

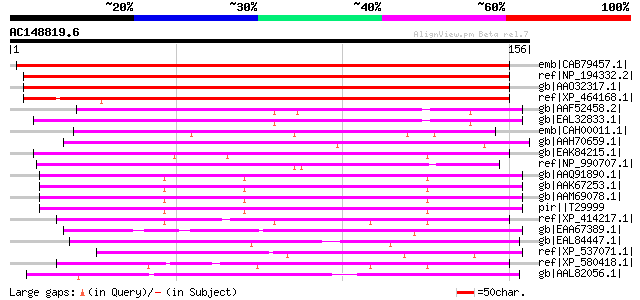
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148819.6 - phase: 0
(156 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAB79457.1| hypothetical protein [Arabidopsis thaliana] gi|4... 155 4e-37
ref|NP_194332.2| expressed protein [Arabidopsis thaliana] 151 5e-36
gb|AAO32317.1| 4/1 protein [Arabidopsis thaliana] 150 1e-35
ref|XP_464168.1| putative 4/1 protein [Oryza sativa (japonica cu... 122 2e-27
gb|AAF52458.2| CG18304-PA [Drosophila melanogaster] gi|24582370|... 54 1e-06
gb|EAL32833.1| GA14882-PA [Drosophila pseudoobscura] 50 2e-05
emb|CAH00011.1| unnamed protein product [Kluyveromyces lactis NR... 50 2e-05
gb|AAH70659.1| LOC431812 protein [Xenopus laevis] 49 4e-05
gb|EAK84215.1| hypothetical protein UM03347.1 [Ustilago maydis 5... 48 9e-05
ref|NP_990707.1| kinectin [Gallus gallus] gi|608483|gb|AAA85818.... 47 1e-04
gb|AAQ91890.1| Lin-5 (five) interacting protein protein 1, isofo... 47 1e-04
gb|AAK67253.1| Lin-5 (five) interacting protein protein 1, isofo... 47 1e-04
gb|AAM69078.1| Lin-5 (five) interacting protein protein 1, isofo... 47 1e-04
pir||T29999 hypothetical protein ZC8.4 - Caenorhabditis elegans 47 1e-04
ref|XP_414217.1| PREDICTED: similar to Growth-arrest-specific pr... 47 2e-04
gb|EAA67389.1| hypothetical protein FG01414.1 [Gibberella zeae P... 47 2e-04
gb|EAL84447.1| vesicle mediated transport protein (Imh1), putati... 46 3e-04
ref|XP_537071.1| PREDICTED: similar to 4932411G06Rik protein [Ca... 46 3e-04
ref|XP_580418.1| PREDICTED: similar to Growth-arrest-specific pr... 46 3e-04
gb|AAL82056.1| hypothetical protein [Pyrococcus furiosus DSM 363... 46 3e-04
>emb|CAB79457.1| hypothetical protein [Arabidopsis thaliana]
gi|4538931|emb|CAB39667.1| hypothetical protein
[Arabidopsis thaliana] gi|7485828|pir||T04257
hypothetical protein F20B18.130 - Arabidopsis thaliana
Length = 273
Score = 155 bits (391), Expect = 4e-37
Identities = 75/148 (50%), Positives = 111/148 (74%)
Query: 3 FSEQGHRRALKLLEEEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILA 62
F++Q HR AL+ L +++ K+ LE +++ L EKA+ + I +L D+ A+K+H+Q ++
Sbjct: 124 FADQEHRNALESLRQKHVTKVEELEYKIRSLLVEKATNDMVIDRLRQDLTANKSHIQAMS 183
Query: 63 NRLDQVHFEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQEM 122
+LD+V EVE KY EI+DLKDCL+ EQ EKND++ K+Q L+KELL+ + + ++Q++
Sbjct: 184 KKLDRVVTEVECKYELEIQDLKDCLLMEQAEKNDISNKLQSLQKELLISRTSIAEKQRDT 243
Query: 123 TSNWQVETLKQKIMKLRKENEVLKRKFS 150
TSN QVETLKQK+MKLRKENE+LKRK S
Sbjct: 244 TSNRQVETLKQKLMKLRKENEILKRKLS 271
>ref|NP_194332.2| expressed protein [Arabidopsis thaliana]
Length = 247
Score = 151 bits (382), Expect = 5e-36
Identities = 74/146 (50%), Positives = 108/146 (73%)
Query: 5 EQGHRRALKLLEEEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANR 64
E HR AL+ L +++ K+ LE +++ L EKA+ + I +L D+ A+K+H+Q ++ +
Sbjct: 100 EHEHRNALESLRQKHVTKVEELEYKIRSLLVEKATNDMVIDRLRQDLTANKSHIQAMSKK 159
Query: 65 LDQVHFEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQEMTS 124
LD+V EVE KY EI+DLKDCL+ EQ EKND++ K+Q L+KELL+ + + ++Q++ TS
Sbjct: 160 LDRVVTEVECKYELEIQDLKDCLLMEQAEKNDISNKLQSLQKELLISRTSIAEKQRDTTS 219
Query: 125 NWQVETLKQKIMKLRKENEVLKRKFS 150
N QVETLKQK+MKLRKENE+LKRK S
Sbjct: 220 NRQVETLKQKLMKLRKENEILKRKLS 245
