

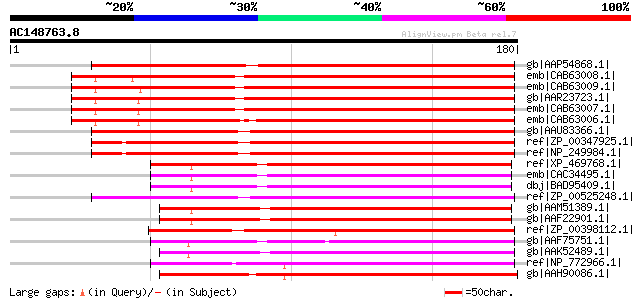
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148763.8 - phase: 1 /pseudo
(180 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAP54868.1| mucin-like protein [Oryza sativa (japonica cultiv... 175 6e-43
emb|CAB63008.1| mucin-like protein [Arabidopsis thaliana] gi|232... 172 4e-42
emb|CAB63009.1| mucin-like protein [Arabidopsis thaliana] gi|235... 168 6e-41
gb|AAR23723.1| At1g73860 [Arabidopsis thaliana] gi|21592926|gb|A... 167 9e-41
emb|CAB63007.1| mucin-like protein [Arabidopsis thaliana] gi|113... 167 9e-41
emb|CAB63006.1| mucin-like protein [Arabidopsis thaliana] gi|215... 166 3e-40
gb|AAU83366.1| conserved hypothetical protein [uncultured archae... 130 1e-29
ref|ZP_00347925.1| COG3386: Gluconolactonase [Pseudomonas aerugi... 125 4e-28
ref|NP_249984.1| hypothetical protein PA1293 [Pseudomonas aerugi... 123 2e-27
ref|XP_469768.1| putative strictosidine synthase [Oryza sativa ... 103 2e-21
emb|CAC34495.1| putative strictosidine synthase-like [Arabidopsi... 101 1e-20
dbj|BAD95409.1| putative strictosidine synthase - like [Arabidop... 101 1e-20
ref|ZP_00525248.1| Strictosidine synthase [Solibacter usitatus E... 99 4e-20
gb|AAM51389.1| unknown protein [Arabidopsis thaliana] gi|1738118... 99 4e-20
gb|AAF22901.1| T27G7.16 [Arabidopsis thaliana] gi|25335889|pir||... 99 4e-20
ref|ZP_00398112.1| Strictosidine synthase [Deinococcus geotherma... 94 2e-18
gb|AAF75751.1| putative strictosidine synthase [Lycopersicon esc... 93 3e-18
gb|AAK52489.1| male fertility protein [Zea mays] 92 7e-18
ref|NP_772966.1| ABC transporter permease protein [Bradyrhizobiu... 91 2e-17
gb|AAH90086.1| LOC548386 protein [Xenopus tropicalis] 90 2e-17
>gb|AAP54868.1| mucin-like protein [Oryza sativa (japonica cultivar-group)]
gi|37536558|ref|NP_922581.1| mucin-like protein [Oryza
sativa (japonica cultivar-group)]
gi|10140763|gb|AAG13594.1| mucin-like protein [Oryza
sativa]
Length = 369
Score = 175 bits (443), Expect = 6e-43
Identities = 80/150 (53%), Positives = 114/150 (75%), Gaps = 4/150 (2%)
Query: 30 GCEDGWIKRITVADSVVENWVHSSGRPLGLALEKTGELIVADAHLGLLRVTQKEGKEPKV 89
GC DGW++R++V+ VE+W + GRPLG+AL G L+VADA +GLL+V+ + V
Sbjct: 84 GCGDGWVRRVSVSSGDVEDWARTGGRPLGVALTADGGLVVADADIGLLKVSPDKA----V 139
Query: 90 EILANEHDGLKFKLTDGVDVGEDGTIYFTEATYKYNLYDFYNDILEGEPHGRFMSYNPAT 149
E+L +E +G+KF LTDGVDV DG IYFT+A++K++L +F D+LE PHGR MS++P+T
