

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148654.3 - phase: 0 /pseudo
(719 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
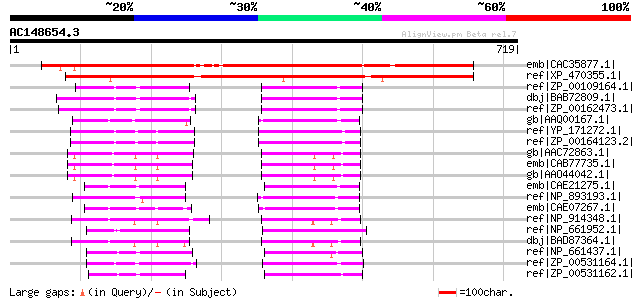
Score E
Sequences producing significant alignments: (bits) Value
emb|CAC35877.1| putative protein [Arabidopsis thaliana] gi|26450... 793 0.0
ref|XP_470355.1| unknown protein [Oryza sativa (japonica cultiva... 717 0.0
ref|ZP_00109164.1| COG2867: Oligoketide cyclase/lipid transport ... 114 1e-23
dbj|BAB72809.1| all0852 [Nostoc sp. PCC 7120] gi|25529736|pir||A... 112 3e-23
ref|ZP_00162473.1| COG2867: Oligoketide cyclase/lipid transport ... 111 7e-23
gb|AAQ00167.1| Oligoketide cyclase family enzyme [Prochlorococcu... 93 3e-17
ref|YP_171272.1| hypothetical protein syc0562_c [Synechococcus e... 93 3e-17
ref|ZP_00164123.2| COG2867: Oligoketide cyclase/lipid transport ... 93 3e-17
gb|AAC72863.1| T15B16.3 gene product [Arabidopsis thaliana] gi|7... 89 5e-16
emb|CAB77735.1| hypothetical protein [Arabidopsis thaliana] gi|1... 89 7e-16
gb|AAO44042.1| At4g01650 [Arabidopsis thaliana] gi|30678933|ref|... 89 7e-16
emb|CAE21275.1| conserved hypothetical protein [Prochlorococcus ... 85 9e-15
ref|NP_893193.1| hypothetical protein PMM1076 [Prochlorococcus m... 84 2e-14
emb|CAE07267.1| conserved hypothetical protein [Synechococcus sp... 80 2e-13
ref|NP_914348.1| P0518C01.12 [Oryza sativa (japonica cultivar-gr... 79 5e-13
ref|NP_661952.1| hypothetical protein CT1061 [Chlorobium tepidum... 78 9e-13
dbj|BAD87364.1| unknown protein [Oryza sativa (japonica cultivar... 77 3e-12
ref|NP_661437.1| hypothetical protein CT0537 [Chlorobium tepidum... 70 2e-10
ref|ZP_00531164.1| Streptomyces cyclase/dehydrase [Chlorobium ph... 65 1e-08
ref|ZP_00531162.1| Streptomyces cyclase/dehydrase [Chlorobium ph... 65 1e-08
>emb|CAC35877.1| putative protein [Arabidopsis thaliana] gi|26450803|dbj|BAC42510.1|
unknown protein [Arabidopsis thaliana]
gi|29029030|gb|AAO64894.1| At5g08720 [Arabidopsis
thaliana] gi|22326677|ref|NP_680157.1| expressed protein
[Arabidopsis thaliana]
Length = 719
Score = 793 bits (2047), Expect = 0.0
Identities = 423/631 (67%), Positives = 482/631 (76%), Gaps = 36/631 (5%)
Query: 46 SKPSFLSLSLFFPRHFHKSIALSST------TQCKPRSH--LGGNLNNGLEED------- 90
++P FLS+ L P ++S++ +C H GG +NGL D
Sbjct: 20 NEPVFLSVLLPSPSRIRVFSSISTSGIGGGVAKCHGTRHSGAGGRGDNGLRRDSGLGFDE 79
Query: 91 -GDREVHCELQVVSWRERRVKAEISINADINSVWNALTDYEHLADFIPNLVWSGRIPCPF 149
G+R+V CE+ V+SWRERR++ EI +++D SVWN LTDYE LADFIPNLVWSGRIPCP
Sbjct: 80 RGERKVRCEVDVISWRERRIRGEIWVDSDSQSVWNVLTDYERLADFIPNLVWSGRIPCPH 139
Query: 150 PGRIWLEQRGFQRAMYWHIEARVVLDLQELLNSEWDRELHFSMVDGDFKKFEGKWSVKSG 209
PGRIWLEQRG QRA+YWHIEARVVLDL E L+S RELHFSMVDGDFKKFEGKWSVKSG
Sbjct: 140 PGRIWLEQRGLQRALYWHIEARVVLDLHECLDSPNGRELHFSMVDGDFKKFEGKWSVKSG 199
Query: 210 TRSSSTNLSYEVNVIPRFNFPAIFLERIVRSDLPVNLRALAYRVERNLLGNQKLPQPEDD 269
RS T LSYEVNVIPRFNFPAIFLERI+RSDLPVNLRA+A + E+ K P +D
Sbjct: 200 IRSVGTVLSYEVNVIPRFNFPAIFLERIIRSDLPVNLRAVARQAEKIYKDCGK-PSIIED 258
Query: 270 LHKTSLVVNGSSVKKINGSLCETDKLAPGQDKEGLDTSISGSLPASSSELNSNWGIFGKV 329
L L + S NG E D LA E S GSL A S+ELN+NWG++GK
Sbjct: 259 L----LGIISSQPAPSNG--IEFDSLAT----ERSVASSVGSL-AHSNELNNNWGVYGKA 307
Query: 330 CSLDKPCVVDEVHLRRFDGLLENGGVHRCVVASITVKAPVRDVWNVMSSYETLPEIVPNL 389
C LDKPC VDEVHLRRFDGLLENGGVHRC VASITVKAPV +VW V++SYE+LPEIVPNL
Sbjct: 308 CKLDKPCTVDEVHLRRFDGLLENGGVHRCAVASITVKAPVCEVWKVLTSYESLPEIVPNL 367
Query: 390 AISKILSRDNNKVRILQEGCKGLLYMVLHARVVLDLCEQLEQEISFEQAEGDFDSFHGKW 449
AISKILSRDNNKVRILQEGCKGLLYMVLHAR VLDL E EQEI FEQ EGDFDS GKW
Sbjct: 368 AISKILSRDNNKVRILQEGCKGLLYMVLHARAVLDLHEIREQEIRFEQVEGDFDSLEGKW 427
Query: 450 TFEQLGNHHTLLKYSVDSKMRRDTFLSEAIMEEVIYEDLPSNLCAIRDYVENQKASQFLE 509
FEQLG+HHTLLKY+V+SKMR+D+FLSEAIMEEVIYEDLPSNLCAIRDY+E ++ + E
Sbjct: 428 IFEQLGSHHTLLKYTVESKMRKDSFLSEAIMEEVIYEDLPSNLCAIRDYIE-KRGEKSSE 486
Query: 510 VCEQNTNSGQQIILSGSGDDNNSS--SADDISDCNVQSSSNQRSRVPGLQRDIEVLKSEL 567
C+ T + S S + + + DD SD + QR R+PGLQRDIEVLKSE+
Sbjct: 487 SCKLETCQVSEETCSSSRAKSVETVYNNDDGSD-----QTKQRRRIPGLQRDIEVLKSEI 541
Query: 568 LKFVAEYGQEGFMPMRKQLRLHGRVDIEKAITRMGGFRKIATIMNLSLAYKYRKPKGYWD 627
