

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149490.9 - phase: 0
(1049 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
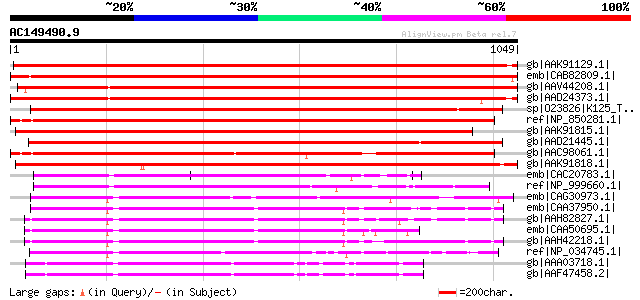
Score E
Sequences producing significant alignments: (bits) Value
gb|AAK91129.1| KRP120-2 [Daucus carota] 1561 0.0
emb|CAB82809.1| kinesin-related protein-like [Arabidopsis thalia... 1558 0.0
gb|AAV44208.1| putative kinesin [Oryza sativa (japonica cultivar... 1410 0.0
gb|AAD24373.1| putative kinesin-like spindle protein [Arabidopsi... 1358 0.0
sp|O23826|K125_TOBAC 125 kDa kinesin-related protein gi|7441387|... 891 0.0
ref|NP_850281.1| kinesin motor protein-related [Arabidopsis thal... 875 0.0
gb|AAK91815.1| kinesin heavy chain [Zea mays] 860 0.0
gb|AAD21445.1| putative kinesin-related cytokinesis protein [Ara... 838 0.0
gb|AAC98061.1| putative kinesin heavy chain [Arabidopsis thalian... 834 0.0
gb|AAK91818.1| kinesin heavy chain [Zea mays] 799 0.0
emb|CAC20783.1| kinesin-like boursin [Paracentrotus lividus] 513 e-143
ref|NP_999660.1| KRP170 [Strongylocentrotus purpuratus] gi|10567... 509 e-142
emb|CAG30973.1| hypothetical protein [Gallus gallus] 501 e-140
emb|CAA37950.1| kinesine [Xenopus laevis] gi|119217|sp|P28025|EG... 492 e-137
gb|AAH82827.1| LOC397908 protein [Xenopus laevis] 490 e-137
emb|CAA50695.1| kinesin like protein [Xenopus laevis] gi|2497521... 489 e-136
gb|AAH42218.1| MGC52588 protein [Xenopus laevis] 488 e-136
ref|NP_034745.1| kinesin family member 11 [Mus musculus] gi|3817... 486 e-135
gb|AAA03718.1| kinesin-like protein 481 e-134
gb|AAF47458.2| CG9191-PA [Drosophila melanogaster] gi|17136642|r... 481 e-134
>gb|AAK91129.1| KRP120-2 [Daucus carota]
Length = 1045
Score = 1561 bits (4041), Expect = 0.0
Identities = 794/1043 (76%), Positives = 915/1043 (87%), Gaps = 9/1043 (0%)
Query: 8 GGLVPLSPSQTPRSTDKPARDLRSADSNSNSHNKYDKEKGVNVQVLVRCRPMNEDEMRLH 67
G V +SPSQTP+S++K RDLRS N++ + EKGVNVQV+VRCRP++EDE++ H
Sbjct: 11 GSFVSISPSQTPKSSEKAIRDLRSEGGNASFKHDSRGEKGVNVQVIVRCRPLSEDEIKAH 70
Query: 68 TPVVISCNEGRREVAAVQSIANKQIDRTFVFDKVFGPNSQQKELYDQAVSPIVYEVLEGY 127
TPVVI+C E RREV AVQ+IA+KQIDR+F+FDKVFGP SQQK+LY+QAVSPIVYEVLEGY
Sbjct: 71 TPVVITCTENRREVCAVQNIASKQIDRSFMFDKVFGPASQQKDLYEQAVSPIVYEVLEGY 130
Query: 128 NCTIFAYGQTGTGKTYTMEGGAIKKNGEFPTDAGVIPRAVKQIFDILEAQSAEYSMKVTF 187
NCTIFAYGQTGTGKTYTMEGG KKNGEFP+DAGVIPRAVKQIF+ILE+Q+AEYSMKVTF
Sbjct: 131 NCTIFAYGQTGTGKTYTMEGGGRKKNGEFPSDAGVIPRAVKQIFNILESQNAEYSMKVTF 190
Query: 188 LELYNEEITDLLAPEETTKFVDEKSKKPIALMEDGKGGVLVRGLEEEIVCTANEIYKILE 247
LELYNEEITDLLAPEE +KF+++KSKKPIALMEDGKGGV VRGLEEEIVCTANEIYKILE
Sbjct: 191 LELYNEEITDLLAPEEFSKFIEDKSKKPIALMEDGKGGVFVRGLEEEIVCTANEIYKILE 250
Query: 248 KGSAKRRTAETLLNKQSSRSHSIFSITIHIKECTPEGEEMIKCGKLNLVDLAGSENISRS 307
KGSAKRRTAETLLNKQSSRSHSIFSITIHIKECTPEGEEMIKCGKLNLVDLAGSENISRS
Sbjct: 251 KGSAKRRTAETLLNKQSSRSHSIFSITIHIKECTPEGEEMIKCGKLNLVDLAGSENISRS 310
Query: 308 GAREGRAREAGEINKSLLTLGRTINALVEHSGHVPYRDSKLTRLLRDSLGGKTKTCIIAT 367
GAREGRAREAGEINKSLLTLGR INALVEHSGHVPYRDSKLTRLLRDSLGGKTKTCIIAT
Sbjct: 311 GAREGRAREAGEINKSLLTLGRVINALVEHSGHVPYRDSKLTRLLRDSLGGKTKTCIIAT 370
Query: 368 VSPSIHCLEETLSTLDYAHRAKNIKNKPEVNQKMMKSAMIKDLYSEIDRLKQEVYAAREK 427
+SPS++ LEETLSTLDYAHRAKNIKNKPE+NQKMMKSAMIKDLYSEIDRLKQEV++AREK
Sbjct: 371 ISPSVYSLEETLSTLDYAHRAKNIKNKPEINQKMMKSAMIKDLYSEIDRLKQEVFSAREK 430
Query: 428 NGIYIPRDRYLNEEAEKKAMAEKIERMELDADSKDKNLVELQELYNSQQLLTAELSAKLE 487
NGIYIP+DRYL +EA+KKAMAEKIERMELD +S+DK +ELQ L+NSQ LTAELS KLE
Sbjct: 431 NGIYIPKDRYLQDEADKKAMAEKIERMELDFESRDKQFMELQGLHNSQLQLTAELSDKLE 490
Query: 488 KTEKSLEETEQTLFDLEERHRQANATIKEKEFLISNLLKSEKELVERAIELRAELENAAS 547
KTEK L ETE L DLEERHRQANATIKEKE+LISNL+KSE+ L+ERA ELRAELE+AA
Sbjct: 491 KTEKKLHETEHALVDLEERHRQANATIKEKEYLISNLIKSERSLIERAFELRAELESAAL 550
Query: 548 DVSNLFSKIERKDKIEEENRVLIQKFQSQLAQQLEALHRTVSASVMHQEQQLKDMEKDMQ 607
DVSNLF+KIERKDKIE NR+LIQKFQ+QL+QQLE LH+TV+ASV QEQQL+ ME+DMQ
Sbjct: 551 DVSNLFTKIERKDKIENGNRILIQKFQAQLSQQLEILHKTVAASVTQQEQQLRAMEEDMQ 610
Query: 608 SFVSTKSEATEDLRVRVVELKNMYGSGIKALDNLAEELKSNNQLTYEDLKSEVAKHSSAL 667
SFVSTK+EATE+LR +++LK MYGSGI ALD++A EL N++ T L +EV+KHSSAL
Sbjct: 611 SFVSTKAEATEELRENLIKLKTMYGSGIGALDDIAGELDENSKSTVGQLNNEVSKHSSAL 670
Query: 668 EDLFKGIALEADSLLNDLQNSLHKQEANVTAFAHQQREAHSRAVETTRSVSKITMKFFET 727
+D FK IA EAD+LLNDLQ SL+ QE +T++A QQREAHSRA+ETTRS+S+IT+ FF T
Sbjct: 671 KDHFKEIASEADTLLNDLQRSLYSQEDKMTSYAQQQREAHSRAMETTRSISQITVNFFNT 730
Query: 728 IDRHASSLTQIVEETQFVNDQKLCELEKKFEECTAYEEKQLLEKVAEMLASSNARKKKLV 787
+D HAS+L+QIVEE Q NDQKL ELEKKFEEC A EE+QLLEKVAE+LASSN+RKKKLV
Sbjct: 731 LDTHASNLSQIVEEAQTDNDQKLSELEKKFEECAANEERQLLEKVAELLASSNSRKKKLV 790
Query: 788 QMAVNDLRESANCRTSKLQREALTMQDSTSFVKAEWMVHMEKTESNYHEDTSSVESGKKD 847
AV LR+SA RT+K Q+E TMQDSTS VK EW ++ K E++Y EDT++VESGKKD
Sbjct: 791 HTAVTSLRDSAASRTNKFQQEMSTMQDSTSLVKVEWSSYIGKAETHYTEDTAAVESGKKD 850
Query: 848 LAEVLQICLNKAEVGSQQWRNAQDSLLSLEKRNAGSVDTIVRGGMEANQALRARFSSSVS 907
+ EVLQ CL KA++G +QW +AQ+SLLSLEK N SVD I+RGGM+ANQ LR+RFS++VS
Sbjct: 851 IEEVLQKCLQKAKMGQKQWSSAQESLLSLEKTNVASVDDIIRGGMDANQILRSRFSTAVS 910
Query: 908 TTLEDAGIANTDINSSIDYSLQLDHEACGNLNSMITPCCGDLTELKGGHYNRIVEITENA 967
+ LEDA IA+ + SSID+SLQLDH+AC NL+S+ITPCCG+L ELK GHY++ VEITE+A
Sbjct: 911 SVLEDANIASRNFISSIDHSLQLDHDACSNLDSIITPCCGELRELKSGHYHKTVEITEDA 970
Query: 968 GKCLLNEYMVDEPSCSTPTRRLFNLPSVSSIEELRTPSFEELLKAFWDAKSQKLANGDVK 1027
GKCLL EY+VD+PSCSTP +R FNLPS++SIEELRTP+FEELLK+FW+AK+ KLANGD K
Sbjct: 971 GKCLLTEYVVDQPSCSTPKKRSFNLPSITSIEELRTPAFEELLKSFWEAKASKLANGDTK 1030
Query: 1028 -HIGSYEETQSVRDSRVPLTTIN 1049
HI + DSR PLT IN
Sbjct: 1031 QHI--------LGDSRAPLTAIN 1045
>emb|CAB82809.1| kinesin-related protein-like [Arabidopsis thaliana]
gi|15231259|ref|NP_190171.1| kinesin motor
protein-related [Arabidopsis thaliana]
gi|11276945|pir||T47525 kinesin-related protein-like -
Arabidopsis thaliana
Length = 1058
Score = 1558 bits (4034), Expect = 0.0
Identities = 795/1057 (75%), Positives = 916/1057 (86%), Gaps = 12/1057 (1%)
Query: 3 VQQKRGGLVPLSPSQTPRSTDKPARDLRSADSNSNSHNKYDKEKGVNVQVLVRCRPMNED 62
+QQ+RGG+V LSP+QTPRS+DK AR+ RS++SNS + N DKEKGVNVQV++RCRP++ED
Sbjct: 4 IQQRRGGIVSLSPAQTPRSSDKSARESRSSESNSTNRN--DKEKGVNVQVILRCRPLSED 61
Query: 63 EMRLHTPVVISCNEGRREVAAVQSIANKQIDRTFVFDKVFGPNSQQKELYDQAVSPIVYE 122
E R+HTPVVISCNE RREVAA QSIA K IDR F FDKVFGP SQQK+LYDQA+ PIV+E
Sbjct: 62 EARIHTPVVISCNENRREVAATQSIAGKHIDRHFAFDKVFGPASQQKDLYDQAICPIVFE 121
Query: 123 VLEGYNCTIFAYGQTGTGKTYTMEGGAIKKNGEFPTDAGVIPRAVKQIFDILEAQSAEYS 182
VLEGYNCTIFAYGQTGTGKTYTMEGGA KKNGEFP+DAGVIPRAVKQIFDILEAQ AEYS
Sbjct: 122 VLEGYNCTIFAYGQTGTGKTYTMEGGARKKNGEFPSDAGVIPRAVKQIFDILEAQGAEYS 181
Query: 183 MKVTFLELYNEEITDLLAPEETTKFVDEKSKKPIALMEDGKGGVLVRGLEEEIVCTANEI 242
MKVTFLELYNEEI+DLLAPEET KFVDEKSKK IALMEDGKG V VRGLEEEIV TANEI
Sbjct: 182 MKVTFLELYNEEISDLLAPEETIKFVDEKSKKSIALMEDGKGSVFVRGLEEEIVSTANEI 241
Query: 243 YKILEKGSAKRRTAETLLNKQSSRSHSIFSITIHIKECTPEGEEMIKCGKLNLVDLAGSE 302
YKILEKGSAKRRTAETLLNKQSSRSHSIFSITIHIKE TPEGEEMIKCGKLNLVDLAGSE
Sbjct: 242 YKILEKGSAKRRTAETLLNKQSSRSHSIFSITIHIKENTPEGEEMIKCGKLNLVDLAGSE 301
Query: 303 NISRSGAREGRAREAGEINKSLLTLGRTINALVEHSGHVPYRDSKLTRLLRDSLGGKTKT 362
NISRSGAREGRAREAGEINKSLLTLGR INALVEHSGH+PYRDSKLTRLLR+SLGGKTKT
Sbjct: 302 NISRSGAREGRAREAGEINKSLLTLGRVINALVEHSGHIPYRDSKLTRLLRESLGGKTKT 361
Query: 363 CIIATVSPSIHCLEETLSTLDYAHRAKNIKNKPEVNQKMMKSAMIKDLYSEIDRLKQEVY 422
C+IAT+SPSIHCLEETLSTLDYAHRAKNIKNKPE+NQKMMKSA++KDLYSEIDRLKQEVY
Sbjct: 362 CVIATISPSIHCLEETLSTLDYAHRAKNIKNKPEINQKMMKSAVMKDLYSEIDRLKQEVY 421
Query: 423 AAREKNGIYIPRDRYLNEEAEKKAMAEKIERMELDADSKDKNLVELQELYNSQQLLTAEL 482
AAREKNGIYIP+DRY+ EEAEKKAMAEKIER+EL ++SKDK +V+LQELYNSQQ+LTAEL
Sbjct: 422 AAREKNGIYIPKDRYIQEEAEKKAMAEKIERLELQSESKDKRVVDLQELYNSQQILTAEL 481
Query: 483 SAKLEKTEKSLEETEQTLFDLEERHRQANATIKEKEFLISNLLKSEKELVERAIELRAEL 542
S KLEKTEK LEETE +LFDLEE++RQANATIKEKEF+ISNLLKSEK LVERA +LR EL
Sbjct: 482 SEKLEKTEKKLEETEHSLFDLEEKYRQANATIKEKEFVISNLLKSEKSLVERAFQLRTEL 541
Query: 543 ENAASDVSNLFSKIERKDKIEEENRVLIQKFQSQLAQQLEALHRTVSASVMHQEQQLKDM 602
E+A+SDVSNLFSKIERKDKIE+ NR LIQKFQSQL QQLE LH+TV++SV QE QLK M
Sbjct: 542 ESASSDVSNLFSKIERKDKIEDGNRFLIQKFQSQLTQQLELLHKTVASSVTQQEVQLKHM 601
Query: 603 EKDMQSFVSTKSEATEDLRVRVVELKNMYGSGIKALDNLAEELKSNNQLTYEDLKSEVAK 662
E+DM+SFVSTKSEATE+LR R+ +LK +YGSGI+ALDN+A +L N+Q T+ L SEV+K
Sbjct: 602 EEDMESFVSTKSEATEELRDRLSKLKRVYGSGIEALDNIAVKLDGNSQSTFSSLNSEVSK 661
Query: 663 HSSALEDLFKGIALEADSLLNDLQNSLHKQEANVTAFAHQQREAHSRAVETTRSVSKITM 722
HS LE++FKG A EAD LL DLQ+SL+KQE + FA QQR+AHSRAV+T RSVSK+T+
Sbjct: 662 HSHELENVFKGFASEADMLLQDLQSSLNKQEEKLITFAQQQRKAHSRAVDTARSVSKVTV 721
Query: 723 KFFETIDRHASSLTQIVEETQFVNDQKLCELEKKFEECTAYEEKQLLEKVAEMLASSNAR 782
+FF+T+D HA+ LT IVEE Q VN +KL E E KFEEC A EE+QLLEKVAE+LA+SNAR
Sbjct: 722 EFFKTLDTHATKLTGIVEEAQTVNHKKLSEFENKFEECAANEERQLLEKVAELLANSNAR 781
Query: 783 KKKLVQMAVNDLRESANCRTSKLQREALTMQDSTSFVKAEWMVHMEKTESNYHEDTSSVE 842
KK LVQMAV+DLRESA+ RT+ LQ E TMQDSTS +KAEW +HMEKTES++HEDTS+VE
Sbjct: 782 KKNLVQMAVHDLRESASTRTTTLQHEMSTMQDSTSSIKAEWSIHMEKTESSHHEDTSAVE 841
Query: 843 SGKKDLAEVLQICLNKAEVGSQQWRNAQDSLLSLEKRNAGSVDTIVRGGMEANQALRARF 902
SGKK + EVL CL K E+ + QWR AQ+SL+SLE+ N SVD+IVRGGM+AN+ LR++F
Sbjct: 842 SGKKAMQEVLLNCLEKTEMSAHQWRKAQESLVSLERNNVASVDSIVRGGMDANENLRSQF 901
Query: 903 SSSVSTTLEDAGIANTDINSSIDYSLQLDHEACGNLNSMITPCCGDLTELKGGHYNRIVE 962
S++VS++L+ AN+ + +SID+SLQLD++AC +NSMI PCC DL ELK H ++I+E
Sbjct: 902 STAVSSSLDVFDAANSSLLTSIDHSLQLDNDACTKVNSMIIPCCEDLIELKSDHNHKIIE 961
Query: 963 ITENAGKCLLNEYMVDEPSCSTPTRRLFNLPSVSSIEELRTPSFEELLKAFWDAKSQKLA 1022
ITENAGKCLL+EY+VDEPSCSTP +R ++PS+ SIEELRTP+ EELL+AF D K K A
Sbjct: 962 ITENAGKCLLDEYVVDEPSCSTPKKRPIDIPSIESIEELRTPASEELLRAFRDEKLSKQA 1021