Sbjct: 140 ELLTDEAEGVKFALTDGVDVAGDGVIYFTDASHKHSLAEFMVDVLEARPHGRLMSFDPST 199
Query: 150 KKVTLLARNLYFANRVAIAPDQKFVVYCET 179
++ T+LAR LYFAN VA++PDQ +V+CET
Sbjct: 200 RRTTVLARGLYFANGVAVSPDQDSLVFCET 229
>emb|CAB63008.1| mucin-like protein [Arabidopsis thaliana]
gi|23296634|gb|AAN13136.1| putative mucin protein
[Arabidopsis thaliana] gi|13430724|gb|AAK25984.1|
putative mucin protein [Arabidopsis thaliana]
gi|15230488|ref|NP_190712.1| strictosidine synthase
family protein [Arabidopsis thaliana]
gi|11358519|pir||T45775 mucin-like protein - Arabidopsis
thaliana
Length = 371
Score = 172 bits (436), Expect = 4e-42
Identities = 85/162 (52%), Positives = 118/162 (72%), Gaps = 8/162 (4%)
Query: 23 EEASYIY-GCEDGWIKRITVA----DSVVENWVHSSGRPLGLALEKTGELIVADAHLGLL 77
E++ +IY GC DGW+KR+ VA DS+VE+ V++ GRPLG+A GE+IVADA+ GLL
Sbjct: 77 EDSGFIYTGCVDGWVKRVKVAESVNDSLVEDLVNTGGRPLGIAFGIHGEVIVADAYKGLL 136
Query: 78 RVTQKEGKEPKVEILANEHDGLKFKLTDGVDVGEDGTIYFTEATYKYNLYDFYNDILEGE 137
++ G K E+L E DG++FKL D V V ++G +YFT+ +YKYNL+ F DILEG+
Sbjct: 137 NIS---GDGKKTELLTEEADGVRFKLPDAVTVADNGVLYFTDGSYKYNLHQFSFDILEGK 193
Query: 138 PHGRFMSYNPATKKVTLLARNLYFANRVAIAPDQKFVVYCET 179
PHGR MS++P TK +L R+LYFAN V+++PDQ +V+CET
Sbjct: 194 PHGRLMSFDPTTKVTRVLLRDLYFANGVSLSPDQTHLVFCET 235
>emb|CAB63009.1| mucin-like protein [Arabidopsis thaliana]
gi|23506011|gb|AAN28865.1| At3g51450/F26O13_90
[Arabidopsis thaliana] gi|21593437|gb|AAM65404.1|
mucin-like protein [Arabidopsis thaliana]
gi|18700143|gb|AAL77683.1| AT3g51450/F26O13_90
[Arabidopsis thaliana] gi|15230490|ref|NP_190713.1|
strictosidine synthase family protein [Arabidopsis
thaliana] gi|51969270|dbj|BAD43327.1| mucin -like
protein [Arabidopsis thaliana]
gi|51968632|dbj|BAD43008.1| mucin -like protein
[Arabidopsis thaliana] gi|11358520|pir||T45776
mucin-like protein - Arabidopsis thaliana
Length = 371
Score = 168 bits (426), Expect = 6e-41
Identities = 82/162 (50%), Positives = 116/162 (70%), Gaps = 8/162 (4%)
Query: 23 EEASYIY-GCEDGWIKRITVADSV----VENWVHSSGRPLGLALEKTGELIVADAHLGLL 77
+E++ IY GC DGW+KR+ VADSV VE+WV++ GRPLG+A GE+IVAD H GLL
Sbjct: 77 KESNLIYTGCVDGWVKRVKVADSVNDSVVEDWVNTGGRPLGIAFGIHGEVIVADVHKGLL 136
Query: 78 RVTQKEGKEPKVEILANEHDGLKFKLTDGVDVGEDGTIYFTEATYKYNLYDFYNDILEGE 137
++ G K E+L +E DG+KFKLTD V V ++G +YFT+A+YKY L D+LEG+
Sbjct: 137 NIS---GDGKKTELLTDEADGVKFKLTDAVTVADNGVLYFTDASYKYTLNQLSLDMLEGK 193
Query: 138 PHGRFMSYNPATKKVTLLARNLYFANRVAIAPDQKFVVYCET 179