LKF++E+GQEGFMPMRKQLRLHGRVDIEKAITRMGGFR+IA +MNLSLAYK+RKPKGYWD
Sbjct: 542 LKFISEHGQEGFMPMRKQLRLHGRVDIEKAITRMGGFRRIALMMNLSLAYKHRKPKGYWD 601
Query: 628 NLENLQDEISRFQRCWGMDPSFMPSRKSFER 658
NLENLQ+EI RFQ+ WGMDPSFMPSRKSFER
Sbjct: 602 NLENLQEEIGRFQQSWGMDPSFMPSRKSFER 632
Score = 42.7 bits (99), Expect = 0.041
Identities = 22/75 (29%), Positives = 41/75 (54%), Gaps = 1/75 (1%)
Query: 555 GLQRDIEVLKSELLKFVAEYGQE-GFMPMRKQLRLHGRVDIEKAITRMGGFRKIATIMNL 613
G ++E L+ E+ +F +G + FMP RK GR DI +A+ + GG +++ ++ L
Sbjct: 598 GYWDNLENLQEEIGRFQQSWGMDPSFMPSRKSFERAGRYDIARALEKWGGLHEVSRLLAL 657
Query: 614 SLAYKYRKPKGYWDN 628
++ + R+ DN
Sbjct: 658 NVRHPNRQLNSRKDN 672
>ref|XP_470355.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|28376712|gb|AAO41142.1| unknown protein [Oryza sativa
(japonica cultivar-group)]
Length = 782
Score = 717 bits (1851), Expect = 0.0
Identities = 377/644 (58%), Positives = 448/644 (69%), Gaps = 80/644 (12%)
Query: 80 GGNLNNGLEEDGDREVHCELQVVSWRERRVKAEISINADINSVWNALTDYEHLADFIPNL 139
GG G + V C++ VVSWRERRV A +++ AD+++VW +TDYE LA+FIPNL
Sbjct: 72 GGGGGRGGKSAAGPGVQCDVDVVSWRERRVFASVAVAADVDTVWRVITDYERLAEFIPNL 131
Query: 140 VW---------------------------------SGRIPCPFPGRIWLEQRGFQRAMYW 166
V SGRIPCP GR+WLEQRG Q+A+YW
Sbjct: 132 VHRSTFHLSFSCVIANSRMLIEGLIGGNLCVVRQRSGRIPCPHQGRVWLEQRGLQQALYW 191
Query: 167 HIEARVVLDLQELLNSEWDRELHFSMVDGDFKKFEGKWSVKSGTRSSSTNLSYEVNVIPR 226
HIEARVVLDL+E+ ++ RELHFSMVDGDFKKFEGKWS++SG RSSS L YEVNVIPR
Sbjct: 192 HIEARVVLDLKEVPDAVNGRELHFSMVDGDFKKFEGKWSIRSGPRSSSAILLYEVNVIPR 251
Query: 227 FNFPAIFLERIVRSDLPVNLRALAYRVERNLLGNQKLPQPEDDLHKTSLVVN-GSSVKKI 285
FNFPAIFLERI+RSDLPVNL ALA R E LGNQ+ H T+ GS
Sbjct: 252 FNFPAIFLERIIRSDLPVNLGALACRAENIYLGNQR--------HGTAKFSGAGSRFHNF 303
Query: 286 NGSLCETDKLAPGQDKEGLDTSISGSLPASSSELNSNWGIFGKVCSLDKPCVVDEVHLRR 345
+ E D +AP + KE + + G L + SELNS WG++G VC LD+PCVVDE+HLRR
Sbjct: 304 RNATTENDAIAPSKFKETPPSGLGGVLASPPSELNSKWGVYGNVCRLDRPCVVDEIHLRR 363
Query: 346 FDGLLENGGVHRCVVASITVKAPVRDVWNVMSSYETLPEIV------------------- 386
FDGLLE+ G HR V ASITVKAPVR+VWN++++YE LPE+V
Sbjct: 364 FDGLLEHEGAHRFVFASITVKAPVREVWNILTAYEKLPELVVLSSFSFYSSDILLYKYEL 423
Query: 387 --------PNLAISKILSRDNNKVRILQEGCKGLLYMVLHARVVLDLCEQLEQEISFEQA 438
PNLAIS+I+ RDNNKVRILQEGCKGLLYMVLHARVV+DL E+LE+EISFEQ
Sbjct: 424 TKLSVRFVPNLAISRIIRRDNNKVRILQEGCKGLLYMVLHARVVMDLREKLEREISFEQV 483
Query: 439 EGDFDSFHGKWTFEQLGNHHTLLKYSVDSKMRRDTFLSEAIMEEVIYEDLPSNLCAIRDY 498
EGDF SF GKW EQLG+ HTLLKY V++KM +DTFLSE+I+EEVIYEDLPSNLCAIRDY
Sbjct: 484 EGDFYSFKGKWRLEQLGDQHTLLKYMVETKMHKDTFLSESILEEVIYEDLPSNLCAIRDY 543
Query: 499 VENQKASQFLEVCEQNTNSGQQIILSGSG----DDNNSSSADDISDCNVQSSSNQRSRVP 554
+E +A E ++ I+ S + D ++ S S QR +VP
Sbjct: 544 IEKAEA-------ESGNSTSSSIVASNADTIAIDYAEGRQSEQASTSCSSSPVKQRPKVP 596
Query: 555 GLQRDIEVLKSELLKFVAEYGQEGFMPMRKQLRLHGRVDIEKAITRMGGFRKIATIMNLS 614
GLQ+DIEVLKSEL KF+A+YGQ+GFMP RK LRLHGRVDIEKAITRMGGFRKIA+IMNLS
Sbjct: 597 GLQKDIEVLKSELEKFIAKYGQDGFMPKRKHLRLHGRVDIEKAITRMGGFRKIASIMNLS 656
Query: 615 LAYKYRKPKGYWDNLENLQDEISRFQRCWGMDPSFMPSRKSFER 658
L+YK RKP+GYWDNLENLQ+EI RFQ+ WGMDP++MPSRKSFER
Sbjct: 657 LSYKNRKPRGYWDNLENLQEEIRRFQKNWGMDPAYMPSRKSFER 700
Score = 42.4 bits (98), Expect = 0.054
Identities = 23/78 (29%), Positives = 44/78 (55%), Gaps = 2/78 (2%)
Query: 546 SSNQRSRVP-GLQRDIEVLKSELLKFVAEYGQE-GFMPMRKQLRLHGRVDIEKAITRMGG 603
S + ++R P G ++E L+ E+ +F +G + +MP RK GR DI +A+ + GG
Sbjct: 656 SLSYKNRKPRGYWDNLENLQEEIRRFQKNWGMDPAYMPSRKSFERAGRYDIARALEKWGG 715
Query: 604 FRKIATIMNLSLAYKYRK 621
+++ +++L L R+
Sbjct: 716 VHEVSRLLSLELRRPRRR 733
>ref|ZP_00109164.1| COG2867: Oligoketide cyclase/lipid transport protein [Nostoc
punctiforme PCC 73102]
Length = 202
Score = 114 bits (284), Expect = 1e-23
Identities = 65/163 (39%), Positives = 95/163 (57%), Gaps = 7/163 (4%)
Query: 94 EVHCELQVVSWRERRVKAEISINADINSVWNALTDYEHLADFIPNLVWSGRIPCPFPGRI 153
+V ++Q ++ R+R++ A++ I + +W LTDYE L DF+PNL S I P G I
Sbjct: 36 KVEVQIQKIAERQRQISAKVQIPQPVEKIWKVLTDYEALPDFLPNLAKSRLIEHP-NGGI 94
Query: 154 WLEQRGFQRAMYWHIEARVVLDLQELLNSEWDRELHFSMVDGDFKKFEGKWSVKSGTRSS 213
LEQ G QR + ++ ARVVLDL+E + RE++F MV+GDFK F G W ++ +
Sbjct: 95 RLEQVGSQRLLNFNFSARVVLDLEEC----FPREINFRMVEGDFKGFSGSWCLEPYSLGE 150
