sbjct: 322 L-LHSKMGHLIAYDLNSNEV 340
>gb|AAR15913.1| S3 self-incompatibility locus-linked putative F-box protein S3-A113
[Petunia integrifolia subsp. inflata]
Length = 376
Score = 106 bits (265), Expect = 1e-21
Identities = 100/366 (27%), Positives = 161/366 (43%), Gaps = 62/366 (16%)
Query: 6 LPPDTLAEIFSRLPVKSLLRFRSTSKSLKSIIDSHNFINLHRKNSLNRSFILRLRSNIYQ 65
LP D + I LPVKSLLRF+ + K+ +II S FI LH ++ N L L ++
Sbjct: 4 LPQDVVIYILVMLPVKSLLRFKCSCKTFCNIIKSSTFIYLHLNHTTNVKDELVLLKRSFK 63
Query: 66 IED-----------------DFSNLTTAVPLNHPFTRNSTNI-ALIGSCNGLLAVSNGEI 107
+D DF +++ V + H T ++ LIG CNGL+A+++
Sbjct: 64 TDDYNFYKSILSFLSSKEGYDFKSISPDVEIPHLTTTSACVFHQLIGPCNGLIALTDSLT 123
Query: 108 ALRHPNAANEITIWNPNIRKHHIIPFLPLPITPRSPSDMNCSLCVHGFGFDPLTGDYKIL 167
+ ++NP RK+ +IP P I P S+ GFGFD DYK++
Sbjct: 124 TI----------VFNPATRKYRLIPPCPFGI----PRGFRRSISGIGFGFDSDANDYKVV 169
Query: 168 RLSWLVSLQNPFYDPHVRLFSLKTNSWKIIPTMPYALVF-AQTMGVLVEDSIHWIMAKKL 226
RLS V + + V ++ +SW+ + +V+ +L + + HW
Sbjct: 170 RLS-EVYKEPCDKEMKVDIYDFSVDSWRELLGQEVPIVYWLPCAEILYKRNFHWFAF--- 225
Query: 227 DGLHPSLIVAFNLTLEIFNEVPLPDEIGEEEVNSNDSVEIDVAALGGCLCMTV------- 279
+I+ F++ E F+ + +PD ++ V + C CMT+
Sbjct: 226 --ADDVVILCFDMNTEKFHNLGMPDACHFDDGKCYGLVIL-------CKCMTLICYPDPM 276
Query: 280 -NYETTKI-DVWVMKQYGLKDSWCKLFTMMKSCVTSHLKSSSPLCYSSDGSKVLIEGIEV 337
+ T K+ D+W+MK+YG K+SW +K C + L SPL D +L I
Sbjct: 277 PSSPTEKLTDIWIMKEYGEKESW------IKRC-SIRLLPESPLAVWMDEILLLQSKIGH 329
Query: 338 LLEVHH 343
L+ H
Sbjct: 330 LIAYDH 335
>gb|AAR15915.1| S2 self-incompatibility locus-linked putative F-box protein S2-A134
[Petunia integrifolia subsp. inflata]
Length = 379
Score = 106 bits (264), Expect = 2e-21
Identities = 108/379 (28%), Positives = 166/379 (43%), Gaps = 70/379 (18%)
Query: 6 LPPDTLAEIFSRLPVKSLLRFRSTSKSLKSIIDSHNFINLHRKNSLN------------- 52
LP D + IF LPVKSLLRF+ T K+ II S FINLH ++ N
Sbjct: 4 LPQDVVIYIFVMLPVKSLLRFKCTCKTFCHIIKSSTFINLHLNHTTNFNDELVLLKRSFE 63
Query: 53 ---RSFILRLRSNIYQIED-DFSNLTTAVPLNHPFTRNSTNI-ALIGSCNGLLAVSNGEI 107
F + S ++ ED DF ++ V + H T + LIG CNGL+ +++
Sbjct: 64 TDEYKFYKSILSFLFAKEDYDFKPISPDVEIPHLTTTAACVCHRLIGPCNGLIVLTDSLT 123
Query: 108 ALRHPNAANEITIWNPNIRKHHIIPFLPLPITPRSPSDMNCSLCVHGFGFDPLTGDYKIL 167
+ ++NP K+ +IP P I P S+ GFGFD DYK++
Sbjct: 124 TI----------VFNPATLKYRLIPPCPFGI----PRGFRRSISGIGFGFDSDANDYKVV 169
