

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149079.6 - phase: 0
(483 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
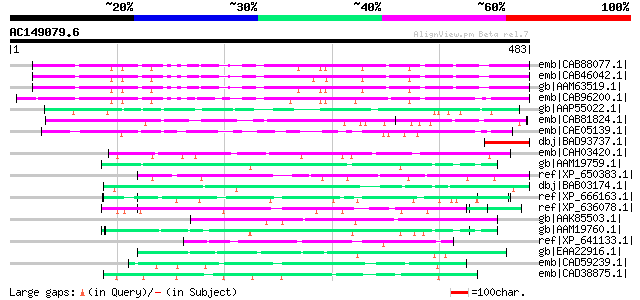
Score E
Sequences producing significant alignments: (bits) Value
emb|CAB88077.1| hypothetical protein [Arabidopsis thaliana] 206 1e-51
emb|CAB46042.1| glycoprotein homolog [Arabidopsis thaliana] gi|7... 206 2e-51
gb|AAM63519.1| glycoprotein homolog [Arabidopsis thaliana] 205 2e-51
emb|CAB96200.1| hypothetical protein [Capsella rubella] 197 8e-49
gb|AAP55022.1| unknown protein [Oryza sativa (japonica cultivar-... 136 2e-30
emb|CAB81824.1| putative protein [Arabidopsis thaliana] gi|15232... 130 7e-29
emb|CAE05139.1| OSJNBa0065H10.11 [Oryza sativa (japonica cultiva... 114 7e-24
dbj|BAD93737.1| glycoprotein homolog [Arabidopsis thaliana] 65 4e-09
emb|CAH03420.1| hypothetical protein [Paramecium tetraurelia] gi... 58 6e-07
gb|AAM19759.1| glutamic acid-rich protein cNBL1500 [Trichinella ... 57 1e-06
ref|XP_650383.1| hypothetical protein 223.t00011 [Entamoeba hist... 56 2e-06
dbj|BAB03174.1| unnamed protein product [Arabidopsis thaliana] g... 55 4e-06
ref|XP_666163.1| myosin heavy chain [Cryptosporidium hominis] gi... 55 5e-06
ref|XP_636078.1| hypothetical protein DDB0188541 [Dictyostelium ... 54 8e-06
gb|AAK85503.1| Hypothetical protein Y55B1BR.3 [Caenorhabditis el... 54 1e-05
gb|AAM19760.1| glutamic acid-rich protein cNBL1700 [Trichinella ... 54 1e-05
ref|XP_641133.1| hypothetical protein DDB0220644 [Dictyostelium ... 53 2e-05
gb|EAA22916.1| erythrocyte membrane-associated giant protein ant... 53 2e-05
emb|CAD59239.1| NAD(+) ADP-ribosyltransferase-3 [Dictyostelium d... 53 2e-05
emb|CAD38875.1| hypothetical protein [Homo sapiens] 52 3e-05
>emb|CAB88077.1| hypothetical protein [Arabidopsis thaliana]
Length = 471
Score = 206 bits (524), Expect = 1e-51
Identities = 185/516 (35%), Positives = 252/516 (47%), Gaps = 112/516 (21%)
Query: 22 QNQQEPNKFHYNFAYKATIVLIFFVILPLLPSQAPEFISQNLLTRNWELLHLLFVGIAIS 81
+ Q P KF+ F +KA I+ + ++P+ SQ PE +Q TR ELLHL+FVGIA+S
Sbjct: 14 KEDQNPRKFYSRFIFKALILTVLCAVVPVFLSQTPELANQ---TRLLELLHLVFVGIAVS 70
Query: 82 YGLFSRRNQEPD----KDNNNNTKFD----NAQSLVSRFLQVSSFFEDEVDHQNQSE--- 130
YGLFSRRN + N+++ K D N+ S V + L+VSS F V H+++SE
Sbjct: 71 YGLFSRRNYDGGGGGGTSNSDHNKADHSNNNSHSYVPKILEVSSVFN--VGHESESEPSD 128
Query: 131 --SEDINKIQTWSNQNQHYRNKPMVIVAPQVQQLQNSVFEDEQSSFNENEKPLLLPVRSL 188
S D K QTW N+ Y K P+V+ + F D SS N EKPLLLPVRSL
Sbjct: 129 DSSGDQRKFQTWKNK---YHMK-----IPEVE----TRFVDRVSSENR-EKPLLLPVRSL 175
Query: 189 K-SRLSDDDCVVQSQSVDGLSKTTTKRFSSNSFNRVRNYAEFEGLVEDKLKQKEEENA-V 246
SR+SD S D +S + +VR+ E LK ++N+ V
Sbjct: 176 NYSRVSDS-------SGD----------NSGRWEKVRSKREL-------LKTLGDDNSDV 211
Query: 247 LPSPIPWRSRSASARMMEPKQ---------------EAIEKALKASMVMEPKQEAN---E 288
LPSPIPWRSRS+S+ K+ + + K L S +++N
Sbjct: 212 LPSPIPWRSRSSSSSSSSSKEVESLPSVKNLTTVESQPLIKNLTPSSSFSSPRKSNPIPN 271
Query: 289 NASK---------------PSMVKTSSIKFTPSESVAKNSEDLIKKKSFYYKSY--PPPP 331
AS+ P+ +SS K P + E + K F + PPPP
Sbjct: 272 LASEFHPSPPPPPPPPPPLPAFYNSSSRKDHPGIYRVERRESSVHKTKFAGGEFHPPPPP 331
Query: 332 PPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEEGTNEK---EDEDEEEKEYSTSSMQEQ 388
PPPPP +YKS K R N R S + ++ +K D E KE T E+
Sbjct: 332 PPPPPVEYYKSPPTKFRLS--NERRKSSEQKMKRNAPKKVWWSDPIVESKEQDT----EK 385
Query: 389 TDKERFIEKRVILEAEETDSDEDEDVGELKQSVKSVKTEEPCSSSLSGGIDTVSDEGPDV 448
D+ + + + E+E + E+ E+ V+ EE S ++ G DV
Sbjct: 386 NDQRSNLGSKAVEESENGEQRRGEN--EIHDEVEKKIVEEEGVSEINNG--------SDV 435
Query: 449 DKKADEFIAKFREQIRLQRIESIKRST-RVARNSSR 483
DKKADEFIAKFREQIRLQRIESIKRST +++ NSSR
Sbjct: 436 DKKADEFIAKFREQIRLQRIESIKRSTNKISANSSR 471
>emb|CAB46042.1| glycoprotein homolog [Arabidopsis thaliana]
gi|7268429|emb|CAB78721.1| glycoprotein homolog
[Arabidopsis thaliana] gi|23506179|gb|AAN31101.1|
At4g16790/dl4420c [Arabidopsis thaliana]
gi|18086583|gb|AAL57715.1| AT4g16790/dl4420c
[Arabidopsis thaliana] gi|25407546|pir||B85187
glycoprotein homolog [imported] - Arabidopsis thaliana
gi|15235884|ref|NP_193412.1| hydroxyproline-rich
glycoprotein family protein [Arabidopsis thaliana]
Length = 473
Score = 206 bits (523), Expect = 2e-51
Identities = 183/516 (35%), Positives = 249/516 (47%), Gaps = 112/516 (21%)
Query: 22 QNQQEPNKFHYNFAYKATIVLIFFVILPLLPSQAPEFISQNLLTRNWELLHLLFVGIAIS 81
+ Q P KF+ F +KA I+ + ++P+ SQ PE +Q TR ELLHL+FVGIA+S
Sbjct: 16 KEDQNPRKFYSRFIFKALILTVLCAVVPVFLSQTPELANQ---TRLLELLHLVFVGIAVS 72
Query: 82 YGLFSRRNQEPD----KDNNNNTKFD----NAQSLVSRFLQVSSFFEDEVDHQNQSE--- 130
YGLFSRRN + N+++ K D N+ S V + L+VSS F V H+++SE
Sbjct: 73 YGLFSRRNYDGGGGGGTSNSDHNKADHSNNNSHSYVPKILEVSSVFN--VGHESESEPSD 130
Query: 131 --SEDINKIQTWSNQNQHYRNKPMVIVAPQVQQLQNSVFEDEQSSFNENEKPLLLPVRSL 188
S D K QTW N+ Y K P+V+ + F D SS N EKPLLLPVRSL
Sbjct: 131 DSSGDQRKFQTWKNK---YHMK-----IPEVE----TRFVDRVSSENR-EKPLLLPVRSL 177
Query: 189 K-SRLSDDDCVVQSQSVDGLSKTTTKRFSSNSFNRVRNYAEFEGLVEDKLKQKEEENA-V 246
SR+SD S D +S + +VR+ E LK ++N+ V
Sbjct: 178 NYSRVSDS-------SGD----------NSGRWEKVRSKREL-------LKTLGDDNSDV 213
Query: 247 LPSPIPWRSRSASARMMEPKQEAIEKALKASMVME----------PKQEANENASKP--- 293
LPSPIPWRSRS+S+ K+ ++K +E P ++ S P
Sbjct: 214 LPSPIPWRSRSSSSSSSSSKEVESLPSVKNLTTVESQPLIKNLTPPSSFSSPRKSNPIPN 273
Query: 294 --------------------SMVKTSSIKFTPSESVAKNSEDLIKKKSFYYKSY--PPPP 331
+ +SS K P + E + K F + PPPP
Sbjct: 274 LASEFHPSPPPPPPPPPPLPAFYNSSSRKDHPGIYRVERRESSVHKTKFAGGEFHPPPPP 333
Query: 332 PPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEEGTNEK---EDEDEEEKEYSTSSMQEQ 388
PPPPP +YKS K R N R S + ++ +K D E KE T E+
Sbjct: 334 PPPPPVEYYKSPPTKFRLS--NERRKSSEQKMKRNAPKKVWWSDPIVESKEQDT----EK 387
Query: 389 TDKERFIEKRVILEAEETDSDEDEDVGELKQSVKSVKTEEPCSSSLSGGIDTVSDEGPDV 448
D+ + + + E+E + E+ E+ V+ EE S ++ G DV
Sbjct: 388 NDQRSNLGSKAVEESENGEQRRGEN--EIHDEVEKKIVEEEGVSEINNG--------SDV 437
Query: 449 DKKADEFIAKFREQIRLQRIESIKRST-RVARNSSR 483
DKKADEFIAKFREQIRLQRIESIKRST +++ NSSR
Sbjct: 438 DKKADEFIAKFREQIRLQRIESIKRSTNKISANSSR 473
>gb|AAM63519.1| glycoprotein homolog [Arabidopsis thaliana]
Length = 473
Score = 205 bits (522), Expect = 2e-51
Identities = 185/516 (35%), Positives = 251/516 (47%), Gaps = 112/516 (21%)
Query: 22 QNQQEPNKFHYNFAYKATIVLIFFVILPLLPSQAPEFISQNLLTRNWELLHLLFVGIAIS 81
+ Q P KF+ F +KA I+ + ++P+ SQ PE +Q TR ELLHL+FVGIA+S
Sbjct: 16 KEDQNPRKFYSRFIFKALILTVLCAVVPVFLSQTPELANQ---TRLLELLHLVFVGIAVS 72
Query: 82 YGLFSRRNQEPD----KDNNNNTKFD----NAQSLVSRFLQVSSFFEDEVDHQNQSE--- 130
YGLFSRRN + N+++ K D N+ S V + L+VSS F V H+++SE
Sbjct: 73 YGLFSRRNYDGGGGGGTSNSDHNKADHSNNNSHSYVPKILEVSSVFN--VGHESESEPSD 130
Query: 131 --SEDINKIQTWSNQNQHYRNKPMVIVAPQVQQLQNSVFEDEQSSFNENEKPLLLPVRSL 188
S D K QTW N+ Y K P+V+ + F D SS N EKPLLLPVRSL
Sbjct: 131 DSSGDQRKFQTWKNK---YHMK-----IPEVE----TRFVDRVSSENR-EKPLLLPVRSL 177
Query: 189 K-SRLSDDDCVVQSQSVDGLSKTTTKRFSSNSFNRVRNYAEFEGLVEDKLKQKEEENA-V 246
SR+SD S D +S + +VR+ E LK ++N+ V
Sbjct: 178 NYSRVSDS-------SGD----------NSGRWEKVRSKREL-------LKTLGDDNSDV 213
Query: 247 LPSPIPWRSRSASARMMEPKQ---------------EAIEKALKASMVMEPKQEAN---E 288
LPSPIPWRSRS+S+ K+ + + K L S +++N
Sbjct: 214 LPSPIPWRSRSSSSSSSSSKEVESLPSVKNLTTVESQPLIKNLTPSSSFSSPRKSNPIPN 273
Query: 289 NASK---------------PSMVKTSSIKFTPSESVAKNSEDLIKKKSFYYKSY--PPPP 331
AS+ P+ +SS K P + E + K F + PPPP
Sbjct: 274 LASEFHPSPPPPPPPPPPLPAFYNSSSRKDHPGIYRVERRESSVHKTKFAGGEFHPPPPP 333
Query: 332 PPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEEGTNEK---EDEDEEEKEYSTSSMQEQ 388
PPPPP +YKS K R N R S + + +K D E KE T E+
Sbjct: 334 PPPPPVEYYKSPPTKFRLS--NERRKSSEQKMXRNAPKKVWWSDPIVESKEQDT----EK 387
Query: 389 TDKERFIEKRVILEAEETDSDEDEDVGELKQSVKSVKTEEPCSSSLSGGIDTVSDEGPDV 448
D+ + + + E+E + E+ E+ V+ EE S ++ G DV
Sbjct: 388 NDQRSNLGSKAVEESENGEQRRGEN--EIHDEVEKKIVEEEGVSEINNG--------SDV 437
Query: 449 DKKADEFIAKFREQIRLQRIESIKRST-RVARNSSR 483
DKKADEFIAKFREQIRLQRIESIKRST +++ NSSR
Sbjct: 438 DKKADEFIAKFREQIRLQRIESIKRSTNKISANSSR 473
>emb|CAB96200.1| hypothetical protein [Capsella rubella]
Length = 470
Score = 197 bits (500), Expect = 8e-49
Identities = 180/527 (34%), Positives = 258/527 (48%), Gaps = 108/527 (20%)
Query: 7 IQITKHQKPTLDNVKQNQQEPNKFHYNFAYKATIVLIFFVILPLLPSQAPEFISQNLLTR 66
++ +KPT K++Q P +F+ F +KA I+ + ++P+ SQ PE +Q TR
Sbjct: 2 VEAKSSKKPTQLGTKEDQN-PTRFYSRFIFKALILTVLCAVVPVFLSQTPELANQ---TR 57
Query: 67 NWELLHLLFVGIAISYGLFSRRNQEPD----KDNNNNTKFD-----NAQSLVSRFLQVSS 117
ELLHL+FVGIA+SYGLFSRRN + N++ K D N+ S V + L+VSS
Sbjct: 58 LLELLHLVFVGIAVSYGLFSRRNYDGGGAGGSSNSDYNKADHHNNNNSHSYVPKLLEVSS 117
Query: 118 FFEDEVDHQNQSE-----SEDINKIQTWSNQNQHYRNKPMVIVAPQVQQLQNSVFEDEQS 172
F VDH+++SE S D K Q W N+ Y K P+V+ + F D S
Sbjct: 118 VFN--VDHESESEPSDDSSGDHRKFQAWRNK---YHMK-----IPEVE----TRFVDRVS 163
Query: 173 SFNENEKPLLLPVRSLKSRLSDDDCVVQSQSVDGLSKTTTKRFSSNSFNRVRNYAEFEGL 232
S EKPLLLPVRSL V S D +S +++VR+ +
Sbjct: 164 S-EIREKPLLLPVRSLNYY------PVPDSSGD----------NSGRWDKVRSKRQL--- 203