P GR +S++P T+ +L ++LYFAN + I+PDQ +++CET
Sbjct: 194 PFGRLLSFDPTTRVTKVLLKDLYFANGITISPDQTHLIFCET 235
>gb|AAR23723.1| At1g73860 [Arabidopsis thaliana] gi|21592926|gb|AAM64876.1|
mucin-like protein [Arabidopsis thaliana]
gi|18087629|gb|AAL58944.1| AT3g51430/F26O13_70
[Arabidopsis thaliana] gi|18086443|gb|AAL57676.1|
AT3g51430/F26O13_70 [Arabidopsis thaliana]
gi|18409339|ref|NP_566951.1| strictosidine synthase,
putative (YLS2) [Arabidopsis thaliana]
gi|13122282|dbj|BAB32882.1| strictosidine synthase-like
protein [Arabidopsis thaliana]
Length = 371
Score = 167 bits (424), Expect = 9e-41
Identities = 81/162 (50%), Positives = 120/162 (74%), Gaps = 8/162 (4%)
Query: 23 EEASYIY-GCEDGWIKRITVADS----VVENWVHSSGRPLGLALEKTGELIVADAHLGLL 77
++++ IY GC DGW+KR++V DS VVE+WV++ GRPLG+A GE+IVADA+ GLL
Sbjct: 77 QDSNLIYTGCIDGWVKRVSVHDSANDSVVEDWVNTGGRPLGIAFGVHGEVIVADAYKGLL 136
Query: 78 RVTQKEGKEPKVEILANEHDGLKFKLTDGVDVGEDGTIYFTEATYKYNLYDFYNDILEGE 137
++ G K E+L ++ +G+KFKLTD V V ++G +YFT+A+YKY L+ DILEG+
Sbjct: 137 NIS---GDGKKTELLTDQAEGVKFKLTDVVAVADNGVLYFTDASYKYTLHQVKFDILEGK 193
Query: 138 PHGRFMSYNPATKKVTLLARNLYFANRVAIAPDQKFVVYCET 179
PHGR MS++P T+ +L ++LYFAN V+++PDQ +++CET
Sbjct: 194 PHGRLMSFDPTTRVTRVLLKDLYFANGVSMSPDQTHLIFCET 235
>emb|CAB63007.1| mucin-like protein [Arabidopsis thaliana] gi|11358518|pir||T45774
mucin-like protein - Arabidopsis thaliana
Length = 367
Score = 167 bits (424), Expect = 9e-41
Identities = 81/162 (50%), Positives = 120/162 (74%), Gaps = 8/162 (4%)
Query: 23 EEASYIY-GCEDGWIKRITVADS----VVENWVHSSGRPLGLALEKTGELIVADAHLGLL 77
++++ IY GC DGW+KR++V DS VVE+WV++ GRPLG+A GE+IVADA+ GLL
Sbjct: 77 QDSNLIYTGCIDGWVKRVSVHDSANDSVVEDWVNTGGRPLGIAFGVHGEVIVADAYKGLL 136
Query: 78 RVTQKEGKEPKVEILANEHDGLKFKLTDGVDVGEDGTIYFTEATYKYNLYDFYNDILEGE 137
++ G K E+L ++ +G+KFKLTD V V ++G +YFT+A+YKY L+ DILEG+
Sbjct: 137 NIS---GDGKKTELLTDQAEGVKFKLTDVVAVADNGVLYFTDASYKYTLHQVKFDILEGK 193
Query: 138 PHGRFMSYNPATKKVTLLARNLYFANRVAIAPDQKFVVYCET 179
PHGR MS++P T+ +L ++LYFAN V+++PDQ +++CET
Sbjct: 194 PHGRLMSFDPTTRVTRVLLKDLYFANGVSMSPDQTHLIFCET 235
>emb|CAB63006.1| mucin-like protein [Arabidopsis thaliana]
gi|21593396|gb|AAM65345.1| mucin-like protein
[Arabidopsis thaliana] gi|15230477|ref|NP_190710.1|
strictosidine synthase family protein [Arabidopsis
thaliana] gi|11358517|pir||T45773 mucin-like protein -
Arabidopsis thaliana
Length = 370
Score = 166 bits (420), Expect = 3e-40
