

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148762.7 + phase: 2 /partial
(212 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
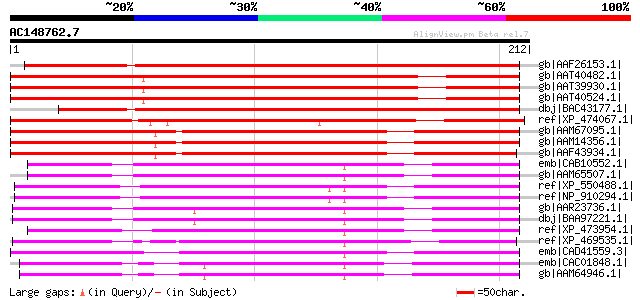
Score E
Sequences producing significant alignments: (bits) Value
gb|AAF26153.1| unknown protein [Arabidopsis thaliana] 218 1e-55
gb|AAT40482.1| hypothetical protein [Solanum demissum] 197 2e-49
gb|AAT39930.1| putative RNA recognition motif containing protein... 196 3e-49
gb|AAT40524.1| putative RNA recognition motif containing protein... 196 4e-49
dbj|BAC43177.1| unknown protein [Arabidopsis thaliana] gi|283729... 194 2e-48
ref|XP_474067.1| OSJNBb0079B02.9 [Oryza sativa (japonica cultiva... 183 2e-45
gb|AAM67095.1| unknown [Arabidopsis thaliana] 177 2e-43
gb|AAM14356.1| unknown protein [Arabidopsis thaliana] gi|1529297... 177 2e-43
gb|AAF43934.1| Contains similarity to Vip1 protein from Schizosa... 175 9e-43
emb|CAB10552.1| hypothetical protein [Arabidopsis thaliana] gi|7... 118 1e-25
gb|AAM65507.1| putative splicing regulatory protein [Arabidopsis... 118 1e-25
ref|XP_550488.1| putative RNA recognition motif (RRM)-containing... 117 2e-25
ref|NP_910294.1| ESTs D15336(C0474),C98053(C0474) correspond to ... 117 2e-25
gb|AAR23736.1| At5g46870 [Arabidopsis thaliana] gi|30695063|ref|... 114 2e-24
dbj|BAA97221.1| unnamed protein product [Arabidopsis thaliana] 114 2e-24
ref|XP_473954.1| OSJNBa0053K19.20 [Oryza sativa (japonica cultiv... 105 7e-22
ref|XP_469535.1| putative splicing regulatory protein [Oryza sat... 105 9e-22
emb|CAD41559.3| OSJNBa0006A01.14 [Oryza sativa (japonica cultiva... 105 1e-21
emb|CAC01848.1| putative protein [Arabidopsis thaliana] gi|15237... 104 1e-21
gb|AAM64946.1| unknown [Arabidopsis thaliana] 104 1e-21
>gb|AAF26153.1| unknown protein [Arabidopsis thaliana]
Length = 249
Score = 218 bits (554), Expect = 1e-55
Identities = 112/202 (55%), Positives = 147/202 (72%), Gaps = 3/202 (1%)
Query: 7 TAYVTFRDAYALETALLLNGSMILDRCISISRWETYTDDSNNWNNLTPNHEDSITYSQMD 66
TAYVTFRDAYAL+ A+LL+G+ I+D+ + IS + Y +SNN L + S+T ++ D
Sbjct: 46 TAYVTFRDAYALDMAVLLSGATIVDQTVWISVYGVYLHESNN---LRQEEDYSVTVTRSD 102
Query: 67 KFVSSPGEALTMAQQVVKTMVAKGFVLSKDAFVMAKAFDESRSVSSTAANKVAELSNKIG 126
F SSPGEA+T+AQQVV+TM+AKG+VLSKDA AKA DES+ SS A K+AE+S+ +G
Sbjct: 103 AFASSPGEAITVAQQVVQTMLAKGYVLSKDAIGKAKALDESQRFSSLVATKLAEISHYLG 162
Query: 127 LTETINSGIETFKSVDEKYHVTDITKSAATVTGTTAIVVATVTGRAAMAAGSAIANSSYF 186
LT+ I S +E +S DEKYH +D TKSA VTGT A+ AT+TG+ A AA +++ NS YF
Sbjct: 163 LTQNIQSSMELVRSADEKYHFSDFTKSAVLVTGTAAVAAATITGKVAAAAATSVVNSRYF 222
Query: 187 AKGALWVSDMLSRAAKSTADLG 208
A GALW SD L RAAK+ A +G
Sbjct: 223 ANGALWFSDALGRAAKAAAHIG 244
>gb|AAT40482.1| hypothetical protein [Solanum demissum]
Length = 244
Score = 197 bits (501), Expect = 2e-49
Identities = 104/210 (49%), Positives = 148/210 (69%), Gaps = 13/210 (6%)
