

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148969.3 + phase: 2 /pseudo
(595 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
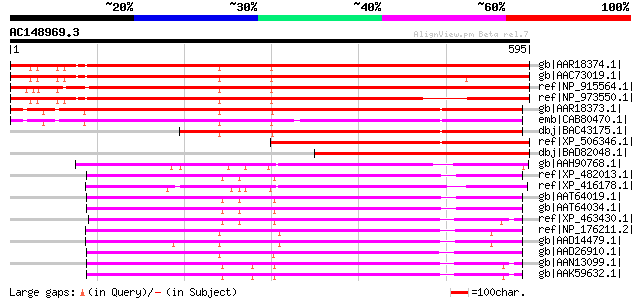
Score E
Sequences producing significant alignments: (bits) Value
gb|AAR18374.1| nucleobase-ascorbate transporter 12 [Arabidopsis ... 742 0.0
gb|AAC73019.1| putative membrane transporter [Arabidopsis thalia... 735 0.0
ref|NP_915564.1| putative permease 1 [Oryza sativa (japonica cul... 666 0.0
ref|NP_973550.1| xanthine/uracil permease family protein [Arabid... 639 0.0
gb|AAR18373.1| nucleobase-ascorbate transporter 11 [Arabidopsis ... 550 e-155
emb|CAB80470.1| putative protein [Arabidopsis thaliana] gi|44671... 520 e-146
dbj|BAC43175.1| unknown protein [Arabidopsis thaliana] gi|289510... 422 e-116
ref|XP_506346.1| PREDICTED P0477A12.37 gene product [Oryza sativ... 348 4e-94
dbj|BAD82048.1| nucleobase-ascorbate transporter-like protein [O... 332 2e-89
gb|AAH90768.1| Zgc:110789 [Danio rerio] gi|61651826|ref|NP_00101... 318 4e-85
ref|XP_482013.1| putative permease 1 [Oryza sativa (japonica cul... 312 2e-83
ref|XP_416178.1| PREDICTED: similar to Solute carrier family 23,... 310 6e-83
gb|AAT64019.1| putative permease [Gossypium hirsutum] 308 2e-82
gb|AAT64034.1| putative permease [Gossypium hirsutum] 307 7e-82
ref|XP_463430.1| putative permease 1 [Oryza sativa (japonica cul... 306 2e-81
ref|NP_176211.2| xanthine/uracil permease family protein [Arabid... 304 6e-81
gb|AAD14479.1| Strong similarity to gi|3337350 F13P17.3 putative... 303 8e-81
gb|AAD26910.1| putative membrane transporter [Arabidopsis thalia... 300 9e-80
gb|AAN13099.1| putative membrane transporter [Arabidopsis thalia... 299 1e-79
gb|AAK59632.1| putative membrane transporter protein [Arabidopsi... 298 4e-79
>gb|AAR18374.1| nucleobase-ascorbate transporter 12 [Arabidopsis thaliana]
gi|20466159|gb|AAM20397.1| putative membrane transporter
[Arabidopsis thaliana] gi|25083906|gb|AAN72132.1|
putative membrane transporter [Arabidopsis thaliana]
gi|30683653|ref|NP_850108.1| xanthine/uracil permease
family protein [Arabidopsis thaliana]
Length = 709
Score = 742 bits (1916), Expect = 0.0
Identities = 403/683 (59%), Positives = 471/683 (68%), Gaps = 91/683 (13%)
Query: 1 WAQKTGFKPVFSAETKAGQSLS-------------RQPDLEAS--------PVVSTPSQA 39
WA+KTGF+P FS ET A S S QPDLEA PV + +
Sbjct: 29 WAKKTGFRPKFSGETTATDSSSGQLSLPVRAKQQETQPDLEAGQTRLRPPPPVSAAVTNG 88
Query: 40 VNDVPHGDKVPPSP------------------SDGVPTNN----------------ARVE 65
D +K PP P SDGV + R+E
Sbjct: 89 ETDKDKKEKPPPPPPGSVAVPVKDQPVKRRRDSDGVVGRSNGPDGANGSGDPVRRPGRIE 148
Query: 66 ERTTRLPVMVDHDDLVLRRRPSPLNYELTDSPALVFLAVYGIQHYLSIIGSLILTPLVIA 125
E LP +D DDLV R + Y L D+P LV + YG+QHYLS++GSLIL PLVI
Sbjct: 149 ETVEVLPQSMD-DDLVARNLH--MKYGLRDTPGLVPIGFYGLQHYLSMLGSLILVPLVIV 205
Query: 126 PAMGASHDETAAMVCTVLLVSGVTTLLHTIFGSRLPLIQGPSFVYLAPVLAIINSPEFQE 185
PAMG SH+E A +V TVL VSG+TTLLHT FGSRLPLIQGPSFV+LAP LAIINSPEFQ
Sbjct: 206 PAMGGSHEEVANVVSTVLFVSGITTLLHTSFGSRLPLIQGPSFVFLAPALAIINSPEFQG 265
Query: 186 LN-ENKFKHIMKELQGAIIIGSAFQTLLGYTGLMSLLVRFINPVVVSSTIAAVGL----- 239
LN N FKHIM+ELQGAIIIGSAFQ +LGY+GLMSL++R +NPVVV+ T+AAVGL
Sbjct: 266 LNGNNNFKHIMRELQGAIIIGSAFQAVLGYSGLMSLILRLVNPVVVAPTVAAVGLSFYSY 325
Query: 240 -FL*LARVLRSVQYRY*CLLFFVLYLRKISVFGHHIFQIYAVPLGLAVTWTFAFLLTENG 298
F + + L + ++ F LYLRKISV H IF IYAVPL LA+TW AFLLTE G
Sbjct: 326 GFPLVGKCLEIGVVQILLVIIFALYLRKISVLSHRIFLIYAVPLSLAITWAAAFLLTETG 385
Query: 299 --------------------------RMKHCQVNTSDTMTSPPWFRFPYPLQWGTPVFNW 332
RMK+C+V+TS ++S PWFRFPYPLQWG P+FNW
Sbjct: 386 AYTYKGCDPNVPVSNVVSTHCRKYMTRMKYCRVDTSHALSSAPWFRFPYPLQWGVPLFNW 445
Query: 333 KMAIVMCVVSLISSVDSVGTYHTSSLLAASGPPTPGVLSRGIGLEGFSSLLAGLWGTGMG 392
KMA VMCVVS+I+SVDSVG+YH SSLL AS PPT GV+SR IGLEGF+S+LAGLWGTG G
Sbjct: 446 KMAFVMCVVSVIASVDSVGSYHASSLLVASRPPTRGVVSRAIGLEGFTSVLAGLWGTGTG 505
Query: 393 STTLTENVHTIAGTKMGSRRPVQLGACLLIVLSLFGKVGGFIASIPEAMVAGLLCIMWAM 452
STTLTENVHTIA TKMGSRR V+LGAC+L++ SL GKVGGF+ASIP+ MVA LLC MWAM
Sbjct: 506 STTLTENVHTIAVTKMGSRRVVELGACVLVIFSLVGKVGGFLASIPQVMVASLLCFMWAM 565
Query: 453 LTALGLSNLRYTETGSSRNIIIVGLSLFFSLSIPAYFQQYESSPESNFSVPSYFQPYIVT 512
TALGLSNLRY+E GSSRNIIIVGLSLFFSLS+PAYFQQY SP SN SVPSY+QPYIV+
Sbjct: 566 FTALGLSNLRYSEAGSSRNIIIVGLSLFFSLSVPAYFQQYGISPNSNLSVPSYYQPYIVS 625
Query: 513 SHGPFRSKYEELNYVLNMIFSLHMVIAFLVALILDNTVPGSKQERELYGWSKPNDAREDP 572
SHGPF+S+Y+ +NYV+N + S+ MVIAF++A+ILDNTVPGSKQER +Y WS A +P
Sbjct: 626 SHGPFKSQYKGMNYVMNTLLSMSMVIAFIMAVILDNTVPGSKQERGVYVWSDSETATREP 685
Query: 573 FIVSEYGLPARVGRCFRWVKWVG 595
+ +Y LP RVGR FRWVKWVG
Sbjct: 686 ALAKDYELPFRVGRFFRWVKWVG 708
>gb|AAC73019.1| putative membrane transporter [Arabidopsis thaliana]
gi|25407923|pir||C84677 probable membrane transporter
[imported] - Arabidopsis thaliana
Length = 721
Score = 735 bits (1897), Expect = 0.0
Identities = 403/695 (57%), Positives = 472/695 (66%), Gaps = 103/695 (14%)
Query: 1 WAQKTGFKPVFSAETKAGQSLS-------------RQPDLEAS--------PVVSTPSQA 39
WA+KTGF+P FS ET A S S QPDLEA PV + +
Sbjct: 29 WAKKTGFRPKFSGETTATDSSSGQLSLPVRAKQQETQPDLEAGQTRLRPPPPVSAAVTNG 88
Query: 40 VNDVPHGDKVPPSP------------------SDGVPTNN----------------ARVE 65
D +K PP P SDGV + R+E
Sbjct: 89 ETDKDKKEKPPPPPPGSVAVPVKDQPVKRRRDSDGVVGRSNGPDGANGSGDPVRRPGRIE 148
Query: 66 ERTTRLPVMVDHDDLVLRRRPSPLNYELTDSPALVFLAVYGIQHYLSIIGSLILTPLVIA 125
E LP +D DDLV R + Y L D+P LV + YG+QHYLS++GSLIL PLVI
Sbjct: 149 ETVEVLPQSMD-DDLVARNLH--MKYGLRDTPGLVPIGFYGLQHYLSMLGSLILVPLVIV 205
Query: 126 PAMGASHDETAAMVCTVLLVSGVTTLLHTIFGSRLPLIQGPSFVYLAPVLAIINSPEFQE 185
PAMG SH+E A +V TVL VSG+TTLLHT FGSRLPLIQGPSFV+LAP LAIINSPEFQ
Sbjct: 206 PAMGGSHEEVANVVSTVLFVSGITTLLHTSFGSRLPLIQGPSFVFLAPALAIINSPEFQG 265
