

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148996.5 + phase: 0
(196 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
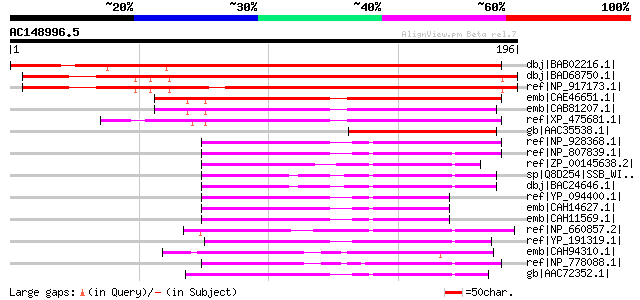
dbj|BAB02216.1| unnamed protein product [Arabidopsis thaliana] g... 215 6e-55
dbj|BAD68750.1| single-strand DNA binding protein-like [Oryza sa... 206 3e-52
ref|NP_917173.1| P0510C12.16 [Oryza sativa (japonica cultivar-gr... 191 1e-47
emb|CAE46651.1| single-stranded DNA binding protein [Arabidopsis... 99 7e-20
emb|CAB81207.1| putative protein [Arabidopsis thaliana] gi|48502... 98 1e-19
ref|XP_475681.1| putative single-strand DNA binding protein [Ory... 97 3e-19
gb|AAC35538.1| contains similarity to the single-strand binding ... 58 1e-07
ref|NP_928368.1| hypothetical protein plu1039 [Photorhabdus lumi... 55 8e-07
ref|NP_807839.1| single strand binding protein [Salmonella enter... 54 2e-06
ref|ZP_00145638.2| COG0629: Single-stranded DNA-binding protein ... 54 2e-06
sp|Q8D254|SSB_WIGBR Single-strand binding protein (SSB) (Helix-d... 54 2e-06
dbj|BAC24646.1| ssb [Wigglesworthia glossinidia endosymbiont of ... 54 2e-06
ref|YP_094400.1| single strand binding protein [Legionella pneum... 53 4e-06
emb|CAH14627.1| Single-strand binding protein (SSB) (Helix-desta... 53 4e-06
emb|CAH11569.1| Single-strand binding protein (SSB) (Helix-desta... 53 4e-06
ref|NP_660857.2| single-strand binding protein [Buchnera aphidic... 52 9e-06
ref|YP_191319.1| Single-strand DNA binding protein [Gluconobacte... 52 9e-06
emb|CAH94310.1| single-strand binding protein, putative [Plasmod... 52 1e-05
ref|NP_778088.1| single-strand binding protein [Buchnera aphidic... 50 3e-05
gb|AAC72352.1| single strand DNA-binding protein; SSB [Vibrio ch... 50 3e-05
>dbj|BAB02216.1| unnamed protein product [Arabidopsis thaliana]
gi|15229675|ref|NP_188488.1| single-strand-binding
family protein [Arabidopsis thaliana]
Length = 217
Score = 215 bits (547), Expect = 6e-55
Identities = 112/199 (56%), Positives = 142/199 (71%), Gaps = 14/199 (7%)
Query: 1 MAKFSISRTSTLYRSLLSTPPPPLHHHRHIPLRFCT---TTTSLSDYDDADSAAPSPSPE 57
MA + + LYRSLLS P + FCT T+ SD+D+ +S + +
Sbjct: 1 MANSMATLSRRLYRSLLSNP-----RISQASMSFCTNNITSPEDSDFDELESPIEPKASD 55
Query: 58 QT------ERTFFDRPLENGLDPGVYRAILVGKAGQKPLQKKLKSGTVVTLLSIGTGGIR 111
ER +RPLENGLD G+++AILVG+ GQ PLQKKLKSG VTL S+GTGGIR
Sbjct: 56 PVSRFSGEERVMEERPLENGLDSGIFKAILVGQVGQLPLQKKLKSGRTVTLFSVGTGGIR 115
Query: 112 NNRRPLDHENPREYANRCAVQWHRVTVYPERLGNLLMKTVLPGSTLYVEGNLETKVFSDP 171
NNRRPL +E+PREYA+R AVQWHRV+VYPERL +L++K V PG+ +Y+EGNLETK+F+DP
Sbjct: 116 NNRRPLINEDPREYASRSAVQWHRVSVYPERLADLVLKNVEPGTVIYLEGNLETKIFTDP 175