Query: 233 VEDKLKQKEEENA-VLPSPIPWRSRSASA-----------RMMEPKQEAIEKALKASMVM 280
LK ++N+ VLPSPIPWRSRS+S+ + + + + K L S
Sbjct: 204 ----LKTLGDDNSDVLPSPIPWRSRSSSSSKEIESPPSIKNLTTVESQPLIKNLTPSSSY 259
Query: 281 EPKQEAN---ENASK---------------PSMVKTSSIKFTPSESVAKNSEDLIKKK-- 320
+++N AS+ P+ K+SS K P + E + K K
Sbjct: 260 SSPRKSNPIPNLASQFHPSPPPPPPPPPPLPAFYKSSSRKVQPGIYRVERRESVHKTKFE 319
Query: 321 SFYYKSYPPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEEGTNEK---EDEDEEE 377
+ + PPPPPPPPP +YKS K R + R + + ++ +K D E
Sbjct: 320 GGEFHNPPPPPPPPPPVEYYKSPPTKFRISSE--RRKTSEQKMKRNFPKKVWWSDPIVES 377
Query: 378 KEYSTSSMQEQTDKERFIEKRVILEAEETDSDEDEDVGELKQSVKSVKTEEPCSSSLSGG 437
KE T E+ D+ ++E + + E+ + E+ + K V+ E C S+
Sbjct: 378 KEQDT----EKNDQRSYLESKEAEDFEKEEQRRRENEMHEEVEKKIVEEERACESN---- 429
Query: 438 IDTVSDEGPDVDKKADEFIAKFREQIRLQRIESIKRS-TRVARNSSR 483
VSD VDKKADEFIAKFREQIRLQRIESIKRS ++++ N+SR
Sbjct: 430 --NVSD----VDKKADEFIAKFREQIRLQRIESIKRSASKISANTSR 470
>gb|AAP55022.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|37536866|ref|NP_922735.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
gi|13569990|gb|AAK31274.1| unknown protein [Oryza
sativa]
Length = 491
Score = 136 bits (342), Expect = 2e-30
Identities = 150/527 (28%), Positives = 211/527 (39%), Gaps = 141/527 (26%)
Query: 33 NFAYKATIVLIFFVILPLLPS-QAPEF---------ISQNLLTRNWELLHLLFVGIAISY 82
+F ++A + ++LPL+PS QAP + L + WELLHLL VGIA+SY
Sbjct: 11 SFPFRAAVFAACVLLLPLVPSPQAPAAGGDGGGGGGRGEAFLAKVWELLHLLVVGIAVSY 70
Query: 83 GLFSRRN----QEPDKDNNNNTKFDNAQSLVSRFLQVSSFFEDEVDHQNQSESEDINKIQ 138
GLFSRRN + +KD K D+A VS+ + S F+D +S N+++
Sbjct: 71 GLFSRRNNAGRRGDEKDAAAQAKADSA-GYVSQMIHDSLVFDDG-GGDVALDSPGGNRVR 128
Query: 139 TWSNQNQHYRNKPMVIVAP-QVQQLQNSVFEDEQSSFNENEKPLLLPVRSLKSRLSDDDC 197
+WS H+ ++P+V+VA ++ E Q + PL LPVR+LK +
Sbjct: 129 SWS--AMHHPDEPVVVVATGGAGGGRSHAVEAAQQA-----PPLSLPVRTLKPQGESSSS 181
Query: 198 VVQSQSVDGLSKTTTKRFSSNSFNRVRNYAEFEGLVEDKLKQKEEENAVLPSPIPWRSRS 257
+ + +R S ++ E VLPSPIPWRSRS
Sbjct: 182 AGYGDGGEPWA-ARPRRISQDTPGGGGGGHE----------------TVLPSPIPWRSRS 224
Query: 258 ASARMMEPKQEAIEKALKASMVMEPKQEANENASKPSMVKTSSIKFTPSESVAKNSEDLI 317
P + PS + S E++AK SED
Sbjct: 225 GRFDASAP-----------------------SPPSPSPKRLSPASSLSKETLAKASEDYS 261
Query: 318 KKKSFYYKSYPPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEEGTNEKEDEDEEE 377
++ YKS PP PPPPPP FL Y + KS +E E+
Sbjct: 262 SRRRSPYKSSPPAPPPPPP------PFLVHGYHPPAAERRTAAKSFKEELQEQTSHSFTT 315
Query: 378 KEYSTSSMQEQTDKERF-------------IEK--------RVILEAEETD--------- 407
E+S SS + K R + K R L+++ +
Sbjct: 316 SEFSRSSSNSSSAKPRISIDSSSSSSSYYPVAKSVRTIRGGRESLQSQSQEQPDVAVAGD 375
Query: 408 -------SDEDEDVGELK--QSVKSVKTEEPCSSSLSGGIDTVSDEGPD----------- 447
SD D+ G + QS+ + E S + G + TVS E D
Sbjct: 376 APALLHGSDSDDPYGGYRAYQSIPRFQYERGSSDPILGNV-TVSSESSDDDDSDVDGDGE 434
Query: 448 --------------------VDKKADEFIAKFREQIRLQRIESIKRS 474
VDKKA+EFIA+FREQIRLQRIESIK+S
Sbjct: 435 LSTRGNSPRRESSPEVDENEVDKKAEEFIARFREQIRLQRIESIKKS 481
>emb|CAB81824.1| putative protein [Arabidopsis thaliana]
gi|15232309|ref|NP_191597.1| expressed protein
[Arabidopsis thaliana] gi|11283517|pir||T47849
hypothetical protein T8B10.40 - Arabidopsis thaliana
Length = 743
Score = 130 bits (328), Expect = 7e-29
Identities = 133/507 (26%), Positives = 215/507 (42%), Gaps = 100/507 (19%)
Query: 34 FAYKATIVLIFFVILPLLPSQAPEFISQNLLTRNWELLHLLFVGIAISYGLFSRRNQEPD 93
F K+ + +F + LPL PSQAP+F+ + +LT+ WEL+HLLFVGIA++YGLFSRRN E
Sbjct: 32 FFCKSVLFALFLLALPLFPSQAPDFVGETVLTKFWELIHLLFVGIAVAYGLFSRRNVESA 91
Query: 94 KDNNNNTKFDNAQSLVSRFLQVSSFFEDEVDH---------------------------- 125
D +++ S VSR QVSS F++E D
Sbjct: 92 VDLRMTRVDESSLSYVSRIFQVSSVFDEEFDDNSCEFVDVRSDESVSARASVVGKSESFV 151
Query: 126 ------QNQSESEDINKIQTWSNQNQHYRNKPMVIVAPQVQQLQNSVFEDEQSSFNENEK 179
+ SE + N+++ W+ +Q+++ K V+VA L V +
Sbjct: 152 VESGELEESSEFGETNEVRAWN--SQYFQGKSKVVVARPAYGLDGHVV----------HQ 199
Query: 180 PLLLPVRSLKSRLSDDDCVVQSQSVDGLSKTTTKRFSSNSFNRVRNYAEFEGLVEDKLKQ 239
PL LP+R L+S L D+ + D AE E L+ D
Sbjct: 200 PLGLPIRRLRSSLRDNAALQDKSFADSCDGAVN--------------AEAESLLADNF-- 243
Query: 240 KEEENAVLPSPIPWRSRSASARMMEPKQEAIEK-ALKASMVMEPKQEANENASKPSMVKT 298
+E A SP+PW++R + + + ++ ++ + ++S+ S
Sbjct: 244 FDEVLAAPASPVPWQARPEMMGIGDNYPSNFQPISVDETLKSISSRSTGSSSSQTSYASQ 303
Query: 299 SSIKFTPSESVA-----KNSEDLIKKKSFYYK------SYPP----PPPPPPPTMFYKST 343
+ +F+PS SV+ N E+L+K+KS S PP P PP P + T
Sbjct: 304 NQNRFSPSRSVSAESLNSNVEELVKEKSRQSSSRSSSPSLPPSPSLSPSPPSPELVPNDT 363
Query: 344 FLKS--RYGEKNGRNMSMGKSIEEGTNEKED-----EDEEEKEYSTSSMQEQTDKERFIE 396
+S + R S + +G+ +ED E+E E E K+
Sbjct: 364 RRRSPELVTDDTPRRASHSRHYSDGSLLEEDVRRGFENELEGSKVRGRKAEFFSKKERGS 423
Query: 397 KRVILEAEETDSDEDEDVGELKQSVKSVKTEEPCS-SSLSGGIDTVSDEGPDVDKKADEF 455
K + L AE + ++ KS ++ P S SS GG D + D+ +K++
Sbjct: 424 KSLNLAAESS-----------RRGNKSRRSYPPESISSPVGGADDSTTRRQDLQQKSNCH 472
Query: 456 IAKFREQIRLQRIESIKRSTRVARNSS 482
+ E IR + +E+ + RV + S
Sbjct: 473 L--LEENIR-KGVEADHNNLRVKKGRS 496
Score = 48.1 bits (113), Expect = 6e-04
Identities = 45/135 (33%), Positives = 64/135 (47%), Gaps = 19/135 (14%)
Query: 360 GKSIEEGTNEKEDEDEEEKEY-----STSSMQEQTD----KERFIEKRVILEAEETDSDE 410
GK ++ + ED E + E S QE+ +E+ E R E EE +E
Sbjct: 612 GKDVKTDGDSSEDRAEAKVESRGRTKSRRPRQEELSIVLHQEKSSETRAKSEPEEVAMEE 671
Query: 411 DEDVGELKQSVKSVKTEEPCSSSLSGGIDTVSDEGPDVDKKADEFIAKFREQIRLQRIES 470
+ E + V + EE S S S + +VD+KA EFIAKFREQIRLQ++ S
Sbjct: 672 PQ--AEQQPEVTFEEEEEAAWESQSNA----SHDHNEVDRKAGEFIAKFREQIRLQKLIS 725
Query: 471 IKR----STRVARNS 481
++ T + RNS
Sbjct: 726 GEQPRGGGTGIFRNS 740
>emb|CAE05139.1| OSJNBa0065H10.11 [Oryza sativa (japonica cultivar-group)]
Length = 412
Score = 114 bits (285), Expect = 7e-24
Identities = 130/474 (27%), Positives = 197/474 (41%), Gaps = 131/474 (27%)
Query: 30 FHYNFAYKATIVLIFFVILPLLP-SQAPEFISQNLLTRNWELLHLLFVGIAISYGLFSRR 88
F N A KA + + +LPLLP SQAP R WEL H+L +G+ ISYG+F +R
Sbjct: 29 FPTNLAAKAVLFAVVVALLPLLPTSQAP---------RIWELPHILLLGLIISYGVFGQR 79
Query: 89 NQEPDKDNNNNTKF---DNAQSLVSRFLQVSSFFEDEVDHQNQSESEDINKIQTWSNQNQ 145
N + + TK ++ +S V++ + FE E D ++++ +Q WS +Q
Sbjct: 80 NADSEVAAVAATKTVDDESVESYVTQMMHGPLVFE-ENDGGGEADAAGKEGVQAWS--SQ 136
Query: 146 HYRNKPMVIVAPQVQQLQNSVFEDEQSSFNENEKPLLLPVRSLKSRLSDDDCVVQSQSVD 205
++ + P+V+VA ++ +E EKPLLLPVR LK +
Sbjct: 137 YFPDDPLVVVA-------DAGAGSNTGKGDEREKPLLLPVRKLKPATEE----------- 178
Query: 206 GLSKTTTKRFSSNSFNRVRNYAEFEGLVEDKLKQKEEENAVLPSPIPWRSRSASARMMEP 265
S T T+ FS +G +E++ +++EEE L AR
Sbjct: 179 --SATLTESFS-------------DGAIEEEEEEEEEETEFL---------LRKARYGGV 214
Query: 266 KQEAIEKALKASMVMEPKQEANENASKPSMVKTSSIKFTPSESVAKNSEDLIKKKSFYYK 325
++ AI PS V + + +P S L+
Sbjct: 215 REHAI--------------------PSPSSVLDADLTLSPC------SPPLLPP------ 242
Query: 326 SYPPPPPPPPPTMFYKSTFLK-------SRYGE----------KNGRNMSMGKSIEEG-- 366
PPPPPPPPP + + L+ + YG G N +I+
Sbjct: 243 --PPPPPPPPPFLDHDRPALRKAKARSFNDYGRVGLQTAAGCGGGGHNFRSKSAIQASRS 300
Query: 367 ---TNEKEDEDEEEK-------EYSTSSMQEQTDKERFIEKRVI-LEAEETDSDE-DEDV 414
T+ +D D EEK +S+ + ++ K + E EE D D D+D
Sbjct: 301 TFPTSPFDDHDLEEKVAASDISSFSSDDVVTDDGEDGDNHKEIYNYEEEEGDVDRLDDDD 360
Query: 415 GELKQSVKSVKTEEPCSSSLSGGIDTVSDEGPDVDKKADEFIAKFREQIRLQRI 468
G + + + T + V DE VD+KADEFIAKFREQIR+QR+
Sbjct: 361 GSCDEELFELATRLAPEEE-----EVVEDE---VDRKADEFIAKFREQIRMQRV 406
>dbj|BAD93737.1| glycoprotein homolog [Arabidopsis thaliana]
Length = 59
Score = 65.5 bits (158), Expect = 4e-09
Identities = 34/42 (80%), Positives = 38/42 (89%), Gaps = 1/42 (2%)
Query: 443 DEGPDVDKKADEFIAKFREQIRLQRIESIKRST-RVARNSSR 483
+ G DVDKKADEFIAKFREQIRLQRIESIKRST +++ NSSR
Sbjct: 18 NNGSDVDKKADEFIAKFREQIRLQRIESIKRSTNKISANSSR 59
>emb|CAH03420.1| hypothetical protein [Paramecium tetraurelia]
gi|50405059|ref|YP_054151.1| hypothetical protein
PTMB.222 [Paramecium tetraurelia]
Length = 2301
Score = 58.2 bits (139), Expect = 6e-07
Identities = 92/413 (22%), Positives = 169/413 (40%), Gaps = 57/413 (13%)
Query: 93 DKDNNNN----TKFDNAQSLVSRFLQVSSFFEDEVDHQNQSES-----EDINKIQTWSNQ 143
D +N NN + +N Q+ L + E EVD + Q+E+ EDIN Q SN
Sbjct: 601 DDENLNNIVEVVEKENTQNQEDMILNDNQNIEQEVDQEIQNETIQNNIEDIN--QDNSNN 658
Query: 144 NQHYRNKPMVIVAPQV---QQLQNSVFEDEQ------SSFNENEKPLLLPVRSLKSRLSD 194
Q ++ + V Q+ Q + V +++Q S+ ++ + + V + +++
Sbjct: 659 KQQILDEQNIQVTEQILNQQDDEQQVMDEQQDMVQDNSNLEQDNQDINNQVETQSNQIKQ 718
Query: 195 DDCVVQSQSVDGLSKTTTKRFSSNSFNRVRNYAEFEGLVEDKLKQKEEENAVLPSPIPWR 254
+ V+ Q DG T + N + N + E + E KL+ EE L I
Sbjct: 719 NPTEVEQQQQDG-----TCEQNENPERILENNKKVEDINEQKLEI--EETGELVQNIQIL 771
Query: 255 SRSASARMMEPKQEAI-------EKALKASMVME-PKQEANENASKPSMVKTSSIKFTPS 306
+S ++ +M+ + E+ EK L+ E KQE N PS++ I+
Sbjct: 772 DQSQNSMLMQDQNESQVQSSPLKEKDLENQNQEEIQKQENNNQGDDPSLLTVEQIQEEQP 831
Query: 307 ES----VAKNSEDL-----IKKKSFYYKSYPPPPPPPPPTMFYKSTFLKSRYGEKNGRNM 357
+ + K ED IK S + P P P ++T + + + N