>gb|AAO32317.1| 4/1 protein [Arabidopsis thaliana]
Length = 242
Score = 150 bits (379), Expect = 1e-35
Identities = 73/146 (50%), Positives = 108/146 (73%)
Query: 5 EQGHRRALKLLEEEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANR 64
E HR AL+ L +++ K+ LE +++ L EKA+ + I +L D+ A+K+H+Q ++ +
Sbjct: 95 EHEHRNALESLRQKHVTKVEELEYKIRSLLVEKATNDMVIDRLRQDLTANKSHIQAMSKK 154
Query: 65 LDQVHFEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQEMTS 124
LD+V EVE KY EI+DLKDCL+ EQ E+ND++ K+Q L+KELL+ + + ++Q++ TS
Sbjct: 155 LDRVVTEVECKYELEIQDLKDCLLMEQAERNDISNKLQSLQKELLISRTSIAEKQRDTTS 214
Query: 125 NWQVETLKQKIMKLRKENEVLKRKFS 150
N QVETLKQK+MKLRKENE+LKRK S
Sbjct: 215 NRQVETLKQKLMKLRKENEILKRKLS 240
>ref|XP_464168.1| putative 4/1 protein [Oryza sativa (japonica cultivar-group)]
gi|45736035|dbj|BAD13062.1| putative 4/1 protein [Oryza
sativa (japonica cultivar-group)]
Length = 256
Score = 122 bits (307), Expect = 2e-27
Identities = 68/157 (43%), Positives = 103/157 (65%), Gaps = 12/157 (7%)
Query: 5 EQGHRRALKLLEEEYNEKIARL-----------EAQVKESLHEKASYEATISQLHGDIAA 53
E+ + R L + EEEY +I +L E ++ ++ ++A+ EA I QL+ ++ A
Sbjct: 94 EKANTRLLSM-EEEYKREIEQLKLGSEMNSNDLENKLSCAVVQQATNEAVIKQLNLELEA 152
Query: 54 HKNHMQILANRLDQVHFEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKA 113
HK H+ +L +RL+QV +V +Y +EI+DLKD ++ EQEEKND++RK+Q E EL + K
Sbjct: 153 HKAHIDMLNSRLEQVTADVHQQYKNEIQDLKDVVIVEQEEKNDMHRKLQNTENELRIMKM 212
Query: 114 KMVDQQQEMTSNWQVETLKQKIMKLRKENEVLKRKFS 150
K +QQ++ S VETLKQK+MK RKENE LKR+ +
Sbjct: 213 KQAEQQRDSISVQHVETLKQKVMKFRKENESLKRRLA 249
>gb|AAF52458.2| CG18304-PA [Drosophila melanogaster] gi|24582370|ref|NP_609083.1|
CG18304-PA [Drosophila melanogaster]
Length = 1833
Score = 54.3 bits (129), Expect = 1e-06
Identities = 40/148 (27%), Positives = 77/148 (52%), Gaps = 16/148 (10%)
Query: 21 EKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANRLDQVHFEVESKYSS-- 78
EK++ LE +++ EK EA ISQL ++ + K + + L++ ++++K S
Sbjct: 1028 EKLSSLEKDMEKQAKEKEKLEAKISQLDAELLSAKKSAEKSKSSLEKEIKDLKTKASKSD 1087
Query: 79 --EIRDLKD-------CLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQEMTSNWQVE 129
+++DLK L AEQ+ DLN + L +E +L +A++ ++Q + + ++
Sbjct: 1088 SKQVQDLKKQVEEVQASLSAEQKRYEDLNNHWEKLSEETILMRAQLTTEKQSLQA--ELN 1145
Query: 130 TLKQKIMK---LRKENEVLKRKFSHIEE 154
KQKI + +R E + RK S ++
Sbjct: 1146 ASKQKIAEMDTIRIERTDMARKLSEAQK 1173
>gb|EAL32833.1| GA14882-PA [Drosophila pseudoobscura]
Length = 1850
Score = 50.1 bits (118), Expect = 2e-05
Identities = 36/161 (22%), Positives = 78/161 (48%), Gaps = 16/161 (9%)
Query: 8 HRRALKLLEEEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANRLDQ 67
++ L+ + EK++ LE +++ EK E+ I QL D+ K + L++
Sbjct: 1017 NKSTLETQSKREKEKLSSLERDLEKQGKEKEKLESKILQLDTDLLTAKKAADKTKSSLEK 1076
Query: 68 VHFEVESKYSS-----------EIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMV 116
++++K S ++ D++ L AEQ+ DLN + L +E +L +A++
Sbjct: 1077 EIKDLKTKASKSDSKQVQELKKQVEDIQTSLSAEQKRNEDLNTHWEKLSEETILMRAQLT 1136
Query: 117 DQQQEMTSNWQVETLKQKIMK---LRKENEVLKRKFSHIEE 154
++Q + + ++ KQK+ + +R E + RK + ++
Sbjct: 1137 TEKQNLQA--ELNASKQKMSEMDTIRIERTDMARKLNEAQK 1175
>emb|CAH00011.1| unnamed protein product [Kluyveromyces lactis NRRL Y-1140]
gi|50309821|ref|XP_454924.1| unnamed protein product