Query: 172 VTGLVRRVREVAVRRNGNL 190
VTGLVRR+REVA+RRNG +
Sbjct: 176 VTGLVRRIREVAIRRNGRV 194
>dbj|BAD68750.1| single-strand DNA binding protein-like [Oryza sativa (japonica
cultivar-group)]
Length = 224
Score = 206 bits (524), Expect = 3e-52
Identities = 115/214 (53%), Positives = 139/214 (64%), Gaps = 33/214 (15%)
Query: 6 ISRTSTLYRSLLSTPPPPLHHHRHIPLRFCTTTTSLSDYDDA---DSAAPS---PSPEQT 59
++R L R +LS+P P F TT +S S + DS A S P PEQ
Sbjct: 9 LARRLLLTRRVLSSPLRP----------FSTTDSSSSSSSSSSSDDSRAGSDAGPDPEQQ 58
Query: 60 E--------------RTFFDRPLENGLDPGVYRAILVGKAGQKPLQKKLKSGTVVTLLSI 105
+ R RPLENGLDPG+Y+AI+VGK GQ+P+QK+L+SG V L S+
Sbjct: 59 QPPPAGQDQQAAARPRAGDTRPLENGLDPGIYKAIMVGKVGQEPIQKRLRSGRTVVLFSL 118
Query: 106 GTGGIRNNRRPLDHENPREYANRCAVQWHRVTVYPERLGNLLMKTVLPGSTLYVEGNLET 165
GTGGIRNNRRPLD E P +YA RC+VQWHRV +YPERLG+L +K V GS LY+EGNLET
Sbjct: 119 GTGGIRNNRRPLDREEPHQYAERCSVQWHRVCIYPERLGSLALKHVKTGSVLYLEGNLET 178
Query: 166 KVFSDPVTGLVRRVREVAVRRNGN---LANSSNA 196
KVFSDP+TGLVRR+RE+AVR NG L N NA
Sbjct: 179 KVFSDPITGLVRRIREIAVRSNGRLLFLGNDCNA 212
>ref|NP_917173.1| P0510C12.16 [Oryza sativa (japonica cultivar-group)]
Length = 218
Score = 191 bits (485), Expect = 1e-47
Identities = 111/214 (51%), Positives = 133/214 (61%), Gaps = 39/214 (18%)
Query: 6 ISRTSTLYRSLLSTPPPPLHHHRHIPLRFCTTTTSLSDYDDA---DSAAPS---PSPEQT 59
++R L R +LS+P P F TT +S S + DS A S P PEQ
Sbjct: 9 LARRLLLTRRVLSSPLRP----------FSTTDSSSSSSSSSSSDDSRAGSDAGPDPEQQ 58
Query: 60 E--------------RTFFDRPLENGLDPGVYRAILVGKAGQKPLQKKLKSGTVVTLLSI 105
+ R RPLENGLDPG+Y K GQ+P+QK+L+SG V L S+
Sbjct: 59 QPPPAGQDQQAAARPRAGDTRPLENGLDPGIY------KVGQEPIQKRLRSGRTVVLFSL 112
Query: 106 GTGGIRNNRRPLDHENPREYANRCAVQWHRVTVYPERLGNLLMKTVLPGSTLYVEGNLET 165
GTGGIRNNRRPLD E P +YA RC+VQWHRV +YPERLG+L +K V GS LY+EGNLET
Sbjct: 113 GTGGIRNNRRPLDREEPHQYAERCSVQWHRVCIYPERLGSLALKHVKTGSVLYLEGNLET 172
Query: 166 KVFSDPVTGLVRRVREVAVRRNGN---LANSSNA 196
KVFSDP+TGLVRR+RE+AVR NG L N NA
Sbjct: 173 KVFSDPITGLVRRIREIAVRSNGRLLFLGNDCNA 206
>emb|CAE46651.1| single-stranded DNA binding protein [Arabidopsis thaliana]
gi|28827408|gb|AAO50548.1| unknown protein [Arabidopsis
thaliana] gi|28393531|gb|AAO42186.1| unknown protein
[Arabidopsis thaliana] gi|30681642|ref|NP_192844.2|
single-strand-binding family protein [Arabidopsis
thaliana] gi|38258307|sp|Q84J78|SSB_ARATH
Single-stranded DNA-binding protein, mitochondrial
precursor
Length = 201
Score = 99.0 bits (245), Expect = 7e-20
Identities = 53/143 (37%), Positives = 86/143 (60%), Gaps = 15/143 (10%)
Query: 57 EQTERTFFDRP--LENGLDP-------GVYRAILVGKAGQKPLQKKLKSGTVVTLLSIGT 107
E E +RP +G+DP GV+RAI+ GK GQ PLQK L++G VT+ ++GT
