

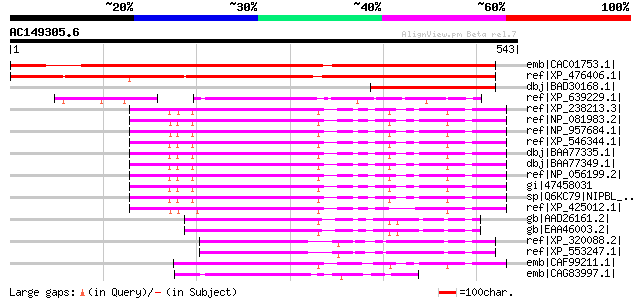
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149305.6 - phase: 0 /pseudo/partial
(543 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAC01753.1| putative protein [Arabidopsis thaliana] gi|15242... 593 e-168
ref|XP_476406.1| Nipped-B gene product-like protein [Oryza sativ... 580 e-164
dbj|BAD30168.1| putative IDN3 protein isoform A [Oryza sativa (j... 167 6e-40
ref|XP_639229.1| hypothetical protein DDB0185354 [Dictyostelium ... 128 4e-28
ref|XP_238213.3| PREDICTED: similar to delangin [Rattus norvegicus] 120 1e-25
ref|NP_081983.2| delangin isoform A [Mus musculus] gi|50400866|s... 119 2e-25
ref|NP_957684.1| delangin isoform B [Mus musculus] gi|48143965|e... 119 2e-25
ref|XP_546344.1| PREDICTED: similar to delangin isoform A [Canis... 119 2e-25
dbj|BAA77335.1| IDN3 [Homo sapiens] 119 2e-25
dbj|BAA77349.1| IDN3-B [Homo sapiens] 119 2e-25
ref|NP_056199.2| delangin isoform B [Homo sapiens] gi|48143963|e... 119 2e-25
gi|47458031 TPA: transcriptional regulator [Homo sapiens] 119 2e-25
sp|Q6KC79|NIPBL_HUMAN Nipped-B-like protein (Delangin) (SCC2 hom... 119 2e-25
ref|XP_425012.1| PREDICTED: similar to transcriptional regulator... 117 1e-24
gb|AAD26161.2| adherin Nipped-B [Drosophila melanogaster] gi|504... 116 2e-24
gb|EAA46003.2| CG17704-PF.3 [Drosophila melanogaster] gi|5195100... 116 2e-24
ref|XP_320088.2| ENSANGP00000010656 [Anopheles gambiae str. PEST... 113 2e-23
ref|XP_553247.1| ENSANGP00000028248 [Anopheles gambiae str. PEST... 113 2e-23
emb|CAF99211.1| unnamed protein product [Tetraodon nigroviridis] 112 4e-23
emb|CAG83997.1| unnamed protein product [Yarrowia lipolytica CLI... 92 3e-17
>emb|CAC01753.1| putative protein [Arabidopsis thaliana] gi|15242325|ref|NP_197058.1|
expressed protein [Arabidopsis thaliana]
gi|11358123|pir||T51532 hypothetical protein T20K14_150 -
Arabidopsis thaliana
Length = 1755
Score = 593 bits (1530), Expect = e-168
Identities = 308/521 (59%), Positives = 376/521 (72%), Gaps = 45/521 (8%)
Query: 1 MAIDILGTIAARLKRDAVICSQEKFWVLQDLLSEDAAPQHYPKDTCCVCLGGRVENLFKC 60
MAI++LGTIAARLKRDAV+CS+++FW L + SE + Q
Sbjct: 596 MAIELLGTIAARLKRDAVLCSKDRFWTLLESDSEISVDQ--------------------- 634
Query: 61 SGCDRLFHADCLDVKENEVPNRNWYCLMCICSKQLLVLQSYCNSQRKDDAKKNRKVS-KD 119
E ++ +RNW+C +C+C +QLLVLQSYC + K K + S ++
Sbjct: 635 ---------------ELDISSRNWHCPLCVCKRQLLVLQSYCKTDTKGTGKLESEESIEN 679
Query: 120 DSTFSNHEIVQQLLLNYFQDVTSADDLHHFICWFYLCSWYKNDPKCQQKPIYYFARMKSR 179
S + E+VQQ+LLNY QDV SADD+H FICWFYLC WYK+ PK Q K YY AR+K++
Sbjct: 680 PSMITKTEVVQQMLLNYLQDVGSADDVHTFICWFYLCLWYKDVPKSQNKFKYYIARLKAK 739
Query: 180 TIVRDSGSVSSMLTRDSIKKITLALGQNSSFCRGFDKIFHTLLVSLKENSPVIRAKALRA 239
+I+R+SG+ +S LTRD+IK+ITLALG NSSF RGFDKI + LL SL+EN+P IRAKALRA
Sbjct: 740 SIIRNSGATTSFLTRDAIKQITLALGMNSSFSRGFDKILNMLLASLRENAPNIRAKALRA 799
Query: 240 VSIIVEADPEVLGDKFVQSSVEGRFCDTAISVREAALELVGRHIASHPDVGFKYFEKITE 299
VSIIVEADPEVL DK VQ +VEGRFCD+AISVREAALELVGRHIASHPDVG KYFEK+ E
Sbjct: 800 VSIIVEADPEVLCDKRVQLAVEGRFCDSAISVREAALELVGRHIASHPDVGIKYFEKVAE 859
Query: 300 RIKDTGVSVRKRAIKIIRDMCCSDGNFLRVYKSLHRDNFTCY***IEYPAYSASDLQDLV 359
RIKDTGVSVRKRAIKIIRDMC S+ NF + + + S +QDLV
Sbjct: 860 RIKDTGVSVRKRAIKIIRDMCTSNPNFSEFTSACAEI--------LSRISDDESSVQDLV 911
Query: 360 CKTFYEFWFEEPSTPQTQVFKDGSTVPLEVAKKTEQIVEMLKRLPNNQLLVTVIKRCLTL 419
CKTFYEFWFEEP TQ D S++PLE+ KKT+Q+V +L R PN QLLVT+IKR L L
Sbjct: 912 CKTFYEFWFEEPPGHHTQFASDASSIPLELEKKTKQMVGLLSRTPNQQLLVTIIKRALAL 971
Query: 420 DFLPQSAKASGVNPVSLVTVRKRCELMCKCLLEKILQVDEMNSNELEKHALPYVLVLHAF 479
DF PQ+AKA+G+NPV+L +VR+RCELMCKCLLEKILQV+EM+ E E LPYVLVLHAF
Sbjct: 972 DFFPQAAKAAGINPVALASVRRRCELMCKCLLEKILQVEEMSREEGEVQVLPYVLVLHAF 1031
Query: 480 CLVDPTLCAPASNPSQFVLTLQPYLKTQVKTNLKTYILGSL 520
CLVDP LC PAS+P++FV+TLQPYLK+Q + +L S+
Sbjct: 1032 CLVDPGLCTPASDPTKFVITLQPYLKSQADSRTGAQLLESI 1072
>ref|XP_476406.1| Nipped-B gene product-like protein [Oryza sativa (japonica
cultivar-group)]
Length = 1755
Score = 580 bits (1495), Expect = e-164
Identities = 303/523 (57%), Positives = 375/523 (70%), Gaps = 15/523 (2%)
Query: 2 AIDILGTIAARLKRDAVICSQEKFWVLQDLLSEDAAPQHYPKDTCCVCLGGRVENLFKCS 61
AID+LG IA+RLKRD+VICS+EK W+LQ+L ++ K+ CC+CLGGR N+ +C
Sbjct: 614 AIDLLGGIASRLKRDSVICSKEKLWILQELTDTESDGSKILKNKCCICLGGRGINM-QCD 672
Query: 62 GCDRLFHADCLDVKENEVPNRNWYCLMCICSKQLLVLQSYCNSQRKDDAKKNRKVSKDDS 121