Query: 1023 NGDVKHIGSYEETQSVR----------DSRVPLTTIN 1049
NGD K ++ +R DSR PL+ +N
Sbjct: 1022 NGDAKQQQQQQQQHLIRASSLYEAAVSDSRYPLSAVN 1058
>gb|AAV44208.1| putative kinesin [Oryza sativa (japonica cultivar-group)]
Length = 1056
Score = 1410 bits (3650), Expect = 0.0
Identities = 741/1049 (70%), Positives = 870/1049 (82%), Gaps = 16/1049 (1%)
Query: 16 SQTPRSTDKPARDLRS-------ADSNSNSHNKYDKEKGVNVQVLVRCRPMNEDEMRLHT 68
S +P+ST+K RDLRS A++NSNS + DKEKGVNVQV++RCRPM+++E + +T
Sbjct: 9 SPSPKSTEKSGRDLRSGGDANGGANTNSNSIPRGDKEKGVNVQVILRCRPMSDEETKSNT 68
Query: 69 PVVISCNEGRREVAAVQSIANKQIDRTFVFDKVFGPNSQQKELYDQAVSPIVYEVLEGYN 128
PVVISCNE RREVAA Q IANKQIDRTF FDKVFGP S+QK+L++Q++SPIV EVLEGYN
Sbjct: 69 PVVISCNERRREVAATQIIANKQIDRTFAFDKVFGPASKQKDLFEQSISPIVNEVLEGYN 128
Query: 129 CTIFAYGQTGTGKTYTMEGGAIKK--NGEFPTDAGVIPRAVKQIFDILEAQSAEYSMKVT 186
CTIFAYGQTGTGKTYTMEGG +K NGE PTDAGVIPRAV+QIFDILEAQ AEYSMKVT
Sbjct: 129 CTIFAYGQTGTGKTYTMEGGGTRKTKNGELPTDAGVIPRAVRQIFDILEAQCAEYSMKVT 188
Query: 187 FLELYNEEITDLLAPEETTKFV---DEKSKKPIALMEDGKGGVLVRGLEEEIVCTANEIY 243
FLELYNEEITDLLAPEE KF ++K+KKPIALMEDGKGGV VRGLEEE+V +A EIY
Sbjct: 189 FLELYNEEITDLLAPEEP-KFPIVPEDKTKKPIALMEDGKGGVFVRGLEEEVVYSAGEIY 247
Query: 244 KILEKGSAKRRTAETLLNKQSSRSHSIFSITIHIKECTPEGEEMIKCGKLNLVDLAGSEN 303
KIL+KGSAKRRTAETLLNKQSSRSHSIFSITIHIKE T EGEEMIK GKLNLVDLAGSEN
Sbjct: 248 KILDKGSAKRRTAETLLNKQSSRSHSIFSITIHIKELTHEGEEMIKIGKLNLVDLAGSEN 307
Query: 304 ISRSGAREGRAREAGEINKSLLTLGRTINALVEHSGHVPYRDSKLTRLLRDSLGGKTKTC 363
ISRSGAR+GRAREAGEINKSLLTLGR INALVEHSGHVPYRDSKLTRLLRDSLGGKTKTC
Sbjct: 308 ISRSGARDGRAREAGEINKSLLTLGRVINALVEHSGHVPYRDSKLTRLLRDSLGGKTKTC 367
Query: 364 IIATVSPSIHCLEETLSTLDYAHRAKNIKNKPEVNQKMMKSAMIKDLYSEIDRLKQEVYA 423
IIAT+SPS++CLEETLSTLDYAHRAKNIKNKPEVNQ+MMKSA+IKDLYSEIDRLKQEV+A
Sbjct: 368 IIATISPSVYCLEETLSTLDYAHRAKNIKNKPEVNQRMMKSAVIKDLYSEIDRLKQEVFA 427
Query: 424 AREKNGIYIPRDRYLNEEAEKKAMAEKIERMELDADSKDKNLVELQELYNSQQLLTAELS 483
AREKNGIYIPR+RYL EEAEKKAM EKIER+ D +++DK LVEL+ELY+++QLL+AELS
Sbjct: 428 AREKNGIYIPRERYLQEEAEKKAMTEKIERLGADLEARDKQLVELKELYDAEQLLSAELS 487
Query: 484 AKLEKTEKSLEETEQTLFDLEERHRQANATIKEKEFLISNLLKSEKELVERAIELRAELE 543
KL KT+K LE+T+ L DLEE++ +A +TIKEKE++I NLLKSEK LV+ A LRAELE
Sbjct: 488 EKLGKTQKDLEDTKNVLHDLEEKYNEAESTIKEKEYVIFNLLKSEKSLVDCAYNLRAELE 547
Query: 544 NAASDVSNLFSKIERKDKIEEENRVLIQKFQSQLAQQLEALHRTVSASVMHQEQQLKDME 603
NAA+DVS LFSKIERKDKIE+ NR L+Q+F+SQL QL+ LH+TVS SVM QE LK+ME
Sbjct: 548 NAAADVSGLFSKIERKDKIEDGNRSLVQRFRSQLTNQLDTLHKTVSTSVMQQENHLKEME 607
Query: 604 KDMQSFVSTKSEATEDLRVRVVELKNMYGSGIKALDNLAEELKSNNQLTYEDLKSEVAKH 663
DMQSFVS+K EA + LR + +LK ++GSGI ALD+LA E+ N+Q T+E L S+V H
Sbjct: 608 DDMQSFVSSKDEAAQGLRESIQKLKLLHGSGITALDSLAGEIDMNSQSTFERLNSQVQSH 667
Query: 664 SSALEDLFKGIALEADSLLNDLQNSLHKQEANVTAFAHQQREAHSRAVETTRSVSKITMK 723
+S+LE F GIA EAD+LLN+LQ SL KQE +T FA +QRE H RAVE +RS+SKIT
Sbjct: 668 TSSLEQCFGGIASEADNLLNELQCSLSKQEERLTQFAKKQREGHLRAVEASRSISKITAG 727
Query: 724 FFETIDRHASSLTQIVEETQFVNDQKLCELEKKFEECTAYEEKQLLEKVAEMLASSNARK 783
FF ++D HAS LT I+EETQ V DQ+L +LEKKFEEC A EEKQLLEKVAEMLASS+ARK
Sbjct: 728 FFSSLDVHASKLTSILEETQSVQDQQLLDLEKKFEECAANEEKQLLEKVAEMLASSHARK 787
Query: 784 KKLVQMAVNDLRESANCRTSKLQREALTMQDSTSFVKAEWMVHMEKTESNYHEDTSSVES 843
KKLVQ AV +LRESA RTS LQ E T QD TS V+ +W +ME+TE NY EDT++V+S
Sbjct: 788 KKLVQTAVGNLRESAVNRTSHLQNEISTAQDFTSSVREKWGFYMEETEKNYIEDTTAVDS 847
Query: 844 GKKDLAEVLQICLNKAEVGSQQWRNAQDSLLSLEKRNAGSVDTIVRGGMEANQALRARFS 903
G+ LAEVL C K +G+QQW+NA+DSL SL K N S D+IVR G EANQ+LR++ S
Sbjct: 848 GRSCLAEVLVECKAKTTMGAQQWKNAEDSLFSLGKGNVESADSIVRTGTEANQSLRSKLS 907
Query: 904 SSVSTTLEDAGIANTDINSSIDYSLQLDHEACGNLNSMITPCCGDLTELKGGHYNRIVEI 963
S+VSTTLE+ IAN + SSID SL+LDH+AC N+ S+I PC +++ELKGGHY+R+VEI
Sbjct: 908 SAVSTTLEEIDIANKALLSSIDSSLKLDHDACANIGSIIKPCHEEISELKGGHYHRVVEI 967
Query: 964 TENAGKCLLNEYMVDEPSCSTPTRRLFNLPSVSSIEELRTPSFEELLKAFWDAK-SQKLA 1022
TENAGKCL EY+VDEPSCSTP RR +LPS+ SIE+LRTP ++ELLK+F +++ S K A
Sbjct: 968 TENAGKCLEEEYLVDEPSCSTPRRRQIDLPSMESIEQLRTPDYDELLKSFRESRASLKQA 1027
Query: 1023 NGDVKHIGSYEET--QSVRDSRVPLTTIN 1049
NGD+KH +E S+ D R PL N
Sbjct: 1028 NGDMKHFLEVQEATPPSITDPRAPLIARN 1056
>gb|AAD24373.1| putative kinesin-like spindle protein [Arabidopsis thaliana]
gi|15226915|ref|NP_180430.1| kinesin motor
protein-related [Arabidopsis thaliana]
gi|25295722|pir||B84687 probable kinesin-like spindle
protein [imported] - Arabidopsis thaliana
Length = 1076
Score = 1358 bits (3514), Expect = 0.0
Identities = 708/1087 (65%), Positives = 859/1087 (78%), Gaps = 49/1087 (4%)
Query: 1 MEVQQKRGGLVPLSPSQTPRSTDKPARDLRSADSNSNSH--NKYDKEKGVNVQVLVRCRP 58
M+ + G SP QTPRST+K RD R DSNSNS+ +K +KEKGVN+QV+VRCRP
Sbjct: 1 MDSNNSKKGSSVKSPCQTPRSTEKSNRDFR-VDSNSNSNPVSKNEKEKGVNIQVIVRCRP 59
Query: 59 MNEDEMRLHTPVVISCNEGRREVAAVQSIANKQIDRTFVFDKVFGPNSQQKELYDQAVSP 118
N +E RL TP V++CN+ ++EVA Q+IA KQID+TF+FDKVFGP SQQK+LY QAVSP
Sbjct: 60 FNSEETRLQTPAVLTCNDRKKEVAVAQNIAGKQIDKTFLFDKVFGPTSQQKDLYHQAVSP 119
Query: 119 IVYEVLEGYNCTIFAYGQTGTGKTYTMEGGAIKKNGEFPTDAGVIPRAVKQIFDILEAQS 178
IV+EVL+GYNCTIFAYGQTGTGKTYTMEGGA KKNGE P+DAGVIPRAVKQIFDILEAQS
Sbjct: 120 IVFEVLDGYNCTIFAYGQTGTGKTYTMEGGARKKNGEIPSDAGVIPRAVKQIFDILEAQS 179
Query: 179 A-EYSMKVTFLELYNEEITDLLAPEETTKFVDEKSKKPIALMEDGKGGVLVRGLEEEIVC 237
A EYS+KV+FLELYNEE+TDLLAPEET KF D+KSKKP+ALMEDGKGGV VRGLEEEIV
Sbjct: 180 AAEYSLKVSFLELYNEELTDLLAPEET-KFADDKSKKPLALMEDGKGGVFVRGLEEEIVS 238
Query: 238 TANEIYKILEKGSAKRRTAETLLNKQSSRSHSIFSITIHIKECTPEGEEMIKCGKLNLVD 297
TA+EIYK+LEKGSAKRRTAETLLNKQSSRSHSIFS+TIHIKECTPEGEE++K GKLNLVD
Sbjct: 239 TADEIYKVLEKGSAKRRTAETLLNKQSSRSHSIFSVTIHIKECTPEGEEIVKSGKLNLVD 298
Query: 298 LAGSENISRSGAREGRAREAGEINKSLLTLGRTINALVEHSGHVPYRDSKLTRLLRDSLG 357
LAGSENISRSGAREGRAREAGEINKSLLTLGR INALVEHSGH+PYR+SKLTRLLRDSLG
Sbjct: 299 LAGSENISRSGAREGRAREAGEINKSLLTLGRVINALVEHSGHIPYRESKLTRLLRDSLG 358
Query: 358 GKTKTCIIATVSPSIHCLEETLSTLDYAHRAKNIKNKPEVNQKMMKSAMIKDLYSEIDRL 417
GKTKTC+IATVSPS+HCLEETLSTLDYAHRAK+IKNKPEVNQKMMKSA++KDLYSEI+RL
Sbjct: 359 GKTKTCVIATVSPSVHCLEETLSTLDYAHRAKHIKNKPEVNQKMMKSAIMKDLYSEIERL 418
Query: 418 KQEVYAAREKNGIYIPRDRYLNEEAEKKAMAEKIERMELDADSKDKNLVELQELYNSQQL 477
KQEVYAAREKNGIYIP++RY EEAEKKAMA+KIE+ME++ ++KDK +++LQELYNS+QL
Sbjct: 419 KQEVYAAREKNGIYIPKERYTQEEAEKKAMADKIEQMEVEGEAKDKQIIDLQELYNSEQL 478
Query: 478 LTAELSAKLEKTEKSLEETEQTLFDLEERHRQANATIKEKEFLISNLLKSEKELVERAIE 537
+TA L KL+KTEK L ETEQ L DLEE+HRQA ATIKEKE+LISNLLKSEK LV+RA+E
Sbjct: 479 VTAGLREKLDKTEKKLYETEQALLDLEEKHRQAVATIKEKEYLISNLLKSEKTLVDRAVE 538
Query: 538 LRAELENAASDVSNLFSKIERKDKIEEENRVLIQKFQSQLAQQLEALHRTVSASVMHQEQ 597
L+AEL NAASDVSNLF+KI RKDKIE+ NR LIQ FQSQL +QLE L+ +V+ SV QE+
Sbjct: 539 LQAELANAASDVSNLFAKIGRKDKIEDSNRSLIQDFQSQLLRQLELLNNSVAGSVSQQEK 598
Query: 598 QLKDMEKDMQSFVSTKSEATEDLRVRVVELKNMYGSGIKALDNLAEELKSNNQLTYEDLK 657
QL+DME M SFVS K++ATE LR + +LK Y +GIK+LD++A L ++Q T DL
Sbjct: 599 QLQDMENVMVSFVSAKTKATETLRGSLAQLKEKYNTGIKSLDDIAGNLDKDSQSTLNDLN 658
Query: 658 SEVAKHSSALEDLFKGIALEADSLLNDLQNSLHKQEANVTAFAHQQREAHSRAVETTRSV 717
SEV KHS ALED+FKG EA +LL LQ SLH QE ++AF QQR+ HSR++++ +SV
Sbjct: 659 SEVTKHSCALEDMFKGFTSEAYTLLEGLQGSLHNQEEKLSAFTQQQRDLHSRSMDSAKSV 718
Query: 718 SKITMKFFETIDRHASSLTQIVEETQFVNDQKLCELEKKFEECTAYEEKQLLEKVAEMLA 777
S + + FF+T+D HA+ LT++ E+ Q VN+QKL KKFEE A EEKQ+LEKVAE+LA
Sbjct: 719 STVMLDFFKTLDTHANKLTKLAEDAQNVNEQKLSAFTKKFEESIANEEKQMLEKVAELLA 778
Query: 778 SSNARKKKLVQMAVNDLRESANCRTSKLQREALTMQDSTSFVKAEWMVHMEKTESNYHED 837
SSNARKK+LVQ+AV D+R+ ++ +T LQ+E MQDS S +K +W H+ + ES++ ++
Sbjct: 779 SSNARKKELVQIAVQDIRQGSSSQTGALQQEMSAMQDSASSIKVQWNSHIVQAESHHLDN 838
Query: 838 TSSVESGKKDLAEVLQICLNKAEVGSQQWRNAQDSLLSLEKRNAGSVDTIVRGGMEANQA 897
S+VE K+D+ ++ CL ++ G+QQW+ AQ+SL+ LEKRN + D+I+RG +E N+
Sbjct: 839 ISAVEVAKEDMQKMHLKCLENSKTGTQQWKTAQESLVDLEKRNVATADSIIRGAIENNEK 898
Query: 898 LRARFSSSVSTTLEDAGIANTDINSSIDYSLQLDHEACGNLNSMITPCCGDLTELKGGHY 957
LR +FSS+VSTTL D +N +I SSID SLQLD +A ++NS I PC +L EL+ H
Sbjct: 899 LRTQFSSAVSTTLSDVDSSNREIISSIDNSLQLDKDASTDVNSTIVPCSENLKELRTHHD 958
Query: 958 NRIVEITENAGKCLLNEY----------------------------------MVDEPSCS 983
+ +VEI +N GKCL +EY VDE + S
Sbjct: 959 DNVVEIKQNTGKCLGHEYKVTRFDPFLYNHHIYMIELDKIVNRKLNSLKTSTQVDEATSS 1018
Query: 984 TPTRRLFNLPSVSSIEELRTPSFEELLKAFWDAKSQK-LANGDVKHIGSYEETQSVRDSR 1042
TP +R +N+P+V SIEEL+TPSFEELLKAF D KS K + NG+ KH V + R
Sbjct: 1019 TPRKREYNIPTVGSIEELKTPSFEELLKAFHDCKSPKQMQNGEAKH---------VSNGR 1069
Query: 1043 VPLTTIN 1049
PLT IN
Sbjct: 1070 PPLTAIN 1076
>sp|O23826|K125_TOBAC 125 kDa kinesin-related protein gi|7441387|pir||T02017
kinesin-related protein TKRP125 - common tobacco
gi|2582971|dbj|BAA23159.1| TKRP125 [Nicotiana tabacum]
Length = 1006
Score = 891 bits (2302), Expect = 0.0
Identities = 477/983 (48%), Positives = 667/983 (67%), Gaps = 10/983 (1%)
Query: 43 DKEKGVNVQVLVRCRPMNEDEMRLHTPVVISCNEGRREVAAVQSIANKQIDRTFVFDKVF 102
+KEKGVNVQVL+RCRP + DE+R + P V++CN+ +REVA Q+IA K IDR F FDKVF
Sbjct: 3 NKEKGVNVQVLLRCRPFSNDELRNNAPQVVTCNDYQREVAVSQNIAGKHIDRIFTFDKVF 62
Query: 103 GPNSQQKELYDQAVSPIVYEVLEGYNCTIFAYGQTGTGKTYTMEGGAIKK----NGEFPT 158
GP++QQ++LYDQA+ PIV EVLEG+NCTIFAYGQTGTGKTYTMEG + NGE P
Sbjct: 63 GPSAQQRDLYDQAIVPIVNEVLEGFNCTIFAYGQTGTGKTYTMEGECKRSKSGPNGELPQ 122
Query: 159 DAGVIPRAVKQIFDILEAQSAEYSMKVTFLELYNEEITDLLAPEETTKFVDEKSKKPIAL 218
+AGVIPRAVKQ+FD LE+Q+AEYS+KVTFLELYNEEITDLLAPE+ ++++ KK + L
Sbjct: 123 EAGVIPRAVKQVFDTLESQNAEYSVKVTFLELYNEEITDLLAPEDLKVALEDRQKKQLPL 182
Query: 219 MEDGKGGVLVRGLEEEIVCTANEIYKILEKGSAKRRTAETLLNKQSSRSHSIFSITIHIK 278
MEDGKGGVLVRGLEEEIV +ANEI+ +LE+GSAKRRTAETLLNKQSSRSHS+FSITIHIK
Sbjct: 183 MEDGKGGVLVRGLEEEIVTSANEIFTLLERGSAKRRTAETLLNKQSSRSHSLFSITIHIK 242
Query: 279 ECTPEGEEMIKCGKLNLVDLAGSENISRSGAREGRAREAGEINKSLLTLGRTINALVEHS 338