Query: 186 LN-ENKFKHIMKELQGAIIIGSAFQTLLGYTGLMSLLVRFINPVVVSSTIAAVGL----- 239
LN N FKHIM+ELQGAIIIGSAFQ +LGY+GLMSL++R +NPVVV+ T+AAVGL
Sbjct: 266 LNGNNNFKHIMRELQGAIIIGSAFQAVLGYSGLMSLILRLVNPVVVAPTVAAVGLSFYSY 325
Query: 240 -FL*LARVLRSVQYRY*CLLFFVLYLRKISVFGHHIFQIYAVPLGLAVTWTFAFLLTENG 298
F + + L + ++ F LYLRKISV H IF IYAVPL LA+TW AFLLTE G
Sbjct: 326 GFPLVGKCLEIGVVQILLVIIFALYLRKISVLSHRIFLIYAVPLSLAITWAAAFLLTETG 385
Query: 299 --------------------------RMKHCQVNTSDTMTSPPWFRFPYPLQWGTPVFNW 332
RMK+C+V+TS ++S PWFRFPYPLQWG P+FNW
Sbjct: 386 AYTYKGCDPNVPVSNVVSTHCRKYMTRMKYCRVDTSHALSSAPWFRFPYPLQWGVPLFNW 445
Query: 333 KMAIVMCVVSLISSVDSVGTYHTSSLLAASGPPTPGVLSRGIGLEGFSSLLAGLWGTGMG 392
KMA VMCVVS+I+SVDSVG+YH SSLL AS PPT GV+SR IGLEGF+S+LAGLWGTG G
Sbjct: 446 KMAFVMCVVSVIASVDSVGSYHASSLLVASRPPTRGVVSRAIGLEGFTSVLAGLWGTGTG 505
Query: 393 STTLTENVHTIAGTKMGSRRPVQLGACLLIVLSLFGKVGGFIASIPEAMVAGLLCIMWAM 452
STTLTENVHTIA TKMGSRR V+LGAC+L++ SL GKVGGF+ASIP+ MVA LLC MWAM
Sbjct: 506 STTLTENVHTIAVTKMGSRRVVELGACVLVIFSLVGKVGGFLASIPQVMVASLLCFMWAM 565
Query: 453 LTALGLSNLRYTETGSSRNIIIVGLSLFFSLSIPAYFQQYESSPESNFSVPSYFQPYIVT 512
TALGLSNLRY+E GSSRNIIIVGLSLFFSLS+PAYFQQY SP SN SVPSY+QPYIV+
Sbjct: 566 FTALGLSNLRYSEAGSSRNIIIVGLSLFFSLSVPAYFQQYGISPNSNLSVPSYYQPYIVS 625
Query: 513 SHGPFRSKYE------------ELNYVLNMIFSLHMVIAFLVALILDNTVPGSKQERELY 560
SHGPF+S+Y+ ++NYV+N + S+ MVIAF++A+ILDNTVPGSKQER +Y
Sbjct: 626 SHGPFKSQYKGDLQFSYLLVYLQMNYVMNTLLSMSMVIAFIMAVILDNTVPGSKQERGVY 685
Query: 561 GWSKPNDAREDPFIVSEYGLPARVGRCFRWVKWVG 595
WS A +P + +Y LP RVGR FRWVKWVG
Sbjct: 686 VWSDSETATREPALAKDYELPFRVGRFFRWVKWVG 720
>ref|NP_915564.1| putative permease 1 [Oryza sativa (japonica cultivar-group)]
Length = 680
Score = 666 bits (1719), Expect = 0.0
Identities = 358/655 (54%), Positives = 440/655 (66%), Gaps = 66/655 (10%)
Query: 1 WAQKTGFKPVFSAETKA-------GQSLSRQP-----DLEASP-------VVSTPSQAVN 41
WA++TGF+ S E+ A GQ +P DLE+ P + P+ A
Sbjct: 31 WAKRTGFQSRVSGESVAIVSASSSGQVSLPRPAEAPSDLESGPPARPNSALPPPPAAAAA 90
Query: 42 DVPHGDKVPPSP---------SDGVPTNNARVEERTTRLPVMVDHDDLVLRRRPSPLNYE 92
+ + PP P SDG N + LP +++ +D RP YE
Sbjct: 91 NAEAKPQPPPPPPPARTRRRDSDGGRPNG---QAAAAPLPQLLEEEDDGAPERPK---YE 144
Query: 93 LTDSPALVFLAVYGIQHYLSIIGSLILTPLVIAPAMGASHDETAAMVCTVLLVSGVTTLL 152
L DSP + +AVYG QHY+S++GS+IL PL++ PAMG S D+ AA+V TVLLVSG+TTLL
Sbjct: 145 LRDSPGVFPIAVYGFQHYISMLGSIILIPLLMVPAMGGSPDDMAAVVSTVLLVSGMTTLL 204
Query: 153 HTIFGSRLPLIQGPSFVYLAPVLAIINSPEFQELNENKFKHIMKELQGAIIIGSAFQTLL 212
HT G+RLPL+QGPSFVYLAP LAII SPEF LN N FKHIMK LQGAIIIG AFQ LL
Sbjct: 205 HTFCGTRLPLVQGPSFVYLAPALAIIYSPEFFGLNHNNFKHIMKHLQGAIIIGGAFQVLL 264
Query: 213 GYTGLMSLLVRFINPVVVSSTIAAVGL------FL*LARVLRSVQYRY*CLLFFVLYLRK 266
GYTGLMSL +R INPVV+S T+AAVGL F + + + ++ F LYLRK
Sbjct: 265 GYTGLMSLFLRLINPVVISPTVAAVGLSFFSYGFTKVGSCIEMGLLQLLIVVMFALYLRK 324
Query: 267 ISVFGHHIFQIYAVPLGLAVTWTFAFLLTENG--------------------------RM 300
+ +FG+ +F IYAVPL L +TW AF+LT G RM
Sbjct: 325 VKLFGYRVFLIYAVPLALGITWAIAFVLTATGVYSYRGCDANIPASNNVSAYCRKHVLRM 384
Query: 301 KHCQVNTSDTMTSPPWFRFPYPLQWGTPVFNWKMAIVMCVVSLISSVDSVGTYHTSSLLA 360
K C+V+TS + S PW RFPYPLQWGTP+F+WKM +VMCV S+I+SVDSVG+YH SSL
Sbjct: 385 KSCRVDTSHALRSSPWLRFPYPLQWGTPIFSWKMGLVMCVASVIASVDSVGSYHASSLFV 444
Query: 361 ASGPPTPGVLSRGIGLEGFSSLLAGLWGTGMGSTTLTENVHTIAGTKMGSRRPVQLGACL 420
A+ PPT GV+SRGIG+EG S++LAGLWGTG+GS T+TENVHTIA TKMG+RR V GA +
Sbjct: 445 ATRPPTAGVVSRGIGVEGVSTVLAGLWGTGVGSATITENVHTIAVTKMGNRRAVGFGAIV 504
Query: 421 LIVLSLFGKVGGFIASIPEAMVAGLLCIMWAMLTALGLSNLRYTETGSSRNIIIVGLSLF 480
LI+LS GKVG FIASIP+ +VA LLC MWAML ALGLSNLRY+ GSSRN I+VGL+LF
Sbjct: 505 LILLSFVGKVGAFIASIPDVLVAALLCFMWAMLCALGLSNLRYSAKGSSRNSIVVGLALF 564
Query: 481 FSLSIPAYFQQYESSPESNFSVPSYFQPYIVTSHGPFRSKYEELNYVLNMIFSLHMVIAF 540
SLS+P+YFQQY P SN SVP+YFQPYIV SHGP + +NY+LN + SL+MVIAF
Sbjct: 565 LSLSVPSYFQQYRLQPNSNSSVPTYFQPYIVASHGPIHTGSSGVNYILNTLLSLNMVIAF 624
Query: 541 LVALILDNTVPGSKQERELYGWSKPNDAREDPFIVSEYGLPARVGRCFRWVKWVG 595
LVALILDNTVPG +QER LY WS+ AR + ++ +Y LP ++G FRWVK VG
Sbjct: 625 LVALILDNTVPGGRQERGLYVWSEAEAARRESAVMKDYELPFKIGHAFRWVKCVG 679
>ref|NP_973550.1| xanthine/uracil permease family protein [Arabidopsis thaliana]
Length = 660
Score = 639 bits (1648), Expect = 0.0
Identities = 365/683 (53%), Positives = 427/683 (62%), Gaps = 140/683 (20%)
Query: 1 WAQKTGFKPVFSAETKAGQSLS-------------RQPDLEAS--------PVVSTPSQA 39
WA+KTGF+P FS ET A S S QPDLEA PV + +
Sbjct: 29 WAKKTGFRPKFSGETTATDSSSGQLSLPVRAKQQETQPDLEAGQTRLRPPPPVSAAVTNG 88
Query: 40 VNDVPHGDKVPPSP------------------SDGVPTNN----------------ARVE 65
D +K PP P SDGV + R+E
Sbjct: 89 ETDKDKKEKPPPPPPGSVAVPVKDQPVKRRRDSDGVVGRSNGPDGANGSGDPVRRPGRIE 148
Query: 66 ERTTRLPVMVDHDDLVLRRRPSPLNYELTDSPALVFLAVYGIQHYLSIIGSLILTPLVIA 125
E LP +D DDLV R + Y L D+P LV + YG+QHYLS++GSLIL PLVI
Sbjct: 149 ETVEVLPQSMD-DDLVARNLH--MKYGLRDTPGLVPIGFYGLQHYLSMLGSLILVPLVIV 205
Query: 126 PAMGASHDETAAMVCTVLLVSGVTTLLHTIFGSRLPLIQGPSFVYLAPVLAIINSPEFQE 185
PAMG SH+E A +V TVL VSG+TTLLHT FGSRLPLIQGPSFV+LAP LAIINSPEFQ
Sbjct: 206 PAMGGSHEEVANVVSTVLFVSGITTLLHTSFGSRLPLIQGPSFVFLAPALAIINSPEFQG 265
Query: 186 LN-ENKFKHIMKELQGAIIIGSAFQTLLGYTGLMSLLVRFINPVVVSSTIAAVGL----- 239
LN N FKHIM+ELQGAIIIGSAFQ +LGY+GLMSL++R +NPVVV+ T+AAVGL
Sbjct: 266 LNGNNNFKHIMRELQGAIIIGSAFQAVLGYSGLMSLILRLVNPVVVAPTVAAVGLSFYSY 325
Query: 240 -FL*LARVLRSVQYRY*CLLFFVLYLRKISVFGHHIFQIYAVPLGLAVTWTFAFLLTENG 298
F + + L + ++ F LYLRKISV H IF IYAVPL LA+TW AFLLTE G
Sbjct: 326 GFPLVGKCLEIGVVQILLVIIFALYLRKISVLSHRIFLIYAVPLSLAITWAAAFLLTETG 385
Query: 299 --------------------------RMKHCQVNTSDTMTSPPWFRFPYPLQWGTPVFNW 332
RMK+C+V+TS ++S PWFRFPYPLQWG P+FNW
Sbjct: 386 AYTYKGCDPNVPVSNVVSTHCRKYMTRMKYCRVDTSHALSSAPWFRFPYPLQWGVPLFNW 445
Query: 333 KMAIVMCVVSLISSVDSVGTYHTSSLLAASGPPTPGVLSRGIGLEGFSSLLAGLWGTGMG 392