Sbjct: 832 QQTDQIIKKEEEDQDTFKQIKNSSEIIEENPIDQPQPRDD---QNTEIIQTHSDVNDTQN 888
Query: 358 SMGKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDKERFIEKRVILEAEETD-SDEDEDVGE 416
+ +++ + ++ DE T ++Q Q D + IE V +ET+ +D E++ E
Sbjct: 889 KSQEEVQDLSKDQILNDE------TLNIQNQKDNQDIIEDVVNQANDETNQNDTQENINE 942
Query: 417 LKQSVKSVKTEEPCSSSLSGGIDTVSDEG---PDVDKKADEFIAKFREQIRLQ 466
KQ + + + ++ + GI ++ D + ++ I K QI Q
Sbjct: 943 KKQIQEDITEKNEENNEENQGISNQLEDNLLQSDKELAEEQNIVKDSSQISKQ 995
Score = 37.0 bits (84), Expect = 1.4
Identities = 20/76 (26%), Positives = 44/76 (57%), Gaps = 5/76 (6%)
Query: 356 NMSMGKSIEEGTNEKEDEDEEEKEYSTSSMQE---QTDKERFIEKRVILEAEETDSDEDE 412
N++ K I+E EK +E+ EE + ++ +++ Q+DKE E+ ++ ++ + DE
Sbjct: 939 NINEKKQIQEDITEKNEENNEENQGISNQLEDNLLQSDKELAEEQNIVKDSSQISKQGDE 998
Query: 413 DVGELKQSVKSVKTEE 428
D + K+ ++ +TE+
Sbjct: 999 D--QNKEIQENFETEQ 1012
Score = 34.7 bits (78), Expect = 6.9
Identities = 29/110 (26%), Positives = 49/110 (44%), Gaps = 18/110 (16%)
Query: 86 SRRNQEPDKDNNNNTKFDNAQSL----VSRFLQ--VSSFFEDEVDHQNQS--------ES 131
+ NQ D NN T D +Q + S LQ V + ED+ + Q Q E
Sbjct: 1143 NEENQNSDNIQNNETSKDTSQQIQVNDPSNVLQTIVQAHIEDQFEFQEQENIESNPLVEP 1202
Query: 132 EDINKIQTWSNQNQHYRNKPMVIVAPQVQQLQNSVFEDEQSSFNENEKPL 181
++ I+ NQNQ+ ++ V Q+ Q Q D++++ + N+K +
Sbjct: 1203 QEEQIIEDKDNQNQNNLDE----VVNQISQNQPETINDDENTKDLNQKEI 1248
>gb|AAM19759.1| glutamic acid-rich protein cNBL1500 [Trichinella spiralis]
Length = 498
Score = 57.4 bits (137), Expect = 1e-06
Identities = 77/376 (20%), Positives = 148/376 (38%), Gaps = 24/376 (6%)
Query: 86 SRRNQEPDKDNNNNTKFDNAQSLVSRFLQVSSFFEDEVDHQNQSESEDINKIQTWSNQNQ 145
S ++E D N+K + S ++E + ++ + ED ++ Q + + +
Sbjct: 128 SEASEEKDVSQEQNSKEEKGASEEDEDTPEEQNSKEE-NGSSEEDDEDASEEQASNEEKE 186
Query: 146 HYRNKPMVIVAPQ-VQQLQNSVFEDEQSSFNENEKPLLLPVRSLKSRLSDDDCVVQSQSV 204
K V + + ++ +D S E++ S D++ + Q+
Sbjct: 187 ASEEKNTVSEERKGASEEEDEEKDDGHESEVESQASEEQTTEEGASEEEDEESASEEQTS 246
Query: 205 DGLSKTTTKRFSSNSFNR----VRNYAEFEGLVEDKLKQKEEENAVLPSPIPWRSRSASA 260
+G K ++ + N V + A E E++ EEE+ S SA
Sbjct: 247 EGEEKGASQEEEEDEGNEQESEVESQASEEQTSEEEESASEEEDEENESKEQTTEEEESA 306
Query: 261 RMMEPKQEAIEKALKASMVMEPKQEANENASKPSMVKTSSIKFTPSESVAKNSEDLIKKK 320
E ++ A E+ K + E + E NE+ + + + S+ + ES ++ E K
Sbjct: 307 SEEEDEESASEREEKNASQEEEEDEGNESKEQTTEEEESASEEEDEESASEGEE----KN 362
Query: 321 SFYYKSYPPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKS--IEEGTNEKEDEDEEEK 378
+ + S S EK G + + EE T+E+E+E E+
Sbjct: 363 ASQEEEEDEGNEQESEVESQASEEQTSEEEEKEGASQEEDEENESEEQTSEEEEEGASEE 422
Query: 379 EYSTSSMQEQTDKERFIEKRVILEAEETDSDEDEDVGELKQSVKSVKTEEPCSSSLSGGI 438
E S+ +EQT +E E++ + EE D + +++ V+S +EE S
Sbjct: 423 EDEESAFEEQTSEEE--EEKGASQEEEEDEENEQE-----SEVESQASEEQTSEE----- 470
Query: 439 DTVSDEGPDVDKKADE 454
+ S+EG D ++ DE
Sbjct: 471 EGASEEGQDASEEEDE 486
Score = 44.7 bits (104), Expect = 0.007
Identities = 63/335 (18%), Positives = 131/335 (38%), Gaps = 41/335 (12%)
Query: 90 QEPDKDNNNNTKFDNAQSLVSRFLQVSSFFEDEVDHQNQSESEDINKIQTWSNQNQHYRN 149
++ +KD+ + ++ ++ S + +S EDE + SE K + + + N
Sbjct: 205 EDEEKDDGHESEVESQASEEQTTEEGASEEEDEESASEEQTSEGEEKGAS-QEEEEDEGN 263
Query: 150 KPMVIVAPQVQQLQNSVFEDEQSSFNENEKPLLLPVRSLKSRLSDDDCVVQSQSVDGLSK 209
+ V Q + Q S E+E +S E+E+ S + +++ + + + S+
Sbjct: 264 EQESEVESQASEEQTSE-EEESASEEEDEEN-----ESKEQTTEEEESASEEEDEESASE 317
Query: 210 TTTKRFSSNSFNRVRNYAEFEGLVEDKLKQKEEENAVLPSPIPWRSRSASARMMEPKQEA 269
K S N ++ E E++ EEE+ S ++ S E ++
Sbjct: 318 REEKNASQEEEEDEGNESK-EQTTEEEESASEEEDEESASEGEEKNASQEEEEDEGNEQE 376
Query: 270 IEKALKASMVMEPKQEANENASKPSMVKTSSIKFTPSESVAKNSEDLIKKKSFYYKSYPP 329
E +AS ++E E AS+ + S + T E SE+ ++ +F ++
Sbjct: 377 SEVESQASEEQTSEEEEKEGASQEEDEENESEEQTSEEEEEGASEEEDEESAFEEQT--- 433
Query: 330 PPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEEGTNEKEDEDEEEKEYSTSSMQEQT 389
+ EE +E+E++EE E + + +
Sbjct: 434 ------------------------------SEEEEEKGASQEEEEDEENEQESEVESQAS 463
Query: 390 DKERFIEKRVILEAEETDSDEDEDVGELKQSVKSV 424
+++ E+ E ++ +EDED E ++S +SV
Sbjct: 464 EEQTSEEEGASEEGQDASEEEDEDESEEEESDESV 498
Score = 43.5 bits (101), Expect = 0.015
Identities = 60/294 (20%), Positives = 116/294 (39%), Gaps = 22/294 (7%)
Query: 194 DDDCVVQSQSVDGLSKT--TTKRFSSNSFNRVRNYAEFEGLVEDKLKQKEE---ENAVLP 248
D+ V++ +S +G+ K T+K S ++ + E +D+ K + E E V
Sbjct: 79 DETTVIEKESENGVDKEKPTSKEESGEKTSQEKESEEKSSQEKDEDKSESEASEEKDVSQ 138
Query: 249 SPIPWRSRSASARMMEPKQEAIEKALKASMVMEPKQEANENASKPSMVKTSSIKFTPSES 308
+ AS + +E K S + + + E AS + S K T SE
Sbjct: 139 EQNSKEEKGASEEDEDTPEEQNSKEENGSSEEDDEDASEEQASNEEK-EASEEKNTVSEE 197
Query: 309 VAKNSEDLIKKKSFYYKSYPPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEEGTN 368
SE+ ++K ++S T ++ E++ + E+G +
Sbjct: 198 RKGASEEEDEEKDDGHESEVESQASEEQTTEEGAS---EEEDEESASEEQTSEGEEKGAS 254
Query: 369 EKEDEDE---EEKEYSTSSMQEQTDKERFI----------EKRVILEAEETDSDEDEDVG 415
++E+EDE +E E + + +EQT +E K E EE+ S+E+++
Sbjct: 255 QEEEEDEGNEQESEVESQASEEQTSEEEESASEEEDEENESKEQTTEEEESASEEEDEES 314
Query: 416 ELKQSVKSVKTEEPCSSSLSGGIDTVSDEGPDVDKKADEFIAKFREQIRLQRIE 469
++ K+ EE T +E +++ +E ++ E+ Q E
Sbjct: 315 ASEREEKNASQEEEEDEGNESKEQTTEEEESASEEEDEESASEGEEKNASQEEE 368
Score = 40.8 bits (94), Expect = 0.096
Identities = 44/228 (19%), Positives = 83/228 (36%), Gaps = 18/228 (7%)
Query: 207 LSKTTTKRFSSNSFNRVRNYAEFEGLVEDKLKQKEEENAVLPSPIPWRSRSASARMM--E 264
L+ T++ S +R+R ++ L L+ E+ W+ R ++ + E
Sbjct: 21 LTVTSSNAIPGRSSSRLRLLERYDSL--PSLRSHSEDRYDDGVDRKWKKREGNSDDICTE 78
Query: 265 PKQEAIEKALKASMVMEPKQEANENASKPSMVKTSSIKFTPSESVAKNSEDLIKKKSFYY 324
+ IEK + + E E+ K S K S K + + K+ + ++K
Sbjct: 79 DETTVIEKESENGVDKEKPTSKEESGEKTSQEKESEEKSSQEKDEDKSESEASEEKDVSQ 138
Query: 325 KSYPPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEEGTNEKEDEDEEEKEYSTSS 384
+ S K E E G++E++DED E++ S
Sbjct: 139 EQ--------------NSKEEKGASEEDEDTPEEQNSKEENGSSEEDDEDASEEQASNEE 184
Query: 385 MQEQTDKERFIEKRVILEAEETDSDEDEDVGELKQSVKSVKTEEPCSS 432
+ +K E+R EE + +D E++ +T E +S
Sbjct: 185 KEASEEKNTVSEERKGASEEEDEEKDDGHESEVESQASEEQTTEEGAS 232
Score = 40.0 bits (92), Expect = 0.16
Identities = 72/394 (18%), Positives = 156/394 (39%), Gaps = 29/394 (7%)
Query: 70 LLHLLFVGIAISYGLFSRRNQEPDKDNNNNTKFDNAQSLVSRFLQVSSFFEDEVDHQ--- 126
LL L F +++ + N P + ++ + SL S ++D VD +
Sbjct: 12 LLQLFF----LTFLTVTSSNAIPGRSSSRLRLLERYDSLPSLRSHSEDRYDDGVDRKWKK 67
Query: 127 NQSESEDI---NKIQTWSNQNQHYRNKPMVIVAPQVQQLQNSVFEDEQSSFNENEKPLLL 183
+ S+DI ++ ++++ +K + + + E E+ S E ++
Sbjct: 68 REGNSDDICTEDETTVIEKESENGVDKEKPTSKEESGEKTSQEKESEEKSSQEKDEDKSE 127
Query: 184 PVRSLKSRLSDDDCVVQSQSVDGLSKTTTKRFSSNSFNRVRNYAEFEGLVEDKLKQKEEE 243
S + +S + + + + T + +S N + + E E++ +E+E
Sbjct: 128 SEASEEKDVSQEQNSKEEKGASEEDEDTPEEQNSKEENG-SSEEDDEDASEEQASNEEKE 186
Query: 244 NAVLPSPIPWRSRSASARMMEPKQEAIEKALKASMVMEPKQEANENASKPSMVKTSSIKF 303
+ + + + AS E K + E +++ E Q E AS+ +++S +
Sbjct: 187 ASEEKNTVSEERKGASEEEDEEKDDGHESEVESQASEE--QTTEEGASEEEDEESASEEQ 244
Query: 304 TPSESVAKNSEDLIKKKSFYYKSYPPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSI 363
T S++ + + +S T S +S E++ N S ++
Sbjct: 245 TSEGEEKGASQEEEEDEGNEQESEVESQASEEQT----SEEEESASEEEDEENESKEQTT 300
Query: 364 EEGTNEKEDEDEE---EKEYSTSSMQEQTDKERFIEKRVILEAEETDSDEDEDVGELKQS 420
EE + E+EDEE E+E +S +E+ D+ +++ E E +EDE+ +
Sbjct: 301 EEEESASEEEDEESASEREEKNASQEEEEDEGNESKEQTTEEEESASEEEDEESASEGEE 360
Query: 421 VKSVKTEEPCSSSLSGGIDTVSDEGPDVDKKADE 454
+ + EE D +++ +V+ +A E
Sbjct: 361 KNASQEEEE---------DEGNEQESEVESQASE 385
>ref|XP_650383.1| hypothetical protein 223.t00011 [Entamoeba histolytica HM-1:IMSS]
gi|56467001|gb|EAL44997.1| hypothetical protein
223.t00011 [Entamoeba histolytica HM-1:IMSS]
Length = 863
Score = 56.2 bits (134), Expect = 2e-06
Identities = 82/385 (21%), Positives = 170/385 (43%), Gaps = 45/385 (11%)
Query: 120 EDEVDHQNQSES----EDINKIQTWSNQNQHYRNKPMVIVAPQVQQLQNSVFEDEQSSFN 175
E EV H NQS + DI+++ +++ K ++++ +N E ++ + +
Sbjct: 146 ETEVLHMNQSSNIAATTDIHELLVQQVDKKNHEIKK----EEKIEKEENEAEESKKDNID 201
Query: 176 EN--EKPLLLPVRSLKSRLSDDDCVVQSQSVDGLSKTTTKRFSSNS-FNRVRNYAEFEGL 232
E E+ L+ R K ++ Q Q + + T S ++ + + F
Sbjct: 202 EEKEEEELVEKQRKQKEIQEQEEAARQKQLEEQQKEAATSSDKSKEKTDKAKEKSLFAAS 261
Query: 233 VED-KLKQKEEENAVLPSPIPWRSRSASARMMEPKQEAIEKALKASMVMEPKQEANEN-- 289
+ D K+K K++++ + KQE I+KA++ ++ E KQ+ ++
Sbjct: 262 MSDVKIKGKKDKDK-----------------KKFKQEDIDKAVEQALA-EKKQKHHKKVA 303
Query: 290 ASKPSMVKTSSIKFTPSESVAKNS----EDLIKKKSFYYKSYPPPPPPPPPTMFYKSTFL 345
A K + + K E K E+L KK K ++ ++T L
Sbjct: 304 ALKAQIEALKAEKDKEIEDAVKEKDIQIEELNKKVQEETKEKEEAKASLAISVAAEAT-L 362
Query: 346 KSRYGEKNGRNMSMGKSIEEGTNE---KEDEDEEEKEYSTSSMQEQTDKERFIEKRVILE 402
K+ +K+ + G+ +E+ E K +E ++EKE T ++E + E+ EK+ + E
Sbjct: 363 KAEVEKKDQELKNKGEELEKEKEEQAKKIEEIQKEKEEQTKKVEE-LEGEKNNEKQKVEE 421
Query: 403 AEETDSDEDEDVGELKQSVKSV--KTEEPCSSSLSGGIDTVSDEGPDVD--KKADEFIAK 458
E+ +D +++ ELK +K + K EE ++ +G + + + ++D KK E ++K
Sbjct: 422 LEKKVNDSEKENNELKGQLKDLQKKLEETEKNAAAGSEELLKQKNEEIDNIKKEKEVLSK 481