Query: 168 RLSWLVSLQNPFYDPHVRLFSLKTNSWKIIPTMPYALVF-AQTMGVLVEDSIHWIMAKKL 226
RLS V + + V ++ +SW+ + VF +L + + HW +
Sbjct: 170 RLS-EVYKEPCDKEMKVDIYDFSVDSWRELLGQDVPFVFWFPCAEILYKRNFHWFAFADV 228
Query: 227 DGLHPSLIVAFNLTLEIFNEVPLPDEIGEEEVNSNDSVEIDVAALGGCLCMTV------N 280
+I+ F++ E F+ + +PD + +D + L C+ + +
Sbjct: 229 -----VVILCFDMNTEKFHNMGMPD-----ACHFDDGKCYGLVILFKCMTLICYPDPKPS 278
Query: 281 YETTKI-DVWVMKQYGLKDSWCKLFTMMKSCVTSHLKSSSPLCYSSDGSKVLIEGIEVLL 339
T K+ D+W+MK+YG K+SW MK C + L SPL D E+LL
Sbjct: 279 SPTEKLTDIWIMKEYGEKESW------MKRC-SIRLLPESPLAVWKD---------EILL 322
Query: 340 EVHHK--KLFWYDLKTEQV 356
+H K L YDL + +V
Sbjct: 323 -LHSKMGHLMAYDLNSNEV 340
>gb|AAU93581.1| putative F-box protein [Solanum demissum]
Length = 383
Score = 104 bits (260), Expect = 4e-21
Identities = 89/326 (27%), Positives = 140/326 (42%), Gaps = 47/326 (14%)
Query: 1 MASDR-----LPPDTLAEIFSRLPVKSLLRFRSTSKSLKSIIDSHNFINLHRKNSLN--- 52
MASD LP + + EI RLP+KSL +F SKS +I S F+ H K + N
Sbjct: 1 MASDSIIPLLLPDELITEILLRLPIKSLSKFMCVSKSWLQLISSPAFVKKHIKLTANDKG 60
Query: 53 ---RSFILRLRSNIYQ---IEDDFSN---LTTAVPLNHPFTRNSTNIALIGSCNGLLAVS 103
I R +N ++ + F+N + + ++ P R + + ++GS NGL+ V+
Sbjct: 61 YIYHRLIFRNTNNDFKFCPLPPLFTNQQLIEEILHIDSPIERTTLSTHIVGSVNGLICVA 120
Query: 104 NGEIALRHPNAANEITIWNPNIRKHHIIPFLPLPITPRSPSDMNCSLCVHGFGFDPLTGD 163
+ E IWNP I K + P+S S++ GFG+D D
Sbjct: 121 HVR--------QREAYIWNPAITKSKEL--------PKSTSNLCSDGIKCGFGYDESRDD 164
Query: 164 YKILRLSWLVSLQNPFYDPH---VRLFSLKTNSWKIIPTMPYALVFAQTMGVLVEDSIHW 220
YK+ V + P H V ++SL+TNSW + + G V ++W
Sbjct: 165 YKV------VFIDYPIRHNHRTVVNIYSLRTNSWTTLHDQLQGIFLLNLHGRFVNGKLYW 218
Query: 221 IMAKKLDGLHPSLIVAFNLTLEIFNEVPLPDEIGEEEVNSNDSVEIDVAALGGCLCMTVN 280
+ ++ I +F+L + + LP G++ N + + V L T
Sbjct: 219 TSSTCINNYKVCNITSFDLADGTWGSLELP-SCGKD----NSYINVGVVGSDLSLLYTCQ 273
Query: 281 YETTKIDVWVMKQYGLKDSWCKLFTM 306
DVW+MK G+ SW KLFT+
Sbjct: 274 LGAATSDVWIMKHSGVNVSWTKLFTI 299
>gb|AAR15912.2| S2 self-incompatibility locus-linked putative F-box protein S2-A113
[Petunia integrifolia subsp. inflata]
gi|38261532|gb|AAR15911.1| S1 self-incompatibility
locus-linked putative F-box protein S1-A113 [Petunia
integrifolia subsp. inflata]
Length = 376
Score = 103 bits (258), Expect = 7e-21
Identities = 93/326 (28%), Positives = 150/326 (45%), Gaps = 57/326 (17%)