Sbjct: 43 EDFEENVTERPELQPHGVDPRKGWGFRGVHRAIICGKVGQAPLQKILRNGRTVTIFTVGT 102
Query: 108 GGIRNNRRPLDHENPREYANRCAVQWHRVTVYPERLGNLLMKTVLPGSTLYVEGNLETKV 167
GG+ + R P+ QWHR+ V+ E LG+ ++ + S++YVEG++ET+V
Sbjct: 103 GGMFDQRLVGATNQPKP------AQWHRIAVHNEVLGSYAVQKLAKNSSVYVEGDIETRV 156
Query: 168 FSDPVTGLVRRVREVAVRRNGNL 190
++D ++ V+ + E+ VRR+G +
Sbjct: 157 YNDSISSEVKSIPEICVRRDGKI 179
>emb|CAB81207.1| putative protein [Arabidopsis thaliana] gi|4850285|emb|CAB43041.1|
putative protein [Arabidopsis thaliana]
gi|7487337|pir||T08185 hypothetical protein T22B4.40 -
Arabidopsis thaliana
Length = 254
Score = 98.2 bits (243), Expect = 1e-19
Identities = 53/141 (37%), Positives = 85/141 (59%), Gaps = 15/141 (10%)
Query: 57 EQTERTFFDRP--LENGLDP-------GVYRAILVGKAGQKPLQKKLKSGTVVTLLSIGT 107
E E +RP +G+DP GV+RAI+ GK GQ PLQK L++G VT+ ++GT
Sbjct: 43 EDFEENVTERPELQPHGVDPRKGWGFRGVHRAIICGKVGQAPLQKILRNGRTVTIFTVGT 102
Query: 108 GGIRNNRRPLDHENPREYANRCAVQWHRVTVYPERLGNLLMKTVLPGSTLYVEGNLETKV 167
GG+ + R P+ QWHR+ V+ E LG+ ++ + S++YVEG++ET+V
Sbjct: 103 GGMFDQRLVGATNQPKP------AQWHRIAVHNEVLGSYAVQKLAKNSSVYVEGDIETRV 156
Query: 168 FSDPVTGLVRRVREVAVRRNG 188
++D ++ V+ + E+ VRR+G
Sbjct: 157 YNDSISSEVKSIPEICVRRDG 177
>ref|XP_475681.1| putative single-strand DNA binding protein [Oryza sativa (japonica
cultivar-group)] gi|48475206|gb|AAT44275.1| putative
single-strand DNA binding protein [Oryza sativa
(japonica cultivar-group)]
Length = 206
Score = 96.7 bits (239), Expect = 3e-19
Identities = 55/163 (33%), Positives = 94/163 (56%), Gaps = 19/163 (11%)
Query: 36 TTTTSLSDYDDADSAAPSPSPEQTERTFFDRPLE-NGLDP-------GVYRAILVGKAGQ 87
++T S SD D+ S + T R LE +DP GV+RAI+ GK GQ
Sbjct: 33 SSTVSFSDIDEK-----SEMGGDDDYTDSRRELEPQSVDPKKGWGFRGVHRAIICGKVGQ 87
Query: 88 KPLQKKLKSGTVVTLLSIGTGGIRNNRRPLDHENPREYANRCAVQWHRVTVYPERLGNLL 147
P+QK L++G VT+ ++GTGG+ + R D + P+ QWHR+ ++ ++LG
Sbjct: 88 VPVQKILRNGRTVTVFTVGTGGMFDQRVVGDADLPKP------AQWHRIAIHNDQLGAFA 141
Query: 148 MKTVLPGSTLYVEGNLETKVFSDPVTGLVRRVREVAVRRNGNL 190
++ ++ S +YVEG++ET+V++D + V+ + E+ +RR+G +
Sbjct: 142 VQKLVKNSAVYVEGDIETRVYNDSINDQVKNIPEICLRRDGKI 184
>gb|AAC35538.1| contains similarity to the single-strand binding proteins family
(Pfam: SSB.hmm, score: 24.02) [Arabidopsis thaliana]
Length = 150
Score = 58.2 bits (139), Expect = 1e-07
Identities = 23/57 (40%), Positives = 42/57 (73%)
Query: 132 QWHRVTVYPERLGNLLMKTVLPGSTLYVEGNLETKVFSDPVTGLVRRVREVAVRRNG 188
QWHR+ V+ E LG+ ++ + S++YVEG++ET+V++D ++ V+ + E+ VRR+G
Sbjct: 17 QWHRIAVHNEVLGSYAVQKLAKNSSVYVEGDIETRVYNDSISSEVKSIPEICVRRDG 73
>ref|NP_928368.1| hypothetical protein plu1039 [Photorhabdus luminescens subsp.