Query: 459 FREQIRLQRIESIKRSTRVARNSSR 483
+Q++ Q + + S + N +
Sbjct: 482 ENKQLKEQISSAEENSNSIIENEKK 506
Score = 37.0 bits (84), Expect = 1.4
Identities = 62/316 (19%), Positives = 129/316 (40%), Gaps = 35/316 (11%)
Query: 130 ESEDINKI--QTWSNQNQHYRNKPMVIVAPQVQQLQNSVFEDEQSSFNENEKPLLLPVRS 187
+ EDI+K Q + + Q + K + A Q++ L+ ++ + + E + + +
Sbjct: 279 KQEDIDKAVEQALAEKKQKHHKKVAALKA-QIEALKAEKDKEIEDAVKEKDIQIEELNKK 337
Query: 188 LKSRLSDDDCVVQSQSVDGLSKTTTKRFSSNSFNRVRNYAEFEGLVEDKLKQKEEENAVL 247
++ + + S ++ ++ T K ++N E +L++++EE A
Sbjct: 338 VQEETKEKEEAKASLAISVAAEATLKAEVEKKDQELKNKGE-------ELEKEKEEQAKK 390
Query: 248 PSPIPWRSRSASARMMEPKQEAIEKALKASMVMEPKQEAN-ENASKPSMVKTSSIKFTPS 306
I + ++ E + E + K + + ++ EN +K K +
Sbjct: 391 IEEIQKEKEEQTKKVEELEGEKNNEKQKVEELEKKVNDSEKENNELKGQLKDLQKKLEET 450
Query: 307 E-SVAKNSEDLIKKKSFYYKSYPPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEE 365
E + A SE+L+K+K+ + + ++ LK + + S+ IE
Sbjct: 451 EKNAAAGSEELLKQKNEEIDNIKKEKE----VLSKENKQLKEQISSAEENSNSI---IEN 503
Query: 366 GTNEKED---EDEEEKEYSTSSMQEQTDKER-FIEKRVILEAEETDSDE----------- 410
EKED ++EE K+ +E KER EK V++ + + S+E
Sbjct: 504 EKKEKEDLKHQNEELKQQIEELKEENNKKERELAEKEVVIVSLQKSSEEVNKKDKSSSSS 563
Query: 411 -DEDVGELKQSVKSVK 425
DE+ E K++ K +K
Sbjct: 564 SDEEENEKKENGKLIK 579
>dbj|BAB03174.1| unnamed protein product [Arabidopsis thaliana]
gi|15228477|ref|NP_189519.1| expressed protein
[Arabidopsis thaliana]
Length = 2081
Score = 55.5 bits (132), Expect = 4e-06
Identities = 79/403 (19%), Positives = 156/403 (38%), Gaps = 31/403 (7%)
Query: 88 RNQEPDKDN--NNNTKFDNAQSLVSRFLQVSSFFEDEVDHQNQSESEDINKIQTWSNQNQ 145
+ +E DK NN K + + S E+ D++ + ESED + +
Sbjct: 957 KKKEEDKKEYVNNELKKQEDNKKETTKSENSKLKEENKDNKEKKESEDSASKNREKKEYE 1016
Query: 146 HYRNKPMVIVAPQVQQLQNSVFEDEQSSFNENEKPLLLPVRSLKSRLSDDDCVVQSQSVD 205
++K + ++ Q+ E++ S +++K R LK++ +++ + +S +
Sbjct: 1017 EKKSKTKEEAKKEKKKSQDKKREEKDSEERKSKKE-KEESRDLKAKKKEEETKEKKESEN 1075
Query: 206 GLSK---TTTKRFSSNSFNRVRNYAEFEGLVEDKLKQKEEENAVLPSPIPWRSRSASARM 262
SK + + S + + E + E K ++KEE+ + S
Sbjct: 1076 HKSKKKEDKKEHEDNKSMKKEEDKKEKKKHEESKSRKKEEDKKDMEKLEDQNSNKKKEDK 1135
Query: 263 MEPKQEAIEKALKASMVMEPKQEANENASKPSMVKTSSIKFTPSESVAKNSEDLIKKKSF 322
E K+ K +K + K+E E + + + S K + K+S+D KKK
Sbjct: 1136 NEKKKSQHVKLVKKESDKKEKKENEEKSETKEIESSKSQKNEVDKKEKKSSKDQQKKKEK 1195
Query: 323 YYKSYPPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEEGTNEKEDEDEEEKEYST 382
K + + +KN + S+EE N+K+ E ++EK
Sbjct: 1196 EMKE------------------SEEKKLKKNEEDRKKQTSVEE--NKKQKETKKEKNKPK 1235
Query: 383 SSMQEQTDKERFIEKRVILEAEETDSDEDEDVGELKQSVKSVKTEEPCSSSLSGGIDTVS 442
+ T + K+ +E+E ++ E++ + S +++ D+ S
Sbjct: 1236 DDKKNTTKQSG--GKKESMESESKEA-ENQQKSQATTQADSDESKNEILMQADSQADSHS 1292
Query: 443 DEGPDVDKKADEFIAKFREQIRLQR--IESIKRSTRVARNSSR 483
D D D+ +E + + Q QR E K+ T VA N +
Sbjct: 1293 DSQADSDESKNEILMQADSQATTQRNNEEDRKKQTSVAENKKQ 1335
Score = 43.9 bits (102), Expect = 0.011
Identities = 68/313 (21%), Positives = 131/313 (41%), Gaps = 44/313 (14%)
Query: 120 EDEVDHQNQSESEDINKIQTWSNQNQHYRNKPMVIVAPQVQQLQNSVF---EDEQSSFNE 176
+DE N+ E++D I T S Q + K + ++ +NS E+++ +
Sbjct: 918 KDEKKEGNKEENKDT--INTSSKQKGKDKKK-------KKKESKNSNMKKKEEDKKEYVN 968
Query: 177 NEKPLLLPVRSLKSRLSDDDCVVQSQSVDGLSKTTTKRFSSNSFNRVRNYAEFEGLVEDK 236
NE L + K ++ ++ ++ D K + S +S ++ R E+E E K
Sbjct: 969 NE--LKKQEDNKKETTKSENSKLKEENKDNKEK----KESEDSASKNREKKEYE---EKK 1019
Query: 237 LKQKEE-ENAVLPSPIPWRSRSASARMMEPKQEAIEKALKASMVMEPKQEANENASKPSM 295
K KEE + S R S K++ + LKA E +E E+ + S
Sbjct: 1020 SKTKEEAKKEKKKSQDKKREEKDSEERKSKKEKEESRDLKAKKKEEETKEKKESENHKSK 1079
Query: 296 VKTSSIKFTPSESVAKNSEDLIKKKSFYYKSYPPPPPPPPPTMFYKSTFLKSRYGEKNGR 355
K + ++S+ K + KKK + +S KSR E++ +
Sbjct: 1080 KKEDKKEHEDNKSMKKEEDKKEKKK--HEES-------------------KSRKKEEDKK 1118
Query: 356 NMSMGKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDKERFIEKRVILEAEETDSDEDEDVG 415
+M + + +KED++E++K +++++DK+ E E +E +S + +
Sbjct: 1119 DMEKLED-QNSNKKKEDKNEKKKSQHVKLVKKESDKKEKKENEEKSETKEIESSKSQKNE 1177
Query: 416 ELKQSVKSVKTEE 428
K+ KS K ++
Sbjct: 1178 VDKKEKKSSKDQQ 1190
Score = 36.2 bits (82), Expect = 2.4
Identities = 64/387 (16%), Positives = 147/387 (37%), Gaps = 38/387 (9%)
Query: 88 RNQEPDKDNNNNTKFDNAQSLVSRFLQVSSFFEDEVDHQNQSESEDINKIQTWSNQNQHY 147
+ Q+ K N K D + + S + + +NQ +S+ + + ++N+
Sbjct: 1222 KKQKETKKEKNKPKDDKKNTTKQSGGKKESMESESKEAENQQKSQATTQADSDESKNE-- 1279
Query: 148 RNKPMVIVAPQVQQLQNSVFEDEQSSFNENEKPLLLPVRSLKS--RLSDDDCVVQSQSVD 205
+++ Q +S D Q+ +E++ +L+ S + R +++D Q+ +
Sbjct: 1280 -----ILMQADSQADSHS---DSQADSDESKNEILMQADSQATTQRNNEEDRKKQTSVAE 1331
Query: 206 GLSKTTTKRFSSNSFNRVRNYAEFEGLVEDKLKQ--KEEENAVLPSPIPWRSRSASARMM 263
+ TK + + +N + G ++ ++ KE EN + A
Sbjct: 1332 NKKQKETKEEKNKPKDDKKNTTKQSGGKKESMESESKEAENQQKSQA------TTQADSD 1385
Query: 264 EPKQEAIEKALKASMVMEPKQEANENASKPSMVKTSSIKFTPSESVAKNSEDLIKKKSFY 323
E K E + +A + Q ++ + +++ S + +N+E+ KK++
Sbjct: 1386 ESKNEILMQADSQADSHSDSQADSDESKNEILMQADS-----QATTQRNNEEDRKKQTSV 1440
Query: 324 YKSYPPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEEGTNEKEDEDEEEKEYSTS 383
++ K K + +KN S GK + KE E++++ + +T
Sbjct: 1441 AENKKQKET--------KEEKNKPKDDKKNTTEQSGGKKESMESESKEAENQQKSQATTQ 1492
Query: 384 SMQEQTDKERFIEKRVILEAE-ETDSDEDEDVGELKQSVKSVKTEEPCSSSLSGGI---- 438
+++ E ++ + + D DE E+ S + S I
Sbjct: 1493 GESDESKNEILMQADSQADTHANSQGDSDESKNEILMQADSQADSQTDSDESKNEILMQA 1552
Query: 439 DTVSDEGPDVDKKADEFIAKFREQIRL 465
D+ +D D D+ +E + + Q ++
Sbjct: 1553 DSQADSQTDSDESKNEILMQADSQAKI 1579
>ref|XP_666163.1| myosin heavy chain [Cryptosporidium hominis]
gi|54657105|gb|EAL35936.1| myosin heavy chain
[Cryptosporidium hominis]
Length = 1604
Score = 55.1 bits (131), Expect = 5e-06
Identities = 82/369 (22%), Positives = 146/369 (39%), Gaps = 59/369 (15%)
Query: 87 RRNQEPDKDNNNNTKFDNAQSLVSRFLQVSSFFEDEVDHQNQSESEDINKIQTWSNQNQH 146
++ E ++ +N+ + D +S S L+ ++V + E ++ +T + H
Sbjct: 1275 KKKDEKEEKDNDEKEDDKEKSKESENLEKKPTISEKVINPESKIDEKDSEKETLN----H 1330
Query: 147 YRNKPMVIVAPQVQQLQNSVFEDEQSSFN----ENEKPLLLPVRSLKSRLSDDDCVVQSQ 202
+ K + + QL+ ++ S N +E L S K + D+
Sbjct: 1331 SKTKE-IAKSKAAAQLKAAILARASSKKNISADNSEDSKLKKSESKKDIIKDEAKDESKT 1389
Query: 203 SVDGLSKTTTKRFSSNSFNRVRNYAEFEGLVEDKLKQKE-----EENAVLPSPIPWRSRS 257
+ DG T + S ++V+ +G V+ K K KE EE++ W S
Sbjct: 1390 NKDGKDLKKTLKEGEGSISKVKG----KGKVKTKAKAKETPPPAEESSWFSG---WFGTS 1442
Query: 258 ASARMMEPKQEAIEKALKASMVMEPKQEANENASKPSMVKTSSIKFTPSESVAKNSEDLI 317
A EP +E +K K P A E++ TS+ + P E AK
Sbjct: 1443 APE---EPPKEEEKKEEKKEDEAPPAPPAEESSWFSGWFGTSAPEEPPKEDPAKK----- 1494
Query: 318 KKKSFYYKSYPPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEEGTNEKEDEDEEE 377
PP + KS+ +K GEK + EE +EKED+ +++
Sbjct: 1495 ---------------PPKKHLVKKSSSVKKDDGEKKEEK----EEKEEKKDEKEDKKDKK 1535
Query: 378 KEYSTSSMQEQTDKERFIEKR---------VILEAEETDSDEDEDV--GELKQSVKSVKT 426
KE + +++ +K+ EK+ I+E E+T +D ++ V + K K+
Sbjct: 1536 KEEIEEAKEKKEEKKESDEKKDETDNKEVKEIVEKEKTGADPEKKVVKAKTKSKAKAKAR 1595
Query: 427 EEPCSSSLS 435
EEP S + S
Sbjct: 1596 EEPTSCTTS 1604
Score = 50.8 bits (120), Expect = 9e-05
Identities = 75/383 (19%), Positives = 154/383 (39%), Gaps = 44/383 (11%)
Query: 88 RNQEPDKDNNNNTKFDNAQSLVSRFLQVSSFFEDEVDHQNQSESEDINKIQTWSNQNQHY 147
+ +E DKD K D + E E + + + E ++ K + ++++
Sbjct: 1004 KEKEEDKDEEKKEKEDKEKK--------EKLKEKEEEGKEKEEGKEKEKEK---DKDEKD 1052
Query: 148 RNKPMVIVAPQVQQLQNSVFED-EQSSFNENEKPLLLPVRSLKSRLSDDDCVVQSQSVDG 206
++K + + + ++L+ + + ++ S +E +K P+ LK+++
Sbjct: 1053 KSKSKIKESQKEEKLEETEKDSSKKESKDEEKKEKEDPIAKLKAKVP--------VKPSP 1104
Query: 207 LSKTTTKRFSSNSFNRVRNYAEFEGLVEDKLKQKEEENAVLPSPIPWRSRSASARMMEPK 266
L K+ +++ ++ + + ++KLK+KEEE + + K
Sbjct: 1105 LLKSKSEKEKEKEEDKDEKKEKEDKEKKEKLKEKEEEGKE-------KEEGKEKEKDKDK 1157
Query: 267 QEAIEKALKASMVMEPKQEANENASKPSMVKTSSIKFTPSESVAKNSEDLIKKKSFYYKS 326
E + K + K + E K + TS + E K ED I K
Sbjct: 1158 DEKDKSKSKTKDFEKEKLKETEKGEKEAEKDTSKKESKDEEK--KEKEDPIAKLK----- 1210
Query: 327 YPPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEEGTNEKEDEDEEEKEYSTSSMQ 386
P P P + KS K + +K+ + K +E EKE+E +E++E
Sbjct: 1211 -AKVPVKPSPLLKSKSEKEKEKEEDKDEKKEKEDKEKKEKLKEKEEEGKEKEE------G 1263
Query: 387 EQTDKERFIEKRVILEAEETDSDEDEDVGELKQSVKSVKTEEPCSSSL---SGGIDTVSD 443
++ +KE+ ++ E EE D+DE ED E + ++++ + S + ID
Sbjct: 1264 KEKEKEKVKAQKKKDEKEEKDNDEKEDDKEKSKESENLEKKPTISEKVINPESKIDEKDS 1323
Query: 444 EGPDVDKKADEFIAKFREQIRLQ 466
E ++ + IAK + +L+
Sbjct: 1324 EKETLNHSKTKEIAKSKAAAQLK 1346
Score = 48.1 bits (113), Expect = 6e-04
Identities = 73/377 (19%), Positives = 148/377 (38%), Gaps = 45/377 (11%)
Query: 120 EDEVDHQNQSESEDINKIQTWSNQNQHYRNKPMV------IVAPQVQQLQNSVFEDEQSS 173
E + + +++ ++ +KI + S +N + K I + + NSV S
Sbjct: 888 ETDAKEKEKTKIKEDSKIDSDSEENSESKEKKSTGLLAKTIAKAKAKSKSNSVLSKLPSK 947
Query: 174 FNENEKPLLLPVRSLKSRLSDDDCVVQSQSVDGLSKTTTKRFSSNSFNRVRNYAEFEGLV 233
N+ L++++ DD + K K S + + + + L+
