

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149130.2 + phase: 0
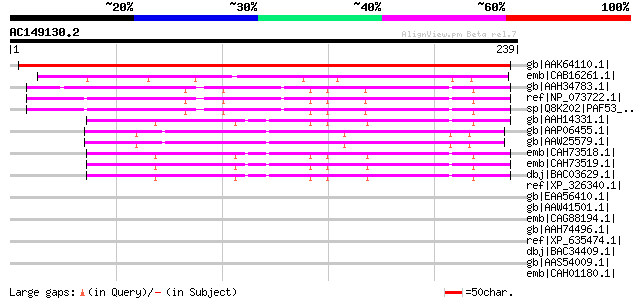
(239 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAK64110.1| unknown protein [Arabidopsis thaliana] gi|1343058... 216 6e-55
emb|CAB16261.1| SPAC2F3.03c [Schizosaccharomyces pombe] gi|19115... 65 1e-09
gb|AAH34783.1| Paf53 protein [Mus musculus] 50 5e-05
ref|NP_073722.1| RNA polymerase I associated factor 53 [Mus musc... 49 9e-05
sp|Q8K202|PAF53_MOUSE DNA-directed RNA polymerase I associated f... 49 9e-05
gb|AAH14331.1| RNA polymerase I associated factor 53 [Homo sapiens] 48 2e-04
gb|AAP06455.1| similar to NM_022811 RNA polymerase I associated ... 48 3e-04
gb|AAW25579.1| unknown [Schistosoma japonicum] 48 3e-04
emb|CAH73518.1| RP11-405L18.3 [Homo sapiens] gi|10436294|dbj|BAB... 47 5e-04
emb|CAH73519.1| RP11-405L18.3 [Homo sapiens] gi|62901107|sp|Q9GZ... 47 5e-04
dbj|BAC03629.1| unnamed protein product [Homo sapiens] 47 5e-04
ref|XP_326340.1| hypothetical protein [Neurospora crassa] gi|289... 44 0.003
gb|EAA56410.1| hypothetical protein MG06381.4 [Magnaporthe grise... 43 0.009
gb|AAW41501.1| expressed protein [Cryptococcus neoformans var. n... 42 0.015
emb|CAG88194.1| unnamed protein product [Debaryomyces hansenii C... 40 0.056
gb|AAH74496.1| MGC84821 protein [Xenopus laevis] gi|62901081|sp|... 40 0.056
ref|XP_635474.1| putative RNA polymerase I subunit [Dictyosteliu... 39 0.095
dbj|BAC34409.1| unnamed protein product [Mus musculus] 39 0.095
gb|AAS54009.1| AFR638Cp [Ashbya gossypii ATCC 10895] gi|45199156... 39 0.12
emb|CAH01180.1| unnamed protein product [Kluyveromyces lactis NR... 39 0.16
>gb|AAK64110.1| unknown protein [Arabidopsis thaliana] gi|13430584|gb|AAK25914.1|
unknown protein [Arabidopsis thaliana]
gi|11994365|dbj|BAB02324.1| unnamed protein product
[Arabidopsis thaliana] gi|15231416|ref|NP_188010.1|
expressed protein [Arabidopsis thaliana]
Length = 442
Score = 216 bits (549), Expect = 6e-55
Identities = 108/232 (46%), Positives = 155/232 (66%)
Query: 5 EEPEAKKNLDEKMKNVEVKETALANTEAHVTRHIPPYNSSATTPQEAYVLDKIILAEEWN 64
++ +K LD K+ + V AL T + V R+IPPYN++ATT EAY L+KII +W+
Sbjct: 211 DDAATQKVLDGKLSELGVNTAALEGTSSTVARNIPPYNTAATTANEAYPLEKIIEKGDWS 270
Query: 65 YLQDIYYSLQKEVAADFSLYPSFIRNRINRLKKIEDDSEKKKLSCILSFINYLVKFKDQH 124
+L+DIY+ LQ+E A YP F+RNR+ RL+ I+DD +K+ + + + +LVKFKD++
Sbjct: 271 FLEDIYWLLQQETEAATEAYPVFVRNRLYRLRDIKDDMKKQTVCGAATLLTHLVKFKDRN 330
Query: 125 SMDGISSSKFEKLPYTLYHRFTTMFDVTESRRLPPEKMNLLISYVLVLTLFSDDFRTDYR 184
SM+G S+K K+P +F +MF +ES R+P +K+NLLISYVLVL+L D+F TD
Sbjct: 331 SMNGYDSAKDHKMPDIFRQKFNSMFKDSESDRIPVDKINLLISYVLVLSLHVDNFMTDPE 390
Query: 185 DIVKDLRMSTLTLRPIFEHLGCKFISSQKVSYATLPIPLTFSQIKKRKRKNK 236
DI KDLR+ST+ LR F LGCKF+ + ATLP PL F ++ +R+R K
Sbjct: 391 DIAKDLRISTVELRKHFLQLGCKFLKQNSTTVATLPTPLNFPEVNRRRRARK 442
>emb|CAB16261.1| SPAC2F3.03c [Schizosaccharomyces pombe]
gi|19115294|ref|NP_594382.1| hypothetical protein