laumondii TTO1] gi|36784450|emb|CAE13334.1| unnamed
protein product [Photorhabdus luminescens subsp.
laumondii TTO1]
Length = 154
Score = 55.5 bits (132), Expect = 8e-07
Identities = 33/116 (28%), Positives = 58/116 (49%), Gaps = 9/116 (7%)
Query: 75 GVYRAILVGKAGQKPLQKKLKSGTVVTLLSIGTGGIRNNRRPLDHENPREYANRCAVQWH 134
GV + IL+G+ GQ+P + G V +LS+ T +R+ + + E WH
Sbjct: 5 GVNKVILIGRLGQEPEVRYRPEGDTVAVLSLATSETWRDRQTGEQKERTE--------WH 56
Query: 135 RVTVYPERLGNLLMKTVLPGSTLYVEGNLETKVFSDPVTGLVRRVREVAVRRNGNL 190
RV ++ +L + + + GS +Y+EG L T+ + D +G R EV + + G +
Sbjct: 57 RVVIF-GKLAEIAGEYLKKGSQVYIEGQLRTRKWFDKASGQDRYSTEVVLNQQGTM 111
>ref|NP_807839.1| single strand binding protein [Salmonella enterica subsp. enterica
serovar Typhi Ty2] gi|16505317|emb|CAD09315.1| single
strand binding protein [Salmonella enterica subsp.
enterica serovar Typhi] gi|29140135|gb|AAO71699.1|
single strand binding protein [Salmonella enterica
subsp. enterica serovar Typhi Ty2]
gi|6513859|gb|AAF14810.1| single strand binding protein
[Salmonella typhi] gi|16763010|ref|NP_458627.1| single
strand binding protein [Salmonella enterica subsp.
enterica serovar Typhi str. CT18]
gi|25294300|pir||AB1027 single strand binding protein
[imported] - Salmonella enterica subsp. enterica serovar
Typhi (strain CT18) gi|37999807|sp|Q9RHF4|SSB2_SALTI
Single-strand binding protein 2 (SSB 2)
(Helix-destabilizing protein 2)
gi|32454454|gb|AAP83026.1| single-strand DNA-binding
protein [Salmonella enterica subsp. enterica serovar
Paratyphi C]
Length = 178
Score = 54.3 bits (129), Expect = 2e-06
Identities = 35/116 (30%), Positives = 60/116 (51%), Gaps = 10/116 (8%)
Query: 75 GVYRAILVGKAGQKPLQKKLKSGTVVTLLSIGTGGIRNNRRPLDHENPREYANRCAVQWH 134
GV + ILVG GQ P + + +G V LS+ T +++ D + E WH
Sbjct: 5 GVNKVILVGNLGQDPEVRYMPNGGAVVNLSLATSDTWTDKQTGDKKERTE--------WH 56
Query: 135 RVTVYPERLGNLLMKTVLPGSTLYVEGNLETKVFSDPVTGLVRRVREVAVRRNGNL 190
RV +Y +L + + + GS +Y+EG L T+ ++D +G+ + EV V ++G +
Sbjct: 57 RVVLY-GKLAEIASEYLRKGSQVYIEGALRTRKWTDQ-SGVEKYTTEVVVSQSGTM 110
>ref|ZP_00145638.2| COG0629: Single-stranded DNA-binding protein [Psychrobacter sp.