C R FH+DC+ E ++ C +C C +QL VLQSY Q K++ K+N + S
Sbjct: 673 VCGRCFHSDCVGAVSQENLQCDYACPLCFCKRQLSVLQSYYELQNKENGKRNAASHRKKS 732
Query: 122 TFSNH----EIVQQLLLNYFQDVTSADDLHHFICWFYLCSWYKNDPKCQQKPIYYFARMK 177
T + +IVQQ+LL Y Q+ DD + F WFYLC WYK+DP Q+K IYY AR+K
Sbjct: 733 TVPDEVTAVDIVQQILLTYIQEGGPQDDGNLFTRWFYLCMWYKDDPHSQEKIIYYLARLK 792
Query: 178 SRTIVRDSGSVSSMLTRDSIKKITLALGQNSSFCRGFDKIFHTLLVSLKENSPVIRAKAL 237
++ I+RDSG+ +L+RD KKI LALGQ +SF RGFDKI LL SL+ENSPVIRAKAL
Sbjct: 793 TKDILRDSGN-GLVLSRDWAKKICLALGQKNSFSRGFDKILSLLLASLRENSPVIRAKAL 851
Query: 238 RAVSIIVEADPEVLGDKFVQSSVEGRFCDTAISVREAALELVGRHIASHPDVGFKYFEKI 297
RAVS IVEADPEVLGDK VQS+VEGRFCD+AISVREAALELVGRHIASHPDVG KY EK+
Sbjct: 852 RAVSSIVEADPEVLGDKRVQSAVEGRFCDSAISVREAALELVGRHIASHPDVGLKYIEKV 911
Query: 298 TERIKDTGVSVRKRAIKIIRDMCCSDGNFLRVYKSLHRDNFTCY***IEYPAYSASDLQD 357
ERIKDTGVSVRKRAIKIIRD+C S+ N D + I S +QD
Sbjct: 912 AERIKDTGVSVRKRAIKIIRDLCASNPN---------TDTTRAFVEIISRVNDEESSVQD 962
Query: 358 LVCKTFYEFWFEEPSTPQTQVFKDGSTVPLEVAKKTEQIVEMLKRLPNNQLLVTVIKRCL 417
LVCKTFYE WFEEP+ + DGS+VP+E+A KTEQIV+ML+++PN+ L+T++KR L
Sbjct: 963 LVCKTFYELWFEEPTGSHKHLVADGSSVPMEIAVKTEQIVDMLRKMPNHLPLITIVKRNL 1022
Query: 418 TLDFLPQSAKASGVNPVSLVTVRKRCELMCKCLLEKILQVDEMNSNELEKHALPYVLVLH 477
LDFLPQSAKA+G+N + ++RKRCEL+CK LLE+ILQV+E ++E E HALPYVL L
Sbjct: 1023 ALDFLPQSAKATGINSSFMASLRKRCELICKRLLERILQVEEGAASETEVHALPYVLALQ 1082
Query: 478 AFCLVDPTLCAPASNPSQFVLTLQPYLKTQVKTNLKTYILGSL 520
AFC+VDPTLC PA+ P QFV TLQPYLK QV +L S+
Sbjct: 1083 AFCIVDPTLCTPATQPFQFVETLQPYLKKQVDNKSTAQLLESI 1125
>dbj|BAD30168.1| putative IDN3 protein isoform A [Oryza sativa (japonica
cultivar-group)]
Length = 764
Score = 167 bits (424), Expect = 6e-40
Identities = 81/134 (60%), Positives = 105/134 (77%)
Query: 387 LEVAKKTEQIVEMLKRLPNNQLLVTVIKRCLTLDFLPQSAKASGVNPVSLVTVRKRCELM 446
+E+A KTEQIV+ML+++PN+ L+T++KR L LDFLPQSAKA+G+N + ++RKRCEL+
Sbjct: 1 MEIAVKTEQIVDMLRKMPNHLPLITIVKRNLALDFLPQSAKATGINSSFMASLRKRCELI 60
Query: 447 CKCLLEKILQVDEMNSNELEKHALPYVLVLHAFCLVDPTLCAPASNPSQFVLTLQPYLKT 506
CK LLE+ILQV+E ++E E HALPYVL L AFC+VDPTLC PA+ P QFV TLQPYLK
Sbjct: 61 CKRLLERILQVEEGAASETEVHALPYVLALQAFCIVDPTLCTPATQPFQFVETLQPYLKK 120
Query: 507 QVKTNLKTYILGSL 520
QV +L S+
Sbjct: 121 QVDNKSTAQLLESI 134
>ref|XP_639229.1| hypothetical protein DDB0185354 [Dictyostelium discoideum]
gi|60467865|gb|EAL65879.1| hypothetical protein
DDB0185354 [Dictyostelium discoideum]
Length = 2063
Score = 128 bits (322), Expect = 4e-28
Identities = 91/319 (28%), Positives = 159/319 (49%), Gaps = 32/319 (10%)
Query: 197 IKKITLALGQNSSFCRGFDKIFHTLLVSLKENSPVIRAKALRAVSIIVEADPEVLGDKFV 256
I+KI +A G S + + + + LL L + S RAKA+++ + IVEADP +L D+ V
Sbjct: 1123 IRKINIASG---SVFQVANNLLYHLLSILFDPSTKARAKAMKSFTTIVEADPSILADEKV 1179
Query: 257 QSSVEGRFCDTAISVREAALELVGRHIASHPDVGFKYFEKITERIKDTGVSVRKRAIKII 316
S+++ RF D +I VRE ++L+G+++ P++ ++Y + I + I D G+SVRKR +K +
Sbjct: 1180 HSAIKNRFLDESILVREHTVDLIGKYVLIKPELTWRYIDLICDLIGDKGISVRKRVVKTL 1239
Query: 317 RDMCCSDGNFLRVYKSLHRDNFTCY***IEYPAYSASDLQDLVCKTFYEFWFEEP----S 372
++C + ++ + C + ++DLV KTF + WF E
Sbjct: 1240 NEICLNHMQHPKI-------PYLCR--VLVTRVSDEESIKDLVLKTFQDLWFSERFYNIE 1290
Query: 373 TPQTQVFKDGSTVPLEVAKKTEQIVEMLKRLPNNQLLVTVIKRCLTLDFLPQSAKASGVN 432
Q +G T+ E+ K +QI+E+ K+L + + +I++ L +A G
Sbjct: 1291 NQQINSNANGCTINPEII-KVKQIIEVSKQLDQDSWFIDLIQKMLNKS--STNATGGGKK 1347
Query: 433 PVSLVTVRKRCEL------MCKCLLEKILQVDEMNSNELEKHALPYVLVLHAFCLVDPTL 486
++ E+ +C L+E ++ +DE S ++ + + LH FC V+PT
Sbjct: 1348 ESKKKLEKEHNEVVQMSNKICSTLIEMLVNLDENKSKNIQSY-MGIFKTLHIFCKVNPTF 1406
Query: 487 CAPASNPSQFVLTLQPYLK 505
P F L PYLK
Sbjct: 1407 LVP------FASILHPYLK 1419
Score = 48.1 bits (113), Expect = 7e-04
Identities = 36/139 (25%), Positives = 61/139 (42%), Gaps = 29/139 (20%)
Query: 49 CLGGRVEN---LFKCSGCDRLFHADCL-DVKENEVPNR--NWYCLMCICSKQLL------ 96
C G+ E+ KC+ C++LFH C D +N N+ +W C+ C +Q++
Sbjct: 894 CNCGKYESDSFTIKCNECEQLFHDGCAPDFTKNGDINQQQSWVCVSCQVKEQVVHYSTTS 953
Query: 97 -VLQSYCNSQRKDDAKKNRKVSKDDS----------------TFSNHEIVQQLLLNYFQD 139
+ + + K+ +KK++K +D S S EI QQL+LNY +
Sbjct: 954 KITPTILEDKLKNQSKKHKKSKRDKSEGGFSEEELPLTMESLVISEKEIYQQLILNYLES 1013
Query: 140 VTSADDLHHFICWFYLCSW 158
S + + F + SW
Sbjct: 1014 KCSTESWINTSKQFLIASW 1032
>ref|XP_238213.3| PREDICTED: similar to delangin [Rattus norvegicus]
Length = 2798
Score = 120 bits (300), Expect = 1e-25