Query: 1 SSDYESTAYVTFRDAYALETALLLNGSMILDRCISISRWETYTDDSNNWNNLT--PNHED 58
+ ++ STAYVTFR+ +ALETA+LL+G+ ILD+ + I+ W Y D+ + WN+ + P +
Sbjct: 40 AGEHASTAYVTFRNPHALETAVLLSGAAILDQRVCITSWGHYQDEFDYWNHSSWKPQEDC 99
Query: 59 SITYSQMDKFVSSPGEALTMAQQVVKTMVAKGFVLSKDAFVMAKAFDESRSVSSTAANKV 118
+ SQ FVSS GEA+T+ Q VVKTM+++G+VL K A AKAFDES +S+TA +KV
Sbjct: 100 HSSDSQGHYFVSSAGEAVTLTQDVVKTMLSQGYVLGKGALGKAKAFDESHGLSATAVSKV 159
Query: 119 AELSNKIGLTETINSGIETFKSVDEKYHVTDITKSAATVTGTTAIVVATVTGRAAMAAGS 178
A+LS +IGLT+ +G+E +SVD++YH++D T+SA + TG TAI AT
Sbjct: 160 ADLSERIGLTDKFCAGVEVARSVDQRYHISDTTRSAVSATGRTAISAAT----------- 208
Query: 179 AIANSSYFAKGALWVSDMLSRAAKSTADLG 208
A+ NSSYF+KGALW+S LS+AA++ ADLG
Sbjct: 209 AVVNSSYFSKGALWMSGALSKAAQAAADLG 238
>gb|AAT39930.1| putative RNA recognition motif containing protein [Solanum
demissum]
Length = 244
Score = 196 bits (499), Expect = 3e-49
Identities = 103/210 (49%), Positives = 148/210 (70%), Gaps = 13/210 (6%)
Query: 1 SSDYESTAYVTFRDAYALETALLLNGSMILDRCISISRWETYTDDSNNWNNLT--PNHED 58
+ ++ STAYVTFR+ +ALETA+LL+G+ ILD+ + I+ W Y D+ + WN+ + P +
Sbjct: 40 AGEHASTAYVTFRNPHALETAVLLSGAAILDQRVCITSWGHYQDEFDYWNHSSWKPQEDC 99
Query: 59 SITYSQMDKFVSSPGEALTMAQQVVKTMVAKGFVLSKDAFVMAKAFDESRSVSSTAANKV 118
+ SQ FVSS GEA+T+ Q VVKTM+++G+VL K A AKAFDES +S+TA +KV
Sbjct: 100 HSSDSQGHYFVSSAGEAVTLTQDVVKTMLSQGYVLGKGALGKAKAFDESHGLSATAVSKV 159
Query: 119 AELSNKIGLTETINSGIETFKSVDEKYHVTDITKSAATVTGTTAIVVATVTGRAAMAAGS 178
A++S +IGLT+ +G+E +SVD++YH++D T+SA + TG TAI AT
Sbjct: 160 ADISERIGLTDKFCAGVEVARSVDQRYHISDTTRSAVSATGRTAISAAT----------- 208
Query: 179 AIANSSYFAKGALWVSDMLSRAAKSTADLG 208
A+ NSSYF+KGALW+S LS+AA++ ADLG
Sbjct: 209 AVVNSSYFSKGALWMSGALSKAAQAAADLG 238
>gb|AAT40524.1| putative RNA recognition motif containing protein [Solanum
demissum]
Length = 244
Score = 196 bits (498), Expect = 4e-49
Identities = 103/210 (49%), Positives = 148/210 (70%), Gaps = 13/210 (6%)
Query: 1 SSDYESTAYVTFRDAYALETALLLNGSMILDRCISISRWETYTDDSNNWNNLT--PNHED 58
+ ++ STAYVTF++ +ALETA+LL+G+ ILD+ + I+ W Y D+ + WN+ + P +
Sbjct: 40 AGEHASTAYVTFKNPHALETAVLLSGAAILDQRVCITSWGHYQDEFDYWNHSSWKPQEDC 99
Query: 59 SITYSQMDKFVSSPGEALTMAQQVVKTMVAKGFVLSKDAFVMAKAFDESRSVSSTAANKV 118
+ SQ FVSS GEA+T+ Q VVKTM+++G+VL K A AKAFDES +S+TA +KV
Sbjct: 100 HSSDSQGHYFVSSAGEAVTLTQDVVKTMLSQGYVLGKGALGKAKAFDESHGLSATAVSKV 159
Query: 119 AELSNKIGLTETINSGIETFKSVDEKYHVTDITKSAATVTGTTAIVVATVTGRAAMAAGS 178
A+LS +IGLT+ +G+E +SVD++YH++D T+SA + TG TAI AT
Sbjct: 160 ADLSERIGLTDKFCAGVEVARSVDQRYHISDTTRSAVSATGRTAISAAT----------- 208
Query: 179 AIANSSYFAKGALWVSDMLSRAAKSTADLG 208
A+ NSSYF+KGALW+S LS+AA++ ADLG
Sbjct: 209 AVVNSSYFSKGALWMSGALSKAAQAAADLG 238
>dbj|BAC43177.1| unknown protein [Arabidopsis thaliana] gi|28372926|gb|AAO39945.1|
At3g01210 [Arabidopsis thaliana]
gi|30678301|ref|NP_186770.2| RNA recognition motif
(RRM)-containing protein [Arabidopsis thaliana]
