

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148762.6 + phase: 0
(93 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
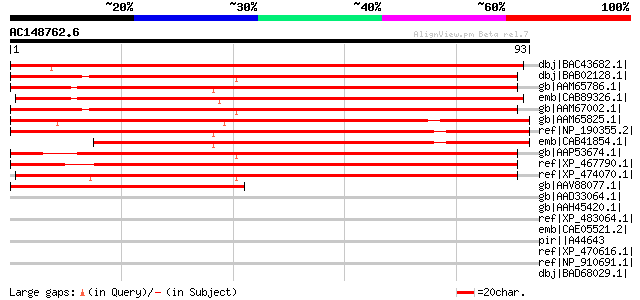
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAC43682.1| unknown protein [Arabidopsis thaliana] gi|284168... 132 3e-30
dbj|BAB02128.1| DNA-binding protein-like [Arabidopsis thaliana] ... 120 6e-27
gb|AAM65786.1| DNA-binding protein-like [Arabidopsis thaliana] g... 120 8e-27
emb|CAB89326.1| putative protein [Arabidopsis thaliana] gi|26452... 118 3e-26
gb|AAM67002.1| DNA-binding protein-like [Arabidopsis thaliana] 118 3e-26
gb|AAM65825.1| putative DNA-binding protein [Arabidopsis thalian... 117 7e-26
ref|NP_190355.2| bHLH family protein [Arabidopsis thaliana] 106 1e-22
emb|CAB41854.1| hypothetical protein [Arabidopsis thaliana] gi|7... 93 1e-18
gb|AAP53674.1| hypothetical protein similar to putative DNA bind... 90 1e-17
ref|XP_467790.1| DNA binding protein-like [Oryza sativa (japonic... 88 6e-17
ref|XP_474070.1| OSJNBb0079B02.12 [Oryza sativa (japonica cultiv... 82 3e-15
gb|AAV88077.1| hypothetical bHLH family protein [Ipomoea batatas] 59 3e-08
gb|AAD33064.1| M protein [Streptococcus pyogenes] 38 0.055
gb|AAH45420.1| Zgc:55668 [Danio rerio] gi|62511041|sp|Q7ZVT3|SAS... 37 0.16
ref|XP_483064.1| bHLH transcription factor-like [Oryza sativa (j... 35 0.36
emb|CAE05521.2| OSJNBa0038P21.14 [Oryza sativa (japonica cultiva... 35 0.36
pir||A44643 M protein precursor - Streptococcus pyogenes (seroty... 35 0.47
ref|XP_470616.1| Hypothetical protein [Oryza sativa (japonica cu... 35 0.61
ref|NP_910691.1| hypothetical protein [Oryza sativa (japonica cu... 34 0.80
dbj|BAD68029.1| putative TA1 protein [Oryza sativa (japonica cul... 34 0.80
>dbj|BAC43682.1| unknown protein [Arabidopsis thaliana] gi|28416803|gb|AAO42932.1|
At1g26948 [Arabidopsis thaliana]
gi|30689711|ref|NP_849712.1| expressed protein
[Arabidopsis thaliana]
Length = 94
Score = 132 bits (331), Expect = 3e-30
Identities = 71/93 (76%), Positives = 80/93 (85%), Gaps = 1/93 (1%)
Query: 1 MSSRRS-RQQSGGSTRISDDQIIDLVCKLRQLVPEIRNRRSDKVPASKVLQETCNYIRNL 59
MSSRRS R + GS+RISDDQI DLV KL+ L+PE+R RRSDKV ASKVLQETCNYIRNL
Sbjct: 1 MSSRRSSRSRQSGSSRISDDQISDLVSKLQHLIPELRRRRSDKVSASKVLQETCNYIRNL 60
Query: 60 QREVDDLSLRLSQLLATIDSDSAEASIIRSLLN 92
REVDDLS RLS+LLA+ D +SAEA+IIRSLLN
