

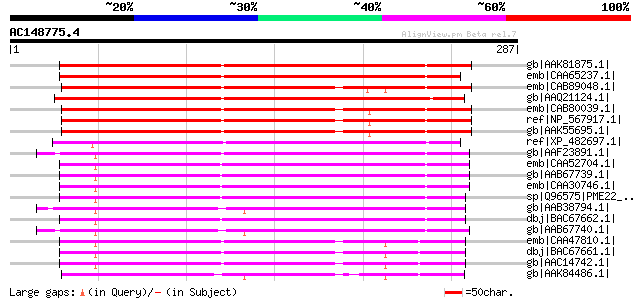
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148775.4 - phase: 0
(287 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAK81875.1| pectin methylesterase PME1 [Vitis vinifera] 213 5e-54
emb|CAA65237.1| pectinesterase [Prunus persica] gi|6093744|sp|Q4... 212 9e-54
emb|CAB89048.1| pectinesterase-like protein [Arabidopsis thalian... 209 6e-53
gb|AAQ21124.1| pectinesterase [Fragaria x ananassa] 203 4e-51
emb|CAB80039.1| pectinesterase-like protein [Arabidopsis thalian... 194 3e-48
ref|NP_567917.1| pectinesterase family protein [Arabidopsis thal... 194 3e-48
gb|AAK55695.1| AT4g33220/F4I10_150 [Arabidopsis thaliana] 194 3e-48
ref|XP_482697.1| putative pectinesterase [Oryza sativa (japonica... 181 2e-44
gb|AAF23891.1| pectin methyl esterase [Solanum tuberosum] 181 3e-44
emb|CAA52704.1| pectin esterase [Lycopersicon esculentum] gi|250... 174 3e-42
gb|AAB67739.1| pectin methylesterase PME2.1 [Lycopersicon escule... 174 3e-42
emb|CAA30746.1| unnamed protein product [Lycopersicon esculentum... 173 6e-42
sp|Q96575|PME22_LYCES Pectinesterase-2 precursor (Pectin methyle... 170 4e-41
gb|AAB38794.1| pectin methylesterase [Lycopersicon esculentum] 169 6e-41
dbj|BAC67662.1| pectin methylesterase [Pisum sativum] 168 1e-40
gb|AAB67740.1| PME1.9 [Lycopersicon esculentum] 167 2e-40
emb|CAA47810.1| pectinesterase [Pisum sativum] gi|7447372|pir||T... 167 3e-40
dbj|BAC67661.1| pectin methylesterase [Pisum sativum] 167 3e-40
gb|AAC14742.1| pectin methylesterase [Pisum sativum] gi|3426335|... 167 3e-40
gb|AAK84486.1| putative thermostable pectinesterase [Citrus sine... 167 4e-40
>gb|AAK81875.1| pectin methylesterase PME1 [Vitis vinifera]
Length = 531
Score = 213 bits (542), Expect = 5e-54
Identities = 119/234 (50%), Positives = 151/234 (63%), Gaps = 3/234 (1%)
Query: 29 GKFPSWVKPGDRKLLQASAVPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKKGV 88
G FPSWV RKLLQAS+V D VA+DG+GNY +MDAV AAP+ S+ YVI+IK+G+
Sbjct: 196 GDFPSWVGKHSRKLLQASSVSPDVTVAADGTGNYTTVMDAVQAAPDYSQNHYVIYIKQGI 255
Query: 89 YNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDISFR 148
Y E+V I K NLMM+GDGMGATVITG+ S+ D T + TF V+G GF A+D++F
Sbjct: 256 YRENVEIKKKKWNLMMVGDGMGATVITGNRSY-IDGWTTYASATFAVKGKGFIARDMTFE 314
Query: 149 NTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLVRQL 208
NTA PE HQAVAL SDSD SV+YRC + G+QD+L + R + +
Sbjct: 315 NTAGPEKHQAVALRSDSDLSVYYRCSMRGYQDTLYPHTNRQFYRECRISGTVDFIFGDAT 374
Query: 209 SSFKT-DILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEFLPFVN 261
F+ ILV+KG Q+NTITAQG + P P GF+ QF N+ AD + L VN
Sbjct: 375 VVFQNCQILVKKGLPNQKNTITAQGRKD-PAQPTGFSIQFSNISADSDLLASVN 427
>emb|CAA65237.1| pectinesterase [Prunus persica] gi|6093744|sp|Q43062|PME_PRUPE
Pectinesterase PPE8B precursor (Pectin methylesterase)
(PE)
Length = 522
Score = 212 bits (540), Expect = 9e-54
Identities = 117/228 (51%), Positives = 147/228 (64%), Gaps = 3/228 (1%)
Query: 29 GKFPSWVKPGDRKLLQASAVPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKKGV 88