SPAC2F3.03c [Schizosaccharomyces pombe 972h-]
gi|12055095|emb|CAC20964.1| Rpa49 subunit specific to
nuclear RNA polymerase I [Schizosaccharomyces pombe]
gi|6094117|sp|O14086|RPA3_SCHPO DNA-directed RNA
polymerase I 49 kDa polypeptide (A49)
gi|7492543|pir||T38536 probable DNA-directed RNA
polymerase (EC 2.7.7.6) I 49K chain - fission yeast
(Schizosaccharomyces pombe)
Length = 425
Score = 65.5 bits (158), Expect = 1e-09
Identities = 66/252 (26%), Positives = 113/252 (44%), Gaps = 32/252 (12%)
Query: 14 DEKMKNVEVKETALANTEAHVT-----RHIPPYNSSATTPQEAYVLDKIILAEEWN--YL 66
D+ + NV+ AL E R IPP N A + ++AY L+ II EE++ Y+
Sbjct: 176 DQLVSNVQKATEALPTQEDIAAAQAQDRPIPPVNVGAESIEDAYKLEDIIPKEEFSAIYI 235
Query: 67 QDIYYSLQKEVAADFSLYPS--FIRNRINRLKKIEDDSEKKKLSCILSFINYLVKFKDQH 124
+ + + + A Y FI R RL IE+ +K+ + IL +I+ L F
Sbjct: 236 KPLLENPDERNWAKLLPYRHSLFINERFQRLLSIEEVDQKR--ARILYYISLLQAFLFSR 293
Query: 125 SMDGISSSKFEKL---PYTLYHRFTTMFDVTE---SRRLPPEKMNLLISYVLVLTLFSDD 178
G + +KL P L F T S ++ +++ +I Y+LVL L D+
Sbjct: 294 RSVGNRETLRKKLADPPEILIDGLIKRFTQTTGIGSVQVSSREVDKIICYILVLCLIVDN 353
Query: 179 FRTDYRDIVKDLRMSTLTLRPIFEHLGCK---FISSQKVSY------------ATLPIPL 223
+ TD + DL + T+ +F +GC+ + +Q+++ A L IPL
Sbjct: 354 YSTDVLTLANDLNVKTMKANELFRTVGCRIMAYTETQRMALGLNKTDAKNHKRAVLKIPL 413
Query: 224 TFSQIKKRKRKN 235
F + ++ + +N
Sbjct: 414 EFPKPRRGRARN 425
>gb|AAH34783.1| Paf53 protein [Mus musculus]
Length = 482
Score = 50.1 bits (118), Expect = 5e-05
Identities = 59/251 (23%), Positives = 117/251 (46%), Gaps = 29/251 (11%)
Query: 9 AKKNLDEKMKNVEVKETALANTEAHVTRHIPPYNSSATTPQEAYVLDKIILAEEWNYLQD 68
A+ +D K N V + A+ + + ++PP + A P++ Y + I+ E++ L+
Sbjct: 235 AESIIDTKGVNALVSD-AMQDDLQYDALYLPPCYADAAKPEDVYRFEDILSPAEYDALES 293
Query: 69 IYYSLQKEVAADF------SLYPSFIRNRINRLKKIE-DDSEKKKLSCILSFINYLVKFK 121
+ +K + D + + S++ I LK + D+ + + + + F++ L++F+
Sbjct: 294 PSEAFRKVTSEDILKMIEENSHCSYV---IEMLKSLPIDEVHRNRQARSIWFLDALIRFR 350
Query: 122 DQHSMDGISSSKFEKLPYT----LYHRFTTM-FDVTESRRLPPEKMNLLI-SYVLVLTLF 175
Q + G + +P+ L +FT + ++ + L M I SY ++L L
Sbjct: 351 AQKVIKG-KRALGPGIPHIINTKLLKQFTCLTYNNGRLQNLISSSMRAKITSYAIILALH 409
Query: 176 SDDFRTDYRDIVKDLRMSTLTLRPIFEHLGCKFISSQKVSYA----------TLPIPLTF 225
++F+ D + KDL++S + I + + K IS QKVS A TL +PL
Sbjct: 410 INNFQVDLTALQKDLKLSEKRIIEIAKAMRLK-ISKQKVSLADGREESHRLGTLSVPLPP 468
Query: 226 SQIKKRKRKNK 236
+Q R+ K +
Sbjct: 469 AQNSDRQSKRR 479
>ref|NP_073722.1| RNA polymerase I associated factor 53 [Mus musculus]
gi|1381029|dbj|BAA03266.1| RNA polymerase I associated
factor (PAF53) [Mus musculus]
Length = 434
Score = 49.3 bits (116), Expect = 9e-05
Identities = 59/251 (23%), Positives = 116/251 (45%), Gaps = 29/251 (11%)
Query: 9 AKKNLDEKMKNVEVKETALANTEAHVTRHIPPYNSSATTPQEAYVLDKIILAEEWNYLQD 68
A+ +D K N V + + + V ++PP + A P++ Y + I+ E++ L+
Sbjct: 187 AESIIDTKGVNALVSDAMQDDLQDGVL-YLPPCYADAAKPEDVYRFEDILSPAEYDALES 245
Query: 69 IYYSLQKEVAADF------SLYPSFIRNRINRLKKIE-DDSEKKKLSCILSFINYLVKFK 121