[Kluyveromyces lactis]
Length = 858
Score = 49.7 bits (117), Expect = 2e-05
Identities = 37/154 (24%), Positives = 79/154 (51%), Gaps = 27/154 (17%)
Query: 20 NEKIARLEAQVKESLHEKASYEATISQLHGDIAA-HKNHMQILANRLDQVHFEVESKYSS 78
NE I LEAQ++ + H++ S E +S L ++++ + H +I+ ++ Q+ E+ SS
Sbjct: 315 NEAIRNLEAQLQRNEHQRQSLEREVSLLQEELSSIRETHQKIITHKDQQIKQLTENLSSS 374
Query: 79 EIRDLK--DCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQ----QQEMTSNW------ 126
+ +K + L +E+ + N+ ++ EK+L + DQ Q+E+ ++
Sbjct: 375 DSEAVKRLNELSSERLRLLETNKSLKFSEKDLKQTVKSLEDQLNSCQEELHASQDKHKEK 434
Query: 127 --------------QVETLKQKIMKLRKENEVLK 146
+VE ++Q+++++RK N++LK
Sbjct: 435 VSELLRKQLPYTSSEVEAMRQELLEMRKSNDILK 468
Score = 31.2 bits (69), Expect = 9.0
Identities = 34/148 (22%), Positives = 68/148 (44%), Gaps = 35/148 (23%)
Query: 9 RRALKLLEEEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANRLDQV 68
++++++ + N +I L + VK+ HE + +S+L + HK +Q L +++
Sbjct: 488 KKSVEVDIQRKNNEIRALRSTVKDLEHELQDVTSNVSKLKEN---HKEEIQKLRKQVNP- 543
Query: 69 HFEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQEMTSNWQV 128
DL ++ EEK+ L ++ LL+ EL K D +++ + W
Sbjct: 544 -------------DLPTDVL---EEKSKLQNRISLLQMELNSIK----DTKEKELAMW-- 581
Query: 129 ETLKQKIMKLRKENEVLKRKFSHIEEGK 156
++K +R+ NE L ++EGK
Sbjct: 582 ---RRKYETIRRSNEEL------LQEGK 600
>gb|AAH70659.1| LOC431812 protein [Xenopus laevis]
Length = 648
Score = 48.9 bits (115), Expect = 4e-05
Identities = 36/148 (24%), Positives = 73/148 (49%), Gaps = 8/148 (5%)
Query: 17 EEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANRLDQVHFEVESKY 76
E + E++ +++ + +E L K E + + ++ A K ++ + DQ +++ +Y
Sbjct: 71 EAFVEELMQMKQRFQEILLVKEEQEDLLRKRERELTALKGALKEEVSTHDQEIDKLKEQY 130
Query: 77 SSEIRDLKDCLMAEQEEKNDL-NRKMQLLEKELLLFKAKMVDQQQEMTSNWQVETLKQKI 135
E++ ++D L E + +D+ N+K + + + + V Q+ W++E L+ KI
Sbjct: 131 DKELKKIRDNLNEEVKNSSDIANQKNEASQLKAVFENKVKVLLQENEELKWKIENLEDKI 190
Query: 136 MKLRKE-------NEVLKRKFSHIEEGK 156
LR+E NE LK K S E+ K
Sbjct: 191 AGLRQEINDLSEENERLKDKLSRSEKEK 218
Score = 31.6 bits (70), Expect = 6.9
Identities = 35/154 (22%), Positives = 67/154 (42%), Gaps = 20/154 (12%)
Query: 6 QGHRRALK-------LLEEEYNEKIARLEAQVK---ESLHEKASYEATISQLHGDIAAHK 55
Q H+RA LL++ + L+A+ + + + S E ISQL ++ K
Sbjct: 367 QDHQRAQDEALTKNHLLQQTIKDLQYELDAKTHVKDDRIRQIKSMEDKISQLEMELDEEK 426
Query: 56 NHMQILANRLDQVHFEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKM 115
N+ +L R+ + ++E S L E+ K DL L+++ K+++
Sbjct: 427 NNSDLLLERVTRCREQIEQTRSE--------LFQERATKQDLECDKISLDRQNKDLKSRI 478
Query: 116 VDQQQEMTSNWQ--VETLKQKIMKLRKENEVLKR 147
V + SN + V ++ +I +L + E +R
Sbjct: 479 VHLENSHRSNKEGLVAQMETRIAELEERLEYEER 512
>gb|EAK84215.1| hypothetical protein UM03347.1 [Ustilago maydis 521]
gi|49073496|ref|XP_400962.1| hypothetical protein
UM03347.1 [Ustilago maydis 521]
Length = 2363
Score = 47.8 bits (112), Expect = 9e-05
Identities = 38/152 (25%), Positives = 75/152 (49%), Gaps = 9/152 (5%)
Query: 8 HRRALKLLEEEYNEKIARLEAQVKESLHEKASYEATISQLH-GDIAAHKNHMQILANR-- 64
H AL ++ + E +A+ + E AS A + H + H+N ++ L
Sbjct: 1016 HASALAESQDRHREAVAKHQNSKSELAAAHASELAALQDGHRAKLDEHRNSIEELVTSHA 1075
Query: 65 -----LDQVHFEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQ 119
L+Q H E + ++++ K + A +E N+++++ KEL KA++ D Q