Sbjct: 61 HREVDDLSDRLSELLASTDDNSAEAAIIRSLLN 93
>dbj|BAB02128.1| DNA-binding protein-like [Arabidopsis thaliana]
gi|42572553|ref|NP_974372.1| expressed protein
[Arabidopsis thaliana]
Length = 92
Score = 120 bits (302), Expect = 6e-27
Identities = 68/92 (73%), Positives = 76/92 (81%), Gaps = 2/92 (2%)
Query: 1 MSSRRSRQQSGGSTRISDDQIIDLVCKLRQLVPEIRNRR-SDKVPASKVLQETCNYIRNL 59
MS+RRSRQ S S RISDDQ+IDLV KLRQ +PEI RR SDKV ASKVLQETCNYIR L
Sbjct: 1 MSNRRSRQTSNAS-RISDDQMIDLVSKLRQFLPEIHERRRSDKVSASKVLQETCNYIRKL 59
Query: 60 QREVDDLSLRLSQLLATIDSDSAEASIIRSLL 91
REVD+LS RLSQLL ++D DS EA++IRSLL
Sbjct: 60 HREVDNLSDRLSQLLDSVDEDSPEAAVIRSLL 91
>gb|AAM65786.1| DNA-binding protein-like [Arabidopsis thaliana]
gi|10176978|dbj|BAB10210.1| DNA-binding protein-like
[Arabidopsis thaliana] gi|15242499|ref|NP_198802.1|
bHLH protein [Arabidopsis thaliana]
Length = 92
Score = 120 bits (301), Expect = 8e-27
Identities = 66/92 (71%), Positives = 78/92 (84%), Gaps = 2/92 (2%)
Query: 1 MSSRRSRQQSGGSTRISDDQIIDLVCKLRQLVPEI-RNRRSDKVPASKVLQETCNYIRNL 59
MS+RRSRQ S + RISD+Q+IDLV KLRQ++PEI + RRSDK ASKVLQETCNYIRNL
Sbjct: 1 MSNRRSRQSSS-APRISDNQMIDLVSKLRQILPEIGQRRRSDKASASKVLQETCNYIRNL 59
Query: 60 QREVDDLSLRLSQLLATIDSDSAEASIIRSLL 91
REVD+LS RLSQLL ++D DS EA++IRSLL
Sbjct: 60 NREVDNLSERLSQLLESVDEDSPEAAVIRSLL 91
>emb|CAB89326.1| putative protein [Arabidopsis thaliana]
gi|26452783|dbj|BAC43472.1| unknown protein
[Arabidopsis thaliana] gi|28973111|gb|AAO63880.1|
putative bHLH protein [Arabidopsis thaliana]
gi|15242227|ref|NP_197020.1| bHLH family protein
[Arabidopsis thaliana] gi|11357892|pir||T49951
hypothetical protein F8M21.50 - Arabidopsis thaliana
Length = 94
Score = 118 bits (296), Expect = 3e-26
Identities = 65/92 (70%), Positives = 77/92 (83%), Gaps = 2/92 (2%)
Query: 2 SSRRSRQQSGGSTRISDDQIIDLVCKLRQLVPEIR-NRRSDKVPASKVLQETCNYIRNLQ 60
SSRRSRQ S S+RISDDQI DL+ KLRQ +PEIR NRRS+ V ASKVLQETCNYIRNL
Sbjct: 3 SSRRSRQASS-SSRISDDQITDLISKLRQSIPEIRQNRRSNTVSASKVLQETCNYIRNLN 61
Query: 61 REVDDLSLRLSQLLATIDSDSAEASIIRSLLN 92
+E DDLS RL+QLL +ID +S +A++IRSL+N
Sbjct: 62 KEADDLSDRLTQLLESIDPNSPQAAVIRSLIN 93
>gb|AAM67002.1| DNA-binding protein-like [Arabidopsis thaliana]
Length = 92
Score = 118 bits (296), Expect = 3e-26
Identities = 67/92 (72%), Positives = 75/92 (80%), Gaps = 2/92 (2%)
Query: 1 MSSRRSRQQSGGSTRISDDQIIDLVCKLRQLVPEIRNRR-SDKVPASKVLQETCNYIRNL 59