Length = 191
Score = 194 bits (492), Expect = 2e-48
Identities = 100/188 (53%), Positives = 134/188 (71%), Gaps = 3/188 (1%)
Query: 21 ALLLNGSMILDRCISISRWETYTDDSNNWNNLTPNHEDSITYSQMDKFVSSPGEALTMAQ 80
A+LL+G+ I+D+ + IS + Y +SNN L + S+T ++ D F SSPGEA+T+AQ
Sbjct: 2 AVLLSGATIVDQTVWISVYGVYLHESNN---LRQEEDYSVTVTRSDAFASSPGEAITVAQ 58
Query: 81 QVVKTMVAKGFVLSKDAFVMAKAFDESRSVSSTAANKVAELSNKIGLTETINSGIETFKS 140
QVV+TM+AKG+VLSKDA AKA DES+ SS A K+AE+S+ +GLT+ I S +E +S
Sbjct: 59 QVVQTMLAKGYVLSKDAIGKAKALDESQRFSSLVATKLAEISHYLGLTQNIQSSMELVRS 118
Query: 141 VDEKYHVTDITKSAATVTGTTAIVVATVTGRAAMAAGSAIANSSYFAKGALWVSDMLSRA 200
DEKYH +D TKSA VTGT A+ AT+TG+ A AA +++ NS YFA GALW SD L RA
Sbjct: 119 ADEKYHFSDFTKSAVLVTGTAAVAAATITGKVAAAAATSVVNSRYFANGALWFSDALGRA 178
Query: 201 AKSTADLG 208
AK+ A +G
Sbjct: 179 AKAAAHIG 186
>ref|XP_474067.1| OSJNBb0079B02.9 [Oryza sativa (japonica cultivar-group)]
gi|38347315|emb|CAE05963.2| OSJNBa0063C18.4 [Oryza
sativa (japonica cultivar-group)]
gi|38344906|emb|CAD41850.2| OSJNBb0079B02.9 [Oryza
sativa (japonica cultivar-group)]
Length = 251
Score = 183 bits (465), Expect = 2e-45
Identities = 106/221 (47%), Positives = 146/221 (65%), Gaps = 24/221 (10%)
Query: 1 SSDYESTAYVTFRDAYALETALLLNGSMILDRCISISRWETYTDDSNNWNNLTPNH--ED 58
S +Y STAYVTF++ Y+LETA+LL+G+ I+D+ + I+RW + N W+ TPN E+
Sbjct: 40 SGEYGSTAYVTFKEPYSLETAVLLSGATIVDQPVCIARWGQPNEPYNFWD--TPNWYTEE 97
Query: 59 SITYS--QMDKFVSSPGEALTMAQQVVKTMVAKGFVLSKDAFVMAKAFDESRSVSSTAAN 116
I Y Q +F S+P EALT+AQ VVKTM+A+G+VLSKDA A+AFDES V++TAA
Sbjct: 98 EIEYRTYQTCQFNSTPQEALTIAQDVVKTMLARGYVLSKDALARARAFDESHQVTATAAA 157
Query: 117 KVAELSNKI-------GLTETINSGIETFKSVDEKYHVTDITKSAATVTGTTAIVVATVT 169
K AELS +I GLT+ +++G+ +SVDE YHV++ TK+ AT TG TA+ V
Sbjct: 158 KAAELSKRIGLTDRVSGLTDRVSAGVGAIRSVDETYHVSETTKTVATATGRTAVKVV--- 214
Query: 170 GRAAMAAGSAIANSSYFAKGALWVSDMLSRAAKSTADLGHH 210
+ I SSYF+ GA+ +SD L RAA++ ADL H
Sbjct: 215 --------NGIMTSSYFSAGAMMLSDALHRAAQAAADLAAH 247
>gb|AAM67095.1| unknown [Arabidopsis thaliana]
Length = 244
Score = 177 bits (449), Expect = 2e-43
Identities = 97/211 (45%), Positives = 136/211 (63%), Gaps = 16/211 (7%)
Query: 1 SSDYESTAYVTFRDAYALETALLLNGSMILDRCISISRWETYTDDSNNWNNLTPNHED-- 58
S + TAYV F+D+Y+ ETA+LL G+ ILD+ + I+RW + ++ + WN + ED
Sbjct: 41 SGEQACTAYVMFKDSYSQETAVLLTGATILDQRVCITRWGQHHEEFDFWNATSRGFEDES 100
Query: 59 -SITYSQMDKFVSSPGEALTMAQQVVKTMVAKGFVLSKDAFVMAKAFDESRSVSSTAANK 117
S Y+Q +F + GEA+T AQ+VVK M+A GFVL KDA AKAFDES VS+ A +
Sbjct: 101 DSHHYAQRSEF--NAGEAVTKAQEVVKIMLATGFVLGKDALSKAKAFDESHGVSAAAVAR 158
Query: 118 VAELSNKIGLTETINSGIETFKSVDEKYHVTDITKSAATVTGTTAIVVATVTGRAAMAAG 177
V++L +IGLT+ I +G+E + D++YHV+D KSA TGR A AA
Sbjct: 159 VSQLEQRIGLTDKIFTGLEAVRMTDQRYHVSDTAKSA-----------VFATGRTAAAAA 207
Query: 178 SAIANSSYFAKGALWVSDMLSRAAKSTADLG 208
+++ NSSYF+ GALW+S L RAAK+ +DLG
Sbjct: 208 TSVVNSSYFSSGALWLSGALERAAKAASDLG 238