Query: 6 LPPDTLAEIFSRLPVKSLLRFRSTSKSLKSIIDSHNFINLHRKNS--LNRSFILRLRS-- 61
LP D + I LPVKSLLRF+ + K+ +II S F+NLH ++ + +L RS
Sbjct: 4 LPQDVVIYILVMLPVKSLLRFKCSCKTFCNIIKSSTFVNLHLNHTTKVKDELVLLKRSFK 63
Query: 62 ----NIYQI--------ED-DFSNLTTAVPLNHPFTRNSTNI--ALIGSCNGLLAVSNGE 106
N Y+ ED DF ++ V + H T S + LIG CNGL+A+++
Sbjct: 64 TDEYNFYKSILSFFSSKEDYDFMPMSPDVEIPH-LTTTSARVFHQLIGPCNGLIALTDSL 122
Query: 107 IALRHPNAANEITIWNPNIRKHHIIPFLPLPITPRSPSDMNCSLCVHGFGFDPLTGDYKI 166
+ ++NP RK+ +IP P I P S+ GFGFD DYK+
Sbjct: 123 TTI----------VFNPATRKYRLIPPCPFGI----PRGFRRSISGIGFGFDSDVNDYKV 168
Query: 167 LRLSWLVSLQNPFYDPHVRLFSLKTNSWKIIPTMPYALVF-AQTMGVLVEDSIHWIMAKK 225
+RLS V + + V ++ +SW+ + +V+ +L + + HW
Sbjct: 169 VRLS-EVYKEPCDKEMKVDIYDFSVDSWRELLGQEVPIVYWLPCADILFKRNFHWFAF-- 225
Query: 226 LDGLHPSLIVAFNLTLEIFNEVPLPDEIGEEEVNSNDSVEIDVAALGGCLCMTV------ 279
+I+ F++ E F+ + +PD ++ V + C CM++
Sbjct: 226 ---ADDVVILCFDMNTEKFHNMGMPDACHFDDGKCYGLVIL-------CKCMSLICYPDP 275
Query: 280 --NYETTKI-DVWVMKQYGLKDSWCK 302
+ T K+ D+W+MK+YG K+SW K
Sbjct: 276 MPSSPTEKLTDIWIMKEYGEKESWIK 301
>gb|AAT38719.1| putative F-Box protein [Solanum demissum]
Length = 327
Score = 102 bits (254), Expect = 2e-20
Identities = 83/324 (25%), Positives = 134/324 (40%), Gaps = 41/324 (12%)
Query: 1 MASDR------LPPDTLAEIFSRLPVKSLLRFRSTSKSLKSIIDSHNFINLHRKNSLNRS 54
MASD LP + + EI +LP+KSLL+F SKS +I S F+ H K + +
Sbjct: 1 MASDSIIPPLLLPDELITEILLKLPIKSLLKFMCVSKSWLQLISSPAFVKNHIKLTADDK 60
Query: 55 FILRLRSNIYQIEDDF------------SNLTTAVPLNHPFTRNSTNIALIGSCNGLLAV 102
+ R DDF + ++ P R + + ++GS NGL+
Sbjct: 61 GYIYHRLIFRNTNDDFKFCPLPPLFTQQQLIKELYHIDSPIERTTLSTHIVGSVNGLICA 120
Query: 103 SNGEIALRHPNAANEITIWNPNIRKHHIIPFLPLPITPRSPSDMNCSLCVHGFGFDPLTG 162
++ E IWNP I K + P+S S++ GFG+D
Sbjct: 121 AHVR--------QREAYIWNPTITKSKEL--------PKSRSNLCSDGIKCGFGYDESRD 164
Query: 163 DYKILRLSWLVSLQNPFYDPHVRLFSLKTNSWKIIPTMPYALVFAQTMGVLVEDSIHWIM 222
DYK++ + + + N + V ++SL+T SW + G V ++W
Sbjct: 165 DYKVVFIDYPIHRHN--HRTVVNIYSLRTKSWTTLHDQLQGFFLLNLHGRFVNGKLYWTS 222
Query: 223 AKKLDGLHPSLIVAFNLTLEIFNEVPLPDEIGEEEVNSNDSVEIDVAALGGCLCMTVNYE 282
+ ++ I +F+L + + LP G++ N + + V L T
Sbjct: 223 SSCINNYKVCNITSFDLADGTWERLELP-SCGKD----NSYINVGVVGSDLSLLYTCQRG 277
Query: 283 TTKIDVWVMKQYGLKDSWCKLFTM 306
DVW+MK G+ SW KLFT+
Sbjct: 278 AATSDVWIMKHSGVNVSWTKLFTI 301
>gb|AAT39293.1| putative F-box-like protein [Solanum demissum]