Query: 214 --STNLSYEVNVIPRFNFPAIFLERIVRSDLPVNLRALAYRVE 254
TNL Y + V P+ P +E + DL +NL A+ RVE
Sbjct: 151 YIGTNLCYTIQVWPKLTMPVGIIENRLSKDLRLNLVAIHQRVE 193
Score = 90.5 bits (223), Expect = 2e-16
Identities = 55/147 (37%), Positives = 80/147 (54%), Gaps = 5/147 (3%)
Query: 357 RCVVASITVKAPVRDVWNVMSSYETLPEIVPNLAISKILSRDNNKVRILQEGCKGLLYMV 416
R + A + + PV +W V++ YE LP+ +PNLA S+++ N +R+ Q G + LL
Sbjct: 49 RQISAKVQIPQPVEKIWKVLTDYEALPDFLPNLAKSRLIEHPNGGIRLEQVGSQRLLNFN 108
Query: 417 LHARVVLDLCEQLEQEISFEQAEGDFDSFHGKWTFE--QLGNH-HTLLKYSVDSKMRRDT 473
ARVVLDL E +EI+F EGDF F G W E LG + T L Y++ ++
Sbjct: 109 FSARVVLDLEECFPREINFRMVEGDFKGFSGSWCLEPYSLGEYIGTNLCYTI--QVWPKL 166
Query: 474 FLSEAIMEEVIYEDLPSNLCAIRDYVE 500
+ I+E + +DL NL AI VE
Sbjct: 167 TMPVGIIENRLSKDLRLNLVAIHQRVE 193
>dbj|BAB72809.1| all0852 [Nostoc sp. PCC 7120] gi|25529736|pir||AB1913 hypothetical
protein all0852 [imported] - Nostoc sp. (strain PCC
7120) gi|17228347|ref|NP_484895.1| hypothetical protein
all0852 [Nostoc sp. PCC 7120]
Length = 202
Score = 112 bits (281), Expect = 3e-23
Identities = 79/203 (38%), Positives = 107/203 (51%), Gaps = 13/203 (6%)
Query: 67 LSSTTQCKPRSHLGGN--LNNGLEEDGDREVHCELQV--VSWRERRVKAEISINADINSV 122
L T Q P L N L+ L D E+QV ++ R+R++ A + I + V
Sbjct: 7 LKVTKQYNPTEDLEFNPDLDGELVAGADNLPSVEIQVEKIADRQRQITARVQIPQPVEQV 66
Query: 123 WNALTDYEHLADFIPNLVWSGRIPCPFPGRIWLEQRGFQRAMYWHIEARVVLDLQELLNS 182
W LT+YE LADFIPNL S + P G I LEQ G QR + + ARVVLDL+E
Sbjct: 67 WQVLTNYEALADFIPNLAKSSLLEHP-NGGIRLEQVGSQRLLNFKFCARVVLDLEEY--- 122
Query: 183 EWDRELHFSMVDGDFKKFEGKWSVKSGTRSS--STNLSYEVNVIPRFNFPAIFLERIVRS 240
+ +E++F MV+GDFK F G W ++ + T+L Y + V P+ P +ER +
Sbjct: 123 -FPKEINFQMVEGDFKGFSGNWCLQPYALGNVIGTDLCYTIQVWPKLTMPITIIERRLSQ 181
Query: 241 DLPVNLRALAYRVERNLLGNQKL 263
DL NL A+ RVE L NQ L
Sbjct: 182 DLRSNLLAIYQRVE--CLANQSL 202
Score = 90.9 bits (224), Expect = 1e-16
Identities = 57/147 (38%), Positives = 81/147 (54%), Gaps = 5/147 (3%)
Query: 357 RCVVASITVKAPVRDVWNVMSSYETLPEIVPNLAISKILSRDNNKVRILQEGCKGLLYMV 416
R + A + + PV VW V+++YE L + +PNLA S +L N +R+ Q G + LL
Sbjct: 51 RQITARVQIPQPVEQVWQVLTNYEALADFIPNLAKSSLLEHPNGGIRLEQVGSQRLLNFK 110
Query: 417 LHARVVLDLCEQLEQEISFEQAEGDFDSFHGKWTFE--QLGN-HHTLLKYSVDSKMRRDT 473
ARVVLDL E +EI+F+ EGDF F G W + LGN T L Y++ ++
Sbjct: 111 FCARVVLDLEEYFPKEINFQMVEGDFKGFSGNWCLQPYALGNVIGTDLCYTI--QVWPKL 168
Query: 474 FLSEAIMEEVIYEDLPSNLCAIRDYVE 500
+ I+E + +DL SNL AI VE
Sbjct: 169 TMPITIIERRLSQDLRSNLLAIYQRVE 195
>ref|ZP_00162473.1| COG2867: Oligoketide cyclase/lipid transport protein [Anabaena
variabilis ATCC 29413]
Length = 194
Score = 111 bits (278), Expect = 7e-23
Identities = 78/200 (39%), Positives = 105/200 (52%), Gaps = 13/200 (6%)
Query: 70 TTQCKPRSHLGGN--LNNGLEEDGDREVHCELQV--VSWRERRVKAEISINADINSVWNA 125
T Q P L N L+ L D E+QV ++ R+R++ A + I + VW
Sbjct: 2 TKQYNPTEDLEFNPDLDGELVAGADNLPSVEIQVEKIADRQRQITARVQIPQPVEQVWQV 61
Query: 126 LTDYEHLADFIPNLVWSGRIPCPFPGRIWLEQRGFQRAMYWHIEARVVLDLQELLNSEWD 185
LT+YE LADFIPNL S + P G I LEQ G QR + + ARVVLDL+E +
Sbjct: 62 LTNYEALADFIPNLAKSSLLEHP-NGGIRLEQVGSQRLLNFKFCARVVLDLEEY----FP 116
Query: 186 RELHFSMVDGDFKKFEGKWSVKSGTRSS--STNLSYEVNVIPRFNFPAIFLERIVRSDLP 243
+E++F MV+GDFK F G W ++ T+L Y + V P+ P +ER + DL
Sbjct: 117 KEINFQMVEGDFKGFSGNWCLQPYALGDVIGTDLCYTIQVWPKLTMPITIIERRLSQDLR 176
Query: 244 VNLRALAYRVERNLLGNQKL 263
NL A+ RVE L NQ L
Sbjct: 177 SNLLAIYQRVE--CLANQSL 194
Score = 89.0 bits (219), Expect = 5e-16
Identities = 56/147 (38%), Positives = 81/147 (55%), Gaps = 5/147 (3%)
Query: 357 RCVVASITVKAPVRDVWNVMSSYETLPEIVPNLAISKILSRDNNKVRILQEGCKGLLYMV 416
R + A + + PV VW V+++YE L + +PNLA S +L N +R+ Q G + LL
Sbjct: 43 RQITARVQIPQPVEQVWQVLTNYEALADFIPNLAKSSLLEHPNGGIRLEQVGSQRLLNFK 102
Query: 417 LHARVVLDLCEQLEQEISFEQAEGDFDSFHGKWTFE--QLGN-HHTLLKYSVDSKMRRDT 473
ARVVLDL E +EI+F+ EGDF F G W + LG+ T L Y++ ++
Sbjct: 103 FCARVVLDLEEYFPKEINFQMVEGDFKGFSGNWCLQPYALGDVIGTDLCYTI--QVWPKL 160
Query: 474 FLSEAIMEEVIYEDLPSNLCAIRDYVE 500
+ I+E + +DL SNL AI VE
Sbjct: 161 TMPITIIERRLSQDLRSNLLAIYQRVE 187
>gb|AAQ00167.1| Oligoketide cyclase family enzyme [Prochlorococcus marinus subsp.
