

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148755.4 + phase: 1 /partial
(441 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
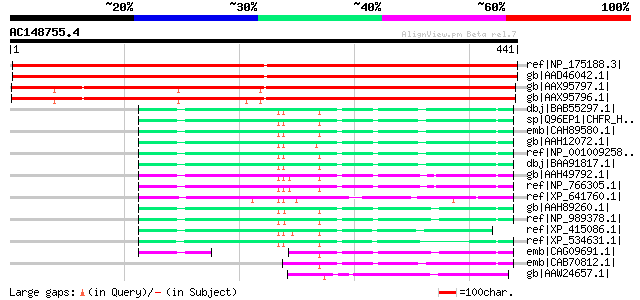
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_175188.3| zinc finger (C3HC4-type RING finger) family pro... 544 e-153
gb|AAD46042.1| F16N3.15 [Arabidopsis thaliana] gi|25405222|pir||... 544 e-153
gb|AAX95797.1| At1g47570 [Oryza sativa (japonica cultivar-group)] 477 e-133
gb|AAX95796.1| At1g47570 [Oryza sativa (japonica cultivar-group)] 461 e-128
dbj|BAB55297.1| unnamed protein product [Homo sapiens] 129 2e-28
sp|Q96EP1|CHFR_HUMAN Ubiquitin ligase protein CHFR (Checkpoint w... 129 2e-28
emb|CAH89580.1| hypothetical protein [Pongo pygmaeus] 128 3e-28
gb|AAH12072.1| CHFR protein [Homo sapiens] 128 4e-28
ref|NP_001009258.1| checkpoint with forkhead and ring finger dom... 128 4e-28
dbj|BAA91817.1| unnamed protein product [Homo sapiens] gi|892267... 127 7e-28
gb|AAH49792.1| Checkpoint with forkhead and ring finger domains ... 125 3e-27
ref|NP_766305.1| checkpoint with forkhead and ring finger domain... 125 3e-27
ref|XP_641760.1| hypothetical protein DDB0205685 [Dictyostelium ... 121 4e-26
gb|AAH89260.1| Unknown (protein for MGC:85038) [Xenopus laevis] 117 5e-25
ref|NP_989378.1| hypothetical protein LOC395009 [Xenopus tropica... 115 2e-24
ref|XP_415086.1| PREDICTED: similar to Ubiquitin ligase protein ... 108 3e-22
ref|XP_534631.1| PREDICTED: similar to Ubiquitin ligase protein ... 92 3e-17
emb|CAG09691.1| unnamed protein product [Tetraodon nigroviridis] 82 3e-14
emb|CAB70812.1| hypothetical protein [Homo sapiens] 81 6e-14
gb|AAW24657.1| unknown [Schistosoma japonicum] 65 5e-09
>ref|NP_175188.3| zinc finger (C3HC4-type RING finger) family protein [Arabidopsis
thaliana] gi|46518399|gb|AAS99681.1| At1g47570
[Arabidopsis thaliana] gi|40823234|gb|AAR92268.1|
At1g47570 [Arabidopsis thaliana]
Length = 466
Score = 544 bits (1402), Expect = e-153
Identities = 252/439 (57%), Positives = 320/439 (72%), Gaps = 2/439 (0%)
Query: 3 DSRYPDVEIRSDKGQIRSEISTASCDKHRWCKIVRNSDLCSATLKNKSSNTILVDGAEVG 62
D+R+ D+EIR + I SEI +S +KH WC+I +N SAT+ NKSS+ ILVD A V
Sbjct: 28 DTRFSDIEIRCNDMVICSEIKPSSLEKHEWCRITKNLGQSSATIHNKSSDAILVDKAVVP 87
Query: 63 KGDTVVIKDRSEIIPGPAKEGFVSYKFQIVSSPEICQTLLKICVDVDHAKCSICLNIWHD 122
K V I SEI+PGP ++G++ Y+F I+ +PE LL+I +D +HAKCSICLNIWHD
Sbjct: 88 KDGAVDIISGSEIVPGPEEQGYLQYRFTIMPAPESRTQLLQISIDPEHAKCSICLNIWHD 147
Query: 123 VVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQFVGKNHFLRTIAEDMVKADS 182
VVT APCLHNFCNGCFSEW+RRS+EK VLCPQCR VQ+VGKNHFL+ I E+++K D+
Sbjct: 148 VVTAAPCLHNFCNGCFSEWMRRSEEKHKHVLCPQCRTTVQYVGKNHFLKNIQEEILKVDA 207
Query: 183 SLRRSHDEVALLDTYASVRSNLVIGSGKKSRKRERANTTMVNQSDGPDHHCSQCVTVVGG 242
+LRR +++A+LD+ AS++SNL+IGS +K R A T + D C QCV +GG
Sbjct: 208 ALRRPAEDIAVLDSSASIQSNLIIGSKRKRRLNMPAPTH--EERDSLRLQCPQCVANIGG 265
Query: 243 FQCDPSTIHLQCQECGGMMPSRTGLGVPQYCLGCDRPFCGAYWSALGVTGSGSYPVCSPD 302
++C+ HLQC C GMMP R L VP +C GCDRPFCGAYWS+ VT S PVC +
Sbjct: 266 YRCEHHGAHLQCHLCQGMMPFRANLQVPLHCKGCDRPFCGAYWSSENVTQGVSGPVCVRE 325
Query: 303 TLRPISEHTISGIPLVAHEKNLHEQNITESCIRQMGRTLQDVISEWIAKLNNREIDTTRL 362
T RPISE TI+ IP + HE N HEQ+IT+ CI M +T+ DV++EW+ NNREID +R+
Sbjct: 326 TFRPISERTITRIPFITHEMNRHEQDITQRCIAHMEKTVPDVVAEWLRLFNNREIDRSRM 385
Query: 363 MLNHAEMMTAGTLVCSDCYQKLVSFFLYWFRISTPKHLLPPEASTREDCWYGYACRTQHR 422
LNHAE +TA T VC+DCY KLV F LYWFRI+ P++ LP + + REDCWYGYACRTQH
Sbjct: 386 PLNHAETITASTHVCNDCYDKLVGFLLYWFRITLPRNHLPADVAAREDCWYGYACRTQHH 445
Query: 423 SEEHARKRNHVCRPTRGSH 441
+E+HARKRNHVCRPTRG+H
Sbjct: 446 NEDHARKRNHVCRPTRGNH 464
>gb|AAD46042.1| F16N3.15 [Arabidopsis thaliana] gi|25405222|pir||C96516 F16N3.15
[imported] - Arabidopsis thaliana
Length = 473
Score = 544 bits (1402), Expect = e-153
Identities = 252/439 (57%), Positives = 320/439 (72%), Gaps = 2/439 (0%)
Query: 3 DSRYPDVEIRSDKGQIRSEISTASCDKHRWCKIVRNSDLCSATLKNKSSNTILVDGAEVG 62
D+R+ D+EIR + I SEI +S +KH WC+I +N SAT+ NKSS+ ILVD A V