Length = 384
Score = 102 bits (253), Expect = 3e-20
Identities = 88/327 (26%), Positives = 138/327 (41%), Gaps = 48/327 (14%)
Query: 1 MASDR------LPPDTLAEIFSRLPVKSLLRFRSTSKSLKSIIDSHNFINLHRKNSLNRS 54
MASD LP + + EI RLP+KSL +F SKS +I S F+ H K + N
Sbjct: 1 MASDSIIPLLLLPDELITEILLRLPIKSLSKFMCVSKSWLQLISSPAFVKNHIKLTANGK 60
Query: 55 FILRLRSNIYQIEDDFS-----NLTTAVPLNH-------PFTRNSTNIALIGSCNGLLAV 102
+ R DDF +L T L P R + + ++GS NGL+
Sbjct: 61 GYIYHRLIFRNTNDDFKFCPLPSLFTKQQLIEELFDIVSPIERTTLSTHIVGSVNGLICA 120
Query: 103 SNGEIALRHPNAANEITIWNPNIRKHHIIPFLPLPITPRSPSDMNCSLCVHGFGFDPLTG 162
++ E IWNP I K + P+S S++ GFG+D
Sbjct: 121 AHVR--------QREAYIWNPTITKSKEL--------PKSRSNLCSDGIKCGFGYDESHD 164
Query: 163 DYKILRLSWLVSLQNPFYDPH---VRLFSLKTNSWKIIPTMPYALVFAQTMGVLVEDSIH 219
DYK++ +++ P + H V ++SL+TNSW + + V++ ++
Sbjct: 165 DYKVVFINY------PSHHNHRSVVNIYSLRTNSWTTLHDQLQGIFLLNLHCRFVKEKLY 218
Query: 220 WIMAKKLDGLHPSLIVAFNLTLEIFNEVPLPDEIGEEEVNSNDSVEIDVAALGGCLCMTV 279
W + ++ I +F+L + + LP G++ N + + V L T
Sbjct: 219 WTSSTCINNYKVCNITSFDLADGTWESLELP-SCGKD----NSYINVGVVGSDLSLLYTC 273
Query: 280 NYETTKIDVWVMKQYGLKDSWCKLFTM 306
DVW+MK G+ SW KLFT+
Sbjct: 274 QRGAANSDVWIMKHSGVNVSWTKLFTI 300
>gb|AAR15916.1| S3 self-incompatibility locus-linked putative F-box protein S3-A134
[Petunia integrifolia subsp. inflata]
Length = 379
Score = 100 bits (250), Expect = 6e-20
Identities = 104/382 (27%), Positives = 165/382 (42%), Gaps = 76/382 (19%)
Query: 6 LPPDTLAEIFSRLPVKSLLRFRSTSKSLKSIIDSHNFINLHRKNSLNRSFILRLRSNIYQ 65
LP D + IF LPVKSLLR + T K+ II S FINLH ++ N + L L ++
Sbjct: 4 LPQDVVIYIFVMLPVKSLLRSKCTCKTFCHIIKSSTFINLHLNHTTNFNDELVLLKRSFE 63
Query: 66 IED-----------------DFSNLTTAVPLNHPFTRNSTNIA--LIGSCNGLLAVSNGE 106
++ DF ++ V + H T + I LIG CNGL+ +++
Sbjct: 64 TDEYNFYKSILSFLFAKKDYDFKPISPDVKIPH-LTTTAACICHRLIGPCNGLIVLTDSL 122
Query: 107 IALRHPNAANEITIWNPNIRKHHIIPFLPLPITPRSPSDMNCSLCVHGFGFDPLTGDYKI 166
+ ++NP K+ +IP P I P S+ GFGFD DYK+
Sbjct: 123 TTI----------VFNPATLKYRLIPPCPFGI----PRGFRRSISGIGFGFDSDANDYKV 168
Query: 167 LRLSWLVSLQNPFYDPHVR--LFSLKTNSWKIIPTMPYALVF-AQTMGVLVEDSIHWIMA 223
+RLS + D ++ ++ +SW+ + VF +L + + HW
Sbjct: 169 VRLS---EVYKGTCDKKMKVDIYDFSVDSWRELLGQDVPFVFWFPCAEILYKRNFHWFAF 225
Query: 224 KKLDGLHPSLIVAFNLTLEIFNEVPLPDEIGEEEVNSNDSVEIDVAALGGCLCMTV---- 279
+I+ F++ E F+ + +PD + +D + L C+ +