E TPEGEE+IKCGKLNLVDLAGSENISRSGAREGRAREAGEINKSLLTLGR INALVEH
Sbjct: 243 EATPEGEELIKCGKLNLVDLAGSENISRSGAREGRAREAGEINKSLLTLGRVINALVEHL 302
Query: 339 GHVPYRDSKLTRLLRDSLGGKTKTCIIATVSPSIHCLEETLSTLDYAHRAKNIKNKPEVN 398
GH+PYRDSKLTRLLRDSLGG+TKTCIIATVSP++HCLEETLSTLDYAHRAKNIKNKPEVN
Sbjct: 303 GHIPYRDSKLTRLLRDSLGGRTKTCIIATVSPAVHCLEETLSTLDYAHRAKNIKNKPEVN 362
Query: 399 QKMMKSAMIKDLYSEIDRLKQEVYAAREKNGIYIPRDRYLNEEAEKKAMAEKIERMELDA 458
QKMMKS +IKDLY EI+RLK EVYAAREKNG+YIP++RY EE E+KAMA++IE+M +
Sbjct: 363 QKMMKSTLIKDLYGEIERLKAEVYAAREKNGVYIPKERYYQEENERKAMADQIEQMGVSI 422
Query: 459 DSKDKNLVELQELYNSQQLLTAELSAKLEKTEKSLEETEQTLFDLEERHRQANATIKEKE 518
++ K ELQ ++SQ ++L+ KL+ T+K L +T + L EE+ RQ+ T+KE++
Sbjct: 423 ENHQKQFEELQSRHDSQVQQCSDLTCKLDVTQKQLNQTSKLLAYTEEQLRQSQYTLKERD 482
Query: 519 FLISNLLKSEKELVERAIELRAELENAASDVSNLFSKIERKDKIEEENRVLIQKFQSQLA 578
F+IS K+E L +A LRA+LE + + ++LF KI R+DK+ +NR L+ FQ++LA
Sbjct: 483 FIISEQKKAENALAHQACVLRADLEKSIQENASLFQKIAREDKLSTDNRSLVNNFQAELA 542
Query: 579 QQLEALHRTVSASVMHQEQQLKDMEKDMQSFVSTKSEATEDLRVRVVELKNMYGSGIKAL 638
+QL +L T++ SV Q + L+ +EK +F+ + +A DL+ ++ +Y S +A+
Sbjct: 543 KQLGSLSSTLATSVCRQTEHLQCVEKFCHNFLDSHDKAVLDLKRKINSSMALYISHFEAM 602
Query: 639 DNLAEELKSNNQLTYEDLKSEVAKHSSALEDLFKGIALEADSLLNDLQNSLHKQEANVTA 698
N+ K+ + T E++ + + +S + ++ A+EA+S+ ++LQ++L + +
Sbjct: 603 QNVVRLHKATSNATLEEVSTLASSNSISTKEFLDAEAVEANSMFDELQSTLSTHQGEMAH 662
Query: 699 FAHQQREAHSRAVETTRSVSKITMKFFETIDRHASSLTQIVEETQFVNDQKLCELEKKFE 758
FA + R+ + + E ++S I +FF+ + + L + + + E EK +E
Sbjct: 663 FARELRQRFNDSTEHLTNISAIIQRFFDKLLDESKRLEKHATTVDEIQTNSIAEFEKAYE 722
Query: 759 ECTAYEEKQLLEKVAEMLASSNARKKKLVQMAVNDLRESANCRTSKLQREALTMQDSTSF 818
E + + ++L+ V ++++ R+K+LV + DLRE+ + + L +M+ T+
Sbjct: 723 EQSKSDAEKLIADVTSLVSNHMRRQKELVGARLVDLRETVSGNRTFLDGHVSSMEGITTD 782
Query: 819 VKAEWMVHMEKTESNYHEDTSSVESGKKDLAEVLQICLNKAEVGSQQWRNAQDSLLSLEK 878
K +W + E E+ + + ++Q C++ AE ++W++ + + +
Sbjct: 783 AKRKWQDFYMQAEGETKENADFSAAKHCRMESLMQKCVSTAETALKRWQSTHELVNDMGN 842
Query: 879 RNAGSVDTIVRGGMEANQALRARFSSSVSTTLEDAGIANTDINSSIDYSLQLDHEACGNL 938
++ ++ ++VR + N+ F S+ + ED + DI SID L E G++
Sbjct: 843 QHVLTMHSVVRNICDNNEQHVTDFDSTRESAEEDVKRNSEDIIKSID---SLSGEERGSI 899
Query: 939 NSMITPCCG---DLTELKGGHYNRIVEITENAGKCLLNEYMVDEPSCSTPTRRLFNLPSV 995
+ ++ L LK H + I + A + +YM EP+ +TP R ++PS
Sbjct: 900 SGVLDTTSAHSETLDVLKKDHCMQSTSIEQIALETFQQKYMDYEPTGATPIRSEPDVPSK 959
Query: 996 SSIEELRTPSFEELLKAFWDAKS 1018
+IE LR E LL+ F + S
Sbjct: 960 VTIESLRAMPMEVLLEEFRENNS 982
>ref|NP_850281.1| kinesin motor protein-related [Arabidopsis thaliana]
Length = 1039
Score = 875 bits (2261), Expect = 0.0
Identities = 456/1002 (45%), Positives = 671/1002 (66%), Gaps = 5/1002 (0%)
Query: 2 EVQQKRGGLVPLSPSQTPRSTDKPARDLRSADSNSNSHNKYDKEKGVNVQVLVRCRPMNE 61
EV ++ G V + PS P T P + R DS SN ++ +KE VNVQV++RC+P++E
Sbjct: 6 EVVSRKSG-VGVIPSPAPFLT--PRLERRRPDSFSNRLDRDNKE--VNVQVILRCKPLSE 60
Query: 62 DEMRLHTPVVISCNEGRREVAAVQSIANKQIDRTFVFDKVFGPNSQQKELYDQAVSPIVY 121
+E + P VISCNE RREV + +IANKQ+DR F FDKVFGP SQQ+ +YDQA++PIV+
Sbjct: 61 EEQKSSVPRVISCNEMRREVNVLHTIANKQVDRLFNFDKVFGPKSQQRSIYDQAIAPIVH 120
Query: 122 EVLEGYNCTIFAYGQTGTGKTYTMEGGAIKKNGEFPTDAGVIPRAVKQIFDILEAQSAEY 181
EVLEG++CT+FAYGQTGTGKTYTMEGG KK G+ P +AGVIPRAV+ IFD LEAQ+A+Y
Sbjct: 121 EVLEGFSCTVFAYGQTGTGKTYTMEGGMRKKGGDLPAEAGVIPRAVRHIFDTLEAQNADY 180
Query: 182 SMKVTFLELYNEEITDLLAPEETTKFVDEKSKKPIALMEDGKGGVLVRGLEEEIVCTANE 241
SMKVTFLELYNEE+TDLLA +++++ ++K +KPI+LMEDGKG V++RGLEEE+V +AN+
Sbjct: 181 SMKVTFLELYNEEVTDLLAQDDSSRSSEDKQRKPISLMEDGKGSVVLRGLEEEVVYSAND 240
Query: 242 IYKILEKGSAKRRTAETLLNKQSSRSHSIFSITIHIKECTPEGEEMIKCGKLNLVDLAGS 301
IY +LE+GS+KRRTA+TLLNK+SSRSHS+F+IT+HIKE + EE+IKCGKLNLVDLAGS
Sbjct: 241 IYALLERGSSKRRTADTLLNKRSSRSHSVFTITVHIKEESMGDEELIKCGKLNLVDLAGS 300
Query: 302 ENISRSGAREGRAREAGEINKSLLTLGRTINALVEHSGHVPYRDSKLTRLLRDSLGGKTK 361
ENI RSGAR+GRAREAGEINKSLLTLGR INALVEHS HVPYRDSKLTRLLRDSLGGKTK
Sbjct: 301 ENILRSGARDGRAREAGEINKSLLTLGRVINALVEHSSHVPYRDSKLTRLLRDSLGGKTK 360
Query: 362 TCIIATVSPSIHCLEETLSTLDYAHRAKNIKNKPEVNQKMMKSAMIKDLYSEIDRLKQEV 421
TCIIAT+SPS H LEETLSTLDYA+RAKNIKNKPE NQK+ K+ ++KDLY E++R+K++V
Sbjct: 361 TCIIATISPSAHSLEETLSTLDYAYRAKNIKNKPEANQKLSKAVLLKDLYLELERMKEDV 420
Query: 422 YAAREKNGIYIPRDRYLNEEAEKKAMAEKIERMELDADSKDKNLVELQELYNSQQLLTAE 481
AAR+KNG+YI +RY EE EKKA E+IE++E + + + + + +LY +++ +
Sbjct: 421 RAARDKNGVYIAHERYTQEEVEKKARIERIEQLENELNLSESEVSKFCDLYETEKEKLLD 480
Query: 482 LSAKLEKTEKSLEETEQTLFDLEERHRQANATIKEKEFLISNLLKSEKELVERAIELRAE 541
+ + L+ +++L + + L DL+E + Q + +KEKE ++S + SE L++RA LR +
Sbjct: 481 VESDLKDCKRNLHNSNKDLLDLKENYIQVVSKLKEKEVIVSRMKASETSLIDRAKGLRCD 540
Query: 542 LENAASDVSNLFSKIERKDKIEEENRVLIQKFQSQLAQQLEALHRTVSASVMHQEQQLKD 601
L++A++D+++LF+++++KDK+E +N+ ++ KF SQL Q L+ LHRTV SV Q+QQL+
Sbjct: 541 LQHASNDINSLFTRLDQKDKLESDNQSMLLKFGSQLDQNLKDLHRTVLGSVSQQQQQLRT 600
Query: 602 MEKDMQSFVSTKSEATEDLRVRVVELKNMYGSGIKALDNLAEELKSNNQLTYEDLKSEVA 661
ME+ SF++ K +AT DL R+ + + Y SGI AL L+E L+ E + +
Sbjct: 601 MEEHTHSFLAHKYDATRDLESRIGKTSDTYTSGIAALKELSEMLQKKASSDLEKKNTSIV 660
Query: 662 KHSSALEDLFKGIALEADSLLNDLQNSLHKQEANVTAFAHQQREAHSRAVETTRSVSKIT 721
A+E A EA ++ D+ N L+ Q+ + A QQ + R++ + + +S T
Sbjct: 661 SQIEAVEKFLTTSATEASAVAQDIHNLLNDQKKLLALAARQQEQGLVRSMRSAQEISNST 720
Query: 722 MKFFETIDRHASSLTQIVEETQFVNDQKLCELEKKFEECTAYEEKQLLEKVAEMLASSNA 781
F I A + + + +Q ++L E KF+E EEKQ L ++ +L+ +
Sbjct: 721 STIFSNIYNQAHDVVEAIRASQAEKSRQLDAFEMKFKEEAEREEKQALNDISLILSKLTS 780
Query: 782 RKKKLVQMAVNDLRESANCRTSKLQREALTMQDSTSFVKAEWMVHMEKTESNYHEDTSSV 841
+K ++ A +++RE +L + MQ + K E +++K ++++ E+T +
Sbjct: 781 KKTAMISDASSNIREHDIQEEKRLYEQMSGMQQVSIGAKEELCDYLKKEKTHFTENTIAS 840
Query: 842 ESGKKDLAEVLQICLNKAEVGSQQWRNAQDSLLSLEKRNAGSVDTIVRGGMEANQALRAR 901
+ L+ CL +A W + + +L + ++ + + N+ ++
Sbjct: 841 AESITVMDSYLEDCLGRANDSKTLWETTETGIKNLNTKYQQELNVTMEDMAKENEKVQDE 900
Query: 902 FSSSVSTTLEDAGIANTDINSSIDYSLQLDHEACGNLNSMITPCCGDLTELKGGHYNRIV 961
F+S+ S+ + +++++++ SL D E +++ C +T L+ H +
Sbjct: 901 FTSTFSSMDANFVSRTNELHAAVNDSLMQDRENKETTEAIVETCMNQVTLLQENHGQAVS 960
Query: 962 EITENAGKCLLNEYMVDEPSCSTPTRRLFNLPSVSSIEELRT 1003
I A + L+ +Y VD+ TP ++ N+PS+ SIEE+RT
Sbjct: 961 NIRNKAEQSLIKDYQVDQHKNETPKKQSINVPSLDSIEEMRT 1002
>gb|AAK91815.1| kinesin heavy chain [Zea mays]
Length = 992
Score = 860 bits (2223), Expect = 0.0
Identities = 466/952 (48%), Positives = 631/952 (65%), Gaps = 7/952 (0%)
Query: 12 PLSPSQTPRSTDKPARDLRSADSNSNSHNKYDKEKGVNVQVLVRCRPMNEDEMRLHTPVV 71
PL +PAR + +++DKEK VNVQVL+RCRP ++DE+R + P V
Sbjct: 15 PLDADILKSKPPQPARPALLSAVACAMSSRHDKEKSVNVQVLLRCRPFSDDELRNNAPQV 74
Query: 72 ISCNEGRREVAAVQSIANKQIDRTFVFDKVFGPNSQQKELYDQAVSPIVYEVLEGYNCTI 131
I+CN+ +REVA Q+IA KQ DR + FDKVFGP ++QKELYDQA+ PIV EVLEG+NCTI
Sbjct: 75 ITCNDYQREVAVTQNIAGKQFDRVYAFDKVFGPTAKQKELYDQAIIPIVNEVLEGFNCTI 134
Query: 132 FAYGQTGTGKTYTMEGGAIKKN----GEFPTDAGVIPRAVKQIFDILEAQSAEYSMKVTF 187
FAYGQTGTGKTYTMEG + G+ P DAGVIPRAVKQIFD LE Q+ EYS+K+TF
Sbjct: 135 FAYGQTGTGKTYTMEGECRRAKSGPKGQLPADAGVIPRAVKQIFDTLERQNTEYSVKITF 194
Query: 188 LELYNEEITDLLAPEETTKFV-DEKSKK--PIALMEDGKGGVLVRGLEEEIVCTANEIYK 244
LELYNEEITDLLAPEE +K ++K KK P+ LMEDGKGGVLVRGLEEEIV A+EI+
Sbjct: 195 LELYNEEITDLLAPEEISKAAFEDKQKKALPLNLMEDGKGGVLVRGLEEEIVTNASEIFS 254
Query: 245 ILEKGSAKRRTAETLLNKQSSRSHSIFSITIHIKECTPEGEEMIKCGKLNLVDLAGSENI 304
+LE+GSAKRRTAETLLNKQSSRSHS+FSITIHIKE TPEGEE+IKCGKLNLVDLAGSENI
Sbjct: 255 LLERGSAKRRTAETLLNKQSSRSHSLFSITIHIKEATPEGEELIKCGKLNLVDLAGSENI 314
Query: 305 SRSGAREGRAREAGEINKSLLTLGRTINALVEHSGHVPYRDSKLTRLLRDSLGGKTKTCI 364
SRSGA+EGRAREAGEINKSLLTLGR I ALVEH GHVPYRDSKLTRLLRDSLGG+TKTCI
Sbjct: 315 SRSGAKEGRAREAGEINKSLLTLGRVITALVEHLGHVPYRDSKLTRLLRDSLGGRTKTCI 374
Query: 365 IATVSPSIHCLEETLSTLDYAHRAKNIKNKPEVNQKMMKSAMIKDLYSEIDRLKQEVYAA 424
IATVSPS+HCLEETLSTLDYAHRAK+IKN+PEVNQKMMKS +IKDLY EIDRLK EVYAA
Sbjct: 375 IATVSPSVHCLEETLSTLDYAHRAKSIKNRPEVNQKMMKSTLIKDLYWEIDRLKAEVYAA 434
Query: 425 REKNGIYIPRDRYLNEEAEKKAMAEKIERMELDADSKDKNLVELQELYNSQQLLTAELSA 484
REK G+YIP+DRY EE E+KAMA++IE+M ++ K + +LQE Y+S+ +A+LS
Sbjct: 435 REKVGVYIPKDRYQQEENERKAMADQIEQMNASLEANQKLISDLQEKYDSELRHSADLSK 494
Query: 485 KLEKTEKSLEETEQTLFDLEERHRQANATIKEKEFLISNLLKSEKELVERAIELRAELEN 544
KLE TEK ++ T L +E +QA +KEK+++IS K+E L +A LR++L+N
Sbjct: 495 KLEVTEKCMDHTSNLLSATKEDLKQAQYNLKEKDYIISEQKKAENALTHQACVLRSDLKN 554
Query: 545 AASDVSNLFSKIERKDKIEEENRVLIQKFQSQLAQQLEALHRTVSASVMHQEQQLKDMEK 604
+ + +L+SKI R DK+ NR ++ FQ+ LA +L+ L T++AS+ Q LK +E
Sbjct: 555 ILAIILSLYSKIARGDKLSATNRSVVNTFQTDLASKLDTLSNTLNASIDQQNMHLKAVED 614
Query: 605 DMQSFVSTKSEATEDLRVRVVELKNMYGSGIKALDNLAEELKSNNQLTYEDLKSEVAKHS 664
QS V + AT +L+ +++ K++Y S ++A N+ K++ T ED+ S A
Sbjct: 615 LCQSCVDSHDRATSELKKKILASKSLYMSHMEAFQNVVLLHKASANATLEDISSLSATSC 674
Query: 665 SALEDLFKGIALEADSLLNDLQNSLHKQEANVTAFAHQQREAHSRAVETTRSVSKITMKF 724
+L+ L + EA ++ ND+ L + +T F Q RE+ +++ T+ +S +
Sbjct: 675 CSLDQLLVCVEGEAQNIFNDIHKLLTTHRSEMTHFTQQLRESFQISLDRTKEMSTYIIGL 734
Query: 725 FETIDRHASSLTQIVEETQFVNDQKLCELEKKFEECTAYEEKQLLEKVAEMLASSNARKK 784
F+ S L T + + + + +EE + EE++LL ++ +++ R++
Sbjct: 735 FDKYVEETSKLQSHSNNTHEAQMKSIEDFQMVYEEQSKSEEQKLLADISSLVSKHITRQR 794
Query: 785 KLVQMAVNDLRESANCRTSKLQREALTMQDSTSFVKAEWMVHMEKTESNYHEDTSSVESG 844
+LV + ++ L ++A + L M+ T K +W E+ E++ +SS
Sbjct: 795 ELVGVRLSSLGDAARGNKAFLDEHTSAMEFVTKDAKRKWETFAEQAENDCKAGSSSSAVK 854
Query: 845 KKDLAEVLQICLNKAEVGSQQWRNAQDSLLSLEKRNAGSVDTIVRGGMEANQALRARFSS 904
+ +LQ C + QQW+ + ++ L ++ V+ +VR +E N+ +S
Sbjct: 855 HCRMETMLQECACTVDSAVQQWKKSHAAVNDLSRKQVAEVEALVRTAIENNEQHEVEVAS 914
Query: 905 SVSTTLEDAGIANTDINSSIDYSLQLDHEACGNLNSMITPCCGDLTELKGGH 956
S + ED ++ DI ++ L+ ++ SM+ +L +L+ H
Sbjct: 915 SRAVAEEDTTNSSKDIAQGVENLLEKAQKSSSRGVSMVEAHFAELQKLQESH 966