+ +K + D + + S++ I LK + D+ + + + + F++ L++F+
Sbjct: 246 PSEAFRKVTSEDILKMIEENSHCSYV---IEMLKSLPIDEVHRNRQARSIWFLDALIRFR 302
Query: 122 DQHSMDGISSSKFEKLPYT----LYHRFTTM-FDVTESRRLPPEKMNLLI-SYVLVLTLF 175
Q + G + +P+ L +FT + ++ + L M I SY ++L L
Sbjct: 303 AQKVIKG-KRALGPGIPHIINTKLLKQFTCLTYNNGRLQNLISSSMRAKITSYAIILALH 361
Query: 176 SDDFRTDYRDIVKDLRMSTLTLRPIFEHLGCKFISSQKVSYA----------TLPIPLTF 225
++F+ D + KDL++S + I + + K IS QKVS A TL +PL
Sbjct: 362 INNFQVDLTALQKDLKLSEKRMIEIAKAMRLK-ISKQKVSLADGREESHRLGTLSVPLPP 420
Query: 226 SQIKKRKRKNK 236
+Q R+ K +
Sbjct: 421 AQNSDRQSKRR 431
>sp|Q8K202|PAF53_MOUSE DNA-directed RNA polymerase I associated factor 53 kDa subunit
Length = 482
Score = 49.3 bits (116), Expect = 9e-05
Identities = 59/251 (23%), Positives = 116/251 (45%), Gaps = 29/251 (11%)
Query: 9 AKKNLDEKMKNVEVKETALANTEAHVTRHIPPYNSSATTPQEAYVLDKIILAEEWNYLQD 68
A+ +D K N V + + + V ++PP + A P++ Y + I+ E++ L+
Sbjct: 235 AESIIDTKGVNALVSDAMQDDLQDGVL-YLPPCYADAAKPEDVYRFEDILSPAEYDALES 293
Query: 69 IYYSLQKEVAADF------SLYPSFIRNRINRLKKIE-DDSEKKKLSCILSFINYLVKFK 121
+ +K + D + + S++ I LK + D+ + + + + F++ L++F+
Sbjct: 294 PSEAFRKVTSEDILKMIEENSHCSYV---IEMLKSLPIDEVHRNRQARSIWFLDALIRFR 350
Query: 122 DQHSMDGISSSKFEKLPYT----LYHRFTTM-FDVTESRRLPPEKMNLLI-SYVLVLTLF 175
Q + G + +P+ L +FT + ++ + L M I SY ++L L
Sbjct: 351 AQKVIKG-KRALGPGIPHIINTKLLKQFTCLTYNNGRLQNLISSSMRAKITSYAIILALH 409
Query: 176 SDDFRTDYRDIVKDLRMSTLTLRPIFEHLGCKFISSQKVSYA----------TLPIPLTF 225
++F+ D + KDL++S + I + + K IS QKVS A TL +PL
Sbjct: 410 INNFQVDLTALQKDLKLSEKRMIEIAKAMRLK-ISKQKVSLADGREESHRLGTLSVPLPP 468
Query: 226 SQIKKRKRKNK 236
+Q R+ K +
Sbjct: 469 AQNSDRQSKRR 479
>gb|AAH14331.1| RNA polymerase I associated factor 53 [Homo sapiens]
Length = 419
Score = 48.1 bits (113), Expect = 2e-04
Identities = 56/221 (25%), Positives = 98/221 (44%), Gaps = 24/221 (10%)
Query: 37 HIPPYNSSATTPQEAYVLDKIILAEEWNYLQ---DIYYSLQKEVAADFSLYPSFIRNRIN 93
++PP A P++ Y + ++ E+ LQ + + ++ E S I
Sbjct: 199 YLPPCYDDAAKPEDVYKFEDLLSPAEYEALQSPSEAFRNVTSEEILKMIEENSHCTFVIE 258
Query: 94 RLKKIEDDSEKK--KLSCILSFINYLVKFKDQHSMDGISSSKFEKLPYT----LYHRFTT 147
LK + D E + + CI F++ L+KF+ H + S+ +P+ L FT
Sbjct: 259 ALKSLPSDVESRDRQARCIW-FLDTLIKFR-AHRVVKRKSALGPGVPHIINTKLLKHFTC 316
Query: 148 M-FDVTESRRLPPEKMNLLIS-YVLVLTLFSDDFRTDYRDIVKDLRMSTLTLRPIFEHLG 205
+ ++ R L + M I+ YV++L L DF+ D + +DL++S + I + +
Sbjct: 317 LTYNNGRLRNLISDSMKAKITAYVIILALHIHDFQIDLTVLQRDLKLSEKRMMEIAKAMR 376
Query: 206 CKFISSQKVSYA----------TLPIPLTFSQIKKRKRKNK 236
K IS +KVS A TL +PL +Q R K +
Sbjct: 377 LK-ISKRKVSVAAGSEEDHKLGTLSLPLPPAQTSDRLAKRR 416
>gb|AAP06455.1| similar to NM_022811 RNA polymerase I associated factor (PAF53) in
Mus musculus [Schistosoma japonicum]
Length = 395
Score = 47.8 bits (112), Expect = 3e-04
Identities = 50/221 (22%), Positives = 94/221 (41%), Gaps = 25/221 (11%)
Query: 36 RHIPPYNSSATTPQEAYVLDKII-------LAEEWNYLQDIYYSLQKEVAADFSLYPSFI 88
R +PP + A+ P++ Y +K++ L E +L + S + S+YP ++
