

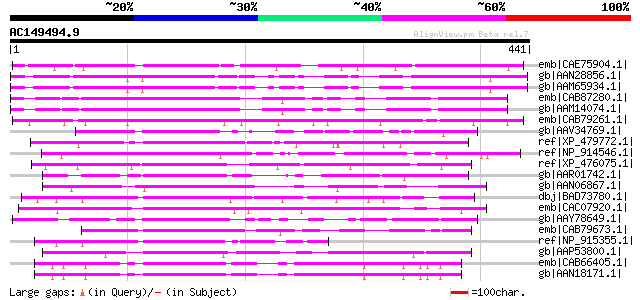
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149494.9 + phase: 0
(441 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAE75904.1| OSJNBa0088I22.17 [Oryza sativa (japonica cultiva... 281 2e-74
gb|AAN28856.1| At5g62070/mtg10_90 [Arabidopsis thaliana] gi|1017... 271 3e-71
gb|AAM65934.1| unknown [Arabidopsis thaliana] 270 9e-71
emb|CAB87280.1| putative protein [Arabidopsis thaliana] gi|29824... 263 1e-68
gb|AAM14074.1| unknown protein [Arabidopsis thaliana] 263 1e-68
emb|CAB79261.1| putative protein [Arabidopsis thaliana] gi|32928... 186 1e-45
gb|AAV34769.1| At3g16490 [Arabidopsis thaliana] gi|9279648|dbj|B... 169 2e-40
ref|XP_479772.1| calmodulin-binding protein family-like [Oryza s... 167 8e-40
ref|NP_914546.1| P0710E05.11 [Oryza sativa (japonica cultivar-gr... 165 2e-39
ref|XP_476075.1| 'unknown protein, contains calmodulin-binding m... 154 7e-36
gb|AAR01742.1| hypothetical protein [Oryza sativa (japonica cult... 139 1e-31
gb|AAN06867.1| Hypothetical protein [Oryza sativa (japonica cult... 137 7e-31
dbj|BAD73780.1| putative SF16 protein [Oryza sativa (japonica cu... 133 1e-29
emb|CAC07920.1| putative protein [Arabidopsis thaliana] gi|28973... 127 9e-28
gb|AAY78649.1| calmodulin-binding family protein [Arabidopsis th... 126 2e-27
emb|CAB79673.1| hypothetical protein [Arabidopsis thaliana] gi|4... 124 8e-27
ref|NP_915355.1| P0460C04.18 [Oryza sativa (japonica cultivar-gr... 123 1e-26
gb|AAP53800.1| unknown protein [Oryza sativa (japonica cultivar-... 119 2e-25
emb|CAB66405.1| SF16-like protein [Arabidopsis thaliana] gi|1135... 115 3e-24
gb|AAN18171.1| At3g49260/F2K15_120 [Arabidopsis thaliana] gi|215... 115 3e-24
>emb|CAE75904.1| OSJNBa0088I22.17 [Oryza sativa (japonica cultivar-group)]
gi|50928045|ref|XP_473550.1| OSJNBa0088I22.17 [Oryza
sativa (japonica cultivar-group)]
Length = 464
Score = 281 bits (720), Expect = 2e-74
Identities = 209/482 (43%), Positives = 263/482 (54%), Gaps = 77/482 (15%)
Query: 3 FLRRLFGAKKPIPPSDGSGKKSDKDNKRRWSFGK---QSSKTKSLPQPPPSAFNQFDSST 59
+LR L G K P+ GSG K+RW FGK + S P PPPSA Q + T
Sbjct: 7 WLRGLLGGGKK--PNSGSGDPKPAREKKRWGFGKSFREKSPAHPPPPPPPSAAVQ-RAVT 63
Query: 60 PL-----------ERNKHAIAVAAATAAVAEAALAAAHAAAEVVRLTSSGVAGSSNKTRG 108
P E++K AIAVAAATAAVAEAA+AAA AAA VVRLTSSG + R
Sbjct: 64 PRRAYTASDEGDDEQSKRAIAVAAATAAVAEAAVAAAQAAAAVVRLTSSGRCAPAAAKR- 122
Query: 109 QLRLPEETAAVKIQSAFRGYLARRALRALKALVKLQALVRGHIVRKKTADMLRRMQTLVR 168
EE AAV+IQ+AFRGYLARRAL+AL+ LVKLQALVRG+IVR++ A+ LR M LVR
Sbjct: 123 -----EEYAAVRIQAAFRGYLARRALKALRGLVKLQALVRGNIVRRQAAETLRCMHALVR 177
Query: 169 LQTKARASRAHLSSDNLHSFKSSLSHYPVPEEYEQPHHVYSTKFGGSSILKRCSSNS--N 226
+Q +ARA RA + S ++ + P PE+Y+Q H K G S LK SS + +
Sbjct: 178 VQRRARACRA-IRSQHVSAHPGP----PTPEKYDQATHEGVPKHGRSGSLKGSSSKTPGS 232
Query: 227 FRKIESEKPRFGSNWLDHWMQENSISQTKNASSKNRHPDEHKSDKILEVDTWKP------ 280
R G NWLD W++E R+ D+ K+ KILEVDT KP
Sbjct: 233 ERLTRERSESCGRNWLDKWVEE-------------RYLDDEKNAKILEVDTGKPGRHASR 279
Query: 281 ---QLNKNENNVNSMSN-----------ESPSKHSTKAQNQSLSVKFHKAKEEVAASRTA 326
+ + ++ +SM++ ESPSK ST AQ S E ++ A
Sbjct: 280 RRSGSHHHHSSCSSMTSEQKSRSYATMPESPSKDSTTAQQSVPSPPSVGMAEALSPLLMA 339
Query: 327 ------DNSPQTFSASSRNGSGVRRNTPFTPTRSECSWSFLGGYSGYPNYMANTESSRAK 380
+SPQ FSA+SR GS R+ FTPT+SECS S GGYS YPNYMANTES RAK
Sbjct: 340 VDIAELCDSPQFFSATSRPGSS--RSRAFTPTKSECSRSLFGGYSDYPNYMANTESFRAK 397
Query: 381 VRSQSAPRQRHEFEEYSSTRRPFQGLWDVGSTNSDNDSDSRS------NKVSPALSRFNR 434
RSQSAP+QR ++E+ SS R+ + GS + S NK P R +R
Sbjct: 398 ARSQSAPKQRPQYEKSSSLRKASAHAFGPGSCAPVAQRTTASLHSKFTNKAYPGSGRLDR 457
Query: 435 IG 436
+G
Sbjct: 458 LG 459
>gb|AAN28856.1| At5g62070/mtg10_90 [Arabidopsis thaliana]
gi|10176925|dbj|BAB10169.1| unnamed protein product
[Arabidopsis thaliana] gi|15241692|ref|NP_201013.1|
calmodulin-binding family protein [Arabidopsis thaliana]
gi|15215590|gb|AAK91340.1| AT5g62070/mtg10_90
[Arabidopsis thaliana]
Length = 403
Score = 271 bits (693), Expect = 3e-71
Identities = 213/465 (45%), Positives = 252/465 (53%), Gaps = 88/465 (18%)
Query: 1 MGFLRRLFGAKKPIPPSDGSGKKSDKDNKRRWSFGKQSSKTKSLPQPPPSAFNQFDSSTP 60
MGF RLFG+KK S K + +KRRWSF +SS + SA
Sbjct: 1 MGFFGRLFGSKKK------SDKAASSRDKRRWSFTTRSSNSSKRAPAVTSA--SVVEQNG 52
Query: 61 LERNKHAIAVAAATAAVAEAALAAAHAAAEVVRLTSSG----VAGSSNKTRGQLR----- 111
L+ +KHAIAVAAATAAVAEAAL AAHAAAEVVRLTS V G N + Q+
Sbjct: 53 LDADKHAIAVAAATAAVAEAALTAAHAAAEVVRLTSGNGGRNVGGGGNSSVFQIGRSNRR 112