>gb|AAD21445.1| putative kinesin-related cytokinesis protein [Arabidopsis thaliana]
gi|15227596|ref|NP_181162.1| kinesin motor
protein-related [Arabidopsis thaliana]
gi|25295721|pir||H84777 probable kinesin-related
cytokinesis protein [imported] - Arabidopsis thaliana
gi|11132972|sp|P82266|K125_ARATH Probable 125 kDa
kinesin-related protein
Length = 1056
Score = 838 bits (2165), Expect = 0.0
Identities = 453/986 (45%), Positives = 645/986 (64%), Gaps = 9/986 (0%)
Query: 40 NKYDKEKGVNVQVLVRCRPMNEDEMRLHTPVVISCNEGRREVAAVQSIANKQIDRTFVFD 99
+++DKEKGVNVQVL+RCRP ++DE+R + P V++CN+ +REVA Q+IA K IDR F FD
Sbjct: 3 SRHDKEKGVNVQVLLRCRPFSDDELRSNAPQVLTCNDLQREVAVSQNIAGKHIDRVFTFD 62
Query: 100 KVFGPNSQQKELYDQAVSPIVYEVLEGYNCTIFAYGQTGTGKTYTMEGGAIKKN----GE 155
KVFGP++QQK+LYDQAV PIV EVLEG+NCTIFAYGQTGTGKTYTMEG + G
Sbjct: 63 KVFGPSAQQKDLYDQAVVPIVNEVLEGFNCTIFAYGQTGTGKTYTMEGECRRSKSAPCGG 122
Query: 156 FPTDAGVIPRAVKQIFDILEAQSAEYSMKVTFLELYNEEITDLLAPEETTKFV-DEKSKK 214
P +AGVIPRAVKQIFD LE Q AEYS+KVTFLELYNEEITDLLAPE+ ++ +EK KK
Sbjct: 123 LPAEAGVIPRAVKQIFDTLEGQQAEYSVKVTFLELYNEEITDLLAPEDLSRVAAEEKQKK 182
Query: 215 PIALMEDGKGGVLVRGLEEEIVCTANEIYKILEKGSAKRRTAETLLNKQSSRSHSIFSIT 274
P+ LMEDGKGGVLVRGLEEEIV +ANEI+ +LE+GS+KRRTAET LNKQSSRSHS+FSIT
Sbjct: 183 PLPLMEDGKGGVLVRGLEEEIVTSANEIFTLLERGSSKRRTAETFLNKQSSRSHSLFSIT 242
Query: 275 IHIKECTPEGEEMIKCGKLNLVDLAGSENISRSGAREGRAREAGEINKSLLTLGRTINAL 334
IHIKE TPEGEE+IKCGKLNLVDLAGSENISRSGAR+GRAREAGEINKSLLTLGR I+AL
Sbjct: 243 IHIKEATPEGEELIKCGKLNLVDLAGSENISRSGARDGRAREAGEINKSLLTLGRVISAL 302
Query: 335 VEHSGHVPYRDSKLTRLLRDSLGGKTKTCIIATVSPSIHCLEETLSTLDYAHRAKNIKNK 394
VEH GHVPYRDSKLTRLLRDSLGG+TKTCIIATVSP++HCLEETLSTLDYAHRAKNI+NK
Sbjct: 303 VEHLGHVPYRDSKLTRLLRDSLGGRTKTCIIATVSPAVHCLEETLSTLDYAHRAKNIRNK 362
Query: 395 PEVNQKMMKSAMIKDLYSEIDRLKQEVYAAREKNGIYIPRDRYLNEEAEKKAMAEKIERM 454
PEVNQKMMKS +IKDLY EI+RLK EVYA+REKNG+Y+P++RY EE+E+K MAE+IE+M
Sbjct: 363 PEVNQKMMKSTLIKDLYGEIERLKAEVYASREKNGVYMPKERYYQEESERKVMAEQIEQM 422
Query: 455 ELDADSKDKNLVELQELYNSQQLLTAELSAKLEKTEKSLEETEQTLFDLEERHRQANATI 514
++ K L ELQ+ Y Q ++L+ KL+ TEK+L +T + L E +++ +
Sbjct: 423 GGQIENYQKQLEELQDKYVGQVRECSDLTTKLDITEKNLSQTCKVLASTNEELKKSQYAM 482
Query: 515 KEKEFLISNLLKSEKELVERAIELRAELENAASDVSNLFSKIERKDKIEEENRVLIQKFQ 574
KEK+F+IS KSE LV++A L++ LE A D S+L KI R+DK+ +NR ++ +Q
Sbjct: 483 KEKDFIISEQKKSENVLVQQACILQSNLEKATKDNSSLHQKIGREDKLSADNRKVVDNYQ 542
Query: 575 SQLAQQLEALHRTVSASVMHQEQQLKDMEKDMQSFVSTKSEATEDLRVRVVELKNMYGSG 634
+L++Q+ L V++ + Q L+ + K QS + ++A +++ +V +++Y S
Sbjct: 543 VELSEQISNLFNRVASCLSQQNVHLQGVNKLSQSRLEAHNKAILEMKKKVKASRDLYSSH 602
Query: 635 IKALDNLAEELKSNNQLTYEDLKSEVAKHSSALEDLFKGIALEADSLLNDLQNSLHKQEA 694
++A+ N+ K+N E++ + + ++++ SL ++LQ++L +
Sbjct: 603 LEAVQNVVRLHKANANACLEEVSALTTSSACSIDEFLASGDETTSSLFDELQSALSSHQG 662
Query: 695 NVTAFAHQQREAHSRAVETTRSVSKITMKFFETIDRHASSLTQIVEETQFVNDQKLCELE 754
+ FA + R+ +E T+ +S+ T FF+ + + + E + + +
Sbjct: 663 EMALFARELRQRFHTTMEQTQEMSEYTSTFFQKLMEESKNAETRAAEANDSQINSIIDFQ 722
Query: 755 KKFEECTAYEEKQLLEKVAEMLASSNARKKKLVQMAVNDLRESANCRTSKLQREALTMQD 814
K +E + + +L+ + +++S R+ +LV +++ +++ + + L + +
Sbjct: 723 KTYEAQSKSDTDKLIADLTNLVSSHIRRQHELVDSRLHNFKDAVSSNKTFLDEHVSAVNN 782
Query: 815 STSFVKAEWMVHMEKTESNYHE--DTSSVESGKKDLAEVLQICLNKAEVGSQQWRNAQDS 872
T K +W + E+ E D S+ + + +L +LQ + AE + + +S
Sbjct: 783 LTKDAKRKWETFSMQAENEAREGADFSAAKHCRMEL--LLQQSVGHAESAFKHCKITHES 840
Query: 873 LLSLEKRNAGSVDTIVRGGMEANQALRARFSSSVSTTLEDAGIANTDINSSIDYSLQLDH 932
L + + V ++VR ++N+ A S+ + +D + DI I+ + +
Sbjct: 841 LKEMTSKQVTDVSSLVRSACDSNEQHDAEVDSARTAAEKDVTKNSDDIIQQIERMSEDEK 900
Query: 933 EACGNLNSMITPCCGDLTELKGGHYNRIVEITENAGKCLLNEYMVDEPSCSTPTRRLFNL 992
+ + + L + + I + A + +YM EP+ +TPT+ +
Sbjct: 901 ASVSKILENVRSHEKTLESFQQDQCCQARCIEDKAQETFQQQYMEYEPTGATPTKNEPEI 960
Query: 993 PSVSSIEELRTPSFEELLKAFWDAKS 1018
P+ ++IE LR E L++ F + S
Sbjct: 961 PTKATIESLRAMPIETLVEEFRENNS 986
>gb|AAC98061.1| putative kinesin heavy chain [Arabidopsis thaliana]
gi|25408550|pir||E84792 probable kinesin heavy chain
[imported] - Arabidopsis thaliana
Length = 1022
Score = 834 bits (2155), Expect = 0.0
Identities = 451/1015 (44%), Positives = 657/1015 (64%), Gaps = 48/1015 (4%)
Query: 2 EVQQKRGGLVPLSPSQTPRSTDKPARDLRSADSNSNSHNKYDKEKGVNVQVLVRCRPMNE 61
EV ++ G V + PS P T P + R DS SN ++ +KE VNVQV++RC+P++E
Sbjct: 6 EVVSRKSG-VGVIPSPAPFLT--PRLERRRPDSFSNRLDRDNKE--VNVQVILRCKPLSE 60
Query: 62 DEMRLHTPVVISCNEGRREVAAVQSIANKQIDRTFVFDKVFGPNSQQKELYDQAVSPIVY 121
+E + P VISCNE RREV + +IANKQ+DR F FDKVFGP SQQ+ +YDQA++PIV+
Sbjct: 61 EEQKSSVPRVISCNEMRREVNVLHTIANKQVDRLFNFDKVFGPKSQQRSIYDQAIAPIVH 120
Query: 122 EVLEGYNCTIFAYGQTGTGKTYTMEGGAIKKNGEFPTDAGVIPRAVKQIFDILEAQSAEY 181
EVLEG++CT+FAYGQTGTGKTYTMEGG KK G+ P +AGVIPRAV+ IFD LEAQ+A+Y
Sbjct: 121 EVLEGFSCTVFAYGQTGTGKTYTMEGGMRKKGGDLPAEAGVIPRAVRHIFDTLEAQNADY 180
Query: 182 SMKVTFLELYNEEITDLLAPEETTKFVDEKSKKPIALMEDGKGGVLVRGLEEEIVCTANE 241
SMKVTFLELYNEE+TDLLA +++++ ++K +KPI+LMEDGKG V++RGLEEE+V +AN+
Sbjct: 181 SMKVTFLELYNEEVTDLLAQDDSSRSSEDKQRKPISLMEDGKGSVVLRGLEEEVVYSAND 240
Query: 242 IYKILEKGSAKRRTAETLLNKQSSRSHSIFSITIHIKECTPEGEEMIKCGKLNLVDLAGS 301
IY +LE+GS+KRRTA+TLLNK+SSRSHS+F+IT+HIKE + EE+IKCGKLNLVDLAGS
Sbjct: 241 IYALLERGSSKRRTADTLLNKRSSRSHSVFTITVHIKEESMGDEELIKCGKLNLVDLAGS 300
Query: 302 ENISRSGAREGRAREAGEINKSLLTLGRTINALVEHSGHVPYRDSKLTRLLRDSLGGKTK 361
ENI RSGAR+GRAREAGEINKSLLTLGR INALVEHS HVPYRDSKLTRLLRDSLGGKTK
Sbjct: 301 ENILRSGARDGRAREAGEINKSLLTLGRVINALVEHSSHVPYRDSKLTRLLRDSLGGKTK 360
Query: 362 TCIIATVSPSIHCLEETLSTLDYAHRAKNIKNKPEVNQKMMKSAMIKDLYSEIDRLKQEV 421
TCIIAT+SPS H LEETLSTLDYA+RAKNIKNKPE NQK+ K+ ++KDLY E++R+K++V
Sbjct: 361 TCIIATISPSAHSLEETLSTLDYAYRAKNIKNKPEANQKLSKAVLLKDLYLELERMKEDV 420
Query: 422 YAAREKNGIYIPRDRYLNEEAEKKAMAEKIERMELDADSKDKNLVELQELYNSQQLLTAE 481
AAR+KNG+YI +RY EE EKKA E+IE++E + + + N ++L + +L
Sbjct: 421 RAARDKNGVYIAHERYTQEEVEKKARIERIEQLENELNLSESN---FRDLVSRLFILLVR 477
Query: 482 LSAKLE--KTEKSLEETEQTLFDLEERHRQANATIKEKEFLISNLLKSEKELVERAIELR 539
+ K + +++L + + L DL+E + Q + +KEKE ++S + SE L++RA LR
Sbjct: 478 VFLKFQTFMIQRNLHNSNKDLLDLKENYIQVVSKLKEKEVIVSRMKASETSLIDRAKGLR 537
Query: 540 AELENAASDVSNLFSKIERKDKIEEENRVLIQKFQSQLAQQLEALHRTVSASVMHQEQQL 599
+L++A++D+++LF+++++KDK+E +N+ ++ KF SQL Q L+ LHRTV SV Q+QQL
Sbjct: 538 CDLQHASNDINSLFTRLDQKDKLESDNQSMLLKFGSQLDQNLKDLHRTVLGSVSQQQQQL 597
Query: 600 KDMEKDMQSFVSTK-----------SEATEDLRVRVVELKNMYGSGIKALDNLAEELKSN 648
+ ME+ SF++ K ++AT DL R+ + + Y SGI AL L+E L+
Sbjct: 598 RTMEEHTHSFLAHKYDLITLVVDLLTQATRDLESRIGKTSDTYTSGIAALKELSEMLQKK 657
Query: 649 NQLTYEDLKSEVAKHSSALEDLFKGIALEADSLLNDLQNSLHKQEANVTAFAHQQREAHS 708
E + + A+E A EA ++ D+ N L+ Q+ + A QQ +
Sbjct: 658 ASSDLEKKNTSIVSQIEAVEKFLTTSATEASAVAQDIHNLLNDQKKLLALAARQQEQGLV 717
Query: 709 RAVETTRSVSKITMKFFETIDRHASSLTQIVEETQFVNDQKLCELEKKFEECTAYEEKQL 768
R++ + + +S T F I +E EEKQ
Sbjct: 718 RSMRSAQEISNSTSTIFSNIYN---------------------------QEEAEREEKQA 750
Query: 769 LEKVAEMLASSNARKKKLVQMAVNDLRESANCRTSKLQREALTMQDSTSFVKAEWMVHME 828
L ++ +L+ ++K ++ A +++RE +L + MQ + K E +++
Sbjct: 751 LNDISLILSKLTSKKTAMISDASSNIREHDIQEEKRLYEQMSGMQQVSIGAKEELCDYLK 810
Query: 829 KTESNYHEDTSSVESGKKDLAEVLQICLNKAEVGSQQWRNAQDSLLSLEKRNAGSVDTIV 888
K ++++ E+T + + L+ CL +A W + + +L + ++ +
Sbjct: 811 KEKTHFTENTIASAESITVMDSYLEDCLGRANDSKTLWETTETGIKNLNTKYQQELNVTM 870
Query: 889 RGGMEANQALRARFSSSVSTTLEDAGIANTDINSSIDYSLQLDHEACGNLNSMITPCCGD 948
+ N+ ++ F+S+ S+ + +++++++ SL D E +++ C
Sbjct: 871 EDMAKENEKVQDEFTSTFSSMDANFVSRTNELHAAVNDSLMQDRENKETTEAIVETCMNQ 930
Query: 949 LTELKGGHYNRIVEITENAGKCLLNEYMVDEPSCSTPTRRLFNLPSVSSIEELRT 1003
+T L+ H + I A + L+ +Y VD+ TP ++ N+PS+ SIEE+RT
Sbjct: 931 VTLLQENHGQAVSNIRNKAEQSLIKDYQVDQHKNETPKKQSINVPSLDSIEEMRT 985
>gb|AAK91818.1| kinesin heavy chain [Zea mays]
Length = 1079
Score = 799 bits (2064), Expect = 0.0
Identities = 464/1070 (43%), Positives = 647/1070 (60%), Gaps = 40/1070 (3%)
Query: 12 PLSPSQTPRSTDKPARDLRSADSNSNSHNKYDKEKGVNVQVLVRCRPMNEDEMRLHTPVV 71
PL +PAR + +++DKEK VNVQVL+RCRP ++DE+R + P V
Sbjct: 18 PLDADILKSKPPQPARPALLSAVACAMSSRHDKEKSVNVQVLLRCRPFSDDELRNNAPQV 77
Query: 72 ISCNEGRREVAAVQSIANKQIDRTFVFDKVFGPNSQQKELYDQAVSPIVYEVLEGYNCTI 131
I+CN+ +REVA Q+IA KQ DR + FDKVFGP ++QKELYDQA+ PIV EVLEG+NCTI
Sbjct: 78 ITCNDYQREVAVTQNIAGKQFDRVYAFDKVFGPTAKQKELYDQAIIPIVNEVLEGFNCTI 137
Query: 132 FAYGQTGTGKTYTMEGGAIKKN----GEFPTDAGVIPRAVKQIFDILEAQSAEYSMKVTF 187
FAYGQTGTGKTYTMEG + G+ P DAGVIPRAVKQIFD LE Q+ EYS+K+TF
Sbjct: 138 FAYGQTGTGKTYTMEGECRRAKSGPKGQLPADAGVIPRAVKQIFDTLERQNTEYSVKITF 197
Query: 188 LELYNEEITDLLAPEETTKFV-DEKSKK--PIALMEDGKGGVLVRGLEEEIVCTANEIYK 244
LELYNEEITDLLAPEE +K ++K KK P+ LMEDGKGGVLVRGLEEEIV A+EI+
Sbjct: 198 LELYNEEITDLLAPEEISKAAFEDKQKKALPLNLMEDGKGGVLVRGLEEEIVTNASEIFS 257
Query: 245 ILEKGSAKRRTAETLLNKQS-SRSHSI---------------FSITI---------HIKE 279
+LE+GSAKRR LNKQS S S S+ F+I +IKE
Sbjct: 258 LLERGSAKRRLQRHYLNKQSRSISASLLLNYTLQQTYVLITTFNILAGPTLYFNHYYIKE 317
Query: 280 CTPEGEEMIKCGKLNLVDLAGSENISRSGAREGRAREAGEINKSLLTLGRTINALVEHSG 339
TPEGEE+IKCGKLNLVDLAGSENISRSGA+EGRAREAGEINKSLLTLGR I ALVEH G
Sbjct: 318 ATPEGEELIKCGKLNLVDLAGSENISRSGAKEGRAREAGEINKSLLTLGRVITALVEHLG 377
Query: 340 HVPYRDSKLTRLLRDSLGGKTKTCIIATVSPSIHCLEETLSTLDYAHRAKNIKNKPEVNQ 399
HVPYRDSKLTRLLRDSLGG+TKTCIIATVSPS+HCLEETLSTLDYAHRAK+IKN+PEVNQ
Sbjct: 378 HVPYRDSKLTRLLRDSLGGRTKTCIIATVSPSVHCLEETLSTLDYAHRAKSIKNRPEVNQ 437
Query: 400 KMMKSAMIKDLYSEIDRLKQEVYAAREKNGIYIPRDRYLNEEAEKKAMAEKIERMELDAD 459
KMMKS +IKDLY EI L+Q +K G P+ + ++ A++IE+M +
Sbjct: 438 KMMKSTLIKDLYGEIADLRQRYTLLEKKLGCTFPKTGISKRKMSERRWADQIEQMNASLE 497
Query: 460 SKDKNLVELQELYNSQQLLTAELSAKLEKTEKSLEETEQTLFDLEERHRQANATIKEKEF 519
K + +LQ+ Y+S+ +A+LS KLE TEK L+ T L +E +QA + EK++
Sbjct: 498 VNHKLISDLQQNYDSELQHSADLSKKLEVTEKCLDHTSNLLSTTKEDLKQAQYNLNEKDY 557