marinus str. CCMP1375] gi|33240572|ref|NP_875514.1|
Oligoketide cyclase family enzyme [Prochlorococcus
marinus subsp. marinus str. CCMP1375]
gi|6469286|emb|CAB61761.1| hypothetical protein
[Prochlorococcus marinus]
Length = 173
Score = 92.8 bits (229), Expect = 3e-17
Identities = 52/171 (30%), Positives = 94/171 (54%), Gaps = 7/171 (4%)
Query: 89 EDGDREVHCELQVVSWRERRVKAEISINADINSVWNALTDYEHLADFIPNLVWSGRIPCP 148
E +R + ++V+ RR+ A+++ + D +S+W LTDY L+DFIPNL+ S +
Sbjct: 7 EKENRTIEQTMEVLPGGTRRLAAQLTTSLDFDSLWKVLTDYNRLSDFIPNLL-SSEVLLK 65
Query: 149 FPGRIWLEQRGFQRAMYWHIEARVVLDLQELLNSEWDRELHFSMVDGDFKKFEGKWSVKS 208
++ L+Q G Q + + A V + +L+ + + L FS++ GDF++FEG W +
Sbjct: 66 TDNQVHLKQVGSQEFLGLNFSAEVCI---KLIEEKENGVLRFSLIKGDFRRFEGSWQIAP 122
Query: 209 GTRSSSTNLSYEVNVIPRFNFPAIFLERIVRSDLPVNLRAL---AYRVERN 256
++ + L+YE+ V F P +E+ ++ +L NL A+ AY + N
Sbjct: 123 SPFNNGSALTYELIVQGCFGMPVALIEKHLKKNLTTNLLAVEKAAYEISSN 173
Score = 60.1 bits (144), Expect = 2e-07
Identities = 41/146 (28%), Positives = 74/146 (50%), Gaps = 7/146 (4%)
Query: 353 GGVHRCVVASITVKAPVRDVWNVMSSYETLPEIVPNLAISKILSRDNNKVRILQEGCKGL 412
GG R + A +T +W V++ Y L + +PNL S++L + +N+V + Q G +
Sbjct: 22 GGTRR-LAAQLTTSLDFDSLWKVLTDYNRLSDFIPNLLSSEVLLKTDNQVHLKQVGSQEF 80
Query: 413 LYMVLHARVVLDLCEQLEQEI-SFEQAEGDFDSFHGKWTFEQLG-NHHTLLKYSVDSKMR 470
L + A V + L E+ E + F +GDF F G W N+ + L Y + +
Sbjct: 81 LGLNFSAEVCIKLIEEKENGVLRFSLIKGDFRRFEGSWQIAPSPFNNGSALTYEL---IV 137
Query: 471 RDTF-LSEAIMEEVIYEDLPSNLCAI 495
+ F + A++E+ + ++L +NL A+
Sbjct: 138 QGCFGMPVALIEKHLKKNLTTNLLAV 163
>ref|YP_171272.1| hypothetical protein syc0562_c [Synechococcus elongatus PCC 6301]
gi|56685530|dbj|BAD78752.1| hypothetical protein
[Synechococcus elongatus PCC 6301]
Length = 204
Score = 92.8 bits (229), Expect = 3e-17
Identities = 59/175 (33%), Positives = 94/175 (53%), Gaps = 6/175 (3%)
Query: 87 LEEDGDREVHCELQVVSWRERRVKAEISINADINSVWNALTDYEHLADFIPNLVWSGRIP 146
L ++V + + R+RR++ +I + I +W LTDY LA+FIPNL S R+P
Sbjct: 35 LHRSPQQDVQIDAHSLGPRQRRIQVQIEVPVAIADLWALLTDYNRLAEFIPNLSISQRLP 94
Query: 147 CPFPGRIWLEQRGFQRAMYWHIEARVVLDLQELLNSEWDRELHFSMVDGDFKKFEGKWSV 206
G I LEQ G Q + + ARVVL +QE S ++ L F M++GDF++F+G W
Sbjct: 95 TS-DGSIRLEQVGSQCFLRFRFCARVVLAMQE---SPYEC-LAFQMIEGDFEQFDGSWRF 149
Query: 207 KSGTRSSSTNLSYEVNVIPRFNFPAIFLERIVRSDLPVNLRALAYRVERNLLGNQ 261
+S + T L+Y+V + P+ P +E + +L NL A+ R + ++
Sbjct: 150 QS-VDADRTQLTYDVTLSPKLPMPIQLIETQLDQNLAANLLAIREEAIRRFVSDR 203
Score = 78.2 bits (191), Expect = 9e-13
Identities = 45/145 (31%), Positives = 75/145 (51%), Gaps = 2/145 (1%)
Query: 353 GGVHRCVVASITVKAPVRDVWNVMSSYETLPEIVPNLAISKILSRDNNKVRILQEGCKGL 412
G R + I V + D+W +++ Y L E +PNL+IS+ L + +R+ Q G +
Sbjct: 51 GPRQRRIQVQIEVPVAIADLWALLTDYNRLAEFIPNLSISQRLPTSDGSIRLEQVGSQCF 110
Query: 413 LYMVLHARVVLDLCEQLEQEISFEQAEGDFDSFHGKWTFEQLGNHHTLLKYSVDSKMRRD 472
L ARVVL + E + ++F+ EGDF+ F G W F+ + T L Y V +
Sbjct: 111 LRFRFCARVVLAMQESPYECLAFQMIEGDFEQFDGSWRFQSVDADRTQLTYDVTLSPKLP 170
Query: 473 TFLSEAIMEEVIYEDLPSNLCAIRD 497
+ ++E + ++L +NL AIR+
Sbjct: 171 --MPIQLIETQLDQNLAANLLAIRE 193
>ref|ZP_00164123.2| COG2867: Oligoketide cyclase/lipid transport protein [Synechococcus
elongatus PCC 7942]
Length = 180
Score = 92.8 bits (229), Expect = 3e-17
Identities = 59/175 (33%), Positives = 94/175 (53%), Gaps = 6/175 (3%)
Query: 87 LEEDGDREVHCELQVVSWRERRVKAEISINADINSVWNALTDYEHLADFIPNLVWSGRIP 146
L ++V + + R+RR++ +I + I +W LTDY LA+FIPNL S R+P
Sbjct: 11 LHRSPQQDVQIDAHSLGPRQRRIQVQIEVPVAIADLWALLTDYNRLAEFIPNLSISQRLP 70
Query: 147 CPFPGRIWLEQRGFQRAMYWHIEARVVLDLQELLNSEWDRELHFSMVDGDFKKFEGKWSV 206
G I LEQ G Q + + ARVVL +QE S ++ L F M++GDF++F+G W
Sbjct: 71 TS-DGSIRLEQVGSQCFLRFRFCARVVLAMQE---SPYEC-LAFQMIEGDFEQFDGSWRF 125
Query: 207 KSGTRSSSTNLSYEVNVIPRFNFPAIFLERIVRSDLPVNLRALAYRVERNLLGNQ 261
+S + T L+Y+V + P+ P +E + +L NL A+ R + ++
Sbjct: 126 QS-VDADRTQLTYDVTLSPKLPMPIQLIETQLDQNLAANLLAIREEAIRRFVSDR 179
Score = 78.2 bits (191), Expect = 9e-13
Identities = 45/145 (31%), Positives = 75/145 (51%), Gaps = 2/145 (1%)
Query: 353 GGVHRCVVASITVKAPVRDVWNVMSSYETLPEIVPNLAISKILSRDNNKVRILQEGCKGL 412
G R + I V + D+W +++ Y L E +PNL+IS+ L + +R+ Q G +