MS+RRSRQ S S RISDDQ+IDLV KLRQ +PEI RR SDKV AS VLQETCNYIR L
Sbjct: 1 MSNRRSRQTSNAS-RISDDQMIDLVSKLRQFLPEIHERRRSDKVSASXVLQETCNYIRKL 59
Query: 60 QREVDDLSLRLSQLLATIDSDSAEASIIRSLL 91
REVD+LS RLSQLL ++D DS EA++IRSLL
Sbjct: 60 HREVDNLSDRLSQLLDSVDEDSPEAAVIRSLL 91
>gb|AAM65825.1| putative DNA-binding protein [Arabidopsis thaliana]
gi|27764980|gb|AAO23611.1| At1g74500 [Arabidopsis
thaliana] gi|15221264|ref|NP_177590.1| bHLH family
protein [Arabidopsis thaliana]
gi|12324788|gb|AAG52350.1| putative DNA-binding
protein; 54988-54618 [Arabidopsis thaliana]
gi|25406349|pir||A96774 probable DNA-binding protein
F1M20.18 [imported] - Arabidopsis thaliana
Length = 93
Score = 117 bits (293), Expect = 7e-26
Identities = 65/95 (68%), Positives = 83/95 (86%), Gaps = 4/95 (4%)
Query: 1 MSSRRSR-QQSGGSTRISDDQIIDLVCKLRQLVPEIRN-RRSDKVPASKVLQETCNYIRN 58
MS RRSR +QS G++RIS+DQI DL+ KL+QL+PE+R+ RRSDKV A++VLQ+TCNYIRN
Sbjct: 1 MSGRRSRSRQSSGTSRISEDQINDLIIKLQQLLPELRDSRRSDKVSAARVLQDTCNYIRN 60
Query: 59 LQREVDDLSLRLSQLLATIDSDSAEASIIRSLLNQ 93
L REVDDLS RLS+LLA +SD+A+A++IRSLL Q
Sbjct: 61 LHREVDDLSERLSELLA--NSDTAQAALIRSLLTQ 93
>ref|NP_190355.2| bHLH family protein [Arabidopsis thaliana]
Length = 92
Score = 106 bits (265), Expect = 1e-22
Identities = 59/94 (62%), Positives = 77/94 (81%), Gaps = 3/94 (3%)
Query: 1 MSSRRSRQQSGGSTRISDDQIIDLVCKLRQLVPEI-RNRRSDKVPASKVLQETCNYIRNL 59
MSSR+SR + G++ I+D+QI DLV +L +L+PE+ NRRS KV AS+VLQETC+YIRNL
Sbjct: 1 MSSRKSRSRQTGASMITDEQINDLVLQLHRLLPELANNRRSGKVSASRVLQETCSYIRNL 60
Query: 60 QREVDDLSLRLSQLLATIDSDSAEASIIRSLLNQ 93
+EVDDLS RLSQLL + +DSA+A++IRSLL Q
Sbjct: 61 SKEVDDLSERLSQLLES--TDSAQAALIRSLLMQ 92
>emb|CAB41854.1| hypothetical protein [Arabidopsis thaliana]
gi|7487374|pir||T07710 hypothetical protein T23J7.40 -
Arabidopsis thaliana
Length = 78
Score = 93.2 bits (230), Expect = 1e-18
Identities = 52/79 (65%), Positives = 66/79 (82%), Gaps = 3/79 (3%)
Query: 16 ISDDQIIDLVCKLRQLVPEI-RNRRSDKVPASKVLQETCNYIRNLQREVDDLSLRLSQLL 74
I+D+QI DLV +L +L+PE+ NRRS KV AS+VLQETC+YIRNL +EVDDLS RLSQLL
Sbjct: 2 ITDEQINDLVLQLHRLLPELANNRRSGKVSASRVLQETCSYIRNLSKEVDDLSERLSQLL 61
Query: 75 ATIDSDSAEASIIRSLLNQ 93
+ +DSA+A++IRSLL Q
Sbjct: 62 ES--TDSAQAALIRSLLMQ 78
>gb|AAP53674.1| hypothetical protein similar to putative DNA binding proteins
[Oryza sativa (japonica cultivar-group)]
gi|37534170|ref|NP_921387.1| hypothetical protein
