

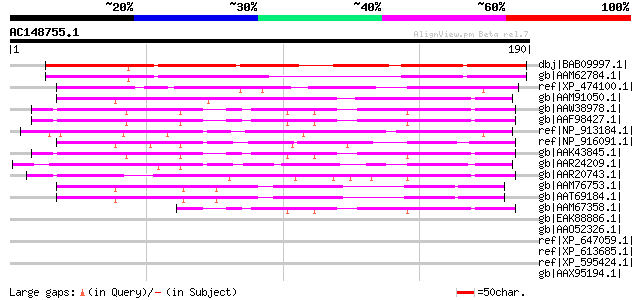
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148755.1 + phase: 0
(190 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAB09997.1| unnamed protein product [Arabidopsis thaliana] g... 174 1e-42
gb|AAM62784.1| unknown [Arabidopsis thaliana] gi|18415830|ref|NP... 134 1e-30
ref|XP_474100.1| OSJNBa0070O11.3 [Oryza sativa (japonica cultiva... 92 6e-18
gb|AAM91050.1| AT5g53830/MGN6_22 [Arabidopsis thaliana] gi|97590... 77 2e-13
gb|AAW38978.1| At1g28280 [Arabidopsis thaliana] gi|56550671|gb|A... 68 1e-10
gb|AAF98427.1| Unknown protein [Arabidopsis thaliana] gi|2551832... 68 1e-10
ref|NP_913184.1| B1015E06.22 [Oryza sativa (japonica cultivar-gr... 67 2e-10
ref|NP_916091.1| P0481E12.14 [Oryza sativa (japonica cultivar-gr... 66 4e-10
gb|AAK43845.1| Unknown protein [Arabidopsis thaliana] 65 1e-09
gb|AAR24209.1| At1g80450 [Arabidopsis thaliana] gi|15220141|ref|... 61 1e-08
gb|AAR20743.1| At3g15300 [Arabidopsis thaliana] gi|9294257|dbj|B... 59 9e-08
gb|AAM76753.1| hypothetical protein [Arabidopsis thaliana] 54 2e-06
gb|AAT69184.1| hypothetical protein At2g33780 [Arabidopsis thali... 54 2e-06
gb|AAM67358.1| unknown [Arabidopsis thaliana] 52 1e-05
gb|EAK88886.1| hypothetical protein with 6 transmembrane domains... 44 0.002
gb|AAO52326.1| similar to Dictyostelium discoideum (Slime mold).... 40 0.034
ref|XP_647059.1| hypothetical protein DDB0202368 [Dictyostelium ... 40 0.034
ref|XP_613685.1| PREDICTED: similar to CBF1 interacting corepres... 40 0.044
ref|XP_595424.1| PREDICTED: similar to CBF1 interacting corepres... 40 0.044
gb|AAX95194.1| hypothetical protein LOC_Os11g12790 [Oryza sativa... 39 0.058
>dbj|BAB09997.1| unnamed protein product [Arabidopsis thaliana]
gi|30682417|ref|NP_850793.1| VQ motif-containing protein
[Arabidopsis thaliana]
Length = 173
Score = 174 bits (441), Expect = 1e-42
Identities = 102/179 (56%), Positives = 120/179 (66%), Gaps = 22/179 (12%)
Query: 14 CKPLTTFVHTNTGAFREVVQRLTGPSEGN---ATKEEQTAKVAPITKRTPPKLHERRKCM 70
CKP+TTFV T+T FRE+VQRLTGP+E N AT E K A I KR KLHERR+CM
Sbjct: 13 CKPVTTFVQTDTNTFREIVQRLTGPTENNAAAATPEATVIKTA-IQKRPTSKLHERRQCM 71
Query: 71 KPKLEIVKPNLQYHKQPGASPSSKSRNSSFPSSPVSGSGSSSLLPSPTTPSTLFSRLTIM 130
+PKLEIVKP L + K G +PSSKS N++ +SPV TPS+LFS L+++
Sbjct: 72 RPKLEIVKPPLSF-KPTGTTPSSKSGNTNLLTSPVG------------TPSSLFSNLSLI 118
Query: 131 EDEKKQGSVINEINIEEEEKAIKERRFYLHPSPRSKASGFNEPELLNLFPLASPKACEK 189
E E + NIEEEEKAIKERRFYLHPSPRSK G+ EPELL LFPL SP + K