>gb|AAM14356.1| unknown protein [Arabidopsis thaliana] gi|15292971|gb|AAK93596.1|
unknown protein [Arabidopsis thaliana]
gi|18394102|ref|NP_563946.1| RNA recognition motif
(RRM)-containing protein [Arabidopsis thaliana]
Length = 244
Score = 177 bits (449), Expect = 2e-43
Identities = 97/211 (45%), Positives = 136/211 (63%), Gaps = 16/211 (7%)
Query: 1 SSDYESTAYVTFRDAYALETALLLNGSMILDRCISISRWETYTDDSNNWNNLTPNHED-- 58
S + TAYV F+D+Y+ ETA+LL G+ ILD+ + I+RW + ++ + WN + ED
Sbjct: 41 SGEQACTAYVMFKDSYSQETAVLLTGATILDQRVCITRWGQHHEEFDFWNATSRGFEDES 100
Query: 59 -SITYSQMDKFVSSPGEALTMAQQVVKTMVAKGFVLSKDAFVMAKAFDESRSVSSTAANK 117
S Y+Q +F + GEA+T AQ+VVK M+A GFVL KDA AKAFDES VS+ A +
Sbjct: 101 DSQHYAQRSEF--NAGEAVTKAQEVVKIMLATGFVLGKDALSKAKAFDESHGVSAAAVAR 158
Query: 118 VAELSNKIGLTETINSGIETFKSVDEKYHVTDITKSAATVTGTTAIVVATVTGRAAMAAG 177
V++L +IGLT+ I +G+E + D++YHV+D KSA TGR A AA
Sbjct: 159 VSQLEQRIGLTDKIFTGLEAVRMTDQRYHVSDTAKSA-----------VFATGRTAAAAA 207
Query: 178 SAIANSSYFAKGALWVSDMLSRAAKSTADLG 208
+++ NSSYF+ GALW+S L RAAK+ +DLG
Sbjct: 208 TSVVNSSYFSSGALWLSGALERAAKAASDLG 238
>gb|AAF43934.1| Contains similarity to Vip1 protein from Schizosaccharomyces pombe
gb|Y13635.1 and contains a RNA recognition motif
PF|00076. EST gb|AI996906 comes from this gene.
[Arabidopsis thaliana] gi|25518024|pir||G86277 F14L17.11
protein - Arabidopsis thaliana
Length = 431
Score = 175 bits (443), Expect = 9e-43
Identities = 96/210 (45%), Positives = 135/210 (63%), Gaps = 16/210 (7%)
Query: 1 SSDYESTAYVTFRDAYALETALLLNGSMILDRCISISRWETYTDDSNNWNNLTPNHED-- 58
S + TAYV F+D+Y+ ETA+LL G+ ILD+ + I+RW + ++ + WN + ED
Sbjct: 41 SGEQACTAYVMFKDSYSQETAVLLTGATILDQRVCITRWGQHHEEFDFWNATSRGFEDES 100
Query: 59 -SITYSQMDKFVSSPGEALTMAQQVVKTMVAKGFVLSKDAFVMAKAFDESRSVSSTAANK 117
S Y+Q +F + GEA+T AQ+VVK M+A GFVL KDA AKAFDES VS+ A +
Sbjct: 101 DSQHYAQRSEF--NAGEAVTKAQEVVKIMLATGFVLGKDALSKAKAFDESHGVSAAAVAR 158
Query: 118 VAELSNKIGLTETINSGIETFKSVDEKYHVTDITKSAATVTGTTAIVVATVTGRAAMAAG 177
V++L +IGLT+ I +G+E + D++YHV+D KSA TGR A AA
Sbjct: 159 VSQLEQRIGLTDKIFTGLEAVRMTDQRYHVSDTAKSA-----------VFATGRTAAAAA 207
Query: 178 SAIANSSYFAKGALWVSDMLSRAAKSTADL 207
+++ NSSYF+ GALW+S L RAAK+ +DL
Sbjct: 208 TSVVNSSYFSSGALWLSGALERAAKAASDL 237
>emb|CAB10552.1| hypothetical protein [Arabidopsis thaliana]
gi|7268525|emb|CAB78775.1| hypothetical protein
[Arabidopsis thaliana] gi|7485085|pir||C71447
hypothetical protein - Arabidopsis thaliana
Length = 321
Score = 118 bits (296), Expect = 1e-25
Identities = 74/205 (36%), Positives = 106/205 (51%), Gaps = 23/205 (11%)
Query: 8 AYVTFRDAYALETALLLNGSMILDRCISISRWETYTDDSNNWNNLTPNHEDSITYSQMDK 67
AYVTF+D ETA+LL+G+ I+D + +S Y L+P S+ +K
Sbjct: 53 AYVTFKDLQGAETAVLLSGATIVDSSVIVSMAPDY--------QLSPEALASLEPKDSNK 104
Query: 68 FVSSPGEALTMAQQVVKTMVAKGFVLSKDAFVMAKAFDESRSVSSTAANKVAELSNKIGL 127