Query: 520 LISNLLKSEKELVERAIELRAELENAASDVSNLFSKIERKDKIEEENRVLIQKFQSQLAQ 579
+IS K+E L + LR++LE D ++L+SKI R DK+ NR ++ FQ+ LA
Sbjct: 558 IISEQKKAENALTHQTYVLRSDLEQYTRDNTSLYSKIARGDKLSATNRSVVNTFQTDLAS 617
Query: 580 QLEALHRTVSASVMHQEQQLKDMEKDMQSFVSTKSEATEDLRVRVVELKNMYGSGIKALD 639
+L+ L T++AS+ Q LK +E QS+V + +AT +L+ +++ K++Y S ++A
Sbjct: 618 KLDILSNTLNASIDQQNMHLKSVEDLCQSYVDSHDKATSELKKKILASKSLYMSHMEAFL 677
Query: 640 NLAEELKSNNQLTYEDLKSEVAKHSSALEDLFKGIALEADSLLNDLQNSLHKQEANVTAF 699
N+ K++ T ED+ S A +L+ L + EA ++ ND+ N L + +T F
Sbjct: 678 NVVLVHKASANGTLEDISSLSAASCCSLDQLLVCVEGEAQNIFNDIHNLLTIHRSEMTHF 737
Query: 700 AHQQREAHSRAVETTRSVSKITMKFFETIDRHASSLTQIVEETQFVNDQKLCELEKKFEE 759
+ RE+ +++ T+ +S + F+ S L T + + + + +EE
Sbjct: 738 TQELRESFQISLDRTKEMSTYIIGLFDKYVEETSKLHSHSNNTHEAQMKSIEDFQMVYEE 797
Query: 760 CTAYEEKQLLEKVAEMLASSNARKKKLVQMAVNDLRESANCRTSKLQREALTMQDSTSFV 819
+ E++LL +++ +++ R+++LV + ++ L ++A + L M+ T
Sbjct: 798 QSKSVEQKLLAEISSLVSKHITRQRELVGVRLSSLGDAARGNKAFLDEHTSAMEFVTKDA 857
Query: 820 KAEWMVHMEKTESNYHEDTSSVESGKKDLAEVLQICLNKAEVGSQQWRNAQDSLLSLEKR 879
K +W E+ E++ +S + + +LQ C + QQW+ + ++ L ++
Sbjct: 858 KRKWETFAEQAENDCKAGSSFSAAKHSRMETMLQECACTVDSAVQQWKKSHAAVNDLSRK 917
Query: 880 NAGSVDTIVRGGMEANQALRARFSSSVSTTLEDAGIANTDINSSIDYSLQLDHEACGNLN 939
V+ +VR +E N+ +SS + EDA + DI ++ L+ + +
Sbjct: 918 QVAEVEALVRTAIENNEQHELEVASSRAVAEEDASNNSKDIAQGVENLLEEARNSSSRVV 977
Query: 940 SMITPCCGDLTELKGGHYNRIVEITENAGKCLLNEYMVDEPSCSTPTRRLFNLPSVSSIE 999
S + G+L L+ H ++ I +A K L Y EPS TP R N+PS SIE
Sbjct: 978 STVEAHFGELQMLQESHSSQAAGINMHADKALQTSYKDYEPSGETPVRSEPNVPSKGSIE 1037
Query: 1000 ELRTPSFEELLKAFWDAKSQKLANGDVKHIGSYEETQSVRDSRVPLTTIN 1049
LR E ++ F + N +H S +E++ + R+PL TIN
Sbjct: 1038 SLRAMPVETMMNEFRE-------NHPYEHESS-KESKLSQIPRLPLATIN 1079
>emb|CAC20783.1| kinesin-like boursin [Paracentrotus lividus]
Length = 1081
Score = 513 bits (1322), Expect = e-143
Identities = 311/814 (38%), Positives = 464/814 (56%), Gaps = 45/814 (5%)
Query: 49 NVQVLVRCRPMNEDEMRLHTPVVISCNEGRREVAAVQSIANKQIDRTFVFDKVFGPNSQQ 108
N+QV+VRCRP++ E + ++ V+ +RE+ +A K +TF FDKVFGP S Q
Sbjct: 12 NIQVVVRCRPVSSSEKKQNSYSVLDVKPAKREIIVGTEVAEKASSKTFSFDKVFGPKSTQ 71
Query: 109 KELYDQAVSPIVYEVLEGYNCTIFAYGQTGTGKTYTMEGGAIKK---NGEFPTDAGVIPR 165
E+Y V+PI+ EVL GYNCT+FAYGQTGTGKT+TMEG + E AG+IPR
Sbjct: 72 IEVYKSVVAPILDEVLMGYNCTVFAYGQTGTGKTFTMEGDRTPDPDLSWEQDPLAGIIPR 131
Query: 166 AVKQIFDILEAQSAEYSMKVTFLELYNEEITDLLAPEETTKFVDEKSKKPIALMEDG--K 223
A+ QIF+ + E+S++V++LELYNEE+ DLL+ +E T+ + + ED K
Sbjct: 132 AMHQIFEKMVGTDVEFSVRVSYLELYNEELFDLLSGQEDTQ--------RMRIFEDSARK 183
Query: 224 GGVLVRGLEEEIVCTANEIYKILEKGSAKRRTAETLLNKQSSRSHSIFSITIHIKECTPE 283
G V+++GLEE V NE+Y ILEKG++KR+TA TL+N SSRSHS+FS+TIHIKE + +
Sbjct: 184 GSVVIQGLEEVTVHNKNEVYAILEKGASKRQTAATLMNAHSSRSHSVFSVTIHIKENSID 243
Query: 284 GEEMIKCGKLNLVDLAGSENISRSGAREGRAREAGEINKSLLTLGRTINALVEHSGHVPY 343
G+E++K GKLNLVDLAGSENI RSGA + RAREAG IN+SLLTLGR I ALVEH+ HVPY
Sbjct: 244 GDELLKTGKLNLVDLAGSENIGRSGAVDKRAREAGNINQSLLTLGRVITALVEHAPHVPY 303
Query: 344 RDSKLTRLLRDSLGGKTKTCIIATVSPSIHCLEETLSTLDYAHRAKNIKNKPEVNQKMMK 403
R+SKLTR+L+DSLGG+TKT IIATVSP+ +EETLSTLDYAHRAKNI N+PE+NQK+ K
Sbjct: 304 RESKLTRILQDSLGGRTKTSIIATVSPASINVEETLSTLDYAHRAKNITNRPEINQKLTK 363
Query: 404 SAMIKDLYSEIDRLKQEVYAAREKNGIYIPRDRYLNEEAEKKAMAEKIERMELDADSKDK 463
A++K+ EI+RL+++++A REKNGI++ + Y + E + +I+ ME + +
Sbjct: 364 KALLKEYTEEIERLRKDLFATREKNGIFLSEEHYRSMETSIASQKAQIKEMEENIEGLTT 423
Query: 464 NLVELQELYNSQQLLTAELSAKLEKTEKSLEETEQTLFDLEERHRQANATIKEKEFLISN 523
+ ++ EL+ Q + + +LE T K+L ET TL E+ R E+ L+S
Sbjct: 424 QMQKVTELFEYTQKELEDRTEELEITTKNLVETTDTLHVTEKDLRVTTQDRDEQRHLVSE 483
Query: 524 LLKSEKELVERAIELRAELENAASDVSNLFSKIERKDKIEEENRVLIQKFQSQLAQQLEA 583
+K+E +L+ A +L + +++ SDV L SK++RK +E N+ + F
Sbjct: 484 HVKTETQLMSEATQLVSTADSSVSDVGGLHSKLDRKRNVEAHNKSAQEVFAESFRSHTSD 543
Query: 584 LHRTVSASVMHQEQQLKDMEKDMQSFVSTKSEATEDLRV----RVVELKNMYGSGIKALD 639
+ +S Q+ + M++ + +S +++ DLR V +K G+ ++
Sbjct: 544 IKSALSQLREEQQGKCASMQQQFEFMISKRTKEAGDLRTALSDMVTSVKGQTGAIMEESQ 603
Query: 640 NLAEELKSNNQLTYEDLKSEVAKHSSALEDLFKGIA-LEADSLLNDLQNSLHKQEANVTA 698
EE K + E+ HS ED+ I ++ L ++ K +
Sbjct: 604 RKREEWKKWSDDACEE-------HSKFKEDVIDRITDFHSNRFLTTMKTLEEKLSVLTES 656
Query: 699 FAHQQREAHSRAVETTRSVSKITMKFFETIDRHASSLTQIVEETQFVNDQKLCELEKKFE 758
+++ S+ + +V + K E + +T VE+ ++ +L + +
Sbjct: 657 LNECRKDIKSQISTQSETVQECIDKQLEQV----KVMTTTVEKFADHQTSRITDLSGQLD 712
Query: 759 ECTAYEEKQLLEKVAEMLASSNARKKKLVQMAVNDLRESANCRTSKLQREALTMQDSTSF 818
E +E + N K +Q + ES TSKLQ + T+
Sbjct: 713 ELKEHERHR------------NQDMMKSIQDLFSKREESFASETSKLQDQFREASTDTAT 760
Query: 819 VKAEWMVHMEKTESNYHEDTSSVESGKKDLAEVL 852
VKA+ ++ E+ E S + KD +E L
Sbjct: 761 VKAD----LQSQETAIQESCESFRTVHKDSSERL 790
Score = 58.2 bits (139), Expect = 1e-06
Identities = 95/484 (19%), Positives = 186/484 (37%), Gaps = 46/484 (9%)
Query: 375 LEETLSTLDYAHRAKNIKNKPEVNQKMMKSAMIK---DLYSEIDRLKQEVYAA-REKNGI 430
L ET TL + + + Q+ + S +K L SE +L ++ + G+
Sbjct: 453 LVETTDTLHVTEKDLRVTTQDRDEQRHLVSEHVKTETQLMSEATQLVSTADSSVSDVGGL 512
Query: 431 YIPRDRYLNEEAEKKAMAEKIERMELDADSKDKNLVELQELYNSQQLLTAELSAKLE--- 487
+ DR N EA K+ E S K+ L +L QQ A + + E
Sbjct: 513 HSKLDRKRNVEAHNKSAQEVFAESFRSHTSDIKSA--LSQLREEQQGKCASMQQQFEFMI 570
Query: 488 -KTEKSLEETEQTLFDLEERHR-QANATIKEKEFLISNLLKSEKELVERAIELRAELENA 545
K K + L D+ + Q A ++E + K + E + + ++ +
Sbjct: 571 SKRTKEAGDLRTALSDMVTSVKGQTGAIMEESQRKREEWKKWSDDACEEHSKFKEDVIDR 630
Query: 546 ASDV-SNLFSKIERKDKIEEENRVLIQKF---QSQLAQQLEALHRTVSASVMHQEQQLKD 601
+D SN F + +EE+ VL + + + Q+ TV + Q +Q+K
Sbjct: 631 ITDFHSNRF--LTTMKTLEEKLSVLTESLNECRKDIKSQISTQSETVQECIDKQLEQVKV 688
Query: 602 MEKDMQSFVSTKSEATEDLRVRVVELKNMYGSGIKALDNLAEELKSNNQLTYEDLKSEVA 661
M ++ F ++ DL ++ ELK + + ++L S + ++ A
Sbjct: 689 MTTTVEKFADHQTSRITDLSGQLDELKEHERHRNQDMMKSIQDLFSKREESF-------A 741
Query: 662 KHSSALEDLFKGIALEADSLLNDLQNSLHKQEANVTAFAHQQREA-----------HSRA 710
+S L+D F+ + + ++ DLQ+ + + +F +++ H +
Sbjct: 742 SETSKLQDQFREASTDTATVKADLQSQETAIQESCESFRTVHKDSSERLHSSADHHHEKT 801
Query: 711 VETTRSVSKITMKFFETIDRHASSLTQIVEETQFVNDQKLCELEK-KFEECTAYEEKQLL 769
V + K E++ H ++L I +T + Q++CE K +E T + KQ
Sbjct: 802 VGQVEDIRKYNASLEESVTSHTNTL--IERQTHYT--QEVCEKVKGHMDEVTQHVTKQ-- 855
Query: 770 EKVAEMLASSNARKKKLVQMAVNDLRESANCRTSKLQREALTMQDSTSFVKAEWMVHMEK 829
E E + + + + + V+ +C + ++ T + + W ME
Sbjct: 856 EADTEAMTNEHLASSETLSTTVDKQLTEMDCTVGQWTEKSKTRSEELT----TWTEEMEG 911
Query: 830 TESN 833
T N
Sbjct: 912 TMEN 915
>ref|NP_999660.1| KRP170 [Strongylocentrotus purpuratus] gi|10567777|gb|AAG18583.1|
KRP170 [Strongylocentrotus purpuratus]
Length = 1081
Score = 509 bits (1310), Expect = e-142
Identities = 334/967 (34%), Positives = 521/967 (53%), Gaps = 54/967 (5%)
Query: 49 NVQVLVRCRPMNEDEMRLHTPVVISCNEGRREVAAVQSIANKQIDRTFVFDKVFGPNSQQ 108
N+QV+VRCRP+N E + ++ V+ ++E+ +A K + F FDKVFGP S Q
Sbjct: 13 NIQVVVRCRPVNSIEKKQNSYSVLDVKPSKKEICVSTEVAEKASSKIFSFDKVFGPKSPQ 72
Query: 109 KELYDQAVSPIVYEVLEGYNCTIFAYGQTGTGKTYTMEGGAIKK---NGEFPTDAGVIPR 165
E+Y V+PI+ EVL GYNCT+FAYGQTGTGKT+TMEG + E AG+IPR
Sbjct: 73 IEVYKSVVAPILDEVLMGYNCTVFAYGQTGTGKTFTMEGERTPDPDLSWEQDPLAGIIPR 132
Query: 166 AVKQIFDILEAQSAEYSMKVTFLELYNEEITDLLAPEETTKFVDEKSKKPIALMEDG--K 223
A+ QIF+ + E+S++V++LELYNEE+ DLL+ +E T+ + + ED K
Sbjct: 133 AMHQIFEKMIGTDIEFSVRVSYLELYNEELFDLLSSQEDTQ--------RLRIFEDSARK 184
Query: 224 GGVLVRGLEEEIVCTANEIYKILEKGSAKRRTAETLLNKQSSRSHSIFSITIHIKECTPE 283
G V+++GLEE V NE+Y ILEKG+AKR+TA TL+N SSRSHS+FS+TIHIKE + E
Sbjct: 185 GSVVIQGLEEVTVHNKNEVYAILEKGAAKRKTAATLMNAHSSRSHSVFSVTIHIKENSIE 244
Query: 284 GEEMIKCGKLNLVDLAGSENISRSGAREGRAREAGEINKSLLTLGRTINALVEHSGHVPY 343
G+E++K GKLNLVDLAGSENI RSGA + RAREAG IN+SLLTLGR I ALVEH+ HVPY
Sbjct: 245 GDELLKTGKLNLVDLAGSENIGRSGAVDKRAREAGNINQSLLTLGRVITALVEHAPHVPY 304
Query: 344 RDSKLTRLLRDSLGGKTKTCIIATVSPSIHCLEETLSTLDYAHRAKNIKNKPEVNQKMMK 403
R+SKLTRLL+DSLGG+TKT IIATVSP+ +EETLSTLDYAHRAK+I N+PE+NQK+ K
Sbjct: 305 RESKLTRLLQDSLGGRTKTSIIATVSPASINVEETLSTLDYAHRAKHITNRPEINQKLTK 364
Query: 404 SAMIKDLYSEIDRLKQEVYAAREKNGIYIPRDRYLNEEAEKKAMAEKIERMELDADSKDK 463
A++K+ EI++L+++++A REKNGIY+ + Y N E +I+ ME + +
Sbjct: 365 KALLKEYTEEIEKLRKDLFATREKNGIYLSEEHYKNMEVSIACQRAQIKEMEENIEDLTT 424
Query: 464 NLVELQELYNSQQLLTAELSAKLEKTEKSLEETEQTLFDLEERHRQANATIKEKEFLISN 523
+ ++ EL+ Q E + +LE T K+LEET +TL E+ R E+ L+S
Sbjct: 425 QMQKVTELFEFTQKELEERTEELEVTTKNLEETTETLHVTEKDLRVTTQDRDEQCHLVSE 484
Query: 524 LLKSEKELVERAIELRAELENAASDVSNLFSKIERKDKIEEENRVLIQKFQSQLAQQLEA 583
+K+E +L+ A +L + +++ +DV L +K++RK +E N+ + F
Sbjct: 485 HVKTETQLMSEATQLLSTADSSVTDVGGLHAKLDRKRTVEAHNKSAQEVFAESFHSYTSD 544
Query: 584 LHRTVSASVMHQEQQLKDMEKDMQSFVSTKSEATEDLRVRVVELKNMYGSGIKALDNLAE 643
+ ++S Q+ + M++ + +S +++ DLR L +M S + E
Sbjct: 545 IKSSLSRLREEQQGKCTSMQQQFEFMISKRTKEAGDLR---ASLSDMVTSVKGHSAGMME 601
Query: 644 ELKSNNQLTYEDLKSEVAKHSSALEDLFKGI-------ALEADSL----LNDLQNSLHKQ 692
E + + + A+HS ED+ + L A ++ L+ L SL++
Sbjct: 602 ESQRKREEWEKWSNDACAEHSKFKEDVIDSMNDFHSNRFLTAMNMLAEKLSSLTESLNEC 661
Query: 693 EANVTAFAHQQREAHSRAVE-TTRSVSKITMKFFETIDRHASSLTQIVEETQFVNDQKLC 751
++ + Q E +E V+ +T + D S ++ + + + DQ+
Sbjct: 662 RRDIKTQINSQSETVQACIEKQLDQVNVMTTTVEKFSDHQRSKISDLSGQLDMLKDQE-- 719
Query: 752 ELEKKFEECTAYEEKQLLEKVAEMLA----SSNARKKKLVQMAVNDLRESANCRTSKLQR 807
+ + +++ + + A S + KL + E+ +TS L +
Sbjct: 720 ----------RHRNQDMMKSIQGLFAQREESFASETNKLQEQLEEASTETERVKTSILSQ 769
Query: 808 EALTMQDSTSFVKAEWMVHMEKTESNYHEDTSSVESGKKDLAEVLQICLNKAEVGSQQWR 867
E + SF VH E +E +S+ +K +A+V I + + +
Sbjct: 770 ETSVQESCESFRN----VHRESSERLI---SSADSHHEKTVAQVEDIRQHNTSL-EESMT 821
Query: 868 NAQDSLLSLEKRNAGSVDTIVRGGM-EANQALRARFSSSVSTTLEDAGIANTDINSSIDY 926
+ ++ + + ++ V +G M E +Q + + + + T+E T ++S++D