Identities = 81/162 (50%), Positives = 119/162 (73%), Gaps = 8/162 (4%)
Query: 23 EEASYIY-GCEDGWIKRITVADS----VVENWVHSSGRPLGLALEKTGELIVADAHLGLL 77
++++ IY GC DGW+KR++V DS +VE+WV++ GRPLG+A GE+IVADA+ GLL
Sbjct: 77 KDSNLIYTGCVDGWVKRVSVHDSANDSIVEDWVNTGGRPLGIAFGLHGEVIVADANKGLL 136
Query: 78 RVTQKEGKEPKVEILANEHDGLKFKLTDGVDVGEDGTIYFTEATYKYNLYDFYNDILEGE 137
++ GK K E+L +E DG++FKLTD V V ++G +YFT+A+ KY+ Y F D LEG+
Sbjct: 137 SISDG-GK--KTELLTDEADGVRFKLTDAVTVADNGVLYFTDASSKYDFYQFIFDFLEGK 193
Query: 138 PHGRFMSYNPATKKVTLLARNLYFANRVAIAPDQKFVVYCET 179
PHGR MS++P T+ +L ++LYFAN ++++PDQ V+CET
Sbjct: 194 PHGRVMSFDPTTRATRVLLKDLYFANGISMSPDQTHFVFCET 235
>gb|AAU83366.1| conserved hypothetical protein [uncultured archaeon GZfos27E7]
Length = 352
Score = 130 bits (328), Expect = 1e-29
Identities = 65/150 (43%), Positives = 94/150 (62%), Gaps = 4/150 (2%)
Query: 30 GCEDGWIKRITVADSVVENWVHSSGRPLGLALEKTGELIVADAHLGLLRVTQKEGKEPKV 89
G EDG I R S E + + GRPLGL + G L+V DA+ GLL +T + +
Sbjct: 69 GMEDGRIMRFQADGSQHEVFADTEGRPLGLHFDAAGNLVVCDAYKGLLSITP----DGSI 124
Query: 90 EILANEHDGLKFKLTDGVDVGEDGTIYFTEATYKYNLYDFYNDILEGEPHGRFMSYNPAT 149
+L+ E G+ F+LTD VD+ DG IYF++A++K+ + D++E P+GR ++YNP+T
Sbjct: 125 IVLSTEQGGVPFRLTDDVDIAADGIIYFSDASFKFTEAEHMADLMEHRPNGRLLAYNPST 184
Query: 150 KKVTLLARNLYFANRVAIAPDQKFVVYCET 179
K L+ NLYFAN VA++PDQ FV+ ET
Sbjct: 185 KTTRLVLNNLYFANGVAVSPDQSFVLVVET 214
>ref|ZP_00347925.1| COG3386: Gluconolactonase [Pseudomonas aeruginosa UCBPP-PA14]
Length = 353
Score = 125 bits (315), Expect = 4e-28
Identities = 66/150 (44%), Positives = 96/150 (64%), Gaps = 5/150 (3%)
Query: 30 GCEDGWIKRITVADSVVENWVHSSGRPLGLALEKTGELIVADAHLGLLRVTQKEGKEPKV 89
G DG + R+ A VE +V + GRPLG+ + G LI+ADA GLLR+ + KV
Sbjct: 77 GLADGRVVRLD-ASGKVETFVDTGGRPLGMDFDAAGNLILADAWKGLLRIDP----QGKV 131
Query: 90 EILANEHDGLKFKLTDGVDVGEDGTIYFTEATYKYNLYDFYNDILEGEPHGRFMSYNPAT 149
E LA E DG+ F TD +D+ DG IYF++A+ K++ D+ D+LE PHGR + Y+P+T
Sbjct: 132 ETLATEADGVPFAFTDDLDIASDGRIYFSDASSKFHQPDYILDLLEARPHGRLLRYDPST 191
Query: 150 KKVTLLARNLYFANRVAIAPDQKFVVYCET 179
K +L ++LYFAN VA++ ++ FV+ ET
Sbjct: 192 GKTEVLLKDLYFANGVALSANEDFVLVNET 221
>ref|NP_249984.1| hypothetical protein PA1293 [Pseudomonas aeruginosa PAO1]
gi|9947229|gb|AAG04682.1| hypothetical protein PA1293
[Pseudomonas aeruginosa PAO1] gi|11349157|pir||H83482
hypothetical protein PA1293 [imported] - Pseudomonas
aeruginosa (strain PAO1)
Length = 353