G+ PSWVK DRKLLQA V DA+VA DG+GN+ + DAV+AAP+ S +RYVI+IK+G
Sbjct: 186 GQIPSWVKTKDRKLLQADGVSVDAIVAQDGTGNFTNVTDAVLAAPDYSMRRYVIYIKRGT 245
Query: 89 YNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDISFR 148
Y E+V I K NLMMIGDGM AT+I+G+ S+ D T + TF V G GF A+DI+F
Sbjct: 246 YKENVEIKKKKWNLMMIGDGMDATIISGNRSF-VDGWTTFRSATFAVSGRGFIARDITFE 304
Query: 149 NTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLVRQL 208
NTA PE HQAVAL SDSD SVFYRC I G+QD+L + R K +
Sbjct: 305 NTAGPEKHQAVALRSDSDLSVFYRCNIRGYQDTLYTHTMRQFYRDCKISGTVDFIFGDAT 364
Query: 209 SSFKT-DILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPE 255
F+ IL +KG Q+N+ITAQG + PN P G + QFCN+ AD +
Sbjct: 365 VVFQNCQILAKKGLPNQKNSITAQGRKD-PNEPTGISIQFCNITADSD 411
>emb|CAB89048.1| pectinesterase-like protein [Arabidopsis thaliana]
gi|20465719|gb|AAM20328.1| putative pectinesterase
[Arabidopsis thaliana] gi|17979141|gb|AAL49828.1|
putative pectinesterase [Arabidopsis thaliana]
gi|30690925|ref|NP_189913.3| pectinesterase family
protein [Arabidopsis thaliana] gi|11279193|pir||T49241
pectinesterase-like protein - Arabidopsis thaliana
Length = 527
Score = 209 bits (533), Expect = 6e-53
Identities = 121/238 (50%), Positives = 158/238 (65%), Gaps = 12/238 (5%)
Query: 30 KFPSWVKPGDRKLLQASAVP-ADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKKGV 88
KFPSWVKPGDRKLLQ + ADAVVA+DG+GN+ I DAV+AAP+ S KRYVIH+K+GV
Sbjct: 192 KFPSWVKPGDRKLLQTDNITVADAVVAADGTGNFTTISDAVLAAPDYSTKRYVIHVKRGV 251
Query: 89 YNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDISFR 148
Y E+V I K N+MM+GDG+ ATVITG+ S+ D T + TF V G GF A+DI+F+
Sbjct: 252 YVENVEIKKKKWNIMMVGDGIDATVITGNRSF-IDGWTTFRSATFAVSGRGFIARDITFQ 310
Query: 149 NTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLT---SYLV 205
NTA PE HQAVA+ SD+D VFYRC + G+QD+L A HS+R E +T ++
Sbjct: 311 NTAGPEKHQAVAIRSDTDLGVFYRCAMRGYQDTLYA----HSMRQFFRECIITGTVDFIF 366
Query: 206 RQLSSF--KTDILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEFLPFVN 261
++ I ++G Q+N+ITAQG + PN P GF QF N+ AD + L +N
Sbjct: 367 GDATAVFQSCQIKAKQGLPNQKNSITAQGRKD-PNEPTGFTIQFSNIAADTDLLLNLN 423
>gb|AAQ21124.1| pectinesterase [Fragaria x ananassa]
Length = 514
Score = 203 bits (517), Expect = 4e-51
Identities = 113/233 (48%), Positives = 145/233 (61%), Gaps = 3/233 (1%)
Query: 26 TQKGKFPSWVKPGDRKLLQASAVPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIK 85
+ G+ P+W K DRKLLQA+ VP D VVA DG+GN+ I A+++AP+ S KRYVI++K
Sbjct: 174 SSNGQTPAWFKAEDRKLLQANGVPVDVVVAQDGTGNFTNITAAILSAPDYSLKRYVIYVK 233
Query: 86 KGVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDI 145
KG+Y E+V I K N+MMIGDGM ATVI+G+ ++ D T + TF V G GF A+DI
Sbjct: 234 KGLYKEYVEIKKKKWNIMMIGDGMDATVISGNHNF-VDGWTTFRSATFAVSGRGFIARDI 292
Query: 146 SFRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLV 205
+F NTA PE H AVAL SDSD S FYRCE G+QD+L + R K +
Sbjct: 293 TFENTAGPEKHMAVALRSDSDLSAFYRCEFRGYQDTLYTHSMRQFYRDCKISGTVDFIFG 352
Query: 206 RQLSSFKT-DILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEFL 257
F+ IL RK Q+N+ITA G K P GF+FQFCN+ A P+ L
Sbjct: 353 DGTVMFQNCQILARKALPNQKNSITAHGRKYKDE-PTGFSFQFCNISAHPDLL 404