Sbjct: 1076 AALSVLEQSHREALATRDNDLQAAKQEVDARTKELAARNQELEMRTKELGAAKAEIDDLQ 1135
Query: 120 QEMTS-NWQVETLKQKIMKLRKENEVLKRKFS 150
+ +T+ ++E LKQ++ +L+K + KR+ S
Sbjct: 1136 KNITALRAEMEQLKQQLAELQKSHTAAKRELS 1167
>ref|NP_990707.1| kinectin [Gallus gallus] gi|608483|gb|AAA85818.1| kinectin
gi|34098657|sp|Q90631|KTN1_CHICK Kinectin
Length = 1364
Score = 47.4 bits (111), Expect = 1e-04
Identities = 36/142 (25%), Positives = 69/142 (48%), Gaps = 5/142 (3%)
Query: 9 RRALKLLEEEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANRLDQV 68
R + LE+E+ +++ +ES K ++ Q+ +I+ K IL + +
Sbjct: 371 RERINALEKEHGTFQSKIHVSYQESQQMKIKFQQRCEQMEAEISHLKQENTILRDAVSTS 430
Query: 69 HFEVESKYSSEIRDLK-DC--LMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQEMTSN 125
++ESK ++E+ L+ DC L+ E EKN ++ +L +K A++ QQQE
Sbjct: 431 TNQMESKQAAELNKLRQDCARLVNELGEKNSKLQQEELQKKNAEQAVAQLKVQQQEAERR 490
Query: 126 WQVETLKQKIMKLRKENEVLKR 147
W E ++ + K E+E ++
Sbjct: 491 W--EEIQVYLRKRTAEHEAAQQ 510
Score = 40.8 bits (94), Expect = 0.011
Identities = 34/144 (23%), Positives = 66/144 (45%), Gaps = 12/144 (8%)
Query: 9 RRALKLLEEEYNEKIARLEAQVKESLHEKASYEATISQLHGDIA---AHKNHMQILANRL 65
+R+++ E ++ K+ + ++K+ AS E + +L +I K + L + L
Sbjct: 1160 QRSVEEEESKWKIKVEESQKELKQMRSSVASLEHEVERLKEEIKEVETLKKEREHLESEL 1219
Query: 66 DQVHFEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKM--------VD 117
++ E S Y SE+R+LKD L Q++ +D + +EL L K K+ VD
Sbjct: 1220 EKAEIE-RSTYVSEVRELKDLLTELQKKLDDSYSEAVRQNEELNLLKMKLSETLSKLKVD 1278
Query: 118 QQQEMTSNWQVETLKQKIMKLRKE 141
Q + + ++ + L +E
Sbjct: 1279 QNERQKVAGDLPKAQESLASLERE 1302
Score = 33.5 bits (75), Expect = 1.8
Identities = 32/137 (23%), Positives = 64/137 (46%), Gaps = 19/137 (13%)
Query: 6 QGHRRALKLLEEEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANRL 65
+G +K +E EK E ++ + + T++QL + ++ L
Sbjct: 873 KGKENQVKTMEALLEEK----EKEIVQKGERLKGQQDTVAQLTSKV------QELEQQNL 922
Query: 66 DQVHFEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQEMTSN 125
Q+ + +S+++DL+ L E+E+ + L ++ E+E+ A V Q Q M S
Sbjct: 923 QQLQ---QVPAASQVQDLESRLRGEEEQISKLKAVLEEKEREI----ASQVKQLQTMQS- 974
Query: 126 WQVETLKQKIMKLRKEN 142
+ E+ K +I +L++EN
Sbjct: 975 -ENESFKVQIQELKQEN 990
>gb|AAQ91890.1| Lin-5 (five) interacting protein protein 1, isoform d [Caenorhabditis
elegans]
Length = 2350
Score = 47.4 bits (111), Expect = 1e-04
Identities = 40/176 (22%), Positives = 82/176 (45%), Gaps = 31/176 (17%)
Query: 10 RALKLLEEEYNEKIARLEAQVKESLHEKASYEATIS--------------QLHGDIAAHK 55
R+L+ EE+++ +LE +++ES + Y+ I+ +L G +
Sbjct: 1793 RSLRDKEEQWDSSRFQLETKMRESDSDTNKYQLQIASFESERQILTEKIKELDGALRLSD 1852
Query: 56 NHMQILANRLDQVH----------------FEVESKYSSEIRDLKDCLMAEQEEKNDLNR 99
+ +Q + + D++ +++SK S E + LKD L+ Q E N N
Sbjct: 1853 SKVQDMKDDTDKLRRDLTKAESVENELRKTIDIQSKTSHEYQLLKDQLLNTQNELNGANN 1912
Query: 100 KMQLLEKELLLFKAKMVDQQQEMTS-NWQVETLKQKIMKLRKENEVLKRKFSHIEE 154
+ Q LE ELL ++++ D +Q + N +V L++++ E ++ +F +E+
Sbjct: 1913 RKQQLENELLNVRSEVRDYKQRVHDVNNRVSELQRQLQDANTEKNRVEDRFLSVEK 1968
Score = 41.6 bits (96), Expect = 0.007
Identities = 40/147 (27%), Positives = 62/147 (41%), Gaps = 5/147 (3%)
Query: 9 RRALKLLEEEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANRLDQV 68
RR L E NE ++ Q K S HE + + ++ N Q L N L V