KMA VMCVVS+I+SVDSVG+YH SSLL AS PPT GV+SR IGLEGF+S+LAGLWGTG G
Sbjct: 446 KMAFVMCVVSVIASVDSVGSYHASSLLVASRPPTRGVVSRAIGLEGFTSVLAGLWGTGTG 505
Query: 393 STTLTENVHTIAGTKMGSRRPVQLGACLLIVLSLFGKVGGFIASIPEAMVAGLLCIMWAM 452
STTLTENVHTIA TKMGSRR V+LGAC+L++ SL GKVGGF+ASIP+ MVA LLC MWAM
Sbjct: 506 STTLTENVHTIAVTKMGSRRVVELGACVLVIFSLVGKVGGFLASIPQVMVASLLCFMWAM 565
Query: 453 LTALGLSNLRYTETGSSRNIIIVGLSLFFSLSIPAYFQQYESSPESNFSVPSYFQPYIVT 512
TALGLSNLRY+E GSSRNIII
Sbjct: 566 FTALGLSNLRYSEAGSSRNIII-------------------------------------- 587
Query: 513 SHGPFRSKYEELNYVLNMIFSLHMVIAFLVALILDNTVPGSKQERELYGWSKPNDAREDP 572
+NYV+N + S+ MVIAF++A+ILDNTVPGSKQER +Y WS A +P
Sbjct: 588 -----------MNYVMNTLLSMSMVIAFIMAVILDNTVPGSKQERGVYVWSDSETATREP 636
Query: 573 FIVSEYGLPARVGRCFRWVKWVG 595
+ +Y LP RVGR FRWVKWVG
Sbjct: 637 ALAKDYELPFRVGRFFRWVKWVG 659
>gb|AAR18373.1| nucleobase-ascorbate transporter 11 [Arabidopsis thaliana]
Length = 709
Score = 550 bits (1418), Expect = e-155
Identities = 301/661 (45%), Positives = 406/661 (60%), Gaps = 83/661 (12%)
Query: 1 WAQKTGFKPVFSAETKAGQSLSRQPDLEASPVVSTPS---QAVNDVPHGDKVPPSPSDGV 57
WA+KTGF +S ET S S + S P Q V H ++ P +
Sbjct: 54 WAKKTGFVSDYSGET----STSTRTKFGESSDFDLPKGRDQVVTGSSHKTEIDPI----L 105
Query: 58 PTNNARVEERTTRLPVMVDHDDLVLRR--------------------------------- 84
N +E T PV + ++ L R
Sbjct: 106 GRNRPEIEHVTGSEPVSREEEERRLNRNEATPETENEGGKINKDLENGFYYPGGGGESSE 165
Query: 85 -----RPSPLNYELTDSPALVFLAVYGIQHYLSIIGSLILTPLVIAPAMGASHDETAAMV 139
+P + + L D+P V L YG+QHYLS++GSL+ PLVI PAM S +TA+++
Sbjct: 166 DGQWPKPILMKFGLRDNPGFVPLIYYGLQHYLSLVGSLVFIPLVIVPAMDGSDKDTASVI 225
Query: 140 CTVLLVSGVTTLLHTIFGSRLPLIQGPSFVYLAPVLAIINSPEFQELNENKFKHIMKELQ 199
T+LL++GVTT+LH FG+RLPL+QG SFVYLAPVL +INS EF+ L E+KF+ M+ELQ
Sbjct: 226 STMLLLTGVTTILHCYFGTRLPLVQGSSFVYLAPVLVVINSEEFRNLTEHKFRDTMRELQ 285
Query: 200 GAIIIGSAFQTLLGYTGLMSLLVRFINPVVVSSTIAAVGL------FL*LARVLRSVQYR 253
GAII+GS FQ +LG++GLMSLL+RFINPVVV+ T+AAVGL F +
Sbjct: 286 GAIIVGSLFQCILGFSGLMSLLLRFINPVVVAPTVAAVGLAFFSYGFPQAGTCVEISVPL 345
Query: 254 Y*CLLFFVLYLRKISVFGHHIFQIYAVPLGLAVTWTFAFLLTENGR-------------- 299
LL F LYLR +S+FGH +F+IYAVPL + WT+AF LT G
Sbjct: 346 ILLLLIFTLYLRGVSLFGHRLFRIYAVPLSALLIWTYAFFLTVGGAYDYRGCNADIPSSN 405
Query: 300 ------------MKHCQVNTSDTMTSPPWFRFPYPLQWGTPVFNWKMAIVMCVVSLISSV 347
MKHC+ + S+ + W R PYP QWG P F+ + +I+M VSL++SV
Sbjct: 406 ILIDECKKHVYTMKHCRTDASNAWRTASWVRIPYPFQWGFPNFHMRTSIIMIFVSLVASV 465
Query: 348 DSVGTYHTSSLLAASGPPTPGVLSRGIGLEGFSSLLAGLWGTGMGSTTLTENVHTIAGTK 407
DSVGTYH++S++ + PT G++SRGI LEGF SLLAG+WG+G GSTTLTEN+HTI TK
Sbjct: 466 DSVGTYHSASMIVNAKRPTRGIVSRGIALEGFCSLLAGIWGSGTGSTTLTENIHTINITK 525
Query: 408 MGSRRPVQLGACLLIVLSLFGKVGGFIASIPEAMVAGLLCIMWAMLTALGLSNLRYTETG 467
+ SRR + +GA LIVLS GK+G +ASIP+A+ A +LC +WA+ +LGLSNLRYT+T
Sbjct: 526 VASRRALVIGAMFLIVLSFLGKLGAILASIPQALAASVLCFIWALTVSLGLSNLRYTQTA 585
Query: 468 SSRNIIIVGLSLFFSLSIPAYFQQYESSPESNFSVPSYFQPYIVTSHGPFRSKYEELNYV 527
S RNI IVG+SLF LSIPAYFQQY+ P S+ +PSY+ P+ S GPF++ E+L++
Sbjct: 586 SFRNITIVGVSLFLGLSIPAYFQQYQ--PLSSLILPSYYIPFGAASSGPFQTGIEQLDFA 643
Query: 528 LNMIFSLHMVIAFLVALILDNTVPGSKQERELYGWSKPNDAREDPFIVSEYGLPARVGRC 587
+N + SL+MV+ FL+A ILDNTVPGSK+ER +Y W++ D + DP + ++Y LP + +
Sbjct: 644 MNAVLSLNMVVTFLLAFILDNTVPGSKEERGVYVWTRAEDMQMDPEMRADYSLPRKFAQI 703
Query: 588 F 588
F
Sbjct: 704 F 704
>emb|CAB80470.1| putative protein [Arabidopsis thaliana] gi|4467111|emb|CAB37545.1|
putative protein [Arabidopsis thaliana]
gi|15233652|ref|NP_195518.1| xanthine/uracil permease
family protein [Arabidopsis thaliana]
gi|7485847|pir||T05632 hypothetical protein F20D10.170 -
Arabidopsis thaliana
Length = 703
Score = 520 bits (1340), Expect = e-146
Identities = 290/661 (43%), Positives = 396/661 (59%), Gaps = 89/661 (13%)
Query: 1 WAQKTGFKPVFSAETKAGQSLSRQPDLEASPVVSTPS---QAVNDVPHGDKVPPSPSDGV 57
WA+KTGF +S ET S S + S P Q V H ++ P +
Sbjct: 54 WAKKTGFVSDYSGET----STSTRTKFGESSDFDLPKGRDQVVTGSSHKTEIDPI----L 105
Query: 58 PTNNARVEERTTRLPVMVDHDDLVLRR--------------------------------- 84
N +E T PV + ++ L R
Sbjct: 106 GRNRPEIEHVTGSEPVSREEEERRLNRNEATPETENEGGKINKDLENGFYYPGGGGESSE 165
Query: 85 -----RPSPLNYELTDSPALVFLAVYGIQHYLSIIGSLILTPLVIAPAMGASHDETAAMV 139
+P + + L D+P V L YG+QHYLS++GSL+ PLVI PAM S +TA+++
Sbjct: 166 DGQWPKPILMKFGLRDNPGFVPLIYYGLQHYLSLVGSLVFIPLVIVPAMDGSDKDTASVI 225
Query: 140 CTVLLVSGVTTLLHTIFGSRLPLIQGPSFVYLAPVLAIINSPEFQELNENKFKHIMKELQ 199
T+LL++GVTT+LH FG+RLPL+QG SFVYLAPVL +INS EF+ L E+KF+ M+ELQ
Sbjct: 226 STMLLLTGVTTILHCYFGTRLPLVQGSSFVYLAPVLVVINSEEFRNLTEHKFRDTMRELQ 285
Query: 200 GAIIIGSAFQTLLGYTGLMSLLVRFINPVVVSSTIAAVGL------FL*LARVLRSVQYR 253
GAII+GS FQ +LG++GLMSLL+RFINPVVV+ T+AAVGL F +
Sbjct: 286 GAIIVGSLFQCILGFSGLMSLLLRFINPVVVAPTVAAVGLAFFSYGFPQAGTCVEISVPL 345
Query: 254 Y*CLLFFVLYLRKISVFGHHIFQIYAVPLGLAVTWTFAFLLTENG--------------- 298
LL F LYLR +S+FGH +F+IYAVPL + WT+AF LT G
Sbjct: 346 ILLLLIFTLYLRGVSLFGHRLFRIYAVPLSALLIWTYAFFLTVGGAYDYRGCNADIPSSN 405
Query: 299 -----------RMKHCQVNTSDTMTSPPWFRFPYPLQWGTPVFNWKMAIVMCVVSLISSV 347
MKHC+ + S+ + W R PYP QWG + + + + ++ +
Sbjct: 406 ILIDECKKHVYTMKHCRTDASNAWRTASWVRIPYPFQWG------GLGMYLFLFAIPVFL 459
Query: 348 DSVGTYHTSSLLAASGPPTPGVLSRGIGLEGFSSLLAGLWGTGMGSTTLTENVHTIAGTK 407
VGTYH++S++ + PT G++SRGI LEGF SLLAG+WG+G GSTTLTEN+HTI TK
Sbjct: 460 LKVGTYHSASMIVNAKRPTRGIVSRGIALEGFCSLLAGIWGSGTGSTTLTENIHTINITK 519
Query: 408 MGSRRPVQLGACLLIVLSLFGKVGGFIASIPEAMVAGLLCIMWAMLTALGLSNLRYTETG 467
+ SRR + +GA LIVLS GK+G +ASIP+A+ A +LC +WA+ +LGLSNLRYT+T
Sbjct: 520 VASRRALVIGAMFLIVLSFLGKLGAILASIPQALAASVLCFIWALTVSLGLSNLRYTQTA 579
Query: 468 SSRNIIIVGLSLFFSLSIPAYFQQYESSPESNFSVPSYFQPYIVTSHGPFRSKYEELNYV 527
S RNI IVG+SLF LSIPAYFQQY+ P S+ +PSY+ P+ S GPF++ E+L++
Sbjct: 580 SFRNITIVGVSLFLGLSIPAYFQQYQ--PLSSLILPSYYIPFGAASSGPFQTGIEQLDFA 637