>emb|CAB80039.1| pectinesterase-like protein [Arabidopsis thaliana]
gi|4455336|emb|CAB36796.1| pectinesterase-like protein
[Arabidopsis thaliana] gi|7447375|pir||T05202
pectinesterase homolog F4I10.150 - Arabidopsis thaliana
Length = 477
Score = 194 bits (492), Expect = 3e-48
Identities = 109/237 (45%), Positives = 152/237 (63%), Gaps = 11/237 (4%)
Query: 30 KFPSWVKPGDRKLLQASAVPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKKGVY 89
+FP WV+P DRKLL+++ D VA DG+GN+ KIMDA+ AP+ S R+VI+IKKG+Y
Sbjct: 143 QFPDWVRPDDRKLLESNGRTYDVSVALDGTGNFTKIMDAIKKAPDYSSTRFVIYIKKGLY 202
Query: 90 NEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDISFRN 149
E+V I K N++M+GDG+ TVI+G+ S+ D T + TF V G GF A+DI+F+N
Sbjct: 203 LENVEIKKKKWNIVMLGDGIDVTVISGNRSF-IDGWTTFRSATFAVSGRGFLARDITFQN 261
Query: 150 TAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTS-----YL 204
TA PE HQAVAL SDSD SVF+RC + G+QD+L H++R E +T +
Sbjct: 262 TAGPEKHQAVALRSDSDLSVFFRCAMRGYQDTLYT----HTMRQFYRECTITGTVDFIFG 317
Query: 205 VRQLSSFKTDILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEFLPFVN 261
+ IL ++G Q+NTITAQG + N P GF+ QF N+ AD + +P++N
Sbjct: 318 DGTVVFQNCQILAKRGLPNQKNTITAQGRKD-VNQPSGFSIQFSNISADADLVPYLN 373
>ref|NP_567917.1| pectinesterase family protein [Arabidopsis thaliana]
Length = 404
Score = 194 bits (492), Expect = 3e-48
Identities = 109/237 (45%), Positives = 152/237 (63%), Gaps = 11/237 (4%)
Query: 30 KFPSWVKPGDRKLLQASAVPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKKGVY 89
+FP WV+P DRKLL+++ D VA DG+GN+ KIMDA+ AP+ S R+VI+IKKG+Y
Sbjct: 70 QFPDWVRPDDRKLLESNGRTYDVSVALDGTGNFTKIMDAIKKAPDYSSTRFVIYIKKGLY 129
Query: 90 NEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDISFRN 149
E+V I K N++M+GDG+ TVI+G+ S+ D T + TF V G GF A+DI+F+N
Sbjct: 130 LENVEIKKKKWNIVMLGDGIDVTVISGNRSF-IDGWTTFRSATFAVSGRGFLARDITFQN 188
Query: 150 TAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTS-----YL 204
TA PE HQAVAL SDSD SVF+RC + G+QD+L H++R E +T +
Sbjct: 189 TAGPEKHQAVALRSDSDLSVFFRCAMRGYQDTLYT----HTMRQFYRECTITGTVDFIFG 244
Query: 205 VRQLSSFKTDILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEFLPFVN 261
+ IL ++G Q+NTITAQG + N P GF+ QF N+ AD + +P++N
Sbjct: 245 DGTVVFQNCQILAKRGLPNQKNTITAQGRKD-VNQPSGFSIQFSNISADADLVPYLN 300
>gb|AAK55695.1| AT4g33220/F4I10_150 [Arabidopsis thaliana]
Length = 525
Score = 194 bits (492), Expect = 3e-48
Identities = 109/237 (45%), Positives = 152/237 (63%), Gaps = 11/237 (4%)
Query: 30 KFPSWVKPGDRKLLQASAVPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKKGVY 89
+FP WV+P DRKLL+++ D VA DG+GN+ KIMDA+ AP+ S R+VI+IKKG+Y
Sbjct: 191 QFPDWVRPDDRKLLESNGRTYDVSVALDGTGNFTKIMDAIKKAPDYSSTRFVIYIKKGLY 250
Query: 90 NEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDISFRN 149
E+V I K N++M+GDG+ TVI+G+ S+ D T + TF V G GF A+DI+F+N
Sbjct: 251 LENVEIKKKKWNIVMLGDGIDVTVISGNRSF-IDGWTTFRSATFAVSGRGFLARDITFQN 309
Query: 150 TAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTS-----YL 204
TA PE HQAVAL SDSD SVF+RC + G+QD+L H++R E +T +