+ L A+ VV +M+AKGF+L KDA AK+ DE ++STA+ KVA KIG
Sbjct: 105 SPKAGDSVLRKAEDVVSSMLAKGFILGKDAIAKAKSVDEKHQLTSTASAKVASFDKKIGF 164
Query: 128 TETINSGI----ETFKSVDEKYHVTDITKSAATVTGTTAIVVATVTGRAAMAAGSAIANS 183
T+ IN+G E + VD+KY V++ TKSA T AGSAI +
Sbjct: 165 TDKINTGTVVVGEKVREVDQKYQVSEKTKSAIAAAEQT-----------VSNAGSAIMKN 213
Query: 184 SYFAKGALWVSDMLSRAAKSTADLG 208
Y GA WV+ ++ AK+ ++G
Sbjct: 214 RYVLTGATWVTGAFNKVAKAAEEVG 238
>gb|AAM65507.1| putative splicing regulatory protein [Arabidopsis thaliana]
gi|28827694|gb|AAO50691.1| putative RRM-containing
protein [Arabidopsis thaliana]
gi|28393291|gb|AAO42073.1| putative RRM-containing
protein [Arabidopsis thaliana]
gi|18414951|ref|NP_567536.1| RNA recognition motif
(RRM)-containing protein [Arabidopsis thaliana]
Length = 313
Score = 118 bits (296), Expect = 1e-25
Identities = 74/205 (36%), Positives = 106/205 (51%), Gaps = 23/205 (11%)
Query: 8 AYVTFRDAYALETALLLNGSMILDRCISISRWETYTDDSNNWNNLTPNHEDSITYSQMDK 67
AYVTF+D ETA+LL+G+ I+D + +S Y L+P S+ +K
Sbjct: 45 AYVTFKDLQGAETAVLLSGATIVDSSVIVSMAPDY--------QLSPEALASLEPKDSNK 96
Query: 68 FVSSPGEALTMAQQVVKTMVAKGFVLSKDAFVMAKAFDESRSVSSTAANKVAELSNKIGL 127
+ L A+ VV +M+AKGF+L KDA AK+ DE ++STA+ KVA KIG
Sbjct: 97 SPKAGDSVLRKAEDVVSSMLAKGFILGKDAIAKAKSVDEKHQLTSTASAKVASFDKKIGF 156
Query: 128 TETINSGI----ETFKSVDEKYHVTDITKSAATVTGTTAIVVATVTGRAAMAAGSAIANS 183
T+ IN+G E + VD+KY V++ TKSA T AGSAI +
Sbjct: 157 TDKINTGTVVVGEKVREVDQKYQVSEKTKSAIAAAEQT-----------VSNAGSAIMKN 205
Query: 184 SYFAKGALWVSDMLSRAAKSTADLG 208
Y GA WV+ ++ AK+ ++G
Sbjct: 206 RYVLTGATWVTGAFNKVAKAAEEVG 230
>ref|XP_550488.1| putative RNA recognition motif (RRM)-containing protein [Oryza
sativa (japonica cultivar-group)]
gi|55295911|dbj|BAD67779.1| putative RNA recognition
motif (RRM)-containing protein [Oryza sativa (japonica
cultivar-group)]
Length = 261
Score = 117 bits (294), Expect = 2e-25
Identities = 79/210 (37%), Positives = 106/210 (49%), Gaps = 23/210 (10%)
Query: 3 DYESTAYVTFRDAYALETALLLNGSMILDRCISISRWETYTDDSNNWNNLTPNHEDSITY 62
++ AYVTF+D ETALLL+G+ I+D + I+ Y P Y
Sbjct: 43 EWSQVAYVTFKDPQGAETALLLSGATIVDLSVIIAPAPEYQPP--------PTSSAPPMY 94
Query: 63 SQMDKFVSSPGEALTMAQQVVKTMVAKGFVLSKDAFVMAKAFDESRSVSSTAANKVAELS 122
S VS + A+ VV TM+AKGF L KDA AKAFDE +STA KVA +
Sbjct: 95 SATSVPVSEDNNVVHKAEDVVSTMLAKGFTLGKDAVGKAKAFDEKHGFTSTAGAKVASID 154
Query: 123 NKIGLTE--TINSGI--ETFKSVDEKYHVTDITKSAATVTGTTAIVVATVTGRAAMAAGS 178
KIGL+E TI + I E K +D+K+ V+D TKSA + AGS
Sbjct: 155 RKIGLSEKFTIGTSIVNEKVKEMDQKFQVSDKTKSA-----------FAAAEQKVSTAGS 203
Query: 179 AIANSSYFAKGALWVSDMLSRAAKSTADLG 208
AI + Y GA WV++ ++ AK+ D+G
Sbjct: 204 AIMKNRYVFTGASWVTNAFNKVAKAATDVG 233
>ref|NP_910294.1| ESTs D15336(C0474),C98053(C0474) correspond to a region of the
predicted gene.~Similar to Arabidopsis thaliana DNA
chromosome 4, ESSA I contig fragment No. 9; hypothetical
protein. (Z97344) [Oryza sativa (japonica