Sbjct: 1866 RRDLTKAESVENELRKTIDIQSKTS-HEYQLLKDQLLNTQNELNGANNRKQQLENELLNV 1924
Query: 69 HFEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQ--EMTSNW 126
EV Y + D+ + + Q + D N + +E L + K+V+ + E
Sbjct: 1925 RSEVRD-YKQRVHDVNNRVSELQRQLQDANTEKNRVEDRFLSVE-KVVNTMRTTETDLRQ 1982
Query: 127 QVETLKQKIMKLRKENEVLKRKFSHIE 153
Q+ET K + KE E LKR+ + +E
Sbjct: 1983 QLETAKNEKRVATKELEDLKRRLAQLE 2009
>gb|AAK67253.1| Lin-5 (five) interacting protein protein 1, isoform b [Caenorhabditis
elegans] gi|17570563|ref|NP_508847.1| major antigen like
(XF611) [Caenorhabditis elegans]
Length = 2315
Score = 47.4 bits (111), Expect = 1e-04
Identities = 40/176 (22%), Positives = 82/176 (45%), Gaps = 31/176 (17%)
Query: 10 RALKLLEEEYNEKIARLEAQVKESLHEKASYEATIS--------------QLHGDIAAHK 55
R+L+ EE+++ +LE +++ES + Y+ I+ +L G +
Sbjct: 1758 RSLRDKEEQWDSSRFQLETKMRESDSDTNKYQLQIASFESERQILTEKIKELDGALRLSD 1817
Query: 56 NHMQILANRLDQVH----------------FEVESKYSSEIRDLKDCLMAEQEEKNDLNR 99
+ +Q + + D++ +++SK S E + LKD L+ Q E N N
Sbjct: 1818 SKVQDMKDDTDKLRRDLTKAESVENELRKTIDIQSKTSHEYQLLKDQLLNTQNELNGANN 1877
Query: 100 KMQLLEKELLLFKAKMVDQQQEMTS-NWQVETLKQKIMKLRKENEVLKRKFSHIEE 154
+ Q LE ELL ++++ D +Q + N +V L++++ E ++ +F +E+
Sbjct: 1878 RKQQLENELLNVRSEVRDYKQRVHDVNNRVSELQRQLQDANTEKNRVEDRFLSVEK 1933
Score = 41.6 bits (96), Expect = 0.007
Identities = 40/147 (27%), Positives = 62/147 (41%), Gaps = 5/147 (3%)
Query: 9 RRALKLLEEEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANRLDQV 68
RR L E NE ++ Q K S HE + + ++ N Q L N L V
Sbjct: 1831 RRDLTKAESVENELRKTIDIQSKTS-HEYQLLKDQLLNTQNELNGANNRKQQLENELLNV 1889
Query: 69 HFEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQ--EMTSNW 126
EV Y + D+ + + Q + D N + +E L + K+V+ + E
Sbjct: 1890 RSEVRD-YKQRVHDVNNRVSELQRQLQDANTEKNRVEDRFLSVE-KVVNTMRTTETDLRQ 1947
Query: 127 QVETLKQKIMKLRKENEVLKRKFSHIE 153
Q+ET K + KE E LKR+ + +E
Sbjct: 1948 QLETAKNEKRVATKELEDLKRRLAQLE 1974
>gb|AAM69078.1| Lin-5 (five) interacting protein protein 1, isoform a [Caenorhabditis
elegans] gi|25151529|ref|NP_508848.2| major antigen like
(273.8 kD) (XF611) [Caenorhabditis elegans]
Length = 2396
Score = 47.4 bits (111), Expect = 1e-04
Identities = 40/176 (22%), Positives = 82/176 (45%), Gaps = 31/176 (17%)
Query: 10 RALKLLEEEYNEKIARLEAQVKESLHEKASYEATIS--------------QLHGDIAAHK 55
R+L+ EE+++ +LE +++ES + Y+ I+ +L G +
Sbjct: 1866 RSLRDKEEQWDSSRFQLETKMRESDSDTNKYQLQIASFESERQILTEKIKELDGALRLSD 1925
Query: 56 NHMQILANRLDQVH----------------FEVESKYSSEIRDLKDCLMAEQEEKNDLNR 99
+ +Q + + D++ +++SK S E + LKD L+ Q E N N
Sbjct: 1926 SKVQDMKDDTDKLRRDLTKAESVENELRKTIDIQSKTSHEYQLLKDQLLNTQNELNGANN 1985
Query: 100 KMQLLEKELLLFKAKMVDQQQEMTS-NWQVETLKQKIMKLRKENEVLKRKFSHIEE 154
+ Q LE ELL ++++ D +Q + N +V L++++ E ++ +F +E+
Sbjct: 1986 RKQQLENELLNVRSEVRDYKQRVHDVNNRVSELQRQLQDANTEKNRVEDRFLSVEK 2041
Score = 41.6 bits (96), Expect = 0.007
Identities = 40/147 (27%), Positives = 62/147 (41%), Gaps = 5/147 (3%)
Query: 9 RRALKLLEEEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANRLDQV 68
RR L E NE ++ Q K S HE + + ++ N Q L N L V
Sbjct: 1939 RRDLTKAESVENELRKTIDIQSKTS-HEYQLLKDQLLNTQNELNGANNRKQQLENELLNV 1997
Query: 69 HFEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQ--EMTSNW 126
EV Y + D+ + + Q + D N + +E L + K+V+ + E