Query: 112 -LPEETAAVKIQSAFRGYLARRALRALKALVKLQALVRGHIVRKKTADMLRRMQTLVRLQ 170
E AA+KIQSAFRGYLARRALRALKALVKLQALVRGHIVRK+TADMLRRMQTLVRLQ
Sbjct: 113 WAQENIAAMKIQSAFRGYLARRALRALKALVKLQALVRGHIVRKQTADMLRRMQTLVRLQ 172
Query: 171 TKARASRAHLSSDNLHSFKSSLSHYPVPEEYEQPHHVYSTKFGGSSILKRCSSNSNFRKI 230
++ARA RA SS + SF SS + P P +++ RC SN+ ++
Sbjct: 173 SQARA-RASRSSHSSASFHSSTA-LLFPSSSSSPRSLHT----------RCVSNA---EV 217
Query: 231 ESEKPRFGSNWLDHWMQENSISQTKNASSKNRHPDEHKSDKILEVDTWKPQLNKNENNVN 290
S R GS LD W E S DKILEVDTWKP + +
Sbjct: 218 SSLDHRGGSKRLD-WQAEES----------------ENGDKILEVDTWKPHYHPKPLR-S 259
Query: 291 SMSNESPSKHSTKAQNQSLSVKFHKAKEEVAASRTADNSPQTFSASSRNGSGVRRNTPFT 350
+NESP K ++ + R+ +NSPQ S SG RR TPFT
Sbjct: 260 ERNNESP----------------RKRQQSLLGPRSTENSPQVGS------SGSRRRTPFT 297
Query: 351 PT-RSECSWSFLG-GYSGY-PNYMANTESSRAKVRSQSAPRQRHEF-EEYSSTRRPFQGL 406
PT RSE SW YSGY PNYMANTES +AKVRSQSAP+QR E E S +R QG
Sbjct: 298 PTSRSEYSWGCNNYYYSGYHPNYMANTESYKAKVRSQSAPKQRVEVSNETSGYKRSVQGQ 357
Query: 407 W-----------DVGSTNSDNDSDSRSNKVSPALSRFNRIGSSNL 440
+ DVGS S++++ S +R+ SS L
Sbjct: 358 YYYYTAVEEESLDVGSAGYYGGGGGDSDRLNRNQSAKSRMHSSFL 402
>gb|AAM65934.1| unknown [Arabidopsis thaliana]
Length = 403
Score = 270 bits (689), Expect = 9e-71
Identities = 212/465 (45%), Positives = 251/465 (53%), Gaps = 88/465 (18%)
Query: 1 MGFLRRLFGAKKPIPPSDGSGKKSDKDNKRRWSFGKQSSKTKSLPQPPPSAFNQFDSSTP 60
MGF RLFG+KK S K + +KRRWSF +SS + SA
Sbjct: 1 MGFFGRLFGSKKK------SDKAASSRDKRRWSFTTRSSNSSKRAPAVTSA--SVVEQNG 52
Query: 61 LERNKHAIAVAAATAAVAEAALAAAHAAAEVVRLTSSG----VAGSSNKTRGQLR----- 111
L+ +KHAIAVAAAT AVAEAAL AAHAAAEVVRLTS V G N + Q+
Sbjct: 53 LDADKHAIAVAAATXAVAEAALTAAHAAAEVVRLTSGNGGRNVGGGGNSSVFQIGRSNRR 112
Query: 112 -LPEETAAVKIQSAFRGYLARRALRALKALVKLQALVRGHIVRKKTADMLRRMQTLVRLQ 170
E AA+KIQSAFRGYLARRALRALKALVKLQALVRGHIVRK+TADMLRRMQTLVRLQ
Sbjct: 113 WAQENIAAMKIQSAFRGYLARRALRALKALVKLQALVRGHIVRKQTADMLRRMQTLVRLQ 172
Query: 171 TKARASRAHLSSDNLHSFKSSLSHYPVPEEYEQPHHVYSTKFGGSSILKRCSSNSNFRKI 230
++ARA RA SS + SF SS + P P +++ RC SN+ ++
Sbjct: 173 SQARA-RASRSSHSSASFHSSTA-LLFPSSSSSPRSLHT----------RCVSNA---EV 217
Query: 231 ESEKPRFGSNWLDHWMQENSISQTKNASSKNRHPDEHKSDKILEVDTWKPQLNKNENNVN 290
S R GS LD W E S DKILEVDTWKP + +
Sbjct: 218 SSLDHRGGSKRLD-WQAEES----------------ENGDKILEVDTWKPHYHPKPLR-S 259
Query: 291 SMSNESPSKHSTKAQNQSLSVKFHKAKEEVAASRTADNSPQTFSASSRNGSGVRRNTPFT 350
+NESP K ++ + R+ +NSPQ S SG RR TPFT
Sbjct: 260 EKNNESP----------------RKRQQSLLGPRSTENSPQVGS------SGSRRRTPFT 297
Query: 351 PT-RSECSWSFLG-GYSGY-PNYMANTESSRAKVRSQSAPRQRHEF-EEYSSTRRPFQGL 406
PT RSE SW YSGY PNYMANTES +AKVRSQSAP+QR E E S +R QG
Sbjct: 298 PTSRSEYSWGCNNYYYSGYHPNYMANTESYKAKVRSQSAPKQRVEVSNETSGYKRSVQGQ 357
Query: 407 W-----------DVGSTNSDNDSDSRSNKVSPALSRFNRIGSSNL 440
+ DVGS S++++ S +R+ SS L
Sbjct: 358 YYYYTAVEEESLDVGSAGYYGGGGGDSDRLNRNQSAKSRMHSSFL 402
>emb|CAB87280.1| putative protein [Arabidopsis thaliana] gi|29824161|gb|AAP04041.1|
unknown protein [Arabidopsis thaliana]
gi|15240730|ref|NP_196341.1| calmodulin-binding family
protein [Arabidopsis thaliana] gi|11358265|pir||T48495
hypothetical protein T28J14.180 - Arabidopsis thaliana
Length = 401
Score = 263 bits (671), Expect = 1e-68
Identities = 192/435 (44%), Positives = 235/435 (53%), Gaps = 82/435 (18%)
Query: 1 MGFLRRLFGAKKPIPPSDGSGKKSDKDNKRRWSFGKQSSKTKSLPQPPPSAFNQFDSSTP 60
MGF RLFG+KK ++ N+RRWSF +SS ++ S+ + D
Sbjct: 1 MGFFGRLFGSKK---------QEKATPNRRRWSFATRSSHPEN-DSSSHSSKRRGDEDV- 49
Query: 61 LERNKHAIAVAAATAAVAEAALAAAHAAAEVVRLTSSGVAGSSNK-TRGQLRLPEE-TAA 118
L +KHAIAVAAATAAVAEAALAAA AAAEVVRLT+ G S + +R R +E AA
Sbjct: 50 LNGDKHAIAVAAATAAVAEAALAAARAAAEVVRLTNGGRNSSVKQISRSNRRWSQEYKAA 109
Query: 119 VKIQSAFRGYLARRALRALKALVKLQALVRGHIVRKKTADMLRRMQTLVRLQTKARASRA 178
+KIQSAFRGYLARRALRALKALVKLQALV+GHIVRK+TADMLRRMQTLVRLQ +ARASR+
Sbjct: 110 MKIQSAFRGYLARRALRALKALVKLQALVKGHIVRKQTADMLRRMQTLVRLQARARASRS 169
Query: 179 HLSSDNLHSFKSSLSHYPVPEEYEQPHHVYSTKFGGSSILKRCSSNSNFRKI-------- 230
SD+ H + P S RC S + + K+
Sbjct: 170 SHVSDSSHPPTLMIPSSP------------------QSFHARCVSEAEYSKVIAMDHHHN 211
Query: 231 ESEKPRFGSNWLDHWMQENSISQTKNASSKNRHPDEHKSDKILEVDTWKPQLNKNENNVN 290
P S LD W E S+ S+ + D+ DKILEVDTWKP
Sbjct: 212 NHRSPMGSSRLLDQWRTEESL-----WSAPKYNEDD---DKILEVDTWKPHF-------- 255
Query: 291 SMSNESPSKHSTKAQNQSLSVKFHKAKEEVAASRTADNSPQTFSASSRNGSGVRRNTPFT 350
ESP K + + + +NSPQ S + + G RR TPFT
Sbjct: 256 ---RESPRKRGS-----------------LVVPTSVENSPQLRSRTGSSSGGSRRKTPFT 295