cultivar-group)]
Length = 306
Score = 117 bits (294), Expect = 2e-25
Identities = 79/210 (37%), Positives = 106/210 (49%), Gaps = 23/210 (10%)
Query: 3 DYESTAYVTFRDAYALETALLLNGSMILDRCISISRWETYTDDSNNWNNLTPNHEDSITY 62
++ AYVTF+D ETALLL+G+ I+D + I+ Y P Y
Sbjct: 88 EWSQVAYVTFKDPQGAETALLLSGATIVDLSVIIAPAPEYQPP--------PTSSAPPMY 139
Query: 63 SQMDKFVSSPGEALTMAQQVVKTMVAKGFVLSKDAFVMAKAFDESRSVSSTAANKVAELS 122
S VS + A+ VV TM+AKGF L KDA AKAFDE +STA KVA +
Sbjct: 140 SATSVPVSEDNNVVHKAEDVVSTMLAKGFTLGKDAVGKAKAFDEKHGFTSTAGAKVASID 199
Query: 123 NKIGLTE--TINSGI--ETFKSVDEKYHVTDITKSAATVTGTTAIVVATVTGRAAMAAGS 178
KIGL+E TI + I E K +D+K+ V+D TKSA + AGS
Sbjct: 200 RKIGLSEKFTIGTSIVNEKVKEMDQKFQVSDKTKSA-----------FAAAEQKVSTAGS 248
Query: 179 AIANSSYFAKGALWVSDMLSRAAKSTADLG 208
AI + Y GA WV++ ++ AK+ D+G
Sbjct: 249 AIMKNRYVFTGASWVTNAFNKVAKAATDVG 278
>gb|AAR23736.1| At5g46870 [Arabidopsis thaliana] gi|30695063|ref|NP_199498.2| RNA
recognition motif (RRM)-containing protein [Arabidopsis
thaliana] gi|45592926|gb|AAS68117.1| At5g46870
[Arabidopsis thaliana]
Length = 295
Score = 114 bits (284), Expect = 2e-24
Identities = 76/215 (35%), Positives = 106/215 (48%), Gaps = 27/215 (12%)
Query: 2 SDYESTAYVTFRDAYALETALLLNGSMILDRCISISRWETYTDDSNNWNNLTPNHEDSIT 61
+D AYVTF+D ETA+LL GS I+D ++++ Y L P+ SI
Sbjct: 39 NDGSKLAYVTFKDLQGAETAVLLTGSTIVDSSVTVTMSPDY--------QLPPDALASIE 90
Query: 62 YSQMDKFVSSPGE----ALTMAQQVVKTMVAKGFVLSKDAFVMAKAFDESRSVSSTAANK 117
+ SSP A+ VV M++KGFVL KDA AK+ DE ++STA+ +
Sbjct: 91 SLKESNKSSSPTREDVSVFRKAEDVVSGMISKGFVLGKDAIAKAKSLDEKHQLTSTASAR 150
Query: 118 VAELSNKIGLTETINSGI----ETFKSVDEKYHVTDITKSAATVTGTTAIVVATVTGRAA 173
V +IG TE IN+G E K VD+K+ VT+ TKSA T
Sbjct: 151 VTSFDKRIGFTEKINTGTTVVSEKVKEVDQKFQVTEKTKSAIAAAEQT-----------V 199
Query: 174 MAAGSAIANSSYFAKGALWVSDMLSRAAKSTADLG 208
AGSAI + Y GA WV+ +R +K+ ++G
Sbjct: 200 SNAGSAIMKNRYVLTGATWVTGAFNRVSKAAEEVG 234
>dbj|BAA97221.1| unnamed protein product [Arabidopsis thaliana]
Length = 293
Score = 114 bits (284), Expect = 2e-24
Identities = 76/215 (35%), Positives = 106/215 (48%), Gaps = 27/215 (12%)
Query: 2 SDYESTAYVTFRDAYALETALLLNGSMILDRCISISRWETYTDDSNNWNNLTPNHEDSIT 61
+D AYVTF+D ETA+LL GS I+D ++++ Y L P+ SI
Sbjct: 37 NDGSKLAYVTFKDLQGAETAVLLTGSTIVDSSVTVTMSPDY--------QLPPDALASIE 88
Query: 62 YSQMDKFVSSPGE----ALTMAQQVVKTMVAKGFVLSKDAFVMAKAFDESRSVSSTAANK 117
+ SSP A+ VV M++KGFVL KDA AK+ DE ++STA+ +
Sbjct: 89 SLKESNKSSSPTREDVSVFRKAEDVVSGMISKGFVLGKDAIAKAKSLDEKHQLTSTASAR 148
Query: 118 VAELSNKIGLTETINSGI----ETFKSVDEKYHVTDITKSAATVTGTTAIVVATVTGRAA 173
V +IG TE IN+G E K VD+K+ VT+ TKSA T
Sbjct: 149 VTSFDKRIGFTEKINTGTTVVSEKVKEVDQKFQVTEKTKSAIAAAEQT-----------V 197
Query: 174 MAAGSAIANSSYFAKGALWVSDMLSRAAKSTADLG 208
AGSAI + Y GA WV+ +R +K+ ++G
Sbjct: 198 SNAGSAIMKNRYVLTGATWVTGAFNRVSKAAEEVG 232
>ref|XP_473954.1| OSJNBa0053K19.20 [Oryza sativa (japonica cultivar-group)]