Sbjct: 35 DTRFSDIEIRCNDMVICSEIKPSSLEKHEWCRITKNLGQSSATIHNKSSDAILVDKAVVP 94
Query: 63 KGDTVVIKDRSEIIPGPAKEGFVSYKFQIVSSPEICQTLLKICVDVDHAKCSICLNIWHD 122
K V I SEI+PGP ++G++ Y+F I+ +PE LL+I +D +HAKCSICLNIWHD
Sbjct: 95 KDGAVDIISGSEIVPGPEEQGYLQYRFTIMPAPESRTQLLQISIDPEHAKCSICLNIWHD 154
Query: 123 VVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQFVGKNHFLRTIAEDMVKADS 182
VVT APCLHNFCNGCFSEW+RRS+EK VLCPQCR VQ+VGKNHFL+ I E+++K D+
Sbjct: 155 VVTAAPCLHNFCNGCFSEWMRRSEEKHKHVLCPQCRTTVQYVGKNHFLKNIQEEILKVDA 214
Query: 183 SLRRSHDEVALLDTYASVRSNLVIGSGKKSRKRERANTTMVNQSDGPDHHCSQCVTVVGG 242
+LRR +++A+LD+ AS++SNL+IGS +K R A T + D C QCV +GG
Sbjct: 215 ALRRPAEDIAVLDSSASIQSNLIIGSKRKRRLNMPAPTH--EERDSLRLQCPQCVANIGG 272
Query: 243 FQCDPSTIHLQCQECGGMMPSRTGLGVPQYCLGCDRPFCGAYWSALGVTGSGSYPVCSPD 302
++C+ HLQC C GMMP R L VP +C GCDRPFCGAYWS+ VT S PVC +
Sbjct: 273 YRCEHHGAHLQCHLCQGMMPFRANLQVPLHCKGCDRPFCGAYWSSENVTQGVSGPVCVRE 332
Query: 303 TLRPISEHTISGIPLVAHEKNLHEQNITESCIRQMGRTLQDVISEWIAKLNNREIDTTRL 362
T RPISE TI+ IP + HE N HEQ+IT+ CI M +T+ DV++EW+ NNREID +R+
Sbjct: 333 TFRPISERTITRIPFITHEMNRHEQDITQRCIAHMEKTVPDVVAEWLRLFNNREIDRSRM 392
Query: 363 MLNHAEMMTAGTLVCSDCYQKLVSFFLYWFRISTPKHLLPPEASTREDCWYGYACRTQHR 422
LNHAE +TA T VC+DCY KLV F LYWFRI+ P++ LP + + REDCWYGYACRTQH
Sbjct: 393 PLNHAETITASTHVCNDCYDKLVGFLLYWFRITLPRNHLPADVAAREDCWYGYACRTQHH 452
Query: 423 SEEHARKRNHVCRPTRGSH 441
+E+HARKRNHVCRPTRG+H
Sbjct: 453 NEDHARKRNHVCRPTRGNH 471
>gb|AAX95797.1| At1g47570 [Oryza sativa (japonica cultivar-group)]
Length = 478
Score = 477 bits (1228), Expect = e-133
Identities = 228/450 (50%), Positives = 308/450 (67%), Gaps = 13/450 (2%)
Query: 2 SDSRYPDVEIRSDKGQIRSEIST-ASCDKHRWCKIVR---NSDLCSATLKNKSSNTILVD 57
SDS +P+VE+ D + S ++ + WC+I R + D SAT++N SS+ I+VD
Sbjct: 30 SDSAFPEVELAEDDAVVCSRVTPDGGGEVAAWCEIRRGGGDGDASSATIRNLSSDAIIVD 89
Query: 58 GAEVGKGDTVVIKDRSEIIPGPAKEGFVSYKFQIVSSPEICQTLLKICVDVDHAKCSICL 117
G + + + V IK SEI+ GP K+G + Y F I + +T +KI +D+++AKCSICL
Sbjct: 90 GRVIQQ-EAVDIKPGSEIVSGPQKDGHLLYTFDITGLNDQDKTNIKIVLDIENAKCSICL 148
Query: 118 NIWHDVVTVAPCLHNFCNGCFSEWLRRS----QEKRSTVLCPQCRAVVQFVGKNHFLRTI 173
N+WHDVVTVAPCLHNFCNGCFSEWLRRS ++K + CPQCR VQ VG+NHFL I
Sbjct: 149 NLWHDVVTVAPCLHNFCNGCFSEWLRRSSANSRDKSQSAACPQCRTAVQSVGRNHFLHNI 208
Query: 174 AEDMVKADSSLRRSHDEVALLDTYASVRSNLVIGSGK-KSRKRE--RANTTMVNQSDGPD 230
E +++A SSL+RS +E+ALL++YASV++N+V+G K +SRKR R+N N ++ D
Sbjct: 209 EEAILQAFSSLQRSDEEIALLESYASVKTNIVLGKQKIQSRKRRLPRSNDE-ANHTNHAD 267
Query: 231 HHCSQCVTVVGGFQCDPSTIHLQCQECGGMMPSRTGLGVPQYCLGCDRPFCGAYWSALGV 290
C QC GGF+C P HL C CGGMMP+R +PQ CLGCDR FCGAYW + GV
Sbjct: 268 FLCPQCGAEFGGFRCSPGAPHLPCNGCGGMMPARPDTSIPQKCLGCDRAFCGAYWCSQGV 327
Query: 291 TGSGSYPVCSPDTLRPISEHTISGIPLVAHEKNLHEQNITESCIRQMGRTLQDVISEWIA 350
S P+C +T + IS+ IS +P H N +E++ITE CI+Q G+ LQ +ISEWI
Sbjct: 328 NSSQHNPICDQETFKMISQRHISSVPDTVHGGNQYEKDITERCIQQSGKALQAIISEWIV 387
Query: 351 KLNNREIDTTRLMLNHAEMMTAGTLVCSDCYQKLVSFFLYWFRISTPKHLLPPEASTRED 410
K +N+E+D +RL LNH + +T+ T VC+ CY K + F LYWFR+S P++LLPP+A+ RE
Sbjct: 388 KFDNKELDRSRLQLNHVDAITSRTYVCNQCYSKFIDFLLYWFRVSMPRNLLPPDAANRES 447
Query: 411 CWYGYACRTQHRSEEHARKRNHVCRPTRGS 440
CWYG+ CRTQH +HA+K NHVCRPTRG+
Sbjct: 448 CWYGFMCRTQHHRPDHAKKLNHVCRPTRGN 477
>gb|AAX95796.1| At1g47570 [Oryza sativa (japonica cultivar-group)]
Length = 508
Score = 461 bits (1187), Expect = e-128
Identities = 228/480 (47%), Positives = 308/480 (63%), Gaps = 43/480 (8%)
Query: 2 SDSRYPDVEIRSDKGQIRSEIST-ASCDKHRWCKIVR---NSDLCSATLKNKSSNTILVD 57
SDS +P+VE+ D + S ++ + WC+I R + D SAT++N SS+ I+VD
Sbjct: 30 SDSAFPEVELAEDDAVVCSRVTPDGGGEVAAWCEIRRGGGDGDASSATIRNLSSDAIIVD 89
Query: 58 GAEVGKGDTVVIKDRSEIIPGPAKEGFVSYKFQIVSSPEICQTLLKICVDVDHAKCSICL 117
G + + + V IK SEI+ GP K+G + Y F I + +T +KI +D+++AKCSICL
Sbjct: 90 GRVIQQ-EAVDIKPGSEIVSGPQKDGHLLYTFDITGLNDQDKTNIKIVLDIENAKCSICL 148
Query: 118 NIWHDVVTVAPCLHNFCNGCFSEWLRRS----QEKRSTVLCPQCRAVVQFVGKNHFLRTI 173
N+WHDVVTVAPCLHNFCNGCFSEWLRRS ++K + CPQCR VQ VG+NHFL I
Sbjct: 149 NLWHDVVTVAPCLHNFCNGCFSEWLRRSSANSRDKSQSAACPQCRTAVQSVGRNHFLHNI 208