Sbjct: 27 GPRQRRIQVQIEVPVAIADLWALLTDYNRLAEFIPNLSISQRLPTSDGSIRLEQVGSQCF 86
Query: 413 LYMVLHARVVLDLCEQLEQEISFEQAEGDFDSFHGKWTFEQLGNHHTLLKYSVDSKMRRD 472
L ARVVL + E + ++F+ EGDF+ F G W F+ + T L Y V +
Sbjct: 87 LRFRFCARVVLAMQESPYECLAFQMIEGDFEQFDGSWRFQSVDADRTQLTYDVTLSPKLP 146
Query: 473 TFLSEAIMEEVIYEDLPSNLCAIRD 497
+ ++E + ++L +NL AIR+
Sbjct: 147 --MPIQLIETQLDQNLAANLLAIRE 169
>gb|AAC72863.1| T15B16.3 gene product [Arabidopsis thaliana] gi|7487023|pir||T02008
hypothetical protein T15B16.3 - Arabidopsis thaliana
Length = 290
Score = 89.0 bits (219), Expect = 5e-16
Identities = 65/198 (32%), Positives = 102/198 (50%), Gaps = 21/198 (10%)
Query: 83 LNNGLEED---GDREVHCELQVVSWRERRVKAEISINADINSVWNALTDYEHLADFIPNL 139
L +G E+ GD V EL+ + RR++++I + A ++SVW+ LTDYE L+DFIP L
Sbjct: 88 LTDGKTEELVVGDDGVLIELKKLEKSSRRIRSKIGMEASLDSVWSVLTDYEKLSDFIPGL 147
Query: 140 VWSGRIPCPFPGRIWLEQRGFQR-AMYWHIEARVVLDLQ----ELLNSEWDRELHFSMVD 194
V S + R+ L Q G Q A+ A+ VLD E+L RE+ F MV+
Sbjct: 148 VVSELVE-KEGNRVRLFQMGQQNLALGLKFNAKAVLDCYEKELEVLPHGRRREIDFKMVE 206
Query: 195 GDFKKFEGKWSVKS------------GTRSSSTNLSYEVNVIPRFNFPAIFLERIVRSDL 242
GDF+ FEGKWS++ + T L+Y V+V P+ P +E + ++
Sbjct: 207 GDFQLFEGKWSIEQLDKGIHGEALDLQFKDFRTTLAYTVDVKPKMWLPVRLVEGRLCKEI 266
Query: 243 PVNLRALAYRVERNLLGN 260
NL ++ ++ + G+
Sbjct: 267 RTNLMSIRDAAQKVIEGS 284
Score = 83.2 bits (204), Expect = 3e-14
Identities = 55/163 (33%), Positives = 92/163 (55%), Gaps = 24/163 (14%)
Query: 357 RCVVASITVKAPVRDVWNVMSSYETLPEIVPNLAISKILSRDNNKVRILQEGCKGL-LYM 415
R + + I ++A + VW+V++ YE L + +P L +S+++ ++ N+VR+ Q G + L L +
Sbjct: 115 RRIRSKIGMEASLDSVWSVLTDYEKLSDFIPGLVVSELVEKEGNRVRLFQMGQQNLALGL 174
Query: 416 VLHARVVLDLCE-QLE-------QEISFEQAEGDFDSFHGKWTFEQL--GNH-------- 457
+A+ VLD E +LE +EI F+ EGDF F GKW+ EQL G H
Sbjct: 175 KFNAKAVLDCYEKELEVLPHGRRREIDFKMVEGDFQLFEGKWSIEQLDKGIHGEALDLQF 234
Query: 458 ---HTLLKYSVDSKMRRDTFLSEAIMEEVIYEDLPSNLCAIRD 497
T L Y+VD K + +L ++E + +++ +NL +IRD
Sbjct: 235 KDFRTTLAYTVDVKPK--MWLPVRLVEGRLCKEIRTNLMSIRD 275
>emb|CAB77735.1| hypothetical protein [Arabidopsis thaliana]
gi|15234259|ref|NP_192074.1| expressed protein
[Arabidopsis thaliana] gi|25361043|pir||C85021
hypothetical protein AT4g01650 [imported] - Arabidopsis
thaliana
Length = 288
Score = 88.6 bits (218), Expect = 7e-16
Identities = 65/197 (32%), Positives = 101/197 (50%), Gaps = 21/197 (10%)
Query: 83 LNNGLEED---GDREVHCELQVVSWRERRVKAEISINADINSVWNALTDYEHLADFIPNL 139
L +G E+ GD V EL+ + RR++++I + A ++SVW+ LTDYE L+DFIP L
Sbjct: 88 LTDGKTEELVVGDDGVLIELKKLEKSSRRIRSKIGMEASLDSVWSVLTDYEKLSDFIPGL 147
Query: 140 VWSGRIPCPFPGRIWLEQRGFQR-AMYWHIEARVVLDLQ----ELLNSEWDRELHFSMVD 194
V S + R+ L Q G Q A+ A+ VLD E+L RE+ F MV+
Sbjct: 148 VVSELVE-KEGNRVRLFQMGQQNLALGLKFNAKAVLDCYEKELEVLPHGRRREIDFKMVE 206
Query: 195 GDFKKFEGKWSVKS------------GTRSSSTNLSYEVNVIPRFNFPAIFLERIVRSDL 242
GDF+ FEGKWS++ + T L+Y V+V P+ P +E + ++
Sbjct: 207 GDFQLFEGKWSIEQLDKGIHGEALDLQFKDFRTTLAYTVDVKPKMWLPVRLVEGRLCKEI 266
Query: 243 PVNLRALAYRVERNLLG 259
NL ++ ++ + G
Sbjct: 267 RTNLMSIRDAAQKVIEG 283
Score = 83.2 bits (204), Expect = 3e-14
Identities = 55/163 (33%), Positives = 92/163 (55%), Gaps = 24/163 (14%)
Query: 357 RCVVASITVKAPVRDVWNVMSSYETLPEIVPNLAISKILSRDNNKVRILQEGCKGL-LYM 415
R + + I ++A + VW+V++ YE L + +P L +S+++ ++ N+VR+ Q G + L L +
Sbjct: 115 RRIRSKIGMEASLDSVWSVLTDYEKLSDFIPGLVVSELVEKEGNRVRLFQMGQQNLALGL 174
Query: 416 VLHARVVLDLCE-QLE-------QEISFEQAEGDFDSFHGKWTFEQL--GNH-------- 457
+A+ VLD E +LE +EI F+ EGDF F GKW+ EQL G H
Sbjct: 175 KFNAKAVLDCYEKELEVLPHGRRREIDFKMVEGDFQLFEGKWSIEQLDKGIHGEALDLQF 234
Query: 458 ---HTLLKYSVDSKMRRDTFLSEAIMEEVIYEDLPSNLCAIRD 497
T L Y+VD K + +L ++E + +++ +NL +IRD
Sbjct: 235 KDFRTTLAYTVDVKPK--MWLPVRLVEGRLCKEIRTNLMSIRD 275
>gb|AAO44042.1| At4g01650 [Arabidopsis thaliana] gi|30678933|ref|NP_849282.1|
expressed protein [Arabidopsis thaliana]
Length = 211
Score = 88.6 bits (218), Expect = 7e-16
Identities = 65/197 (32%), Positives = 101/197 (50%), Gaps = 21/197 (10%)
Query: 83 LNNGLEED---GDREVHCELQVVSWRERRVKAEISINADINSVWNALTDYEHLADFIPNL 139
L +G E+ GD V EL+ + RR++++I + A ++SVW+ LTDYE L+DFIP L
Sbjct: 11 LEDGKTEELVVGDDGVLIELKKLEKSSRRIRSKIGMEASLDSVWSVLTDYEKLSDFIPGL 70