Sbjct: 948 VNKETSKA---ENDLENKIEKDD--------ESNKKEIKKEEDSIAKLKAKVPVKPSPLL 996
Query: 234 EDKL-KQKEEENAVLPSPIPWRSRSASARMMEPKQEAIEKALKASMVMEPKQEANENASK 292
+ K K+KE+E + ++ E ++E EK + E +++ +E
Sbjct: 997 KSKSEKEKEKEEDKDEEKKEKEDKEKKEKLKEKEEEGKEK--EEGKEKEKEKDKDEKDKS 1054
Query: 293 PSMVKTSS----IKFTPSESVAKNSEDLIKKKSF--YYKSYPPPPPPPPPTMFYKSTFLK 346
S +K S ++ T +S K S+D KK+ K P P P + KS K
Sbjct: 1055 KSKIKESQKEEKLEETEKDSSKKESKDEEKKEKEDPIAKLKAKVPVKPSPLLKSKSEKEK 1114
Query: 347 SRYGEKNGRNMSMGKSIEEGTNEKEDED-------EEEKEYSTSSMQEQTDKERFIEKRV 399
+ +K+ + K +E EKE+E E+EK+ + K + EK
Sbjct: 1115 EKEEDKDEKKEKEDKEKKEKLKEKEEEGKEKEEGKEKEKDKDKDEKDKSKSKTKDFEKEK 1174
Query: 400 ILEAEETDSD----------EDEDVGELKQSVKSVKTEEPCSSS--LSGGIDTVSDEGPD 447
+ E E+ + + +DE+ E + + +K + P S L + ++ D
Sbjct: 1175 LKETEKGEKEAEKDTSKKESKDEEKKEKEDPIAKLKAKVPVKPSPLLKSKSEKEKEKEED 1234
Query: 448 VDKKADEFIAKFREQIR 464
D+K ++ + +E+++
Sbjct: 1235 KDEKKEKEDKEKKEKLK 1251
Score = 45.4 bits (106), Expect = 0.004
Identities = 58/260 (22%), Positives = 108/260 (41%), Gaps = 40/260 (15%)
Query: 238 KQKEEENAVLPSPIPWRSRSASARMMEPKQEAIEK---ALKASMVMEPKQEANENA---S 291
K+KEE +A + S E E+ EK L A + + K ++ N+
Sbjct: 884 KEKEETDAKEKEKTKIKEDSKIDSDSEENSESKEKKSTGLLAKTIAKAKAKSKSNSVLSK 943
Query: 292 KPSMV--KTSSIKFTPSESVAK----NSEDLIKKKSFYYKSYPPPPPPPPPTMFYKSTFL 345
PS V +TS + + K N +++ K++ K P P P + KS
Sbjct: 944 LPSKVNKETSKAENDLENKIEKDDESNKKEIKKEEDSIAKLKAKVPVKPSPLLKSKSEKE 1003
Query: 346 KSRYGEKNGRNMSM---------------GKSIEEGTNEKEDEDEEEKEYSTSSMQEQTD 390
K + +K+ GK EEG +++++D++EK+ S S ++E
Sbjct: 1004 KEKEEDKDEEKKEKEDKEKKEKLKEKEEEGKEKEEGKEKEKEKDKDEKDKSKSKIKESQK 1063
Query: 391 KERFIEKRVILEAEETDS----DEDEDVGELKQSVKSVKTEEPCSSS--LSGGIDTVSDE 444
+E+ LE E DS +DE+ E + + +K + P S L + ++
Sbjct: 1064 EEK-------LEETEKDSSKKESKDEEKKEKEDPIAKLKAKVPVKPSPLLKSKSEKEKEK 1116
Query: 445 GPDVDKKADEFIAKFREQIR 464
D D+K ++ + +E+++
Sbjct: 1117 EEDKDEKKEKEDKEKKEKLK 1136
Score = 38.5 bits (88), Expect = 0.47
Identities = 67/372 (18%), Positives = 152/372 (40%), Gaps = 36/372 (9%)
Query: 132 EDINKIQTWSNQNQHYRNKPMVIVAPQVQQLQ--NSVFEDEQSSFNENEKPLLLPVRSLK 189
+ INK + ++ YR + + ++ QL+ N + E E+ E L + +
Sbjct: 381 DTINKKNSKLDEALTYRIESAITAEREINQLKEKNQIIEFEKKMMEEK-----LEISNNN 435
Query: 190 SRLSDDDCVVQSQSVDGLSKTTTKRFSSNSFNRVRNYAEFEGLVEDKLKQKEEENAVLPS 249
++ DD+ ++ L++ K N + ++ + E VE+ + + ++ S
Sbjct: 436 IKIKDDEIQQLKTTIQDLNRQNIKL--ENELHSLQESSRRES-VENTFRSSRIGSNIVSS 492
Query: 250 PIPWRSRSA--------SARMMEPKQEAIEKALKASMVMEPKQEANENASKPSMVKTSSI 301
I S S S+ + E +EK + A + + + + + + VK S
Sbjct: 493 KINASSGSIVNSSENTISSNIATSNAETVEKVVNADAIADTSIQDSPINAASTTVKMLSS 552
Query: 302 KFTPSESVAKNSEDLIKKKSFYYKSYPPPPPPPPPTMFYKSTF--LKSRYGEKN------ 353
K S+S A +S K KS + T+ ++ +KS+ G K+
Sbjct: 553 KSISSKS-AISSLPKSKAKSAVKSAPATEENNGDQTISEPASISEVKSKAGSKSMPVSKS 611
Query: 354 -----GRNMSMGKS-IEEGTNEKEDEDEEEKEYSTSSMQEQTDKERFIEKRVILEAEETD 407
++ + KS ++ G KED + ++ E + ++ Q +KE E + + E EE +
Sbjct: 612 KSGIKAKSQVVAKSDMDSGKIVKEDNETKDNEDEAAKVRAQKEKEE-AEAKALKEKEEAE 670
Query: 408 SDEDEDVGELKQSVKSVKTEEPCSSSLSGGIDTVSDEGPDVDKKADEFIAKFREQIRLQR 467
+ +++ E + K++K +E + + + ++A+ K +E+ +
Sbjct: 671 AKANKEKEEAE--AKALKEKEEAEAKAKKEKEEAEAKAKKEKEEAEAKAKKEKEEAEAKA 728
Query: 468 IESIKRSTRVAR 479
++ + + A+
Sbjct: 729 LKEKEEAEAKAK 740
Score = 37.0 bits (84), Expect = 1.4
Identities = 50/223 (22%), Positives = 82/223 (36%), Gaps = 26/223 (11%)
Query: 237 LKQKEEENAVLPSPIPWRSRSASARMMEPKQEAIEKALKASMVMEPKQEANENASKPSMV 296
LK+KEE A A A+ + K+EA KALK K+EA A K
Sbjct: 729 LKEKEEAEAKAKK----EKEEAEAKAKKEKEEAEAKALKE------KEEAEAKAKKEKEE 778
Query: 297 KTSSIKFTPSESVAKNSEDLIKKKSFYYKSYPPPPPPPPPTMFYKSTFLKSRYGEKNGRN 356
+ K E+ AK ++ + ++ K K EK
Sbjct: 779 TEAKAKKEKEEAEAKAKKEKEEAEAKAKKE--------------KEEAEAKALKEKEEAE 824
Query: 357 MSMGKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDKERFIEKRVILEAEETDSDEDEDVGE 416
K EE K +++EE E +E+ + + EK + + +E E +
Sbjct: 825 AKAKKEKEEA-EAKAKKEKEEAEAKAKKEKEEAEAKALKEKEEAEAKGKKEKEEAEAKAK 883
Query: 417 LKQSVKSVKTEEPCSSSLSGGIDTVSDEGPD-VDKKADEFIAK 458
++ K +E ID+ S+E + +KK+ +AK
Sbjct: 884 KEKEETDAKEKEKTKIKEDSKIDSDSEENSESKEKKSTGLLAK 926
>ref|XP_636078.1| hypothetical protein DDB0188541 [Dictyostelium discoideum]
gi|60464424|gb|EAL62571.1| hypothetical protein
DDB0188541 [Dictyostelium discoideum]
Length = 1419
Score = 54.3 bits (129), Expect = 8e-06
Identities = 75/402 (18%), Positives = 152/402 (37%), Gaps = 47/402 (11%)
Query: 86 SRRNQEPDKDNNNNTKFDNA-----------QSLVSRFLQVSSFFEDEVDHQNQSESEDI 134
+R N+ D +NNNN +NA Q L + ++ +D+ + + + E E
Sbjct: 300 NRVNETDDDNNNNNNHNNNANNEFKSITDKKQKLSNNIIKKKEESDDDDEKEKEKEKEKN 359
Query: 135 NKIQTWSNQNQHYRNKPMVIVAPQVQQLQNSVFEDEQSSFNENEKPLLLPVRSLKSRLSD 194
K++ + + + + +P Q +QS N ++K S S+
Sbjct: 360 KKLKNSNINKNNSKESQSIKKSPTKSQ-------PKQSKKNASDKITKTIEESQSESESE 412
Query: 195 DDCVVQSQSVDGLSKTTTKRFSSNSFNRVRNYAEFEGLVEDKLKQKEEENAVLPSPIPWR 254
+ + +S D +K+ S S E + +QK+ N PI +
Sbjct: 413 EPIEISKKSTDKPILKQSKKKPSESEE------------ESEKEQKKIVNKSSTKPISKQ 460
Query: 255 SRSASARMMEPKQEAIEKALKASMVMEPKQEANENASKPSMVKTSSIKFTPSESVAKNSE 314
S+ + +E E + E K+ N+ ++KP + K S K SE +++
Sbjct: 461 SKKKAVE--SESEEESEPGSEEESEEESKKAVNKASAKP-ISKQSKKKAVESEPESESES 517
Query: 315 DLIKKKSFYYKSYPPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEEGTNEKEDED 374
D ++ + KST + +K K IE + K +E+
Sbjct: 518 DEESEEE------EESEEEETNKVVTKSTSKPLKQSKK--------KEIESESESKSEEE 563
Query: 375 EEEKEYSTSSMQEQTDKERFIEKRVILEAEETDSDEDEDVGELKQSVKSVKTEEPCSSSL 434
EEE+E ++ + +K+V E++ + +E+ + K++VK +++
Sbjct: 564 EEEEEEEKEEESKKKSSSKQSKKKVTKSKFESEHESEEESKDSKKNVKKSASKQSKKKVT 623
Query: 435 SGGIDTVSDEGPDVDKKADEFIAKFREQIRLQRIESIKRSTR 476
+ + E + KK K ++ +L+ E + R
Sbjct: 624 QESDEELKSESDEEIKKEKSKKVKKEKENKLKEKEKKEEEKR 665
Score = 50.4 bits (119), Expect = 1e-04
Identities = 57/348 (16%), Positives = 146/348 (41%), Gaps = 36/348 (10%)
Query: 86 SRRNQEPDKDNNNNTKFDNAQSLVSRFLQVSSFFEDEVDHQNQSESEDINKIQTWSNQNQ 145
+ +N +K+NNNN +N + + ++ + ++ N + + + N +++ N
Sbjct: 44 NNKNNSKNKNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNYNSNNN 103
Query: 146 HYRNKPMVIVAPQVQQLQNSVFEDEQSSFNENEKPLLLPVRSLKSRLSDDDC---VVQSQ 202
+ + V+ L++ + ++ + +++ L ++ K + + D V+ +
Sbjct: 104 INDVRVQIDPFESVELLKDVQRDKKKHRISNDKEELKDKTKNNKEKEKEKDKEKEKVKEK 163
Query: 203 SVDGLSKTTTKRFSSNSFNRVRNYAEFEGLVEDKLKQKEEENAVLPSPIPWRSRSASARM 262
L + T + N N ++N E E +V++ + ++E + R +
Sbjct: 164 DDTELKEETKENEKLNRKNNLKNKKEQEEIVKENKEIDKKE----------KKRKRDEKE 213
Query: 263 MEPKQEAIEKALKASMVMEPKQEANENASKPSMVKTSSIKFTPSESVAKNSEDLIKKKSF 322
+ + +EK + + K++ +E V+ +K + +K ++ +KK
Sbjct: 214 KDKSNDTVEKEKHNNKNILKKKKPDEEEEVEEEVEEEVVKEDKVDKKSKKKKEKAEKKQ- 272
Query: 323 YYKSYPPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSI--EEGTNEKEDEDEEEKEY 380
T +T + G+ N +GK++ E NE +D++ +
Sbjct: 273 ------------TTTTTTTTTATTTTKGKSN-----VGKNLINENRVNETDDDNNNNNNH 315
Query: 381 STSSMQE---QTDKERFIEKRVILEAEETDSDEDEDVGELKQSVKSVK 425
+ ++ E TDK++ + +I + EE+D D++++ + K+ K +K
Sbjct: 316 NNNANNEFKSITDKKQKLSNNIIKKKEESDDDDEKEKEKEKEKNKKLK 363
Score = 48.5 bits (114), Expect = 5e-04
Identities = 65/312 (20%), Positives = 121/312 (37%), Gaps = 33/312 (10%)
Query: 120 EDEVDHQNQSESEDINKIQTWSNQNQHYRNKPMVIVAPQVQQLQNSVFEDEQSSFNENEK 179
++E + + +SE E+ NK+ T S ++K I + + + E+E+ E++K
Sbjct: 518 DEESEEEEESEEEETNKVVTKSTSKPLKQSKKKEIESESESKSEEEEEEEEEEKEEESKK 577
Query: 180 PLLLPVRSLKSRLSDDDCVVQSQSVDGLSKTTTKRFSSN-SFNRVRNYAEFEGLVEDKLK 238
K S + +S+ SK K+ +S S +V ++ E E +
Sbjct: 578 KSSSKQSKKKVTKSKFESEHESEEESKDSKKNVKKSASKQSKKKVTQESDEELKSESDEE 637
Query: 239 QKEEENAVLPSPIPWRSRSASARMMEPKQEAIEKALKASMVMEPKQEANENASKPSMVKT 298
K+E++ + + + + E K+E K K + E K++ E + K
Sbjct: 638 IKKEKSKKVK-----KEKENKLKEKEKKEEEKRKEEKEKLEREKKEKEKEKEKEKEKEKE 692
Query: 299 SSIKFTPSES--VAKNSEDLIKKKSFYYKSYPPPPPPPPPTMFYKSTFLKSRYGEKNGRN 356
K E + +N E K+K K + EK+ +
Sbjct: 693 KEKKRIEKEKKKIRENEEKERKQKE-------------------KDEKKRKEKEEKDRKE 733
Query: 357 MSMGKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDKERFIEKRVILEAEETDSDEDEDVGE 416
K +E KE+E+ E KE +E+ KE+ E++ E EE + E E+
Sbjct: 734 ----KEEKEEKERKENEENERKEKEEKKRKEKERKEK--EEKERKEKEEKEIKEKEEKKR 787
Query: 417 LKQSVKSVKTEE 428
++ K K +E
Sbjct: 788 KEKEEKDRKEKE 799
Score = 47.8 bits (112), Expect = 8e-04
Identities = 72/368 (19%), Positives = 146/368 (39%), Gaps = 34/368 (9%)
Query: 86 SRRNQEPDKDNNNN--TKFDNAQSLVSRFLQVSSFFE---DEVDHQNQSESEDINKIQTW 140
S +E +++ N TK + S+ ++ S E +E + + + E E+ +K ++
Sbjct: 521 SEEEEESEEEETNKVVTKSTSKPLKQSKKKEIESESESKSEEEEEEEEEEKEEESKKKSS 580
Query: 141 SNQNQHYRNKPMVIVAPQVQQLQNSVFEDEQSSFNENEKPLLLPVRSLKSRLSDDDCVVQ 200
S Q++ K + ++ +E+S ++ +S K + D ++
Sbjct: 581 SKQSKKKVTK---------SKFESEHESEEESKDSKKNVKKSASKQSKKKVTQESDEELK 631
Query: 201 SQSVDGLSKTTTKRFSSNSFNRVRNYAEFEGLVEDKLKQKEEENAVLPSPIPWRSRSASA 260
S+S + + K +K+ N+++ E E E+K K+++E+