Query: 528 LNMIFSLHMVIAFLVALILDNTVPGSKQERELYGWSKPNDAREDPFIVSEYGLPARVGRC 587
+N + SL+MV+ FL+A ILDNTVPGSK+ER +Y W++ D + DP + ++Y LP + +
Sbjct: 638 MNAVLSLNMVVTFLLAFILDNTVPGSKEERGVYVWTRAEDMQMDPEMRADYSLPRKFAQI 697
Query: 588 F 588
F
Sbjct: 698 F 698
>dbj|BAC43175.1| unknown protein [Arabidopsis thaliana] gi|28951001|gb|AAO63424.1|
At4g38050 [Arabidopsis thaliana]
Length = 429
Score = 422 bits (1085), Expect = e-116
Identities = 216/426 (50%), Positives = 289/426 (67%), Gaps = 34/426 (7%)
Query: 195 MKELQGAIIIGSAFQTLLGYTGLMSLLVRFINPVVVSSTIAAVGL------FL*LARVLR 248
M+ELQGAII+GS FQ +LG++GLMSLL+RFINPVVV+ T+AAVGL F +
Sbjct: 1 MRELQGAIIVGSLFQCILGFSGLMSLLLRFINPVVVAPTVAAVGLAFFSYGFPQAGTCVE 60
Query: 249 SVQYRY*CLLFFVLYLRKISVFGHHIFQIYAVPLGLAVTWTFAFLLTENGR--------- 299
LL F LYLR +S+FGH +F+IYAVPL + WT+AF LT G
Sbjct: 61 ISVPLILLLLIFTLYLRGVSLFGHRLFRIYAVPLSALLIWTYAFFLTVGGAYDYRGCNAD 120
Query: 300 -----------------MKHCQVNTSDTMTSPPWFRFPYPLQWGTPVFNWKMAIVMCVVS 342
MKHC+ + S+ + W R PYP QWG P F+ + +I+M VS
Sbjct: 121 IPSSNILIDECKKHVYTMKHCRTDASNAWRTASWVRIPYPFQWGFPNFHMRTSIIMIFVS 180
Query: 343 LISSVDSVGTYHTSSLLAASGPPTPGVLSRGIGLEGFSSLLAGLWGTGMGSTTLTENVHT 402
L++SVDSVGTYH++S++ + PT G++SRGI LEGF SLLAG+WG+G GSTTLTEN+HT
Sbjct: 181 LVASVDSVGTYHSASMIVNAKRPTRGIVSRGIALEGFCSLLAGIWGSGTGSTTLTENIHT 240
Query: 403 IAGTKMGSRRPVQLGACLLIVLSLFGKVGGFIASIPEAMVAGLLCIMWAMLTALGLSNLR 462
I TK+ SRR + +GA LIVLS GK+G +ASIP+A+ A +LC +WA+ +LGLSNLR
Sbjct: 241 INITKVASRRALVIGAMFLIVLSFLGKLGAILASIPQALAASVLCFIWALTVSLGLSNLR 300
Query: 463 YTETGSSRNIIIVGLSLFFSLSIPAYFQQYESSPESNFSVPSYFQPYIVTSHGPFRSKYE 522
YT+T S RNI IVG+SLF LSIPAYFQQY+ P S+ +PSY+ P+ S GPF++ E
Sbjct: 301 YTQTASFRNITIVGVSLFLGLSIPAYFQQYQ--PLSSLILPSYYIPFGAASSGPFQTGIE 358
Query: 523 ELNYVLNMIFSLHMVIAFLVALILDNTVPGSKQERELYGWSKPNDAREDPFIVSEYGLPA 582
+L++ +N + SL+MV+ FL+A ILDNTVPGSK+ER +Y W++ D + DP + ++Y LP
Sbjct: 359 QLDFAMNAVLSLNMVVTFLLAFILDNTVPGSKEERGVYVWTRAEDMQMDPEMRADYSLPR 418
Query: 583 RVGRCF 588
+ + F
Sbjct: 419 KFAQIF 424
>ref|XP_506346.1| PREDICTED P0477A12.37 gene product [Oryza sativa (japonica
cultivar-group)] gi|50937285|ref|XP_478170.1| putative
permease [Oryza sativa (japonica cultivar-group)]
gi|33146998|dbj|BAC80070.1| putative permease [Oryza
sativa (japonica cultivar-group)]
Length = 305
Score = 348 bits (892), Expect = 4e-94
Identities = 163/296 (55%), Positives = 212/296 (71%), Gaps = 2/296 (0%)
Query: 300 MKHCQVNTSDTMTSPPWFRFPYPLQWGTPVFNWKMAIVMCVVSLISSVDSVGTYHTSSLL 359
M+ C+ + S+ + W R PYP QWG P F++K +I+M +VSL++SVDS+ +YH +SLL
Sbjct: 11 MRRCRTDASNAWRTAAWVRVPYPFQWGPPTFHFKTSIIMVIVSLVASVDSLSSYHATSLL 70
Query: 360 AASGPPTPGVLSRGIGLEGFSSLLAGLWGTGMGSTTLTENVHTIAGTKMGSRRPVQLGAC 419
PPT GV+SRGIG EG S+L+AG+WGTG GSTTLTEN+HT+ TKM SRR +Q GA
Sbjct: 71 VNLSPPTRGVVSRGIGFEGISTLIAGVWGTGTGSTTLTENIHTLENTKMASRRALQFGAV 130
Query: 420 LLIVLSLFGKVGGFIASIPEAMVAGLLCIMWAMLTALGLSNLRYTETGSSRNIIIVGLSL 479
LL++ S FGK+G +ASIP A+ A +LC WA++ ALGLS LRYT+ SSRN+IIVG +L
Sbjct: 131 LLVIFSFFGKIGALLASIPVALAASVLCFTWALIVALGLSTLRYTQAASSRNMIIVGFTL 190
Query: 480 FFSLSIPAYFQQYESSPESNFSVPSYFQPYIVTSHGPFRSKYEELNYVLNMIFSLHMVIA 539
F S+S+PAYFQQYE P +N +PSY PY S GP RS LN+ +N + S+++V+A
Sbjct: 191 FISMSVPAYFQQYE--PSTNLILPSYLLPYAAASSGPVRSGSNGLNFAVNALLSINVVVA 248
Query: 540 FLVALILDNTVPGSKQERELYGWSKPNDAREDPFIVSEYGLPARVGRCFRWVKWVG 595
LVALILDNTVPGS+QER +Y WS PN DP + Y LP ++ FRW K VG
Sbjct: 249 LLVALILDNTVPGSRQERGVYIWSDPNSLEMDPASLEPYRLPEKISCWFRWAKCVG 304
>dbj|BAD82048.1| nucleobase-ascorbate transporter-like protein [Oryza sativa
(japonica cultivar-group)]
Length = 253
Score = 332 bits (852), Expect = 2e-89
Identities = 162/246 (65%), Positives = 194/246 (78%)
Query: 350 VGTYHTSSLLAASGPPTPGVLSRGIGLEGFSSLLAGLWGTGMGSTTLTENVHTIAGTKMG 409
VG+YH SSL A+ PPT GV+SRGIG+EG S++LAGLWGTG+GS T+TENVHTIA TKMG
Sbjct: 7 VGSYHASSLFVATRPPTAGVVSRGIGVEGVSTVLAGLWGTGVGSATITENVHTIAVTKMG 66
Query: 410 SRRPVQLGACLLIVLSLFGKVGGFIASIPEAMVAGLLCIMWAMLTALGLSNLRYTETGSS 469
+RR V GA +LI+LS GKVG FIASIP+ +VA LLC MWAML ALGLSNLRY+ GSS
Sbjct: 67 NRRAVGFGAIVLILLSFVGKVGAFIASIPDVLVAALLCFMWAMLCALGLSNLRYSAKGSS 126
Query: 470 RNIIIVGLSLFFSLSIPAYFQQYESSPESNFSVPSYFQPYIVTSHGPFRSKYEELNYVLN 529
RN I+VGL+LF SLS+P+YFQQY P SN SVP+YFQPYIV SHGP + +NY+LN
Sbjct: 127 RNSIVVGLALFLSLSVPSYFQQYRLQPNSNSSVPTYFQPYIVASHGPIHTGSSGVNYILN 186
Query: 530 MIFSLHMVIAFLVALILDNTVPGSKQERELYGWSKPNDAREDPFIVSEYGLPARVGRCFR 589
+ SL+MVIAFLVALILDNTVPG +QER LY WS+ AR + ++ +Y LP ++G FR
Sbjct: 187 TLLSLNMVIAFLVALILDNTVPGGRQERGLYVWSEAEAARRESAVMKDYELPFKIGHAFR 246
Query: 590 WVKWVG 595
WVK VG
Sbjct: 247 WVKCVG 252
>gb|AAH90768.1| Zgc:110789 [Danio rerio] gi|61651826|ref|NP_001013353.1|
hypothetical protein LOC503757 [Danio rerio]
Length = 619
Score = 318 bits (814), Expect = 4e-85
Identities = 189/570 (33%), Positives = 298/570 (52%), Gaps = 73/570 (12%)
Query: 76 DHDDLVLRRRPSPLNYELTDSPALVFLAVYGIQHYLSIIGSLILTPLVIAPAMGASHD-- 133
+ D R P+ L Y +TD P GIQHYL+ G +I PL+++ + HD
Sbjct: 32 ESDGFSERGDPNKLAYCVTDIPPWYLCIFLGIQHYLTAFGGIIAIPLILSQGLCLQHDGL 91
Query: 134 ETAAMVCTVLLVSGVTTLLHTIFGSRLPLIQGPSFVYLAPVLAIINSPEFQ--------- 184
+ ++ T+ VSGV TLL FG RLP++QG +F L+P +A+++ PE+
Sbjct: 92 TQSHLISTIFFVSGVCTLLQVTFGVRLPILQGGTFTLLSPTMALLSMPEWTCPAWTQNAS 151
Query: 185 --ELNENKFKHI----MKELQGAIIIGSAFQTLLGYTGLMSLLVRFINPVVVSSTIAAVG 238
+F H+ M+ LQG+I++GS FQ L+G++GL+ L +RFI P+ ++ TI+ +G
Sbjct: 152 LVNTTSPEFIHVWQSRMQMLQGSIMVGSLFQVLVGFSGLIGLFMRFIGPLTIAPTISLIG 211
Query: 239 LFL*LARVLRS-----VQYRY*CLL-FFVLYLRKISV-------------FGHHIFQIYA 279
L L + + + + CL+ F YLR I++ IFQI
Sbjct: 212 LSLFDSAGMNAGHHWGISAMTTCLIVIFSQYLRHIAIPVPKYSRAKKFHTTRIFIFQILP 271
Query: 280 VPLGLAVTWTFAFLLT----------ENGRMKHCQVNTSDTMTSPPWFRFPYPLQWGTPV 329