273-4]
Length = 213
Score = 54.3 bits (129), Expect = 2e-06
Identities = 31/108 (28%), Positives = 58/108 (53%), Gaps = 10/108 (9%)
Query: 75 GVYRAILVGKAGQKPLQKKLKSGTVVTLLSIGTGGIRNNRRPLDHENPREYANRCAVQWH 134
GV + I++G G P ++ +G VT +S+ T +++ + R A +WH
Sbjct: 3 GVNKVIIIGNLGADPEARQFNNGGSVTNISVATSEQWTDKQSGE--------KREATEWH 54
Query: 135 RVTVYPERLGNLLMKTVLPGSTLYVEGNLETKVFSDPVTGLVRRVREV 182
R++++ RLG + + + GS +Y+EG+L T+ + DP G R + E+
Sbjct: 55 RISLF-NRLGEIAAQYLRKGSKVYIEGSLRTRKYQDP-NGQDRYITEI 100
>sp|Q8D254|SSB_WIGBR Single-strand binding protein (SSB) (Helix-destabilizing protein)
Length = 163
Score = 53.9 bits (128), Expect = 2e-06
Identities = 35/114 (30%), Positives = 59/114 (51%), Gaps = 10/114 (8%)
Query: 75 GVYRAILVGKAGQKPLQKKLKSGTVVTLLSIGTGGIRNNRRPLDHENPREYANRCAVQWH 134
G+ + IL+G GQ P + + +G +T LS+ T N R + +E +WH
Sbjct: 5 GINKVILIGNLGQDPEVRYIPNGNAITNLSLATS---ENWRDKNTGESKE-----KTEWH 56
Query: 135 RVTVYPERLGNLLMKTVLPGSTLYVEGNLETKVFSDPVTGLVRRVREVAVRRNG 188
RV ++ +L + + + GS +Y+EGNL+T+ + D G R + E+ V NG
Sbjct: 57 RVILF-GKLAEVAGEYLKKGSQVYIEGNLQTRKWQDQ-NGQDRYITEIVVGING 108
>dbj|BAC24646.1| ssb [Wigglesworthia glossinidia endosymbiont of Glossina
brevipalpis] gi|32491249|ref|NP_871503.1| hypothetical
protein WGLp500 [Wigglesworthia glossinidia endosymbiont
of Glossina brevipalpis]
Length = 167
Score = 53.9 bits (128), Expect = 2e-06
Identities = 35/114 (30%), Positives = 59/114 (51%), Gaps = 10/114 (8%)
Query: 75 GVYRAILVGKAGQKPLQKKLKSGTVVTLLSIGTGGIRNNRRPLDHENPREYANRCAVQWH 134
G+ + IL+G GQ P + + +G +T LS+ T N R + +E +WH
Sbjct: 9 GINKVILIGNLGQDPEVRYIPNGNAITNLSLATS---ENWRDKNTGESKE-----KTEWH 60
Query: 135 RVTVYPERLGNLLMKTVLPGSTLYVEGNLETKVFSDPVTGLVRRVREVAVRRNG 188
RV ++ +L + + + GS +Y+EGNL+T+ + D G R + E+ V NG
Sbjct: 61 RVILF-GKLAEVAGEYLKKGSQVYIEGNLQTRKWQDQ-NGQDRYITEIVVGING 112
>ref|YP_094400.1| single strand binding protein [Legionella pneumophila subsp.
pneumophila str. Philadelphia 1]
gi|52627712|gb|AAU26453.1| single strand binding protein
[Legionella pneumophila subsp. pneumophila str.