Sbjct: 226 -----ADDVVILCFDMNTEKFHNMGMPD-----ACHFDDGKSYGLVILFKCMTLICYPDP 275
Query: 280 --NYETTKI-DVWVMKQYGLKDSWCKLFTMMKSCVTSHLKSSSPLCYSSDGSKVLIEGIE 336
+ T K+ D+W+MK+YG K+SW +K C + L SPL D E
Sbjct: 276 MPSSPTEKLTDIWIMKEYGEKESW------IKRC-SIRLLPESPLAVWKD---------E 319
Query: 337 VLLEVHHK--KLFWYDLKTEQV 356
+LL +H K L YDL + +V
Sbjct: 320 ILL-LHSKMGHLIAYDLNSNEV 340
>gb|AAX11682.1| non-S F-box protein 1 [Petunia axillaris subsp. axillaris]
Length = 389
Score = 100 bits (249), Expect = 8e-20
Identities = 91/337 (27%), Positives = 153/337 (45%), Gaps = 45/337 (13%)
Query: 5 RLPPDTLAEIFSRLPVKSLLRFRSTSKSLKSIIDSHNFINLH--RKNSLNRSFILRLRSN 62
+LP D + I PVKSL+RF+ SKS +I S FIN H RK + FIL R+
Sbjct: 8 KLPEDLVFLILLTFPVKSLMRFKCISKSWSFLIQSTGFINRHVNRKTNTKDEFILFKRA- 66
Query: 63 IYQIEDDFSNLTTAVP-----LNHPF----------TRNSTNIALIGSCNGLLAVSNGEI 107
I E++F N+ + LN F N T LIG C+GL+A+++ I
Sbjct: 67 IKDEEEEFINILSFFSGYDDVLNPLFPDIDVSYMTSNCNCTFNPLIGPCDGLIALTDTII 126
Query: 108 ALRHPNAANEITIWNPNIRKHHIIPFLPLPITPRSPSDMNCSLCVHGFGFDPLTGDYKIL 167
+ + NP R ++P P P P + S+ GFG D ++ YK++
Sbjct: 127 TI----------LLNPATRNFRLLP--PSPFA--CPKGYHRSIEGVGFGLDTISNYYKVV 172
Query: 168 RLSWLVSLQNPFY----DPHVRLFSLKTNSWKIIPTMPYALVF-AQTMGVLVEDSIHWIM 222
R+S + + Y D + + L T+SW+ + + L++ G+L + +HW
Sbjct: 173 RISEVYCEEADGYPGPKDSKIDVCDLVTDSWRELDHIQLPLIYWVPCSGMLYMEMVHWFA 232
Query: 223 AKKLDGLHPSLIVAFNLTLEIFNEVPLPDEIGE--EEVNSNDSVEIDVAALGGCL--CMT 278
+ +I+ F+++ E+F + +PD E+ + D L G +
Sbjct: 233 TTDIS----MVILCFDMSTEVFRNMKMPDTCTRITHELYYGLVILCDSFTLIGYSNPIGS 288
Query: 279 VNYETTKIDVWVMKQYGLKDSWCKLFTMMKSCVTSHL 315
++ K+ +WVM +YG+ +SW +T+ + S L
Sbjct: 289 IDSARDKMHIWVMMEYGVSESWIMKYTIKPLSIESPL 325
>gb|AAU14836.1| S3 putative F-box protein SLF-S3B [Petunia x hybrida]
Length = 379
Score = 100 bits (249), Expect = 8e-20
Identities = 105/379 (27%), Positives = 161/379 (41%), Gaps = 70/379 (18%)
Query: 6 LPPDTLAEIFSRLPVKSLLRFRSTSKSLKSIIDSHNFINLHRKNSLN------------- 52
LP D + IF LPVKSLLRF+ T K+ II S FINLH ++ N
Sbjct: 4 LPQDVVIYIFVMLPVKSLLRFKCTCKTFCHIIKSSTFINLHLNHTTNFNDELVLLKRSFE 63
Query: 53 ---RSFILRLRSNIYQIED-DFSNLTTAVPLNHPFTRNSTNI-ALIGSCNGLLAVSNGEI 107
+F + S ++ ED DF ++ V + H T + LIG CNGL+ +++
Sbjct: 64 TDEYNFYKSILSFLFAKEDYDFKPISPDVEIPHLTTTAACVCHRLIGPCNGLIVLTDSLT 123
Query: 108 ALRHPNAANEITIWNPNIRKHHIIPFLPLPITPRSPSDMNCSLCVHGFGFDPLTGDYKIL 167