V LG+ ++W +LLT + G + + D + PWFRFPYP QWG P
Sbjct: 272 VLLGITLSWLICYLLTIYNVLPSDPDKYGYLARTDIK-GDVTSKAPWFRFPYPGQWGVPS 330
Query: 330 FNWKMAIVMCVVSLISSVDSVGTYHTSSLLAASGPPTPGVLSRGIGLEGFSSLLAGLWGT 389
+ + + S ++SVG YH + L+ + PP ++RGIG+EG LLAG WGT
Sbjct: 331 VSLAGVFGILAGVISSMIESVGDYHACARLSGAPPPPRHAINRGIGIEGIGCLLAGAWGT 390
Query: 390 GMGSTTLTENVHTIAGTKMGSRRPVQLGACLLIVLSLFGKVGGFIASIPEAMVAGLLCIM 449
G G+T+ +ENV + TK+GSR + ++I++ +FGK+G +IP ++ G+ +M
Sbjct: 391 GNGTTSYSENVGALGITKVGSRMVIVASGFIMIIMGMFGKIGAIFTTIPTPVIGGMFLVM 450
Query: 450 WAMLTALGLSNLRYTETGSSRNIIIVGLSLFFSLSIPAYFQQYESSPESNFSVPSYFQPY 509
+ ++TA G+SNL+YT+ SSRNI I G S+F L+IP +
Sbjct: 451 FGVITAAGISNLQYTDMNSSRNIFIFGFSMFTGLTIP---------------------NW 489
Query: 510 IVTSHGPFRSKYEELNYVLNMIFSLHMVIAFLVALILDNTVPGSKQERELYGWSKPNDAR 569
I+ + + EL++VL ++ + M + +LDNTVPG+K+ER + W+K +
Sbjct: 490 IIKNPTSIATGVVELDHVLQVLLTTSMFVGGFFGFLLDNTVPGTKRERGITAWNKAHQDD 549
Query: 570 EDPFIVSE--YGLPARVGRC---FRWVKWV 594
+ S+ YGLP R+ C RW K+V
Sbjct: 550 SHNTLESDEVYGLPFRINSCLSSLRWTKYV 579
>ref|XP_482013.1| putative permease 1 [Oryza sativa (japonica cultivar-group)]
gi|38637273|dbj|BAD03537.1| putative permease 1 [Oryza
sativa (japonica cultivar-group)]
gi|38637220|dbj|BAD03486.1| putative permease 1 [Oryza
sativa (japonica cultivar-group)]
Length = 524
Score = 312 bits (799), Expect = 2e-83
Identities = 177/518 (34%), Positives = 284/518 (54%), Gaps = 34/518 (6%)
Query: 89 LNYELTDSPALVFLAVYGIQHYLSIIGSLILTPLVIAPAMGASHDETAAMVCTVLLVSGV 148
L Y + +P+ G QHY+ +G+ ++ P ++ P MG + + A +V T+L V+G+
Sbjct: 20 LEYCIDSNPSWGEAIALGFQHYILCLGTAVMIPTLLVPLMGGNAHDKAKVVQTMLFVTGI 79
Query: 149 TTLLHTIFGSRLPLIQGPSFVYLAPVLAIINSPEFQELNENKFKHIM--KELQGAIIIGS 206
T+L T+FG+RLP I G S+ ++ PV++II P ++ ++ + IM + +QGA+II S
Sbjct: 80 NTMLQTLFGTRLPTIIGGSYAFVIPVISIIKDPSLAQITDDHTRFIMTMRAIQGALIISS 139
Query: 207 AFQTLLGYTGLMSLLVRFINPVVVSSTIAAVGLFL*---LARVLRSVQYRY*CLLFFVL- 262
Q +LGY+ L + RF +P+ + +A VGL L + R V+ L+ FV
Sbjct: 140 CIQIILGYSQLWGICSRFFSPLGMVPVVALVGLGLFERGFPVIGRCVEIGLPMLVLFVAL 199
Query: 263 --YLRKISVFGHHIFQIYAVPLGLAVTWTFAFLLTENGRMKH--------CQVNTSDTMT 312
YL+ + V I + ++V + +A+ W +A +LT +G KH C+ + ++ +T
Sbjct: 200 SQYLKHVQVRHFPILERFSVLISIALVWVYAHILTASGTYKHTSLLTQINCRTDRANLIT 259
Query: 313 SPPWFRFPYPLQWGTPVFNWKMAIVMCVVSLISSVDSVGTYHTSSLLAASGPPTPGVLSR 372
S W PYPLQWG P F+ A M ++S ++S G + ++ LA++ PP P VLSR
Sbjct: 260 SADWIDIPYPLQWGPPTFSADHAFGMMAAVVVSLIESAGAFKAAARLASATPPPPYVLSR 319
Query: 373 GIGLEGFSSLLAGLWGTGMGSTTLTENVHTIAGTKMGSRRPVQLGACLLIVLSLFGKVGG 432
GIG +G L GL+GTG GST EN+ + T++GSRR +Q+ A +I S+ G+ G
Sbjct: 320 GIGWQGIGLLFDGLFGTGTGSTVSVENIGLLGSTRIGSRRVIQISAGFMIFFSILGRFGA 379
Query: 433 FIASIPEAMVAGLLCIMWAMLTALGLSNLRYTETGSSRNIIIVGLSLFFSLSIPAYFQQY 492
ASIP M A + C+M+ + A+GLS +++T S R++ I+G+SLF +SIP YF +Y
Sbjct: 380 LFASIPFTMFAAIYCVMFGYVGAVGLSFMQFTNMNSMRSLFIIGVSLFLGISIPEYFFRY 439
Query: 493 ESSPESNFSVPSYFQPYIVTSHGPFRSKYEELNYVLNMIFSLHMVIAFLVALILDNT--V 550
S HGP ++ N +N +FS + +VA+ILDNT V
Sbjct: 440 TMS----------------ALHGPAHTRAGWFNDYINTVFSSPPTVGLIVAVILDNTLEV 483
Query: 551 PGSKQERELYGWSKPNDAREDPFIVSEYGLPARVGRCF 588
+ ++R + W++ R D Y LP + R F
Sbjct: 484 RDAARDRGMPWWARFRTFRGDSRNEEFYTLPFNLNRFF 521
>ref|XP_416178.1| PREDICTED: similar to Solute carrier family 23, member 1
(Sodium-dependent vitamin C transporter 1) (hSVCT1)
(Na(+)/L-ascorbic acid transporter 1) (Yolk sac
permease-like molecule 3) [Gallus gallus]
Length = 633
Score = 310 bits (795), Expect = 6e-83
Identities = 184/559 (32%), Positives = 298/559 (52%), Gaps = 78/559 (13%)
Query: 87 SPLNYELTDSPALVFLAVYGIQHYLSIIGSLILTPLVIAPAMGASHD--ETAAMVCTVLL 144
S L Y +TD P + GIQH+L+ +G L+ PL+++ + HD + ++ T+
Sbjct: 46 SKLAYTVTDMPPWYLCILLGIQHFLTAMGGLVAIPLILSKELCLQHDLLTQSHLISTIFF 105
Query: 145 VSGVTTLLHTIFGSRLPLIQGPSFVYLAPVLAIIN-------------------SPEFQE 185
VSG+ TLL +FG RLP+IQG +F +L P LA+++ SPEF E
Sbjct: 106 VSGICTLLQVLFGVRLPIIQGGTFAFLTPTLAMLSLPKWKCPAWTENATLVNTSSPEFIE 165
Query: 186 LNENKFKHIMKELQGAIIIGSAFQTLLGYTGLMSLLVRFINPVVVSSTIAAVGLFL*LAR 245
+ + + M+E+QGAI++ S FQ L+G++G++ L+RFI P+ ++ TI V L L +
Sbjct: 166 VWQTR----MREVQGAIMVASCFQILVGFSGIIGFLMRFIGPLTIAPTITLVALPLFDSA 221
Query: 246 VLRSVQY---RY*CLLFFVL---YLRKISV-------------FGHHIFQIYAVPLGLAV 286
++ Q+ + + F VL YL+ + V ++FQI+ V LGL++
Sbjct: 222 GDKAGQHWGIAFMTIFFIVLFSQYLKDVPVPLPSFRRGKKCHFSPIYVFQIFPVLLGLSL 281
Query: 287 TWTFAFLLTEN----------GRMKHCQVNTSDTMTSPPWFRFPYPLQWGTPVFNWKMAI 336
+W ++LT G + D ++ PWFR PYP QWGTP +
Sbjct: 282 SWLLCYVLTVTDVLPTDPTAYGHLARTDTR-GDVLSQAPWFRLPYPGQWGTPTVSLAGIF 340
Query: 337 VMCVVSLISSVDSVGTYHTSSLLAASGPPTPGVLSRGIGLEGFSSLLAGLWGTGMGSTTL 396
+ + S ++S+G Y+ + L+ + PP ++RGIG+EG LLAG WGTG G+T+
Sbjct: 341 GILAGVISSMLESMGDYYACARLSGAPPPPKHAINRGIGVEGIGCLLAGAWGTGNGTTSY 400
Query: 397 TENVHTIAGTKMGSRRPVQLGACLLIVLSLFGKVGGFIASIPEAMVAGLLCIMWAMLTAL 456
+ENV + TK+GSR + GAC +++ +FGKVG +ASIP ++ G+ +M+ ++TA+
Sbjct: 401 SENVGALGITKVGSRMVIIAGACAMLLSGVFGKVGAMLASIPTPVIGGMFLVMFGIITAV 460
Query: 457 GLSNLRYTETGSSRNIIIVGLSLFFSLSIPAYFQQYESSPESNFSVPSYFQPYIVTSHGP 516
G+SNL+YT+ SSRNI I G S+F L++P + + + E+
Sbjct: 461 GISNLQYTDMNSSRNIFIFGFSVFAGLTVPNWANKNNTLLETEII--------------- 505
Query: 517 FRSKYEELNYVLNMIFSLHMVIAFLVALILDNTVPGSKQERELYGW--SKPNDAREDPFI 574
+L+ V+ ++ + M + L+ ILDNT+PG+++ER L W S +A I
Sbjct: 506 ------QLDQVIQVLLTTGMFVGGLLGFILDNTIPGTQEERGLLAWKHSHKGEADNSQLI 559
Query: 575 VSEYGLPARVGRCFRWVKW 593
Y LP +G + V W
Sbjct: 560 SKVYDLPFGIGTKYCAVSW 578
>gb|AAT64019.1| putative permease [Gossypium hirsutum]
Length = 524
Score = 308 bits (790), Expect = 2e-82
Identities = 171/518 (33%), Positives = 283/518 (54%), Gaps = 34/518 (6%)
Query: 89 LNYELTDSPALVFLAVYGIQHYLSIIGSLILTPLVIAPAMGASHDETAAMVCTVLLVSGV 148