Identities = 116/453 (25%), Positives = 189/453 (41%), Gaps = 93/453 (20%)
Query: 129 VQQLLLNYFQDVTSADDLHHFICWFYLCSWYKNDPKCQQKP------------------- 169
+Q+ LL+Y + T D F FY+ W+++ +K
Sbjct: 1645 LQKALLDYLDENTETDPSLVFSRKFYIAQWFRDTTLETEKAMKSQKDEESSDGAHHAKEI 1704
Query: 170 -----IYYFARMKSR---TIVRDSGSVSSMLTR-------DSIKKITLALGQNSSFCRGF 214
I + A + R +I++ + S S L D I L F + F
Sbjct: 1705 ETTGQIMHRAESRKRFLRSIIKTTPSQFSTLKMNSDTVDYDDACLIVRYLASMRPFAQSF 1764
Query: 215 DKIFHTLLVSLKENSPVIRAKALRAVSIIVEADPEVLGDKFVQSSVEGRFCDTAISVREA 274
D +L L EN+ +R KA++ +S +V DP +L +Q V GR D + SVREA
Sbjct: 1765 DIYLTQILRVLGENAIAVRTKAMKCLSEVVAVDPSILARLDMQRGVHGRLMDNSTSVREA 1824
Query: 275 ALELVGRHIASHPDVGFKYFEKITERIKDTGVSVRKRAIKIIRDMCCSDGNFLRV----Y 330
A+EL+GR + P + +Y++ + ERI DTG+SVRKR IKI+RD+C F ++
Sbjct: 1825 AVELLGRFVLCRPQLAEQYYDMLIERILDTGISVRKRVIKILRDICIEQPTFPKITEMCV 1884
Query: 331 KSLHRDNFTCY***IEYPAYSASDLQDLVCKTFYEFWFEEPSTPQTQVFKDGSTVPLEVA 390
K + R N ++ LV +TF + WF TP K+ T
Sbjct: 1885 KMIRRVN-------------DEEGIKKLVNETFQKLWF----TPTPHNDKEAMT------ 1921
Query: 391 KKTEQIVEMLKRLPN------NQLLVTVIKRCLTLDFLPQSAKASGVNPVSLVTVRKRCE 444
+K I +++ + QLL ++K S + S P V+K C
Sbjct: 1922 RKILNITDVVAACRDTGYDWFEQLLQNLLK----------SEEDSSYKP-----VKKACT 1966
Query: 445 LMCKCLLEKILQVDEMNSNELEK-----HALPYVLVLHAFCLVDPTLCAPASNPSQFVLT 499
+ L+E IL+ +E ++ K + + L F + P L + +T
Sbjct: 1967 QLVDNLVEHILKYEESLADSDNKGVNSGRLVACITTLFLFSKIRPQLMV------KHAMT 2020
Query: 500 LQPYLKTQVKTNLKTYILGSLQCYYALCIELLE 532
+QPYL T+ T ++ ++ L + L+E
Sbjct: 2021 MQPYLTTKCSTQNDFMVICNVAKILELVVPLME 2053
>ref|NP_081983.2| delangin isoform A [Mus musculus] gi|50400866|sp|Q6KCD5|NPBL_MOUSE
Nipped-B-like protein (Delangin homolog) (SCC2 homolog)
gi|48143960|emb|CAF25291.1| delangin [Mus musculus]
Length = 2798
Score = 119 bits (299), Expect = 2e-25
Identities = 114/453 (25%), Positives = 191/453 (41%), Gaps = 93/453 (20%)
Query: 129 VQQLLLNYFQDVTSADDLHHFICWFYLCSWYKNDPKCQQKPI------------YYFARM 176
+Q+ LL+Y + T D F FY+ W+++ +K + ++ +
Sbjct: 1645 LQKALLDYLDENTETDPSLVFSRKFYIAQWFRDTTLETEKAMKSQKDEESSDATHHAKEL 1704
Query: 177 KS---------------RTIVRDSGSVSSMLTR-------DSIKKITLALGQNSSFCRGF 214
++ R+I++ + S S L D I L F + F
Sbjct: 1705 ETTGQIMHRAENRKKFLRSIIKTTPSQFSTLKMNSDTVDYDDACLIVRYLASMRPFAQSF 1764
Query: 215 DKIFHTLLVSLKENSPVIRAKALRAVSIIVEADPEVLGDKFVQSSVEGRFCDTAISVREA 274
D +L L EN+ +R KA++ +S +V DP +L +Q V GR D + SVREA
Sbjct: 1765 DIYLTQILRVLGENAIAVRTKAMKCLSEVVAVDPSILARLDMQRGVHGRLMDNSTSVREA 1824
Query: 275 ALELVGRHIASHPDVGFKYFEKITERIKDTGVSVRKRAIKIIRDMCCSDGNFLRV----Y 330
A+EL+GR + P + +Y++ + ERI DTG+SVRKR IKI+RD+C F ++
Sbjct: 1825 AVELLGRFVLCRPQLAEQYYDMLIERILDTGISVRKRVIKILRDICIEQPTFPKITEMCV 1884
Query: 331 KSLHRDNFTCY***IEYPAYSASDLQDLVCKTFYEFWFEEPSTPQTQVFKDGSTVPLEVA 390
K + R N ++ LV +TF + WF TP K+ T
Sbjct: 1885 KMIRRVN-------------DEEGIKKLVNETFQKLWF----TPTPHNDKEAMT------ 1921
Query: 391 KKTEQIVEMLKRLPN------NQLLVTVIKRCLTLDFLPQSAKASGVNPVSLVTVRKRCE 444
+K I +++ + QLL ++K S + S P V+K C
Sbjct: 1922 RKILNITDVVAACRDTGYDWFEQLLQNLLK----------SEEDSSYKP-----VKKACT 1966
Query: 445 LMCKCLLEKILQVDEMNSNELEK-----HALPYVLVLHAFCLVDPTLCAPASNPSQFVLT 499
+ L+E IL+ +E ++ K + + L F + P L + +T
Sbjct: 1967 QLVDNLVEHILKYEESLADSDNKGVNSGRLVACITTLFLFSKIRPQLMV------KHAMT 2020
Query: 500 LQPYLKTQVKTNLKTYILGSLQCYYALCIELLE 532
+QPYL T+ T ++ ++ L + L+E
Sbjct: 2021 MQPYLTTKCSTQNDFMVICNVAKILELVVPLME 2053
>ref|NP_957684.1| delangin isoform B [Mus musculus] gi|48143965|emb|CAG26692.1|
delangin [Mus musculus]
Length = 2691
Score = 119 bits (299), Expect = 2e-25
Identities = 114/453 (25%), Positives = 191/453 (41%), Gaps = 93/453 (20%)
Query: 129 VQQLLLNYFQDVTSADDLHHFICWFYLCSWYKNDPKCQQKPI------------YYFARM 176
+Q+ LL+Y + T D F FY+ W+++ +K + ++ +
Sbjct: 1645 LQKALLDYLDENTETDPSLVFSRKFYIAQWFRDTTLETEKAMKSQKDEESSDATHHAKEL 1704
Query: 177 KS---------------RTIVRDSGSVSSMLTR-------DSIKKITLALGQNSSFCRGF 214
++ R+I++ + S S L D I L F + F
Sbjct: 1705 ETTGQIMHRAENRKKFLRSIIKTTPSQFSTLKMNSDTVDYDDACLIVRYLASMRPFAQSF 1764
Query: 215 DKIFHTLLVSLKENSPVIRAKALRAVSIIVEADPEVLGDKFVQSSVEGRFCDTAISVREA 274
D +L L EN+ +R KA++ +S +V DP +L +Q V GR D + SVREA
Sbjct: 1765 DIYLTQILRVLGENAIAVRTKAMKCLSEVVAVDPSILARLDMQRGVHGRLMDNSTSVREA 1824
Query: 275 ALELVGRHIASHPDVGFKYFEKITERIKDTGVSVRKRAIKIIRDMCCSDGNFLRV----Y 330
A+EL+GR + P + +Y++ + ERI DTG+SVRKR IKI+RD+C F ++