Sbjct: 1998 RSEVRD-YKQRVHDVNNRVSELQRQLQDANTEKNRVEDRFLSVE-KVVNTMRTTETDLRQ 2055
Query: 127 QVETLKQKIMKLRKENEVLKRKFSHIE 153
Q+ET K + KE E LKR+ + +E
Sbjct: 2056 QLETAKNEKRVATKELEDLKRRLAQLE 2082
>pir||T29999 hypothetical protein ZC8.4 - Caenorhabditis elegans
Length = 2288
Score = 47.4 bits (111), Expect = 1e-04
Identities = 40/176 (22%), Positives = 82/176 (45%), Gaps = 31/176 (17%)
Query: 10 RALKLLEEEYNEKIARLEAQVKESLHEKASYEATIS--------------QLHGDIAAHK 55
R+L+ EE+++ +LE +++ES + Y+ I+ +L G +
Sbjct: 1758 RSLRDKEEQWDSSRFQLETKMRESDSDTNKYQLQIASFESERQILTEKIKELDGALRLSD 1817
Query: 56 NHMQILANRLDQVH----------------FEVESKYSSEIRDLKDCLMAEQEEKNDLNR 99
+ +Q + + D++ +++SK S E + LKD L+ Q E N N
Sbjct: 1818 SKVQDMKDDTDKLRRDLTKAESVENELRKTIDIQSKTSHEYQLLKDQLLNTQNELNGANN 1877
Query: 100 KMQLLEKELLLFKAKMVDQQQEMTS-NWQVETLKQKIMKLRKENEVLKRKFSHIEE 154
+ Q LE ELL ++++ D +Q + N +V L++++ E ++ +F +E+
Sbjct: 1878 RKQQLENELLNVRSEVRDYKQRVHDVNNRVSELQRQLQDANTEKNRVEDRFLSVEK 1933
Score = 41.6 bits (96), Expect = 0.007
Identities = 40/147 (27%), Positives = 62/147 (41%), Gaps = 5/147 (3%)
Query: 9 RRALKLLEEEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANRLDQV 68
RR L E NE ++ Q K S HE + + ++ N Q L N L V
Sbjct: 1831 RRDLTKAESVENELRKTIDIQSKTS-HEYQLLKDQLLNTQNELNGANNRKQQLENELLNV 1889
Query: 69 HFEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQ--EMTSNW 126
EV Y + D+ + + Q + D N + +E L + K+V+ + E
Sbjct: 1890 RSEVRD-YKQRVHDVNNRVSELQRQLQDANTEKNRVEDRFLSVE-KVVNTMRTTETDLRQ 1947
Query: 127 QVETLKQKIMKLRKENEVLKRKFSHIE 153
Q+ET K + KE E LKR+ + +E
Sbjct: 1948 QLETAKNEKRVATKELEDLKRRLAQLE 1974
>ref|XP_414217.1| PREDICTED: similar to Growth-arrest-specific protein 8 (Growth
arrest-specific 11) [Gallus gallus]
Length = 480
Score = 47.0 bits (110), Expect = 2e-04
Identities = 38/142 (26%), Positives = 73/142 (50%), Gaps = 8/142 (5%)
Query: 15 LEEEYNEKIARLEAQVKESLHEKASYEATIS-QLHGDIAAHKNHMQILANRLDQVHFEVE 73
+EE N +I+ L +++ H+ +Y ++ Q I+ K M+ + R + H E E
Sbjct: 213 VEERKNNQISELMKNHEKAFHDFKNYHDDVTFQNLALISLLKEQMEEMKKR--ETHLEKE 270
Query: 74 SKYSS-EIRDLKDCLMAEQEEKNDLNRKMQLLEKE---LLLFKAKMVDQQQEMTS-NWQV 128
+ + + LKD L QE+ +L +K+ +K+ L+ KA + Q+E+ W+
Sbjct: 271 KADALLQNKQLKDSLQQTQEQVFELQKKLAHYDKDKEALMNTKAHLKVTQKELKDLRWEH 330
Query: 129 ETLKQKIMKLRKENEVLKRKFS 150
E L+Q+ K++ E + L +KF+
Sbjct: 331 EVLEQRFSKVQAERDELYQKFT 352
>gb|EAA67389.1| hypothetical protein FG01414.1 [Gibberella zeae PH-1]
gi|46109064|ref|XP_381590.1| hypothetical protein
FG01414.1 [Gibberella zeae PH-1]
Length = 774
Score = 46.6 bits (109), Expect = 2e-04
Identities = 39/143 (27%), Positives = 68/143 (47%), Gaps = 12/143 (8%)
Query: 17 EEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANRLDQVHFEVESKY 76
E+ EKI LEAQ +L E A I+ L ++ K + L E K
Sbjct: 56 EDLREKIKELEAQSSLALDET---HARIAILQDEL---KKGGDSTSEELRSTKEAAEQK- 108
Query: 77 SSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQ-----EMTSNWQVETL 131
+ E+ D K L A +E+ L ++ Q + EL KA++V+ ++ E +++TL
Sbjct: 109 AKELEDAKSSLTATEEKLKGLEQERQSIADELATLKAELVEAKEAREALEAALTKEIDTL 168
Query: 132 KQKIMKLRKENEVLKRKFSHIEE 154
K +I + ++++ L + S +EE
Sbjct: 169 KTQISEAEQKHQALTKAHSTLEE 191
>gb|EAL84447.1| vesicle mediated transport protein (Imh1), putative [Aspergillus
fumigatus Af293]
Length = 1001