Sbjct: 119 EGEPDSCTT----NIEEEEKAIKERRFYLHPSPRSK-PGYTEPELLTLFPLTSPNSSGK 172
>gb|AAM62784.1| unknown [Arabidopsis thaliana] gi|18415830|ref|NP_568198.1| VQ
motif-containing protein [Arabidopsis thaliana]
Length = 142
Score = 134 bits (337), Expect = 1e-30
Identities = 84/179 (46%), Positives = 94/179 (51%), Gaps = 53/179 (29%)
Query: 14 CKPLTTFVHTNTGAFREVVQRLTGPSEGN---ATKEEQTAKVAPITKRTPPKLHERRKCM 70
CKP+TTFV T+T FRE+VQRLTGP+E N AT E K A I KR KLHERR+CM
Sbjct: 13 CKPVTTFVQTDTNTFREIVQRLTGPTENNAAAATPEATVIKTA-IQKRPTSKLHERRQCM 71
Query: 71 KPKLEIVKPNLQYHKQPGASPSSKSRNSSFPSSPVSGSGSSSLLPSPTTPSTLFSRLTIM 130
+PKLEIVKP L + G S +
Sbjct: 72 RPKLEIVKPPLSFKPTEGEPDSCTT----------------------------------- 96
Query: 131 EDEKKQGSVINEINIEEEEKAIKERRFYLHPSPRSKASGFNEPELLNLFPLASPKACEK 189
NIEEEEKAIKERRFYLHPSPRSK G+ EPELL LFPL SP + K
Sbjct: 97 -------------NIEEEEKAIKERRFYLHPSPRSK-PGYTEPELLTLFPLTSPNSSGK 141
>ref|XP_474100.1| OSJNBa0070O11.3 [Oryza sativa (japonica cultivar-group)]
gi|32487350|emb|CAE03172.1| OSJNBa0070O11.3 [Oryza
sativa (japonica cultivar-group)]
gi|5852187|emb|CAB55425.1| zhb0017.1 [Oryza sativa
(indica cultivar-group)]
Length = 202
Score = 92.4 bits (228), Expect = 6e-18
Identities = 71/186 (38%), Positives = 94/186 (50%), Gaps = 39/186 (20%)
Query: 18 TTFVHTNTGAFREVVQRLTGPSEGNATKEEQTAKVAPITKRTPPKLHERRKCMKPKLEIV 77
TTFV + FR +VQ+LTG +K A AP+ +R PKL ERR+ +LE+
Sbjct: 33 TTFVQADPATFRALVQKLTGAPGSGGSKP---APAAPVMRR--PKLQERRRAAPARLELA 87
Query: 78 KPNLQY----HKQPGASP------------SSKSRNSSFPSSPVSGSGSSSLLPSPTTPS 121
+P Y H + SP SS S +SS PSS SSSL PSP S
Sbjct: 88 RPQPLYYSHHHHRLMHSPVSPMDYAYVMASSSSSSSSSLPSS------SSSLSPSPPASS 141
Query: 122 TLFSRLTIMEDEKKQGSVINEINIEEEEKAIKERRFYLHPSPRSKASGFNE-PELLNLFP 180
+ + I ++E+ E EEKAI + FYLH SPRS +G E P+LL LFP
Sbjct: 142 SSCGVVVITKEEE-----------EREEKAIASKGFYLHSSPRSGGAGDGERPKLLPLFP 190
Query: 181 LASPKA 186
+ SP++
Sbjct: 191 VHSPRS 196
>gb|AAM91050.1| AT5g53830/MGN6_22 [Arabidopsis thaliana] gi|9759077|dbj|BAB09555.1|
unnamed protein product [Arabidopsis thaliana]
gi|15238851|ref|NP_200194.1| VQ motif-containing protein
[Arabidopsis thaliana] gi|13926342|gb|AAK49634.1|
AT5g53830/MGN6_22 [Arabidopsis thaliana]
Length = 243
Score = 77.0 bits (188), Expect = 2e-13
Identities = 63/198 (31%), Positives = 87/198 (43%), Gaps = 38/198 (19%)
Query: 18 TTFVHTNTGAFREVVQRLTG-------------PSEGNATKEEQTAKVAPITKRTPPKLH 64
TTFV +T F++VVQ LTG PS N + + + PI K KL+
Sbjct: 49 TTFVQADTSTFKQVVQMLTGSSTDTTTGKHHEAPSPVNNNNKGSSFSIPPIKKTNSFKLY 108