gi|38344181|emb|CAE03512.2| OSJNBa0053K19.20 [Oryza
sativa (japonica cultivar-group)]
Length = 376
Score = 105 bits (263), Expect = 7e-22
Identities = 70/205 (34%), Positives = 98/205 (47%), Gaps = 27/205 (13%)
Query: 8 AYVTFRDAYALETALLLNGSMILDRCISISRWETYTDDSNNWNNLTPNHEDSITYSQMDK 67
AY+TF D E A+LL G+ I+D + I+ Y + ++ K
Sbjct: 44 AYITFEDDEGAERAMLLTGATIVDMSVIITPATNYQLPA------------AVLADIESK 91
Query: 68 FVSSPGEALTMAQQVVKTMVAKGFVLSKDAFVMAKAFDESRSVSSTAANKVAELSNKIGL 127
AL A+ V +M+AKGFVL KDA AK+FDE ++STA KV L K+GL
Sbjct: 92 NAGGMESALRKAEDAVVSMLAKGFVLGKDALERAKSFDEKHQLTSTATAKVTSLDRKMGL 151
Query: 128 TETINSGI----ETFKSVDEKYHVTDITKSAATVTGTTAIVVATVTGRAAMAAGSAIANS 183
++ ++G E K +D+KY V + TKSA T AGSAI ++
Sbjct: 152 SQKFSTGTLVVNEKMKEMDQKYQVAEKTKSALAAAEQT-----------VSTAGSAIMSN 200
Query: 184 SYFAKGALWVSDMLSRAAKSTADLG 208
Y GA WV+D S+ A + D G
Sbjct: 201 RYILTGAAWVTDAYSKVATTATDAG 225
>ref|XP_469535.1| putative splicing regulatory protein [Oryza sativa (japonica
cultivar-group)] gi|18071362|gb|AAL58221.1| putative
splicing regulatory protein [Oryza sativa (japonica
cultivar-group)]
Length = 284
Score = 105 bits (262), Expect = 9e-22
Identities = 72/210 (34%), Positives = 109/210 (51%), Gaps = 27/210 (12%)
Query: 2 SDYESTAYVTFRDAYALETALLLNGSMILDRCISISRWETYTDDSNNWNNLTPNHEDSIT 61
S+ AYVTF+D+ +TA+LL+G+ I+DR + I+ Y L P D+
Sbjct: 39 SERSQLAYVTFKDSQGADTAVLLSGATIVDRSVIITPVVNY--------QLPP---DARK 87
Query: 62 YSQMDKFVSSPGEALTMAQQVVKTMVAKGFVLSKDAFVMAKAFDESRSVSSTAANKVAEL 121
S +K SS + A+ VV +M+AKGFVLSKDA +A++FDE ++ S A VA L
Sbjct: 88 QSAGEKS-SSAESVVRKAEDVVSSMLAKGFVLSKDALNVARSFDERHNILSNATATVASL 146
Query: 122 SNKIGLTETINSGI----ETFKSVDEKYHVTDITKSAATVTGTTAIVVATVTGRAAMAAG 177
+ G++E I+ G K VD++Y V+++TKSA A + A
Sbjct: 147 DRQYGVSEKISLGRAIVGSKVKEVDDRYQVSELTKSALAAAEQKASI-----------AS 195
Query: 178 SAIANSSYFAKGALWVSDMLSRAAKSTADL 207
SAI N+ Y + GA W++ K+ D+
Sbjct: 196 SAIMNNQYVSAGASWLTSAFGMVTKAAGDM 225
>emb|CAD41559.3| OSJNBa0006A01.14 [Oryza sativa (japonica cultivar-group)]
gi|32489377|emb|CAE04149.1| OSJNBa0009P12.34 [Oryza
sativa (japonica cultivar-group)]
Length = 314
Score = 105 bits (261), Expect = 1e-21
Identities = 67/212 (31%), Positives = 104/212 (48%), Gaps = 29/212 (13%)
Query: 1 SSDYESTAYVTFRDAYALETALLLNGSMILDRCISISRWETYTDDSNNWNNLTPNHEDSI 60
S + AY+TF+D ETA+LL G+ I+D + ++ Y ++ L P
Sbjct: 41 SDELSQVAYITFKDNQGSETAMLLTGATIVDMAVIVTPATDYELPASVLAALEPK----- 95
Query: 61 TYSQMDKFVSSPGEALTMAQQVVKTMVAKGFVLSKDAFVMAKAFDESRSVSSTAANKVAE 120
S AL A+ +V TM+AKGF+L +DA AKA DE ++STA +V+
Sbjct: 96 ---------DSKPSALQKAEDIVGTMLAKGFILGRDALDRAKALDEKHQLTSTATARVSS 146
Query: 121 LSNKIGLTETINSGI----ETFKSVDEKYHVTDITKSAATVTGTTAIVVATVTGRAAMAA 176
K+GL+E I+ G + K +D+KY V++ T+SA ++ A
Sbjct: 147 FDKKMGLSEKISVGTSAVNDKVKEMDQKYQVSEKTRSA-----------LAAAEQSVSTA 195
Query: 177 GSAIANSSYFAKGALWVSDMLSRAAKSTADLG 208