similar to putative DNA binding proteins [Oryza sativa
(japonica cultivar-group)] gi|21671920|gb|AAM74282.1|
Hypothetical protein similar to putative DNA binding
proteins [Oryza sativa (japonica cultivar-group)]
Length = 88
Score = 90.1 bits (222), Expect = 1e-17
Identities = 50/93 (53%), Positives = 70/93 (74%), Gaps = 8/93 (8%)
Query: 1 MSSRRSRQQSGGSTRISDDQIIDLVCKLRQLVPEIRNRR--SDKVPASKVLQETCNYIRN 58
MS RR+ S RI+DD+I +L+ KL+ L+PE RR + + PA+K+L+E C+YI++
Sbjct: 1 MSGRRA------SGRITDDEINELISKLQSLLPESSRRRGATSRSPATKLLKEMCSYIKS 54
Query: 59 LQREVDDLSLRLSQLLATIDSDSAEASIIRSLL 91
L REVDDLS RLS+L+AT+DS+S +A IIRSLL
Sbjct: 55 LHREVDDLSERLSELMATMDSNSPQADIIRSLL 87
>ref|XP_467790.1| DNA binding protein-like [Oryza sativa (japonica cultivar-group)]
gi|46390936|dbj|BAD16450.1| DNA binding protein-like
[Oryza sativa (japonica cultivar-group)]
Length = 88
Score = 87.8 bits (216), Expect = 6e-17
Identities = 48/91 (52%), Positives = 66/91 (71%), Gaps = 5/91 (5%)
Query: 1 MSSRRSRQQSGGSTRISDDQIIDLVCKLRQLVPEIRNRRSDKVPASKVLQETCNYIRNLQ 60
MSSRRS + S IS+++I +L+ KL+ L+P R R S + +K+L+ETCNYI++L
Sbjct: 1 MSSRRSSRGS-----ISEEEINELISKLQSLLPNSRRRGSSQASTTKLLKETCNYIKSLH 55
Query: 61 REVDDLSLRLSQLLATIDSDSAEASIIRSLL 91
REVDDLS RLS L+AT+D +S A IIRS+L
Sbjct: 56 REVDDLSDRLSDLMATMDHNSPGAEIIRSIL 86
>ref|XP_474070.1| OSJNBb0079B02.12 [Oryza sativa (japonica cultivar-group)]
gi|32490047|emb|CAE05966.1| OSJNBa0063C18.7 [Oryza
sativa (japonica cultivar-group)]
gi|38344909|emb|CAD41853.2| OSJNBb0079B02.12 [Oryza
sativa (japonica cultivar-group)]
gi|10241623|emb|CAC09463.1| putative DNA binding protein
[Oryza sativa (indica cultivar-group)]
Length = 104
Score = 82.4 bits (202), Expect = 3e-15
Identities = 50/101 (49%), Positives = 70/101 (68%), Gaps = 11/101 (10%)
Query: 2 SSRRSRQQSGGS----------TRISDDQIIDLVCKLRQLVPEIRNRR-SDKVPASKVLQ 50
SSRRSR + GS T IS+DQI +L+ KL+ L+PE + R + + A++VLQ
Sbjct: 3 SSRRSRSRRAGSSVPSSSSSSRTSISEDQIAELLSKLQALLPESQARNGAHRGSAARVLQ 62
Query: 51 ETCNYIRNLQREVDDLSLRLSQLLATIDSDSAEASIIRSLL 91
ETC+YIR+L +EVD+LS L+QLLA+ D S +A++IRSLL
Sbjct: 63 ETCSYIRSLHQEVDNLSETLAQLLASPDVTSDQAAVIRSLL 103
>gb|AAV88077.1| hypothetical bHLH family protein [Ipomoea batatas]
Length = 198
Score = 58.9 bits (141), Expect = 3e-08
Identities = 29/42 (69%), Positives = 35/42 (83%)
Query: 1 MSSRRSRQQSGGSTRISDDQIIDLVCKLRQLVPEIRNRRSDK 42