Score = 123 bits (309), Expect = 2e-27
Identities = 65/150 (43%), Positives = 95/150 (63%), Gaps = 5/150 (3%)
Query: 30 GCEDGWIKRITVADSVVENWVHSSGRPLGLALEKTGELIVADAHLGLLRVTQKEGKEPKV 89
G DG + R+ A VE +V + GRPLG+ + G LI+ADA GLLR+ + KV
Sbjct: 77 GLADGRVVRLD-ASGKVETFVDTGGRPLGMDFDAAGNLILADAWKGLLRIDP----QGKV 131
Query: 90 EILANEHDGLKFKLTDGVDVGEDGTIYFTEATYKYNLYDFYNDILEGEPHGRFMSYNPAT 149
E LA E D + F TD +D+ DG IYF++A+ K++ D+ D+LE PHGR + Y+P+T
Sbjct: 132 ETLATEADSVPFAFTDDLDIASDGRIYFSDASSKFHQPDYILDLLEARPHGRLLRYDPST 191
Query: 150 KKVTLLARNLYFANRVAIAPDQKFVVYCET 179
K +L ++LYFAN VA++ ++ FV+ ET
Sbjct: 192 GKTEVLLKDLYFANGVALSANEDFVLVNET 221
>ref|XP_469768.1| putative strictosidine synthase [Oryza sativa (japonica
cultivar-group)] gi|40538997|gb|AAR87254.1| putative
strictosidine synthase [Oryza sativa (japonica
cultivar-group)]
Length = 480
Score = 103 bits (257), Expect = 2e-21
Identities = 52/129 (40%), Positives = 79/129 (60%), Gaps = 4/129 (3%)
Query: 51 HSSGRPLGLALEK-TGELIVADAHLGLLRVTQKEGKEPKVEILANEHDGLKFKLTDGVDV 109
H GR LGL ++ TG+L +ADA+ GLL+V G LA E +G++F T+ +D+
Sbjct: 214 HICGRALGLRFDRRTGDLYIADAYFGLLKVGPDGGLATP---LATEAEGVRFNFTNDLDL 270
Query: 110 GEDGTIYFTEATYKYNLYDFYNDILEGEPHGRFMSYNPATKKVTLLARNLYFANRVAIAP 169
+DG +YFT+++ Y F + G+P GR + Y+P TKK T+L RN+ F N V+++
Sbjct: 271 DDDGNVYFTDSSIHYQRRHFMQLVFSGDPSGRLLKYDPNTKKATVLHRNIQFPNGVSMSK 330
Query: 170 DQKFVVYCE 178
D F V+CE
Sbjct: 331 DGLFFVFCE 339
>emb|CAC34495.1| putative strictosidine synthase-like [Arabidopsis thaliana]
gi|22326950|ref|NP_680189.1| strictosidine synthase
family protein [Arabidopsis thaliana]
gi|48525339|gb|AAT44971.1| At5g22020 [Arabidopsis
thaliana]
Length = 395
Score = 101 bits (251), Expect = 1e-20
Identities = 51/129 (39%), Positives = 77/129 (59%), Gaps = 4/129 (3%)
Query: 51 HSSGRPLGLALEK-TGELIVADAHLGLLRVTQKEGKEPKVEILANEHDGLKFKLTDGVDV 109
H GRPLGL +K TG+L +ADA++GLL+V + G L E +G+ T+ +D+
Sbjct: 133 HICGRPLGLRFDKRTGDLYIADAYMGLLKVGPEGGLATP---LVTEAEGVPLGFTNDLDI 189
Query: 110 GEDGTIYFTEATYKYNLYDFYNDILEGEPHGRFMSYNPATKKVTLLARNLYFANRVAIAP 169
+DGT+YFT+++ Y +F + G+ GR + Y+P KK +L NL F N V+I+
Sbjct: 190 ADDGTVYFTDSSISYQRRNFLQLVFSGDNTGRVLKYDPVAKKAVVLVSNLQFPNGVSISR 249
Query: 170 DQKFVVYCE 178
D F V+CE
Sbjct: 250 DGSFFVFCE 258
>dbj|BAD95409.1| putative strictosidine synthase - like [Arabidopsis thaliana]
Length = 394
Score = 101 bits (251), Expect = 1e-20
Identities = 51/129 (39%), Positives = 77/129 (59%), Gaps = 4/129 (3%)