Query: 351 PTRSECSWSFLGGYSGY-PNYMANTESSRAKVRSQSAPRQR-HEFEEYSSTRRPFQGLWD 408
P RSE + YSGY PNYMANTES +AKVRSQSAPRQR + S +R QG +
Sbjct: 296 PARSEYEY-----YSGYHPNYMANTESYKAKVRSQSAPRQRLQDLPSESGYKRSIQGQYY 350
Query: 409 VGSTNSDNDSDSRSN 423
+ ++ D RS+
Sbjct: 351 YYTPAAERSFDQRSD 365
>gb|AAM14074.1| unknown protein [Arabidopsis thaliana]
Length = 437
Score = 263 bits (671), Expect = 1e-68
Identities = 192/435 (44%), Positives = 235/435 (53%), Gaps = 82/435 (18%)
Query: 1 MGFLRRLFGAKKPIPPSDGSGKKSDKDNKRRWSFGKQSSKTKSLPQPPPSAFNQFDSSTP 60
MGF RLFG+KK ++ N+RRWSF +SS ++ S+ + D
Sbjct: 37 MGFFGRLFGSKK---------QEKATPNRRRWSFATRSSHPEN-DSSSHSSKRRGDEDV- 85
Query: 61 LERNKHAIAVAAATAAVAEAALAAAHAAAEVVRLTSSGVAGSSNK-TRGQLRLPEE-TAA 118
L +KHAIAVAAATAAVAEAALAAA AAAEVVRLT+ G S + +R R +E AA
Sbjct: 86 LNGDKHAIAVAAATAAVAEAALAAARAAAEVVRLTNGGRNSSVKQISRSNRRWSQEYKAA 145
Query: 119 VKIQSAFRGYLARRALRALKALVKLQALVRGHIVRKKTADMLRRMQTLVRLQTKARASRA 178
+KIQSAFRGYLARRALRALKALVKLQALV+GHIVRK+TADMLRRMQTLVRLQ +ARASR+
Sbjct: 146 MKIQSAFRGYLARRALRALKALVKLQALVKGHIVRKQTADMLRRMQTLVRLQARARASRS 205
Query: 179 HLSSDNLHSFKSSLSHYPVPEEYEQPHHVYSTKFGGSSILKRCSSNSNFRKI-------- 230
SD+ H + P S RC S + + K+
Sbjct: 206 SHVSDSSHPPTLMIPSSP------------------QSFHARCVSEAEYSKVIAMDHHHN 247
Query: 231 ESEKPRFGSNWLDHWMQENSISQTKNASSKNRHPDEHKSDKILEVDTWKPQLNKNENNVN 290
P S LD W E S+ S+ + D+ DKILEVDTWKP
Sbjct: 248 NHRSPMGSSRLLDQWRTEESL-----WSAPKYNEDD---DKILEVDTWKPHF-------- 291
Query: 291 SMSNESPSKHSTKAQNQSLSVKFHKAKEEVAASRTADNSPQTFSASSRNGSGVRRNTPFT 350
ESP K + + + +NSPQ S + + G RR TPFT
Sbjct: 292 ---RESPRKRGS-----------------LVVPTSVENSPQLRSRTGSSSGGSRRKTPFT 331
Query: 351 PTRSECSWSFLGGYSGY-PNYMANTESSRAKVRSQSAPRQR-HEFEEYSSTRRPFQGLWD 408
P RSE + YSGY PNYMANTES +AKVRSQSAPRQR + S +R QG +
Sbjct: 332 PARSEYEY-----YSGYHPNYMANTESYKAKVRSQSAPRQRLQDLPSESGYKRSIQGQYY 386
Query: 409 VGSTNSDNDSDSRSN 423
+ ++ D RS+
Sbjct: 387 YYTPAAERSFDQRSD 401
>emb|CAB79261.1| putative protein [Arabidopsis thaliana] gi|3292832|emb|CAA19822.1|
putative protein [Arabidopsis thaliana]
gi|15235911|ref|NP_194037.1| calmodulin-binding family
protein [Arabidopsis thaliana] gi|7486576|pir||T05138
hypothetical protein F7H19.250 - Arabidopsis thaliana
Length = 543
Score = 186 bits (472), Expect = 1e-45
Identities = 179/537 (33%), Positives = 239/537 (44%), Gaps = 110/537 (20%)
Query: 3 FLRRLFGAKKPIP-----PSDGSGKKSDKDNKRRWSFGKQSSKTKSLP------------ 45
+ R LFG KKP P + + + + KRRWSF K + +S P
Sbjct: 7 WFRSLFGVKKPDPGYPDLSVETPSRSTSSNLKRRWSFVKSKREKESTPINQVPHTPSLPN 66
Query: 46 -QPPPSAFNQFDSSTPLER---------------NKHAIAVAAATAAVAEAALAAAHAAA 89
PPP + +Q S+P R +KHAIAVAAATAAVAEAA+AAA+AAA
Sbjct: 67 STPPPPSHHQ---SSPRRRRKQKPMWEDEGSEDSDKHAIAVAAATAAVAEAAVAAANAAA 123
Query: 90 EVVRLTSSG------------------VAGSSNKTRGQLRLPEETAAVKIQSAFRGYLAR 131
VVRLTS+ V +K G R E A +KIQS FRGYLA+
Sbjct: 124 AVVRLTSTSGRSTRSPVKARFSDGFDDVVAHGSKFYGHGRDSCELAVIKIQSIFRGYLAK 183
Query: 132 RALRALKALVKLQALVRGHIVRKKTADMLRRMQTLVRLQTKARASRAHLS--SDNLHSFK 189
RALRALK LV+LQA+VRGHI RK+ + LRRM LVR Q + RA+R ++ S + S
Sbjct: 184 RALRALKGLVRLQAIVRGHIERKRMSVHLRRMHALVRAQARVRATRVIVTPESSSSQSNN 243
Query: 190 SSLSHY---------------------PVPEEYEQPHHVYSTKFGGSSILKRCSSNSNF- 227
+ SH+ P PE+ E S+K S + K F
Sbjct: 244 TKSSHFQNPVSLVKFPMIVPFNLKHGPPTPEKLEHSISSRSSKLAHSHLFKVLHFQLLFV 303
Query: 228 -------RKIESEKPRFGSNWLDHWMQENSISQTKN---ASSKNRHPDEHK--------S 269
I S+ R ++Q S + N AS NR H+
Sbjct: 304 SSVFVACGPISSKFQRLYKLLTLLYVQNKSNLKNWNGSKASDNNRLYPAHRETFSATDEE 363
Query: 270 DKILEVDTWKPQLNKNENNVNSMSNESPSKHSTKAQNQSLSVKF--HKAKEEVAAS-RTA 326
+KIL++D N + + + + F + EE+ + TA
Sbjct: 364 EKILQIDRKHISSYTRRNRPDMFYSSHLILDNAGLSEPVFATPFSPSSSHEEITSQFCTA 423
Query: 327 DNSPQTFSASSRNGSGVRRNTPFTPTRSECSWSFLGGYSGYPNYMANTESSRAKVRSQSA 386
+NSPQ +SA+SR+ + P S+C+ S G +P+YMA TESSRAK RS SA
Sbjct: 424 ENSPQLYSATSRSKRSAFTASSIAP--SDCTKSCCDG--DHPSYMACTESSRAKARSASA 479
Query: 387 PRQRHE--FEEYSSTRRPFQGLWDVGSTNSDNDSDSR-----SNKVSPALSRFNRIG 436
P+ R + +E SS R F L G T S S NK P R +R+G
Sbjct: 480 PKSRPQLFYERPSSKRFGFVDLPYCGDTKSGPQKGSALHTSFMNKAYPGSGRLDRLG 536
>gb|AAV34769.1| At3g16490 [Arabidopsis thaliana] gi|9279648|dbj|BAB01148.1| unnamed
protein product [Arabidopsis thaliana]
gi|58531348|gb|AAW78596.1| At3g16490 [Arabidopsis
thaliana] gi|15228224|ref|NP_188270.1|
calmodulin-binding family protein [Arabidopsis thaliana]
Length = 389
Score = 169 bits (427), Expect = 2e-40