Query: 140 VWSGRIPCPFPGRIWLEQRGFQR-AMYWHIEARVVLDLQ----ELLNSEWDRELHFSMVD 194
V S + R+ L Q G Q A+ A+ VLD E+L RE+ F MV+
Sbjct: 71 VVSELVE-KEGNRVRLFQMGQQNLALGLKFNAKAVLDCYEKELEVLPHGRRREIDFKMVE 129
Query: 195 GDFKKFEGKWSVKS------------GTRSSSTNLSYEVNVIPRFNFPAIFLERIVRSDL 242
GDF+ FEGKWS++ + T L+Y V+V P+ P +E + ++
Sbjct: 130 GDFQLFEGKWSIEQLDKGIHGEALDLQFKDFRTTLAYTVDVKPKMWLPVRLVEGRLCKEI 189
Query: 243 PVNLRALAYRVERNLLG 259
NL ++ ++ + G
Sbjct: 190 RTNLMSIRDAAQKVIEG 206
Score = 83.2 bits (204), Expect = 3e-14
Identities = 55/163 (33%), Positives = 92/163 (55%), Gaps = 24/163 (14%)
Query: 357 RCVVASITVKAPVRDVWNVMSSYETLPEIVPNLAISKILSRDNNKVRILQEGCKGL-LYM 415
R + + I ++A + VW+V++ YE L + +P L +S+++ ++ N+VR+ Q G + L L +
Sbjct: 38 RRIRSKIGMEASLDSVWSVLTDYEKLSDFIPGLVVSELVEKEGNRVRLFQMGQQNLALGL 97
Query: 416 VLHARVVLDLCE-QLE-------QEISFEQAEGDFDSFHGKWTFEQL--GNH-------- 457
+A+ VLD E +LE +EI F+ EGDF F GKW+ EQL G H
Sbjct: 98 KFNAKAVLDCYEKELEVLPHGRRREIDFKMVEGDFQLFEGKWSIEQLDKGIHGEALDLQF 157
Query: 458 ---HTLLKYSVDSKMRRDTFLSEAIMEEVIYEDLPSNLCAIRD 497
T L Y+VD K + +L ++E + +++ +NL +IRD
Sbjct: 158 KDFRTTLAYTVDVKPK--MWLPVRLVEGRLCKEIRTNLMSIRD 198
>emb|CAE21275.1| conserved hypothetical protein [Prochlorococcus marinus str. MIT
9313] gi|33863371|ref|NP_894931.1| hypothetical protein
PMT1100 [Prochlorococcus marinus str. MIT 9313]
Length = 190
Score = 84.7 bits (208), Expect = 9e-15
Identities = 48/143 (33%), Positives = 78/143 (53%), Gaps = 4/143 (2%)
Query: 107 RRVKAEISINADINSVWNALTDYEHLADFIPNLVWSGRIPCPFPGRIWLEQRGFQRAMYW 166
RR+ ++ + +W+ LTDY+ L++FIPNL S + R+WL Q G Q+ +
Sbjct: 41 RRLAVQLRTPIKESLLWDVLTDYDKLSEFIPNLA-SSTVLERTGNRVWLNQVGSQQLLGL 99
Query: 167 HIEARVVLDLQELLNSEWDRELHFSMVDGDFKKFEGKWSVKSGTRSSSTNLSYEVNVIPR 226
A+V L+L E + +L F ++ GDF++FEG W ++ +ST+L YE+ V
Sbjct: 100 RFSAQVQLELVEY---RAEGKLQFHLLKGDFRRFEGSWIMRELAEGTSTSLLYELTVQGC 156
Query: 227 FNFPAIFLERIVRSDLPVNLRAL 249
P +E+ +R DL NL A+
Sbjct: 157 IGMPVALIEQRLRDDLTANLLAV 179
Score = 71.6 bits (174), Expect = 8e-11
Identities = 43/138 (31%), Positives = 79/138 (57%), Gaps = 6/138 (4%)
Query: 362 SITVKAPVRD--VWNVMSSYETLPEIVPNLAISKILSRDNNKVRILQEGCKGLLYMVLHA 419
++ ++ P+++ +W+V++ Y+ L E +PNLA S +L R N+V + Q G + LL + A
Sbjct: 44 AVQLRTPIKESLLWDVLTDYDKLSEFIPNLASSTVLERTGNRVWLNQVGSQQLLGLRFSA 103
Query: 420 RVVLDLCE-QLEQEISFEQAEGDFDSFHGKWTFEQLG-NHHTLLKYSVDSKMRRDTFLSE 477
+V L+L E + E ++ F +GDF F G W +L T L Y + ++ +
Sbjct: 104 QVQLELVEYRAEGKLQFHLLKGDFRRFEGSWIMRELAEGTSTSLLYEL--TVQGCIGMPV 161
Query: 478 AIMEEVIYEDLPSNLCAI 495
A++E+ + +DL +NL A+
Sbjct: 162 ALIEQRLRDDLTANLLAV 179
>ref|NP_893193.1| hypothetical protein PMM1076 [Prochlorococcus marinus subsp.
pastoris str. CCMP1986] gi|33634209|emb|CAE19535.1|
conserved hypothetical protein [Prochlorococcus marinus
subsp. pastoris str. CCMP1986]
Length = 178
Score = 83.6 bits (205), Expect = 2e-14
Identities = 55/165 (33%), Positives = 91/165 (54%), Gaps = 11/165 (6%)
Query: 89 EDGD-REVHCELQVVSWRERRVKAEISINADINSVWNALTDYEHLADFIPNLVWSGRIPC 147
++GD R + ++ +S RR+ A+++ +A NS+WN LTDY+ L +IPNL+ S +I
Sbjct: 12 QNGDYRTIEQTMEKLSGGTRRLAAQLTTSATFNSLWNVLTDYDRLNLYIPNLLSSRKI-Y 70
Query: 148 PFPGRIWLEQRGFQRAMYWHIEARVVLDLQELLNSEWDRE---LHFSMVDGDFKKFEGKW 204
+ L+Q G Q + A V +DL E ++E L FS++ GDF++FEG W
Sbjct: 71 KNNNNVHLKQVGAQDFLGMKFSAEVTIDLFE------EKELGLLKFSLIKGDFRRFEGSW 124
Query: 205 SVKSGTRSSSTNLSYEVNVIPRFNFPAIFLERIVRSDLPVNLRAL 249
+K +S +L Y++ V P +E+ ++ DL NL A+
Sbjct: 125 KIKKIKDTSKNSLIYDLTVQGCQWMPIGMIEKRLKKDLSENLIAV 169
Score = 69.3 bits (168), Expect = 4e-10
Identities = 43/147 (29%), Positives = 78/147 (52%), Gaps = 7/147 (4%)
Query: 352 NGGVHRCVVASITVKAPVRDVWNVMSSYETLPEIVPNLAISKILSRDNNKVRILQEGCKG 411
+GG R + A +T A +WNV++ Y+ L +PNL S+ + ++NN V + Q G +
Sbjct: 27 SGGTRR-LAAQLTTSATFNSLWNVLTDYDRLNLYIPNLLSSRKIYKNNNNVHLKQVGAQD 85
Query: 412 LLYMVLHARVVLDLCEQLEQE-ISFEQAEGDFDSFHGKWTFEQL--GNHHTLLKYSVDSK 468
L M A V +DL E+ E + F +GDF F G W +++ + ++L+ D
Sbjct: 86 FLGMKFSAEVTIDLFEEKELGLLKFSLIKGDFRRFEGSWKIKKIKDTSKNSLI---YDLT 142
Query: 469 MRRDTFLSEAIMEEVIYEDLPSNLCAI 495
++ ++ ++E+ + +DL NL A+
Sbjct: 143 VQGCQWMPIGMIEKRLKKDLSENLIAV 169