Query: 51 HSSGRPLGLALEK-TGELIVADAHLGLLRVTQKEGKEPKVEILANEHDGLKFKLTDGVDV 109
H GRPLGL +K TG+L +ADA++GLL+V + G L E +G+ T+ +D+
Sbjct: 132 HICGRPLGLRFDKRTGDLYIADAYMGLLKVGPEGGLATP---LVTEAEGVPLGFTNDLDI 188
Query: 110 GEDGTIYFTEATYKYNLYDFYNDILEGEPHGRFMSYNPATKKVTLLARNLYFANRVAIAP 169
+DGT+YFT+++ Y +F + G+ GR + Y+P KK +L NL F N V+I+
Sbjct: 189 ADDGTVYFTDSSISYQRRNFLQLVFSGDNTGRVLKYDPVAKKAVVLVSNLQFPNGVSISR 248
Query: 170 DQKFVVYCE 178
D F V+CE
Sbjct: 249 DGSFFVFCE 257
>ref|ZP_00525248.1| Strictosidine synthase [Solibacter usitatus Ellin6076]
gi|67860629|gb|EAM55673.1| Strictosidine synthase
[Solibacter usitatus Ellin6076]
Length = 384
Score = 99.4 bits (246), Expect = 4e-20
Identities = 51/150 (34%), Positives = 88/150 (58%), Gaps = 4/150 (2%)
Query: 30 GCEDGWIKRITVADSVVENWVHSSGRPLGLALEKTGELIVADAHLGLLRVTQKEGKEPKV 89
G +DG + R+ S E +V + GRPLG+ + G L+V DA GLL ++ E K+
Sbjct: 105 GLQDGRVMRMLPDGSGRETFVQTGGRPLGMKFDTAGNLVVGDAFRGLLSISP----ERKI 160
Query: 90 EILANEHDGLKFKLTDGVDVGEDGTIYFTEATYKYNLYDFYNDILEGEPHGRFMSYNPAT 149
+LA+ +G + T+ + + DG+++F++A+ +++ + + D LEG GR + Y P T
Sbjct: 161 SVLADSVNGERMLFTNDLAIAADGSVWFSDASRRFDQHHWTLDFLEGRATGRLLHYEPRT 220
Query: 150 KKVTLLARNLYFANRVAIAPDQKFVVYCET 179
+VT++ L FAN VA+ P ++V+ ET
Sbjct: 221 GQVTVVLDRLMFANGVALGPGDQYVLVNET 250
>gb|AAM51389.1| unknown protein [Arabidopsis thaliana] gi|17381182|gb|AAL36403.1|
unknown protein [Arabidopsis thaliana]
gi|18390900|ref|NP_563818.1| strictosidine synthase
family protein [Arabidopsis thaliana]
gi|16930481|gb|AAL31926.1| At1g08470/T27G7_9
[Arabidopsis thaliana]
Length = 390
Score = 99.4 bits (246), Expect = 4e-20
Identities = 49/126 (38%), Positives = 77/126 (60%), Gaps = 4/126 (3%)
Query: 54 GRPLGLALEK-TGELIVADAHLGLLRVTQKEGKEPKVEILANEHDGLKFKLTDGVDVGED 112
GRPLGL +K G+L +ADA+LG+++V + G V NE DG+ + T+ +D+ ++
Sbjct: 130 GRPLGLRFDKKNGDLYIADAYLGIMKVGPEGGLATSV---TNEADGVPLRFTNDLDIDDE 186
Query: 113 GTIYFTEATYKYNLYDFYNDILEGEPHGRFMSYNPATKKVTLLARNLYFANRVAIAPDQK 172
G +YFT+++ + F I+ GE GR + YNP TK+ T L RNL F N +++ D
Sbjct: 187 GNVYFTDSSSFFQRRKFMLLIVSGEDSGRVLKYNPKTKETTTLVRNLQFPNGLSLGKDGS 246
Query: 173 FVVYCE 178
F ++CE
Sbjct: 247 FFIFCE 252
>gb|AAF22901.1| T27G7.16 [Arabidopsis thaliana] gi|25335889|pir||H86217 protein
T27G7.16 [imported] - Arabidopsis thaliana
Length = 421
Score = 99.4 bits (246), Expect = 4e-20
Identities = 49/126 (38%), Positives = 77/126 (60%), Gaps = 4/126 (3%)