Query: 174 AEDMVKADSSLRRSHDEVALLDTYASVRSNL----------------------------- 204
E +++A SSL+RS +E+ALL++YASV++N+
Sbjct: 209 EEAILQAFSSLQRSDEEIALLESYASVKTNIAWHAFIPFPWSYAGAMILRSTMGINICQR 268
Query: 205 -VIGSGK-KSRKRE--RANTTMVNQSDGPDHHCSQCVTVVGGFQCDPSTIHLQCQECGGM 260
V+G K +SRKR R+N N ++ D C QC GGF+C P HL C CGGM
Sbjct: 269 EVLGKQKIQSRKRRLPRSNDE-ANHTNHADFLCPQCGAEFGGFRCSPGAPHLPCNGCGGM 327
Query: 261 MPSRTGLGVPQYCLGCDRPFCGAYWSALGVTGSGSYPVCSPDTLRPISEHTISGIPLVAH 320
MP+R +PQ CLGCDR FCGAYW + GV S P+C +T + IS+ IS +P H
Sbjct: 328 MPARPDTSIPQKCLGCDRAFCGAYWCSQGVNSSQHNPICDQETFKMISQRHISSVPDTVH 387
Query: 321 EKNLHEQNITESCIRQMGRTLQDVISEWIAKLNNREIDTTRLMLNHAEMMTAGTLVCSDC 380
N +E++ITE CI+Q G+ LQ +ISEWI K +N+E+D +RL LNH + +T+ T VC+ C
Sbjct: 388 GGNQYEKDITERCIQQSGKALQAIISEWIVKFDNKELDRSRLQLNHVDAITSRTYVCNQC 447
Query: 381 YQKLVSFFLYWFRISTPKHLLPPEASTREDCWYGYACRTQHRSEEHARKRNHVCRPTRGS 440
Y K + F LYWFR+S P++LLPP+A+ RE CWYG+ CRTQH +HA+K NHVCRPTRG+
Sbjct: 448 YSKFIDFLLYWFRVSMPRNLLPPDAANRESCWYGFMCRTQHHRPDHAKKLNHVCRPTRGN 507
>dbj|BAB55297.1| unnamed protein product [Homo sapiens]
Length = 652
Score = 129 bits (323), Expect = 2e-28
Identities = 94/378 (24%), Positives = 151/378 (39%), Gaps = 72/378 (19%)
Query: 113 CSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQFVGKNHFLRT 172
C IC ++ HD V++ PC+H FC C+S W+ RS LCP CR V+ + KNH L
Sbjct: 292 CIICQDLLHDCVSLQPCMHTFCAACYSGWMERSS------LCPTCRCPVERICKNHILNN 345
Query: 173 IAEDMVKADSSLRRSHDEVALLDTYASVRSNLVIGSGKKSRKRERANTTMVNQSDGPDHH 232
+ E + RS ++V +D + +++ ++S E ++ + + D
Sbjct: 346 LVEAYLIQHPDKSRSEEDVQSMDARNKITQDMLQPKVRRSFSDEEGSSEDLLELSDVDSE 405
Query: 233 ----------CSQC------------------------------------VTVVGGFQCD 246
C QC T V + C
Sbjct: 406 SSDISQPYVVCRQCPEYRRQAAQPPHCPAPEGEPGAPQALGDAPSTSVSLTTAVQDYVCP 465
Query: 247 PSTIHLQCQECGGMMPSRTGLG------VPQYCLGCDRPFCGAYWSALGVTGSGSYPVCS 300
H C C MP R PQ C C +PFC YW G T +G Y +
Sbjct: 466 LQGSHALCTCCFQPMPDRRAEREQDPRVAPQQCAVCLQPFCHLYW---GCTRTGCYGCLA 522
Query: 301 PDTLRPISEHTISGIPLVAHEKNLHEQNITESCIRQMGRTLQDVISEWIAKLNNREIDTT 360
P + + + G+ N +E +I ++ + G T +++++E + L
Sbjct: 523 PFCELNLGDKCLDGVL----NNNSYESDILKNYLATRGLTWKNMLTESLVALQRG----- 573
Query: 361 RLMLNHAEMMTAGTLVCSDCYQKLVSFFLYWFRISTPKHLLPPEASTREDCWYGYACRTQ 420
+ L +T T++C C + Y +R + P LP ++R DC++G CRTQ
Sbjct: 574 -VFLLSDYRVTGDTVLCYCCGLRSFRELTYQYRQNIPASELPVAVTSRPDCYWGRNCRTQ 632
Query: 421 HRSEEHARKRNHVCRPTR 438
++ HA K NH+C TR
Sbjct: 633 VKA-HHAMKFNHICEQTR 649
>sp|Q96EP1|CHFR_HUMAN Ubiquitin ligase protein CHFR (Checkpoint with forkhead and RING
finger domains protein) gi|9651170|gb|AAF91084.1| cell
cycle checkpoint protein CHFR [Homo sapiens]
Length = 664
Score = 129 bits (323), Expect = 2e-28
Identities = 94/378 (24%), Positives = 151/378 (39%), Gaps = 72/378 (19%)
Query: 113 CSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQFVGKNHFLRT 172
C IC ++ HD V++ PC+H FC C+S W+ RS LCP CR V+ + KNH L
Sbjct: 304 CIICQDLLHDCVSLQPCMHTFCAACYSGWMERSS------LCPTCRCPVERICKNHILNN 357
Query: 173 IAEDMVKADSSLRRSHDEVALLDTYASVRSNLVIGSGKKSRKRERANTTMVNQSDGPDHH 232
+ E + RS ++V +D + +++ ++S E ++ + + D
Sbjct: 358 LVEAYLIQHPDKSRSEEDVQSMDARNKITQDMLQPKVRRSFSDEEGSSEDLLELSDVDSE 417
Query: 233 ----------CSQC------------------------------------VTVVGGFQCD 246
C QC T V + C
Sbjct: 418 SSDISQPYVVCRQCPEYRRQAAQPPHCPAPEGEPGAPQALGDAPSTSVSLTTAVQDYVCP 477
Query: 247 PSTIHLQCQECGGMMPSRTGLG------VPQYCLGCDRPFCGAYWSALGVTGSGSYPVCS 300
H C C MP R PQ C C +PFC YW G T +G Y +
Sbjct: 478 LQGSHALCTCCFQPMPDRRAEREQDPRVAPQQCAVCLQPFCHLYW---GCTRTGCYGCLA 534
Query: 301 PDTLRPISEHTISGIPLVAHEKNLHEQNITESCIRQMGRTLQDVISEWIAKLNNREIDTT 360
P + + + G+ N +E +I ++ + G T +++++E + L
Sbjct: 535 PFCELNLGDKCLDGVL----NNNSYESDILKNYLATRGLTWKNMLTESLVALQRG----- 585
Query: 361 RLMLNHAEMMTAGTLVCSDCYQKLVSFFLYWFRISTPKHLLPPEASTREDCWYGYACRTQ 420
+ L +T T++C C + Y +R + P LP ++R DC++G CRTQ
Sbjct: 586 -VFLLSDYRVTGDTVLCYCCGLRSFRELTYQYRQNIPASELPVAVTSRPDCYWGRNCRTQ 644
Query: 421 HRSEEHARKRNHVCRPTR 438
++ HA K NH+C TR
Sbjct: 645 VKA-HHAMKFNHICEQTR 661
>emb|CAH89580.1| hypothetical protein [Pongo pygmaeus]