GSAI + Y GA WV+ ++ A + D+G
Sbjct: 196 GSAIMKNRYVLTGAAWVTGAFNKVANAANDVG 227
>emb|CAC01848.1| putative protein [Arabidopsis thaliana]
gi|15237419|ref|NP_197186.1| RNA recognition motif
(RRM)-containing protein [Arabidopsis thaliana]
gi|11357808|pir||T51516 hypothetical protein F5E19_180 -
Arabidopsis thaliana
Length = 259
Score = 104 bits (260), Expect = 1e-21
Identities = 70/209 (33%), Positives = 106/209 (50%), Gaps = 32/209 (15%)
Query: 5 ESTAYVTFRDAYALETALLLNGSMILDRCISISRWETYTDDSNNWNNLTPNHEDSITYSQ 64
E +AYVTF++ ETA+LL+G+ I D+ + I Y+ + P+ E
Sbjct: 41 EHSAYVTFKETQGAETAVLLSGASIADQSVIIELAPNYSPPA------APHAETQ----- 89
Query: 65 MDKFVSSPGEALTM-AQQVVKTMVAKGFVLSKDAFVMAKAFDESRSVSSTAANKVAELSN 123
SS E++ A+ VV +M+AKGF+L KDA AKAFDE +STA VA L
Sbjct: 90 -----SSGAESVVQKAEDVVSSMLAKGFILGKDAVGKAKAFDEKHGFTSTATAGVASLDQ 144
Query: 124 KIGLTETINSGI----ETFKSVDEKYHVTDITKSAATVTGTTAIVVATVTGRAAMAAGSA 179
KIGL++ + +G E K+VD+ + VT+ TKS V + +AGSA
Sbjct: 145 KIGLSQKLTAGTSLVNEKIKAVDQNFQVTERTKS-----------VYAAAEQTVSSAGSA 193
Query: 180 IANSSYFAKGALWVSDMLSRAAKSTADLG 208
+ + Y G W + +R A++ ++G
Sbjct: 194 VMKNRYVLTGVSWAAGAFNRVAQAAGEVG 222
>gb|AAM64946.1| unknown [Arabidopsis thaliana]
Length = 259
Score = 104 bits (260), Expect = 1e-21
Identities = 70/209 (33%), Positives = 106/209 (50%), Gaps = 32/209 (15%)
Query: 5 ESTAYVTFRDAYALETALLLNGSMILDRCISISRWETYTDDSNNWNNLTPNHEDSITYSQ 64
E +AYVTF++ ETA+LL+G+ I D+ + I Y+ + P+ E
Sbjct: 41 EHSAYVTFKETQGAETAVLLSGASIADQSVIIELAPNYSPPA------APHAETQ----- 89
Query: 65 MDKFVSSPGEALTM-AQQVVKTMVAKGFVLSKDAFVMAKAFDESRSVSSTAANKVAELSN 123
SS E++ A+ VV +M+AKGF+L KDA AKAFDE +STA VA L
Sbjct: 90 -----SSGAESVVQKAEDVVSSMLAKGFILGKDAVGKAKAFDEKHGFTSTATAGVASLDQ 144
Query: 124 KIGLTETINSGI----ETFKSVDEKYHVTDITKSAATVTGTTAIVVATVTGRAAMAAGSA 179
KIGL++ + +G E K+VD+ + VT+ TKS V + +AGSA
Sbjct: 145 KIGLSQKLTAGTSLVNEKIKAVDQNFQVTERTKS-----------VYAAAEQTVSSAGSA 193
Query: 180 IANSSYFAKGALWVSDMLSRAAKSTADLG 208
+ + Y G W + +R A++ ++G
Sbjct: 194 VMKNRYVLTGVSWAAGAFNRVAQAAGEVG 222
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.312 0.122 0.333
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 292,756,752
Number of Sequences: 2540612
Number of extensions: 9672071
Number of successful extensions: 31675
Number of sequences better than 10.0: 112
Number of HSP's better than 10.0 without gapping: 40
Number of HSP's successfully gapped in prelim test: 73
Number of HSP's that attempted gapping in prelim test: 31449
Number of HSP's gapped (non-prelim): 210
length of query: 212
length of database: 863,360,394
effective HSP length: 122
effective length of query: 90
effective length of database: 553,405,730
effective search space: 49806515700
effective search space used: 49806515700
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 72 (32.3 bits)
Medicago: description of AC148762.7