Identities = 134/344 (38%), Positives = 181/344 (51%), Gaps = 63/344 (18%)
Query: 57 SSTPLERNKHAIAVAAATAAVAEAALAAAHAAAEVVRLTSSGVAGSSNKTRGQLRLPEET 116
+ T E+NKHAIAVAAATAA A+AA+AAA AA VVRLTS+G +G + E
Sbjct: 54 AETDKEQNKHAIAVAAATAAAADAAVAAAQAAVAVVRLTSNGRSGGYSGNA-----MERW 108
Query: 117 AAVKIQSAFRGYLARRALRALKALVKLQALVRGHIVRKKTADMLRRMQTLVRLQTKARAS 176
AAVKIQS F+GYLAR+ALRALK LVKLQALVRG++VRK+ A+ L MQ L+R QT R+
Sbjct: 109 AAVKIQSVFKGYLARKALRALKGLVKLQALVRGYLVRKRAAETLHSMQALIRAQTSVRSQ 168
Query: 177 RAHLSSDNLHSFKSSLSHYPVPEEYEQPHHVYSTKFGGSSILKRCSSNSNFRKIESEKPR 236
R + ++++ H P H + +++ +
Sbjct: 169 RIN---------RNNMFH---------PRH-------------------SLERLDDSRSE 191
Query: 237 FGSNWLDHWMQENSISQTKNASSKNRHPDEHKSDKILEVDTWKPQLNKNENNVNSMSNES 296
S + SIS K ++ N DE S KI+E+DT+K + NV +E
Sbjct: 192 IHSKRI-------SISVEKQSNHNNNAYDE-TSPKIVEIDTYKTKSRSKRMNV--AVSEC 241
Query: 297 PSKHSTKAQNQSLSVKFHKAKEEVAASRTADNSPQTFSASSRNGSGVRRNTPFTPTRSEC 356
+A++ S K K TA N+P+ FS+S N + TP +P +S C
Sbjct: 242 GDDFIYQAKDFEWSFPGEKCK-----FPTAQNTPR-FSSSMANNN--YYYTPPSPAKSVC 293
Query: 357 -SWSFLGGYSGY--PNYMANTESSRAKVRSQSAPRQRHEFEEYS 397
F Y G P+YMANT+S +AKVRS SAPRQR + + S
Sbjct: 294 RDACFRPSYPGLMTPSYMANTQSFKAKVRSHSAPRQRPDRKRLS 337
>ref|XP_479772.1| calmodulin-binding protein family-like [Oryza sativa (japonica
cultivar-group)] gi|42409298|dbj|BAD10560.1|
calmodulin-binding protein family-like [Oryza sativa
(japonica cultivar-group)]
Length = 543
Score = 167 bits (422), Expect = 8e-40
Identities = 150/436 (34%), Positives = 207/436 (47%), Gaps = 77/436 (17%)
Query: 18 DGSGKKSDKDNKRRWSFGKQSSKTKSLPQPPPSAFNQFDS--STPLERNKHAIAVAAATA 75
D + K +RRW F K SS + + P PPP Q S + E +HAIA+A ATA
Sbjct: 36 DDTDDDKGKRERRRWLFRKSSSPSPAPPTPPPPQQQQQQSRAAAVTEEQRHAIALAVATA 95
Query: 76 AVAEAALAAAHAAAEVVRLTSSGVAGSSNKTRGQLRLPEETAAVKIQSAFRGYLARRALR 135
A AEAA+A A AAAEVVRLT SS+ R E AA+ +Q+AFRGYLARRALR
Sbjct: 96 ATAEAAVATAQAAAEVVRLTRP----SSSFVR------EHYAAIVVQTAFRGYLARRALR 145
Query: 136 ALKALVKLQALVRGHIVRKKTADMLRRMQTLVRLQTKARASRAHLSSDNLHSFKSSLSHY 195
ALK LVKLQALVRGH VRK+ LR MQ LVR+Q + R R LS D++ ++ S
Sbjct: 146 ALKGLVKLQALVRGHNVRKQANMTLRCMQALVRVQARVRDQRMRLSQDSISLSAAAASAA 205
Query: 196 PVPEEYEQPHHVYSTKFGGSSILKRCSSNSNFRKIESEKPRFGSNWL--DHWMQENSISQ 253
P + + V ++ F S ++ ++ R I E+ R GS++ D W +
Sbjct: 206 PCGSS-KSSYSVDTSTFWDSKYTHDFAA-ADRRSI--ERSRDGSSFAAGDDWDDRPRTIE 261
Query: 254 TKNASSKNRHPDEHKSDKIL-----------------EVDT-----W-------KPQLNK 284
A + R K ++ L E+D W + +
Sbjct: 262 EIQAMLQTRKDAALKRERALSYAFSHQIWRNPAPSVEEMDVDGQPRWAERWMASRASFDT 321
Query: 285 NENNVNSMSNESPSKHSTKAQNQSLSVKFHKA--------KEEVAASRTADNSP------ 330
+ + V + + +P + ST ++Q +++ A + AS A +SP
Sbjct: 322 SRSTVRASAAAAPGRASTDHRDQVKTLEIDTARPFSYSTPRRHGNASYHASSSPMHRAHH 381
Query: 331 ---------------QTFSASSRNGSGVRRNTPFTPT-RSECSWSFLGGYSGYPNYMANT 374
Q SAS R G +TP+ S + GG + PNYMA T
Sbjct: 382 HSPVTPSPSKARPPIQVRSASPRVERGGGGGGSYTPSLHSHRHHASSGGAAAVPNYMAAT 441
Query: 375 ESSRAKVRSQSAPRQR 390
ES++A+VRSQSAPRQR
Sbjct: 442 ESAKARVRSQSAPRQR 457
>ref|NP_914546.1| P0710E05.11 [Oryza sativa (japonica cultivar-group)]
gi|9049461|dbj|BAA99426.1| unknown protein [Oryza sativa
(japonica cultivar-group)]
Length = 465
Score = 165 bits (418), Expect = 2e-39
Identities = 154/442 (34%), Positives = 209/442 (46%), Gaps = 65/442 (14%)
Query: 28 NKRRWSFGKQSSKTK------------SLPQPPPSAF-NQFDSSTPLERNKHAIAVAAAT 74
+++RWSF K S + ++ + +A+ S T E++KHAIAVAAAT
Sbjct: 36 DRKRWSFAKSSRDSTEGEAAAAVGGNAAIAKAAEAAWLKSMYSDTEREQSKHAIAVAAAT 95
Query: 75 AAVAEAALAAAHAAAEVVRLTSSGVAGSSNKTRGQLRLPE-ETAAVKIQSAFRGYLARRA 133
AA A+AA+AAA AA EVVRLTS G SS G + P AAVKIQ+AFRG+LA++A
Sbjct: 96 AAAADAAVAAAQAAVEVVRLTSQGPPTSSVFVCGGVLDPRGRAAAVKIQTAFRGFLAKKA 155
Query: 134 LRALKALVKLQALVRGHIVRKKTADMLRRMQTLVRLQTKARASR----AHLSSDNLHSFK 189
LRALKALVKLQALVRG++VR++ A L+ MQ LVR Q RA+R A L +LH
Sbjct: 156 LRALKALVKLQALVRGYLVRRQAAATLQSMQALVRAQAAVRAARSSRGAALPPLHLHHHP 215
Query: 190 SSLSHYPVPEEYEQPHHVYSTKFGGSSILKRCSSNSNFRKIESEKPRFGSNWLDHWMQEN 249
Y + E Y ++ G ++ +R S++ IES +G + +
Sbjct: 216 PVRPRYSLQERYMDDTR---SEHGVAAYSRRLSAS-----IESSS--YG------YDRSP 259
Query: 250 SISQTKNASSKNRHPDEHKSDKILEVDTWKPQLNKNENNVNSMSNESPSKHSTKAQNQSL 309
I + K+R S +++ E NS+S+ H +
Sbjct: 260 KIVEMDTGRPKSRSSSVRTSPPVVDAGA------AEEWYANSVSSPLLPFHQLPGAPPRI 313
Query: 310 SVKFHKAKEEVAASRTADNSPQTFSASSRNGSGVRRNTPFTPTRSECSWSFLGGYSGYPN 369
S + E P T ++ R + P TPT+S C GGY PN
Sbjct: 314 SAPSARHFPEYDWCPLEKPRPATAQSTPR-----LAHMPVTPTKSVCGG---GGYGASPN 365