>emb|CAE07267.1| conserved hypothetical protein [Synechococcus sp. WH 8102]
gi|33865286|ref|NP_896845.1| hypothetical protein
SYNW0752 [Synechococcus sp. WH 8102]
Length = 180
Score = 80.5 bits (197), Expect = 2e-13
Identities = 54/152 (35%), Positives = 78/152 (50%), Gaps = 10/152 (6%)
Query: 107 RRVKAEISINADINSVWNALTDYEHLADFIPNLVWSGRIPCPFPGRIWLEQRGFQRAMYW 166
RR+ ++ + W+ LTDY HLADFIPNL S + + L+Q G Q+ +
Sbjct: 34 RRLAVQLKSSIPAELFWDVLTDYAHLADFIPNLS-SSELVMRDGETVRLQQVGSQQLLGM 92
Query: 167 HIEARVVLDLQELLNSEWDRELHFSMVDGDFKKFEGKWSVKSGTRSSSTNLSYEVNVIPR 226
A+V+L+L+E + D L F M+ GDF++FEG W V+ T + L YE+ V
Sbjct: 93 RFSAQVLLELREF---KPDGVLRFQMLKGDFRRFEGSWQVR--TLPEGSTLLYELMVQGC 147
Query: 227 FNFPAIFLERIVRSDLPVNLRALAYRVERNLL 258
P +E +R DL NL + VER L
Sbjct: 148 LGMPIGLIEERLRDDLSSNL----FAVEREAL 175
Score = 70.5 bits (171), Expect = 2e-10
Identities = 46/144 (31%), Positives = 75/144 (51%), Gaps = 5/144 (3%)
Query: 353 GGVHRCVVASITVKAPVRDVWNVMSSYETLPEIVPNLAISKILSRDNNKVRILQEGCKGL 412
GG R V + P W+V++ Y L + +PNL+ S+++ RD VR+ Q G + L
Sbjct: 31 GGARRLAV-QLKSSIPAELFWDVLTDYAHLADFIPNLSSSELVMRDGETVRLQQVGSQQL 89
Query: 413 LYMVLHARVVLDLCE-QLEQEISFEQAEGDFDSFHGKWTFEQLGNHHTLLKYSVDSKMRR 471
L M A+V+L+L E + + + F+ +GDF F G W L TLL + ++
Sbjct: 90 LGMRFSAQVLLELREFKPDGVLRFQMLKGDFRRFEGSWQVRTLPEGSTLL---YELMVQG 146
Query: 472 DTFLSEAIMEEVIYEDLPSNLCAI 495
+ ++EE + +DL SNL A+
Sbjct: 147 CLGMPIGLIEERLRDDLSSNLFAV 170
>ref|NP_914348.1| P0518C01.12 [Oryza sativa (japonica cultivar-group)]
Length = 873
Score = 79.0 bits (193), Expect = 5e-13
Identities = 70/213 (32%), Positives = 104/213 (47%), Gaps = 22/213 (10%)
Query: 88 EEDGDREVHCELQV--VSWRERR-VKAEISINADINSVWNALTDYEHLADFIPNLVWSGR 144
+E+ D E+QV + R RR V+A + ++A +++VW LTDYE LA FIP L R
Sbjct: 74 KEERDERYGFEIQVRKLPKRNRRLVRARVRVDAPLDAVWATLTDYEGLAGFIPGLS-ECR 132
Query: 145 IPCPFPGRIWLEQRGFQ-RAMYWHIEARVVLD-----LQELLNSEWDRELHFSMVDGDFK 198
+ L Q G Q A+ + AR +D LQ L RE+ F+M+DGDFK
Sbjct: 133 LLDQSDCFARLYQVGEQDLALGFKFNARGTIDCYEGELQLLPAGARRREIAFNMIDGDFK 192
Query: 199 KFEGKWSVK--------SGTRSSSTNLSYEVNVIPRFNFPAIFLERIVRSDLPVNLRALA 250
FEG WSV+ S + T LSY V + P+ P LE + +++ NL ++
Sbjct: 193 VFEGNWSVQEEVDGGEISADQEFQTILSYVVELEPKLWVPVRLLEGRICNEIKTNLFSIR 252
Query: 251 YRVERNLLGNQKLPQPEDDLHKTSLVVNGSSVK 283
+R Q+L + TSL++ + K
Sbjct: 253 EEAQR----IQRLQDKASSQYYTSLILKNTLQK 281
Score = 54.3 bits (129), Expect = 1e-05
Identities = 44/160 (27%), Positives = 76/160 (47%), Gaps = 21/160 (13%)
Query: 357 RCVVASITVKAPVRDVWNVMSSYETLPEIVPNLAISKILSRDNNKVRILQEGCKGL-LYM 415
R V A + V AP+ VW ++ YE L +P L+ ++L + + R+ Q G + L L
Sbjct: 96 RLVRARVRVDAPLDAVWATLTDYEGLAGFIPGLSECRLLDQSDCFARLYQVGEQDLALGF 155
Query: 416 VLHARVVLDLCE---QL------EQEISFEQAEGDFDSFHGKWTFEQL---------GNH 457
+AR +D E QL +EI+F +GDF F G W+ ++
Sbjct: 156 KFNARGTIDCYEGELQLLPAGARRREIAFNMIDGDFKVFEGNWSVQEEVDGGEISADQEF 215
Query: 458 HTLLKYSVDSKMRRDTFLSEAIMEEVIYEDLPSNLCAIRD 497
T+L Y V+ + ++ ++E I ++ +NL +IR+
Sbjct: 216 QTILSYVVE--LEPKLWVPVRLLEGRICNEIKTNLFSIRE 253
>ref|NP_661952.1| hypothetical protein CT1061 [Chlorobium tepidum TLS]
gi|21647024|gb|AAM72294.1| conserved hypothetical
protein [Chlorobium tepidum TLS]
Length = 235
Score = 78.2 bits (191), Expect = 9e-13
Identities = 46/146 (31%), Positives = 76/146 (51%), Gaps = 5/146 (3%)
Query: 109 VKAEISINADINSVWNALTDYEHLADFIPNLVWSGRIPCPFPGRIWLEQRGFQRAMYWHI 168
V ++ I A VW A+TDY + F+P L+ SG I GR EQ F+R
Sbjct: 67 VVGKVYIEASPKHVWAAITDYNNHKSFVPKLIDSGLISDN--GR---EQVMFERGKTGIF 121
Query: 169 EARVVLDLQELLNSEWDRELHFSMVDGDFKKFEGKWSVKSGTRSSSTNLSYEVNVIPRFN 228
R + ++ L E+ + L F ++GDFK +EG W ++ + + L++ + P F
Sbjct: 122 LFRKTVYIKLSLQGEYPKRLDFHQIEGDFKVYEGDWLIERASDGKGSILTFRAKIKPDFF 181
Query: 229 FPAIFLERIVRSDLPVNLRALAYRVE 254
PA+F+ ++ ++DLP+ L A+ R E
Sbjct: 182 APAMFVRKVQQNDLPMVLAAMKKRAE 207
Score = 58.2 bits (139), Expect = 9e-07
Identities = 35/147 (23%), Positives = 66/147 (44%), Gaps = 1/147 (0%)
Query: 359 VVASITVKAPVRDVWNVMSSYETLPEIVPNLAISKILSRDNNKVRILQEGCKGLLYMVLH 418
VV + ++A + VW ++ Y VP L S ++S + + + + G G+
Sbjct: 67 VVGKVYIEASPKHVWAAITDYNNHKSFVPKLIDSGLISDNGREQVMFERGKTGIFLFRKT 126