Query: 65 ERRKCMK------------------PKLEIVKPNLQYHKQPGASPSSKSRNSSFPSSPVS 106
ERR+ +L N +H+ P SP + S + + SP
Sbjct: 109 ERRQNNNNMFAKNDLMINTLRLQNSQRLMFTGGNSSHHQSPRFSPRNSSSSENILLSPSM 168
Query: 107 GSGSSSLLPSPTTPSTLFSRLTIMEDEKKQGSVINEINIEEEEKAIKERRFYLHPSPRSK 166
L SP TP L +D + S ++ N EE+KAI ++ FYLHPSP S
Sbjct: 169 LDFPKLGLNSPVTP------LRSNDDPFNKSSPLSLGNSSEEDKAIADKGFYLHPSPVST 222
Query: 167 ASGFNEPELLNLFPLASP 184
++P LL LFP+ASP
Sbjct: 223 PRD-SQPLLLPLFPVASP 239
>gb|AAW38978.1| At1g28280 [Arabidopsis thaliana] gi|56550671|gb|AAV97789.1|
At1g28280 [Arabidopsis thaliana]
gi|30690315|ref|NP_564303.2| VQ motif-containing protein
[Arabidopsis thaliana]
Length = 247
Score = 68.2 bits (165), Expect = 1e-10
Identities = 70/209 (33%), Positives = 94/209 (44%), Gaps = 52/209 (24%)
Query: 9 STTGDCKPLTTFVHTNTGAFREVVQRLTGPSEG---------NATKEEQTAKVAPITKRT 59
S +G+ P TTFV +T +F++VVQ LTG +E N T + + P +
Sbjct: 49 SESGNPYP-TTFVQADTSSFKQVVQMLTGSAERPKHGSSLKPNPTHHQPDPRSTPSSFSI 107
Query: 60 PP-----------------KLHERRKCMKPKLEIVKPNLQYHKQPGASPSSKSRNSSF-P 101
PP +L+ERR MK NL+ + +P NS+F P
Sbjct: 108 PPIKAVPNKKQSSSSASGFRLYERRNSMK--------NLKINP---LNPVFNPVNSAFSP 156
Query: 102 SSPVSGSGS----SSLLPSPTTPSTLFSRLTIMEDEKKQGSVINEIN-IEEEEKAIKERR 156
P S S SL+ SP TP I + + GS N + EEKA+KER
Sbjct: 157 RKPEILSPSILDFPSLVLSPVTP-------LIPDPFDRSGSSNQSPNELAAEEKAMKERG 209
Query: 157 FYLHPSPRSKASGFNEPELLNLFPLASPK 185
FYLHPSP + EP LL LFP+ SP+
Sbjct: 210 FYLHPSPATTPMD-PEPRLLPLFPVTSPR 237
>gb|AAF98427.1| Unknown protein [Arabidopsis thaliana] gi|25518323|pir||A86409
hypothetical protein F3H9.7 [imported] - Arabidopsis
thaliana
Length = 259
Score = 68.2 bits (165), Expect = 1e-10
Identities = 70/209 (33%), Positives = 94/209 (44%), Gaps = 52/209 (24%)
Query: 9 STTGDCKPLTTFVHTNTGAFREVVQRLTGPSEG---------NATKEEQTAKVAPITKRT 59
S +G+ P TTFV +T +F++VVQ LTG +E N T + + P +
Sbjct: 61 SESGNPYP-TTFVQADTSSFKQVVQMLTGSAERPKHGSSLKPNPTHHQPDPRSTPSSFSI 119
Query: 60 PP-----------------KLHERRKCMKPKLEIVKPNLQYHKQPGASPSSKSRNSSF-P 101
PP +L+ERR MK NL+ + +P NS+F P
Sbjct: 120 PPIKAVPNKKQSSSSASGFRLYERRNSMK--------NLKINP---LNPVFNPVNSAFSP 168
Query: 102 SSPVSGSGS----SSLLPSPTTPSTLFSRLTIMEDEKKQGSVINEIN-IEEEEKAIKERR 156
P S S SL+ SP TP I + + GS N + EEKA+KER
Sbjct: 169 RKPEILSPSILDFPSLVLSPVTP-------LIPDPFDRSGSSNQSPNELAAEEKAMKERG 221
Query: 157 FYLHPSPRSKASGFNEPELLNLFPLASPK 185
FYLHPSP + EP LL LFP+ SP+
Sbjct: 222 FYLHPSPATTPMD-PEPRLLPLFPVTSPR 249
>ref|NP_913184.1| B1015E06.22 [Oryza sativa (japonica cultivar-group)]