Query: 370 ---YMANTESSRAKVRSQSAPRQRHEFEEYSST------RRPFQ--------GLWDVGST 412
YM++T+SS AKVRSQSAP+QR E T R P L VG
Sbjct: 366 CRGYMSSTQSSEAKVRSQSAPKQRPEPGVAGGTGGGARKRVPLSEVTLEARASLSGVGMQ 425
Query: 413 NSDNDSDSRSNKVSPALSRFNR 434
S N N + LSRF+R
Sbjct: 426 RSCNRVQEAFNFKTAVLSRFDR 447
>ref|XP_476075.1| 'unknown protein, contains calmodulin-binding motif' [Oryza sativa
(japonica cultivar-group)] gi|46275850|gb|AAS86400.1|
hypothetical protein [Oryza sativa (japonica
cultivar-group)]
Length = 497
Score = 154 bits (388), Expect = 7e-36
Identities = 141/439 (32%), Positives = 204/439 (46%), Gaps = 91/439 (20%)
Query: 19 GSGKKSDKDNK-----RRWSFGKQSSKTKSLPQPPPSAFNQFD----------------- 56
G G+K K +RWSFGK S + +A +
Sbjct: 13 GGGRKEQKGEAPASGGKRWSFGKSSRDSAEAAAAAAAAAAEASGGNAAIARAAEAAWLRS 72
Query: 57 --SSTPLERNKHAIAVAAATAAVAEAALAAAHAAAEVVRLTSSGVAGS--SNKTRGQLRL 112
+ T E++KHAIAVAAATAA A+AA+AAA AA VVRLTS G + + G R
Sbjct: 73 VYADTEREQSKHAIAVAAATAAAADAAVAAAQAAVAVVRLTSKGRSAPVLAATVAGDTR- 131
Query: 113 PEETAAVKIQSAFRGYLARRALRALKALVKLQALVRGHIVRKKTADMLRRMQTLVRLQTK 172
AAV+IQ+AFRG+LA++ALRALKALVKLQALVRG++VR++ A L+ MQ LVR Q
Sbjct: 132 SLAAAAVRIQTAFRGFLAKKALRALKALVKLQALVRGYLVRRQAAATLQSMQALVRAQAT 191
Query: 173 ARASRAHL-SSDNLHSFKSSLSHYPVPEEYEQPHHVYSTKFGGSSILKRCSSNSN----- 226
RA R+ ++ NL PH ++ + S+++R + ++
Sbjct: 192 VRAHRSGAGAAANL------------------PHLHHAPFWPRRSLVRRWLNLADDIAMY 233
Query: 227 --------FRKIESEKPRFGSNWLDHWMQENSISQTKNASSKNRHPDEHKSDKILEVDTW 278
+R ++ E+ +H + + S+ +AS ++ +S KI+EVDT
Sbjct: 234 MFDVDVVCWRWMQQERCAGDDTRSEHGVA--AYSRRLSASIESSSYGYDRSPKIVEVDTG 291
Query: 279 KPQLNKNENN---------------------VNSMSNESPSKHSTKAQNQSLSVKFHKAK 317
+P+ + + NSMS+ P A ++V +
Sbjct: 292 RPKSRSSSSRRASSPLLLDAAGCASGGEDWCANSMSSPLPCYLPGGAPPPRIAVPTSRHF 351
Query: 318 EEVAASRTADNSPQTFSASSRNGSGVRRNTPFTPTRSECSWSFLGGYS----GYPNYMAN 373
+ P T ++ R + P TPT+S C GG PNYM+N
Sbjct: 352 PDYDWCALEKARPATAQSTPRYA-----HAPPTPTKSVCGGGGGGGIHSSPLNCPNYMSN 406
Query: 374 TESSRAKVRSQSAPRQRHE 392
T+S AKVRSQSAP+QR E
Sbjct: 407 TQSFEAKVRSQSAPKQRPE 425
>gb|AAR01742.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|50917185|ref|XP_468989.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 447
Score = 139 bits (351), Expect = 1e-31
Identities = 131/374 (35%), Positives = 177/374 (47%), Gaps = 80/374 (21%)
Query: 29 KRRWSFGKQSSKTKSLPQPPPSAFNQFDSSTPLERNKHAIAVAAATAAVAEAALAAAHAA 88
KRRWSF + + K P ++ ++HA+AVA ATAA AEAA+AA AA
Sbjct: 41 KRRWSFRRPAVKDGGGGFLEPR----------VDPDQHAVAVAIATAAAAEAAVAAKQAA 90
Query: 89 AEVVRLTSSGVAGSSNKTRGQLRLPEETAAVKIQSAFRGYLARRALRALKALVKLQALVR 148
A VVRL AGSS + + + EE AA+KIQ FR YLAR+AL AL+ LVKLQALVR
Sbjct: 91 AAVVRL-----AGSSRRGVVVVGI-EEAAAIKIQCVFRSYLARKALCALRGLVKLQALVR 144
Query: 149 GHIVRKKTADMLRRMQTLVRLQTKARASRAHLSSDNLHSFKSSLSHYP-VPEEYEQPHHV 207
GH+VR++ ++ LR MQ LV Q +ARA+R L D+ + L H P + PHH
Sbjct: 145 GHLVRRQASNTLRCMQALVAAQHRARAARLRLLDDDK---EKPLLHTPRMMPTRRSPHH- 200
Query: 208 YSTKFGGSSILKRCSSNSNFRKIESEKPRFGSNWLDHWMQENSISQTKNASSKNRHPDEH 267
PRF H Q+ +
Sbjct: 201 ---------------------------PRF-----RHQQQQQ---------------EAE 213
Query: 268 KSDKILEVDT-WKPQLNKNENNVN-SMSNESPSKHSTKAQNQSLSVKFHKAKEEVAASRT 325
++ KI+EVDT + E + S+ S ++T V+ ++ K S
Sbjct: 214 ENVKIVEVDTGFGGGGGSGEAHCTPRTSSRRSSCYATPLCRTPSKVELYQ-KVSPTPSAL 272
Query: 326 ADNSPQTFS--------ASSRNGSGVRRNTPFTPTRSECSWSFLGGYS-GYPNYMANTES 376
D S +T+S +++RN + P + G + +PNYMANTES
Sbjct: 273 TDASARTYSGRYDDFSFSTARNSPWHHHHASDAPCKPHHPHHGNGDHPLFFPNYMANTES 332
Query: 377 SRAKVRSQSAPRQR 390
SRAK RSQSAPRQR
Sbjct: 333 SRAKARSQSAPRQR 346
>gb|AAN06867.1| Hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 417
Score = 137 bits (345), Expect = 7e-31
Identities = 125/394 (31%), Positives = 175/394 (43%), Gaps = 108/394 (27%)
Query: 29 KRRWSFGKQSSKTKSLPQPPPSA-----------FNQFDSSTPLERNKHAIAVAAATAAV 77
++RWSF + K+ + +A F+ S + ++ +HA+AVA ATAA
Sbjct: 36 EKRWSFRRPVHGEKAAAEAAAAADGVVVGEAEAGFDLSASESEFDQKRHAMAVAVATAAA 95
Query: 78 AEAALAAAHAAAEVVRLTSSGVAGSSNKTRGQLRLP----EETAAVKIQSAFRGYLARRA 133
A+AA+AAAHAAA VRL+S R +LP EE AAV+IQ+ FRGYLAR A
Sbjct: 96 ADAAVAAAHAAAAAVRLSS----------RKAHQLPASAVEEAAAVRIQATFRGYLARTA 145
Query: 134 LRALKALVKLQALVRGHIVRKKTADMLRRMQTLVRLQTKARASRAHLSSDNLHSFKSSLS 193
L AL+ +VKLQALVRG +VRK+ LR MQ L+ Q++ RA + + + H
Sbjct: 146 LCALRGIVKLQALVRGQLVRKQATATLRCMQALLAAQSQLRAQAQRVRALHEHHRTPPRP 205
Query: 194 HYPVPEEYEQPHHVYSTKFGGSSILKRCSSNSNFRKIESEKPRFGSNWLDHWMQENSISQ 253
P P ++ + Y +D +EN+