MSSRRSR + ++RISDDQI DLV KL++L+PEIR RRSDK
Sbjct: 1 MSSRRSRSRQPETSRISDDQIADLVSKLQRLIPEIRGRRSDK 42
>gb|AAD33064.1| M protein [Streptococcus pyogenes]
Length = 270
Score = 38.1 bits (87), Expect = 0.055
Identities = 20/79 (25%), Positives = 38/79 (47%)
Query: 5 RSRQQSGGSTRISDDQIIDLVCKLRQLVPEIRNRRSDKVPASKVLQETCNYIRNLQREVD 64
++ +QS + QI DLV K +L E+ D+ ++V++ + LQ ++D
Sbjct: 187 KAEEQSNEQLEFKNQQIADLVGKKAELEMELAKAEEDRATYNRVIEGAKRELTELQAKLD 246
Query: 65 DLSLRLSQLLATIDSDSAE 83
+ L+ A +D+ AE
Sbjct: 247 ETKQELANQQAQLDASKAE 265
>gb|AAH45420.1| Zgc:55668 [Danio rerio] gi|62511041|sp|Q7ZVT3|SAS6_BRARE Spindle
assembly abnormal protein 6 homolog
gi|47087417|ref|NP_998603.1| hypothetical protein
LOC406747 [Danio rerio]
Length = 627
Score = 36.6 bits (83), Expect = 0.16
Identities = 19/48 (39%), Positives = 29/48 (59%)
Query: 24 LVCKLRQLVPEIRNRRSDKVPASKVLQETCNYIRNLQREVDDLSLRLS 71
L L+ L+ +I+ + S VP K+LQET + ++ QRE+ D RLS
Sbjct: 394 LQADLKALLGKIKVKNSVTVPQEKILQETSDKLQRQQRELQDTQQRLS 441
>ref|XP_483064.1| bHLH transcription factor-like [Oryza sativa (japonica
cultivar-group)] gi|42408258|dbj|BAD09414.1| bHLH
transcription factor-like [Oryza sativa (japonica
cultivar-group)]
Length = 365
Score = 35.4 bits (80), Expect = 0.36
Identities = 26/78 (33%), Positives = 43/78 (54%), Gaps = 10/78 (12%)
Query: 15 RISDDQIIDLVCKLRQLVPEIRNRRSDKVPASKVLQETCNYIRNLQREVDDLSLRLSQLL 74
R+ ++I + + L+ LVP S + +L E NY+++LQR+V+ LS++LS +
Sbjct: 192 RLRREKISERMKLLQDLVPGC----SKVTGKALMLDEIINYVQSLQRQVEFLSMKLSAVN 247
Query: 75 ATIDSDSAEASIIRSLLN 92
ID D I SL+N
Sbjct: 248 PRIDLD------IESLVN 259
>emb|CAE05521.2| OSJNBa0038P21.14 [Oryza sativa (japonica cultivar-group)]
Length = 263
Score = 35.4 bits (80), Expect = 0.36
Identities = 22/67 (32%), Positives = 42/67 (61%), Gaps = 4/67 (5%)
Query: 15 RISDDQIIDLVCKLRQLVPEIRNRRSDKVPASKVLQETCNYIRNLQREVDDLSLRLSQLL 74
R+ ++I + + L+ LVP N+ + K + +L E NY+++LQR+V+ LS++L+ +
Sbjct: 97 RVRRERISERMKLLQSLVPGC-NKITGK---ALMLDEIINYVQSLQRQVEFLSMKLATMN 152
Query: 75 ATIDSDS 81
+D DS
Sbjct: 153 PQLDFDS 159
>pir||A44643 M protein precursor - Streptococcus pyogenes (serotype M57)
(fragment)
Length = 501
Score = 35.0 bits (79), Expect = 0.47
Identities = 18/79 (22%), Positives = 38/79 (47%)
Query: 5 RSRQQSGGSTRISDDQIIDLVCKLRQLVPEIRNRRSDKVPASKVLQETCNYIRNLQREVD 64
++ +QS + QI DL+ K +L ++ D+ ++V++ + LQ ++D