Sbjct: 175 RLLPPIDLKASRPEDVYPYNKLVPPKIVEALNSEVQHLLSLDTS-SVDTLLTKSIYPQWV 233
Query: 89 RNRINRLKKIEDDSEKKKLSCILSFINYLVKFKDQHSMDGISSSKFEKLPYTLYHRFTTM 148
+++ L + K ++ ++ + L F D + P + T
Sbjct: 234 MDKLCCLCSNKSADYSKHVTSLVLLSHMLALFH-LPKRDLMRKVPLPDTPNNVSKHLLTQ 292
Query: 149 FDVTESRR---------LPPEKMNLLISYVLVLTLFSDDFRTDYRDIVKDLRMSTLTLRP 199
F + SR+ + P + LI+++LVL L D+F+T ++ DL++ L
Sbjct: 293 FTASISRQGMGRLQVRAMSPMLRDKLITHILVLILHCDNFKTIIDNLTVDLKIGRDRLAQ 352
Query: 200 IFEHLGC---KFISSQKVS----YATLPIPLTFSQIKKRKR 233
F++LGC K ++QK S A L P+ K +R
Sbjct: 353 FFQYLGCRCTKVENAQKKSESNIIAELVTPVKLLDFKPSRR 393
>gb|AAW25579.1| unknown [Schistosoma japonicum]
Length = 277
Score = 47.8 bits (112), Expect = 3e-04
Identities = 50/221 (22%), Positives = 94/221 (41%), Gaps = 25/221 (11%)
Query: 36 RHIPPYNSSATTPQEAYVLDKII-------LAEEWNYLQDIYYSLQKEVAADFSLYPSFI 88
R +PP + A+ P++ Y +K++ L E +L + S + S+YP ++
Sbjct: 57 RLLPPIDLKASRPEDVYPYNKLVPPKIVEALNSEVQHLLSLDAS-SVDTLLTKSIYPQWV 115
Query: 89 RNRINRLKKIEDDSEKKKLSCILSFINYLVKFKDQHSMDGISSSKFEKLPYTLYHRFTTM 148
+++ L + K ++ ++ + L F D + P + T
Sbjct: 116 MDKLCCLCSNKSADYSKHVTSLVLLSHMLALFH-LPKRDLMRKVPLPDTPNNVSKHLLTQ 174
Query: 149 FDVTESRR---------LPPEKMNLLISYVLVLTLFSDDFRTDYRDIVKDLRMSTLTLRP 199
F + SR+ + P + LI+++LVL L D+F+T ++ DL++ L
Sbjct: 175 FTASISRQGMGRLQVRAMSPMLRDKLITHILVLILHCDNFKTIIDNLTVDLKIGRDRLAQ 234
Query: 200 IFEHLGC---KFISSQKVS----YATLPIPLTFSQIKKRKR 233
F++LGC K ++QK S A L P+ K +R
Sbjct: 235 FFQYLGCRCTKVENAQKKSESNIIAELVTPVKLLDFKPSRR 275
>emb|CAH73518.1| RP11-405L18.3 [Homo sapiens] gi|10436294|dbj|BAB14791.1| unnamed
protein product [Homo sapiens]
gi|10435392|dbj|BAB14579.1| unnamed protein product
[Homo sapiens] gi|12654979|gb|AAH01337.1| RNA polymerase
I associated factor 53 [Homo sapiens]
gi|48146743|emb|CAG33594.1| PAF53 [Homo sapiens]
gi|11968047|ref|NP_071935.1| RNA polymerase I associated
factor 53 [Homo sapiens]
Length = 419
Score = 47.0 bits (110), Expect = 5e-04
Identities = 55/221 (24%), Positives = 98/221 (43%), Gaps = 24/221 (10%)
Query: 37 HIPPYNSSATTPQEAYVLDKIILAEEWNYLQ---DIYYSLQKEVAADFSLYPSFIRNRIN 93
++PP A P++ Y + ++ E+ LQ + + ++ E S I
Sbjct: 199 YLPPCYDDAAKPEDVYKFEDLLSPAEYEALQSPSEAFRNVTSEEILKMIEENSHCTFVIE 258
Query: 94 RLKKIEDDSEKK--KLSCILSFINYLVKFKDQHSMDGISSSKFEKLPYT----LYHRFTT 147
LK + D E + + CI F++ L+KF+ H + S+ +P+ L FT
Sbjct: 259 ALKSLPSDVESRDRQARCIW-FLDTLIKFR-AHRVVKRKSALGPGVPHIINTKLLKHFTC 316
Query: 148 M-FDVTESRRLPPEKMNLLIS-YVLVLTLFSDDFRTDYRDIVKDLRMSTLTLRPIFEHLG 205
+ ++ R L + M I+ YV++L L DF+ D + +DL++S + I + +
Sbjct: 317 LTYNNGRLRNLISDSMKAKITAYVIILALHIHDFQIDLTVLQRDLKLSEKRMMEIAKAMR 376
Query: 206 CKFISSQKVSYA----------TLPIPLTFSQIKKRKRKNK 236
K IS ++VS A TL +PL +Q R K +
Sbjct: 377 LK-ISKRRVSVAAGSEEDHKLGTLSLPLPPAQTSDRLAKRR 416
>emb|CAH73519.1| RP11-405L18.3 [Homo sapiens] gi|62901107|sp|Q9GZS1|RPF53_HUMAN
DNA-directed RNA polymerase I associated factor 53 kDa
subunit
Length = 481