Sbjct: 632 SESDEEIKKEKSKKVKKEKENKLK---EKEKKEEEKRKEEKEKLEREKKEKEKEKEKEKE 688
Query: 261 RMMEPKQEAIEKALKASMVMEPKQ----EANENASKPSMVKTSSIKFTPSESVAKNSEDL 316
+ E +++ IEK K E K+ E +E K K K E K +E+
Sbjct: 689 KEKEKEKKRIEKEKKKIRENEEKERKQKEKDEKKRKEKEEKDRKEKEEKEEKERKENEEN 748
Query: 317 IKKKSFYYKSYPPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEEGTNEKEDEDEE 376
+K+ K K K K + + E+ EKE++D +
Sbjct: 749 ERKEKEEKKRKE------------KERKEKEEKERKEKEEKEIKEKEEKKRKEKEEKDRK 796
Query: 377 EKEYSTSSMQEQTDKERFIEKRVILEAEETDSDEDEDVGELKQSVKSVKTEEPCSSSLSG 436
EKE + +++ +KE E++ E E+ + + +E + K+ + + EE S
Sbjct: 797 EKERKENEEKKRKEKEE-KERKEKEEREKQEKEREEKEKQEKEERERKEKEENESIDCII 855
Query: 437 GIDTVSDE 444
DT+ E
Sbjct: 856 CTDTIKKE 863
Score = 41.2 bits (95), Expect = 0.073
Identities = 64/283 (22%), Positives = 108/283 (37%), Gaps = 29/283 (10%)
Query: 148 RNKPMVIVAPQVQ-------QLQNSVFEDEQSSFNENEKPLLLPVRSLKSRLSDDDCVVQ 200
R K VIV+ + Q +L N + ED + FN+ E+ + S++S L DD +
Sbjct: 908 RKKNSVIVSAKDQIAEYTEEELVNLLGEDYEQHFNDEEEGVDADALSVESSLDSDDSLND 967
Query: 201 ------SQSVDGLSKTTTKRFSSNSFNRVRNYAEFEGLVEDKLKQKEEENAVLPSPIPWR 254
S + D TT S+ F R R F V+D +++ +
Sbjct: 968 FIVGDDSFASDEEPFRTTNHRVSSQFPRFRR--SFFNKVDDDDDDDDDDYHDDDDDVS-E 1024
Query: 255 SRSASARMMEPKQEAIEKALKASMVMEPKQEANENASKPSMVKTSSIKFTPSESVAKNSE 314
S S + +E + + S + + + T++ T S+ +N+
Sbjct: 1025 SESNLSDFIEETNSLVTPVTRNSS----RNPTTTTTTTTTTTTTTTSPITRRSSLRRNTI 1080
Query: 315 DLIKKKSFYYKSYPPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEEGTNEKEDED 374
I +S S PP P + T + N N K+ ++G E + E
Sbjct: 1081 T-ISDESIGIPSTPPITRRNPIRISDDLTTTTTTTRSNNNNN----KNDDDGAIELDSET 1135
Query: 375 EEEKEYSTSSMQ----EQTDKERFIEKRVILEAEETDSDEDED 413
EE+K++ Q T+ E IEK E E D +++ED
Sbjct: 1136 EEDKKHFNRIFQIESDSDTENENEIEKEEECEEVEEDEEDEED 1178
Score = 34.7 bits (78), Expect = 6.9
Identities = 28/103 (27%), Positives = 44/103 (42%), Gaps = 8/103 (7%)
Query: 345 LKSRYGEKNGRNMSMGKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDKERFIEKRVILEAE 404
++S +N + + EE ++EDE++EE + E + + E V EAE
Sbjct: 1148 IESDSDTENENEIEKEEECEEVEEDEEDEEDEEDD-------EDDEDDEQEEDGVYEEAE 1200
Query: 405 ETDSDEDEDVGELKQSVKSVKTEEPCSSSLSGGIDTVSDEGPD 447
E D DE+E L + C L G+D SD+ D
Sbjct: 1201 EDDRDEEELEAHLNVPFGECEDCGECDYCLD-GLDLHSDDDED 1242
>gb|AAK85503.1| Hypothetical protein Y55B1BR.3 [Caenorhabditis elegans]
gi|17556242|ref|NP_497198.1| chromo domain containing
protein (77.9 kD) (3B49) [Caenorhabditis elegans]
Length = 679
Score = 53.9 bits (128), Expect = 1e-05
Identities = 62/295 (21%), Positives = 125/295 (42%), Gaps = 13/295 (4%)
Query: 169 DEQSSFNENEKPLLLPVRSLKSRLSDDDCVVQSQSVDGLSKTTTKRFS-SNSFNRVRNYA 227
D S F++ + RS + SDDD ++ + +K+F S R N +
Sbjct: 224 DSDSDFDKKKWKKNKAKRSKRDDSSDDDSEMERRRKKSKKSKKSKKFKKSEKRKRAVNDS 283
Query: 228 EFEGLVEDKL---KQKEEENAVLPSPIP--WRSRSASARMMEPKQEAIEKALKASMVMEP 282
+ E++ + K+ + AV+ S +S+ R + K+E+ E+ +AS E
Sbjct: 284 SSDDEDEEEKPEKRSKKSKKAVIDSSSEDEEEEKSSKKRSKKSKKESDEEQ-QASDSEEE 342
Query: 283 KQEANENASKPSMV-KTSSIKFTPSESVAKNSEDLIKKK--SFYYKSYPPPPPPPPPTMF 339
E +N+ P K +++K ES E+++KKK S K
Sbjct: 343 VVEVKKNSKSPKKTPKKTAVKEESEESSGDEEEEVVKKKKSSKINKRKAKESSSDEEEEV 402
Query: 340 YKSTFLKSRYGEKNGRNMSMGKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDKERFIEKRV 399
+S K++ K+ + + + EE E D +EEE+ + + ++ K+ +
Sbjct: 403 EESPKKKTKSPRKSSKKSA---AKEESEEESSDNEEEEEVDYSPKKKVKSPKKSSKKPAA 459
Query: 400 ILEAEETDSDEDEDVGELKQSVKSVKTEEPCSSSLSGGIDTVSDEGPDVDKKADE 454
+E+EE +E+E+ + +K KT S + +++ G + +++ +E
Sbjct: 460 KVESEEPSDNEEEEEEVEESPIKKDKTPRKYSRKSAAKVESTESSGNEEEEEVEE 514
Score = 41.2 bits (95), Expect = 0.073
Identities = 55/294 (18%), Positives = 111/294 (37%), Gaps = 18/294 (6%)
Query: 172 SSFNENEKPLLLPVRSLKSRLSDDDCVVQSQSVDGLSKTTTKRFSSNSFNRVRNYAEFEG 231
SS + E+ RS KS+ D+ + Q+ D + + +S S + +
Sbjct: 308 SSSEDEEEEKSSKKRSKKSKKESDE---EQQASDSEEEVVEVKKNSKSPKKTPKKTAVKE 364
Query: 232 LVEDKLKQKEEENAVLPSPIPWRSRSASARMMEPKQEAIEKALKASMVMEPKQEANENAS 291
E+ +EEE R A + ++E E K + P++ + ++A+
Sbjct: 365 ESEESSGDEEEEVVKKKKSSKINKRKAKESSSDEEEEVEESPKKKTK--SPRKSSKKSAA 422
Query: 292 KPSMVKTSS-------IKFTPSESVAKNSEDLIKKKSFYYKSYPPPPPPPPPTMFYKSTF 344
K + SS + ++P + V K+ + KK + +S P +S
Sbjct: 423 KEESEEESSDNEEEEEVDYSPKKKV-KSPKKSSKKPAAKVESEEPSDNEEEEEEVEESPI 481
Query: 345 LKSRYGEKNGRNMSMGKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDKERFIEKRVILEAE 404
K + K R KS + + + +EEE+E S ++ + +K +E
Sbjct: 482 KKDKTPRKYSR-----KSAAKVESTESSGNEEEEEVEESPKKKGKTPRKSSKKSAAVEES 536
Query: 405 ETDSDEDEDVGELKQSVKSVKTEEPCSSSLSGGIDTVSDEGPDVDKKADEFIAK 458
+ + ++ E+ + + S + + D +E D KK D+ + K
Sbjct: 537 DNEEEDVEESPKKRTSPRKSSKKRAAKEESEESSDNEVEEVEDSPKKNDKTLRK 590
>gb|AAM19760.1| glutamic acid-rich protein cNBL1700 [Trichinella spiralis]
Length = 571
Score = 53.5 bits (127), Expect = 1e-05
Identities = 76/374 (20%), Positives = 146/374 (38%), Gaps = 28/374 (7%)
Query: 90 QEPDKDNNNNTKFDNAQSLVSRFLQVSSFFEDEVDHQNQSESEDINKIQTWSNQNQHYRN 149
++ +KD+ + ++ ++ S + +S EDE + SE K + + + N
Sbjct: 205 EDEEKDDGHESEVESQASEEQTTEEGASEEEDEESASEEQTSEGEEKGAS-QEEEEDEGN 263
Query: 150 KPMVIVAPQVQQLQNSVFEDEQSSFNENEKPLLLPVRSLKSRLSDDDCVVQSQSVDGLSK 209
+ V Q + Q S E+E +S E+E+ S + +++ + + + S+
Sbjct: 264 EQESEVESQASEEQTSE-EEESASEEEDEEN-----ESKEQTTEEEESASEEEDEESASE 317
Query: 210 TTTKRFSSNSFNRVRNYAEFEGLVEDKLKQKEEENAVLPSPIPWRSRSASARMMEPKQEA 269
K S N ++ + E++ +EE+ + A E + E
Sbjct: 318 REEKNASQEEEEDEGNESKEQTTEEEESASEEEDEESVSEEQTSEGEEKGASQEEEEDEG 377
Query: 270 IEKALKA-SMVMEPKQEANENASKPSMVKTSSIKFTPSESVAKNSEDLI------KKKSF 322
++ + S E + E AS+ + S + T E A ED +K +
Sbjct: 378 NDQESEVESQASEEQTSEEEGASEEEDEENESEEQTTEEESASEEEDEESASEGEEKNAS 437
Query: 323 YYKSYPPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKS--IEEGTNEKEDEDEEEKEY 380
+ S S EK G + + EE T+E+E+E E+E
Sbjct: 438 QEEEEDEGNEQESEVESQASEEQTSEEEEKEGASQEEDEENESEEQTSEEEEEGASEEED 497
Query: 381 STSSMQEQTDKERFIEKRVILEAEETDSDEDEDVGELKQSVKSVKTEEPCSSSLSGGIDT 440
S+ +EQT +E E++ + EE D + +++ V+S +EE S +
Sbjct: 498 EESAFEEQTSEEE--EEKGASQEEEEDEENEQE-----SEVESQASEEQTSEE-----EG 545
Query: 441 VSDEGPDVDKKADE 454
S+EG D ++ DE
Sbjct: 546 ASEEGQDASEEEDE 559
Score = 52.8 bits (125), Expect = 2e-05
Identities = 75/382 (19%), Positives = 146/382 (37%), Gaps = 14/382 (3%)
Query: 86 SRRNQEPDKDNNNNTKFDNAQSLVSRFLQVSSFFEDEVDHQNQSESEDINKIQTWSNQNQ 145
S ++E D N+K + S ++E + ++ + ED ++ Q + + +
Sbjct: 128 SEASEEKDVSQEQNSKEEKGASEEDEDTPEEQNSKEE-NGSSEEDDEDASEEQASNEEKE 186
Query: 146 HYRNKPMVIVAPQ-VQQLQNSVFEDEQSSFNENEKPLLLPVRSLKSRLSDDDCVVQSQSV 204
K V + + ++ +D S E++ S D++ + Q+
Sbjct: 187 ASEEKNTVSEERKGASEEEDEEKDDGHESEVESQASEEQTTEEGASEEEDEESASEEQTS 246
Query: 205 DGLSKTTTKRFSSNSFNR----VRNYAEFEGLVEDKLKQKEEENAVLPSPIPWRSRSASA 260
+G K ++ + N V + A E E++ EEE+ S SA
Sbjct: 247 EGEEKGASQEEEEDEGNEQESEVESQASEEQTSEEEESASEEEDEENESKEQTTEEEESA 306
Query: 261 RMMEPKQEAIEKALKASMVMEPKQEANENASKPSMVKTSSIKFTPSESVAKNSEDLIKKK 320
E ++ A E+ K + E + E NE+ + + + S+ + ESV++ ++K
Sbjct: 307 SEEEDEESASEREEKNASQEEEEDEGNESKEQTTEEEESASEEEDEESVSEEQTSEGEEK 366
Query: 321 SFYYKSYPPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEEGTNEKEDEDEEEKEY 380
+ + +++ ++ E EE T E+E EEE E
Sbjct: 367 GASQEEEEDEGNDQESEVESQASEEQTSEEEGASEEEDEENESEEQTTEEESASEEEDEE 426
Query: 381 STS-------SMQEQTDKERFIEKRVILEA-EETDSDEDEDVGELKQSVKSVKTEEPCSS 432
S S S +E+ D+ E V +A EE S+E+E G ++ + ++EE S
Sbjct: 427 SASEGEEKNASQEEEEDEGNEQESEVESQASEEQTSEEEEKEGASQEEDEENESEEQTSE 486
Query: 433 SLSGGIDTVSDEGPDVDKKADE 454
G DE +++ E
Sbjct: 487 EEEEGASEEEDEESAFEEQTSE 508
Score = 51.6 bits (122), Expect = 5e-05
Identities = 70/352 (19%), Positives = 135/352 (37%), Gaps = 57/352 (16%)
Query: 89 NQEPDKDNNNNTKFD-NAQSLVSRFLQVSSFFEDEVDHQNQSESEDINKIQTWSNQNQHY 147
+QE ++D N + + +Q+ + + +E D +N+S+ + + ++ S +
Sbjct: 254 SQEEEEDEGNEQESEVESQASEEQTSEEEESASEEEDEENESKEQTTEEEESASEEEDEE 313
Query: 148 RNKPMVIVAPQVQQLQNSVFEDEQSSFNENEKPLLLPVRSLKSRLSDDDCVVQSQSVDGL 207
+ ++ +N+ E+E+ NE+++ S S D++ V + Q+ +G
Sbjct: 314 --------SASEREEKNASQEEEEDEGNESKEQTTEEEESA-SEEEDEESVSEEQTSEGE 364
Query: 208 SKTTTKRFSSNSFN----RVRNYAEFEGLVEDKLKQKEEENAVLPSPIPWRSRSASARMM 263
K ++ + N V + A E E++ +EE+ SAS
Sbjct: 365 EKGASQEEEEDEGNDQESEVESQASEEQTSEEEGASEEEDEENESEEQTTEEESASEE-- 422
Query: 264 EPKQEAIEKALKASMVMEPKQEANENASKPSMVKTSSIKFTPSESVAKNSEDLIKKKSFY 323
E ++ A E K + E + E NE S+ V++ + + SE K
Sbjct: 423 EDEESASEGEEKNASQEEEEDEGNEQESE---VESQASEEQTSEEEEKEGAS-------- 471
Query: 324 YKSYPPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEEGTNEKEDEDE-------E 376
E+N + EEG +E+EDE+ E
Sbjct: 472 -----------------------QEEDEENESEEQTSEEEEEGASEEEDEESAFEEQTSE 508
Query: 377 EKEYSTSSMQEQTDKERFIEKRVILEAEETDSDEDEDVGELKQSVKSVKTEE 428
E+E +S +E+ D+E E V +A E + E+E E Q + E+
Sbjct: 509 EEEEKGASQEEEEDEENEQESEVESQASEEQTSEEEGASEEGQDASEEEDED 560
Score = 43.5 bits (101), Expect = 0.015
Identities = 57/283 (20%), Positives = 113/283 (39%), Gaps = 22/283 (7%)