Sbjct: 1825 AVELLGRFVLCRPQLAEQYYDMLIERILDTGISVRKRVIKILRDICIEQPTFPKITEMCV 1884
Query: 331 KSLHRDNFTCY***IEYPAYSASDLQDLVCKTFYEFWFEEPSTPQTQVFKDGSTVPLEVA 390
K + R N ++ LV +TF + WF TP K+ T
Sbjct: 1885 KMIRRVN-------------DEEGIKKLVNETFQKLWF----TPTPHNDKEAMT------ 1921
Query: 391 KKTEQIVEMLKRLPN------NQLLVTVIKRCLTLDFLPQSAKASGVNPVSLVTVRKRCE 444
+K I +++ + QLL ++K S + S P V+K C
Sbjct: 1922 RKILNITDVVAACRDTGYDWFEQLLQNLLK----------SEEDSSYKP-----VKKACT 1966
Query: 445 LMCKCLLEKILQVDEMNSNELEK-----HALPYVLVLHAFCLVDPTLCAPASNPSQFVLT 499
+ L+E IL+ +E ++ K + + L F + P L + +T
Sbjct: 1967 QLVDNLVEHILKYEESLADSDNKGVNSGRLVACITTLFLFSKIRPQLMV------KHAMT 2020
Query: 500 LQPYLKTQVKTNLKTYILGSLQCYYALCIELLE 532
+QPYL T+ T ++ ++ L + L+E
Sbjct: 2021 MQPYLTTKCSTQNDFMVICNVAKILELVVPLME 2053
>ref|XP_546344.1| PREDICTED: similar to delangin isoform A [Canis familiaris]
Length = 2936
Score = 119 bits (299), Expect = 2e-25
Identities = 116/453 (25%), Positives = 189/453 (41%), Gaps = 93/453 (20%)
Query: 129 VQQLLLNYFQDVTSADDLHHFICWFYLCSWYKNDPKCQQKP------------------- 169
+Q+ LL+Y + T D F FY+ W+++ +K
Sbjct: 1783 LQKALLDYLDENTETDPSLVFSRKFYIAQWFRDTTLETEKAMKSQKDEESSEGTHHAKEV 1842
Query: 170 -----IYYFARMKS---RTIVRDSGSVSSMLTR-------DSIKKITLALGQNSSFCRGF 214
I + A + R+I++ + S S L D I L F + F
Sbjct: 1843 ETTGQIMHRAENRKKFLRSIIKTTPSQFSTLKMNSDTVDYDDACLIVRYLASMRPFAQSF 1902
Query: 215 DKIFHTLLVSLKENSPVIRAKALRAVSIIVEADPEVLGDKFVQSSVEGRFCDTAISVREA 274
D +L L EN+ +R KA++ +S +V DP +L +Q V GR D + SVREA
Sbjct: 1903 DIYLTQILRVLGENAIAVRTKAMKCLSEVVAVDPSILARLDMQRGVHGRLMDNSTSVREA 1962
Query: 275 ALELVGRHIASHPDVGFKYFEKITERIKDTGVSVRKRAIKIIRDMCCSDGNFLRV----Y 330
A+EL+GR + P + +Y++ + ERI DTG+SVRKR IKI+RD+C F ++
Sbjct: 1963 AVELLGRFVLCRPQLAEQYYDMLIERILDTGISVRKRVIKILRDICIEQPTFPKITEMCV 2022
Query: 331 KSLHRDNFTCY***IEYPAYSASDLQDLVCKTFYEFWFEEPSTPQTQVFKDGSTVPLEVA 390
K + R N ++ LV +TF + WF TP K+ T
Sbjct: 2023 KMIRRVN-------------DEEGIKKLVNETFQKLWF----TPTPHNDKEAMT------ 2059
Query: 391 KKTEQIVEMLKRLPN------NQLLVTVIKRCLTLDFLPQSAKASGVNPVSLVTVRKRCE 444
+K I +++ + QLL ++K S + S P V+K C
Sbjct: 2060 RKILNITDVVAACRDTGYDWFEQLLQNLLK----------SEEDSSYKP-----VKKACT 2104
Query: 445 LMCKCLLEKILQVDEMNSNELEK-----HALPYVLVLHAFCLVDPTLCAPASNPSQFVLT 499
+ L+E IL+ +E ++ K + + L F + P L + +T
Sbjct: 2105 QLVDNLVEHILKYEESLADSDNKGVNSGRLVACITTLFLFSKIRPQLMV------KHAMT 2158
Query: 500 LQPYLKTQVKTNLKTYILGSLQCYYALCIELLE 532
+QPYL T+ T ++ ++ L + L+E
Sbjct: 2159 MQPYLTTKCSTQNDFMVICNVAKILELVVPLME 2191
>dbj|BAA77335.1| IDN3 [Homo sapiens]
Length = 2265
Score = 119 bits (299), Expect = 2e-25
Identities = 116/453 (25%), Positives = 189/453 (41%), Gaps = 93/453 (20%)
Query: 129 VQQLLLNYFQDVTSADDLHHFICWFYLCSWYKNDPKCQQKP------------------- 169
+Q+ LL+Y + T D F FY+ W+++ +K
Sbjct: 1112 LQKALLDYLDENTETDPSLVFSRKFYIAQWFRDTTLETEKAMKSQKDEESSEGTHHAKEI 1171
Query: 170 -----IYYFARMKS---RTIVRDSGSVSSMLTR-------DSIKKITLALGQNSSFCRGF 214
I + A + R+I++ + S S L D I L F + F
Sbjct: 1172 ETTGQIMHRAENRKKFLRSIIKTTPSQFSTLKMNSDTVDYDDACLIVRYLASMRPFAQSF 1231
Query: 215 DKIFHTLLVSLKENSPVIRAKALRAVSIIVEADPEVLGDKFVQSSVEGRFCDTAISVREA 274
D +L L EN+ +R KA++ +S +V DP +L +Q V GR D + SVREA
Sbjct: 1232 DIYLTQILRVLGENAIAVRTKAMKCLSEVVAVDPSILARLDMQRGVHGRLMDNSTSVREA 1291
Query: 275 ALELVGRHIASHPDVGFKYFEKITERIKDTGVSVRKRAIKIIRDMCCSDGNFLRV----Y 330
A+EL+GR + P + +Y++ + ERI DTG+SVRKR IKI+RD+C F ++
Sbjct: 1292 AVELLGRFVLCRPQLAEQYYDMLIERILDTGISVRKRVIKILRDICIEQPTFPKITEMCV 1351
Query: 331 KSLHRDNFTCY***IEYPAYSASDLQDLVCKTFYEFWFEEPSTPQTQVFKDGSTVPLEVA 390
K + R N ++ LV +TF + WF TP K+ T
Sbjct: 1352 KMIRRVN-------------DEEGIKKLVNETFQKLWF----TPTPHNDKEAMT------ 1388
Query: 391 KKTEQIVEMLKRLPN------NQLLVTVIKRCLTLDFLPQSAKASGVNPVSLVTVRKRCE 444
+K I +++ + QLL ++K S + S P V+K C
Sbjct: 1389 RKILNITDVVAACRDTGYDWFEQLLQNLLK----------SEEDSSYKP-----VKKACT 1433
Query: 445 LMCKCLLEKILQVDEMNSNELEK-----HALPYVLVLHAFCLVDPTLCAPASNPSQFVLT 499
+ L+E IL+ +E ++ K + + L F + P L + +T
Sbjct: 1434 QLVDNLVEHILKYEESLADSDNKGVNSGRLVACITTLFLFSKIRPQLMV------KHAMT 1487
Query: 500 LQPYLKTQVKTNLKTYILGSLQCYYALCIELLE 532
+QPYL T+ T ++ ++ L + L+E
Sbjct: 1488 MQPYLTTKCSTQNDFMVICNVAKILELVVPLME 1520
>dbj|BAA77349.1| IDN3-B [Homo sapiens]
Length = 2158
Score = 119 bits (299), Expect = 2e-25
Identities = 116/453 (25%), Positives = 189/453 (41%), Gaps = 93/453 (20%)
Query: 129 VQQLLLNYFQDVTSADDLHHFICWFYLCSWYKNDPKCQQKP------------------- 169