Sbjct: 310 TAGPEKHQAVALRSDSDLSVFFRCAMRGYQDTLYT----HTMRQFYRECTITGTVDFIFG 365
Query: 205 VRQLSSFKTDILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEFLPFVN 261
+ IL ++G Q+NTITAQG + N P GF+ QF N+ AD + +P++N
Sbjct: 366 DGTVVFQNCQILAKRGLPNQKNTITAQGRKD-VNQPSGFSIQFSNISADADLVPYLN 421
>ref|XP_482697.1| putative pectinesterase [Oryza sativa (japonica cultivar-group)]
gi|42407616|dbj|BAD08731.1| putative pectinesterase
[Oryza sativa (japonica cultivar-group)]
Length = 557
Score = 181 bits (459), Expect = 2e-44
Identities = 106/236 (44%), Positives = 138/236 (57%), Gaps = 7/236 (2%)
Query: 25 ITQKGKFPSWVKPGDRKLLQA----SAVPADAVVASDGSGNYMKIMDAVMAAPNGSKKRY 80
+ + G P W+ +R+LLQ +P DAVVA DGSGNY + AV AAP S RY
Sbjct: 204 LAEGGGAPHWLGARERRLLQMPLGPGGMPVDAVVAKDGSGNYTTVSAAVDAAPTESASRY 263
Query: 81 VIHIKKGVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGF 140
VI++KKGVY E V I K NLM++GDGMG TVI+G ++ D T + T V G GF
Sbjct: 264 VIYVKKGVYKETVDIKKKKWNLMLVGDGMGVTVISGHRNYV-DGYTTFRSATVAVNGKGF 322
Query: 141 SAQDISFRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARL 200
A+D++F NTA P HQAVAL DSD SVFYRC G+QD+L A+ R + +
Sbjct: 323 MARDVTFENTAGPSKHQAVALRCDSDLSVFYRCGFEGYQDTLYAHSLRQFYRDCRVSGTV 382
Query: 201 TSYLVRQLSSFKTDILVRKGPT-GQQNTITAQGGPEKPNLPFGFAFQFCNVCADPE 255
+ F+ L + P Q+N++TAQG + N+ GFAFQFCNV AD +
Sbjct: 383 DFVFGNAAAVFQNCTLAARLPLPDQKNSVTAQGRLDG-NMTTGFAFQFCNVTADDD 437
>gb|AAF23891.1| pectin methyl esterase [Solanum tuberosum]
Length = 530
Score = 181 bits (458), Expect = 3e-44
Identities = 104/248 (41%), Positives = 145/248 (57%), Gaps = 7/248 (2%)
Query: 16 TSSSDILDTITQKGKFPSWVKPGDRKLLQASA--VPADAVVASDGSGNYMKIMDAVMAAP 73
T + D+ T+ GK PSWV DRKL+++S + A+AVVA DG+G+Y + +AV AAP
Sbjct: 182 TQNEDVFRTVL--GKMPSWVSSRDRKLMESSGKDIKANAVVAQDGTGDYQTLAEAVAAAP 239
Query: 74 NGSKKRYVIHIKKGVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTF 133
+ SK RYVI++K G+Y E+V + + K NLM++GDGM AT+ITG L++ D T + T
Sbjct: 240 DKSKTRYVIYVKMGIYKENVEVTSRKMNLMIVGDGMNATIITGSLNY-VDGTTTFRSATL 298
Query: 134 GVEGLGFSAQDISFRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRI 193
G GF QDI +NTA PE HQAVAL D SV RC I +QD+L A+ + R
Sbjct: 299 AAVGQGFILQDICIQNTAGPEKHQAVALRVGGDMSVINRCPIDAYQDTLYAHSQRQFYRD 358
Query: 194 AKSEARLTSYLVRQLSSF-KTDILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCA 252
+ + F K ++ RK Q+N +TAQG + PN G + QFC++ A
Sbjct: 359 SYVSGTIDFIFGNAAVVFQKCQLVARKPSKNQKNMVTAQGRTD-PNQATGTSIQFCDIIA 417
Query: 253 DPEFLPFV 260
P+ P V
Sbjct: 418 SPDLEPVV 425
>emb|CAA52704.1| pectin esterase [Lycopersicon esculentum]
gi|2507165|sp|P09607|PME21_LYCES Pectinesterase-2
precursor (Pectin methylesterase 2) (PE 2)
Length = 550
Score = 174 bits (441), Expect = 3e-42
Identities = 102/235 (43%), Positives = 139/235 (58%), Gaps = 5/235 (2%)
Query: 29 GKFPSWVKPGDRKLLQASA--VPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKK 86
GK PSWV DRKL+++S + A+AVVA DG+G Y + +AV AAP+ SK RYVI++K+
Sbjct: 213 GKMPSWVSSRDRKLMESSGKDIGANAVVAKDGTGKYRTLAEAVAAAPDKSKTRYVIYVKR 272
Query: 87 GVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDIS 146