Sbjct: 206 RPPSPPQHPRHRRSYE--------------------------------MDRSCEENA--- 230
Query: 254 TKNASSKNRHPDEHKSDKILEVDT--WKPQLNKNENNVNSMSNESPSKHSTKAQNQSLSV 311
KI+EVD+ +P E + + +PS + ++ S
Sbjct: 231 -----------------KIVEVDSGAGEPARRGGEYGHHGRWSPAPSAMTEVMSPRAYSG 273
Query: 312 KFHKAKEEVAASRTADNSPQTFSASSRNGSGVRRNTPFTPTRSECSWSFLGGYSGYPNYM 371
F E++A + TA +SP SASS P+YM
Sbjct: 274 HF----EDMAFAATAHSSPHHASASSE-------------------------LLCCPSYM 304
Query: 372 ANTESSRAKVRSQSAPRQRHEFEEYSSTRRPFQG 405
ANTESSRAK RSQSAPRQR + E +RR G
Sbjct: 305 ANTESSRAKARSQSAPRQRTDALERQPSRRKSGG 338
>dbj|BAD73780.1| putative SF16 protein [Oryza sativa (japonica cultivar-group)]
Length = 500
Score = 133 bits (334), Expect = 1e-29
Identities = 131/457 (28%), Positives = 200/457 (43%), Gaps = 93/457 (20%)
Query: 11 KKPIPPSDGSGK-----KSDKDNKRR-WSFGKQS------------SKTKSLPQPPPSAF 52
KK SD G+ K+DK RR W FGK + T PQPPP
Sbjct: 12 KKVFSSSDPDGREAKIEKADKSRSRRKWPFGKSKKSDPWTSTVAVPTSTAPPPQPPPPPP 71
Query: 53 NQFDSSTP------------LERNKHAIAVAAATAAVAEAALAAAHAAAEVVRLTSSGVA 100
P E+NKHA +VA A+A AEAA AA AAAEVVRLT++ A
Sbjct: 72 THPIQPQPEEIKDVKAVETDSEQNKHAYSVALASAVAAEAAAVAAQAAAEVVRLTTATTA 131
Query: 101 GSSNKTRGQLRLPEETAAVKIQSAFRGYLARRALRALKALVKLQALVRGHIVRKKTADML 160
+ + +E AA+KIQ+AFRGYLARRALRAL+ LV+L++LV G+ V+++TA L
Sbjct: 132 VPKSPVSSK----DELAAIKIQTAFRGYLARRALRALRGLVRLKSLVDGNAVKRQTAHTL 187
Query: 161 RRMQTLVRLQTKARASRAHLSSDNLH-------SFKSSLSHYPVPEEYEQPHHVYSTKFG 213
QT+ R+QT+ + R + + + L + E+++ H +
Sbjct: 188 HCTQTMTRVQTQIYSRRVKMEEEKQALQRQLQLKHQRELEKMKIDEDWDHSHQ-SKEQVE 246
Query: 214 GSSILKRCSSNSNFRKI--------------------ESEKPRFGSNWLDHWMQENSISQ 253
S ++K+ ++ R + + P +G +W++ WM
Sbjct: 247 TSLMMKQEAALRRERALAYAFSHQWKNSGRTITPTFTDQGNPNWGWSWMERWMTSR---P 303
Query: 254 TKNASSKNRHPDEHKSDKILEVD---TWKPQ-LNKNENNVNSMSNESPSKHSTK---AQN 306
++ ++ P +H S K T+ P+ ++ + S+ PS+ S ++
Sbjct: 304 WESRVISDKDPKDHYSTKNPSTSASRTYVPRAISIQRPATPNKSSRPPSRQSPSTPPSRV 363
Query: 307 QSLSVKFHKA--------KEEVAASRTADNSPQTFSASSRNGSGVRRNTPFTPTRSECSW 358
S++ K A KE+ S T+ S + S G+ VR + T T
Sbjct: 364 PSVTGKIRPASPRDSWLYKEDDLRSITSIRSERP-RRQSTGGASVRDDASLTST------ 416
Query: 359 SFLGGYSGYPNYMANTESSRAKVRSQSAPRQRHEFEE 395
P+YM +TES+RAK R +S R E E
Sbjct: 417 ------PALPSYMQSTESARAKSRYRSLLTDRFEVPE 447
>emb|CAC07920.1| putative protein [Arabidopsis thaliana] gi|28973469|gb|AAO64059.1|
unknown protein [Arabidopsis thaliana]
gi|27754608|gb|AAO22750.1| unknown protein [Arabidopsis
thaliana] gi|15231175|ref|NP_190797.1|
calmodulin-binding family protein [Arabidopsis thaliana]
gi|11292165|pir||T46099 hypothetical protein T25B15.60 -
Arabidopsis thaliana
Length = 430
Score = 127 bits (318), Expect = 9e-28
Identities = 122/439 (27%), Positives = 184/439 (41%), Gaps = 71/439 (16%)
Query: 8 FGAKKPIPPSDGSGKKSDKDNKRRWSFGKQSS---KTKSLPQPPPSAFNQFDSSTPLERN 64
F A K + KK K +K + FGK P + + +++
Sbjct: 6 FSAVKKALSPEPKQKKEQKPHKSKKWFGKSKKLDVTNSGAAYSPRTVKDAKLKEIEEQQS 65
Query: 65 KHAIAVAAATAAVAEAALAAAHAAAEVVRLTS-SGVAGSSNKTRGQLRLPEETAAVKIQS 123
+HA +VA ATAA AEAA+AAA AAAEVVRL++ S G S EE AA+KIQ+
Sbjct: 66 RHAYSVAIATAAAAEAAVAAAQAAAEVVRLSALSRFPGKSM---------EEIAAIKIQT 116
Query: 124 AFRGYLARRALRALKALVKLQALVRGHIVRKKTADMLRRMQTLVRLQTKARASRAHLSSD 183
AFRGY+ARRALRAL+ LV+L++LV+G VR++ L+ MQTL R+Q + R R LS D
Sbjct: 117 AFRGYMARRALRALRGLVRLKSLVQGKCVRRQATSTLQSMQTLARVQYQIRERRLRLSED 176
Query: 184 NLHSFK-------------------SSLSHYPVPEEY-------EQPHHVYSTKFGGSSI 217
+ S+LS V + + F +
Sbjct: 177 KQALTRQLQQKHNKDFDKTGENWNDSTLSREKVEANMLNKQVATMRREKALAYAFSHQNT 236
Query: 218 LKRCSSNSNFRKIESEKPRFGSNWLDHWM---QENSISQTKNASSKNRHPDEHKSDKILE 274
K + + ++ P +G +WL+ WM + S T + + K+ S + E
Sbjct: 237 WKNSTKMGSQTFMDPNNPHWGWSWLERWMAARPNENHSLTPDNAEKDSSARSVASRAMSE 296
Query: 275 VDTWKPQLNKNENNVNSMSNESPSKHSTKAQNQSLSVKFHKAKEEVAASRTADNSPQTFS 334
+ L+ NS SP +++ + V F + T + P T
Sbjct: 297 MIPRGKNLSPRGKTPNSRRGSSPRVRQVPSEDSNSIVSFQSEQPCNRRHSTCGSIPST-- 354
Query: 335 ASSRNGSGVRRNTPFTPTRSECSWSFLGGYSGYPNYMANTESSRAKVR--------SQSA 386
R + FT + S+ P YMA T++++A+ R S+
Sbjct: 355 ---------RDDESFTSSFSQ----------SVPGYMAPTQAAKARARFSNLSPLSSEKT 395
Query: 387 PRQRHEFEEYSSTRRPFQG 405
++R F T R F G
Sbjct: 396 AKKRLSFSGSPKTVRRFSG 414
>gb|AAY78649.1| calmodulin-binding family protein [Arabidopsis thaliana]
gi|15218082|ref|NP_175608.1| calmodulin-binding family
protein [Arabidopsis thaliana] gi|4220443|gb|AAD12670.1|
Similar to gb|X74772 SF16 protein from Helianthus annuus
and contains calmodulin-binding motif PF|00612.