Score = 46.2 bits (108), Expect = 3e-04
Identities = 36/160 (22%), Positives = 71/160 (43%), Gaps = 30/160 (18%)
Query: 19 YNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANRLDQVHFE------V 72
++ ++ RLE+++KE E + ++ + L D+A + + + L+ E +
Sbjct: 117 FDNELPRLESELKEKQEEVETLKSQVESLKRDLAVARESTEGMVQNLESATRELVELRDI 176
Query: 73 ESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKA------------------- 113
K SEI+ LKD AE ++ K +L E E + KA
Sbjct: 177 RDKQDSEIKKLKDTRQAEVDDV-----KAKLAESENTIAKAGEEAEKLKAELKQKTEEIE 231
Query: 114 KMVDQQQEMTSNWQVETLKQKIMKLRKENEVLKRKFSHIE 153
K+ DQ + N Q + L++K+ ++ KE + ++K ++
Sbjct: 232 KLQDQVTQQKDNDQQKELQEKLEEVAKEKDASEKKLGVLQ 271
Score = 33.9 bits (76), Expect = 1.4
Identities = 27/141 (19%), Positives = 64/141 (45%), Gaps = 7/141 (4%)
Query: 10 RALKLLEEEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANRLDQVH 69
+ L + ++ E +++LE ++ + AS A ++H D+ ++++ L+
Sbjct: 477 KELNVEKKALEETVSKLEKELADIRTSHASKSADSEKMHSDLKEDYENLKVKLTNLETEL 536
Query: 70 FEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQEMTSNWQVE 129
+ ++ +DL + Q+ + +L + +++ EL K + ++ E+
Sbjct: 537 SAAQQLAATRFKDLTELRETLQKLQPEL-KSLRVESSELKSTKEALASKESEL------R 589
Query: 130 TLKQKIMKLRKENEVLKRKFS 150
TL+ K +LR E + LK S
Sbjct: 590 TLEGKHEELRAEVKTLKSTIS 610
>ref|XP_537071.1| PREDICTED: similar to 4932411G06Rik protein [Canis familiaris]
Length = 1382
Score = 46.2 bits (108), Expect = 3e-04
Identities = 36/147 (24%), Positives = 68/147 (45%), Gaps = 20/147 (13%)
Query: 27 EAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANRLDQVHFEVESKYSSEIRD---- 82
E ++ E+L ++ + E+ + HG++ + +Q L + L + +E KYS+ I+D
Sbjct: 865 ETELTEALQKREALESELQNAHGELKSTLRQLQELRDVLQKAQLSLEEKYST-IKDLTAE 923
Query: 83 LKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVD-QQQEMTSNWQVETLKQKIMKL--- 138
L++C M +++K +L Q L++ K + +MT ++QKI+KL
Sbjct: 924 LRECKMENEDKKQELLEMDQALKERNWELKQRAAQVTHLDMTIREHRGEMEQKIIKLEGT 983
Query: 139 -----------RKENEVLKRKFSHIEE 154
K+ E L K H +E
Sbjct: 984 LEKSELELKECNKQIESLNEKLQHSKE 1010
Score = 39.3 bits (90), Expect = 0.033
Identities = 28/138 (20%), Positives = 66/138 (47%), Gaps = 6/138 (4%)
Query: 17 EEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANRLDQVHFEVESKY 76
+E ++I RLE+Q+++ + E T+S L DI ++H++ LD++ E + +
Sbjct: 584 KEMEKQIERLESQLEKKDQQFKEQEKTMSILQQDIICKQHHLE----SLDRLLTESKGEM 639
Query: 77 SSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQEMTSNWQVETLKQKIM 136
E + L Q + L+ +++ ++EL L ++ +++ Q++ +++
Sbjct: 640 EKENMKKDEALRTLQNQVRQLDSALEICKEELALHLNQLEGNKEKFEK--QLKKKSEEVY 697
Query: 137 KLRKENEVLKRKFSHIEE 154
L+KE ++ E
Sbjct: 698 CLQKELKIKNHSLQETAE 715
Score = 36.6 bits (83), Expect = 0.21
Identities = 42/179 (23%), Positives = 73/179 (40%), Gaps = 36/179 (20%)
Query: 1 MTFSEQGHRRALKLLEEEY---------NEKIARLEAQVKESLHEKASYEATISQLHGDI 51
M S Q ++ +LEE+ +K+ R E Q E+L+ +
Sbjct: 141 MLESVQQQAASVPILEEQIINLEAEVSAQDKVLRAERQRNEALYNAEELSKAFQHYKEKV 200
Query: 52 AAHKNHMQILANRLDQ--VHFEVESK--------YSSEIRDLKDCLMAEQEEKNDLNRKM 101
A +Q+ L++ + E E+K Y S++ LK+ L +EE N+ K+
Sbjct: 201 AEKLEKVQVEEEILEKNLISCEKENKRLQEKCGLYKSDLEILKEKLRQLKEENNNGKEKL 260
Query: 102 QLLEKELLLFKAKMVDQQQEMTSNWQVETLKQKIMKLRKE----NEVLKRKFSHIEEGK 156
+++ V + M Q+ +Q I+KL E EVL+ KFS + E +