+ ++NP K+ +IP P I P S+ GFGFD DYK++
Sbjct: 124 TI----------VFNPATLKYRLIPPCPFGI----PRGFRRSISGIGFGFDSDANDYKVV 169
Query: 168 RLSWLVSLQNPFYDPHVRLFSLKTNSWKIIPTMPYALVF-AQTMGVLVEDSIHWIMAKKL 226
RLS V + + V ++ +SW+ + VF +L + + HW +
Sbjct: 170 RLS-EVYKEPCDKEMKVDIYDFSVDSWRELLGQDVPFVFWFPCAEILYKRNFHWFAFADV 228
Query: 227 DGLHPSLIVAFNLTLEIFNEVPLPDEIGEEEVNSNDSVEIDVAALGGCLC-------MTV 279
+I+ F + E F+ + +PD + D + L C+ M
Sbjct: 229 -----VVILCFEMNTEKFHNMGMPD-----ACHFADGKCYGLVILFKCMTLICYPDPMPS 278
Query: 280 NYETTKIDVWVMKQYGLKDSWCKLFTMMKSCVTSHLKSSSPLCYSSDGSKVLIEGIEVLL 339
+ D+W+MK+YG K+SW +K C + L SPL D E+LL
Sbjct: 279 SPTEXWTDIWIMKEYGEKESW------IKRC-SIRLLPESPLAVWKD---------EILL 322
Query: 340 EVHHK--KLFWYDLKTEQV 356
+H K L YD + +V
Sbjct: 323 -LHSKTGHLIAYDFNSNEV 340
>gb|AAX11681.1| S19-locus linked F-box protein [Petunia axillaris subsp. axillaris]
Length = 389
Score = 100 bits (248), Expect = 1e-19
Identities = 88/343 (25%), Positives = 152/343 (43%), Gaps = 57/343 (16%)
Query: 5 RLPPDTLAEIFSRLPVKSLLRFRSTSKSLKSIIDSHNFINLH--RKNSLNRSFILRLRSN 62
+LP D + I PVKSLLRF+ SK+ +I S FIN H RK + FIL RS
Sbjct: 8 KLPEDLVFLILLTFPVKSLLRFKCISKAWSILIQSTTFINRHINRKTNTKAEFILFKRS- 66
Query: 63 IYQIEDDFSNLTTAVPLNHPFTR---------------NSTNIALIGSCNGLLAVSNGEI 107
I E++F N+ + N + T LIG C+GL+A+++ I
Sbjct: 67 IKDEEEEFINILSFFSGNDDVLNPLFPDIDVSYMTSKCDCTFTPLIGPCDGLIALTDTII 126
Query: 108 ALRHPNAANEITIWNPNIRKHHIIPFLPLPITPRSPSDMNCSLCVH------GFGFDPLT 161
+ + NP R ++P PS C H GFGFD ++
Sbjct: 127 TI----------VLNPATRNFRVLP----------PSPFGCPKGYHRSVEGVGFGFDTIS 166
Query: 162 GDYKILRLSWLVSLQNPFY----DPHVRLFSLKTNSWKIIPTMPY-ALVFAQTMGVLVED 216
YK++R+S + + Y D + + L T+SW+ + + ++ + G+L ++
Sbjct: 167 YYYKVVRISEVYCEEADGYPGPKDSKIDVCDLSTDSWRELDHVQLPSIYWVPCAGMLYKE 226
Query: 217 SIHWIMAKKLDGLHPSLIVAFNLTLEIFNEVPLPDEIGE--EEVNSNDSVEIDVAALGGC 274
+HW +I+ F+++ E+F+++ +PD E+ + + L G
Sbjct: 227 MVHWFATTDTS----MVILCFDMSTEMFHDMKMPDTCSRITHELYYGLVILCESFTLIGY 282
Query: 275 L--CMTVNYETTKIDVWVMKQYGLKDSWCKLFTMMKSCVTSHL 315
+++ K+ +WVM +YG+ +SW +T+ + S L
Sbjct: 283 SNPISSIDPVEDKMHIWVMMEYGVSESWIMKYTIRPLSIESPL 325
>gb|AAX11680.1| S17-locus linked F-box protein [Petunia axillaris subsp. axillaris]
Length = 388
Score = 100 bits (248), Expect = 1e-19
Identities = 89/343 (25%), Positives = 154/343 (43%), Gaps = 58/343 (16%)