Length = 212
Score = 67.4 bits (163), Expect = 2e-10
Identities = 69/203 (33%), Positives = 88/203 (42%), Gaps = 32/203 (15%)
Query: 5 PVHGSTTGD-CKPL-TTFVHTNTGAFREVVQRLTGPSE----------GNATKEEQTAKV 52
P+ + GD P TTFV +T +F++VVQ LTG E A AK
Sbjct: 16 PLPAAAAGDPSNPFPTTFVQADTTSFKQVVQILTGTPETAAAAAAGGAAAAAPPPAAAKP 75
Query: 53 APIT---KRTPPKLHERRKCMKPKLEIVKPNLQYHKQPGASPSSKSRNSSFPSSPVS--- 106
AP K+ KL+ERR MK L+++ P L P A+ + F SP
Sbjct: 76 APAPPGPKKPAFKLYERRSSMK-SLKMLCPLL-----PAAAAFAAGGGGGFSPSPRGFSP 129
Query: 107 -GSGSSSLLPSPTTPSTLFSRLTIMEDEKKQGSVINEINIEEEEKAIKERRFYLHPSPRS 165
G G L PS +L + GS + E++AI E+ FYLHPSPR
Sbjct: 130 RGGGMEVLSPSMLDFPSLALGSPVTPLPPLPGS---DEAAAAEDRAIAEKGFYLHPSPRG 186
Query: 166 KASGFNE----PELLNLFPLASP 184
A E P LL LFPL SP
Sbjct: 187 NAGAAGELQPPPRLLPLFPLQSP 209
>ref|NP_916091.1| P0481E12.14 [Oryza sativa (japonica cultivar-group)]
gi|14209533|dbj|BAB56029.1| VQ motif-containing
protein-like [Oryza sativa (japonica cultivar-group)]
Length = 231
Score = 66.2 bits (160), Expect = 4e-10
Identities = 67/189 (35%), Positives = 83/189 (43%), Gaps = 44/189 (23%)
Query: 18 TTFVHTNTGAFREVVQRLTG---PSEGNATKEEQTA-------------KVAPITKRTPP 61
TTFV +T +F++VVQ LTG PS+ AT A P + P
Sbjct: 60 TTFVQADTASFKQVVQMLTGAEQPSKNAATAATAAAGNSSAAGIGGGQGANGPCRPKKPA 119
Query: 62 -KLHERRKCMKPKLEIVKPNLQYHKQPGASPSSKSRNSSFPS--SPVSGSGSSSLLPSPT 118
KL+ERR +K L+++ P GA PS R P SP S SL SP
Sbjct: 120 FKLYERRSSLK-NLKMIAPLAM-----GALPSPTGRKVGTPEILSP-SVLDFPSLKLSPV 172
Query: 119 TPST--LFSRLTIMEDEKKQGSVINEINIEEEEKAIKERRFYLHPSPRSKASGFNEPELL 176
TP T F+R E + E AI ER F+LHPSPR G P LL
Sbjct: 173 TPLTGEPFNRSPASSSE------------DAERAAISERGFFLHPSPR----GAEPPRLL 216
Query: 177 NLFPLASPK 185
LFP+ SP+
Sbjct: 217 PLFPVTSPR 225
>gb|AAK43845.1| Unknown protein [Arabidopsis thaliana]
Length = 247
Score = 65.1 bits (157), Expect = 1e-09
Identities = 69/209 (33%), Positives = 92/209 (44%), Gaps = 52/209 (24%)
Query: 9 STTGDCKPLTTFVHTNTGAFREVVQRLTGPSEG---------NATKEEQTAKVAPITKRT 59
S +G+ P TTF +T +F++VVQ LTG E N T + + P +
Sbjct: 49 SESGNPYP-TTFDQADTSSFKQVVQMLTGSGERPKHGSSIKPNPTHHQPDPRSTPSSFSI 107
Query: 60 PP-----------------KLHERRKCMKPKLEIVKPNLQYHKQPGASPSSKSRNSSF-P 101
PP +L+ERR MK NL+ + +P NS+F P
Sbjct: 108 PPIKAVPNKKQSSSSASGFRLYERRNSMK--------NLKINP---LNPVFNPVNSAFSP 156
Query: 102 SSPVSGSGS----SSLLPSPTTPSTLFSRLTIMEDEKKQGSVINEIN-IEEEEKAIKERR 156
P S S SL+ SP TP I + + GS N + EEKA+KER
Sbjct: 157 RKPEILSPSILDFPSLVLSPVTP-------LIPDPFDRSGSSNQSPNELAAEEKAMKERG 209