Score = 47.0 bits (110), Expect = 5e-04
Identities = 55/221 (24%), Positives = 98/221 (43%), Gaps = 24/221 (10%)
Query: 37 HIPPYNSSATTPQEAYVLDKIILAEEWNYLQ---DIYYSLQKEVAADFSLYPSFIRNRIN 93
++PP A P++ Y + ++ E+ LQ + + ++ E S I
Sbjct: 261 YLPPCYDDAAKPEDVYKFEDLLSPAEYEALQSPSEAFRNVTSEEILKMIEENSHCTFVIE 320
Query: 94 RLKKIEDDSEKK--KLSCILSFINYLVKFKDQHSMDGISSSKFEKLPYT----LYHRFTT 147
LK + D E + + CI F++ L+KF+ H + S+ +P+ L FT
Sbjct: 321 ALKSLPSDVESRDRQARCIW-FLDTLIKFR-AHRVVKRKSALGPGVPHIINTKLLKHFTC 378
Query: 148 M-FDVTESRRLPPEKMNLLIS-YVLVLTLFSDDFRTDYRDIVKDLRMSTLTLRPIFEHLG 205
+ ++ R L + M I+ YV++L L DF+ D + +DL++S + I + +
Sbjct: 379 LTYNNGRLRNLISDSMKAKITAYVIILALHIHDFQIDLTVLQRDLKLSEKRMMEIAKAMR 438
Query: 206 CKFISSQKVSYA----------TLPIPLTFSQIKKRKRKNK 236
K IS ++VS A TL +PL +Q R K +
Sbjct: 439 LK-ISKRRVSVAAGSEEDHKLGTLSLPLPPAQTSDRLAKRR 478
>dbj|BAC03629.1| unnamed protein product [Homo sapiens]
Length = 481
Score = 47.0 bits (110), Expect = 5e-04
Identities = 55/221 (24%), Positives = 98/221 (43%), Gaps = 24/221 (10%)
Query: 37 HIPPYNSSATTPQEAYVLDKIILAEEWNYLQ---DIYYSLQKEVAADFSLYPSFIRNRIN 93
++PP A P++ Y + ++ E+ LQ + + ++ E S I
Sbjct: 261 YLPPCYDDAAKPEDVYKFEDLLSPAEYEALQSPSEAFRNVTSEEILKMIEENSHCTFVIE 320
Query: 94 RLKKIEDDSEKK--KLSCILSFINYLVKFKDQHSMDGISSSKFEKLPYT----LYHRFTT 147
LK + D E + + CI F++ L+KF+ H + S+ +P+ L FT
Sbjct: 321 ALKSLPSDVESRDRQARCIW-FLDTLIKFR-AHRVVKRKSALGPGVPHIINTKLLKHFTC 378
Query: 148 M-FDVTESRRLPPEKMNLLIS-YVLVLTLFSDDFRTDYRDIVKDLRMSTLTLRPIFEHLG 205
+ ++ R L + M I+ YV++L L DF+ D + +DL++S + I + +
Sbjct: 379 LTYNNGRLRNLISDSMKAKITAYVIILALHIHDFQIDLTVLQRDLKLSEKRMMEIAKAMR 438
Query: 206 CKFISSQKVSYA----------TLPIPLTFSQIKKRKRKNK 236
K IS ++VS A TL +PL +Q R K +
Sbjct: 439 LK-ISKRRVSVAAGSEEDHKLGTLSLPLPPAQTSDRLAKRR 478
>ref|XP_326340.1| hypothetical protein [Neurospora crassa] gi|28918211|gb|EAA27889.1|
hypothetical protein [Neurospora crassa]
Length = 436
Score = 44.3 bits (103), Expect = 0.003
Identities = 52/240 (21%), Positives = 98/240 (40%), Gaps = 21/240 (8%)
Query: 8 EAKKNLDEKMKNVEVKETALANTEAHVTRHIPPYNSSATTPQEAYVLDKIILAEEWNYLQ 67
+A K + M E + A+ N R +P N +A + YV +II +E N +
Sbjct: 204 DAIKTATKDMSTKEDLQAAVDNA-----RPVPRGNFNADEIADVYVPSQIIGSEVLNAIP 258
Query: 68 DIYYSLQKEVAADFSLYPSFIRNRINRLKKIEDDSEKKKLSCILSFINYLVKFKDQHSMD 127
+ + + D + F+ R++ + +D + ++ L F+ Q
Sbjct: 259 VMDWQESVKNLEDIQVPSRFVAKRVSNVASSDDAVHRLRVLRYLLFVIIFWNATSQGRER 318
Query: 128 GISS-SKFEKL-------PYTLYHRFTTMFDVTESRRLPPEKMNLLISYVLVLTLFSDDF 179
G S K +KL P + F ++ + ++LL+++ D F
Sbjct: 319 GTRSIGKRDKLREVMAPAPEIVIENIRRKF--SDGGVMRKTHVDLLMTHCCAFAAIIDGF 376
Query: 180 RTDYRDIVKDLRMSTLTLRPIFEHLGCKFISSQKV-----SYATLPIPLTFSQIKKRKRK 234
R + D+ +DL++ L F +G + + QKV ++A L +PL F +I+ RK
Sbjct: 377 RVNTIDLREDLKLEQKQLNQYFIEIGAR-VKQQKVADKMENFAVLQLPLQFPKIRLPARK 435
>gb|EAA56410.1| hypothetical protein MG06381.4 [Magnaporthe grisea 70-15]
gi|39976957|ref|XP_369866.1| hypothetical protein
MG06381.4 [Magnaporthe grisea 70-15]