Length = 571
Score = 128 bits (322), Expect = 3e-28
Identities = 93/377 (24%), Positives = 152/377 (39%), Gaps = 71/377 (18%)
Query: 113 CSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQFVGKNHFLRT 172
C IC ++ HD V++ PC+H FC C+S W+ RS LCP CR V+ + KNH L
Sbjct: 212 CIICQDLLHDCVSLQPCMHTFCAACYSGWMERSS------LCPTCRCPVERICKNHILNN 265
Query: 173 IAEDMVKADSSLRRSHDEVALLDTYASVRSNLVIGSGKKSRKRERANTTMVNQSDGPDHH 232
+ E + RS ++V +D + +++ ++S E ++ + + D
Sbjct: 266 LVEAYLIQHPDKSRSEEDVQSMDARNKITQDMLQPKVRRSFSDEEGSSEDLLELSDVDSE 325
Query: 233 ----------CSQC-----------------------------------VTVVGGFQCDP 247
C QC +T V + C
Sbjct: 326 SSDISQPYVVCRQCPEYRRQAAQPPPCPAPEGEPGVPQALVDAPSTSVSLTTVQDYVCPL 385
Query: 248 STIHLQCQECGGMMPSRTGLG------VPQYCLGCDRPFCGAYWSALGVTGSGSYPVCSP 301
H C C MP R PQ C C +PFC YW G T +G + +P
Sbjct: 386 QGSHALCTCCFQPMPDRRAEREQNPRVAPQQCAVCLQPFCHLYW---GCTRTGCFGCLAP 442
Query: 302 DTLRPISEHTISGIPLVAHEKNLHEQNITESCIRQMGRTLQDVISEWIAKLNNREIDTTR 361
+ + + G+ N +E +I ++ + G T +++++E + L
Sbjct: 443 FCELNLGDKCLDGVL----NNNSYESDILKNYLATRGLTWKNMLTESLVALQRG------ 492
Query: 362 LMLNHAEMMTAGTLVCSDCYQKLVSFFLYWFRISTPKHLLPPEASTREDCWYGYACRTQH 421
+ L +T T++C C + Y +R + P LP ++R DC++G CRTQ
Sbjct: 493 VFLLSDYRVTGDTILCYCCGLRSFRELTYQYRQNIPASELPVAVTSRPDCYWGRNCRTQV 552
Query: 422 RSEEHARKRNHVCRPTR 438
++ HA K NH+C TR
Sbjct: 553 KA-HHAMKFNHICEQTR 568
>gb|AAH12072.1| CHFR protein [Homo sapiens]
Length = 652
Score = 128 bits (321), Expect = 4e-28
Identities = 94/378 (24%), Positives = 151/378 (39%), Gaps = 72/378 (19%)
Query: 113 CSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQFVGKNHFLRT 172
C IC ++ HD V++ PC+H FC C+S W+ RS LCP CR V+ + KNH L
Sbjct: 292 CIICQDLLHDCVSLQPCMHTFCAACYSGWMERSS------LCPTCRCPVERICKNHILNN 345
Query: 173 IAEDMVKADSSLRRSHDEVALLDTYASVRSNLVIGSGKKSRKRERANTTMVNQSDGPDHH 232
+ E + RS ++V +D + +++ ++S E ++ + + D
Sbjct: 346 LVEAYLIQHPDKSRSEEDVQSMDARNKITQDMLQPKVRRSFSDEEGSSEDLLELSDVDSE 405
Query: 233 ----------CSQC------------------------------------VTVVGGFQCD 246
C QC T V + C
Sbjct: 406 SSDISQPYVVCRQCPEYRRQAAQPPHCPAPEGEPGAPQALGDAPSTSVSLTTAVQDYVCP 465
Query: 247 PSTIHLQCQECGGMMPSRT------GLGVPQYCLGCDRPFCGAYWSALGVTGSGSYPVCS 300
H C C MP R PQ C C +PFC YW G T +G Y +
Sbjct: 466 LQGSHALCTCCFQPMPDRRVEREQDPRVAPQQCAVCLQPFCHLYW---GCTRTGCYGCLA 522
Query: 301 PDTLRPISEHTISGIPLVAHEKNLHEQNITESCIRQMGRTLQDVISEWIAKLNNREIDTT 360
P + + + G+ N +E +I ++ + G T +++++E + L
Sbjct: 523 PFCELNLGDKCLDGVL----NNNSYESDILKNYLATRGLTWKNMLTESLVALQRG----- 573
Query: 361 RLMLNHAEMMTAGTLVCSDCYQKLVSFFLYWFRISTPKHLLPPEASTREDCWYGYACRTQ 420
+ L +T T++C C + Y +R + P LP ++R DC++G CRTQ
Sbjct: 574 -VFLLSDYRVTGDTVLCYCCGLRSFRELTYQYRQNIPASELPVAVTSRPDCYWGRNCRTQ 632
Query: 421 HRSEEHARKRNHVCRPTR 438
++ HA K NH+C TR
Sbjct: 633 VKA-HHAMKFNHICEQTR 649
>ref|NP_001009258.1| checkpoint with forkhead and ring finger domains (predicted)
[Rattus norvegicus] gi|56268938|gb|AAH87162.1|
Checkpoint with forkhead and ring finger domains
(predicted) [Rattus norvegicus]
Length = 663
Score = 128 bits (321), Expect = 4e-28
Identities = 92/378 (24%), Positives = 152/378 (39%), Gaps = 72/378 (19%)
Query: 113 CSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQFVGKNHFLRT 172
C IC ++ HD V++ PC+H FC C+S W+ RS LCP CR V+ + KNH L
Sbjct: 303 CIICQDLLHDCVSLQPCMHTFCAACYSGWMERSS------LCPTCRCPVERICKNHILNN 356
Query: 173 IAEDMVKADSSLRRSHDEVALLDTYASVRSNLVIGSGKKSRKRERANTTMVNQSDGPDHH 232
+ E + RS ++V +D + +++ ++S E ++ + + D
Sbjct: 357 LVEAYLLQHPDKSRSEEDVRSMDARNKITQDMLQPKVRRSFSDEEGSSEDLLELSDVDSE 416
Query: 233 ----------CSQC------------------------------------VTVVGGFQCD 246
C QC +T G+ C
Sbjct: 417 SSDISQPYIVCRQCPEYRRQAVQSLPCPVPESELGATQALGGEAPSTSASLTTAPGYTCP 476
Query: 247 PSTIHLQCQECGGMMPSRTGLG------VPQYCLGCDRPFCGAYWSALGVTGSGSYPVCS 300
H C C MP R PQ C C +PFC YW G T +G + +
Sbjct: 477 LQGSHAICTCCFQPMPDRRAEHEQDSRIAPQQCAVCLQPFCHLYW---GCTRTGCFGCLA 533
Query: 301 PDTLRPISEHTISGIPLVAHEKNLHEQNITESCIRQMGRTLQDVISEWIAKLNNREIDTT 360
P + + + G+ N +E +I ++ + G T +++++E + L