Query: 157 FYLHPSPRSKASGFNEPELLNLFPLASPK 185
FYLHPSP + EP LL LFP+ SP+
Sbjct: 210 FYLHPSPATTPMD-PEPRLLPLFPVTSPR 237
>gb|AAR24209.1| At1g80450 [Arabidopsis thaliana] gi|15220141|ref|NP_178160.1| VQ
motif-containing protein [Arabidopsis thaliana]
gi|46931258|gb|AAT06433.1| At1g80450 [Arabidopsis
thaliana] gi|25406665|pir||C96836 unknown protein
T21F11.22 [imported] - Arabidopsis thaliana
gi|6730734|gb|AAF27124.1| unknown protein; 73942-74475
[Arabidopsis thaliana]
Length = 177
Score = 61.2 bits (147), Expect = 1e-08
Identities = 62/188 (32%), Positives = 85/188 (44%), Gaps = 34/188 (18%)
Query: 2 EKPPVHGSTTGDCKPLTTFVHTNTGAFREVVQRLTGPSEGNATKEEQTAKVA----PITK 57
++PP + + +P T FV + FR +VQ+LTG ++ A P+T
Sbjct: 4 QQPPSYAT-----EPNTMFVQADPSNFRNIVQKLTGAPPDISSSSFSAVSAAHQKLPLTP 58
Query: 58 RTPP-KLHERRKCMKPKLEIVKPNLQYHKQPGASPSSKSRNSSFPSSPVSGSGSSSLLPS 116
+ P KLHERR+ K K+E+ N+ P + S R F SPVS S
Sbjct: 59 KKPAFKLHERRQSSK-KMELKVNNIT---NPNDAFSHFHRG--FLVSPVSHLDPFWARVS 112
Query: 117 PTTPSTLFSRLTIMEDEKKQGSVINEINIEEEEKAIKERRFYLHPSPRSKASGFNEPELL 176
P + E+ Q +EE+KAI E+ FY PSPRS + PELL
Sbjct: 113 PHSAR---------EEHHAQPD-------KEEQKAIAEKGFYFLPSPRSGSE--PAPELL 154
Query: 177 NLFPLASP 184
LFPL SP
Sbjct: 155 PLFPLRSP 162
>gb|AAR20743.1| At3g15300 [Arabidopsis thaliana] gi|9294257|dbj|BAB02159.1| unnamed
protein product [Arabidopsis thaliana]
gi|15232547|ref|NP_188148.1| VQ motif-containing protein
[Arabidopsis thaliana] gi|41349916|gb|AAS00343.1|
At3g15300 [Arabidopsis thaliana]
Length = 219
Score = 58.5 bits (140), Expect = 9e-08
Identities = 59/200 (29%), Positives = 94/200 (46%), Gaps = 33/200 (16%)
Query: 7 HGSTTGDCKPLTTFVHTNTGAFREVVQRLTGPSEGNATKEEQTAKVAPITKRTPPKLHER 66
H T D P TTFV ++ +F++VVQ LTG S +P + R P +
Sbjct: 27 HIITRSDHYP-TTFVQADSSSFKQVVQMLTGSSSPR----------SPDSPRPPTTPSGK 75
Query: 67 RKCMKPKLEIVKP-----NLQYHKQPGASPSSKSRNSSFPSS-PVSGSGSSSLLPSP--- 117
+ P ++ +P N Y ++ ++ +NS ++ + G G+ S SP
Sbjct: 76 GNFVIPPIKTAQPKKHSGNKLYERRSHGGFNNNLKNSLMINTLMIGGGGAGSPRFSPRNQ 135
Query: 118 --TTPSTL-FSRLTIME--DEKKQGSVINEIN-------IEEEEKAIKERRFYLHPSPRS 165
+PS L F +L + KQG+ NE + + EEE+ I ++ +YLH SP S
Sbjct: 136 EILSPSCLDFPKLALNSPVTPLKQGTNGNEGDPFDKMSPLSEEERGIADKGYYLHRSPIS 195
Query: 166 KASGFNEPELLNLFPLASPK 185
+EP+LL LFP+ SP+
Sbjct: 196 TPRD-SEPQLLPLFPVTSPR 214
>gb|AAM76753.1| hypothetical protein [Arabidopsis thaliana]
Length = 204
Score = 53.9 bits (128), Expect = 2e-06
Identities = 55/184 (29%), Positives = 76/184 (40%), Gaps = 48/184 (26%)