+Q+ LL+Y + T D F FY+ W+++ +K
Sbjct: 1112 LQKALLDYLDENTETDPSLVFSRKFYIAQWFRDTTLETEKAMKSQKDEESSEGTHHAKEI 1171
Query: 170 -----IYYFARMKS---RTIVRDSGSVSSMLTR-------DSIKKITLALGQNSSFCRGF 214
I + A + R+I++ + S S L D I L F + F
Sbjct: 1172 ETTGQIMHRAENRKKFLRSIIKTTPSQFSTLKMNSDTVDYDDACLIVRYLASMRPFAQSF 1231
Query: 215 DKIFHTLLVSLKENSPVIRAKALRAVSIIVEADPEVLGDKFVQSSVEGRFCDTAISVREA 274
D +L L EN+ +R KA++ +S +V DP +L +Q V GR D + SVREA
Sbjct: 1232 DIYLTQILRVLGENAIAVRTKAMKCLSEVVAVDPSILARLDMQRGVHGRLMDNSTSVREA 1291
Query: 275 ALELVGRHIASHPDVGFKYFEKITERIKDTGVSVRKRAIKIIRDMCCSDGNFLRV----Y 330
A+EL+GR + P + +Y++ + ERI DTG+SVRKR IKI+RD+C F ++
Sbjct: 1292 AVELLGRFVLCRPQLAEQYYDMLIERILDTGISVRKRVIKILRDICIEQPTFPKITEMCV 1351
Query: 331 KSLHRDNFTCY***IEYPAYSASDLQDLVCKTFYEFWFEEPSTPQTQVFKDGSTVPLEVA 390
K + R N ++ LV +TF + WF TP K+ T
Sbjct: 1352 KMIRRVN-------------DEEGIKKLVNETFQKLWF----TPTPHNDKEAMT------ 1388
Query: 391 KKTEQIVEMLKRLPN------NQLLVTVIKRCLTLDFLPQSAKASGVNPVSLVTVRKRCE 444
+K I +++ + QLL ++K S + S P V+K C
Sbjct: 1389 RKILNITDVVAACRDTGYDWFEQLLQNLLK----------SEEDSSYKP-----VKKACT 1433
Query: 445 LMCKCLLEKILQVDEMNSNELEK-----HALPYVLVLHAFCLVDPTLCAPASNPSQFVLT 499
+ L+E IL+ +E ++ K + + L F + P L + +T
Sbjct: 1434 QLVDNLVEHILKYEESLADSDNKGVNSGRLVACITTLFLFSKIRPQLMV------KHAMT 1487
Query: 500 LQPYLKTQVKTNLKTYILGSLQCYYALCIELLE 532
+QPYL T+ T ++ ++ L + L+E
Sbjct: 1488 MQPYLTTKCSTQNDFMVICNVAKILELVVPLME 1520
>ref|NP_056199.2| delangin isoform B [Homo sapiens] gi|48143963|emb|CAG26691.1|
delangin [Homo sapiens]
Length = 2697
Score = 119 bits (299), Expect = 2e-25
Identities = 116/453 (25%), Positives = 189/453 (41%), Gaps = 93/453 (20%)
Query: 129 VQQLLLNYFQDVTSADDLHHFICWFYLCSWYKNDPKCQQKP------------------- 169
+Q+ LL+Y + T D F FY+ W+++ +K
Sbjct: 1651 LQKALLDYLDENTETDPSLVFSRKFYIAQWFRDTTLETEKAMKSQKDEESSEGTHHAKEI 1710
Query: 170 -----IYYFARMKS---RTIVRDSGSVSSMLTR-------DSIKKITLALGQNSSFCRGF 214
I + A + R+I++ + S S L D I L F + F
Sbjct: 1711 ETTGQIMHRAENRKKFLRSIIKTTPSQFSTLKMNSDTVDYDDACLIVRYLASMRPFAQSF 1770
Query: 215 DKIFHTLLVSLKENSPVIRAKALRAVSIIVEADPEVLGDKFVQSSVEGRFCDTAISVREA 274
D +L L EN+ +R KA++ +S +V DP +L +Q V GR D + SVREA
Sbjct: 1771 DIYLTQILRVLGENAIAVRTKAMKCLSEVVAVDPSILARLDMQRGVHGRLMDNSTSVREA 1830
Query: 275 ALELVGRHIASHPDVGFKYFEKITERIKDTGVSVRKRAIKIIRDMCCSDGNFLRV----Y 330
A+EL+GR + P + +Y++ + ERI DTG+SVRKR IKI+RD+C F ++
Sbjct: 1831 AVELLGRFVLCRPQLAEQYYDMLIERILDTGISVRKRVIKILRDICIEQPTFPKITEMCV 1890
Query: 331 KSLHRDNFTCY***IEYPAYSASDLQDLVCKTFYEFWFEEPSTPQTQVFKDGSTVPLEVA 390
K + R N ++ LV +TF + WF TP K+ T
Sbjct: 1891 KMIRRVN-------------DEEGIKKLVNETFQKLWF----TPTPHNDKEAMT------ 1927
Query: 391 KKTEQIVEMLKRLPN------NQLLVTVIKRCLTLDFLPQSAKASGVNPVSLVTVRKRCE 444
+K I +++ + QLL ++K S + S P V+K C
Sbjct: 1928 RKILNITDVVAACRDTGYDWFEQLLQNLLK----------SEEDSSYKP-----VKKACT 1972
Query: 445 LMCKCLLEKILQVDEMNSNELEK-----HALPYVLVLHAFCLVDPTLCAPASNPSQFVLT 499
+ L+E IL+ +E ++ K + + L F + P L + +T
Sbjct: 1973 QLVDNLVEHILKYEESLADSDNKGVNSGRLVACITTLFLFSKIRPQLMV------KHAMT 2026
Query: 500 LQPYLKTQVKTNLKTYILGSLQCYYALCIELLE 532
+QPYL T+ T ++ ++ L + L+E
Sbjct: 2027 MQPYLTTKCSTQNDFMVICNVAKILELVVPLME 2059
>gi|47458031 TPA: transcriptional regulator [Homo sapiens]
Length = 2804
Score = 119 bits (299), Expect = 2e-25
Identities = 116/453 (25%), Positives = 189/453 (41%), Gaps = 93/453 (20%)
Query: 129 VQQLLLNYFQDVTSADDLHHFICWFYLCSWYKNDPKCQQKP------------------- 169
+Q+ LL+Y + T D F FY+ W+++ +K
Sbjct: 1651 LQKALLDYLDENTETDPSLVFSRKFYIAQWFRDTTLETEKAMKSQKDEESSEGTHHAKEI 1710
Query: 170 -----IYYFARMKS---RTIVRDSGSVSSMLTR-------DSIKKITLALGQNSSFCRGF 214
I + A + R+I++ + S S L D I L F + F
Sbjct: 1711 ETTGQIMHRAENRKKFLRSIIKTTPSQFSTLKMNSDTVDYDDACLIVRYLASMRPFAQSF 1770
Query: 215 DKIFHTLLVSLKENSPVIRAKALRAVSIIVEADPEVLGDKFVQSSVEGRFCDTAISVREA 274
D +L L EN+ +R KA++ +S +V DP +L +Q V GR D + SVREA
Sbjct: 1771 DIYLTQILRVLGENAIAVRTKAMKCLSEVVAVDPSILARLDMQRGVHGRLMDNSTSVREA 1830
Query: 275 ALELVGRHIASHPDVGFKYFEKITERIKDTGVSVRKRAIKIIRDMCCSDGNFLRV----Y 330
A+EL+GR + P + +Y++ + ERI DTG+SVRKR IKI+RD+C F ++
Sbjct: 1831 AVELLGRFVLCRPQLAEQYYDMLIERILDTGISVRKRVIKILRDICIEQPTFPKITEMCV 1890
Query: 331 KSLHRDNFTCY***IEYPAYSASDLQDLVCKTFYEFWFEEPSTPQTQVFKDGSTVPLEVA 390
K + R N ++ LV +TF + WF TP K+ T
Sbjct: 1891 KMIRRVN-------------DEEGIKKLVNETFQKLWF----TPTPHNDKEAMT------ 1927