Length = 458
Score = 42.7 bits (99), Expect = 0.009
Identities = 44/212 (20%), Positives = 90/212 (41%), Gaps = 21/212 (9%)
Query: 38 IPPYNSSATTPQEAYVLDKIILAEEWNYLQDIYYSLQKEVAADFSLYPSFIRNRINRLKK 97
+P N A Q+ Y D+II ++ N + + + + L ++ NR+ R+ +
Sbjct: 253 VPKGNYDAAEIQDVYDPDQIIGSDVLNAIPVRDWQTANKNDENVQLTSRYVANRLPRVGE 312
Query: 98 IEDDSEKKKLSCILSFI-----------NYLVKFKDQHSMDGISSSKFEKLPYTLYHRFT 146
+ ++ ++ L F+ N + K + + S + + +L H+F
Sbjct: 313 HAESLKRLRILRYLYFLMTFIKIAKPGKNGVRKLPPRDKLREALSPAPDVVLDSLRHKFA 372
Query: 147 TMFDVTESRRLPPEKMNLLISYVLVLTLFSDDFRTDYRDIVKDLRMSTLTLRPIFEHLGC 206
D T+ R++ + L+++ VL D F D D+ +DL++ L F +G
Sbjct: 373 ---DGTQMRKIHAD---LIMTTCCVLASIIDSFEVDTFDLREDLQLEQKQLNSYFAEIGA 426
Query: 207 K----FISSQKVSYATLPIPLTFSQIKKRKRK 234
+ +K + A L +PL F ++ +RK
Sbjct: 427 RTKPVVNGDKKTTIAKLALPLQFPKVSMGRRK 458
>gb|AAW41501.1| expressed protein [Cryptococcus neoformans var. neoformans JEC21]
gi|50259891|gb|EAL22559.1| hypothetical protein CNBB4360
[Cryptococcus neoformans var. neoformans B-3501A]
gi|58262796|ref|XP_568808.1| expressed protein
[Cryptococcus neoformans var. neoformans JEC21]
Length = 427
Score = 42.0 bits (97), Expect = 0.015
Identities = 45/254 (17%), Positives = 101/254 (39%), Gaps = 39/254 (15%)
Query: 8 EAKKNLDEKMKNVEVKETALANTEAHVTRHIPPYNSSATTPQEAYVLDKIILAEEWNYLQ 67
+ K +L E + +E + ++A +E IP N+ P E Y + II +EW +
Sbjct: 178 DVKGHLMESIGELEEENDSVAPSE-----FIPNPNTDTADPTEVYTRESIITTQEWAAID 232
Query: 68 DIYYSLQKEVAADFSLYP----SFIRNRINRLKKIEDDSEKKK-------LSCILSFINY 116
++ S P S+++ + + I+D + +K LSC+L+ +++
Sbjct: 233 ASRLLAIEDDKERASALPYKRSSWLQYKSRAVANIKDKTARKTQMKYLYYLSCLLTLLDF 292
Query: 117 LVKFKDQ--HSMDGISSSKFEKLPYTLYHRFTTMFDVTESRR--LPPEKMNLLISYVLVL 172
+ H + + F ++P + + F + + + L+ ++ +L
Sbjct: 293 SPRLSKTPVHKL----APAFPQVPRQIIDGMLSRFAEPNGLKHNVTEKTKTKLLVWICLL 348
Query: 173 TLFSDDFRTDYRDIVKDLRMSTLTLRPIFEHLGCK---------------FISSQKVSYA 217
L + + + + K+L+M + +++ LGC + A
Sbjct: 349 YLILEGYSVEVAKVAKELKMEPAKVATLYKQLGCNVKLASPAEREAQGITLAQANAARRA 408
Query: 218 TLPIPLTFSQIKKR 231
L P+ F ++K+R
Sbjct: 409 VLVAPVRFPKLKRR 422
>emb|CAG88194.1| unnamed protein product [Debaryomyces hansenii CBS767]
gi|50422743|ref|XP_459948.1| unnamed protein product
[Debaryomyces hansenii]
Length = 415
Score = 40.0 bits (92), Expect = 0.056
Identities = 53/229 (23%), Positives = 90/229 (39%), Gaps = 32/229 (13%)
Query: 36 RHIPPYNSSATTPQEAYVLDKIILAEEWNYLQDIYYSLQKEVAADFSLYP----SFIRNR 91
R P N AT ++ Y L II EW+YL+ +++ L P ++
Sbjct: 186 RPTPIANVEATNVEDIYELYNIIPKREWSYLRVNSLFTEEDPKQRLELLPYDKSIYVSKH 245
Query: 92 INRLKKIEDDSEKKKLSCILSFI-----NYLVKFKDQHSMDGISSSKFEKLPYTLYHRFT 146
+ L +++EK +L S + N VK K Q M + + E L + RFT
Sbjct: 246 LAPLIS-SNNTEKLQLLYYASLLFGVYENRRVKDK-QTLMLKLQNKPSEILVDGILERFT 303
Query: 147 ----TMFDVTESRR--LPPEKMNLLISYVLVLTLFSDDFRTDYRDIVKDLRMSTLTLRPI 200
+ F ++ R + P + L+ Y+L + D+F + + +L M L +
Sbjct: 304 IPRASQFGKSKDRSFVIDPHHEDKLLCYLLAIIFHIDNFMVELPPLAHELNMKPTRLVSL 363