Query: 194 DDDCVVQSQSVDGLSKT--TTKRFSSNSFNRVRNYAEFEGLVEDKLKQKEE---ENAVLP 248
D+ V++ +S +G+ K T+K S ++ + E +D+ K + E E V
Sbjct: 79 DETTVIEKESENGVDKEKPTSKEESGEKTSQEKESEEKSSQEKDEDKSESEASEEKDVSQ 138
Query: 249 SPIPWRSRSASARMMEPKQEAIEKALKASMVMEPKQEANENASKPSMVKTSSIKFTPSES 308
+ AS + +E K S + + + E AS + S K T SE
Sbjct: 139 EQNSKEEKGASEEDEDTPEEQNSKEENGSSEEDDEDASEEQASNEEK-EASEEKNTVSEE 197
Query: 309 VAKNSEDLIKKKSFYYKSYPPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEEGTN 368
SE+ ++K ++S T ++ E++ + E+G +
Sbjct: 198 RKGASEEEDEEKDDGHESEVESQASEEQTTEEGAS---EEEDEESASEEQTSEGEEKGAS 254
Query: 369 EKEDEDE---EEKEYSTSSMQEQTDKERFI----------EKRVILEAEETDSDEDEDVG 415
++E+EDE +E E + + +EQT +E K E EE+ S+E+++
Sbjct: 255 QEEEEDEGNEQESEVESQASEEQTSEEEESASEEEDEENESKEQTTEEEESASEEEDEES 314
Query: 416 ELKQSVKSVKTEEPCSSSLSGGIDTVSDEGPDVDKKADEFIAK 458
++ K+ EE T +E +++ +E +++
Sbjct: 315 ASEREEKNASQEEEEDEGNESKEQTTEEEESASEEEDEESVSE 357
Score = 40.8 bits (94), Expect = 0.096
Identities = 44/228 (19%), Positives = 83/228 (36%), Gaps = 18/228 (7%)
Query: 207 LSKTTTKRFSSNSFNRVRNYAEFEGLVEDKLKQKEEENAVLPSPIPWRSRSASARMM--E 264
L+ T++ S +R+R ++ L L+ E+ W+ R ++ + E
Sbjct: 21 LTVTSSNAIPGRSSSRLRLLERYDSL--PSLRSHSEDRYDDGVDRKWKKREGNSDDICTE 78
Query: 265 PKQEAIEKALKASMVMEPKQEANENASKPSMVKTSSIKFTPSESVAKNSEDLIKKKSFYY 324
+ IEK + + E E+ K S K S K + + K+ + ++K
Sbjct: 79 DETTVIEKESENGVDKEKPTSKEESGEKTSQEKESEEKSSQEKDEDKSESEASEEKDVSQ 138
Query: 325 KSYPPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEEGTNEKEDEDEEEKEYSTSS 384
+ S K E E G++E++DED E++ S
Sbjct: 139 EQ--------------NSKEEKGASEEDEDTPEEQNSKEENGSSEEDDEDASEEQASNEE 184
Query: 385 MQEQTDKERFIEKRVILEAEETDSDEDEDVGELKQSVKSVKTEEPCSS 432
+ +K E+R EE + +D E++ +T E +S
Sbjct: 185 KEASEEKNTVSEERKGASEEEDEEKDDGHESEVESQASEEQTTEEGAS 232
>ref|XP_641133.1| hypothetical protein DDB0220644 [Dictyostelium discoideum]
gi|60469158|gb|EAL67154.1| CHD gene family protein
containing chromodomain, helicase domain, and
DNA-binding domain [Dictyostelium discoideum]
Length = 2373
Score = 53.1 bits (126), Expect = 2e-05
Identities = 56/255 (21%), Positives = 111/255 (42%), Gaps = 26/255 (10%)
Query: 162 LQNSVFEDEQSSFNENEKPLLLPVRSLKSRLSDDDCVVQSQSVDGLSKTTTKRFSSNSFN 221
L+N+ D ++ N+N+K S KS LS +D V+ K + N+
Sbjct: 49 LENNNDSDSDNNNNKNKKSTKSK-NSKKSNLSGEDSENDDNDVE-------KDENENNDV 100
Query: 222 RVRNYAEFEGLVEDKLKQKEEENAVLPSPIPWRSRSA-SARMMEPKQEAIEKALKASMVM 280
+++ E E E+ +++EE+++ + +SR A + +P K K+ +
Sbjct: 101 EMKDEEEEEEEEEELEEEEEEDDSEDSDDLSKKSRGRPKASVSKPPPP---KVPKSKPIA 157
Query: 281 EPKQEANENASKPSMVKTSSIKFTPSESVAKNSEDLIKKKSFYYKSYPPPPPPPPPTMFY 340
+ + N N + + + S I T + + + I PPP P P
Sbjct: 158 KTNNKKNSNTKEVIIKEKSKITTTTTTTTTATTTATITP--------PPPKPVSKPVGKP 209
Query: 341 KSTFLK--SRYGEKNGRNMSMGKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDKERFIEKR 398
K + K + +K S + EE +E E+++EEE+E + +++ +KE ++
Sbjct: 210 KGSVSKKAAALAKKKNNVQSESEESEESESESEEDEEEEEEKGANKNKKKNEKENQVKG- 268
Query: 399 VILEAEETDSDEDED 413
E E+ D++++ED
Sbjct: 269 ---EGEDDDNEKEED 280
>gb|EAA22916.1| erythrocyte membrane-associated giant protein antigen 332 [Plasmodium
yoelii yoelii]
Length = 3404
Score = 53.1 bits (126), Expect = 2e-05
Identities = 67/376 (17%), Positives = 151/376 (39%), Gaps = 41/376 (10%)
Query: 120 EDEVDHQNQSESE-DINKIQTWSNQNQHYRNKPMVIVAPQVQQLQNSVFEDEQSSFNENE 178
E+E + + + SE D+ I+ S+ + + V ++N ++E S E
Sbjct: 885 EEEEEEEEIARSEIDVGSIKNQSDAESEEEEEEITRTKVDVGSIKNQS-DNEIESEEEEM 943
Query: 179 KPLLLPVRSLKSRLSDDDCVVQSQSVDGLSKTTTKRFSSNSFNRVRNYAEFEGLVEDKLK 238
+ + V S+K++ D + +++S + ++++ + + N + +E E E+++
Sbjct: 944 ARIEIDVESIKNQ-HDAESESENESEEEIARSKIDTATIKNQNDTESESESE---EEEIT 999
Query: 239 QKEEENAVLPSPIPWRSRSASARMMEPKQEAIEKALKASMVMEPKQEANENASKPSMVKT 298
+ E + + + S + + + +E ++K E + E E A +
Sbjct: 1000 RSEINVSSIKNQNDSESENEEEEEIARSEVNVE-SIKNQNYAESESEEKEIARNE--IDV 1056
Query: 299 SSIKFTPSESVAKNSEDLIKKKSF--------YYKSYPPPPPPPPPTMFYKSTFLKSRYG 350
+SIK + E +I+ K + YY+S + +T Y
Sbjct: 1057 NSIKNQSDNEIESEEEKIIRTKVYVSRIKSQSYYESESEEEEEIARSDIDAATIKNQNYA 1116
Query: 351 EKNGRNMSMGKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDKERF----IEKRVILEAE-- 404
E N S +S E +E E E+E E E S +E+ + + I+ + E+E
Sbjct: 1117 ENESENESENESESENESENESENESENESENESEEEEIARSKIDVGTIKNQNYAESEEE 1176
Query: 405 ------------------ETDSDEDEDVGELKQSVKSVKTEEPCSSSLSGGIDTVSDEGP 446
E++++E+E++ + +V+S+K + S + ++
Sbjct: 1177 GITRNEIDVNSIKNQNDNESENEEEEEIARSEVNVESIKNQHDAESDNEIEEEEIAQSEI 1236
Query: 447 DVDKKADEFIAKFREQ 462
DVD ++ ++ E+
Sbjct: 1237 DVDSIENQSDSEIEEE 1252
Score = 46.2 bits (108), Expect = 0.002
Identities = 71/365 (19%), Positives = 146/365 (39%), Gaps = 28/365 (7%)
Query: 120 EDEVDHQNQSESEDINKIQTWSNQNQHYRNKPMVIVAPQVQQLQNSVFEDEQSSFNENEK 179
E+E Q++ + E I ++++ ++ + V ++N ++E S E
Sbjct: 606 EEEEIAQSEIDVESIKNQNDTESESEIEEDEEITRTKVDVGSIKNQS-DNEIESEEEEMA 664
Query: 180 PLLLPVRSLKSRL-------SDDDCVVQSQ-SVDGLSKTTTKRFSSNSFNRVRNYAEFEG 231
+ V S+K++ S+++ + QS+ VD + + R +
Sbjct: 665 RSEIDVESIKNQHDAESESESEEEEIAQSEIDVDSIENQSDSEIEEEEEEITRTKVDVNT 724
Query: 232 LVEDKLKQKEEENAVLPSPIPWRS-RSASARMMEPKQEAIEKALKASMVMEPKQEANENA 290
+ + EEE ++ S I S ++ S +E ++E I + + + E
Sbjct: 725 IKNQSDTESEEEEGIIRSEIDADSIKNQSDSEIEEEEEIIRTKVDVNTIKNQSDNGIEEE 784
Query: 291 SKPSMVKTSSIKFTPSESVAKNSEDLIKKKSFYYKSYPPPPPPPPPTMFYKSTFLKSRYG 350
S + S+K S+S ++ E I + S + +S
Sbjct: 785 IARSKINVGSVK-NQSDSEIESEEGEIDRSEIDIDSIKNQNDTESES--------ESESE 835
Query: 351 EKNGRNMSMGKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDKERFIEKRVILEAEETDSDE 410
E+ R+ +SI+ N+ E E+E E+E + S + T I+ + E+EE + +E
Sbjct: 836 EEIARSKINVESIKN-KNDAESENESEEEIARSKIDVGT-----IKNQNYAESEEEEEEE 889
Query: 411 DEDVGELKQSVKSVKTEEPCSSSLSGG--IDTVSDEGPDVDKKADEFIAKFREQIRLQ-R 467
+E++ + V S+K + S T D G ++ +E ++ E R++
Sbjct: 890 EEEIARSEIDVGSIKNQSDAESEEEEEEITRTKVDVGSIKNQSDNEIESEEEEMARIEID 949
Query: 468 IESIK 472
+ESIK
Sbjct: 950 VESIK 954
Score = 43.5 bits (101), Expect = 0.015
Identities = 68/400 (17%), Positives = 166/400 (41%), Gaps = 49/400 (12%)
Query: 96 NNNNTKFDNAQSLVSRFLQVSSFFEDEVDHQNQSESED--------INKIQTWSNQNQHY 147
N N+T+ ++ + ++R + +++ D +++S+ E+ ++ I+ S+
Sbjct: 1625 NQNDTESESEEDEITRTKLDVNSIKNQNDTESESKEEEEIARSEVNVDPIKNQSDNEIES 1684
Query: 148 RNKPMVIVAPQVQQLQN-SVFEDEQSSFNENEKPLLLPVRSLKSRLSDDDCVVQSQSVDG 206
+ ++ V +++N S +E E+ + + V +K++ + +S+ +
Sbjct: 1685 EEEKIIRTKVYVSRIKNQSYYESEEEEITRTK----IDVDPIKNQNYAES---ESEEEEE 1737
Query: 207 LSKTTTKRFSSNSFNRVRNYAEFEGLVEDKLKQKEEENAVLPSPIPWRSRSASARMMEPK 266
+++ S + +R++N +E E E ++ + + + A + + S + S K
Sbjct: 1738 ITR------SEVNVSRIQNQSESE---EGEIARSDMDAATIKNQNDTESENESEEEEVAK 1788
Query: 267 QEAIEKALK--ASMVMEPKQEANENASKPSMVKTSSIKFTPSESVAKNSEDLIKKKSFYY 324
E ++K + +E ++E E + V SSIK S+S ++ E+ I +
Sbjct: 1789 SEIDVSSIKKQSDNEIESEEEEEEEGITRNEVDVSSIK-NQSDSENESEEEEIARSEVNV 1847
Query: 325 KSYPPPPPPPPP-----------TMFYKSTFLKSRYGEKNGRNMSMGKSIEEGTNEKEDE 373
+S + + +S E +++G ++ NE E+E
Sbjct: 1848 ESIKNQNDTESEEDEITRNEIDVSSIKNQSESESEEEEITRSEIAVGSIKKQSDNEIEEE 1907
Query: 374 DEEEKEYSTSSMQEQTDKERFIEKRVILEAE----------ETDSDEDEDVGELKQSVKS 423
+ + + S++ + D E E+ I +E +T+S+E+E++ + + V S
Sbjct: 1908 EIARTKINAESIKNKNDAESEEEEEGITRSEIDVNNIKNQNDTESEEEEEIAQSEIDVGS 1967
Query: 424 VKTEEPCSSSLSGGIDTVSDEGPDVDKKADEFIAKFREQI 463
+K + S I + + ++D I E+I
Sbjct: 1968 IKNQSDAESEEEEEITRTKVDVNTIKNQSDNEIESEEEKI 2007
Score = 41.2 bits (95), Expect = 0.073
Identities = 67/374 (17%), Positives = 142/374 (37%), Gaps = 71/374 (18%)
Query: 96 NNNNTKFDNAQSLVSRFLQVSSFFEDEVDH--------QNQSESEDINK-------IQTW 140
N N+T+ +N E+E+D QNQS++E + I
Sbjct: 1269 NQNDTESENESE------------EEEIDRNEIDVGSIQNQSDAESEEEEEIARSDIDAA 1316
Query: 141 SNQNQHYRNKPMVIVAPQVQQLQNSVFEDEQSSFNENEKPLL----LPVRSLKSRLSDDD 196
+ +NQ+Y ++ + + V + S E+EK + + V S+K++ +D
Sbjct: 1317 TTKNQNYAESENESEEEEIDKSEIDVGSIQNQSNAESEKEEITRSEINVNSIKNQ---ND 1373
Query: 197 CVVQSQSVDGLSKTTTKRFSSNSFNRVRNYAEFEGLVEDKLK----QKEEENAVLPSPIP 252
+++ + ++++ S + N + +E E + ++ +K+ +N +
Sbjct: 1374 SESENEEEEEIARSEVNVESIKNQNDTESESEEEEIARYEIAVGSIKKQSDNEI------ 1427
Query: 253 WRSRSASARMMEPKQEAIEKALKASMVMEPKQEANENASKPSMVKTSSIKFTPSESVAKN 312
S + E ++K +E + E+ E S + +SIK ++S ++
Sbjct: 1428 ---ESEEEEEGITRNEIDVSSIKNQSDVESENESEEKGITKSEIDVNSIK---NQSYYES 1481
Query: 313 SEDLIKKKSFYYKSYPPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEEGT----- 367
E+ I + Y +Y+S + E + + EEG
Sbjct: 1482 DEEKIIRTKVYVSRIKNQS-------YYESEEEEITRSEVDVSRIQNQSESEEGEIARSD 1534
Query: 368 ---------NEKEDEDEEEKEYSTSSMQEQTDKERFIEKRVILEAEETDSDEDEDVGELK 418
N+ E EDE E E S +E+ + + +++ E+E++ K
Sbjct: 1535 IDTATINNQNDTESEDESENESENESEEEEITRSEIAVGSIKKQSDNEIESEEEEITRTK 1594
Query: 419 QSVKSVKTEEPCSS 432
+V+S+K + S
Sbjct: 1595 INVESIKNKNDAES 1608
Score = 39.7 bits (91), Expect = 0.21
Identities = 86/460 (18%), Positives = 180/460 (38%), Gaps = 72/460 (15%)
Query: 80 ISYGLFSRRNQEPDKDNNNNTKFDNAQSLVSRFLQVSSFFEDEVDHQNQSESEDIN--KI 137