Query: 391 KKTEQIVEMLKRLPN------NQLLVTVIKRCLTLDFLPQSAKASGVNPVSLVTVRKRCE 444
+K I +++ + QLL ++K S + S P V+K C
Sbjct: 1928 RKILNITDVVAACRDTGYDWFEQLLQNLLK----------SEEDSSYKP-----VKKACT 1972
Query: 445 LMCKCLLEKILQVDEMNSNELEK-----HALPYVLVLHAFCLVDPTLCAPASNPSQFVLT 499
+ L+E IL+ +E ++ K + + L F + P L + +T
Sbjct: 1973 QLVDNLVEHILKYEESLADSDNKGVNSGRLVACITTLFLFSKIRPQLMV------KHAMT 2026
Query: 500 LQPYLKTQVKTNLKTYILGSLQCYYALCIELLE 532
+QPYL T+ T ++ ++ L + L+E
Sbjct: 2027 MQPYLTTKCSTQNDFMVICNVAKILELVVPLME 2059
>sp|Q6KC79|NIPBL_HUMAN Nipped-B-like protein (Delangin) (SCC2 homolog)
gi|47578105|ref|NP_597677.2| delangin isoform A [Homo
sapiens] gi|48143958|emb|CAF25290.1| delangin [Homo
sapiens]
Length = 2804
Score = 119 bits (299), Expect = 2e-25
Identities = 116/453 (25%), Positives = 189/453 (41%), Gaps = 93/453 (20%)
Query: 129 VQQLLLNYFQDVTSADDLHHFICWFYLCSWYKNDPKCQQKP------------------- 169
+Q+ LL+Y + T D F FY+ W+++ +K
Sbjct: 1651 LQKALLDYLDENTETDPSLVFSRKFYIAQWFRDTTLETEKAMKSQKDEESSEGTHHAKEI 1710
Query: 170 -----IYYFARMKS---RTIVRDSGSVSSMLTR-------DSIKKITLALGQNSSFCRGF 214
I + A + R+I++ + S S L D I L F + F
Sbjct: 1711 ETTGQIMHRAENRKKFLRSIIKTTPSQFSTLKMNSDTVDYDDACLIVRYLASMRPFAQSF 1770
Query: 215 DKIFHTLLVSLKENSPVIRAKALRAVSIIVEADPEVLGDKFVQSSVEGRFCDTAISVREA 274
D +L L EN+ +R KA++ +S +V DP +L +Q V GR D + SVREA
Sbjct: 1771 DIYLTQILRVLGENAIAVRTKAMKCLSEVVAVDPSILARLDMQRGVHGRLMDNSTSVREA 1830
Query: 275 ALELVGRHIASHPDVGFKYFEKITERIKDTGVSVRKRAIKIIRDMCCSDGNFLRV----Y 330
A+EL+GR + P + +Y++ + ERI DTG+SVRKR IKI+RD+C F ++
Sbjct: 1831 AVELLGRFVLCRPQLAEQYYDMLIERILDTGISVRKRVIKILRDICIEQPTFPKITEMCV 1890
Query: 331 KSLHRDNFTCY***IEYPAYSASDLQDLVCKTFYEFWFEEPSTPQTQVFKDGSTVPLEVA 390
K + R N ++ LV +TF + WF TP K+ T
Sbjct: 1891 KMIRRVN-------------DEEGIKKLVNETFQKLWF----TPTPHNDKEAMT------ 1927
Query: 391 KKTEQIVEMLKRLPN------NQLLVTVIKRCLTLDFLPQSAKASGVNPVSLVTVRKRCE 444
+K I +++ + QLL ++K S + S P V+K C
Sbjct: 1928 RKILNITDVVAACRDTGYDWFEQLLQNLLK----------SEEDSSYKP-----VKKACT 1972
Query: 445 LMCKCLLEKILQVDEMNSNELEK-----HALPYVLVLHAFCLVDPTLCAPASNPSQFVLT 499
+ L+E IL+ +E ++ K + + L F + P L + +T
Sbjct: 1973 QLVDNLVEHILKYEESLADSDNKGVNSGRLVACITTLFLFSKIRPQLMV------KHAMT 2026
Query: 500 LQPYLKTQVKTNLKTYILGSLQCYYALCIELLE 532
+QPYL T+ T ++ ++ L + L+E
Sbjct: 2027 MQPYLTTKCSTQNDFMVICNVAKILELVVPLME 2059
>ref|XP_425012.1| PREDICTED: similar to transcriptional regulator [Gallus gallus]
Length = 2380
Score = 117 bits (292), Expect = 1e-24
Identities = 113/447 (25%), Positives = 183/447 (40%), Gaps = 99/447 (22%)
Query: 129 VQQLLLNYFQDVTSADDLHHFICWFYLCSWYKNDPKCQQKPI------------------ 170
+Q+ LL+Y + T D F FY+ W+++ +K I
Sbjct: 1239 LQKALLDYLDENTETDASLVFSRKFYIAQWFRDTTMETEKAIKSQKDEDSSEGTHHAKDV 1298
Query: 171 ----YYFARMKSR-----TIVRDSGSVSSMLT--RDSIKK-----ITLALGQNSSFCRGF 214
R +SR +I++ + S S L D++ I L F + F
Sbjct: 1299 ETTGQIMHRAESRKKFLRSIIKTAPSQFSTLKIYSDTVDYEDACLIVRYLASMRPFAQSF 1358
Query: 215 DKIFHTLLVSLKENSPVIRAKALRAVSIIVEADPEVLGDKFVQSSVEGRFCDTAISVREA 274
D +L L EN+ +R KA++ +S +V DP +L +Q V GR D + SVREA
Sbjct: 1359 DIYLTQILRVLGENAIAVRTKAMKCLSEVVAVDPSILARPDMQRGVHGRLMDNSTSVREA 1418
Query: 275 ALELVGRHIASHPDVGFKYFEKITERIKDTGVSVRKRAIKIIRDMCCSDGNFLRV----Y 330
A+EL+GR + P + +Y++ + ERI DTG+SVRKR IKI+RD+C F ++
Sbjct: 1419 AVELLGRFVLCRPQLAEQYYDMLIERILDTGISVRKRVIKILRDICIEQPTFPKITEMCV 1478
Query: 331 KSLHRDNFTCY***IEYPAYSASDLQDLVCKTFYEFWFEEPSTPQTQVFKDGSTVPLEVA 390
K + R N ++ LV +TF + WF TP K+ T
Sbjct: 1479 KMIRRVN-------------DEEGIKKLVNETFQKLWF----TPTPHHDKEAMT------ 1515
Query: 391 KKTEQIVEMLKRLPNNQLLVTVIKRCLTLDFLPQSAKASGVNPVSLVTVRKRCELMCKCL 450
+K I ++L + + S V+K C + L
Sbjct: 1516 RKILNITDVLLKSEED---------------------------ASYKPVKKACTQLVDNL 1548
Query: 451 LEKILQVDEMNSNELEK-----HALPYVLVLHAFCLVDPTLCAPASNPSQFVLTLQPYLK 505
+E IL+ +E S+ K + + L F + P L + +T+QPYL
Sbjct: 1549 VEHILKYEESLSDSDNKGVNSGRLVACITTLFLFSKIRPQLMV------KHAMTMQPYLA 1602
Query: 506 TQVKTNLKTYILGSLQCYYALCIELLE 532
T+ T ++ ++ L + L+E
Sbjct: 1603 TKCSTQNDFMVICNVAKILELVVPLME 1629
>gb|AAD26161.2| adherin Nipped-B [Drosophila melanogaster]
gi|50400943|sp|Q7PLI2|NIPB_DROME Nipped-B protein (SCC2
homolog)
Length = 2077
Score = 116 bits (291), Expect = 2e-24
Identities = 87/331 (26%), Positives = 164/331 (49%), Gaps = 50/331 (15%)
Query: 188 VSSMLTRDSIKKITLALGQNSSFCRGFDKIFHTLLVSLKENSPVIRAKALRAVSIIVEAD 247
+ + + ++ + I L F + FD +++ + E S +R +A++ ++ IVE D