Sbjct: 534 PFCELNLGDKCLDGVL----NNNNYESDILKNYLTTRGLTWKNMLTESLLALQRG----- 584
Query: 361 RLMLNHAEMMTAGTLVCSDCYQKLVSFFLYWFRISTPKHLLPPEASTREDCWYGYACRTQ 420
+ + +T T++C C + Y +R + P LP ++R DC++G CRTQ
Sbjct: 585 -VFILSDYRITGNTVLCYCCGLRSFRELTYQYRQNIPASELPVTVTSRPDCYWGRNCRTQ 643
Query: 421 HRSEEHARKRNHVCRPTR 438
++ HA K NH+C TR
Sbjct: 644 VKA-HHAMKFNHICEQTR 660
>dbj|BAA91817.1| unnamed protein product [Homo sapiens] gi|8922675|ref|NP_060693.1|
checkpoint with forkhead and ring finger domains [Homo
sapiens]
Length = 623
Score = 127 bits (319), Expect = 7e-28
Identities = 93/378 (24%), Positives = 151/378 (39%), Gaps = 72/378 (19%)
Query: 113 CSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQFVGKNHFLRT 172
C IC ++ HD V++ PC+H FC C+S W+ RS LCP CR V+ + KNH L
Sbjct: 263 CIICQDLLHDCVSLQPCMHTFCAACYSGWMERSS------LCPTCRCPVERICKNHILNN 316
Query: 173 IAEDMVKADSSLRRSHDEVALLDTYASVRSNLVIGSGKKSRKRERANTTMVNQSDGPDHH 232
+ E + RS ++V +D + +++ ++S E ++ + + D
Sbjct: 317 LVEAYLIQHPDKSRSEEDVQSMDARNKITQDMLQPKVRRSFSDEEGSSEDLLELSDVDSE 376
Query: 233 ----------CSQC------------------------------------VTVVGGFQCD 246
C QC T V + C
Sbjct: 377 SSDISQPYVVCRQCPEYRRQAAQPPHCPAPEGEPGAPQALGDAPPTSVSLTTAVQDYVCP 436
Query: 247 PSTIHLQCQECGGMMPSRTGLG------VPQYCLGCDRPFCGAYWSALGVTGSGSYPVCS 300
H C C MP R PQ C C +PFC YW G T +G Y +
Sbjct: 437 LQGSHALCTCCFQPMPDRRAEREQDPRVAPQQCAVCLQPFCHLYW---GCTRTGCYGCLA 493
Query: 301 PDTLRPISEHTISGIPLVAHEKNLHEQNITESCIRQMGRTLQDVISEWIAKLNNREIDTT 360
P + + + G+ N +E +I ++ + G T +++++E + L
Sbjct: 494 PFCELNLGDKCLDGVL----NNNSYESDILKNYLATRGLTWKNMLTESLVALQRG----- 544
Query: 361 RLMLNHAEMMTAGTLVCSDCYQKLVSFFLYWFRISTPKHLLPPEASTREDCWYGYACRTQ 420
+ L +T T++C C + Y ++ + P LP ++R DC++G CRTQ
Sbjct: 545 -VFLLSDYRVTGDTVLCYCCGLRSFRELTYQYQQNIPASELPVAVTSRPDCYWGRNCRTQ 603
Query: 421 HRSEEHARKRNHVCRPTR 438
++ HA K NH+C TR
Sbjct: 604 VKA-HHAMKFNHICEQTR 620
>gb|AAH49792.1| Checkpoint with forkhead and ring finger domains [Mus musculus]
gi|41688490|sp|Q810L3|CHFR_MOUSE Ubiquitin ligase
protein CHFR (Checkpoint with forkhead and RING finger
domains protein)
Length = 664
Score = 125 bits (313), Expect = 3e-27
Identities = 95/379 (25%), Positives = 152/379 (40%), Gaps = 73/379 (19%)
Query: 113 CSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQFVGKNHFLRT 172
C IC ++ HD V++ PC+H FC C+S W+ RS LCP CR V+ + KNH L
Sbjct: 303 CIICQDLLHDCVSLQPCMHTFCAACYSGWMERSS------LCPTCRCPVERICKNHILNN 356
Query: 173 IAEDMVKADSSLRRSHDEVALLDTYASVRSNLVIGSGKKSRKRERANTTMVNQSDGPDHH 232
+ E + RS ++V +D + +++ ++S E ++ + + D
Sbjct: 357 LVEAYLIQHPDKSRSEEDVRSMDARNKITQDMLQPKVRRSFSDEEGSSEDLLELSDVDSE 416
Query: 233 ----------CSQC----------------------VTVVGG---------------FQC 245
C QC +GG + C
Sbjct: 417 SSDISQPYIVCRQCPEYRRQAVQSLPCPVPESELGATLALGGEAPSTSASLPTAAPDYMC 476
Query: 246 DPSTIHLQCQECGGMMPSRTGLG------VPQYCLGCDRPFCGAYWSALGVTGSGSYPVC 299
H C C MP R PQ C C +PFC YW G T +G +
Sbjct: 477 PLQGSHAICTCCFQPMPDRRAEREQDPRVAPQQCAVCLQPFCHLYW---GCTRTGCFGCL 533
Query: 300 SPDTLRPISEHTISGIPLVAHEKNLHEQNITESCIRQMGRTLQDVISEWIAKLNNREIDT 359
+P + + + G+ N +E +I ++ + G T + V++E + L
Sbjct: 534 APFCELNLGDKCLDGVL----NNNNYESDILKNYLATRGLTWKSVLTESLLALQRGVF-- 587
Query: 360 TRLMLNHAEMMTAGTLVCSDCYQKLVSFFLYWFRISTPKHLLPPEASTREDCWYGYACRT 419
ML+ + T T++C C + Y +R + P LP ++R DC++G CRT
Sbjct: 588 ---MLSDYRI-TGNTVLCYCCGLRSFRELTYQYRQNIPASELPVTVTSRPDCYWGRNCRT 643
Query: 420 QHRSEEHARKRNHVCRPTR 438
Q ++ HA K NH+C TR
Sbjct: 644 QVKA-HHAMKFNHICEQTR 661
>ref|NP_766305.1| checkpoint with forkhead and ring finger domains [Mus musculus]
gi|26346522|dbj|BAC36912.1| unnamed protein product [Mus
musculus]
Length = 663
Score = 125 bits (313), Expect = 3e-27
Identities = 95/378 (25%), Positives = 152/378 (40%), Gaps = 72/378 (19%)
Query: 113 CSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQFVGKNHFLRT 172
C IC ++ HD V++ PC+H FC C+S W+ RS LCP CR V+ + KNH L
Sbjct: 303 CIICQDLLHDCVSLQPCMHTFCAACYSGWMERSS------LCPTCRCPVERICKNHILNN 356
Query: 173 IAEDMVKADSSLRRSHDEVALLDTYASVRSNLVIGSGKKSRKRERANTTMVNQSDGPDHH 232