Query: 201 FEHLGCKF---------------ISSQKVSYATLPIPLTFSQIKKRKRK 234
+ LG +S+ ATL +P ++ KR RK
Sbjct: 364 CKALGAIIKSATVAQAEAFGIPKVSASTYKVATLKVPFKLPEMSKRVRK 412
>gb|AAH74496.1| MGC84821 protein [Xenopus laevis] gi|62901081|sp|Q6GLI9|RPF53_XENLA
DNA-directed RNA polymerase I associated factor 53 kDa
subunit
Length = 419
Score = 40.0 bits (92), Expect = 0.056
Identities = 45/250 (18%), Positives = 102/250 (40%), Gaps = 25/250 (10%)
Query: 8 EAKKNLDEKMKNVEVKETALANTEAHVTRHIPPYNSSATTPQEAYVLDKIILAEEWNYLQ 67
+A + + E E+ + A E + +PP + +A P+ AY D +I E+ L+
Sbjct: 169 KAAEEIIESRGTTELIKDAAEKREQDTSLFLPPCDFNADKPENAYKFDNLISPVEYAALE 228
Query: 68 DIYYSLQKEVAADFSLYPS------FIRNRINRLKKIEDDSEKKKLSCILSFINYLVKFK 121
+L+ + F+ ++ L++I+D+ + L +++ L+K
Sbjct: 229 TASAALRNITSEGLQQMVEEKKSGLFVLQELHGLREIKDEKALDHQARCLWYLDALIKLS 288
Query: 122 DQHSM---DGISSSKFEKLPYTLYHRFT--TMFDVTESRRLPPEKMNLLISYVLVLTLFS 176
++ D ++ + + L FT T + + +++Y++ + L
Sbjct: 289 QLRTVKRKDILTPECPSVVCWKLMKNFTVETYKNGRIQNAISGTTKTKIVAYIIAIALHI 348
Query: 177 DDFRTDYRDIVKDLRMSTLTLRPIFEHLGCKFISSQKVSYAT---------LPIPLTF-- 225
DF+ D + +D+++ + I + +G K S ++ L IPLT
Sbjct: 349 CDFQVDLTLLQRDMKLKESRILEIAKVMGLKIKKRMMYSESSIEEGHKIGLLTIPLTVYK 408
Query: 226 ---SQIKKRK 232
++K++K
Sbjct: 409 PSGGELKRKK 418
>ref|XP_635474.1| putative RNA polymerase I subunit [Dictyostelium discoideum]
gi|60463794|gb|EAL61970.1| putative RNA polymerase I
subunit [Dictyostelium discoideum]
Length = 375
Score = 39.3 bits (90), Expect = 0.095
Identities = 45/224 (20%), Positives = 103/224 (45%), Gaps = 34/224 (15%)
Query: 6 EPEAKKNLD--EKMKNVEVKETALANTEAHVTRH-IPPYNSSATTPQEAYVLDKIILAEE 62
E E +NLD + K+++ + LA+ E++ +P +++ ++ Y ++ I+
Sbjct: 142 EMENVENLDYEKTTKSIDAVKNELADIESNEKPFDLPDFDAETNDVRKIYNIEAILPPR- 200
Query: 63 WNYLQDIYYSLQKEVAADF--------SLYPSFIRNRINRLKKIEDDSEKKKLSC-ILSF 113
I++ L + D S P + ++++ R + I+D+ EK +C +L+F
Sbjct: 201 ------IFFGLNHQPFLDIISKTSEAPSYLPEYFKDQLKRFEDIKDE-EKVVYNCKLLTF 253
Query: 114 INY---LVKFKDQHS---MDGISSSKFEKLPYTLYHRFT-TMFDVTESRRLPPEKMNLLI 166
+ Y L+K K H +G +S L + +++++ +L +
Sbjct: 254 MWYSMILMKSKGGHEPLIKEGCTSEVINFLVSIFGSKINHKKYNISQDDKLK------IY 307
Query: 167 SYVLVLTLFSDDFR-TDYRDIVKDLRMSTLTLRPIFEHLGCKFI 209
+Y+ + L +DF+ TD + + + L +++ + F +GCK +
Sbjct: 308 NYLAIFALKLEDFQITDIKSLAQSLSLTSSSFEKHFTRVGCKVV 351
>dbj|BAC34409.1| unnamed protein product [Mus musculus]
Length = 393
Score = 39.3 bits (90), Expect = 0.095
Identities = 43/198 (21%), Positives = 93/198 (46%), Gaps = 18/198 (9%)
Query: 9 AKKNLDEKMKNVEVKETALANTEAHVTRHIPPYNSSATTPQEAYVLDKIILAEEWNYLQD 68
A+ +D K N V + + + V ++PP + A P++ Y + I+ E++ L+
Sbjct: 187 AESIIDTKGVNALVSDAMQDDLQDGVL-YLPPCYADAAKPEDVYRFEDILSPAEYDALES 245
Query: 69 IYYSLQKEVAADF------SLYPSFIRNRINRLKKIE-DDSEKKKLSCILSFINYLVKFK 121
+ +K + D + + S++ I LK + D+ + + + + F++ L++F+
Sbjct: 246 PSEAFRKVTSEDILKMIEENSHCSYV---IEMLKSLPIDEVHRNRQARSIWFLDALIRFR 302
Query: 122 DQHSMDGISSSKFEKLPYT----LYHRFTTM-FDVTESRRLPPEKMNLLI-SYVLVLTLF 175