G Y E+V +++ K NLM+IGDGM AT+ITG L+ D T ++ T G GF QDI
Sbjct: 273 GTYKENVEVSSRKMNLMIIGDGMYATIITGSLN-VVDGSTTFHSATLAAVGKGFILQDIC 331
Query: 147 FRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLVR 206
+NTA P HQAVAL +D SV RC I +QD+L A+ + R + +
Sbjct: 332 IQNTAGPAKHQAVALRVGADKSVINRCRIDAYQDTLYAHSQRQFYRDSYVTGTIDFIFGN 391
Query: 207 QLSSF-KTDILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEFLPFV 260
F K ++ RK QQN +TAQG + PN G + QFC++ A P+ P V
Sbjct: 392 AAVVFQKCQLVARKPGKYQQNMVTAQGRTD-PNQATGTSIQFCDIIASPDLKPVV 445
>gb|AAB67739.1| pectin methylesterase PME2.1 [Lycopersicon esculentum]
Length = 550
Score = 174 bits (441), Expect = 3e-42
Identities = 102/235 (43%), Positives = 139/235 (58%), Gaps = 5/235 (2%)
Query: 29 GKFPSWVKPGDRKLLQASA--VPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKK 86
GK PSWV DRKL+++S + A+AVVA DG+G Y + +AV AAP+ SK RYVI++K+
Sbjct: 213 GKMPSWVSSRDRKLMESSGKDIGANAVVAKDGTGKYRTLAEAVAAAPDKSKTRYVIYVKR 272
Query: 87 GVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDIS 146
G Y E+V +++ K NLM+IGDGM AT+ITG L+ D T ++ T G GF QDI
Sbjct: 273 GTYKENVEVSSRKMNLMIIGDGMYATIITGSLN-VVDGSTTFHSATLAAVGKGFILQDIC 331
Query: 147 FRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLVR 206
+NTA P HQAVAL +D SV RC I +QD+L A+ + R + +
Sbjct: 332 IQNTAGPAKHQAVALRVGADKSVINRCRIDAYQDTLYAHSQRQFYRDSYVTGTIDFIFGN 391
Query: 207 QLSSF-KTDILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEFLPFV 260
F K ++ RK QQN +TAQG + PN G + QFC++ A P+ P V
Sbjct: 392 AAVVFQKCQLVARKPGKYQQNMVTAQGRTD-PNQATGTSIQFCDIIASPDLKPVV 445
>emb|CAA30746.1| unnamed protein product [Lycopersicon esculentum]
gi|82097|pir||S00629 pectinesterase (EC 3.1.1.11)
precursor (clone PE1) - tomato
Length = 389
Score = 173 bits (438), Expect = 6e-42
Identities = 101/235 (42%), Positives = 139/235 (58%), Gaps = 5/235 (2%)
Query: 29 GKFPSWVKPGDRKLLQASA--VPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKK 86
GK PSWV DRKL+++S + A+AVVA DG+G Y + +AV AAP+ SK RYVI++K+
Sbjct: 37 GKMPSWVSSRDRKLMESSGKDIGANAVVAKDGTGKYRTLAEAVAAAPDKSKTRYVIYVKR 96
Query: 87 GVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDIS 146
G Y E+V +++ K NLM+IGDGM AT+ITG L+ D T ++ T G GF QDI
Sbjct: 97 GTYKENVEVSSRKMNLMIIGDGMYATIITGSLN-VVDGSTTFHSATLAAVGKGFILQDIC 155
Query: 147 FRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLVR 206
+NTA P HQAVAL +D SV RC I +QD+L A+ + + + +
Sbjct: 156 IQNTAGPAKHQAVALRVGADKSVINRCRIDAYQDTLYAHSQRQFYQSSYVTGTIDFIFGN 215
Query: 207 QLSSF-KTDILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEFLPFV 260
F K ++ RK QQN +TAQG + PN G + QFC++ A P+ P V
Sbjct: 216 AAVVFQKCQLVARKPGKYQQNMVTAQGRTD-PNQATGTSIQFCDIIASPDLKPVV 269
>sp|Q96575|PME22_LYCES Pectinesterase-2 precursor (Pectin methylesterase 2) (PE 2)
gi|1617584|gb|AAB38792.1| pectin methylesterase
[Lycopersicon esculentum]
Length = 550
Score = 170 bits (431), Expect = 4e-41
Identities = 99/233 (42%), Positives = 138/233 (58%), Gaps = 5/233 (2%)
Query: 29 GKFPSWVKPGDRKLLQASA--VPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKK 86
GK PSWV DRKL+++S + A+AVVA DG+G Y + +AV AAP+ SK RYVI++K+