Query: 54 GRPLGLALEK-TGELIVADAHLGLLRVTQKEGKEPKVEILANEHDGLKFKLTDGVDVGED 112
GRPLGL +K G+L +ADA+LG+++V + G V NE DG+ + T+ +D+ ++
Sbjct: 130 GRPLGLRFDKKNGDLYIADAYLGIMKVGPEGGLATSV---TNEADGVPLRFTNDLDIDDE 186
Query: 113 GTIYFTEATYKYNLYDFYNDILEGEPHGRFMSYNPATKKVTLLARNLYFANRVAIAPDQK 172
G +YFT+++ + F I+ GE GR + YNP TK+ T L RNL F N +++ D
Sbjct: 187 GNVYFTDSSSFFQRRKFMLLIVSGEDSGRVLKYNPKTKETTTLVRNLQFPNGLSLGKDGS 246
Query: 173 FVVYCE 178
F ++CE
Sbjct: 247 FFIFCE 252
>ref|ZP_00398112.1| Strictosidine synthase [Deinococcus geothermalis DSM 11300]
gi|66780263|gb|EAL81274.1| Strictosidine synthase
[Deinococcus geothermalis DSM 11300]
Length = 363
Score = 93.6 bits (231), Expect = 2e-18
Identities = 54/131 (41%), Positives = 79/131 (60%), Gaps = 5/131 (3%)
Query: 50 VHSSGRPLGLALEKTGELIVADAHLGLLRVTQKEGKEPKVEILANEHDGLKFKLTDGVDV 109
V++ GRPLGL + L+VADA GLLRV G + VE+LA E +G+ F+ TD +DV
Sbjct: 99 VNTGGRPLGLRVHPDSTLLVADALRGLLRV----GLDGAVEVLATEAEGVPFRFTDDLDV 154
Query: 110 GEDGT-IYFTEATYKYNLYDFYNDILEGEPHGRFMSYNPATKKVTLLARNLYFANRVAIA 168
G +YFT+A+ KY D+LE HGR + ++ T + T+LAR L F N V +
Sbjct: 155 DRAGRFVYFTDASSKYGWPHELLDLLEHGGHGRVLRHDLQTGETTVLARGLNFPNGVTLG 214
Query: 169 PDQKFVVYCET 179
P +++++ ET
Sbjct: 215 PGEEYLLVTET 225
>gb|AAF75751.1| putative strictosidine synthase [Lycopersicon esculentum]
Length = 351
Score = 93.2 bits (230), Expect = 3e-18
Identities = 55/130 (42%), Positives = 76/130 (58%), Gaps = 5/130 (3%)
Query: 51 HSSGRPLGLALE-KTGELIVADAHLGLLRVTQKEGKEPKVEILANEHDGLKFKLTDGVDV 109
H GRPLGL + KTGEL +ADA+LGL V K G L + +G T+ VD+
Sbjct: 100 HICGRPLGLRFDTKTGELYIADAYLGLQVVGPKGGLATP---LVQKFEGKPLVFTNDVDI 156
Query: 110 GEDGTIYFTEATYKYNLYDFYNDILEGEPHGRFMSYNPATKKVTLLARNLYFANRVAIAP 169
+D IYFT+ + KY + F G+ GR M Y+ +TKKVT+L +L FAN VA++
Sbjct: 157 DDD-VIYFTDTSTKYQRWQFLTSFSSGDTTGRLMKYDKSTKKVTVLLGDLAFANGVALSK 215
Query: 170 DQKFVVYCET 179
++ FV+ ET
Sbjct: 216 NKSFVLVTET 225
>gb|AAK52489.1| male fertility protein [Zea mays]
Length = 412
Score = 92.0 bits (227), Expect = 7e-18
Identities = 51/127 (40%), Positives = 76/127 (59%), Gaps = 4/127 (3%)
Query: 54 GRPLGLALE-KTGELIVADAHLGLLRVTQKEGKEPKVEILANEHDGLKFKLTDGVDVGED 112
GRPLGL +TGEL VADA+ GL+ V Q G V A E DG + + +DV +
Sbjct: 150 GRPLGLRFHGETGELYVADAYYGLMVVGQSGGVASSV---AREADGDPIRFANDLDVHRN 206
Query: 113 GTIYFTEATYKYNLYDFYNDILEGEPHGRFMSYNPATKKVTLLARNLYFANRVAIAPDQK 172
G+++FT+ + +Y+ D N +LEGE GR + Y+P T V ++ + L F N V I+ D +