Sbjct: 1101 IKTYIDYNNAQLIAQYLATKRPFSQSFDGCLKKIILVVNEPSIAVRTRAMKCLANIVEVD 1160
Query: 248 PEVLGDKFVQSSVEGRFCDTAISVREAALELVGRHIASHPDVGFKYFEKITERIKDTGVS 307
P VL K +Q V +F DTAISVREAA++LVG+ + S+ D+ +Y++ ++ RI DTGVS
Sbjct: 1161 PLVLKRKDMQMGVNQKFLDTAISVREAAVDLVGKFVLSNQDLIDQYYDMLSTRILDTGVS 1220
Query: 308 VRKRAIKIIRDMCCSDGNFLRV----YKSLHRDNFTCY***IEYPAYSASDLQDLVCKTF 363
VRKR IKI+RD+C +F ++ K + R + +Q LV + F
Sbjct: 1221 VRKRVIKILRDICLEYPDFSKIPEICVKMIRR-------------VHDEEGIQKLVTEVF 1267
Query: 364 YEFWFEEPSTPQTQVFKDGSTVPLEVAKKTEQIVEMLKRLPN------NQLLVTVIK--- 414
+ WF TP T+ K G + +K I++++ + LL+++ K
Sbjct: 1268 MKMWF----TPCTKNDKIG------IQRKINHIIDVVNTAHDTGTTWLEGLLMSIFKPRD 1317
Query: 415 -RCLTLDFLPQSAKASGVNPVSLVTVRKRCELMCKCLLEKILQVDEMNSNELEKHALPYV 473
+ + + K + P+ +V C+ + L+++++++++ +++ + L +
Sbjct: 1318 NMLRSEGCVQEFIKKNSEPPMDIVIA---CQQLADGLVDRLIELEDTDNSRM----LGCI 1370
Query: 474 LVLHAFCLVDPTLCAPASNPSQFVLTLQPYL 504
LH V P L + +T++PYL
Sbjct: 1371 TTLHLLAKVRPQLLV------KHAITIEPYL 1395
>gb|EAA46003.2| CG17704-PF.3 [Drosophila melanogaster] gi|51951003|gb|EAA46004.2|
CG17704-PE.3 [Drosophila melanogaster]
gi|62862616|ref|NP_001015455.1| CG17704-PF.3 [Drosophila
melanogaster] gi|62862614|ref|NP_001015454.1|
CG17704-PE.3 [Drosophila melanogaster]
Length = 2069
Score = 116 bits (291), Expect = 2e-24
Identities = 87/331 (26%), Positives = 164/331 (49%), Gaps = 50/331 (15%)
Query: 188 VSSMLTRDSIKKITLALGQNSSFCRGFDKIFHTLLVSLKENSPVIRAKALRAVSIIVEAD 247
+ + + ++ + I L F + FD +++ + E S +R +A++ ++ IVE D
Sbjct: 1093 IKTYIDYNNAQLIAQYLATKRPFSQSFDGCLKKIILVVNEPSIAVRTRAMKCLANIVEVD 1152
Query: 248 PEVLGDKFVQSSVEGRFCDTAISVREAALELVGRHIASHPDVGFKYFEKITERIKDTGVS 307
P VL K +Q V +F DTAISVREAA++LVG+ + S+ D+ +Y++ ++ RI DTGVS
Sbjct: 1153 PLVLKRKDMQMGVNQKFLDTAISVREAAVDLVGKFVLSNQDLIDQYYDMLSTRILDTGVS 1212
Query: 308 VRKRAIKIIRDMCCSDGNFLRV----YKSLHRDNFTCY***IEYPAYSASDLQDLVCKTF 363
VRKR IKI+RD+C +F ++ K + R + +Q LV + F
Sbjct: 1213 VRKRVIKILRDICLEYPDFSKIPEICVKMIRR-------------VHDEEGIQKLVTEVF 1259
Query: 364 YEFWFEEPSTPQTQVFKDGSTVPLEVAKKTEQIVEMLKRLPN------NQLLVTVIK--- 414
+ WF TP T+ K G + +K I++++ + LL+++ K
Sbjct: 1260 MKMWF----TPCTKNDKIG------IQRKINHIIDVVNTAHDTGTTWLEGLLMSIFKPRD 1309
Query: 415 -RCLTLDFLPQSAKASGVNPVSLVTVRKRCELMCKCLLEKILQVDEMNSNELEKHALPYV 473
+ + + K + P+ +V C+ + L+++++++++ +++ + L +
Sbjct: 1310 NMLRSEGCVQEFIKKNSEPPMDIVIA---CQQLADGLVDRLIELEDTDNSRM----LGCI 1362
Query: 474 LVLHAFCLVDPTLCAPASNPSQFVLTLQPYL 504
LH V P L + +T++PYL
Sbjct: 1363 TTLHLLAKVRPQLLV------KHAITIEPYL 1387
>ref|XP_320088.2| ENSANGP00000010656 [Anopheles gambiae str. PEST]
gi|55235638|gb|EAA15100.2| ENSANGP00000010656 [Anopheles
gambiae str. PEST]
Length = 1518
Score = 113 bits (282), Expect = 2e-23
Identities = 89/334 (26%), Positives = 152/334 (44%), Gaps = 63/334 (18%)
Query: 204 LGQNSSFCRGFDKIFHTLLVSLKENSPVIRAKALRAVSIIVEADPEVLGDKFVQSSVEGR 263
L SF + +DK +++ ++E IR +A++ ++ IVE D VL K +Q V+ +
Sbjct: 659 LASKRSFSQSYDKYLQKIILVVREPVVAIRTRAMKCLANIVEVDQLVLARKDMQMGVQQK 718
Query: 264 FCDTAISVREAALELVGRHIASHPDVGFKYFEKITERIKDTGVSVRKRAIKIIRDMCCSD 323
DTAISVREAA++LVG++I S P++ +Y+E I++RI DTGVSVRKR IKI+RD+C
Sbjct: 719 LLDTAISVREAAVDLVGKYILSDPELIDQYYEMISQRILDTGVSVRKRVIKILRDIC--- 775
Query: 324 GNFLRVYKSLHRDNFTCY***IEYPAY---------------SASDLQDLVCKTFYEFWF 368
IEYPA+ +Q LV F WF
Sbjct: 776 ---------------------IEYPAHEKIPDICVKMIRRVNDEEGIQKLVMDVFMTMWF 814
Query: 369 EEPSTPQTQVFKDGSTVPLEVAKKTEQIVEMLKRLPNNQLLVTVIKRCLTLDFLPQSAKA 428
P + D +K QI++++ +++ L F P+ +K
Sbjct: 815 -TPCNDNDKAAMD---------RKITQIIDVV--CSSHETGTQGFDALLKTIFEPKESKD 862
Query: 429 SGVNPVSLV--TVRKRCELMCKCLLEKILQVDEMNSNELEKHALPYVLVLHAFCLVDPTL 486
+ T+ K C+ + L++ ++++ + L + + LH F + P L
Sbjct: 863 DNKKLKKEIPKTLIKACQQIVDGLVDATMRLEGAENTRL----VGCITALHLFAKIQPQL 918
Query: 487 CAPASNPSQFVLTLQPYLKTQVKTNLKTYILGSL 520
++L+PYL + + + + + S+
Sbjct: 919 LV------NHAMSLEPYLNMRCQNQIISKFISSI 946
>ref|XP_553247.1| ENSANGP00000028248 [Anopheles gambiae str. PEST]
gi|55235637|gb|EAL39093.1| ENSANGP00000028248 [Anopheles
gambiae str. PEST]
Length = 1425
Score = 113 bits (282), Expect = 2e-23
Identities = 89/334 (26%), Positives = 152/334 (44%), Gaps = 63/334 (18%)
Query: 204 LGQNSSFCRGFDKIFHTLLVSLKENSPVIRAKALRAVSIIVEADPEVLGDKFVQSSVEGR 263
L SF + +DK +++ ++E IR +A++ ++ IVE D VL K +Q V+ +
Sbjct: 659 LASKRSFSQSYDKYLQKIILVVREPVVAIRTRAMKCLANIVEVDQLVLARKDMQMGVQQK 718