Sbjct: 822 SQTNTQIDRQTQHMEQVCENFKGHMEEISQHVTKQKADKEAMTIEHEASDET-LSSTVDK 880
Query: 927 SLQLDHEACGNLNSMITPCCGDLTELKGGHYNRIVEITENAGKCLLNEYMVDEPSCSTPT 986
L D+T + + L + D P+ +TP
Sbjct: 881 QLTEMTTTVTEWTEDSKARSEDMTSWTAEMEETVESGLKRVEGFLTRDLKEDVPTGTTPQ 940
Query: 987 RRLFNLP 993
R+ F+ P
Sbjct: 941 RKHFSYP 947
>emb|CAG30973.1| hypothetical protein [Gallus gallus]
Length = 1067
Score = 501 bits (1290), Expect = e-140
Identities = 340/1042 (32%), Positives = 543/1042 (51%), Gaps = 98/1042 (9%)
Query: 44 KEKGVNVQVLVRCRPMNEDEMRLHTPVVISCNEGRREVAA-VQSIANKQIDRTFVFDKVF 102
+EKG N+QV+VRCRP N E + ++ V+ C++ R+EV+ + +K + +T+ FD VF
Sbjct: 15 EEKGKNIQVVVRCRPFNASERKANSYAVVDCDQARKEVSVRTGGVTDKMLKKTYTFDMVF 74
Query: 103 GPNSQQKELYDQAVSPIVYEVLEGYNCTIFAYGQTGTGKTYTMEGGAIKKNGEFPTD--- 159
G ++Q ++Y V PI+ EV+ GYNCT+FAYGQTGTGKT+TMEG N E+ +
Sbjct: 75 GAQAKQIDVYRSVVCPILDEVIMGYNCTVFAYGQTGTGKTFTMEGER-SPNEEYTWEEDP 133
Query: 160 -AGVIPRAVKQIFDILEAQSAEYSMKVTFLELYNEEITDLLAP-----EETTKFVDEKSK 213
AG+IPR + QIF+ L E+S+KV+ LE+YNEE+ DLL P E F D ++K
Sbjct: 134 LAGIIPRTLHQIFEKLTENGTEFSVKVSLLEIYNEELFDLLNPAPDVGERLQMFDDPRNK 193
Query: 214 KPIALMEDGKGGVLVRGLEEEIVCTANEIYKILEKGSAKRRTAETLLNKQSSRSHSIFSI 273
+ GV+++GLEE V NE+Y+ILE+G+AKR TA T +N SSRSHS+FSI
Sbjct: 194 R----------GVIIKGLEEVTVHNKNEVYQILERGAAKRTTAATYMNAYSSRSHSVFSI 243
Query: 274 TIHIKECTPEGEEMIKCGKLNLVDLAGSENISRSGAREGRAREAGEINKSLLTLGRTINA 333
TIH+KE T +GEE++K GKLNLVDLAGSENI RSGA + RAREAG IN+SLLTLGR I+A
Sbjct: 244 TIHMKETTVDGEELVKIGKLNLVDLAGSENIGRSGAVDKRAREAGNINQSLLTLGRVISA 303
Query: 334 LVEHSGHVPYRDSKLTRLLRDSLGGKTKTCIIATVSPSIHCLEETLSTLDYAHRAKNIKN 393
LVE + H+PYR+SKLTR+L+DSLGG+TKT IIATVSP+ LEETLSTL+YAHRAK+I N
Sbjct: 304 LVERAPHIPYRESKLTRILQDSLGGRTKTSIIATVSPASINLEETLSTLEYAHRAKHIMN 363
Query: 394 KPEVNQKMMKSAMIKDLYSEIDRLKQEVYAAREKNGIYIPRDRY--LNEE--AEKKAMAE 449
KPEVNQK+ K A+IK+ EI+RLK+++ A REKNG+YI + + LN + +++ +AE
Sbjct: 364 KPEVNQKLTKKALIKEYTEEIERLKRDLAATREKNGVYISLENFEALNGKLTVQEEQIAE 423
Query: 450 KIERMELDADSKDKNLVELQELYNSQQLLTAELSAKLEKTEKSLEETEQTLFDLEERHRQ 509
I+++ + ++ + + EL+ + + LE EK LEET++ L ++
Sbjct: 424 YIDKISV----MEEEMKRVTELFTVNKNELKQCKTDLEIKEKELEETQKDL-------QE 472
Query: 510 ANATIKEKEFLISNLLKSEKELVERAIELRAELENAASDVSNLFSKIERKDKIEEENRVL 569
+ E+E+++S L +E++L A +L +E DVS L +K++RK +++ N ++
Sbjct: 473 TKVHLAEEEYVVSVLENAEQKLHGTASKLLNTVEETTKDVSGLHAKLDRKKAVDQHNAIV 532
Query: 570 IQKFQSQLAQQLEALHRTVSASVMHQEQQLKDMEKDMQSFVSTKSEA----TEDLRVRVV 625
F Q+ + +V+ + + Q+Q L + +ST S A T +
Sbjct: 533 QNTFAQQMTDLFNKIQDSVNENSVKQQQMLTSYTNFIDDILSTTSSASNILTSVVSASFA 592
Query: 626 ELKNMYGSGIKALDNLAEELKSNNQLTYE---DLKSEVAKHSSALEDLFKGIALEADSLL 682
LK + + + ++E++ + L+ + +L + +H+S L + + +L
Sbjct: 593 SLKELVSTEVSC---MSEKVLQHENLSLDHKAELLRLIEEHASGLGSALNSLTPMVEFVL 649
Query: 683 N---DLQNSLHKQEANVTAFAHQQREAHSRAVETTRSVSKI---TMKFFETIDRHASSLT 736
Q ++ K A ++E + + + ++ K+ T + +L
Sbjct: 650 GLNCQFQTNVKKYSAVADKMNGHKKEMDTFFEDLSLTLKKLQEETASVLAQLQNDCENLR 709
Query: 737 QIVEETQFVNDQKLCELEKKFEECTAYEEKQLLEKVAEMLASSNARKKKL------VQMA 790
+ VE T+ + + EL + ++ + +A++LA + + K + V +
Sbjct: 710 EEVEMTRQAHTKNAAELMSSLQRQLDLFAQETQKNLADVLAKNGSLKTTITAVQENVHLK 769
Query: 791 VNDLRESANCRTSKLQREALTMQDSTSFVKAEWMVHMEKTESNYHEDTSSVESGKKDLAE 850
DL S S+ + +E +E + + ++ + +D +
Sbjct: 770 TTDLVSSTASNHSRFLASVDNFSKELRTINSENKRMLEDSTDRCQQLLINLRNVSQDADK 829
Query: 851 VLQICLNK-AEVGSQQ---WRNAQDSLLSLEKRNAGSVDTIVRGGMEANQALRARFSSSV 906
++ + + A QQ +R+ + L+ +K+N G+ + +
Sbjct: 830 CAELTIAQIASFTDQQLLSFRDEKQQLMCFQKKNRGNCEKAI------------------ 871
Query: 907 STTLEDAGIANTDINSSIDYSLQLDHEACGNLNSMITPCCGDLTELKGGHYNRIVEITEN 966
+I ID + + L + G L E K
Sbjct: 872 -----------AEIADHIDTQKSAEEKVLNGLLDQMKVDQGVLLEQKLALNEEAQRGLAQ 920
Query: 967 AGKCLLNEYMVDEPSCSTPTRRLFNLPSVSSIEELRTPSFEEL------LKAFWDAKSQK 1020
L + VD P+ +TP RR ++ P E R E+L L A D+ ++
Sbjct: 921 VNGFLKEDLKVDIPTGTTPQRRDYSYPVTLVKTEPRELLLEQLRQKQLKLDAMLDSVVKE 980
Query: 1021 L-ANGDVKHIGSYEETQSVRDS 1041
N D +G EE Q +S
Sbjct: 981 AEENADEDLLGVEEEMQESSES 1002
>emb|CAA37950.1| kinesine [Xenopus laevis] gi|119217|sp|P28025|EG51_XENLA
Kinesin-related motor protein Eg5 1
Length = 1060
Score = 492 bits (1266), Expect = e-137
Identities = 339/1016 (33%), Positives = 532/1016 (51%), Gaps = 96/1016 (9%)
Query: 44 KEKGVNVQVLVRCRPMNEDEMRLHTPVVISCNEGRREVAAVQSIANKQIDR-TFVFDKVF 102
++KG N+QV+VRCRP N+ E + + V+ C+ R+EV N ++ + T+ FD VF
Sbjct: 6 EDKGKNIQVVVRCRPFNQLERKASSHSVLECDSQRKEVYVRTGEVNDKLGKKTYTFDMVF 65
Query: 103 GPNSQQKELYDQAVSPIVYEVLEGYNCTIFAYGQTGTGKTYTMEGGAIKKNGEFPTD--- 159
GP ++Q E+Y V PI+ EV+ GYNCTIFAYGQTGTGKT+TMEG + EF +
Sbjct: 66 GPAAKQIEVYRSVVCPILDEVIMGYNCTIFAYGQTGTGKTFTMEGER-SSDEEFTWEQDP 124
Query: 160 -AGVIPRAVKQIFDILEAQSAEYSMKVTFLELYNEEITDLLAP-----EETTKFVDEKSK 213
AG+IPR + QIF+ L E+S+KV+ LE+YNEE+ DLL+P E F D ++K
Sbjct: 125 LAGIIPRTLHQIFEKLSENGTEFSVKVSLLEIYNEELFDLLSPSPDVGERLQMFDDPRNK 184
Query: 214 KPIALMEDGKGGVLVRGLEEEIVCTANEIYKILEKGSAKRRTAETLLNKQSSRSHSIFSI 273
+ GV+++GLEE V +E+Y ILE+G+A+R+TA TL+N SSRSHS+FS+
Sbjct: 185 R----------GVIIKGLEEISVHNKDEVYHILERGAARRKTASTLMNAYSSRSHSVFSV 234
Query: 274 TIHIKECTPEGEEMIKCGKLNLVDLAGSENISRSGAREGRAREAGEINKSLLTLGRTINA 333
TIH+KE T +GEE++K GKLNLVDLAGSENI RSGA + RAREAG IN+SLLTLGR I A
Sbjct: 235 TIHMKETTVDGEELVKIGKLNLVDLAGSENIGRSGAVDKRAREAGNINQSLLTLGRVITA 294
Query: 334 LVEHSGHVPYRDSKLTRLLRDSLGGKTKTCIIATVSPSIHCLEETLSTLDYAHRAKNIKN 393
LVE + H+PYR+SKLTR+L+DSLGG+TKT IIATVSP+ LEET+STLDYA+RAK+I N
Sbjct: 295 LVERTPHIPYRESKLTRILQDSLGGRTKTSIIATVSPASINLEETVSTLDYANRAKSIMN 354
Query: 394 KPEVNQKMMKSAMIKDLYSEIDRLKQEVYAAREKNGIYIPRDRYLNEEAEKKAMAEKIER 453
KPEVNQK+ K A+IK+ EI+RLK+E+ AAREKNG+Y+ + Y + + + E I
Sbjct: 355 KPEVNQKLTKKALIKEYTEEIERLKRELAAAREKNGVYLSSENYEQLQGKVLSQEEMITE 414
Query: 454 MELDADSKDKNLVELQELYNSQQLLTAELSAKLEKTEKSLEETEQTLFDLEERHRQANAT 513
+ ++ L + EL+ + E + L+ EK LEET+ L + +E+
Sbjct: 415 YTEKITAMEEELKSISELFADNKKELEECTTILQCKEKELEETQNHLQESKEQ------- 467
Query: 514 IKEKEFLISNLLKSEKELVERAIELRAELENAASDVSNLFSKIERKDKIEEENRVLIQKF 573
+ ++ F++S +EK+L A +L + + DVS L K++RK +++ N + + F
Sbjct: 468 LAQESFVVSAFETTEKKLHGTANKLLSTVRETTRDVSGLHEKLDRKKAVDQHNFQVHENF 527
Query: 574 QSQLAQQLEALHRTVSASVMHQEQQLKDMEKDMQSFVSTKSEATEDLRVRVVE-LKNMYG 632
Q+ ++ + RTV + Q+ L + + S V + ++
Sbjct: 528 AEQMDRRFSVIQRTVDDYSVKQQGMLDFYTNSIDDLLGASSSRLSATASAVAKSFASVQE 587
Query: 633 SGIKALDNLAEELKSNNQLTYE---DLKSEVAKHSSALEDLFKGIALEADSLLNDLQNSL 689
+ K + + EE+ L+ + DL+ +A H + LE+ + L + + DL + L
Sbjct: 588 TVTKQVSHSVEEILKQETLSSQAKGDLQQLMAAHRTGLEEALRSDLLPVVTAVLDLNSHL 647
Query: 690 ----------------HKQEANVTAFAHQQREAHSRAVETTRSVSKITMKFFETIDRHAS 733
HK++ N + F R H ++++ ++S I ++
Sbjct: 648 SHCLQNFLIVADKIDSHKEDMN-SFFTEHSRSLHKLRLDSSSALSSIQSEY--------E 698
Query: 734 SLTQIVEETQFVNDQKLCELEKKFEECTAYEEKQLLEKVAEMLASSNARKKKLVQMAVND 793
SL + + Q ++ + + L + + ++ + L S + +K V D
Sbjct: 699 SLKEDIATAQSMHSEGVNNLISSLQNQLNLLGMETQQQFSGFL-SKGGKLQKSVGSLQQD 757
Query: 794 L----RESANCRTSKLQREALTMQDSTSFVKAEWMVHMEKTESNYHEDTSSVESGKKDLA 849
L E+ C +S ++ A QD ++ +M E ES K+
Sbjct: 758 LDLVSSEAIECISSHHKKLAEQSQDVAVEIRQLAGSNMSTLE----------ESSKQ--C 805
Query: 850 EVLQICLNKAEVGSQQWRNAQDSLLSLEKRNAGSVDTIVRGGMEANQALRARFSSSVSTT 909
E L +N SQQW + +D+++ + + + +
Sbjct: 806 EKLTSSINTISQESQQWCESAGQ----------KIDSVLEEQVCYLHSSKKHLQNLHKGV 855
Query: 910 LEDAGIANTDINSSIDYSLQLDHEACGNLNSMITPCCGDLTELKGGHYNRIVEITEN--- 966
+ G + +I ++ Q + +A +L + D E+ G + E ++
Sbjct: 856 EDSCGSSVVEITDRVNAQRQAEEKALTSLVEQVR----DDQEMVGEQRLELQEQVQSGLN 911
Query: 967 -AGKCLLNEYMVDEPSCSTPTRRLFNLPSVSSIEELRTPSFEELLKAFWDAKSQKL 1021
L E D P+ +TP RR + PS+ ++T + LL+ F + + L
Sbjct: 912 KVHSYLKEELRNDVPTGTTPQRRDYAYPSLL----VKTKPRDVLLEQFRQQQQEYL 963
>gb|AAH82827.1| LOC397908 protein [Xenopus laevis]
Length = 1067
Score = 490 bits (1262), Expect = e-137
Identities = 339/1030 (32%), Positives = 536/1030 (51%), Gaps = 101/1030 (9%)
Query: 32 ADSNSNSHNKYDKEKGVNVQVLVRCRPMNEDEMRLHTPVVISCNEGRREV-AAVQSIANK 90
+ NS +K D +KG N+QV+VRCRP N+ E + + V+ C+ R+EV I +K
Sbjct: 2 SSQNSFMASKKD-DKGKNIQVVVRCRPFNQLERKASSHSVLECDSQRKEVYVRTGGINDK 60
Query: 91 QIDRTFVFDKVFGPNSQQKELYDQAVSPIVYEVLEGYNCTIFAYGQTGTGKTYTMEGGAI 150
+T+ FD VFGP ++Q ++Y V PI+ EV+ GYNCTIFAYGQTGTGKT+TMEG
Sbjct: 61 LGKKTYTFDMVFGPAAKQIDVYRSVVCPILDEVIMGYNCTIFAYGQTGTGKTFTMEGER- 119
Query: 151 KKNGEFPTD----AGVIPRAVKQIFDILEAQSAEYSMKVTFLELYNEEITDLLAP----- 201
+ EF + AG+IPR + QIF+ L E+S+KV+ LE+YNEE+ DLL+P
Sbjct: 120 SSDEEFTWEQDPLAGIIPRTLHQIFEKLSENGTEFSVKVSLLEIYNEELFDLLSPSPDVG 179
Query: 202 EETTKFVDEKSKKPIALMEDGKGGVLVRGLEEEIVCTANEIYKILEKGSAKRRTAETLLN 261
E F D ++K+ GV+++GLEE V +E+Y ILE+G+A+R+TA TL+N
Sbjct: 180 ERLQMFDDPRNKR----------GVIIKGLEEISVHNKDEVYHILERGAARRKTASTLMN 229
Query: 262 KQSSRSHSIFSITIHIKECTPEGEEMIKCGKLNLVDLAGSENISRSGAREGRAREAGEIN 321
SSRSHS+F++TIH+KE T +GEE++K GKLNLVDLAGSENI RSGA + RAREAG IN
Sbjct: 230 AYSSRSHSVFAVTIHMKETTVDGEELVKIGKLNLVDLAGSENIGRSGAVDKRAREAGNIN 289
Query: 322 KSLLTLGRTINALVEHSGHVPYRDSKLTRLLRDSLGGKTKTCIIATVSPSIHCLEETLST 381
+SLLTLGR I ALVE + H+PYR+SKLTR+L+DSLGG+TKT IIATVSP+ LEET+ST
Sbjct: 290 QSLLTLGRVITALVERTPHIPYRESKLTRILQDSLGGRTKTSIIATVSPASINLEETVST 349
Query: 382 LDYAHRAKNIKNKPEVNQKMMKSAMIKDLYSEIDRLKQEVYAAREKNGIYIPRDRYLNEE 441
LDYA+RAK+I NKPEVNQK+ K A+IK+ EI+RLK+E+ AAREKNG+Y+ + Y +
Sbjct: 350 LDYANRAKSIMNKPEVNQKLTKKALIKEYTEEIERLKRELAAAREKNGVYLSSENYEQLQ 409
Query: 442 AEKKAMAEKIERMELDADSKDKNLVELQELYNSQQLLTAELSAKLEKTEKSLEETEQTLF 501
+ + E I + ++ L + EL+ + E + L+ EK LEET+ L
Sbjct: 410 GKVLSQEEMITEYTEKITAMEEELKSISELFADNKKELEECTTILQCKEKELEETQNHLQ 469
Query: 502 DLEERHRQANATIKEKEFLISNLLKSEKELVERAIELRAELENAASDVSNLFSKIERKDK 561
+ +E+ + ++ F++S +EK+L A +L + + DVS L K++RK
Sbjct: 470 ESKEQ-------LAQESFVVSAFETTEKKLHGTANKLLSTVRETTRDVSGLHEKLDRKKA 522