L Y + +P+ G QHY+ +G+ ++ P + P MG + D+ +V T+L V G+
Sbjct: 20 LEYCIDSNPSWGEAIALGFQHYILALGTAVMIPSFLVPLMGGTDDDKVRVVQTLLFVEGI 79
Query: 149 TTLLHTIFGSRLPLIQGPSFVYLAPVLAIINSPEFQELNEN--KFKHIMKELQGAIIIGS 206
TLL T+FG+RLP + G S+ ++ P+++II+ + +N +F + M+ +QGA+I+ S
Sbjct: 80 NTLLQTLFGTRLPTVIGGSYAFMVPIISIIHDTTLLNIEDNHMRFLYTMRAVQGALIVAS 139
Query: 207 AFQTLLGYTGLMSLLVRFINPVVVSSTIAAVGLFL*---LARVLRSVQYRY*CLLFFVL- 262
+ Q +LGY+ + ++ RF +P+ + IA VG L V R V+ L+ F+
Sbjct: 140 SIQIILGYSQMWAICTRFFSPLGMIPVIALVGFGLFDKGFPVVGRCVEIGIPMLILFIAF 199
Query: 263 --YLRKISVFGHHIFQIYAVPLGLAVTWTFAFLLTENGRMKH--------CQVNTSDTMT 312
YL+ I + +A+ + + V W +A LLT +G KH C+ + ++ ++
Sbjct: 200 SQYLKNFHTKQLPILERFALIISITVIWAYAHLLTASGAYKHRPELTQLNCRTDKANLIS 259
Query: 313 SPPWFRFPYPLQWGTPVFNWKMAIVMCVVSLISSVDSVGTYHTSSLLAASGPPTPGVLSR 372
S PW + PYPLQWG P F+ A M L+S ++S G+Y ++ LA++ PP +LSR
Sbjct: 260 SAPWIKIPYPLQWGAPTFDAGHAFGMMAAVLVSLIESTGSYKAAARLASATPPPAHILSR 319
Query: 373 GIGLEGFSSLLAGLWGTGMGSTTLTENVHTIAGTKMGSRRPVQLGACLLIVLSLFGKVGG 432
GIG +G LL GL+GT GST ENV + T++GSRR +Q+ A +I S+ GK G
Sbjct: 320 GIGWQGIGILLDGLFGTLTGSTVSVENVGLLGSTRVGSRRVIQISAGFMIFFSILGKFGA 379
Query: 433 FIASIPEAMVAGLLCIMWAMLTALGLSNLRYTETGSSRNIIIVGLSLFFSLSIPAYFQQY 492
ASIP + A + C+++ ++ ++GLS +++T S RN+ I+G++LF LS+P Y+++Y
Sbjct: 380 LFASIPFTIFAAVYCVLFGIVASVGLSFMQFTNMNSMRNLFIIGVALFLGLSVPEYYREY 439
Query: 493 ESSPESNFSVPSYFQPYIVTSHGPFRSKYEELNYVLNMIFSLHMVIAFLVALILDNTV-- 550
+ HGP ++ N LN IF +A +VA++LDNT+
Sbjct: 440 TAK----------------ALHGPAHTRAVWFNDFLNTIFFSSPTVALIVAVLLDNTLDY 483
Query: 551 PGSKQERELYGWSKPNDAREDPFIVSEYGLPARVGRCF 588
S ++R + W+ + D Y LP + R F
Sbjct: 484 KDSARDRGMPWWANFRTFKGDSRSEEFYSLPFNLNRFF 521
>gb|AAT64034.1| putative permease [Gossypium hirsutum]
Length = 524
Score = 307 bits (786), Expect = 7e-82
Identities = 170/518 (32%), Positives = 283/518 (53%), Gaps = 34/518 (6%)
Query: 89 LNYELTDSPALVFLAVYGIQHYLSIIGSLILTPLVIAPAMGASHDETAAMVCTVLLVSGV 148
L Y + +P+ G QHY+ +G+ ++ P + P MG + D+ +V T+L V G+
Sbjct: 20 LEYCIDSNPSWGEAIALGFQHYILALGTAVMIPSFLVPLMGGTDDDKVRVVQTLLFVEGI 79
Query: 149 TTLLHTIFGSRLPLIQGPSFVYLAPVLAIINSPEFQELNEN--KFKHIMKELQGAIIIGS 206
TLL T+FG+RLP + G S+ ++ P+++II+ + +N +F + M+ +QGA+I+ S
Sbjct: 80 NTLLQTLFGTRLPTVIGGSYAFMVPIISIIHDTTLLSIEDNHMRFLYTMRAVQGALIVAS 139
Query: 207 AFQTLLGYTGLMSLLVRFINPVVVSSTIAAVGLFL*---LARVLRSVQYRY*CLLFFVL- 262
+ Q +LGY+ + ++ RF +P+ + IA VG L V R V+ L F+
Sbjct: 140 SIQIILGYSQMWAICTRFFSPLGMIPVIALVGFGLFDKGFPVVGRCVEIGIPMLFLFIAF 199
Query: 263 --YLRKISVFGHHIFQIYAVPLGLAVTWTFAFLLTENGRMKH--------CQVNTSDTMT 312
YL+ I + +A+ + + V W +A LLT++G KH C+ + ++ ++
Sbjct: 200 SQYLKNFLTKQLPILERFALIISITVIWAYAHLLTKSGAYKHRPELTQLNCRTDKANLIS 259
Query: 313 SPPWFRFPYPLQWGTPVFNWKMAIVMCVVSLISSVDSVGTYHTSSLLAASGPPTPGVLSR 372
S PW + PYPLQWG P F+ A M L+S ++S G+Y ++ LA++ PP +LSR
Sbjct: 260 SAPWIKIPYPLQWGAPTFDAGHAFGMMAAVLVSLIESTGSYKAAARLASATPPPAHILSR 319
Query: 373 GIGLEGFSSLLAGLWGTGMGSTTLTENVHTIAGTKMGSRRPVQLGACLLIVLSLFGKVGG 432
GIG +G LL GL+GT GST ENV + T++GSRR +Q+ A +I S+ GK G
Sbjct: 320 GIGWQGIGILLDGLFGTLTGSTVSVENVGLLGSTRVGSRRVIQISAGFMIFFSILGKFGA 379
Query: 433 FIASIPEAMVAGLLCIMWAMLTALGLSNLRYTETGSSRNIIIVGLSLFFSLSIPAYFQQY 492
ASIP + A + C+++ ++ ++GLS +++T S RN+ I+G+++F LS+P Y+++Y
Sbjct: 380 LFASIPFTIFAAVYCVLFGIVASVGLSFMQFTNMNSMRNLFIIGVAMFLGLSVPEYYREY 439
Query: 493 ESSPESNFSVPSYFQPYIVTSHGPFRSKYEELNYVLNMIFSLHMVIAFLVALILDNTV-- 550
+ HGP ++ N LN IF +A +VA++LDNT+
Sbjct: 440 TAK----------------ALHGPAHTRAVWFNDFLNTIFFSSPTVALIVAVLLDNTLDY 483
Query: 551 PGSKQERELYGWSKPNDAREDPFIVSEYGLPARVGRCF 588
S ++R + W+ + D Y LP + R F
Sbjct: 484 KDSARDRGMPWWANFRTFKGDGRSEEFYSLPFNLNRFF 521
>ref|XP_463430.1| putative permease 1 [Oryza sativa (japonica cultivar-group)]
gi|20804662|dbj|BAB92350.1| putative permease 1 [Oryza
sativa (japonica cultivar-group)]
gi|14587294|dbj|BAB61205.1| putative permease 1 [Oryza
sativa (japonica cultivar-group)]
Length = 524
Score = 306 bits (783), Expect = 2e-81
Identities = 174/520 (33%), Positives = 284/520 (54%), Gaps = 42/520 (8%)
Query: 91 YELTDSPALVFLAVYGIQHYLSIIGSLILTPLVIAPAMGASHDETAAMVCTVLLVSGVTT 150
Y + +P + G QHY+ +G+ ++ P V+ P MG S + +V T+L V+G+ T
Sbjct: 22 YCIDSNPPWGEAIILGFQHYILALGTAVMIPAVLVPMMGGSDGDRVRVVQTLLFVTGINT 81
Query: 151 LLHTIFGSRLPLIQGPSFVYLAPVLAIINSPEFQELNEN--KFKHIMKELQGAIIIGSAF 208
LL ++FG+RLP + G S+ ++ P++AII + ++ +F M+ +QGA+I+ S+
Sbjct: 82 LLQSLFGTRLPTVIGGSYAFVVPIMAIIQDSSLAAIPDDHERFLQTMRAIQGALIVSSSI 141
Query: 209 QTLLGYTGLMSLLVRFINPVVVSSTIAAVGLFL*---LARVLRSVQYRY*CLLFFVL--- 262
Q +LGY+ L + RF +P+ ++ +A +G L V R V+ L+ FV+
Sbjct: 142 QIILGYSQLWGIFSRFFSPLGMAPVVALLGFGLFERGFPVVGRCVEVGLPMLILFVVLSQ 201
Query: 263 YLRKISVFGHHIFQIYAVPLGLAVTWTFAFLLTENGRMKH--------CQVNTSDTMTSP 314
YL+ + + I + +++ + +A+ W +A +LT G KH C+ + ++ ++S
Sbjct: 202 YLKNVQIRDIPILERFSLFICIALVWAYAQILTAGGAYKHSPEVTQINCRTDRANLISSA 261
Query: 315 PWFRFPYPLQWGTPVFNWKMAIVMCVVSLISSVDSVGTYHTSSLLAASGPPTPGVLSRGI 374
PW + P+PLQWG P F+ + M L+S V+S +Y ++ LA++ PP +LSRGI
Sbjct: 262 PWIKIPFPLQWGAPTFSAGQSFGMVSAVLVSLVESTASYKAAARLASATPPPAHILSRGI 321
Query: 375 GLEGFSSLLAGLWGTGMGSTTLTENVHTIAGTKMGSRRPVQLGACLLIVLSLFGKVGGFI 434
G +G LL GL+GTG GST ENV + T++GSRR +Q+ A +I S+ GK G
Sbjct: 322 GWQGIGILLDGLFGTGTGSTVSVENVGLLGSTRIGSRRVIQISAGFMIFFSMLGKFGALF 381
Query: 435 ASIPEAMVAGLLCIMWAMLTALGLSNLRYTETGSSRNIIIVGLSLFFSLSIPAYFQQYES 494
ASIP + A + C+++ ++ A+GLS L++T S RN+ IVG+S+F LS+P YF +Y
Sbjct: 382 ASIPFTIFAAVYCVLFGLVAAVGLSFLQFTNMNSMRNLFIVGVSIFLGLSVPEYFFRYS- 440
Query: 495 SPESNFSVPSYFQPYIVTSHGPFRSKYEELNYVLNMIFSLHMVIAFLVALILDNT--VPG 552
+ GP +K N +N IFS + +VA+ LDNT V
Sbjct: 441 ---------------MAAQRGPAHTKAGWFNDYINTIFSSPPTVGLIVAVFLDNTLEVKN 485
Query: 553 SKQERELYGW----SKPNDAREDPFIVSEYGLPARVGRCF 588
+ ++R + W S DAR + F Y LP + R F