Query: 264 FCDTAISVREAALELVGRHIASHPDVGFKYFEKITERIKDTGVSVRKRAIKIIRDMCCSD 323
DTAISVREAA++LVG++I S P++ +Y+E I++RI DTGVSVRKR IKI+RD+C
Sbjct: 719 LLDTAISVREAAVDLVGKYILSDPELIDQYYEMISQRILDTGVSVRKRVIKILRDIC--- 775
Query: 324 GNFLRVYKSLHRDNFTCY***IEYPAY---------------SASDLQDLVCKTFYEFWF 368
IEYPA+ +Q LV F WF
Sbjct: 776 ---------------------IEYPAHEKIPDICVKMIRRVNDEEGIQKLVMDVFMTMWF 814
Query: 369 EEPSTPQTQVFKDGSTVPLEVAKKTEQIVEMLKRLPNNQLLVTVIKRCLTLDFLPQSAKA 428
P + D +K QI++++ +++ L F P+ +K
Sbjct: 815 -TPCNDNDKAAMD---------RKITQIIDVV--CSSHETGTQGFDALLKTIFEPKESKD 862
Query: 429 SGVNPVSLV--TVRKRCELMCKCLLEKILQVDEMNSNELEKHALPYVLVLHAFCLVDPTL 486
+ T+ K C+ + L++ ++++ + L + + LH F + P L
Sbjct: 863 DNKKLKKEIPKTLIKACQQIVDGLVDATMRLEGAENTRL----VGCITALHLFAKIQPQL 918
Query: 487 CAPASNPSQFVLTLQPYLKTQVKTNLKTYILGSL 520
++L+PYL + + + + + S+
Sbjct: 919 LV------NHAMSLEPYLNMRCQNQIISKFISSI 946
>emb|CAF99211.1| unnamed protein product [Tetraodon nigroviridis]
Length = 1794
Score = 112 bits (279), Expect = 4e-23
Identities = 98/373 (26%), Positives = 165/373 (43%), Gaps = 60/373 (16%)
Query: 176 MKSRTIVRDS-GSVSSMLTRDSIKKITLALGQNSSFCRGFDKIFHTLLVSLKENSPVIRA 234
MK+ ++ S GS S + + I L F + FD +L L E++ +R
Sbjct: 779 MKASSVRSHSQGSNSETVEYEDSCLIVRYLASMRPFAQSFDIYLSQILRVLGESAIAVRT 838
Query: 235 KALRAVSIIVEADPEVLGDKFVQSSVEGRFCDTAISVREAALELVGRHIASHPDVGFKYF 294
KA++ +S +V D +L +Q V GR D + SVREAA+ELVGR + S P++ +Y+
Sbjct: 839 KAMKCLSEVVAVDSSILARVDMQRGVHGRLMDNSTSVREAAVELVGRFVLSRPELIEQYY 898
Query: 295 EKITERIKDTGVSVRKRAIKIIRDMCCSDGNFLRV----YKSLHRDNFTCY***IEYPAY 350
+ + ERI DTG+SVRKR IKI+RD+C F ++ K + R N
Sbjct: 899 DMLIERILDTGISVRKRVIKILRDICLEQPGFRKITEMCVKMIRRVN------------- 945
Query: 351 SASDLQDLVCKTFYEFWFEEPSTPQTQVFKDGSTVPLEVAKKTEQIVEMLKRLPNN---- 406
++ LV +TF + WF T+ + +K I +++ ++
Sbjct: 946 DEEGIKKLVNETFQKLWFTPTPGQDTEA----------MTRKILNITDVVLACKDSGYDW 995
Query: 407 --QLLVTVIKRCLTLDFLPQSAKASGVNPVSLVTVRKRCELMCKCLLEKILQVDEMNSNE 464
QLL ++K S + + P +K C + L+E IL+ ++ ++
Sbjct: 996 FEQLLQNLLK----------SEEQASYKP-----TKKACVQLVDNLVEHILKYEDSLADC 1040
Query: 465 LEK-----HALPYVLVLHAFCLVDPTLCAPASNPSQFVLTLQPYLKTQVKTNLKTYILGS 519
EK + LH F + L + +T+QPYL T+ T ++ +
Sbjct: 1041 EEKGFNSDRLVSCTTTLHLFSKIRAQLMV------KHAMTIQPYLTTKCNTQNDFMVICN 1094
Query: 520 LQCYYALCIELLE 532
+ L + L+E
Sbjct: 1095 VAKILELVVPLME 1107
>emb|CAG83997.1| unnamed protein product [Yarrowia lipolytica CLIB99]
gi|50545033|ref|XP_500068.1| hypothetical protein
[Yarrowia lipolytica]
Length = 1644
Score = 92.4 bits (228), Expect = 3e-17
Identities = 70/265 (26%), Positives = 129/265 (48%), Gaps = 32/265 (12%)
Query: 177 KSRTIVRDSGSVSSMLTRDSIKKITLALGQNSSFCRGFDKIFHTLLVSLKENSPVIRAKA 236
KS T S + S+ + + I +I L + ++ I T+ SL+++ IR++A
Sbjct: 699 KSMTDEISSRTNDSLDSANDISRILLHM----PLAESYNTILATICHSLEQSKIQIRSRA 754
Query: 237 LRAVSIIVEADPEVLGDKFVQSSVEGRFCDTAISVREAALELVGRHIASHPDVGFKYFEK 296
L++++ ++ DP+++ FV +S++ R D + SVR+AA+E+VG+++ P +YFE
Sbjct: 755 LKSLTTLLSYDPKIMRLPFVANSLKSRLVDASASVRDAAVEIVGKYVLLQPTKTTQYFES 814
Query: 297 ITERIKDTGVSVRKRAIKIIRDMCCSDGNFLRVYKSLHRDNFTCY***IEYPAYSASD-- 354
+ ER+ D + VRKR ++++RD+ ++ V S+ + IE Y D
Sbjct: 815 LFERVSDMSLGVRKRVLRLLRDLYATN-----VDNSVKLEI-------IEKLLYRTEDEE 862
Query: 355 --LQDLVCKTFYEFWFEEPSTPQTQVFKDGSTVPLEVAKKTEQIVEMLKRLPNNQLLVTV 412
+Q+L T E WF Q + KK+EQ+ + L R L ++
Sbjct: 863 ESVQELATSTLKEMWFMVAKLEDAQSM-------VATLKKSEQLAQYLAR-----FLDSI 910
Query: 413 IKRCLTLDFLPQSAKASGVNPVSLV 437
L D +P++ G +SL+
Sbjct: 911 FNPFLHKDPIPEATLQVGTQYISLL 935
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.327 0.139 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 880,077,616
Number of Sequences: 2540612
Number of extensions: 35125124
Number of successful extensions: 105047
Number of sequences better than 10.0: 748
Number of HSP's better than 10.0 without gapping: 349
Number of HSP's successfully gapped in prelim test: 399
Number of HSP's that attempted gapping in prelim test: 104095
Number of HSP's gapped (non-prelim): 1273
length of query: 543
length of database: 863,360,394
effective HSP length: 133
effective length of query: 410
effective length of database: 525,458,998
effective search space: 215438189180
effective search space used: 215438189180
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 78 (34.7 bits)
Medicago: description of AC149305.6