Sbjct: 213 GKMPSWVSSRDRKLMESSGKDIGANAVVAKDGTGKYRTLAEAVAAAPDKSKTRYVIYVKR 272
Query: 87 GVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDIS 146
G+Y E+V +++ K LM++GDGM AT+ITG+L+ D T ++ T G GF QDI
Sbjct: 273 GIYKENVEVSSRKMKLMIVGDGMHATIITGNLN-VVDGSTTFHSATLAAVGKGFILQDIC 331
Query: 147 FRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLVR 206
+NTA P HQAVAL +D SV RC I +QD+L A+ + R + +
Sbjct: 332 IQNTAGPAKHQAVALRVGADKSVINRCRIDAYQDTLYAHSQRQFYRDSYVTGTIDFIFGN 391
Query: 207 QLSSF-KTDILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEFLP 258
F K ++ RK QQN +TAQG + PN G + QFCN+ A + P
Sbjct: 392 AAVVFQKCKLVARKPGKYQQNMVTAQGRTD-PNQATGTSIQFCNIIASSDLEP 443
>gb|AAB38794.1| pectin methylesterase [Lycopersicon esculentum]
Length = 439
Score = 169 bits (429), Expect = 6e-41
Identities = 104/249 (41%), Positives = 143/249 (56%), Gaps = 13/249 (5%)
Query: 16 TSSSDILDTITQKGKFPSWVKPGDRKLLQASA--VPADAVVASDGSGNYMKIMDAVMAAP 73
T D+ +T GK PSWV DRKL+++S + A+ VVA DG+G Y + +AV AAP
Sbjct: 91 TQDEDVF--MTGLGKMPSWVSSMDRKLMESSGKDIIANRVVAQDGTGKYRTLAEAVAAAP 148
Query: 74 NGSKKRYVIHIKKGVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTY-- 131
N SKKRYVI++K+G+Y E+V ++++K NLM++GDGM AT ITG L + +D S T+
Sbjct: 149 NRSKKRYVIYVKRGIYKENVEVSSNKMNLMIVGDGMYATTITGSL----NVVDGSTTFRS 204
Query: 132 -TFGVEGLGFSAQDISFRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHS 190
T G GF QDI +NTA P QAVAL +D SV RC I +QD+L A+ +
Sbjct: 205 ATLAAVGQGFILQDICIQNTAGPAKDQAVALRVGADMSVINRCRIDAYQDTLYAHSQRQF 264
Query: 191 IRIAKSEARLTSYLVRQLSSF-KTDILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCN 249
R + + F K ++ RK QQN +TAQG + PN G + QFCN
Sbjct: 265 YRDSYVTGTVDFIFGNAAVVFQKCQLVARKPGKYQQNMVTAQGTTD-PNQATGTSIQFCN 323
Query: 250 VCADPEFLP 258
+ A + P
Sbjct: 324 IIASSDLEP 332
>dbj|BAC67662.1| pectin methylesterase [Pisum sativum]
Length = 553
Score = 168 bits (426), Expect = 1e-40
Identities = 98/233 (42%), Positives = 136/233 (58%), Gaps = 5/233 (2%)
Query: 29 GKFPSWVKPGDRKLLQASA--VPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKK 86
G+FPSWV DR+LL++S + A+ VVA DGSG + + +AV + PN K RYVI++KK
Sbjct: 217 GEFPSWVTSKDRRLLESSVGDITANVVVAKDGSGKFKTVAEAVASVPNKGKTRYVIYVKK 276
Query: 87 GVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDIS 146
G Y E+V I++ K+N+M++GDGM AT+ITG L+ D T + T G GF AQDI
Sbjct: 277 GTYKENVEISSQKTNVMLVGDGMDATIITGSLN-VVDGTGTFQSATVAAVGDGFIAQDIG 335
Query: 147 FRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLVR 206
F+NTA PE HQAVAL SD SV RC I FQD+L A+ R +
Sbjct: 336 FKNTAGPEKHQAVALRVGSDQSVINRCRIDAFQDTLYAHSNRQFYRDCFITGTIDFIFGN 395
Query: 207 QLSSF-KTDILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEFLP 258
+ F K+ ++ RK + Q+N +TAQG + PN + Q C++ + P
Sbjct: 396 AAAVFQKSKLVARKPMSNQKNMVTAQGRLD-PNQNTATSIQQCDIIPSTDLKP 447
>gb|AAB67740.1| PME1.9 [Lycopersicon esculentum]
Length = 430
Score = 167 bits (424), Expect = 2e-40
Identities = 103/251 (41%), Positives = 145/251 (57%), Gaps = 13/251 (5%)
Query: 16 TSSSDILDTITQKGKFPSWVKPGDRKLLQASA--VPADAVVASDGSGNYMKIMDAVMAAP 73