Sbjct: 486 AAKDRGMPWWVPFRSFKGDARSEEF----YSLPFNLNRFF 521
>ref|NP_176211.2| xanthine/uracil permease family protein [Arabidopsis thaliana]
Length = 538
Score = 304 bits (778), Expect = 6e-81
Identities = 180/522 (34%), Positives = 274/522 (52%), Gaps = 36/522 (6%)
Query: 87 SPLNYELTDSPALVFLAVYGIQHYLSIIGSLILTPLVIAPAMGASHDETAAMVCTVLLVS 146
S ++Y +T P + G QHYL ++G+ +L P + P MG ++E A MV T+L VS
Sbjct: 30 SSISYCITSPPPWPEAILLGFQHYLVMLGTTVLIPTYLVPQMGGGNEEKAKMVQTLLFVS 89
Query: 147 GVTTLLHTIFGSRLPLIQGPSFVYLAPVLAIINSPEFQEL--NENKFKHIMKELQGAIII 204
G+ TLL + FG+RLP + G S+ Y+ L+II + + ++ + KFK IM+ +QGA+I+
Sbjct: 90 GLNTLLQSFFGTRLPAVIGGSYTYVPTTLSIILAGRYSDILDPQEKFKRIMRGIQGALIV 149
Query: 205 GSAFQTLLGYTGLMSLLVRFINPVVVSSTIAAVGL------FL*LARVLRSVQYRY*CLL 258
S Q ++G++GL +VR ++P+ +A G F LA+ + LL
Sbjct: 150 ASILQIVVGFSGLWRNVVRLLSPLSAVPLVALAGFGLYEHGFPLLAKCIEIGLPEIILLL 209
Query: 259 FFVLYLRKISVFGHHIFQIYAVPLGLAVTWTFAFLLTENGRMKHCQVNT--------SDT 310
F Y+ + +F +AV + + W +A LLT G K+ VNT S
Sbjct: 210 LFSQYIPHLIRGERQVFHRFAVIFSVVIVWIYAHLLTVGGAYKNTGVNTQTSCRTDRSGL 269
Query: 311 MTSPPWFRFPYPLQWGTPVFNWKMAIVMCVVSLISSVDSVGTYHTSSLLAASGPPTPGVL 370
++ PW R PYP QWG P F+ A M VS +S ++S GTY S A++ PP P VL
Sbjct: 270 ISGSPWIRVPYPFQWGPPTFHAGEAFAMMAVSFVSLIESTGTYIVVSRFASATPPPPSVL 329
Query: 371 SRGIGLEGFSSLLAGLWGTGMGSTTLTENVHTIAGTKMGSRRPVQLGACLLIVLSLFGKV 430
SRG+G +G LL GL+G G G++ EN +A T++GSRR VQ+ A +I S+ GK
Sbjct: 330 SRGVGWQGVGVLLCGLFGAGNGASVSVENAGLLALTRVGSRRVVQISAGFMIFFSILGKF 389
Query: 431 GGFIASIPEAMVAGLLCIMWAMLTALGLSNLRYTETGSSRNIIIVGLSLFFSLSIPAYFQ 490
G ASIP +VA L C+ +A + A GLS L++ S R I+G S+F LSIP YF
Sbjct: 390 GAIFASIPAPVVAALHCLFFAYVGAGGLSLLQFCNLNSFRTKFILGFSVFMGLSIPQYFN 449
Query: 491 QYESSPESNFSVPSYFQPYIVTSHGPFRSKYEELNYVLNMIFSLHMVIAFLVALILDNTV 550
QY + V +GP + N ++N+ FS +A ++A LD T+
Sbjct: 450 QYTA----------------VNKYGPVHTHARWFNDMINVPFSSKAFVAGILAFFLDVTM 493
Query: 551 ----PGSKQERELYGWSKPNDAREDPFIVSEYGLPARVGRCF 588
++++R ++ W + + D Y LP + + F
Sbjct: 494 SSKDSATRKDRGMFWWDRFMSFKSDTRSEEFYSLPFNLNKYF 535
>gb|AAD14479.1| Strong similarity to gi|3337350 F13P17.3 putative permease from
Arabidopsis thaliana BAC gb|AC004481
gi|25309278|pir||F96624 hypothetical protein T2K10.8
[imported] - Arabidopsis thaliana
Length = 543
Score = 303 bits (777), Expect = 8e-81
Identities = 181/527 (34%), Positives = 274/527 (51%), Gaps = 41/527 (7%)
Query: 87 SPLNYELTDSPALVFLAVYGIQHYLSIIGSLILTPLVIAPAMGASHDETAAMVCTVLLVS 146
S ++Y +T P + G QHYL ++G+ +L P + P MG ++E A MV T+L VS
Sbjct: 30 SSISYCITSPPPWPEAILLGFQHYLVMLGTTVLIPTYLVPQMGGGNEEKAKMVQTLLFVS 89
Query: 147 GVTTLLHTIFGSRLPLIQGPSFVYLAPVLAIINSPEFQEL-------NENKFKHIMKELQ 199
G+ TLL + FG+RLP + G S+ Y+ L+II + + ++ N KFK IM+ +Q
Sbjct: 90 GLNTLLQSFFGTRLPAVIGGSYTYVPTTLSIILAGRYSDILDPQESENMQKFKRIMRGIQ 149
Query: 200 GAIIIGSAFQTLLGYTGLMSLLVRFINPVVVSSTIAAVGL------FL*LARVLRSVQYR 253
GA+I+ S Q ++G++GL +VR ++P+ +A G F LA+ +
Sbjct: 150 GALIVASILQIVVGFSGLWRNVVRLLSPLSAVPLVALAGFGLYEHGFPLLAKCIEIGLPE 209
Query: 254 Y*CLLFFVLYLRKISVFGHHIFQIYAVPLGLAVTWTFAFLLTENGRMKHCQVNT------ 307
LL F Y+ + +F +AV + + W +A LLT G K+ VNT
Sbjct: 210 IILLLLFSQYIPHLIRGERQVFHRFAVIFSVVIVWIYAHLLTVGGAYKNTGVNTQTSCRT 269
Query: 308 --SDTMTSPPWFRFPYPLQWGTPVFNWKMAIVMCVVSLISSVDSVGTYHTSSLLAASGPP 365
S ++ PW R PYP QWG P F+ A M VS +S ++S GTY S A++ PP
Sbjct: 270 DRSGLISGSPWIRVPYPFQWGPPTFHAGEAFAMMAVSFVSLIESTGTYIVVSRFASATPP 329
Query: 366 TPGVLSRGIGLEGFSSLLAGLWGTGMGSTTLTENVHTIAGTKMGSRRPVQLGACLLIVLS 425
P VLSRG+G +G LL GL+G G G++ EN +A T++GSRR VQ+ A +I S
Sbjct: 330 PPSVLSRGVGWQGVGVLLCGLFGAGNGASVSVENAGLLALTRVGSRRVVQISAGFMIFFS 389
Query: 426 LFGKVGGFIASIPEAMVAGLLCIMWAMLTALGLSNLRYTETGSSRNIIIVGLSLFFSLSI 485
+ GK G ASIP +VA L C+ +A + A GLS L++ S R I+G S+F LSI
Sbjct: 390 ILGKFGAIFASIPAPVVAALHCLFFAYVGAGGLSLLQFCNLNSFRTKFILGFSVFMGLSI 449
Query: 486 PAYFQQYESSPESNFSVPSYFQPYIVTSHGPFRSKYEELNYVLNMIFSLHMVIAFLVALI 545
P YF QY + V +GP + N ++N+ FS +A ++A
Sbjct: 450 PQYFNQYTA----------------VNKYGPVHTHARWFNDMINVPFSSKAFVAGILAFF 493
Query: 546 LDNTV----PGSKQERELYGWSKPNDAREDPFIVSEYGLPARVGRCF 588
LD T+ ++++R ++ W + + D Y LP + + F
Sbjct: 494 LDVTMSSKDSATRKDRGMFWWDRFMSFKSDTRSEEFYSLPFNLNKYF 540
>gb|AAD26910.1| putative membrane transporter [Arabidopsis thaliana]
gi|66792650|gb|AAY56427.1| At2g05760 [Arabidopsis
thaliana] gi|15224977|ref|NP_178636.1| xanthine/uracil
permease family protein [Arabidopsis thaliana]
gi|25309276|pir||D84471 probable membrane transporter
[imported] - Arabidopsis thaliana
Length = 520
Score = 300 bits (768), Expect = 9e-80
Identities = 177/519 (34%), Positives = 277/519 (53%), Gaps = 35/519 (6%)
Query: 89 LNYELTDSPALVFLAVYGIQHYLSIIGSLILTPLVIAPAMGASHDETAAMVCTVLLVSGV 148
L Y + +P + Q+Y+ ++G+ P ++ PAMG S + A ++ T+L V+G+
Sbjct: 15 LEYCIDSNPPWPETVLLAFQNYILMLGTSAFIPALLVPAMGGSDGDRARVIQTLLFVAGI 74
Query: 149 TTLLHTIFGSRLPLIQGPSFVYLAPVLAIINSPEFQELNEN--KFKHIMKELQGAIIIGS 206
TLL +FG+RLP + G S Y+ P+ IIN Q+++ + +F H M+ +QGA+I+ S
Sbjct: 75 KTLLQALFGTRLPAVVGGSLAYVVPIAYIINDSSLQKISNDHERFIHTMRAIQGALIVAS 134
Query: 207 AFQTLLGYTGLMSLLVRFINPVVVSSTIAAVGL------FL*LARVLRSVQYRY*CLLFF 260
+ Q +LGY+ + L RF +P+ ++ + VGL F L + ++
Sbjct: 135 SIQIILGYSQVWGLFSRFFSPLGMAPVVGLVGLGMFQRGFPQLGNCIEIGLPMLLLVIGL 194
Query: 261 VLYLRKISVFGH-HIFQIYAVPLGLAVTWTFAFLLTENGRMK--------HCQVNTSDTM 311
YL+ + F IF+ + + + + + W +A +LT +G + C+ + ++ +
Sbjct: 195 TQYLKHVRPFKDVPIFERFPILICVTIVWIYAVILTASGAYRGKPSLTQHSCRTDKANLI 254
Query: 312 TSPPWFRFPYPLQWGTPVFNWKMAIVMCVVSLISSVDSVGTYHTSSLLAASGPPTPGVLS 371
++ PWF+FPYPLQWG P F+ + M L+S V+S G Y +S LA + PP VLS
Sbjct: 255 STAPWFKFPYPLQWGPPTFSVGHSFAMMSAVLVSMVESTGAYIAASRLAIATPPPAYVLS 314
Query: 372 RGIGLEGFSSLLAGLWGTGMGSTTLTENVHTIAGTKMGSRRPVQLGACLLIVLSLFGKVG 431