[Arabidopsis thaliana] gi|25405512|pir||A96559
hypothetical protein F5F19.1 [imported] - Arabidopsis
thaliana
Length = 351
Score = 126 bits (316), Expect = 2e-27
Identities = 126/395 (31%), Positives = 174/395 (43%), Gaps = 81/395 (20%)
Query: 3 FLRRLFGAKKPIPPSDGSGKKSDKDNKRRWSFGKQSSKTKSLPQPPPSAFNQFDSSTPLE 62
+ + +FG KK S SG S K F + T +
Sbjct: 7 WFKGMFGTKKSKDRSHVSGGDSVKGGDHSGDFNVPRDSV---------LLGTILTDTEKD 57
Query: 63 RNKHAIAVAAATAAVAEAALAAAHAAAEVVRLTSSGVAGSSNKTRGQLRLPEETAAVKIQ 122
+NK+AIAVA ATA A+AA++AA VVRLTS G AG T+ E AAVKIQ
Sbjct: 58 QNKNAIAVATATATAADAAVSAA-----VVRLTSEGRAGDIIITK-----EERWAAVKIQ 107
Query: 123 SAFRGYLARRALRALKALVKLQALVRGHIVRKKTADMLRRMQTLVRLQTKARASRAHLSS 182
FRG LAR+ALRALK +VKLQALVRG++VRK+ A ML+ +QTL+R+QT R+ R + S
Sbjct: 108 KVFRGSLARKALRALKGIVKLQALVRGYLVRKRAAAMLQSIQTLIRVQTAMRSKRINRSL 167
Query: 183 DNLHSFKSSLSHYPVPEEYEQPHHVYSTKFGGSSILKRCSSNSNFRKIESEKPRFGSNWL 242
+ ++ QP + KF ++ R + KI + R+
Sbjct: 168 NKEYN------------NMFQPRQSFD-KFDEATFDDRRT------KIVEKDDRY----- 203
Query: 243 DHWMQENSISQTKNASSKNRHPDEHKSDKILEVDTWKPQLNKNENNVNSMSNESPSKHST 302
+ +SS++R H NV SMS+ +
Sbjct: 204 -----------MRRSSSRSRSRQVH--------------------NVVSMSD---YEGDF 229
Query: 303 KAQNQSLSVKFHKAKEEVAASRTADNSPQTFSASSRNGSGVRRNTPFTPTRSECSWSFLG 362
+ L + F K + A TA N+P+ S N +P + +
Sbjct: 230 VYKGNDLELCFSDEKWKFA---TAQNTPRLLHHHSANNRYYVMQSPAKSVGGKALCDYES 286
Query: 363 GYSGYPNYMANTESSRAKVRSQSAPRQRHEFEEYS 397
S P YM T+S +AKVRS SAPRQR E + S
Sbjct: 287 SVS-TPGYMEKTKSFKAKVRSHSAPRQRSERQRLS 320
>emb|CAB79673.1| hypothetical protein [Arabidopsis thaliana]
gi|4972061|emb|CAB43929.1| hypothetical protein
[Arabidopsis thaliana] gi|28393019|gb|AAO41944.1|
unknown protein [Arabidopsis thaliana]
gi|15233461|ref|NP_194644.1| calmodulin-binding family
protein [Arabidopsis thaliana] gi|7485745|pir||T08970
hypothetical protein F19B15.180 - Arabidopsis thaliana
Length = 383
Score = 124 bits (310), Expect = 8e-27
Identities = 114/334 (34%), Positives = 151/334 (45%), Gaps = 61/334 (18%)
Query: 62 ERNKHAIAVAAATAAVAEAALAAAHAAAEVVRLTSSGVAGSSNKTRGQLRLPEETAAVKI 121
ER HAIAVAAATAA A+AA+AAA AAA VVRL G +G + + E AA++I
Sbjct: 66 ERRTHAIAVAAATAAAADAAVAAAKAAAAVVRLQGQGKSGPLGGGKSR----EHRAAMQI 121
Query: 122 QSAFRGYLARRALRALKALVKLQALVRGHIVRKKTADMLRRMQTLVRLQTKARASRAHLS 181
Q AFRGYLAR+ALRAL+ +VK+QALVRG +VR + A LR M+ LVR Q + RA
Sbjct: 122 QCAFRGYLARKALRALRGVVKIQALVRGFLVRNQAAATLRSMEALVRAQKTVKIQRALRR 181
Query: 182 SDNLHSFKSSLSHYPVPEEYEQPHHVYSTKFGGSSILKRCSSNSNFR--KIESEKPRF-G 238
+ N + S + E + G K ++ R P G
Sbjct: 182 NGNAAPARKSTERFSGSLE---------NRNNGEETAKIVEVDTGTRPGTYRIRAPVLSG 232
Query: 239 SNWLDHWMQENSISQTKNASSKNRHPDEHKSDKILEVDTWKPQLNKNENNVNSMSNESPS 298
S++LD N +T ++ R P P+L+ + S++ P+
Sbjct: 233 SDFLD-----NPFRRTLSSPLSGRVP---------------PRLSMPKPEWEECSSKFPT 272
Query: 299 KHSTKAQNQSLSVKFHKAKEEVAASRTADNSPQTFSASSRNGSGVRRNTPFTPTRSECSW 358
ST R + SP S G +T R +
Sbjct: 273 AQST--------------------PRFSGGSPARSVCCSGGGVEAEVDTEADANR----F 308
Query: 359 SFLGGYSGYPNYMANTESSRAKVRSQSAPRQRHE 392
FL G YMA+T S RAK+RS SAPRQR E
Sbjct: 309 CFLSGEFN-SGYMADTTSFRAKLRSHSAPRQRPE 341
>ref|NP_915355.1| P0460C04.18 [Oryza sativa (japonica cultivar-group)]
Length = 302
Score = 123 bits (309), Expect = 1e-26
Identities = 92/275 (33%), Positives = 136/275 (49%), Gaps = 46/275 (16%)
Query: 22 KKSDKDNKRR-WSFGKQS------------SKTKSLPQPPPSAFNQFDSSTP-------- 60
+K+DK RR W FGK + T PQPPP P
Sbjct: 4 EKADKSRSRRKWPFGKSKKSDPWTSTVAVPTSTAPPPQPPPPPPTHPIQPQPEEIKDVKA 63
Query: 61 ----LERNKHAIAVAAATAAVAEAALAAAHAAAEVVRLTSSGVAGSSNKTRGQLRLPEET 116
E+NKHA +VA A+A AEAA AA AAAEVVRLT++ A + + +E
Sbjct: 64 VETDSEQNKHAYSVALASAVAAEAAAVAAQAAAEVVRLTTATTAVPKSPVSSK----DEL 119
Query: 117 AAVKIQSAFRGYLARRALRALKALVKLQALVRGHIVRKKTADMLRRMQTLVRLQTKARAS 176
AA+KIQ+AFRGYLARRALRAL+ LV+L++LV G+ V+++TA L QT+ R+QT+ +
Sbjct: 120 AAIKIQTAFRGYLARRALRALRGLVRLKSLVDGNAVKRQTAHTLHCTQTMTRVQTQIYSR 179
Query: 177 RAHLSSDNLHSFKSSLSHYPVPEEYEQPHHVYSTKFGGSSILKRCSSNSNFRKIESEKPR 236
R + + K +L + ++++ K G +I + N P
Sbjct: 180 RVKMEEE-----KQALQR-QLQLKHQRELEKMKWKNSGRTITPTFTDQGN--------PN 225
Query: 237 FGSNWLDHWMQENSISQTKNASSKNRHPDEHKSDK 271
+G +W++ WM ++ ++ P +H S K
Sbjct: 226 WGWSWMERWMTSR---PWESRVISDKDPKDHYSTK 257
>gb|AAP53800.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|37534422|ref|NP_921513.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
Length = 485
Score = 119 bits (298), Expect = 2e-25
Identities = 112/379 (29%), Positives = 164/379 (42%), Gaps = 62/379 (16%)
Query: 29 KRRWSFGKQSSKTKSLPQPPPSAFNQFDSSTPLERNKHAIAVAAATAAVAEA------AL 82
K+RWSF + S+ + P+A + P +++ A A E A+
Sbjct: 48 KKRWSFRRSSASASAAAMGKPAAVTAPSTPEPSVSGLASVSERARDVADLEGQSKHAMAV 107
Query: 83 AAAHAAAEVVRLTSSGVAGSSNKTRGQLRLPEETAAVKIQSAFRGYLARRALRALKALVK 142
AA AAE +++S V E AAV IQ+ +RGYLAR+AL AL+ LVK
Sbjct: 108 AAVATAAEGDDVSASAV--------------EVVAAVMIQATYRGYLARKALCALRGLVK 153
Query: 143 LQALVRGHIVRKKTADMLRRMQTLVRLQTKARASRAHL----SSDNLHSFKSSLSHYPVP 198
LQAL+RG++VRK+ LRRMQ L+ Q + RA R + D++H H