Q + G + +P+ L +FT + ++ + L M I SY ++L L
Sbjct: 303 AQKVIKG-KRALGPGIPHIINTKLLKQFTCLTYNNGRLQNLISSSMRAKITSYAIILALH 361
Query: 176 SDDFRTDYRDIVKDLRMS 193
++F+ D + KDL++S
Sbjct: 362 INNFQVDLTALQKDLKLS 379
>gb|AAS54009.1| AFR638Cp [Ashbya gossypii ATCC 10895] gi|45199156|ref|NP_986185.1|
AFR638Cp [Eremothecium gossypii]
Length = 409
Score = 38.9 bits (89), Expect = 0.12
Identities = 45/224 (20%), Positives = 90/224 (40%), Gaps = 29/224 (12%)
Query: 36 RHIPPYNSSATTPQEAYVLDKIILAEEWNYLQDIYYSLQKEVAADFSLYP----SFIRNR 91
R IPP N AT ++ Y + II EWN+++ +K+ + P ++I +
Sbjct: 181 RPIPPPNVDATDVEQIYPVRSIIPQHEWNFIRVGPIMKEKDAQQRLQMLPYTKSAYITKK 240
Query: 92 INRLKKIEDDSEKKKLSCILSFINYLVKFKDQHSMDGI---SSSKFEKLPYTLYHRFTTM 148
+ L + EK +L LS + + + + + + +S + L + RFT +
Sbjct: 241 LPTLTQ-SVQMEKLQLLYYLSLLLGVYNNRRVSNKENLLEKLNSPADTLVDGILERFTVV 299
Query: 149 ----FDVTESR--RLPPEKMNLLISYVLVLTLFSDDFRTDYRDIVKDLRMSTLTLRPIFE 202
F ++ R + P++ + L+ Y+L L + D+F + +L + L +
Sbjct: 300 RPGQFGRSKDRSFAIDPQREDKLLCYILALIMHLDNFIVEISPFAHELGLKPSKLVNLLR 359
Query: 203 HLGCKF---------------ISSQKVSYATLPIPLTFSQIKKR 231
+G ++ ATL +P ++ KR
Sbjct: 360 TMGATVKGATVAQAQAFGIPKSAASTYKIATLKVPFKLPELNKR 403
>emb|CAH01180.1| unnamed protein product [Kluyveromyces lactis NRRL Y-1140]
gi|50304747|ref|XP_452329.1| unnamed protein product
[Kluyveromyces lactis]
Length = 412
Score = 38.5 bits (88), Expect = 0.16
Identities = 45/216 (20%), Positives = 92/216 (41%), Gaps = 25/216 (11%)
Query: 4 DEEPEAKKNLDEKMKNVEVKETALANTEAHVTRHIPPYNSSATTPQEAYVLDKIILAEEW 63
D A K L K E++ET ++ R P + AT ++ Y L II +EW
Sbjct: 161 DTVKTASKELPSKE---ELQETVSSD------RPTPLADLDATDVEQIYPLYNIIPKKEW 211
Query: 64 NYLQ--DIYYSLQKEVAADFSLYPSFIRNRINRLKKIEDDSEKKKLSCILSFINYLVKFK 121
++++ I E ++ Y F + N+L + ++ +KL +L +++ L
Sbjct: 212 SFIRVGSIMKEQDPEKRLEYFPYTKF-QYVANKLPTLTQAAQMQKLQ-LLYYLSLLAGVY 269
Query: 122 DQHSMDGISS--SKFEKLPYTLYHRFTTMFDVT----------ESRRLPPEKMNLLISYV 169
D ++ + + P L + F ++ +S + P++ + L+ Y+
Sbjct: 270 DNRRVNNKTKLLERINNPPDALLNGILERFTISRPGQFGRSKDKSFFIDPQREDKLLCYI 329
Query: 170 LVLTLFSDDFRTDYRDIVKDLRMSTLTLRPIFEHLG 205
LV+ + D+F + + + L + L +F+ LG
Sbjct: 330 LVIIMHLDNFIVEVPPLAQQLNLKPSKLVSLFKILG 365
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.319 0.134 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 384,378,561
Number of Sequences: 2540612
Number of extensions: 14855897
Number of successful extensions: 44866
Number of sequences better than 10.0: 69
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 61
Number of HSP's that attempted gapping in prelim test: 44831
Number of HSP's gapped (non-prelim): 75
length of query: 239
length of database: 863,360,394
effective HSP length: 124
effective length of query: 115
effective length of database: 548,324,506
effective search space: 63057318190
effective search space used: 63057318190
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 73 (32.7 bits)
Medicago: description of AC149130.2