T D+L T+ GK PSWV DRKL+++S + A+AVVA DG+G+Y + +AV AAP
Sbjct: 82 TQDEDVLMTVL--GKMPSWVSSMDRKLMESSGKDIIANAVVAQDGTGDYQTLAEAVAAAP 139
Query: 74 NGSKKRYVIHIKKGVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTY-- 131
+ SK RYVI++K+G Y E+V ++++K NLM++GDGM AT ITG L + +D S T+
Sbjct: 140 DKSKTRYVIYVKRGTYKENVEVSSNKMNLMIVGDGMYATTITGSL----NVVDGSTTFRS 195
Query: 132 -TFGVEGLGFSAQDISFRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHS 190
T G GF QDI +NTA P QAVAL +D SV RC I +QD+L A+ +
Sbjct: 196 ATLAAVGQGFILQDICIQNTAGPAKDQAVALRVGADMSVINRCRIDAYQDTLYAHSQRQF 255
Query: 191 IRIAKSEARLTSYLVRQLSSF-KTDILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCN 249
R + + F K ++ RK QQN +TAQG + PN G + QFC+
Sbjct: 256 YRDSYVTGTVDFIFGNAAVVFQKCQLVARKPGKYQQNMVTAQGRTD-PNQATGTSIQFCD 314
Query: 250 VCADPEFLPFV 260
+ A + P +
Sbjct: 315 IIASSDLEPLL 325
>emb|CAA47810.1| pectinesterase [Pisum sativum] gi|7447372|pir||T06468
pectinesterase (EC 3.1.1.11) precursor - garden pea
Length = 554
Score = 167 bits (423), Expect = 3e-40
Identities = 100/237 (42%), Positives = 141/237 (59%), Gaps = 13/237 (5%)
Query: 29 GKFPSWVKPGDRKLLQASA--VPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKK 86
G+FPSWV DR+LL+++ + A+ VVA DGSG + + +AV +AP+ K RYVI++K+
Sbjct: 217 GEFPSWVTSKDRRLLESTVGDIKANVVVAKDGSGKFKTVAEAVASAPDNGKARYVIYVKR 276
Query: 87 GVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDIS 146
G Y E V I K+N+M++GDGM AT+ITG+L++ D T + T G GF AQDI
Sbjct: 277 GTYKEKVEIGKKKTNVMLVGDGMDATIITGNLNF-IDGTTTFNSATVAAVGDGFIAQDIG 335
Query: 147 FRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLVR 206
F+NTA PE HQAVAL +D SV RC+I FQD+L A HS R ++ +T +
Sbjct: 336 FQNTAGPEKHQAVALRVGADQSVINRCKIDAFQDTLYA----HSNRQFYRDSFITGTVDF 391
Query: 207 QLSSF-----KTDILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEFLP 258
+ K+ ++ RK + Q+N +TAQ G E PN + Q CNV + P
Sbjct: 392 IFGNAGVVFQKSKLVARKPMSNQKNMVTAQ-GREDPNQNTATSIQQCNVIPSSDLKP 447
>dbj|BAC67661.1| pectin methylesterase [Pisum sativum]
Length = 554
Score = 167 bits (423), Expect = 3e-40
Identities = 100/237 (42%), Positives = 141/237 (59%), Gaps = 13/237 (5%)
Query: 29 GKFPSWVKPGDRKLLQASA--VPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKK 86
G+FPSWV DR+LL+++ + A+ VVA DGSG + + +AV +AP+ K RYVI++K+
Sbjct: 217 GEFPSWVTSKDRRLLESTVGDIKANVVVAKDGSGKFKTVAEAVASAPDNGKARYVIYVKR 276
Query: 87 GVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDIS 146
G Y E V I K+N+M++GDGM AT+ITG+L++ D T + T G GF AQDI
Sbjct: 277 GTYKEKVEIGKKKTNVMLVGDGMDATIITGNLNF-IDGTTTFNSATVAAVGDGFIAQDIG 335
Query: 147 FRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLVR 206
F+NTA PE HQAVAL +D SV RC+I FQD+L A HS R ++ +T +
Sbjct: 336 FQNTAGPEKHQAVALRVGADQSVINRCKIDAFQDTLYA----HSNRQFYRDSFITGTVDF 391
Query: 207 QLSSF-----KTDILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEFLP 258
+ K+ ++ RK + Q+N +TAQ G E PN + Q CNV + P
Sbjct: 392 IFGNAGVVFQKSKLVARKPMSNQKNMVTAQ-GREDPNQNTATSIQQCNVIPSSDLKP 447
>gb|AAC14742.1| pectin methylesterase [Pisum sativum] gi|3426335|gb|AAC32273.1|
pectin methylesterase [Pisum sativum]