Query: 18 TTFVHTNTGAFREVVQRLTG----PSEGNATKEEQTAKVAPITKRTPPK------LHERR 67
TTF+ T+ +F++VVQ LTG P+ + + PI T K L ERR
Sbjct: 36 TTFIRTDPSSFKQVVQLLTGIPKNPTHQPDPRFPPFHSIPPIKAVTNKKQSSSFRLSERR 95
Query: 68 KCMKPKL----------EIVKPNLQYHKQPGASPSSKSRNSSFPSSPVSGSGSSSLLPSP 117
MK L EI+ P + SP ++ S P GS S PS
Sbjct: 96 NSMKHYLNINPTHSGPPEILTPTILNFPALDLSP-----DTPLMSDPFYXPGSFSQSPSX 150
Query: 118 TTPSTLFSRLTIMEDEKKQGSVINEINIEEEEKAIKERRFYLHPSPRSKASGFNEPELLN 177
+ PS +++E++IKE+ FYL PSP S EP LL+
Sbjct: 151 SKPSF----------------------DDDQERSIKEKGFYLRPSP-STTPRDTEPRLLS 187
Query: 178 LFPL 181
LFP+
Sbjct: 188 LFPM 191
>gb|AAT69184.1| hypothetical protein At2g33780 [Arabidopsis thaliana]
gi|2253579|gb|AAC69144.1| hypothetical protein
[Arabidopsis thaliana] gi|25408308|pir||E84749
hypothetical protein At2g33780 [imported] - Arabidopsis
thaliana gi|15226165|ref|NP_180934.1| VQ
motif-containing protein [Arabidopsis thaliana]
Length = 204
Score = 53.9 bits (128), Expect = 2e-06
Identities = 55/184 (29%), Positives = 76/184 (40%), Gaps = 48/184 (26%)
Query: 18 TTFVHTNTGAFREVVQRLTG----PSEGNATKEEQTAKVAPITKRTPPK------LHERR 67
TTF+ T+ +F++VVQ LTG P+ + + PI T K L ERR
Sbjct: 36 TTFIRTDPSSFKQVVQLLTGIPKNPTHQPDPRFPPFHSIPPIKAVTNKKQSSSFRLSERR 95
Query: 68 KCMKPKL----------EIVKPNLQYHKQPGASPSSKSRNSSFPSSPVSGSGSSSLLPSP 117
MK L EI+ P + SP ++ S P GS S PS
Sbjct: 96 NSMKHYLNINPTHSGPPEILTPTILNFPALDLSP-----DTPLMSDPFYRPGSFSQSPSD 150
Query: 118 TTPSTLFSRLTIMEDEKKQGSVINEINIEEEEKAIKERRFYLHPSPRSKASGFNEPELLN 177
+ PS +++E++IKE+ FYL PSP S EP LL+
Sbjct: 151 SKPSF----------------------DDDQERSIKEKGFYLRPSP-STTPRDTEPRLLS 187
Query: 178 LFPL 181
LFP+
Sbjct: 188 LFPM 191
>gb|AAM67358.1| unknown [Arabidopsis thaliana]
Length = 141
Score = 51.6 bits (122), Expect = 1e-05
Identities = 49/130 (37%), Positives = 63/130 (47%), Gaps = 25/130 (19%)
Query: 62 KLHERRKCMKPKLEIVKPNLQYHKQPGASPSSKSRNSSF-PSSPVSGSGS----SSLLPS 116
+L+ERR MK NL+ + +P NS+F P P S S SL+ S
Sbjct: 21 RLYERRNSMK--------NLKINP---LNPVFNPVNSAFSPRKPEILSPSILDFPSLVLS 69
Query: 117 PTTPSTLFSRLTIMEDEKKQGSVINEIN-IEEEEKAIKERRFYLHPSPRSKASGFNEPEL 175
P TP I + + GS N + EEKA+KER FYLHPSP + EP L
Sbjct: 70 PVTP-------LIPDPFDRSGSSNQSPNELAAEEKAMKERGFYLHPSPATTPMD-PEPRL 121
Query: 176 LNLFPLASPK 185
L LFP+ SP+
Sbjct: 122 LPLFPVTSPR 131
>gb|EAK88886.1| hypothetical protein with 6 transmembrane domains [Cryptosporidium
parvum] gi|66358226|ref|XP_626291.1| hypothetical
protein cgd2_540 [Cryptosporidium parvum]
Length = 572
Score = 44.3 bits (103), Expect = 0.002