Query: 562 IEEENRVLIQKFQSQLAQQLEALHRTVSASVMHQEQQLKDMEKDMQSFVSTKSEATEDLR 621
+++ N + + F Q+ ++ + RTV + Q+ L + + S A
Sbjct: 523 VDQHNFQVHENFAEQMDRRFSVIQRTVDDYSVKQQGMLDFYTNSIDDLLGASSSALSATA 582
Query: 622 VRVVE-LKNMYGSGIKALDNLAEELKSNNQLTYE---DLKSEVAKHSSALEDLFKGIALE 677
V + ++ + K + + EE+ L+ + D++ + H + LE+ + L
Sbjct: 583 SAVAKSFASVQETVTKQVSHSVEEILKQETLSSQAKGDMQQLMTAHRTGLEEALRSDLLP 642
Query: 678 ADSLLNDLQNSL----------------HKQEANVTAFAHQQREAHSRAVETTRSVSKIT 721
+ + DL + L HK++ N + F R H ++++ ++S I
Sbjct: 643 VVTAVLDLNSHLSHCLQSFLIVADKIDSHKEDMN-SFFTEHSRSLHKLRLDSSSALSSIQ 701
Query: 722 MKFFETIDRHASSLTQIVEETQFVNDQKLCELEKKFEECTAYEEKQLLEKVAEMLASSNA 781
++ SL + + Q ++ + + L + + LL + S
Sbjct: 702 SEY--------ESLKEDIATAQSMHSEGVNNLISSLQ-----NQLNLLGMETQQQFSGFL 748
Query: 782 RKKKLVQMAVNDLRESANCRTSKL------QREALTMQDSTSFVKAEWMVHMEKTESNYH 835
K +Q +V L++ + +S+ + LT Q V+ +
Sbjct: 749 SKGGKLQKSVGSLQQDLDLVSSEAIECISSHHKKLTEQSQDVAVEIRQLA---------G 799
Query: 836 EDTSSVESGKKDLAEVLQICLNKAEVGSQQWRNAQDSLLSLEKRNAGSVDTIVRGGMEAN 895
+ S++E K E L +N SQQW + +D+++ +
Sbjct: 800 SNMSTLEESSKQ-CEKLTSSINTISQESQQWCESAGQ----------KIDSVLEEQVCYL 848
Query: 896 QALRARFSSSVSTTLEDAGIANTDINSSIDYSLQLDHEACGNLNSMITPCCGDLTELKGG 955
+ + + + G + +I ++ Q + +A +L + D E+ G
Sbjct: 849 HSSKKHLQNLHKGVEDSCGSSVVEITDHVNVQRQAEEKALTSLVEQVR----DDQEMVGE 904
Query: 956 HYNRIVEITEN----AGKCLLNEYMVDEPSCSTPTRRLFNLPSVSSIEELRTPSFEELLK 1011
+ E ++ L E D P+ +TP RR + PS+ ++T + LL+
Sbjct: 905 QRLELQEQVQSGLNKVHSYLKEELRNDVPTGTTPQRRDYAYPSLL----VKTKPRDVLLE 960
Query: 1012 AFWDAKSQKL 1021
F + + L
Sbjct: 961 QFRQQQQEYL 970
>emb|CAA50695.1| kinesin like protein [Xenopus laevis]
gi|2497521|sp|Q91783|EG52_XENLA Kinesin-related motor
protein Eg5 2
Length = 1067
Score = 489 bits (1259), Expect = e-136
Identities = 311/876 (35%), Positives = 478/876 (54%), Gaps = 81/876 (9%)
Query: 32 ADSNSNSHNKYDKEKGVNVQVLVRCRPMNEDEMRLHTPVVISCNEGRREVAAVQSIANKQ 91
+ NS +K D +KG N+QV+VRCRP N+ E + + V+ C R+EV N +
Sbjct: 2 SSQNSFMSSKKD-DKGKNIQVVVRCRPFNQLERKASSHSVLECESQRKEVCVRTGEVNDK 60
Query: 92 IDR-TFVFDKVFGPNSQQKELYDQAVSPIVYEVLEGYNCTIFAYGQTGTGKTYTMEGGAI 150
+ + T+ FD VFGP ++Q ++Y V PI+ EV+ GYNCTIFAYGQTGTGKT+TMEG
Sbjct: 61 LGKKTYTFDMVFGPAAKQIDVYRSVVCPILDEVIMGYNCTIFAYGQTGTGKTFTMEGER- 119
Query: 151 KKNGEFPTD----AGVIPRAVKQIFDILEAQSAEYSMKVTFLELYNEEITDLLAP----- 201
+ EF + AG+IPR + QIF+ L E+S+KV+ LE+YNEE+ DLL+P
Sbjct: 120 SSDEEFTWEQDPLAGIIPRTLHQIFEKLSEIGTEFSVKVSLLEIYNEELFDLLSPSPDVG 179
Query: 202 EETTKFVDEKSKKPIALMEDGKGGVLVRGLEEEIVCTANEIYKILEKGSAKRRTAETLLN 261
E F D ++K+ GV+++GLEE V +E+Y+ILE+G+AKR+TA TL+N
Sbjct: 180 ERLQMFDDPRNKR----------GVIIKGLEEISVHNKDEVYQILERGAAKRKTASTLMN 229
Query: 262 KQSSRSHSIFSITIHIKECTPEGEEMIKCGKLNLVDLAGSENISRSGAREGRAREAGEIN 321
SSRSHS+FS+TIH+KE T +GEE++K GKLNLVDLAGSENI RSGA + RAREAG IN
Sbjct: 230 AYSSRSHSVFSVTIHMKETTIDGEELVKIGKLNLVDLAGSENIGRSGAVDKRAREAGNIN 289
Query: 322 KSLLTLGRTINALVEHSGHVPYRDSKLTRLLRDSLGGKTKTCIIATVSPSIHCLEETLST 381
+SLLTLGR I ALVE + H+PYR+SKLTR+L+DSLGG+TKT IIATVSP+ LEET+ST
Sbjct: 290 QSLLTLGRVITALVERAPHIPYRESKLTRILQDSLGGRTKTSIIATVSPASINLEETMST 349
Query: 382 LDYAHRAKNIKNKPEVNQKMMKSAMIKDLYSEIDRLKQEVYAAREKNGIYIPRDRYLNEE 441
LDYA RAKNI NKPEVNQK+ K A+IK+ EI+RLK+E+ AREKNG+Y+ + Y +
Sbjct: 350 LDYASRAKNIMNKPEVNQKLTKKALIKEYTEEIERLKRELATAREKNGVYLSNENYEQLQ 409
Query: 442 AEKKAMAEKIERMELDADSKDKNLVELQELYNSQQLLTAELSAKLEKTEKSLEETEQTLF 501
+ + E I + ++ + + EL+ + E + L+ EK LE T+ L
Sbjct: 410 GKVLSQEEMITEYSEKIAAMEEEIKRIGELFADNKKELEECTTILQCKEKELEATQNNLQ 469
Query: 502 DLEERHRQANATIKEKEFLISNLLKSEKELVERAIELRAELENAASDVSNLFSKIERKDK 561
+ +E+ + ++ F++S + +EK+L A +L + + DVS L K++RK
Sbjct: 470 ESKEQ-------LAQEAFVVSAMETTEKKLHGTANKLLSTVRETTRDVSGLHEKLDRKRA 522
Query: 562 IEEENRVLIQKFQSQLAQQLEALHRTVSASVMHQEQQLKDMEKDMQSFVSTKSEATEDLR 621
+E+ N + + F Q+ ++ + +TV + Q+ L + + S A
Sbjct: 523 VEQHNSQVHENFAEQINRRFSVIQQTVDEYSVKQQGMLDFYTNSIDDLLGASSSALSATA 582
Query: 622 VRVVE-LKNMYGSGIKALDNLAEELKSNNQLT---YEDLKSEVAKHSSALEDLFKGIALE 677
V + ++ + K + + EE+ L+ +DL+ + H + LE + L
Sbjct: 583 TAVAKSFASVQETVSKQVSHSVEEILKQETLSSQAKDDLQKLMTAHRTGLEQALRTDLLP 642
Query: 678 ADSLLNDLQNSL----------------HKQEANVTAFAHQQREAHSRAVETTRSVSKIT 721
+ + DL + L HK++ N + F R H ++++ ++S I
Sbjct: 643 VVTAVLDLNSHLSHCLQSFLGVADKIDSHKEDMN-SFFTEHSRSLHKLRLDSSSALSSIQ 701
Query: 722 MKFFETIDR-------HASSLTQIVEETQFVNDQKLCELEK------------KFEE--- 759
++ + H+ + ++ Q N L +E K +E
Sbjct: 702 SEYESLKEEIATAQSTHSEGVNNLISSLQ--NQLNLLAMETRQQFSGFLSKGGKLQESVG 759
Query: 760 CTAYEEKQLLEKVAEMLASSNARKKKLVQMAVNDLRESANCRTSKLQREALTMQDSTSFV 819
C + + E ++S +++ + Q ++R+ A S L+ + + T+ +
Sbjct: 760 CLQQDLDLVSSDAIECISSHHSKFTEQSQAVTVEIRQLAGSNMSTLEESSKQCEKLTNSI 819
Query: 820 KA------EWMVHM-EKTESNYHEDTSSVESGKKDL 848
+W +K +S E + S KK +
Sbjct: 820 NTICQESQQWCESAGQKMDSLLEEQVCYLHSSKKQI 855
>gb|AAH42218.1| MGC52588 protein [Xenopus laevis]
Length = 1067
Score = 488 bits (1255), Expect = e-136
Identities = 339/1028 (32%), Positives = 529/1028 (50%), Gaps = 97/1028 (9%)
Query: 32 ADSNSNSHNKYDKEKGVNVQVLVRCRPMNEDEMRLHTPVVISCNEGRREVAA-VQSIANK 90
+ NS +K D +KG N+QV+VRCRP N+ E + + V+ C R+EV I +K
Sbjct: 2 SSQNSFMSSKKD-DKGKNIQVVVRCRPFNQLERKASSHSVLECESQRKEVCVRTGGINDK 60
Query: 91 QIDRTFVFDKVFGPNSQQKELYDQAVSPIVYEVLEGYNCTIFAYGQTGTGKTYTMEGGAI 150
+T+ FD VFGP ++Q ++Y V PI+ EV+ GYNCTIFAYGQTGTGKT+TMEG
Sbjct: 61 LGKKTYTFDMVFGPAAKQIDVYRSVVCPILDEVIMGYNCTIFAYGQTGTGKTFTMEGER- 119
Query: 151 KKNGEFPTD----AGVIPRAVKQIFDILEAQSAEYSMKVTFLELYNEEITDLLAP----- 201
+ EF + AG+IPR + QIF+ L E+S+KV+ LE+YNEE+ DLL+P
Sbjct: 120 SSDEEFTWEQDPLAGIIPRTLHQIFEKLSENGTEFSVKVSLLEIYNEELFDLLSPSPDVG 179
Query: 202 EETTKFVDEKSKKPIALMEDGKGGVLVRGLEEEIVCTANEIYKILEKGSAKRRTAETLLN 261
E F D ++K+ GV+++GLEE V +E+Y+ILE+G+AKR+TA TL+N
Sbjct: 180 ERLQMFDDPRNKR----------GVIIKGLEEISVHNKDEVYQILERGAAKRKTASTLMN 229
Query: 262 KQSSRSHSIFSITIHIKECTPEGEEMIKCGKLNLVDLAGSENISRSGAREGRAREAGEIN 321
SSRSHS+FS+TIH+KE T +GEE++K GKLNLVDLAGSENI RSGA + RAREAG IN
Sbjct: 230 AYSSRSHSVFSVTIHMKETTIDGEELVKIGKLNLVDLAGSENIGRSGAVDKRAREAGNIN 289
Query: 322 KSLLTLGRTINALVEHSGHVPYRDSKLTRLLRDSLGGKTKTCIIATVSPSIHCLEETLST 381
+SLLTLGR I ALVE + H+PYR+SKLTR+L+DSLGG+TKT IIATVSP+ LEET+ST
Sbjct: 290 QSLLTLGRVITALVERAPHIPYRESKLTRILQDSLGGRTKTSIIATVSPASINLEETMST 349
Query: 382 LDYAHRAKNIKNKPEVNQKMMKSAMIKDLYSEIDRLKQEVYAAREKNGIYIPRDRYLNEE 441
L+YA RAKNI NKPEVNQK+ K A+IK+ EI+RLK+E+ AREKNG+Y+ + Y +
Sbjct: 350 LEYASRAKNIMNKPEVNQKLTKKALIKEYTEEIERLKRELATAREKNGVYLSNENYEQLQ 409
Query: 442 AEKKAMAEKIERMELDADSKDKNLVELQELYNSQQLLTAELSAKLEKTEKSLEETEQTLF 501
+ + E I + ++ + + EL+ + E + L+ EK LE T+ L
Sbjct: 410 GKVLSQEEVITEYSEKIAAMEEEIKRIGELFADNKKELEECTTILQCKEKELEATQNNLQ 469
Query: 502 DLEERHRQANATIKEKEFLISNLLKSEKELVERAIELRAELENAASDVSNLFSKIERKDK 561
+ +E+ + ++ F++S + +EK+L A +L + + DVS L K++RK
Sbjct: 470 ESKEQ-------LAQEAFVVSAMETTEKKLHGTANKLLSTVRETTRDVSGLHEKLDRKRA 522
Query: 562 IEEENRVLIQKFQSQLAQQLEALHRTVSASVMHQEQQLKDMEKDMQSFVSTKSEATEDLR 621
+E+ N + + F Q+ ++ + +TV + Q+ L + + S A
Sbjct: 523 VEQHNSQVHENFAEQINRRFSVIQQTVDEYSVKQQGMLDFYTNSIDDLLGASSSALSATA 582
Query: 622 VRVVE-LKNMYGSGIKALDNLAEELKSNNQLT---YEDLKSEVAKHSSALEDLFKGIALE 677
V + ++ + K + + EE+ L+ +DL+ + + LE + L
Sbjct: 583 TAVAKSFASVQETVSKQVSHSVEEILRQETLSSQAKDDLQKLMIAQRTGLEQALRTDLLP 642
Query: 678 ADSLLNDLQNSL----------------HKQEANVTAFAHQQREAHSRAVETTRSVSKIT 721
+ + DL + L HK++ N + F R H ++++ ++S I
Sbjct: 643 VVTAVLDLNSHLSHCLQSFLGVADKIDSHKEDMN-SFFTEHSRSLHKLRLDSSSALSSIQ 701
Query: 722 MKFFETIDRHASSLTQIVEETQFVNDQKLCELEKKFEECTAYEEKQLLEKVAEMLASSNA 781
++ SL + + Q + + + L SS
Sbjct: 702 SEY--------ESLKEEIATAQSTHSEGV-----------------------NNLISSLQ 730
Query: 782 RKKKLVQMAVNDLRESANCRTSKLQREALTMQDSTSFVKAEWMVHMEKTESNYHEDTSSV 841
+ L+ M + KLQ +Q V ++ + + S + E + +V
Sbjct: 731 NQLNLLAMETRQQFSGFLSKGGKLQESVGCLQQDLDLVSSDAIECISSHHSKFTEQSQAV 790
Query: 842 ESGKKDLAEVLQICLNKAEVGSQQWRNAQDSLLSLEKRNAGSVDTIVRGGMEANQALRAR 901
+ LA L ++ ++ N+ +++ + S + +E
Sbjct: 791 AVEIRQLAGSNMSTLEESSKQCEKLTNSINTICQESQHWCESASQKMDSLLEEQVCYLHS 850
Query: 902 FSSSVSTTLED----AGIANTDINSSIDYSLQLDHEACGNLNSMITPCCGDLTELKGG-- 955
+ T +D G + +I+ ++ Q +A +L + D E+ G
Sbjct: 851 SKKQIQTLHKDVEDGCGSSVVEISDRVNVQRQAQEKALTSLVEQVK----DDKEMLGEQR 906
Query: 956 -HYNRIVEITENAGKCLLNEYM-VDEPSCSTPTRRLFNLPSVSSIEELRTPSFEELLKAF 1013
N V+I+ N L E + D P+ +TP RR + PS+ +RT + LL+ F
Sbjct: 907 LELNEQVQISLNKVHVYLKEELRNDVPTGTTPQRRDYVYPSLL----IRTKPRDVLLEQF 962
Query: 1014 WDAKSQKL 1021
+ + L
Sbjct: 963 KQQQQEYL 970
>ref|NP_034745.1| kinesin family member 11 [Mus musculus] gi|38174473|gb|AAH60670.1|
Kinesin family member 11 [Mus musculus]
Length = 1052
Score = 486 bits (1250), Expect = e-135
Identities = 351/1008 (34%), Positives = 533/1008 (52%), Gaps = 101/1008 (10%)
Query: 41 KYDKEKGVNVQVLVRCRPMNEDEMRLHTPVVISCNEGRREVAA-VQSIANKQIDRTFVFD 99
K +EKG N+QV+VRCRP N E + + V+ C+ R+EV+ + +K +T+ FD
Sbjct: 9 KKKEEKGRNIQVVVRCRPFNLAERKANAHSVVECDHARKEVSVRTAGLTDKTSKKTYTFD 68
Query: 100 KVFGPNSQQKELYDQAVSPIVYEVLEGYNCTIFAYGQTGTGKTYTMEGGAIKKN---GEF 156
VFG +++Q ++Y V PI+ EV+ GYNCTIFAYGQTGTGKT+TMEG E
Sbjct: 69 MVFGASTKQIDVYRSVVCPILDEVIMGYNCTIFAYGQTGTGKTFTMEGERSPNEVYTWEE 128
Query: 157 PTDAGVIPRAVKQIFDILEAQSAEYSMKVTFLELYNEEITDLLAP-----EETTKFVDEK 211
AG+IPR + QIF+ L E+S+KV+ LE+YNEE+ DLL+P E F D +
Sbjct: 129 DPLAGIIPRTLHQIFEKLTDNGTEFSVKVSLLEIYNEELFDLLSPSSDVSERLQMFDDPR 188
Query: 212 SKKPIALMEDGKGGVLVRGLEEEIVCTANEIYKILEKGSAKRRTAETLLNKQSSRSHSIF 271
+K+ GV+++GLEE V +E+Y+ILEKG+AKR TA TL+N SSRSHS+F
Sbjct: 189 NKR----------GVIIKGLEEITVHNKDEVYQILEKGAAKRTTAATLMNAYSSRSHSVF 238
Query: 272 SITIHIKECTPEGEEMIKCGKLNLVDLAGSENISRSGAREGRAREAGEINKSLLTLGRTI 331
S+TIH+KE T +GEE++K GKLNLVDLAGSENI RSGA + RAREAG IN+SLLTLGR I