Sbjct: 207 GSVFFTDTSMRYSRKDHLNILLEGEGTGRLLRYDPETSGVHVVLKGLVFPNGVQISEDHQ 266
Query: 173 FVVYCET 179
F+++ ET
Sbjct: 267 FLLFSET 273
>ref|NP_772966.1| ABC transporter permease protein [Bradyrhizobium japonicum USDA
110] gi|27354605|dbj|BAC51591.1| ABC transporter
permease protein [Bradyrhizobium japonicum USDA 110]
Length = 601
Score = 90.5 bits (223), Expect = 2e-17
Identities = 45/134 (33%), Positives = 78/134 (57%), Gaps = 6/134 (4%)
Query: 51 HSSGRPLGLALEKTGELIVADAHLGLLRVTQKEGKEPKVEILANEH-----DGLKFKLTD 105
H G PLG+AL++ G L + A +GL +V + G ++ N D KL D
Sbjct: 426 HIGGSPLGMALDRQGHLNICVAGMGLYQV-EMNGTVRRLTAETNRSLLSIIDDSNMKLAD 484
Query: 106 GVDVGEDGTIYFTEATYKYNLYDFYNDILEGEPHGRFMSYNPATKKVTLLARNLYFANRV 165
+D+ DGTIYF+EAT ++ ++D+Y+D LE +GR + Y+P T++ + +NL F N +
Sbjct: 485 DLDIAPDGTIYFSEATTRFEMHDWYSDALESRGNGRIIRYDPKTRRTRTVLKNLVFPNGI 544
Query: 166 AIAPDQKFVVYCET 179
++ D + +++ E+
Sbjct: 545 CMSYDNESLLFAES 558
>gb|AAH90086.1| LOC548386 protein [Xenopus tropicalis]
Length = 431
Score = 90.1 bits (222), Expect = 2e-17
Identities = 47/129 (36%), Positives = 79/129 (60%), Gaps = 4/129 (3%)
Query: 54 GRPLGLALEKTGELIVADAHLGLLRVTQKEGKEPKVEILANEH--DGLKFKLTDGVDVGE 111
GRP G+ + G LIVAD++ GL RV G+ K +++NE D + F+ +G++V +
Sbjct: 166 GRPHGIRMAPDGYLIVADSYFGLYRVQPHTGE--KSLLISNEAGLDQIPFRFLNGLEVSK 223
Query: 112 DGTIYFTEATYKYNLYDFYNDILEGEPHGRFMSYNPATKKVTLLARNLYFANRVAIAPDQ 171
+GTIYFT+++ K+ ++LE GR + Y+P T+K L LY AN +A++P++
Sbjct: 224 NGTIYFTDSSSKWGRRHHRYEVLETNHLGRLLQYDPVTQKAKSLLDKLYMANGIALSPEE 283
Query: 172 KFVVYCETA 180
F++ ET+
Sbjct: 284 DFILVAETS 292
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.330 0.148 0.475
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 309,336,919
Number of Sequences: 2540612
Number of extensions: 12605226
Number of successful extensions: 35908
Number of sequences better than 10.0: 221
Number of HSP's better than 10.0 without gapping: 132
Number of HSP's successfully gapped in prelim test: 89
Number of HSP's that attempted gapping in prelim test: 35524
Number of HSP's gapped (non-prelim): 232
length of query: 180
length of database: 863,360,394
effective HSP length: 120
effective length of query: 60
effective length of database: 558,486,954
effective search space: 33509217240
effective search space used: 33509217240
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 71 (32.0 bits)
Medicago: description of AC148763.8