+ E + RS ++V +D + +++ ++S E ++ + + D
Sbjct: 357 LVEAYLIQHPDKSRSEEDVRSMDARNKITQDMLQPKVRRSFSDEEGSSEDLLELSDVDSE 416
Query: 233 ----------CSQC----------------------VTVVGG--------------FQCD 246
C QC +GG + C
Sbjct: 417 SSDISQPYIVCRQCPEYRRQAVQSLPCPVPESELGATLALGGEAPSTSASLPTAPDYMCP 476
Query: 247 PSTIHLQCQECGGMMPSRTGLG------VPQYCLGCDRPFCGAYWSALGVTGSGSYPVCS 300
H C C MP R PQ C C +PFC YW G T +G + +
Sbjct: 477 LQGSHAICTCCFQPMPDRRAEREQDPRVAPQQCAVCLQPFCHLYW---GCTRTGCFCCLA 533
Query: 301 PDTLRPISEHTISGIPLVAHEKNLHEQNITESCIRQMGRTLQDVISEWIAKLNNREIDTT 360
P + + + G+ N +E +I ++ + G T + V++E + L
Sbjct: 534 PFCELNLGDKCLDGVL----NNNNYESDILKNYLATRGLTWKSVLTESLLALQRGVF--- 586
Query: 361 RLMLNHAEMMTAGTLVCSDCYQKLVSFFLYWFRISTPKHLLPPEASTREDCWYGYACRTQ 420
ML+ + T T++C C + Y +R + P LP ++R DC++G CRTQ
Sbjct: 587 --MLSDYRI-TGNTVLCYCCGLRSFRELTYQYRQNIPASELPVTVTSRPDCYWGRNCRTQ 643
Query: 421 HRSEEHARKRNHVCRPTR 438
++ HA K NH+C TR
Sbjct: 644 VKA-HHAMKFNHICEQTR 660
>ref|XP_641760.1| hypothetical protein DDB0205685 [Dictyostelium discoideum]
gi|60469793|gb|EAL67780.1| hypothetical protein
DDB0205685 [Dictyostelium discoideum]
Length = 742
Score = 121 bits (304), Expect = 4e-26
Identities = 90/361 (24%), Positives = 152/361 (41%), Gaps = 58/361 (16%)
Query: 113 CSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQFVGKNHFLRT 172
C IC +I + +T+ PC+HNFC C+ +W S + CPQCR V+ KNH +
Sbjct: 404 CGICQDIIYKCLTLIPCMHNFCVCCYGDWRANSMD------CPQCRQSVKSAQKNHAINN 457
Query: 173 IAEDMVKADSSLRRSHDEVALLDTYASVRSNLVIGSG--KKSRKRERANTTMVNQSDGPD 230
+ E +K + R +E+ +D + ++ K SRK + ++ D
Sbjct: 458 LIELYLKKNPEKVRDPEELESMDKRCKITDEMLKNGSLNKSSRKYDNDEYEDEDEDYSED 517
Query: 231 HH--------------CSQC----VTVVGGFQCDPS---------TIHLQCQECGGMMPS 263
C+ C + G+QC S T HL C C + P
Sbjct: 518 EEYYSSEEDDKFIPSKCTFCHPNPPNQITGYQCPESPANATYATQTKHLNCSNCYKIFPK 577
Query: 264 RTGLGVPQYCLGCDRPFCGAYWSALGVTGSGSYPVCSPDTLRPISEHTISGIPLVAHEKN 323
V C C FC Y + G + + ++ +P A N
Sbjct: 578 EPVEPVGIRCGFCHNAFCNLYQACSKGGNFGK-----------MKDLPVNNLPSDAFGGN 626
Query: 324 LHEQNITESCIRQMGRTLQDVISEWIAKLNNREIDTTRLMLNHAEMMTAGTLVCSDCYQK 383
E I + + +T++ + + + +ID+ +L++ + + T CS+ Y+
Sbjct: 627 QFEVKILNDYLTKNVKTVRQLYLDIL-----EDIDSKKLIIPTSSNIIMPTSPCSEQYRC 681
Query: 384 L------VSFFLYWFRISTPKHLLPPEASTREDCWYGYACRTQHRSEEHARKRNHVCRPT 437
L S L+++R+S PK LP A+ R++C+YG +CRTQ +HA+K NH+C T
Sbjct: 682 LGCASHFFSKTLFYYRLSIPKEKLPDYAN-RDNCYYGKSCRTQFSKFDHAKKLNHICEQT 740
Query: 438 R 438
+
Sbjct: 741 K 741
>gb|AAH89260.1| Unknown (protein for MGC:85038) [Xenopus laevis]
Length = 625
Score = 117 bits (294), Expect = 5e-25
Identities = 89/377 (23%), Positives = 145/377 (37%), Gaps = 71/377 (18%)
Query: 113 CSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQFVGKNHFLRT 172
C IC + HD V++ PC+H FC C+S W+ RS LCP CR V+ + KNH L
Sbjct: 266 CIICQELLHDCVSLQPCMHTFCAACYSGWMERSS------LCPTCRCPVERICKNHILNN 319
Query: 173 IAEDMVKADSSLRRSHDEVALLDTYASVRSNLVIGSGKKSRKRERANTTMVNQSDGPDHH 232
+ E + RS ++ +D + +++ ++S E ++ + + D
Sbjct: 320 LVEAYLIQHPEKCRSEEDRCSMDARNKITQDMLQPKVRRSFSDEEGSSEDLLELSDVDSE 379
Query: 233 ----------CSQCV-----------------------------------TVVGGFQCDP 247
C QC T + C
Sbjct: 380 SSDISQPYTVCRQCPGFVRHSMQPPPYPPPSDTETSRTQGDAPSTSTNFPTATQEYVCPS 439
Query: 248 STIHLQCQECGGMMPSRTGLG------VPQYCLGCDRPFCGAYWSALGVTGSGSYPVCSP 301
H+ C C MP R PQ C C PFC YW G G + +P
Sbjct: 440 HGSHVICTCCFQPMPDRRAEREHNSHVAPQQCTICLEPFCHMYW---GCNRMGCFGCLAP 496
Query: 302 DTLRPISEHTISGIPLVAHEKNLHEQNITESCIRQMGRTLQDVISEWIAKLNNREIDTTR 361
+ + + G+ N +E +I ++ + G T +D+++E +A +
Sbjct: 497 FCELNLGDKCLDGVL----NNNNYESDILKNYLASRGLTWKDMLNESLAAVQRGVFMLPD 552
Query: 362 LMLNHAEMMTAGTLVCSDCYQKLVSFFLYWFRISTPKHLLPPEASTREDCWYGYACRTQH 421
+N T++C C + Y +R + P LP ++R +C++G CRTQ
Sbjct: 553 YRINGT------TVLCYFCGLRNFRILTYQYRQNIPASELPVTVTSRPNCYWGRNCRTQV 606
Query: 422 RSEEHARKRNHVCRPTR 438
++ HA K NH+C TR
Sbjct: 607 KA-HHAMKFNHICEQTR 622
>ref|NP_989378.1| hypothetical protein LOC395009 [Xenopus tropicalis]
gi|40353026|gb|AAH64721.1| Hypothetical protein MGC75959
[Xenopus tropicalis]
Length = 626