Query: 5 RLPPDTLAEIFSRLPVKSLLRFRSTSKSLKSIIDSHNFINLHRKNSLNR--SFILRLRSN 62
+LP D + I VKSL+RF+ SK+ +I S FIN H + +N+ FIL R+
Sbjct: 8 KLPEDLVFLILLTFSVKSLMRFKCISKAFSILIQSTTFINRHVNHEINKEDEFILFKRA- 66
Query: 63 IYQIEDDFSNLTTAVP-----LNHPFTR----------NSTNIALIGSCNGLLAVSNGEI 107
I E++F N+ + LN F N T LIG C+GL+A+++ I
Sbjct: 67 IKDEEEEFINILSFFSGHDDVLNPLFPDIDVSYMTSKFNCTFNPLIGPCDGLIALTDSII 126
Query: 108 ALRHPNAANEITIWNPNIRKHHIIPFLPLPITPRSPSDMNCSLCVH------GFGFDPLT 161
+ I NP R ++P PS C H GFG D ++
Sbjct: 127 TI----------ILNPATRNFRVLP----------PSPFGCPKGYHRSVEGVGFGLDTIS 166
Query: 162 GDYKILRLSWLVSLQNPFY----DPHVRLFSLKTNSWKIIPTMPYALVF-AQTMGVLVED 216
YK++R+S + + Y D + +F L+T++WK + + L++ G+L +
Sbjct: 167 NYYKVVRISEVYCEEAGGYPGPKDSKIDVFDLRTDTWKELDHVQLPLIYWLPCSGMLYKQ 226
Query: 217 SIHWIMAKKLDGLHPSLIVAFNLTLEIFNEVPLPDE--IGEEEVNSNDSVEIDVAALGGC 274
+HW + +I+ F+++ E+F + +PD + E+ + + L G
Sbjct: 227 MVHWFATTDM-----MVILCFDISTEMFRNMKMPDTCCLITHELYYGLVILCESFTLIGY 281
Query: 275 L--CMTVNYETTKIDVWVMKQYGLKDSWCKLFTMMKSCVTSHL 315
+++ K+ +WVM +YG+ +SW +T+ + S L
Sbjct: 282 SNPISSIDPARDKMHIWVMMEYGVSESWIMKYTIRPISIKSPL 324
>gb|AAT38720.1| putative F-Box protein [Solanum demissum]
Length = 372
Score = 99.4 bits (246), Expect = 2e-19
Identities = 99/398 (24%), Positives = 163/398 (40%), Gaps = 64/398 (16%)
Query: 6 LPPDTLAEIFSRLPVKSLLRFRSTSKSLKSIIDSHNFINLHRKNSLNRSFILRLRSNIYQ 65
LP + + EI ++P KSLL+F SK+ +I S FI H + N R +
Sbjct: 9 LPHEIIIEILLKVPPKSLLKFMCVSKTWLELISSAKFIKTHLELIANDKEYSHHRIIFQE 68
Query: 66 IEDDF-----------SNLTTAVPLNHPFTRNSTNIALIGSCNGLLAVSNGEIALRHPNA 114
+F T + P + ++GS NGL+ + +
Sbjct: 69 SACNFKVCCLPSMLNKERSTELFDIGSPMENPTIYTWIVGSVNGLICLY---------SK 119
Query: 115 ANEITIWNPNIRKHHIIPFLPLPITPRSPSDMNCSLCV-HGFGFDPLTGDYKILRLSWLV 173
E +WNP ++K +P L + CS + +GFG+D DYK++ + +
Sbjct: 120 IEETVLWNPAVKKSKKLPTLGAKLR------NGCSYYLKYGFGYDETRDDYKVVVIQCIY 173
Query: 174 SLQNPFYDPHVRLFSLKTNSWKIIPTMPYALVFAQTMGVLVEDSIHWIMAKKLDGLHPSL 233
+ D V ++SLK +SW+ I + G V I+W ++ +D +
Sbjct: 174 E-DSGSCDSVVNIYSLKADSWRTINKF-QGNFLVNSPGKFVNGKIYWALSADVDTFNMCN 231
Query: 234 IVAFNLTLEIFNEVPLPDEIGEEEVNSNDSVEIDVAALG---GCLCMTVNYETTKIDVWV 290
I++ +L E + + LPD G+ S + + +G LC+ E T DVW+
Sbjct: 232 IISLDLADETWRRLELPDSYGK------GSYPLALGVVGSHLSVLCLNC-IEGTNSDVWI 284
Query: 291 MKQYGLKDSWCKLFTMMKSCVTSHLKS----------SSPLCYSSDGSKVLIEGIEVLLE 340