gi|7447371|pir||T06374 probable pectinesterase (EC
3.1.1.11) precursor - garden pea
Length = 554
Score = 167 bits (423), Expect = 3e-40
Identities = 100/237 (42%), Positives = 141/237 (59%), Gaps = 13/237 (5%)
Query: 29 GKFPSWVKPGDRKLLQASA--VPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKK 86
G+FPSWV DR+LL+++ + A+ VVA DGSG + + +AV +AP+ K RYVI++K+
Sbjct: 217 GEFPSWVTSKDRRLLESTVGDIKANVVVAKDGSGKFKTVAEAVASAPDNGKARYVIYVKR 276
Query: 87 GVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDIS 146
G Y E V I K+N+M++GDGM AT+ITG+L++ D T + T G GF AQDI
Sbjct: 277 GTYKEKVEIGKKKTNVMLVGDGMDATIITGNLNF-IDGTTTFNSATVAAVGDGFIAQDIG 335
Query: 147 FRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLVR 206
F+NTA PE HQAVAL +D SV RC+I FQD+L A HS R ++ +T +
Sbjct: 336 FQNTAGPEKHQAVALRVGADQSVINRCKIDAFQDTLYA----HSNRQFYRDSFITGTVDF 391
Query: 207 QLSSF-----KTDILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEFLP 258
+ K+ ++ RK + Q+N +TAQ G E PN + Q CNV + P
Sbjct: 392 IFGNAGVVFQKSKLVARKPMSNQKNMVTAQ-GREDPNQNTATSIQQCNVIPSSDLKP 447
>gb|AAK84486.1| putative thermostable pectinesterase [Citrus sinensis]
gi|24250746|gb|AAK84485.1| putative thermostable
pectinesterase [Citrus sinensis]
Length = 631
Score = 167 bits (422), Expect = 4e-40
Identities = 103/241 (42%), Positives = 134/241 (54%), Gaps = 27/241 (11%)
Query: 30 KFPSWVKPGDRKLLQASAVPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKKGVY 89
K+P W+ GDR+LLQA+ V D VA+DGSGNY+ + AV AAP GS +RY+I IK G Y
Sbjct: 299 KWPEWLSAGDRRLLQATTVVPDVTVAADGSGNYLTVAAAVAAAPEGSSRRYIIRIKAGEY 358
Query: 90 NEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTY---TFGVEGLGFSAQDIS 146
E+V + K NLM IGDG T+ITG R+ +D S T+ T V G GF A+DI+
Sbjct: 359 RENVEVPKKKINLMFIGDGRTTTIITG----SRNVVDGSTTFNSATVAVVGDGFLARDIT 414
Query: 147 FRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLVR 206
F+NTA P HQAVAL SD S FYRC++ +QD+L HS+R TS ++
Sbjct: 415 FQNTAGPSKHQAVALRVGSDLSAFYRCDMLAYQDTLYV----HSLR-----QFYTSCIIA 465
Query: 207 QLSSF----------KTDILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEF 256
F DI R+ Q+N +TAQ G + PN G Q C + A +
Sbjct: 466 GTVDFIFGNAAAVFQNCDIHARRPNPNQRNMVTAQ-GRDDPNQNTGIVIQKCRIGATSDL 524
Query: 257 L 257
L
Sbjct: 525 L 525
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.320 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 481,757,985
Number of Sequences: 2540612
Number of extensions: 19754686
Number of successful extensions: 45616
Number of sequences better than 10.0: 306
Number of HSP's better than 10.0 without gapping: 246
Number of HSP's successfully gapped in prelim test: 60
Number of HSP's that attempted gapping in prelim test: 44808
Number of HSP's gapped (non-prelim): 325
length of query: 287
length of database: 863,360,394
effective HSP length: 127
effective length of query: 160
effective length of database: 540,702,670
effective search space: 86512427200
effective search space used: 86512427200
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 74 (33.1 bits)
Medicago: description of AC148775.4