Identities = 32/119 (26%), Positives = 55/119 (45%), Gaps = 2/119 (1%)
Query: 29 REVVQRLTGPSEGNATKEE-QTAKVAPITKRTPPKLHERRKCMKPKLEIVKPNLQYHKQP 87
+EV+Q + P EG T+ + +A P ++++PPK + P+ + +
Sbjct: 109 KEVIQVPSVPKEGGNTQSQTSSAPSMPSSQQSPPKQSSSQPAPPPQQSSSQQSSSQQSSS 168
Query: 88 GASPSSKSRNSSFPSSPVSGSGSSSLLPSPTTPSTLFSRLTIMEDEKKQGSVINEINIE 146
SP +S PS P +SS PS +TPS+ S+ T M+ +K I+ I+
Sbjct: 169 QQSPPKQSPPQQSPSQPAPPPRNSSSQPSSSTPSSKPSQ-TQMDSQKSNSQKISPQKIQ 226
>gb|AAO52326.1| similar to Dictyostelium discoideum (Slime mold). Protein kinase
gi|66820326|ref|XP_643791.1| putative LISK family
protein kinase [Dictyostelium discoideum]
gi|60471909|gb|EAL69863.1| putative tyrosine kinase-like
(TKL) protein [Dictyostelium discoideum]
Length = 921
Score = 40.0 bits (92), Expect = 0.034
Identities = 28/114 (24%), Positives = 49/114 (42%), Gaps = 4/114 (3%)
Query: 8 GSTTGDCKPLTTFVHTNTGAFREVVQRLTGPSEGNATKEEQTAKVAPITKRTPPKLHERR 67
G T + + + +T A + + LT S + Q + + TK+ P +
Sbjct: 802 GDTVSHRRSRSLDIKNSTKAIIKKFEELTRFSSQQSNLVHQPSSSSSSTKQPPTSQFLQN 861
Query: 68 KCMKPKLEIVKPNLQYHKQPGASPSSKSRNSSFPSSPVSGSGSSSLLPSPTTPS 121
M+ KL + L + K P SP S P+S S S+S +P+P++P+
Sbjct: 862 PRMEAKLSFAQETLVFEKSPSTSPKS----LDIPTSSSLTSNSNSSIPAPSSPT 911
>ref|XP_647059.1| hypothetical protein DDB0202368 [Dictyostelium discoideum]
gi|60475169|gb|EAL73105.1| hypothetical protein
DDB0202368 [Dictyostelium discoideum]
Length = 1141
Score = 40.0 bits (92), Expect = 0.034
Identities = 37/136 (27%), Positives = 61/136 (44%), Gaps = 16/136 (11%)
Query: 15 KPLTTFVHTNTGAFREVVQRL-TGPSEGNATKEEQTAKVAPITKRTPPKLHERRKCMKPK 73
+P+T F+ T ++++L T + + + Q + H ++
Sbjct: 967 EPITNFISTLFFESHNIIKKLKTTTTSSSQNYQSQNYQ------------HTVKEFNDNS 1014
Query: 74 LEIVKPNLQYHKQPGASPSSKSRNSSFPSSPVSGSGSSSLLPSPTTPSTLFSRLTIMEDE 133
+E +K QY +S SS S +SS SS S S SSS L SP+ S L +I E+
Sbjct: 1015 IESMKEERQYCSSSSSSSSSSSSSSSSSSSSSSSSSSSSTLSSPS--SKLIGE-SISENR 1071
Query: 134 KKQGSVINEINIEEEE 149
KK ++ EI +E+
Sbjct: 1072 KKNEQLLQEIEKLKEQ 1087
>ref|XP_613685.1| PREDICTED: similar to CBF1 interacting corepressor isoform 1,
partial [Bos taurus]
Length = 424
Score = 39.7 bits (91), Expect = 0.044
Identities = 35/122 (28%), Positives = 57/122 (46%), Gaps = 4/122 (3%)
Query: 45 KEEQTAKVAPITKRTPPKLHERRKCMKPKLEIVKPNLQYHKQPGASPSSKSRNSSFPSSP 104
K++ K+ + K+ K +R+K KL+ + + HK +SPSS S SS SP
Sbjct: 192 KQKLLRKLDRLEKKKKKKKKDRKK---KKLQKSRSKHKKHKNKPSSPSSSSTTSSSSPSP 248
Query: 105 VSGSGSSSLLPSPTTPST-LFSRLTIMEDEKKQGSVINEINIEEEEKAIKERRFYLHPSP 163