Sbjct: 261 RIM----------AVKNSEVMA---QLTESRQSILKLENELEDKKEVLREKFSLMNENR 306
Score = 35.0 bits (79), Expect = 0.63
Identities = 42/189 (22%), Positives = 72/189 (37%), Gaps = 41/189 (21%)
Query: 3 FSEQGHRRALKLLEEEYNEKIARLEAQVKESLHEKASYEATISQL-------HGDIAAHK 55
FS R LKL NE++ + +++ S E S E +SQL + K
Sbjct: 299 FSLMNENRELKLRVAAQNERLDLCQQEIESSRVELRSLEKVMSQLPLKREMFGYKSSLSK 358
Query: 56 NHMQILANRLDQVHFEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAK- 114
+ M L+N+ D E SE+R +K + + +K N L + L+ + K
Sbjct: 359 HQMSSLSNKEDIGCCEANKMVISELR-IKLAIKEAEIQKLQANLAANQLSQSLIGYNDKQ 417
Query: 115 -----------------------MVDQQQEMTSNWQ---------VETLKQKIMKLRKEN 142
+ D+Q M+ ++ +E L+ K+M++ EN
Sbjct: 418 ESGKLNSLETEPVKLGGNQLAERIKDRQHTMSKQYEKERQRLASGIEELRTKLMQIEAEN 477
Query: 143 EVLKRKFSH 151
LK +H
Sbjct: 478 SDLKVNMAH 486
>ref|XP_580418.1| PREDICTED: similar to Growth-arrest-specific protein 8 (Growth
arrest-specific 11), partial [Bos taurus]
Length = 468
Score = 46.2 bits (108), Expect = 3e-04
Identities = 39/143 (27%), Positives = 79/143 (54%), Gaps = 10/143 (6%)
Query: 15 LEEEYNEKIARLEAQVKESLHEKASY--EATISQLHGDIAAHKNHMQILANRLDQVHFEV 72
+EE N +I L + +E+ + +Y + T++ L I + K M+ + R + H E
Sbjct: 201 VEERKNGQITTLMQRHEEAFTDIKNYYNDITLNNL-ALINSLKEQMEDM--RKKEEHLEK 257
Query: 73 E-SKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKE---LLLFKAKMVDQQQEMTS-NWQ 127
E ++ + + R L D L +EE +D+ +K+ E++ L+ KA++ ++E+ S W+
Sbjct: 258 EMTEVAMQNRRLADPLQKAREEMSDMQKKLGSYERDKQILVCTKARLKVTEKELKSLRWE 317
Query: 128 VETLKQKIMKLRKENEVLKRKFS 150
E L+Q+ +K+++E + L KF+
Sbjct: 318 HEVLEQRFIKVQQERDDLYHKFT 340
>gb|AAL82056.1| hypothetical protein [Pyrococcus furiosus DSM 3638]
gi|18978304|ref|NP_579661.1| hypothetical protein PF1932
[Pyrococcus furiosus DSM 3638]
Length = 269
Score = 46.2 bits (108), Expect = 3e-04
Identities = 39/156 (25%), Positives = 71/156 (45%), Gaps = 16/156 (10%)
Query: 6 QGHRRALKLLEEEY--------NEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNH 57
+G +A +L+ E Y +E I +E K L E + E +L + K
Sbjct: 84 KGESKAGQLVRETYELIKQGKLDELIKTIEMIEKGGLREVVAKEE-YEKLLQEYEKLKQE 142
Query: 58 MQILANRLDQVHFEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVD 117
+ + +++ E K EI +LK L +EEK +L EKEL K K+++
Sbjct: 143 YEAIKEKVEAAELESLEKAKEEIENLKKKLQKLEEEKKEL-------EKELKEQKVKLIE 195
Query: 118 QQQEMTSNWQVETLKQKIMKLRKENEVLKRKFSHIE 153
+ + ++E +++ +L KE+E LK+K ++
Sbjct: 196 YEAKAKRAEELENKVKELEQLAKESEELKKKLEEVQ 231
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.313 0.127 0.334
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 223,646,993
Number of Sequences: 2540612
Number of extensions: 7905391
Number of successful extensions: 65089
Number of sequences better than 10.0: 4348
Number of HSP's better than 10.0 without gapping: 294
Number of HSP's successfully gapped in prelim test: 4118
Number of HSP's that attempted gapping in prelim test: 58015
Number of HSP's gapped (non-prelim): 10368
length of query: 156
length of database: 863,360,394
effective HSP length: 117
effective length of query: 39
effective length of database: 566,108,790
effective search space: 22078242810
effective search space used: 22078242810
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 69 (31.2 bits)
Medicago: description of AC148819.6