Sbjct: 239 SVTIHMKETTIDGEELVKIGKLNLVDLAGSENIGRSGAVDKRAREAGNINQSLLTLGRVI 298
Query: 332 NALVEHSGHVPYRDSKLTRLLRDSLGGKTKTCIIATVSPSIHCLEETLSTLDYAHRAKNI 391
ALVE + H+PYR+SKLTR+L+DSLGG+T+T IIAT+SP+ LEETLSTL+YAHRAKNI
Sbjct: 299 TALVERTPHIPYRESKLTRILQDSLGGRTRTSIIATISPASFNLEETLSTLEYAHRAKNI 358
Query: 392 KNKPEVNQKMMKSAMIKDLYSEIDRLKQEVYAAREKNGIYIPRDRYLNEEAEKKAMAEKI 451
NKPEVNQK+ K A+IK+ EI+RLK+++ AAREKNG+YI + + +AM K+
Sbjct: 359 MNKPEVNQKLTKKALIKEYTEEIERLKRDLAAAREKNGVYISEESF-------RAMNGKV 411
Query: 452 ERMELDADSKDKNLVELQELYNSQQLLTAELSAKLEKTEKSLEETEQTLFDLEERHRQAN 511
E + + L+E + L + +L++ + L+ Q L ++ ++
Sbjct: 412 TVQEEQIVELVEKIAVLEEELSKATELFMDSKNELDQCKSDLQTKTQELETTQKHLQETK 471
Query: 512 ATIKEKEFLISNLLKSEKELVERAIELRAELENAASDVSNLFSKIERKDKIEEENRVLIQ 571
+ ++E++ S L ++EK L + A +L ++ VS L SK++RK I+E N +
Sbjct: 472 LQLVKEEYVSSALERTEKTLHDTASKLLNTVKETTRAVSGLHSKLDRKRAIDEHNAEAQE 531
Query: 572 KFQSQLAQQLEALHRTVSASVMHQEQQLKDMEKDMQSFVSTKSEATEDLRVRVVELKNMY 631
F L + + Q+ L + + +S+ A + + +E
Sbjct: 532 SFGKNLNSLFNNMEELIKDGSAKQKAMLDVHKTLFGNLMSSSVSALDTITTTALE----- 586
Query: 632 GSGIKALDNLAEELKSNNQLTYEDLKSEVAKHSSALEDLFKGIALEADSLLNDLQNSLHK 691
S + +N++ + + + E+ +S A+ S L+ L D L+ DL SL
Sbjct: 587 -SLVSIPENVSARVSQISDMILEE-QSLAAQSKSVLQGLI-------DELVTDLFTSLKT 637
Query: 692 QEA-------NVTAFAHQQREAHSRAVETTRSVSKITMKFFETIDRHASSLTQIVEET-- 742
A N+ A S E + F + ++L ++ E T
Sbjct: 638 IVAPSVVSILNINKQLQHIFRASSTVAEKVEDQKREIDSFLSIL---CNNLHELRENTVS 694
Query: 743 QFVNDQKLC----ELEKKFEECTAYEEKQLLEKVAEMLASSNARKKKLVQMAVNDLRESA 798
V QKLC E K +E + E QL AE + +K + +Q +N ++E+
Sbjct: 695 SLVESQKLCGDLTEDLKTIKETHSQELCQLSSSWAERFCALE-KKYENIQKPLNSIQENT 753
Query: 799 NCRTSKLQREALTMQDSTSFVKAEWMV----HMEKTESNYHEDT----SSVESGKKDLA- 849
R++ + + T+ +++ ++ H + + E++ SS+ S + ++
Sbjct: 754 ELRSTDIINKT-TVHSKKILAESDGLLQELRHFNQEGTQLVEESVGHCSSLNSNLETVSQ 812
Query: 850 EVLQIC--LNKAEVG-SQQWRNAQDSLLSLEKRNAGSVDTIVRGGMEANQALRARFSSSV 906
E+ Q C LN + V S QW S LS K ++ V G +A+ SS +
Sbjct: 813 EITQKCGTLNTSTVHFSDQWA----SCLSKRKEELENLMEFVNGCCKAS-------SSEI 861
Query: 907 STTLEDAGIANTDINSSIDYSLQLDHEAC--GNLNSMITPCCGDLTELKGGHYNRIVEIT 964
+ + + A + +SS + D E+C G+L T G LT+L
Sbjct: 862 TKKVREQSAAVANQHSSFVAQMTSDEESCKAGSLELDKTIKTG-LTKL------------ 908
Query: 965 ENAGKCLLNEYM-VDEPSCSTPTRRLFNLPSVSSIEELRTPSFEELLK 1011
C L + + +D P+ TP R+ + P+ E R ++L K
Sbjct: 909 ----NCFLKQDLKLDIPTGMTPERKKYLYPTTLVRTEPREQLLDQLQK 952
>gb|AAA03718.1| kinesin-like protein
Length = 1066
Score = 481 bits (1238), Expect = e-134
Identities = 307/844 (36%), Positives = 496/844 (58%), Gaps = 61/844 (7%)
Query: 34 SNSNSHNKYDKEKGVNVQVLVRCRPMNEDEMRLHTPVVISCNEGRREVAAVQSIANKQID 93
S N+ + K+ N+QV VR RP+N E + + V+ G REV ++ +K +
Sbjct: 4 SGGNTSRQPQKKSNQNIQVYVRVRPLNSRERCIRSAEVVDV-VGPREVVTRHTLDSK-LT 61
Query: 94 RTFVFDKVFGPNSQQKELYDQAVSPIVYEVLEGYNCTIFAYGQTGTGKTYTMEGGA---I 150
+ F FD+ FGP S+Q ++Y VSP++ EVL GYNCT+FAYGQTGTGKT+TM G +
Sbjct: 62 KKFTFDRSFGPESKQCDVYSVVVSPLIEEVLNGYNCTVFAYGQTGTGKTHTMVGNETAEL 121
Query: 151 KKNGEFPTDAGVIPRAVKQIFDILEAQSAEYSMKVTFLELYNEEITDLLAPEETTKF--V 208
K + E +D G+IPRA+ +FD L EY+M++++LELYNEE+ DLL+ ++TTK
Sbjct: 122 KSSWEDDSDIGIIPRALSHLFDELRMMEVEYTMRISYLELYNEELCDLLSTDDTTKIRIF 181
Query: 209 DEKSKKPIALMEDGKGGVLVRGLEEEIVCTANEIYKILEKGSAKRRTAETLLNKQSSRSH 268
D+ +KK G V+++GLEE V + +++YK+LEKG +R+TA TL+N QSSRSH
Sbjct: 182 DDSTKK---------GSVIIQGLEEIPVHSKDDVYKLLEKGKERRKTATTLMNAQSSRSH 232
Query: 269 SIFSITIHIKECTPEGEEMIKCGKLNLVDLAGSENISRSGAREG-RAREAGEINKSLLTL 327
++FSI +HI+E EGE+M+K GKLNLVDLAGSEN+S++G +G R RE IN+SLLTL
Sbjct: 233 TVFSIVVHIRENGIEGEDMLKIGKLNLVDLAGSENVSKAGNEKGIRVRETVNINQSLLTL 292
Query: 328 GRTINALVEHSGHVPYRDSKLTRLLRDSLGGKTKTCIIATVSPSIHCLEETLSTLDYAHR 387
GR I ALV+ + HVPYR+SKLTRLL++SLGG+TKT IIAT+SP +EETLSTL+YAHR
Sbjct: 293 GRVITALVDRAPHVPYRESKLTRLLQESLGGRTKTSIIATISPGHKDIEETLSTLEYAHR 352
Query: 388 AKNIKNKPEVNQKMMKSAMIKDLYSEIDRLKQEVYAAREKNGIYIPRDRY----LNEEAE 443
AKNI+NKPEVNQK+ K ++K+ EID+LK+++ AAR+KNGIY+ + Y L E++
Sbjct: 353 AKNIQNKPEVNQKLTKKTVLKEYTEEIDKLKRDLMAARDKNGIYLAEETYGEITLKLESQ 412
Query: 444 KKAMAEKIERMELDADSKDKNLVELQELYNSQQLLTAELSAKLEKTEKSLEETEQTLFDL 503
+ + EK+ ++ D L +++++ + E + +L+KTE++L T+ TL
Sbjct: 413 NRELNEKMLLLKALKD----ELQNKEKIFSEVSMSLVEKTQELKKTEENLLNTKGTLLLT 468
Query: 504 EERHRQANATIKEKEFLISNLLKSEKELVERAIELRAELENAASDVSNLFSKIERKDKIE 563
++ + KEK+ L+++ +K+E+ L +A E+ A + A D L IER+ +++
Sbjct: 469 KKVLTKTKRRYKEKKELVASHMKTEQVLTTQAQEILAAADLATDDTHQLHGTIERRRELD 528
Query: 564 EENRVLIQKFQSQLAQQLEALHRTVSASVMHQEQQLKDMEKDMQSFVSTKSEATEDLRVR 623
E+ R +F+ ++ LE + +++ ++Q+QQ E+ Q V++ S ++ L +
Sbjct: 529 EKIRRSCDQFKDRMQDNLEMIGGSLN---LYQDQQAALKEQLSQEMVNS-SYVSQRLALN 584
Query: 624 ----VVELKNMYGSGIKALDNLAEELKSNNQLTYEDLKSEVAKHSSALEDLFKGIALEAD 679
+ LK M ++ NL +N+L E +K + + L + + +
Sbjct: 585 SSKSIEMLKEMCAQSLQDQTNL------HNKLIGEVMKISDQHSQAFVAKLMEQMQQQQL 638
Query: 680 SLLNDLQNSLHKQEANVTAFAHQQREAHSRAVETTRSVSKITMKFFETIDRHASSLTQIV 739
+ ++Q +L E N +Q+ +A +++ + I ++++ HA + + +
Sbjct: 639 LMSKEIQTNLQVIEEN-----NQRHKAMLDSMQ--EKFATIIDSSLQSVEEHAKQMHKKL 691
Query: 740 EE---TQFVNDQKLCELEKKF--EECTAYEEKQLLEK-VAEMLASSNARKKKLVQMAV-- 791
E+ + ++L L+++ E A +E LLE + +M N R K + M+V
Sbjct: 692 EQLGAMSLPDAEELQNLQEELANERALAQQEDALLESMMMQMEQIKNLRSKNSISMSVHL 751
Query: 792 NDLRESANCRTSKLQREALTMQDSTSFVKAEWMVHMEKTESNYHEDTSSVESGKKDLAEV 851
N + ES R ++ D S ++ + +E ++S E TS +E+G L +
Sbjct: 752 NKMEESRLTRNHRI-------DDIKSGIQDYQKLGIEASQSAQAELTSQMEAGMLCLDQG 804
Query: 852 LQIC 855
+ C
Sbjct: 805 VANC 808
>gb|AAF47458.2| CG9191-PA [Drosophila melanogaster] gi|17136642|ref|NP_476818.1|
CG9191-PA [Drosophila melanogaster]
gi|17862220|gb|AAL39587.1| LD15641p [Drosophila
melanogaster] gi|26006996|sp|P46863|KL61_DROME Bipolar
kinesin KRP-130 (Kinesin-like protein Klp61F)
Length = 1066
Score = 481 bits (1238), Expect = e-134
Identities = 307/844 (36%), Positives = 496/844 (58%), Gaps = 61/844 (7%)
Query: 34 SNSNSHNKYDKEKGVNVQVLVRCRPMNEDEMRLHTPVVISCNEGRREVAAVQSIANKQID 93
S N+ + K+ N+QV VR RP+N E + + V+ G REV ++ +K +
Sbjct: 4 SGGNTSRQPQKKSNQNIQVYVRVRPLNSRERCIRSAEVVDV-VGPREVVTRHTLDSK-LT 61
Query: 94 RTFVFDKVFGPNSQQKELYDQAVSPIVYEVLEGYNCTIFAYGQTGTGKTYTMEGGA---I 150
+ F FD+ FGP S+Q ++Y VSP++ EVL GYNCT+FAYGQTGTGKT+TM G +
Sbjct: 62 KKFTFDRSFGPESKQCDVYSVVVSPLIEEVLNGYNCTVFAYGQTGTGKTHTMVGNETAEL 121
Query: 151 KKNGEFPTDAGVIPRAVKQIFDILEAQSAEYSMKVTFLELYNEEITDLLAPEETTKF--V 208
K + E +D G+IPRA+ +FD L EY+M++++LELYNEE+ DLL+ ++TTK
Sbjct: 122 KSSWEDDSDIGIIPRALSHLFDELRMMEVEYTMRISYLELYNEELCDLLSTDDTTKIRIF 181
Query: 209 DEKSKKPIALMEDGKGGVLVRGLEEEIVCTANEIYKILEKGSAKRRTAETLLNKQSSRSH 268
D+ +KK G V+++GLEE V + +++YK+LEKG +R+TA TL+N QSSRSH
Sbjct: 182 DDSTKK---------GSVIIQGLEEIPVHSKDDVYKLLEKGKERRKTATTLMNAQSSRSH 232
Query: 269 SIFSITIHIKECTPEGEEMIKCGKLNLVDLAGSENISRSGAREG-RAREAGEINKSLLTL 327
++FSI +HI+E EGE+M+K GKLNLVDLAGSEN+S++G +G R RE IN+SLLTL
Sbjct: 233 TVFSIVVHIRENGIEGEDMLKIGKLNLVDLAGSENVSKAGNEKGIRVRETVNINQSLLTL 292
Query: 328 GRTINALVEHSGHVPYRDSKLTRLLRDSLGGKTKTCIIATVSPSIHCLEETLSTLDYAHR 387
GR I ALV+ + HVPYR+SKLTRLL++SLGG+TKT IIAT+SP +EETLSTL+YAHR
Sbjct: 293 GRVITALVDRAPHVPYRESKLTRLLQESLGGRTKTSIIATISPGHKDIEETLSTLEYAHR 352
Query: 388 AKNIKNKPEVNQKMMKSAMIKDLYSEIDRLKQEVYAAREKNGIYIPRDRY----LNEEAE 443
AKNI+NKPEVNQK+ K ++K+ EID+LK+++ AAR+KNGIY+ + Y L E++
Sbjct: 353 AKNIQNKPEVNQKLTKKTVLKEYTEEIDKLKRDLMAARDKNGIYLAEETYGEITLKLESQ 412
Query: 444 KKAMAEKIERMELDADSKDKNLVELQELYNSQQLLTAELSAKLEKTEKSLEETEQTLFDL 503
+ + EK+ ++ D L +++++ + E + +L+KTE++L T+ TL
Sbjct: 413 NRELNEKMLLLKALKD----ELQNKEKIFSEVSMSLVEKTQELKKTEENLLNTKGTLLLT 468
Query: 504 EERHRQANATIKEKEFLISNLLKSEKELVERAIELRAELENAASDVSNLFSKIERKDKIE 563
++ + KEK+ L+++ +K+E+ L +A E+ A + A D L IER+ +++
Sbjct: 469 KKVLTKTKRRYKEKKELVASHMKTEQVLTTQAQEILAAADLATDDTHQLHGTIERRRELD 528
Query: 564 EENRVLIQKFQSQLAQQLEALHRTVSASVMHQEQQLKDMEKDMQSFVSTKSEATEDLRVR 623
E+ R +F+ ++ LE + +++ ++Q+QQ E+ Q V++ S ++ L +
Sbjct: 529 EKIRRSCDQFKDRMQDNLEMIGGSLN---LYQDQQAALKEQLSQEMVNS-SYVSQRLALN 584
Query: 624 ----VVELKNMYGSGIKALDNLAEELKSNNQLTYEDLKSEVAKHSSALEDLFKGIALEAD 679
+ LK M ++ NL +N+L E +K + + L + + +
Sbjct: 585 SSKSIEMLKEMCAQSLQDQTNL------HNKLIGEVMKISDQHSQAFVAKLMEQMQQQQL 638
Query: 680 SLLNDLQNSLHKQEANVTAFAHQQREAHSRAVETTRSVSKITMKFFETIDRHASSLTQIV 739
+ ++Q +L E N +Q+ +A +++ + I ++++ HA + + +
Sbjct: 639 LMSKEIQTNLQVIEEN-----NQRHKAMLDSMQ--EKFATIIDSSLQSVEEHAKQMHKKL 691
Query: 740 EE---TQFVNDQKLCELEKKF--EECTAYEEKQLLEK-VAEMLASSNARKKKLVQMAV-- 791
E+ + ++L L+++ E A +E LLE + +M N R K + M+V
Sbjct: 692 EQLGAMSLPDAEELQNLQEELANERALAQQEDALLESMMMQMEQIKNLRSKNSISMSVHL 751
Query: 792 NDLRESANCRTSKLQREALTMQDSTSFVKAEWMVHMEKTESNYHEDTSSVESGKKDLAEV 851
N + ES R ++ D S ++ + +E ++S E TS +E+G L +
Sbjct: 752 NKMEESRLTRNHRI-------DDIKSGIQDYQKLGIEASQSAQAELTSQMEAGMLCLDQG 804
Query: 852 LQIC 855
+ C
Sbjct: 805 VANC 808
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.311 0.127 0.341
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,561,690,831
Number of Sequences: 2540612
Number of extensions: 63987572
Number of successful extensions: 279087
Number of sequences better than 10.0: 12722
Number of HSP's better than 10.0 without gapping: 2261
Number of HSP's successfully gapped in prelim test: 10845
Number of HSP's that attempted gapping in prelim test: 221295
Number of HSP's gapped (non-prelim): 33967
length of query: 1049
length of database: 863,360,394
effective HSP length: 138
effective length of query: 911
effective length of database: 512,755,938
effective search space: 467120659518
effective search space used: 467120659518
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 81 (35.8 bits)
Medicago: description of AC149490.9