+S S SSS ++ S +++ +K + S N + EE K+ K +R H
Sbjct: 249 LSSSSSSSDSSESSSESDGKGTKIQKRRRKKNKRSGHNNSDSEERGKSKKRKRDEEHSCS 308
Query: 164 RS 165
S
Sbjct: 309 HS 310
>ref|XP_595424.1| PREDICTED: similar to CBF1 interacting corepressor isoform 1,
partial [Bos taurus]
Length = 291
Score = 39.7 bits (91), Expect = 0.044
Identities = 35/122 (28%), Positives = 57/122 (46%), Gaps = 4/122 (3%)
Query: 45 KEEQTAKVAPITKRTPPKLHERRKCMKPKLEIVKPNLQYHKQPGASPSSKSRNSSFPSSP 104
K++ K+ + K+ K +R+K KL+ + + HK +SPSS S SS SP
Sbjct: 59 KQKLLRKLDRLEKKKKKKKKDRKK---KKLQKSRSKHKKHKNKPSSPSSSSTTSSSSPSP 115
Query: 105 VSGSGSSSLLPSPTTPST-LFSRLTIMEDEKKQGSVINEINIEEEEKAIKERRFYLHPSP 163
+S S SSS ++ S +++ +K + S N + EE K+ K +R H
Sbjct: 116 LSSSSSSSDSSESSSESDGKGTKIQKRRRKKNKRSGHNNSDSEERGKSKKRKRDEEHSCS 175
Query: 164 RS 165
S
Sbjct: 176 HS 177
>gb|AAX95194.1| hypothetical protein LOC_Os11g12790 [Oryza sativa (japonica
cultivar-group)] gi|62734542|gb|AAX96651.1| hypothetical
protein [Oryza sativa (japonica cultivar-group)]
Length = 164
Score = 39.3 bits (90), Expect = 0.058
Identities = 48/169 (28%), Positives = 70/169 (41%), Gaps = 37/169 (21%)
Query: 28 FREVVQRLTG--PSEGNATKEEQTAKVAPITKRTPPKLHE----RRKCMKPKLEIVKPNL 81
F+E+VQRLTG P+E A A VA +R +H + +PKL ++P
Sbjct: 14 FKELVQRLTGQPPAEAAAVAAPAPAPVAGAPRRGRLGVHNPPAFKPTPHRPKLPTIRPEH 73
Query: 82 QYHKQPGASPSSKSRNSSFPSSPVSGSG------SSSLLPSPTT-PSTLFSRLTIMEDEK 134
ASP S S + SG+G L PSPT+ STL + + E
Sbjct: 74 PRLLAGFASPPSPPSLSPW----CSGAGQCVQNMQDELPPSPTSASSTLAEEVVVGETVS 129
Query: 135 KQGSVINEINIEEEEKAIKERRFYLHPSPRSKASGFNEPELLNLFPLAS 183
++G + ++H P + E +LLNLFPL +
Sbjct: 130 EEG-----------------KPDHMHQPPPVRT---GEAKLLNLFPLTA 158
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.309 0.127 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 359,943,731
Number of Sequences: 2540612
Number of extensions: 15797425
Number of successful extensions: 67224
Number of sequences better than 10.0: 455
Number of HSP's better than 10.0 without gapping: 56
Number of HSP's successfully gapped in prelim test: 402
Number of HSP's that attempted gapping in prelim test: 66556
Number of HSP's gapped (non-prelim): 850
length of query: 190
length of database: 863,360,394
effective HSP length: 121
effective length of query: 69
effective length of database: 555,946,342
effective search space: 38360297598
effective search space used: 38360297598
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 71 (32.0 bits)
Medicago: description of AC148755.1