K G++ SW K+FT+ H K S CY S+ ++L+ V+L
Sbjct: 285 RKDCGVEVSWTKIFTV------DHPKDLGEFIFFTSIFSVPCYQSNKDEILLLLPPVILT 338
Query: 341 VHHKKLFWYDLKTEQVS-YEEIPDFNGAKICVGSLVPP 377
Y+ T QV + + A+I V SLV P
Sbjct: 339 --------YNGSTRQVEVVDRFEECAAAEIYVESLVDP 368
>gb|AAT38695.1| putative F-box protein [Solanum demissum]
Length = 427
Score = 98.2 bits (243), Expect = 4e-19
Identities = 83/327 (25%), Positives = 139/327 (42%), Gaps = 46/327 (14%)
Query: 1 MASDR------LPPDTLAEIFSRLPVKSLLRFRSTSKSLKSIIDSHNFINLHRKNSLNRS 54
MASD LP + + EI +LPVKSL +F SKS +I S F+ H K + +
Sbjct: 42 MASDSIIPLLLLPDELITEILLKLPVKSLSKFMCVSKSWLQLISSPTFVKNHIKLTADDK 101
Query: 55 FILRLRSNIYQIEDDF------------SNLTTAVPLNHPFTRNSTNIALIGSCNGLLAV 102
+ R I+ +F + ++ P R++ + ++GS NGL+ V
Sbjct: 102 GYIHHRLIFRNIDGNFKFCSLPPLFTKQQHTEELFHIDSPIERSTLSTHIVGSVNGLICV 161
Query: 103 SNGEIALRHPNAANEITIWNPNIRKHHIIPFLPLPITPRSPSDMNCSLCVHGFGFDPLTG 162
+G+ E IWNP I K + P+ S+M S +GFG+D
Sbjct: 162 VHGQ---------KEAYIWNPTITKSKEL--------PKFTSNMCSSSIKYGFGYDESRD 204
Query: 163 DYKILRLSWLVS-LQNPFYDPHVRLFSLKTNSWKIIPTMPYALVFAQTMGVLVEDSIHWI 221
DYK++ + + + + V ++SL+ NSW + G V ++W
Sbjct: 205 DYKVVFIHYPYNHSSSSNMTTVVHIYSLRNNSWTTFRDQLQCFL-VNHYGRFVNGKLYWT 263
Query: 222 MAKKLDGLHPSLIVAFNLTLEIFNEVPLPDEIGEEEVNSNDSVEIDVAALGGCLCM--TV 279
+ ++ I +F+L + + LP + D+ +I++ +G L + T
Sbjct: 264 SSTCINKYKVCNITSFDLADGTWGSLDLP-------ICGKDNFDINLGVVGSDLSLLYTC 316
Query: 280 NYETTKIDVWVMKQYGLKDSWCKLFTM 306
DVW+MK + SW KLFT+
Sbjct: 317 QRGAATSDVWIMKHSAVNVSWTKLFTI 343
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.322 0.138 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 723,200,583
Number of Sequences: 2540612
Number of extensions: 30814423
Number of successful extensions: 65519
Number of sequences better than 10.0: 419
Number of HSP's better than 10.0 without gapping: 254
Number of HSP's successfully gapped in prelim test: 165
Number of HSP's that attempted gapping in prelim test: 64810
Number of HSP's gapped (non-prelim): 520
length of query: 412
length of database: 863,360,394
effective HSP length: 130
effective length of query: 282
effective length of database: 533,080,834
effective search space: 150328795188
effective search space used: 150328795188
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 76 (33.9 bits)
Medicago: description of AC149495.3