I G +N ++ + + A+S + V S +++ D +++ E E+I K+
Sbjct: 869 IDVGTIKNQNYAESEEEEEEEEEEIARSEID----VGSI-KNQSDAESEEEEEEITRTKV 923
Query: 138 QTWSNQNQ-----HYRNKPMVIVAPQVQQLQNSVFEDEQSSFNENEKPLLLPVRSLKSRL 192
S +NQ + M + V+ ++N + E S NE+E+ + +
Sbjct: 924 DVGSIKNQSDNEIESEEEEMARIEIDVESIKNQ-HDAESESENESEEEIARSKIDTATIK 982
Query: 193 SDDDCVVQSQSVDGLSKTTTKRFSSNSFNRVRNYAEFEGLVEDKLKQKE----------- 241
+ +D +S+S + + T + +S +N +E E E+++ + E
Sbjct: 983 NQNDTESESESEE--EEITRSEINVSSIKN-QNDSESENEEEEEIARSEVNVESIKNQNY 1039
Query: 242 -----EENAVLPSPIPWRS-RSASARMMEPKQEAIEKA------LKASMVMEPKQEANEN 289
EE + + I S ++ S +E ++E I + +K+ E + E E
Sbjct: 1040 AESESEEKEIARNEIDVNSIKNQSDNEIESEEEKIIRTKVYVSRIKSQSYYESESEEEEE 1099
Query: 290 ASKPSM----VKTSSIKFTPSESVAKN-------SEDLIKKKSFYYKSYPPPPPPPPPTM 338
++ + +K + SE+ ++N SE+ + +S +
Sbjct: 1100 IARSDIDAATIKNQNYAENESENESENESESENESENESENESENESENESEEEEIARSK 1159
Query: 339 FYKSTFLKSRYGEKNGRNMSMGK----SIE-EGTNEKEDEDEEE---KEYSTSSMQEQTD 390
T Y E ++ + SI+ + NE E+E+EEE E + S++ Q D
Sbjct: 1160 IDVGTIKNQNYAESEEEGITRNEIDVNSIKNQNDNESENEEEEEIARSEVNVESIKNQHD 1219
Query: 391 KE--RFIEKRVILEAE------------ETDSDEDEDVGELKQSVKSVKTEEPCSSSLSG 436
E IE+ I ++E E + +E+E++ K V ++K + S
Sbjct: 1220 AESDNEIEEEEIAQSEIDVDSIENQSDSEIEEEEEEEITRTKVDVNTIKNQNDTESENES 1279
Query: 437 GIDTVSDEGPDVDKKADEFIAKFREQIRLQRIESIKRSTR 476
+ + DV ++ A+ E+ + R + +T+
Sbjct: 1280 EEEEIDRNEIDVGSIQNQSDAESEEEEEIARSDIDAATTK 1319
Score = 39.3 bits (90), Expect = 0.28
Identities = 58/336 (17%), Positives = 136/336 (40%), Gaps = 50/336 (14%)
Query: 120 EDEVDHQNQSESE-DINKIQTWSNQNQHYRNKPMVIVAPQVQQLQNSVFEDEQSSFNENE 178
E E + + + SE D+N I+ NQN + I ++ D +S E
Sbjct: 1926 ESEEEEEGITRSEIDVNNIK---NQNDTESEEEEEIAQSEIDVGSIKNQSDAESEEEEEI 1982
Query: 179 KPLLLPVRSLKSRLSDDDCVVQSQSVDGLSKTTTKRFSSNSFNRVRNYAEFEGLVEDKLK 238
+ V ++K++ SD++ + + + TK + NR++N + +E E+++
Sbjct: 1983 TRTKVDVNTIKNQ-SDNEIESEEEKI-----IRTKVY----VNRIKNQSYYESESEEEIT 2032
Query: 239 QKEEENAVLPSPIPWRSRSASARMMEPKQEAIEKALKASMVMEPKQEANENASKPSMVKT 298
+ E + + + + S + S E ++E I ++ + + KQ NE+ K
Sbjct: 2033 RSEIDVSSIKNQNDTESENESESESESEEEEITRS-EIDVGSIKKQSDNESEEKEITRTK 2091
Query: 299 SSIKFTPSESVAKNSEDLIKKKSFYYKSYPPPPPPPPPTMFYKSTFLKSRYGEKNGRNMS 358
+++ +++ A++ E+ + ++ + +N +
Sbjct: 2092 INVESIKNKNDAESEEE-------------------------EEGITRNEIDVSSIKNQN 2126
Query: 359 MGKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDKE----RFIEKRVILEA------EETDS 408
+S E +E E+E+ E S+++Q+D E ++ +E+ E++
Sbjct: 2127 DTESENESESESEEEEIARSEIDVGSIKKQSDNESEEKEITRTKINVESIKNKNDAESEE 2186
Query: 409 DEDEDVGELKQSVKSVKTEEPCSSSLSGGIDTVSDE 444
+E+E + + V S+K + S ++ +E
Sbjct: 2187 EEEEGITRNEIDVSSIKNQNDTESENENESESEEEE 2222
Score = 38.9 bits (89), Expect = 0.36
Identities = 66/379 (17%), Positives = 141/379 (36%), Gaps = 37/379 (9%)
Query: 86 SRRNQEPDKDNNNNTKFDNAQSLVSRFLQVSSFFEDEVDHQNQSESEDINK-------IQ 138
SR + + + + D + ++ S E E + +N+SE E+I + I+
Sbjct: 1517 SRIQNQSESEEGEIARSDIDTATINNQNDTESEDESENESENESEEEEITRSEIAVGSIK 1576
Query: 139 TWSNQNQHYRNKPMVIVAPQVQQLQN---SVFEDEQSSFNENEKPLLLPVRSLKSRLSDD 195
S+ + + V+ ++N + E+E+ + +E + +S +D
Sbjct: 1577 KQSDNEIESEEEEITRTKINVESIKNKNDAESEEEEIARSEVNVESIKNQNDTESESEED 1636
Query: 196 DCVVQSQSVDGL-SKTTTKRFSSNSFNRVRNYAEFEGLVEDKLKQKEEENAVLPSPIPWR 254
+ V+ + ++ T+ S R+ + + + E E + +
Sbjct: 1637 EITRTKLDVNSIKNQNDTESESKEEEEIARSEVNVDPIKNQSDNEIESEEEKIIRTKVYV 1696
Query: 255 SRSASARMMEPKQEAIEKALKASMVMEPKQEANENASKPSMVKTSSIKFTPSESVAKNSE 314
SR + E ++E I + ++ + A + + + S + + ++ +++ E
Sbjct: 1697 SRIKNQSYYESEEEEITRTKIDVDPIKNQNYAESESEEEEEITRSEVNVSRIQNQSESEE 1756
Query: 315 DLIKKKSFYYKSYPPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEEGTNEKEDED 374
I + + + KS ++S K + E E+E+
Sbjct: 1757 GEIARSDMDAATIKNQNDTESENESEEEEVAKSEI------DVSSIKKQSDNEIESEEEE 1810
Query: 375 EEE----KEYSTSSMQEQTDKERFIEKRVILEAE----------ETDSDEDE------DV 414
EEE E SS++ Q+D E E+ I +E +T+S+EDE DV
Sbjct: 1811 EEEGITRNEVDVSSIKNQSDSENESEEEEIARSEVNVESIKNQNDTESEEDEITRNEIDV 1870
Query: 415 GELKQSVKSVKTEEPCSSS 433
+K +S EE + S
Sbjct: 1871 SSIKNQSESESEEEEITRS 1889
>emb|CAD59239.1| NAD(+) ADP-ribosyltransferase-3 [Dictyostelium discoideum]
Length = 2536
Score = 52.8 bits (125), Expect = 2e-05
Identities = 80/351 (22%), Positives = 132/351 (36%), Gaps = 62/351 (17%)
Query: 111 RFLQVSSFFEDEVDHQNQSESEDIN----------KIQTWSNQNQHYRNKPMVI--VAPQ 158
R+L VS EDE +++ ES N ++ S Q YR + + V
Sbjct: 54 RWLTVSD--EDETEYKVGPESSSSNIPIESIIVTTTLKANSKQESSYRISKIELRRVNTI 111
Query: 159 VQQLQNSVFEDEQSSFNENEKP----------------LLLPVRSLKSRLSDDDCVVQSQ 202
V+ +S E+E+ S E EK +PV+ K +L ++ Q +
Sbjct: 112 VENESSSEEEEEEESNGEEEKEGNAQESGSESEESESESEVPVKKTKKQLVEE----QKK 167
Query: 203 SVDGLSKTTTKRFSSNSFNRVRNYAEFEGLVEDKLKQKEEENAVLPSPIPWRSRSASARM 262
S K ++ + + + E E E KQKE+E +++ +
Sbjct: 168 SAAAAKKLQKEKEKEKEKEKAKKFKEKEKEKEKAKKQKEKEKE--------KAKKLKEKE 219
Query: 263 MEPKQEAIEKALKASMVMEPKQEANENASKPSMVKTSSIKFTPSESVAKNSEDLIKKKSF 322
E K++ E KQE EN S+ S + SE + + KKK+
Sbjct: 220 KEKKKKNSESEKPKKSNKSKKQEDTENESESESESESESSSSESEKEESSKKSSSKKKTT 279
Query: 323 YYKSYPPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEEGTNEKEDEDEEEKEYST 382
+ P KS ++ E++G + S G E+ ++ DEDE EK+ S
Sbjct: 280 TKEKSKPTK---------KSKKQETEDEEESGSDESSGDDDEKSSS---DEDEREKKSSK 327
Query: 383 SSMQEQTDKERFIE--------KRVILEAEETDSDEDEDVGELKQSVKSVK 425
S K + I+ KR E + S ++E+ E K + K K
Sbjct: 328 KSSTAANGKPQEIKPIVKKSTGKRKKNEDSDASSGDEEEEEEDKTTTKKSK 378
Score = 38.9 bits (89), Expect = 0.36
Identities = 63/307 (20%), Positives = 109/307 (34%), Gaps = 36/307 (11%)
Query: 175 NENEKPLLLPVRSLKSRLSDDDCVVQSQSVDGLSKTTTKRFSSNSFNRVRNYAEFEGLVE 234
+E+E + S S + + +V + + ++ R S RV E E E
Sbjct: 60 DEDETEYKVGPESSSSNIPIESIIVTTTLKANSKQESSYRISKIELRRVNTIVENESSSE 119
Query: 235 DKLKQKEEENAVLPSPIPWRSRSASARMMEPKQEAIEKALKASMVMEPKQEANENASKPS 294
++ ++EE N + + + E + E K K +V E K+ A
Sbjct: 120 EE--EEEESNGEEEKEGNAQESGSESEESESESEVPVKKTKKQLVEEQKKSA-------- 169
Query: 295 MVKTSSIKFTPSESVAKNSEDLIKKKSFYYKSYPPPPPPPPPTMFYKSTFLKSRYGEKNG 354
++ K + K E K K F K K + EK
Sbjct: 170 ---AAAKKLQKEKEKEKEKE---KAKKFKEKE--------------KEKEKAKKQKEKEK 209
Query: 355 RNMSMGKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDKERFIEKRVILEAEETDSDEDEDV 414
K E+ +K E E+ K+ + S QE T+ E E E+ ++S+++E
Sbjct: 210 EKAKKLKEKEKEKKKKNSESEKPKKSNKSKKQEDTENESESESESESESSSSESEKEES- 268
Query: 415 GELKQSVKSVKTEE---PCSSSLSGGIDTVSDEGPDVDKKADEFIAKFREQIRLQRIESI 471
+ S K T+E P S + + G D D+ + E R ++ S
Sbjct: 269 SKKSSSKKKTTTKEKSKPTKKSKKQETEDEEESGSDESSGDDDEKSSSDEDEREKK--SS 326
Query: 472 KRSTRVA 478
K+S+ A
Sbjct: 327 KKSSTAA 333
>emb|CAD38875.1| hypothetical protein [Homo sapiens]
Length = 819
Score = 52.4 bits (124), Expect = 3e-05
Identities = 78/375 (20%), Positives = 150/375 (39%), Gaps = 52/375 (13%)
Query: 88 RNQEPDKDNNN---NTKFDNAQSLVSRFLQVSSFFEDEV--DHQNQSESEDINKIQTWSN 142
R + P D+ N N +++S +R L S +E+ H + SE+ED+ K +
Sbjct: 100 RAEPPASDSENEDVNQHGSDSESEETRKLPGSDSENEELLNGHASDSENEDVGKHPASDS 159
Query: 143 QNQHYRNKPMV------IVAPQVQQLQNSVFEDEQSSFNENEKPLLLPVRSLKSRLSDDD 196
+ + + P + PQ+ ++ Q+S +ENE+P K R+SD +
Sbjct: 160 EIEELQKSPASDSETEDALKPQISDSESEEPPRHQASDSENEEPP-------KPRMSDSE 212
Query: 197 C--VVQSQSVDGLSKTTTKRFSSNSFNRVR---NYAEFEGLVEDKLKQKEEENAVLPSPI 251
+ + Q D S+ + +S+S N ++ E + + + EN LP P
Sbjct: 213 SEELPKPQVSDSESEEPPRHQASDSENEELPKPRISDSESEDPPRHQASDSENEELPKPR 272
Query: 252 --------PWRSRSASARMME-PKQEAIEKALKASMVMEPKQEANENASKPSMVKTSSIK 302
P R++++ + E PK + + E+AS+ S
Sbjct: 273 ISDSESEDPPRNQASDSENEELPKPRVSDSESEGPQKGPASDSETEDASRHKQKPESD-- 330
Query: 303 FTPSESVAKNSEDLIKKKSFYYKSYPPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKS 362
S+ K + ++ SF+ S+ + F S E+ + M
Sbjct: 331 -DDSDRENKGEDTEMQNDSFHSDSHMD-----------RKKFHSSDSEEEEHKKQKM--- 375
Query: 363 IEEGTNEKEDEDEEEKEYSTSSMQEQTDKERFIEK--RVILEAEETDSDEDEDVGELKQS 420
+ +EKE E+E+ + + + + D+E+ K RV+ +A+++DSD D ++
Sbjct: 376 -DSDEDEKEGEEEKVAKRKAAVLSDSEDEEKASAKKSRVVSDADDSDSDAVSDKSGKREK 434
Query: 421 VKSVKTEEPCSSSLS 435
+ +EE LS
Sbjct: 435 TIASDSEEEAGKELS 449
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.309 0.126 0.342
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 817,400,024
Number of Sequences: 2540612
Number of extensions: 37362431
Number of successful extensions: 407478
Number of sequences better than 10.0: 4108
Number of HSP's better than 10.0 without gapping: 702
Number of HSP's successfully gapped in prelim test: 3673
Number of HSP's that attempted gapping in prelim test: 353799
Number of HSP's gapped (non-prelim): 21847
length of query: 483
length of database: 863,360,394
effective HSP length: 132
effective length of query: 351
effective length of database: 527,999,610
effective search space: 185327863110
effective search space used: 185327863110
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 77 (34.3 bits)
Medicago: description of AC149079.6