RGIG +G LL GL+GTG GST L ENV + T++GSRR VQ+ A +IV S GK G
Sbjct: 315 RGIGWQGIGVLLDGLFGTGTGSTVLVENVGLLGLTRVGSRRVVQVSAGFMIVFSTLGKFG 374
Query: 432 GFIASIPEAMVAGLLCIMWAMLTALGLSNLRYTETGSSRNIIIVGLSLFFSLSIPAYFQQ 491
ASIP + A L CI++ ++ A+GLS L++T S RN++I GLSLF +SIP +F Q
Sbjct: 375 AVFASIPVPIYAALHCILFGLVAAVGLSFLQFTNMNSMRNLMITGLSLFLGISIPQFFAQ 434
Query: 492 YESSPESNFSVPSYFQPYIVTSHGPFRSKYEELNYVLNMIFSLHMVIAFLVALILDNT-- 549
Y + +G + N LN +F + ++A+ +DNT
Sbjct: 435 Y----------------WDARHYGLVHTNAGWFNAFLNTLFMSPATVGLIIAVFMDNTME 478
Query: 550 VPGSKQERELYGWSKPNDAREDPFIVSEYGLPARVGRCF 588
V SK++R + W K R D Y LP + R F
Sbjct: 479 VERSKKDRGMPWWVKFRTFRGDNRNEEFYTLPFNLNRFF 517
>gb|AAN13099.1| putative membrane transporter [Arabidopsis thaliana]
gi|3337350|gb|AAC27395.1| putative membrane transporter
[Arabidopsis thaliana] gi|15226243|ref|NP_180966.1|
xanthine/uracil permease family protein [Arabidopsis
thaliana] gi|7488005|pir||T02307 probable membrane
transporter At2g34190 [imported] - Arabidopsis thaliana
Length = 524
Score = 299 bits (766), Expect = 1e-79
Identities = 171/522 (32%), Positives = 282/522 (53%), Gaps = 42/522 (8%)
Query: 89 LNYELTDSPALVFLAVYGIQHYLSIIGSLILTPLVIAPAMGASHDETAAMVCTVLLVSGV 148
L Y + +P G +HY+ +G+ ++ P ++ P MG + +V T+L + GV
Sbjct: 20 LEYCIDSNPPWGEAIALGFEHYILALGTAVMIPSILVPMMGGDDGDKVRVVQTLLFLQGV 79
Query: 149 TTLLHTIFGSRLPLIQGPSFVYLAPVLAIINSPEFQELNENKFKHI--MKELQGAIIIGS 206
TLL T+FG+RLP + G S+ ++ P+++II+ + + + + + M+ +QGAII+ S
Sbjct: 80 NTLLQTLFGTRLPTVIGGSYAFMVPIISIIHDSSLTRIEDPQLRFLSTMRAVQGAIIVAS 139
Query: 207 AFQTLLGYTGLMSLLVRFINPVVVSSTIAAVGLFL*---LARVLRSVQYRY*CLLFFVLY 263
+ Q +LG++ + ++ RF +P+ + IA G L V V+ L+ FV++
Sbjct: 140 SVQIILGFSQMWAICSRFFSPIGMVPVIALTGFGLFNRGFPVVGNCVEIGLPMLILFVIF 199
Query: 264 LRKISVFGHHIFQI---YAVPLGLAVTWTFAFLLTENGRMKH--------CQVNTSDTMT 312
+ + F F + +A+ + L + W +A +LT +G KH C+ + S+ ++
Sbjct: 200 SQYLKNFQFRQFPVVERFALIIALIIVWAYAHVLTASGAYKHRPHQTQLNCRTDMSNLIS 259
Query: 313 SPPWFRFPYPLQWGTPVFNWKMAIVMCVVSLISSVDSVGTYHTSSLLAASGPPTPGVLSR 372
S PW + PYPLQWG P F+ A M L+S ++S G + ++ LA++ PP P VLSR
Sbjct: 260 SAPWIKIPYPLQWGAPSFDAGHAFAMMAAVLVSLIESTGAFKAAARLASATPPPPHVLSR 319
Query: 373 GIGLEGFSSLLAGLWGTGMGSTTLTENVHTIAGTKMGSRRPVQLGACLLIVLSLFGKVGG 432
GIG +G LL GL+GT GS+ EN+ + T++GSRR +Q+ A +I S+ GK G
Sbjct: 320 GIGWQGIGILLNGLFGTLSGSSVSVENIGLLGSTRVGSRRVIQISAGFMIFFSMLGKFGA 379
Query: 433 FIASIPEAMVAGLLCIMWAMLTALGLSNLRYTETGSSRNIIIVGLSLFFSLSIPAYFQQY 492
ASIP + A + C+++ ++ ++GLS L++T S RN+ IVG+SLF LSIP YF+ +
Sbjct: 380 LFASIPFTIFAAVYCVLFGLVASVGLSFLQFTNMNSLRNLFIVGVSLFLGLSIPEYFRDF 439
Query: 493 ESSPESNFSVPSYFQPYIVTSHGPFRSKYEELNYVLNMIFSLHMVIAFLVALILDNTV-- 550
+ HGP + N LN IF ++A +VA+ LDNT+
Sbjct: 440 S----------------MKALHGPAHTNAGWFNDFLNTIFLSSPMVALMVAVFLDNTLDY 483
Query: 551 PGSKQERELYGWSK----PNDAREDPFIVSEYGLPARVGRCF 588
+ ++R L W+K D+R + F Y LP + R F
Sbjct: 484 KETARDRGLPWWAKFRTFKGDSRNEEF----YTLPFNLNRFF 521
>gb|AAK59632.1| putative membrane transporter protein [Arabidopsis thaliana]
Length = 524
Score = 298 bits (762), Expect = 4e-79
Identities = 170/522 (32%), Positives = 281/522 (53%), Gaps = 42/522 (8%)
Query: 89 LNYELTDSPALVFLAVYGIQHYLSIIGSLILTPLVIAPAMGASHDETAAMVCTVLLVSGV 148
L Y + +P G +HY+ +G+ ++ P ++ P MG + +V T+L + GV
Sbjct: 20 LEYCIDSNPPWGEAIALGFEHYILALGTAVMIPSILVPMMGGDDGDKVRVVQTLLFLQGV 79
Query: 149 TTLLHTIFGSRLPLIQGPSFVYLAPVLAIINSPEFQELNENKFKHI--MKELQGAIIIGS 206
TLL T+FG+RLP + G S+ ++ P+++II+ + + + + + M+ +QGAII+ S
Sbjct: 80 NTLLQTLFGTRLPTVIGGSYAFMVPIISIIHDSSLTRIEDPQLRFLSTMRAVQGAIIVAS 139
Query: 207 AFQTLLGYTGLMSLLVRFINPVVVSSTIAAVGLFL*---LARVLRSVQYRY*CLLFFVLY 263
+ Q +LG++ + ++ RF +P+ + IA G L V V+ + FV++
Sbjct: 140 SVQIILGFSQMWAICSRFFSPIGMVPVIALTGFGLFNRGFPVVGNCVEIGLPMFILFVIF 199
Query: 264 LRKISVFGHHIFQI---YAVPLGLAVTWTFAFLLTENGRMKH--------CQVNTSDTMT 312
+ + F F + +A+ + L + W +A +LT +G KH C+ + S+ ++
Sbjct: 200 SQYLKNFQFRQFPVVERFALIIALIIVWAYAHVLTASGAYKHRPHQTQLNCRTDMSNLIS 259
Query: 313 SPPWFRFPYPLQWGTPVFNWKMAIVMCVVSLISSVDSVGTYHTSSLLAASGPPTPGVLSR 372
S PW + PYPLQWG P F+ A M L+S ++S G + ++ LA++ PP P VLSR
Sbjct: 260 SAPWIKIPYPLQWGAPSFDAGHAFAMMAAVLVSLIESTGAFKAAARLASATPPPPHVLSR 319
Query: 373 GIGLEGFSSLLAGLWGTGMGSTTLTENVHTIAGTKMGSRRPVQLGACLLIVLSLFGKVGG 432
GIG +G LL GL+GT GS+ EN+ + T++GSRR +Q+ A +I S+ GK G
Sbjct: 320 GIGWQGIGILLNGLFGTLSGSSVSVENIGLLGSTRVGSRRVIQISAGFMIFFSMLGKFGA 379
Query: 433 FIASIPEAMVAGLLCIMWAMLTALGLSNLRYTETGSSRNIIIVGLSLFFSLSIPAYFQQY 492
ASIP + A + C+++ ++ ++GLS L++T S RN+ IVG+SLF LSIP YF+ +
Sbjct: 380 LFASIPFTIFAAVYCVLFGLVASVGLSFLQFTNMNSLRNLFIVGVSLFLGLSIPEYFRDF 439
Query: 493 ESSPESNFSVPSYFQPYIVTSHGPFRSKYEELNYVLNMIFSLHMVIAFLVALILDNTV-- 550
+ HGP + N LN IF ++A +VA+ LDNT+
Sbjct: 440 S----------------MKALHGPAHTNAGWFNDFLNTIFLSSPMVALMVAVFLDNTLDY 483
Query: 551 PGSKQERELYGWSK----PNDAREDPFIVSEYGLPARVGRCF 588
+ ++R L W+K D+R + F Y LP + R F
Sbjct: 484 KETARDRGLPWWAKFRTFKGDSRNEEF----YTLPFNLNRFF 521
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.325 0.140 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,029,547,378
Number of Sequences: 2540612
Number of extensions: 45105388
Number of successful extensions: 164616
Number of sequences better than 10.0: 567
Number of HSP's better than 10.0 without gapping: 452
Number of HSP's successfully gapped in prelim test: 115
Number of HSP's that attempted gapping in prelim test: 162748
Number of HSP's gapped (non-prelim): 873
length of query: 595
length of database: 863,360,394
effective HSP length: 134
effective length of query: 461
effective length of database: 522,918,386
effective search space: 241065375946
effective search space used: 241065375946
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 78 (34.7 bits)
Medicago: description of AC148969.3