Score = 115 bits (289), Expect = 2e-24
Identities = 88/377 (23%), Positives = 144/377 (37%), Gaps = 71/377 (18%)
Query: 113 CSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQFVGKNHFLRT 172
C IC + HD V++ PC+H FC C+S W+ RS LCP CR V+ + KNH L
Sbjct: 267 CIICQELLHDCVSLQPCMHTFCAACYSGWMERSS------LCPTCRCPVERICKNHILNN 320
Query: 173 IAEDMVKADSSLRRSHDEVALLDTYASVRSNLVIGSGKKSRKRERANTTMVNQSDGPDHH 232
+ E + RS ++ +D + +++ ++S E ++ + + D
Sbjct: 321 LVEAYLIQHPEKCRSEEDRCSMDARNKITQDMLQPKVRRSFSDEEGSSEDLLELSDVDSE 380
Query: 233 ----------CSQCV-----------------------------------TVVGGFQCDP 247
C QC T + C
Sbjct: 381 SSDISQPYTVCRQCPGYVRHNIQPPPYPPPSDTEASRTQGDAPSTSTNFPTATQEYVCPS 440
Query: 248 STIHLQCQECGGMMPSRTGLG------VPQYCLGCDRPFCGAYWSALGVTGSGSYPVCSP 301
H+ C C MP R PQ C C PFC YW G G + +P
Sbjct: 441 HGSHVICTCCFQPMPDRRAEREHNSHVAPQQCTICLEPFCHMYW---GCNRMGCFGCLAP 497
Query: 302 DTLRPISEHTISGIPLVAHEKNLHEQNITESCIRQMGRTLQDVISEWIAKLNNREIDTTR 361
+ + + G+ N +E +I ++ + G T +D+++E ++
Sbjct: 498 FCELNLGDKCLDGVL----NNNNYESDILKNYLASRGLTWKDMLNESLSAAQRGVFMLPD 553
Query: 362 LMLNHAEMMTAGTLVCSDCYQKLVSFFLYWFRISTPKHLLPPEASTREDCWYGYACRTQH 421
+N T++C C + Y +R + P LP ++R +C++G CRTQ
Sbjct: 554 YRINGT------TVLCYFCGLRNFRILTYQYRQNIPASELPVTVTSRPNCYWGRNCRTQV 607
Query: 422 RSEEHARKRNHVCRPTR 438
++ HA K NH+C TR
Sbjct: 608 KA-HHAMKFNHICEQTR 623
>ref|XP_415086.1| PREDICTED: similar to Ubiquitin ligase protein CHFR (Checkpoint
with forkhead and RING finger domains protein) [Gallus
gallus]
Length = 667
Score = 108 bits (270), Expect = 3e-22
Identities = 85/360 (23%), Positives = 138/360 (37%), Gaps = 71/360 (19%)
Query: 113 CSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQFVGKNHFLRT 172
C IC + HD V++ PC+H FC C+S W+ RS LCP CR V+ + KNH L
Sbjct: 320 CIICQELLHDCVSLQPCMHTFCAACYSGWMERSS------LCPTCRCPVERICKNHILNN 373
Query: 173 IAEDMVKADSSLRRSHDEVALLDTYASVRSNLVIGSGKKSRKRERANTTMVNQSDGPDHH 232
+ E + R+ D+V +D + +++ ++S E ++ + + D
Sbjct: 374 LVEAYLIQHPDKCRNEDDVRSMDARNKITQDMLQPKVRRSFSDEEGSSEDLLELSDVDSE 433
Query: 233 ----------CSQC----------------VTVVGGFQ--------------------CD 246
C QC T GG Q C
Sbjct: 434 SSDISQPYIVCRQCPGYRRHPVPALPGTGQETEAGGMQALGDAPSTSANFPAAVQEYVCP 493
Query: 247 PSTIHLQCQECGGMMPSRTGLG------VPQYCLGCDRPFCGAYWSALGVTGSGSYPVCS 300
H+ C C MP R PQ C C +PFC YW G T + +
Sbjct: 494 AQGSHVICTCCFQPMPDRRAEREQNPHVAPQQCTVCLQPFCHLYW---GCTRMACFGCLA 550
Query: 301 PDTLRPISEHTISGIPLVAHEKNLHEQNITESCIRQMGRTLQDVISEWIAKLNNREIDTT 360
P + + + G+ N +E +I + + G T +++++E + L
Sbjct: 551 PFCEINLGDKCLDGV----LNNNHYESDILKDYLASRGLTWKNMLNESLLALQR------ 600
Query: 361 RLMLNHAEMMTAGTLVCSDCYQKLVSFFLYWFRISTPKHLLPPEASTREDCWYGYACRTQ 420
+ + +T T++C C + Y +R + P LP ++R DC++G CRTQ
Sbjct: 601 GVFMLSDYRITGNTVLCYCCGLRSFRELAYQYRQNIPVAELPVTVTSRPDCYWGRNCRTQ 660
>ref|XP_534631.1| PREDICTED: similar to Ubiquitin ligase protein CHFR (Checkpoint with
forkhead and RING finger domains protein) [Canis
familiaris]
Length = 1039
Score = 92.0 bits (227), Expect = 3e-17
Identities = 80/377 (21%), Positives = 131/377 (34%), Gaps = 107/377 (28%)
Query: 113 CSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQFVGKNHFLRT 172
C IC ++ HD V++ PC+H FC C+S W+ RS CP CR V+ + KNH L
Sbjct: 716 CIICQDLLHDCVSLQPCMHTFCAACYSGWMERS------TWCPTCRCPVERICKNHILNN 769
Query: 173 IAEDMVKADSSLRRSHDEVALLDTYASVRSNLVIGSGKKSRKRERANTTMVNQSDGPDHH 232
+ E + RS ++V + + +++ ++S E ++ + + D
Sbjct: 770 LVEAYLLQHPDKSRSEEDVRSMAARNKITQDMLQPKVRRSFSDEEGSSEDLLELSDVDSE 829
Query: 233 ----------CSQC-----------------------------------VTVVGGFQCDP 247
C QC V + C
Sbjct: 830 SSDVSQPYIVCRQCPEYRRQGGQALPCPGPNSEPGVPQAAGDAPSTSTSVKTAQDYVCAL 889
Query: 248 STIHLQCQECGGMMPSRTGLG------VPQYCLGCDRPFCGAYWSALGVTGSGSYPVCSP 301
H C C MP R PQ C C +PFC YW G +G +P
Sbjct: 890 QGSHAICTCCFQPMPDRRAEREQDPRIAPQQCAVCLQPFCHLYW---GCARAGCLGCLAP 946
Query: 302 DTLRPISEHTISGIPLVAHEKNLHEQNITESCIRQMGRTLQDVISEWIAKLNNREIDTTR 361
+ + + G+ N +E ++ ++ + G T +++++E + L
Sbjct: 947 FCELDLGDRCLDGV----LSNNHYESDVLKNYLATRGLTWKNMLTESLVALQRGAF---- 998