Sbjct: 185 KAEEQSNEQLEFKNQQIADLIGKKAELEMKLAKAEEDRATYNRVIEGAKRELTELQAKLD 244
Query: 65 DLSLRLSQLLATIDSDSAE 83
+ L+ A +D+ AE
Sbjct: 245 ETKQELANQQAQLDAYKAE 263
>ref|XP_470616.1| Hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|27311243|gb|AAO00689.1| Hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 776
Score = 34.7 bits (78), Expect = 0.61
Identities = 22/66 (33%), Positives = 41/66 (61%), Gaps = 4/66 (6%)
Query: 15 RISDDQIIDLVCKLRQLVPEIRNRRSDKVPASKVLQETCNYIRNLQREVDDLSLRLSQLL 74
R ++I + + L+ LVP N+ + K + +L E NY+++LQR+V+ LS++LS +
Sbjct: 644 RFRREKINERMKLLQDLVPGC-NKITGK---AMMLDEIINYVQSLQRQVEFLSMKLSTIS 699
Query: 75 ATIDSD 80
++SD
Sbjct: 700 PELNSD 705
>ref|NP_910691.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 569
Score = 34.3 bits (77), Expect = 0.80
Identities = 20/64 (31%), Positives = 37/64 (57%), Gaps = 4/64 (6%)
Query: 15 RISDDQIIDLVCKLRQLVPEIRNRRSDKVPASKVLQETCNYIRNLQREVDDLSLRLSQLL 74
R+ ++I + + L+ LVP V +L E NY+++LQR+V+ LS++L+ +
Sbjct: 408 RVRREKISERMKYLQDLVPGCSKVTGKAV----MLDEIINYVQSLQRQVEFLSMKLASVN 463
Query: 75 ATID 78
T+D
Sbjct: 464 PTLD 467
>dbj|BAD68029.1| putative TA1 protein [Oryza sativa (japonica cultivar-group)]
Length = 437
Score = 34.3 bits (77), Expect = 0.80
Identities = 20/64 (31%), Positives = 37/64 (57%), Gaps = 4/64 (6%)
Query: 15 RISDDQIIDLVCKLRQLVPEIRNRRSDKVPASKVLQETCNYIRNLQREVDDLSLRLSQLL 74
R+ ++I + + L+ LVP V +L E NY+++LQR+V+ LS++L+ +
Sbjct: 276 RVRREKISERMKYLQDLVPGCSKVTGKAV----MLDEIINYVQSLQRQVEFLSMKLASVN 331
Query: 75 ATID 78
T+D
Sbjct: 332 PTLD 335
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.316 0.130 0.339
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 120,735,733
Number of Sequences: 2540612
Number of extensions: 3683104
Number of successful extensions: 15696
Number of sequences better than 10.0: 108
Number of HSP's better than 10.0 without gapping: 64
Number of HSP's successfully gapped in prelim test: 44
Number of HSP's that attempted gapping in prelim test: 15600
Number of HSP's gapped (non-prelim): 123
length of query: 93
length of database: 863,360,394
effective HSP length: 69
effective length of query: 24
effective length of database: 688,058,166
effective search space: 16513395984
effective search space used: 16513395984
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 68 (30.8 bits)
Medicago: description of AC148762.6