Sbjct: 154 LQALIRGNLVRKQATATLRRMQALLVAQARLRAQRMRMLEEEEDDDVH------GHGHHH 207
Query: 199 EEYEQPHHVYSTKFGGSSILKRCSSNSNFRKIESEKPRFGSNWLDHWMQENSISQTKNAS 258
PHH + S NF + H +QE S + A
Sbjct: 208 HRRSSPHHPRHRRSYVSRARLPSRRRGNFSYV-------------HALQEMDRSGEEQA- 253
Query: 259 SKNRHPDEHKSDKILEVDTWKP-QLNKNENNVNSMSNESPSKHSTKAQNQSLSVKFHKAK 317
KI+EVD +P + ++ + ++ES + + + +
Sbjct: 254 ------------KIVEVDVGEPAPPRRGRSSCSVAASESRERRMAEYGYYAQCSPAPSSS 301
Query: 318 EEVAASRTADNSPQTFSASSRNGS----GVRRNTPFTPTRSECSWSFLGGYSGYPNYMAN 373
AA+ + ++S + S R++P+ P S GG +PNYMAN
Sbjct: 302 AFTAAAAASPPRDASYSGHFDDFSPFEPATARSSPYIPP-SPGGGGGGGGGEFFPNYMAN 360
Query: 374 TESSRAKVRSQSAPRQRHE 392
T+SSRAK RSQSAPRQR E
Sbjct: 361 TQSSRAKARSQSAPRQRTE 379
>emb|CAB66405.1| SF16-like protein [Arabidopsis thaliana] gi|11358844|pir||T45831
SF16-like protein - Arabidopsis thaliana
Length = 535
Score = 115 bits (288), Expect = 3e-24
Identities = 123/412 (29%), Positives = 184/412 (43%), Gaps = 85/412 (20%)
Query: 22 KKSDKDNKRRWSFG-------KQSSKTKSL---------PQPPPSAFNQFDSSTPLER-- 63
K S KD+KR + G +Q T+ + P S + +STP
Sbjct: 19 KSSPKDSKRENNIGSNNADIWQQQHDTQEVVSFEHFPAESSPEISHDVESTASTPATNVG 78
Query: 64 -NKHAIAVAAATAAVAEAALAAAHAAAEVVRLTSSGVAGSSNKTRGQLRLPEETAAVKIQ 122
KHA+AVA ATAA AEAA+AAA AAA+VVRL AG + +T E++AAV IQ
Sbjct: 79 DRKHAMAVAIATAAAAEAAVAAAQAAAKVVRL-----AGYNRQTE------EDSAAVLIQ 127
Query: 123 SAFRGYLARRALRALKALVKLQALVRGHIVRKKTADMLRRMQTLVRLQTKARASRAHLSS 182
S +RGYLARRALRALK LV+LQALVRG+ VRK+ ++ MQ LVR+Q + RA R ++
Sbjct: 128 SHYRGYLARRALRALKGLVRLQALVRGNHVRKQAQMTMKCMQALVRVQGRVRARRLQVAH 187
Query: 183 DNLHSFKSSLSHYPVPEEYEQPHHVYSTKFGGSSILKRCSSNSNFRKIESEKPRFGSNWL 242
D FK E+P+ ++ K E EKP+
Sbjct: 188 DR---FKKQFEEEEKRSGMEKPNKGFAN-----------------LKTEREKPK-----K 222
Query: 243 DHWMQENSISQTKNASSKNRHPDEHKSDKILEVDTWKPQL-NKNENNVNSMSNESPSKH- 300
H + S+ QT+ + + + T++ Q+ + N +S+ P ++
Sbjct: 223 LHEVNRTSLYQTQGKEKERSEGMMKRERALAYAYTYQRQMQHTNSEEGIGLSSNGPDRNQ 282
Query: 301 -STKAQNQSLSVKFHKAKEEVAASRTADNSPQTF-----SASSRNGSGVRRNT----PFT 350
+ + +S + + ++ +P + +A++ V T T
Sbjct: 283 WAWNWLDHWMSSQPYTGRQTGPGPGPGQYNPPPYPPFPTAAATTTSDDVSEKTVEMDVTT 342
Query: 351 PTR--------SECSWSFLGGY----------SGYPNYMANTESSRAKVRSQ 384
PT + + LG Y + P+YMA T S++AKVR Q
Sbjct: 343 PTSLKGNIIGLIDREYIDLGSYRQGHKQRKSPTHIPSYMAPTASAKAKVRDQ 394
>gb|AAN18171.1| At3g49260/F2K15_120 [Arabidopsis thaliana]
gi|21593446|gb|AAM65413.1| SF16-like protein
[Arabidopsis thaliana] gi|13605619|gb|AAK32803.1|
AT3g49260/F2K15_120 [Arabidopsis thaliana]
gi|42572619|ref|NP_974405.1| calmodulin-binding family
protein [Arabidopsis thaliana]
gi|18408809|ref|NP_566917.1| calmodulin-binding family
protein [Arabidopsis thaliana]
gi|39980284|gb|AAR32943.1| guard cell associated protein
[Arabidopsis thaliana]
Length = 471
Score = 115 bits (288), Expect = 3e-24
Identities = 123/412 (29%), Positives = 184/412 (43%), Gaps = 85/412 (20%)
Query: 22 KKSDKDNKRRWSFG-------KQSSKTKSL---------PQPPPSAFNQFDSSTPLER-- 63
K S KD+KR + G +Q T+ + P S + +STP
Sbjct: 19 KSSPKDSKRENNIGSNNADIWQQQHDTQEVVSFEHFPAESSPEISHDVESTASTPATNVG 78
Query: 64 -NKHAIAVAAATAAVAEAALAAAHAAAEVVRLTSSGVAGSSNKTRGQLRLPEETAAVKIQ 122
KHA+AVA ATAA AEAA+AAA AAA+VVRL AG + +T E++AAV IQ
Sbjct: 79 DRKHAMAVAIATAAAAEAAVAAAQAAAKVVRL-----AGYNRQTE------EDSAAVLIQ 127
Query: 123 SAFRGYLARRALRALKALVKLQALVRGHIVRKKTADMLRRMQTLVRLQTKARASRAHLSS 182
S +RGYLARRALRALK LV+LQALVRG+ VRK+ ++ MQ LVR+Q + RA R ++
Sbjct: 128 SHYRGYLARRALRALKGLVRLQALVRGNHVRKQAQMTMKCMQALVRVQGRVRARRLQVAH 187
Query: 183 DNLHSFKSSLSHYPVPEEYEQPHHVYSTKFGGSSILKRCSSNSNFRKIESEKPRFGSNWL 242
D FK E+P+ ++ K E EKP+
Sbjct: 188 DR---FKKQFEEEEKRSGMEKPNKGFAN-----------------LKTEREKPK-----K 222
Query: 243 DHWMQENSISQTKNASSKNRHPDEHKSDKILEVDTWKPQL-NKNENNVNSMSNESPSKH- 300
H + S+ QT+ + + + T++ Q+ + N +S+ P ++
Sbjct: 223 LHEVNRTSLYQTQGKEKERSEGMMKRERALAYAYTYQRQMQHTNSEEGIGLSSNGPDRNQ 282
Query: 301 -STKAQNQSLSVKFHKAKEEVAASRTADNSPQTF-----SASSRNGSGVRRNT----PFT 350
+ + +S + + ++ +P + +A++ V T T
Sbjct: 283 WAWNWLDHWMSSQPYTGRQTGPGPGPGQYNPPPYPPFPTAAATTTSDDVSEKTVEMDVTT 342
Query: 351 PTR--------SECSWSFLGGY----------SGYPNYMANTESSRAKVRSQ 384
PT + + LG Y + P+YMA T S++AKVR Q
Sbjct: 343 PTSLKGNIIGLIDREYIDLGSYRQGHKQRKSPTHIPSYMAPTASAKAKVRDQ 394
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.310 0.123 0.348
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 727,456,841
Number of Sequences: 2540612
Number of extensions: 29499869
Number of successful extensions: 143575
Number of sequences better than 10.0: 819
Number of HSP's better than 10.0 without gapping: 155
Number of HSP's successfully gapped in prelim test: 701
Number of HSP's that attempted gapping in prelim test: 138811
Number of HSP's gapped (non-prelim): 3093
length of query: 441
length of database: 863,360,394
effective HSP length: 131
effective length of query: 310
effective length of database: 530,540,222
effective search space: 164467468820
effective search space used: 164467468820
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 77 (34.3 bits)
Medicago: description of AC149494.9