Query: 362 LMLNHAEMMTAGTLVCSDCYQKLVSFFLYWFRISTPKHLLPPEASTREDCWYGYACRTQH 421
LL ++R DC++G CRTQ
Sbjct: 999 --------------------------------------LLSVAVTSRPDCYWGRNCRTQV 1020
Query: 422 RSEEHARKRNHVCRPTR 438
++ HA K NH+C TR
Sbjct: 1021 KA-HHAMKFNHICEQTR 1036
>emb|CAG09691.1| unnamed protein product [Tetraodon nigroviridis]
Length = 709
Score = 82.0 bits (201), Expect = 3e-14
Identities = 55/200 (27%), Positives = 91/200 (45%), Gaps = 18/200 (9%)
Query: 243 FQCDPSTIHLQCQECGGMMPSRTG-LG---VPQYCLGCDRPFCGAYWSALGVTGSGSYPV 298
++C P H+ C C MP R LG V Q C C R FC YW G G
Sbjct: 521 YRCPPRGFHVFCTCCLQPMPDRRAELGSQQVAQQCGLCQRSFCHMYW---GCRRIGCRGC 577
Query: 299 CSPDTLRPISEHTISGIPLVAHEKNLHEQNITESCIRQMGRTLQDVISEWIAKLNNREID 358
+P + ++ + G+ N +E + ++ + G++ +D+ E + + + D
Sbjct: 578 LAPFSELNFTDKCLDGVL----NNNSYESEVLQNYLSSRGKSWKDLRQEALQSIEQGKYD 633
Query: 359 TTRLMLNHAEMMTAGTLVCSDCYQKLVSFFLYWFRISTPKHLLPPEASTREDCWYGYACR 418
T ++ + T++C C + ++ Y +R S P LP + R DC++G CR
Sbjct: 634 LTDRRISRS------TIMCIYCGLRALADLAYKYRQSIPPSELPAAVTARPDCYWGRNCR 687
Query: 419 TQHRSEEHARKRNHVCRPTR 438
TQ ++ HA K NH+C TR
Sbjct: 688 TQVKA-HHAMKFNHICEQTR 706
Score = 64.7 bits (156), Expect = 5e-09
Identities = 26/63 (41%), Positives = 37/63 (58%), Gaps = 6/63 (9%)
Query: 113 CSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQFVGKNHFLRT 172
C IC ++ HD +++ PC+H FC C+S W+ RS LCP CR V+ + KNH L
Sbjct: 268 CVICQDLLHDCISLQPCMHVFCAACYSGWMERSS------LCPTCRCPVERIRKNHILNN 321
Query: 173 IAE 175
+ E
Sbjct: 322 LVE 324
>emb|CAB70812.1| hypothetical protein [Homo sapiens]
Length = 306
Score = 81.3 bits (199), Expect = 6e-14
Identities = 57/207 (27%), Positives = 90/207 (42%), Gaps = 20/207 (9%)
Query: 238 TVVGGFQCDPSTIHLQCQECGGMMPSRTGLG------VPQYCLGCDRPFCGAYWSALGVT 291
T V + C H C C MP R PQ C C +PFC YW G T
Sbjct: 111 TAVQDYVCPLQGSHALCTCCFQPMPDRRAEREQDPRVAPQQCAVCLQPFCHLYW---GCT 167
Query: 292 GSGSYPVCSPDTLRPISEHTISGIPLVAHEKNLHEQNITESCIRQMGRTLQDVISEWIAK 351
+G Y +P + + + G+ N +E +I ++ + G T +++++E +
Sbjct: 168 RTGCYGCLAPFCELNLGDKCLDGVL----NNNSYESDILKNYLATRGLTWKNMLTESLVA 223
Query: 352 LNNREIDTTRLMLNHAEMMTAGTLVCSDCYQKLVSFFLYWFRISTPKHLLPPEASTREDC 411
L + L +T T++C C + Y +R + P LP ++R DC
Sbjct: 224 LQRG------VFLLSDYRVTGDTVLCYCCGLRSFRELTYQYRQNIPASELPVAVTSRPDC 277
Query: 412 WYGYACRTQHRSEEHARKRNHVCRPTR 438
++G CRTQ ++ HA K NH+C TR
Sbjct: 278 YWGRNCRTQVKA-HHAMKFNHICEQTR 303
>gb|AAW24657.1| unknown [Schistosoma japonicum]
Length = 227
Score = 64.7 bits (156), Expect = 5e-09
Identities = 51/198 (25%), Positives = 84/198 (41%), Gaps = 18/198 (9%)
Query: 242 GFQCDPSTIHLQCQECGGMMPSRTGLGVPQY----CLGCDRPFCGAYWSALGVTGSGSYP 297
G + + + H C C MP +T L + C C FC S + G +
Sbjct: 39 GPRINECSAHCYCACCNRPMPFQTTLQLHGMNRTECEVCRHTFC----SLINPHGCST-- 92
Query: 298 VCSPDTLRPISEHT-ISGIPLVAHEKNLHEQNITESCIRQMGRTLQDVISEWIAKLNNRE 356
C L + + T + IP N E I + + G ++ IS+ ++ N
Sbjct: 93 -CDGYCLSRVQDLTRYNSIPRTLLLNNRIETRILDDYLTSQGISIPIFISQCLSLYINTT 151
Query: 357 IDTTRLMLNHAEMMTAGTLVCSDCYQKLVSFFLYWFRISTPKHLLPPEASTREDCWYGYA 416
+ +LN +L+C +C + L+S Y +R+S LP + T+ +C+YG
Sbjct: 152 VIYPSCILNE------NSLICKNCGEYLLSQLAYQYRVSICSDELPNDVITKPNCYYGRY 205
Query: 417 CRTQHRSEEHARKRNHVC 434
CR Q + HAR+ NH+C
Sbjct: 206 CRYQSYNYHHARRFNHIC 223
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.321 0.134 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 778,940,511
Number of Sequences: 2540612
Number of extensions: 33033861
Number of successful extensions: 74636
Number of sequences better than 10.0: 1443
Number of HSP's better than 10.0 without gapping: 275
Number of HSP's successfully gapped in prelim test: 1169
Number of HSP's that attempted gapping in prelim test: 73817
Number of HSP's gapped (non-prelim): 1633
length of query: 441
length of database: 863,360,394
effective HSP length: 131
effective length of query: 310
effective length of database: 530,540,222
effective search space: 164467468820
effective search space used: 164467468820
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 77 (34.3 bits)
Medicago: description of AC148755.4