Philadelphia 1]
Length = 162
Score = 53.1 bits (126), Expect = 4e-06
Identities = 32/96 (33%), Positives = 49/96 (50%), Gaps = 9/96 (9%)
Query: 75 GVYRAILVGKAGQKPLQKKLKSGTVVTLLSIGTGGIRNNRRPLDHENPREYANRCAVQWH 134
G+ + ILVG G P + L +G VT LS+ T ++ + ++ E WH
Sbjct: 7 GINKVILVGNVGADPDVRYLPNGNAVTTLSVATSETWKDKTTGEKQDRTE--------WH 58
Query: 135 RVTVYPERLGNLLMKTVLPGSTLYVEGNLETKVFSD 170
RV + RLG + + + GS LYVEG+L T+ + D
Sbjct: 59 RVVCF-NRLGEIAGEYIRKGSKLYVEGSLRTRKWQD 93
>emb|CAH14627.1| Single-strand binding protein (SSB) (Helix-destabilizing protein)
[Legionella pneumophila str. Lens]
gi|54293348|ref|YP_125763.1| Single-strand binding
protein (SSB) (Helix-destabilizing protein) [Legionella
pneumophila str. Lens]
Length = 159
Score = 53.1 bits (126), Expect = 4e-06
Identities = 32/96 (33%), Positives = 49/96 (50%), Gaps = 9/96 (9%)
Query: 75 GVYRAILVGKAGQKPLQKKLKSGTVVTLLSIGTGGIRNNRRPLDHENPREYANRCAVQWH 134
G+ + ILVG G P + L +G VT LS+ T ++ + ++ E WH
Sbjct: 4 GINKVILVGNVGADPDVRYLPNGNAVTTLSVATSETWKDKTTGEKQDRTE--------WH 55
Query: 135 RVTVYPERLGNLLMKTVLPGSTLYVEGNLETKVFSD 170
RV + RLG + + + GS LYVEG+L T+ + D
Sbjct: 56 RVVCF-NRLGEIAGEYIRKGSKLYVEGSLRTRKWQD 90
>emb|CAH11569.1| Single-strand binding protein (SSB) (Helix-destabilizing protein)
[Legionella pneumophila str. Paris]
gi|54296392|ref|YP_122761.1| Single-strand binding
protein (SSB) (Helix-destabilizing protein) [Legionella
pneumophila str. Paris]
Length = 159
Score = 53.1 bits (126), Expect = 4e-06
Identities = 32/96 (33%), Positives = 49/96 (50%), Gaps = 9/96 (9%)
Query: 75 GVYRAILVGKAGQKPLQKKLKSGTVVTLLSIGTGGIRNNRRPLDHENPREYANRCAVQWH 134
G+ + ILVG G P + L +G VT LS+ T ++ + ++ E WH
Sbjct: 4 GINKVILVGNVGADPDVRYLPNGNAVTTLSVATSETWKDKTTGEKQDRTE--------WH 55
Query: 135 RVTVYPERLGNLLMKTVLPGSTLYVEGNLETKVFSD 170
RV + RLG + + + GS LYVEG+L T+ + D
Sbjct: 56 RVVCF-NRLGEIAGEYIRKGSKLYVEGSLRTRKWQD 90
>ref|NP_660857.2| single-strand binding protein [Buchnera aphidicola str. Sg
(Schizaphis graminum)]
Length = 176
Score = 52.0 bits (123), Expect = 9e-06
Identities = 36/132 (27%), Positives = 64/132 (48%), Gaps = 14/132 (10%)
Query: 68 LENGL----DPGVYRAILVGKAGQKPLQKKLKSGTVVTLLSIGTGGIRNNRRPLDHENPR 123
LENG+ GV + IL+G GQ P + + +G V +++ T + ++
Sbjct: 8 LENGVLIMASRGVNKVILIGHLGQDPEVRYMPNGNAVVNMTLATSE--------NWKDKN 59
Query: 124 EYANRCAVQWHRVTVYPERLGNLLMKTVLPGSTLYVEGNLETKVFSDPVTGLVRRVREVA 183
N+ +WHR+ ++ +L + + + GS +Y+EG+L+T+ + D GL R EV
Sbjct: 60 TGENKEKTEWHRIVLF-GKLAEIAGEYLRKGSQVYIEGSLQTRKWQDQ-NGLERYTTEVI 117
Query: 184 VRRNGNLANSSN 195
V G + N
Sbjct: 118 VNIGGTMQMLGN 129
>ref|YP_191319.1| Single-strand DNA binding protein [Gluconobacter oxydans 621H]
gi|58001769|gb|AAW60663.1| Single-strand DNA binding
protein [Gluconobacter oxydans 621H]
Length = 188
Score = 52.0 bits (123), Expect = 9e-06
Identities = 30/111 (27%), Positives = 56/111 (50%), Gaps = 9/111 (8%)
Query: 76 VYRAILVGKAGQKPLQKKLKSGTVVTLLSIGTGGIRNNRRPLDHENPREYANRCAVQWHR 135
V + ILVG G+ P + +SG + L++ T N+++ + + E WHR
Sbjct: 5 VNKVILVGNLGKDPEVRNTQSGAKIVNLTVATSDTWNDKQSGERKERTE--------WHR 56
Query: 136 VTVYPERLGNLLMKTVLPGSTLYVEGNLETKVFSDPVTGLVRRVREVAVRR 186
V ++ ER ++ + + G +Y+EG L T+ ++D +G + EV + R
Sbjct: 57 VVIFNERTADVAERYLRKGRKVYIEGELRTRKWTDQ-SGQEKYTTEVVIDR 106
>emb|CAH94310.1| single-strand binding protein, putative [Plasmodium berghei]
Length = 276
Score = 51.6 bits (122), Expect = 1e-05
Identities = 34/129 (26%), Positives = 64/129 (49%), Gaps = 11/129 (8%)
Query: 60 ERTFFDRPLENGLDPGVYRAILVGKAGQKPLQKKLKSGTVVTLLSIGTGGIRNNRRPLDH 119
+R + +RP+ N + + + L+G+ G +P K L G V S+ T +R
Sbjct: 63 KRFYNNRPVMN--EKSLNKITLIGRVGCEPDIKILNGGDKVATFSLATNEFWRDR----- 115
Query: 120 ENPREYANRCAVQWHRVTVYPERLGNLLMKTVLPGSTLYVEGNLET-KVFSDPVTGLVRR 178
N E ++ WHR+ VY + + +L+ K + G +Y++G+L T K F + + R+
Sbjct: 116 -NSNELKSK--TDWHRIVVYDQNIVDLIDKYLRKGRRVYIQGSLHTRKWFGNDLNNQPRQ 172
Query: 179 VREVAVRRN 187
+ E+ + N
Sbjct: 173 ITEIVLSYN 181
>ref|NP_778088.1| single-strand binding protein [Buchnera aphidicola str. Bp
(Baizongia pistaciae)] gi|27904360|gb|AAO27193.1|
single-strand binding protein [Buchnera aphidicola str.
Bp (Baizongia pistaciae)]
gi|32699636|sp|Q89A53|SSB_BUCBP Single-strand binding
protein (SSB) (Helix-destabilizing protein)
Length = 166
Score = 50.4 bits (119), Expect = 3e-05
Identities = 31/116 (26%), Positives = 62/116 (52%), Gaps = 10/116 (8%)
Query: 75 GVYRAILVGKAGQKPLQKKLKSGTVVTLLSIGTGGIRNNRRPLDHENPREYANRCAVQWH 134
G+ + IL+G GQ P + +++G+ VT +++ T ++ N E + +WH
Sbjct: 5 GINKVILIGYLGQDPDVRYMQNGSAVTNITLATSETWKDK------NNGEIKEK--TEWH 56
Query: 135 RVTVYPERLGNLLMKTVLPGSTLYVEGNLETKVFSDPVTGLVRRVREVAVRRNGNL 190
R+ V+ +L + + + GS +Y+EG+L+T+ + D G+ R + E+ V G +
Sbjct: 57 RI-VFFNKLAEIAGEYLKKGSQVYIEGSLQTRKWKDQ-NGIERYITEIIVSVGGTM 110
>gb|AAC72352.1| single strand DNA-binding protein; SSB [Vibrio cholerae]
Length = 192
Score = 50.4 bits (119), Expect = 3e-05
Identities = 35/117 (29%), Positives = 58/117 (48%), Gaps = 10/117 (8%)
Query: 69 ENGLDPGVYRAILVGKAGQKPLQKKLKSGTVVTLLSIGTGGIRNNRRPLDHENPREYANR 128
+N GV + IL+G GQ P + + SG V ++I T ++ + + E
Sbjct: 14 QNMASRGVNKVILIGNLGQDPEVRYMPSGGAVANITIATSETWRDKATGEQKEKTE---- 69
Query: 129 CAVQWHRVTVYPERLGNLLMKTVLPGSTLYVEGNLETKVFSDPVTGLVRRVREVAVR 185
WHRVT+Y +L + + + GS +Y+EG L+T+ + D +G R EV V+
Sbjct: 70 ----WHRVTLY-GKLAEVAGEYLRKGSQVYIEGQLQTRKWQDQ-SGQDRYSTEVVVQ 120
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.317 0.134 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 362,758,904
Number of Sequences: 2540612
Number of extensions: 16187302
Number of successful extensions: 42532
Number of sequences better than 10.0: 261
Number of HSP's better than 10.0 without gapping: 78
Number of HSP's successfully gapped in prelim test: 183
Number of HSP's that attempted gapping in prelim test: 42334
Number of HSP's gapped (non-prelim): 277
length of query: 196
length of database: 863,360,394
effective HSP length: 121
effective length of query: 75
effective length of database: 555,946,342
effective search space: 41695975650
effective search space used: 41695975650
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 71 (32.0 bits)
Medicago: description of AC148996.5