Query: 419 ARVVLDLCEQLEQEISFEQAEGDFDSFHGKWTFEQLGNHHTLLKYSVDSKMRRDTFLSEA 478
+ L L + + + F Q EGDF + G W E+ + + + +K++ D F
Sbjct: 127 VYIKLSLQGEYPKRLDFHQIEGDFKVYEGDWLIERASDGKGSI-LTFRAKIKPDFFAPAM 185
Query: 479 IMEEVIYEDLPSNLCAIRDYVENQKAS 505
+ +V DLP L A++ E+ + S
Sbjct: 186 FVRKVQQNDLPMVLAAMKKRAESAEGS 212
>dbj|BAD87364.1| unknown protein [Oryza sativa (japonica cultivar-group)]
Length = 268
Score = 76.6 bits (187), Expect = 3e-12
Identities = 67/188 (35%), Positives = 95/188 (49%), Gaps = 21/188 (11%)
Query: 88 EEDGDREVHCELQV--VSWRERR-VKAEISINADINSVWNALTDYEHLADFIPNLVWSGR 144
+E+ D E+QV + R RR V+A + ++A +++VW LTDYE LA FIP L R
Sbjct: 74 KEERDERYGFEIQVRKLPKRNRRLVRARVRVDAPLDAVWATLTDYEGLAGFIPGLS-ECR 132
Query: 145 IPCPFPGRIWLEQRGFQ-RAMYWHIEARVVLD-----LQELLNSEWDRELHFSMVDGDFK 198
+ L Q G Q A+ + AR +D LQ L RE+ F+M+DGDFK
Sbjct: 133 LLDQSDCFARLYQVGEQDLALGFKFNARGTIDCYEGELQLLPAGARRREIAFNMIDGDFK 192
Query: 199 KFEGKWSVK--------SGTRSSSTNLSYEVNVIPRFNFPAIFLERIVRSDLPVNL---R 247
FEG WSV+ S + T LSY V + P+ P LE + +++ NL R
Sbjct: 193 VFEGNWSVQEEVDGGEISADQEFQTILSYVVELEPKLWVPVRLLEGRICNEIKTNLFSIR 252
Query: 248 ALAYRVER 255
A R++R
Sbjct: 253 EEAQRIQR 260
Score = 54.3 bits (129), Expect = 1e-05
Identities = 44/160 (27%), Positives = 76/160 (47%), Gaps = 21/160 (13%)
Query: 357 RCVVASITVKAPVRDVWNVMSSYETLPEIVPNLAISKILSRDNNKVRILQEGCKGL-LYM 415
R V A + V AP+ VW ++ YE L +P L+ ++L + + R+ Q G + L L
Sbjct: 96 RLVRARVRVDAPLDAVWATLTDYEGLAGFIPGLSECRLLDQSDCFARLYQVGEQDLALGF 155
Query: 416 VLHARVVLDLCE---QL------EQEISFEQAEGDFDSFHGKWTFEQL---------GNH 457
+AR +D E QL +EI+F +GDF F G W+ ++
Sbjct: 156 KFNARGTIDCYEGELQLLPAGARRREIAFNMIDGDFKVFEGNWSVQEEVDGGEISADQEF 215
Query: 458 HTLLKYSVDSKMRRDTFLSEAIMEEVIYEDLPSNLCAIRD 497
T+L Y V+ + ++ ++E I ++ +NL +IR+
Sbjct: 216 QTILSYVVE--LEPKLWVPVRLLEGRICNEIKTNLFSIRE 253
>ref|NP_661437.1| hypothetical protein CT0537 [Chlorobium tepidum TLS]
gi|21646468|gb|AAM71779.1| hypothetical protein CT0537
[Chlorobium tepidum TLS]
Length = 259
Score = 70.1 bits (170), Expect = 2e-10
Identities = 43/142 (30%), Positives = 70/142 (49%), Gaps = 5/142 (3%)
Query: 362 SITVKAPVRDVWNVMSSYETLPEIVPNLAISKILSRDNNKVRILQEGCKGLLYMVLHARV 421
S+ V+A VW +++ Y+ L E +P + S++L +N I Q G G+
Sbjct: 115 SVFVEAEPPVVWRMLTDYDHLHETMPKVISSRLLETNNQTRIIAQSGKSGIFIFEKTVNF 174
Query: 422 VLDLCEQLEQEISFEQAEGDFDSFHGKWTFEQL---GNHHTLLKYSVDSKMRRDTFLSEA 478
L + E + + F Q GDF + G+W E + H TLL Y ++++ D F +
Sbjct: 175 TLKVEEVFPEHLWFSQIGGDFQVYEGEWQLEAVEGKNGHATLLSY--QAEIKPDFFAPQF 232
Query: 479 IMEEVIYEDLPSNLCAIRDYVE 500
++ V +DLP+ L AIR Y E
Sbjct: 233 VVSFVQSQDLPTILRAIRSYCE 254
Score = 66.6 bits (161), Expect = 3e-09
Identities = 43/152 (28%), Positives = 71/152 (46%), Gaps = 7/152 (4%)
Query: 110 KAEISINADINSVWNALTDYEHLADFIPNLVWSGRIPCPFPGRIWLEQRGFQRAMYWHIE 169
K + + A+ VW LTDY+HL + +P ++ S + RI + Q G +
Sbjct: 113 KGSVFVEAEPPVVWRMLTDYDHLHETMPKVISSRLLETNNQTRI-IAQSGKSGIFIFEKT 171
Query: 170 ARVVLDLQELLNSEWDRELHFSMVDGDFKKFEGKWSVKS--GTRSSSTNLSYEVNVIPRF 227
L ++E+ + L FS + GDF+ +EG+W +++ G +T LSY+ + P F
Sbjct: 172 VNFTLKVEEV----FPEHLWFSQIGGDFQVYEGEWQLEAVEGKNGHATLLSYQAEIKPDF 227
Query: 228 NFPAIFLERIVRSDLPVNLRALAYRVERNLLG 259
P + + DLP LRA+ E G
Sbjct: 228 FAPQFVVSFVQSQDLPTILRAIRSYCEARAKG 259
>ref|ZP_00531164.1| Streptomyces cyclase/dehydrase [Chlorobium phaeobacteroides BS1]
gi|67915112|gb|EAM64438.1| Streptomyces
cyclase/dehydrase [Chlorobium phaeobacteroides BS1]
Length = 175
Score = 64.7 bits (156), Expect = 1e-08
Identities = 39/145 (26%), Positives = 70/145 (47%), Gaps = 5/145 (3%)
Query: 359 VVASITVKAPVRDVWNVMSSYETLPEIVPNLAISKILSRDNNKVRILQEGCKGLLYMVLH 418
V I + A +VWN +++Y+ L + +P + S ++ +N + + Q G G+L
Sbjct: 26 VTGKIFIDARPEEVWNTLTNYDNLSKTLPKVLESHLIENENGHIILEQTGRTGILIFEKT 85
Query: 419 ARVVLDLCEQLEQEISFEQAEGDFDSFHGKWTFEQLGNHH-TLLKYSVDSKMRRDTFLSE 477
R L + E+ ++FEQ GDF + G+W E L + T L+Y + + F +
Sbjct: 86 VRFRLKIQEEYLHRVTFEQISGDFHVYSGQWLLETLPDQRGTFLQYHA---LIKPLFFAP 142
Query: 478 AIMEEVIY-EDLPSNLCAIRDYVEN 501
I+ + +DLP L A + E+
Sbjct: 143 PILVSFVQRQDLPGILSAHKQQAES 167
Score = 63.9 bits (154), Expect = 2e-08
Identities = 48/156 (30%), Positives = 73/156 (46%), Gaps = 6/156 (3%)
Query: 109 VKAEISINADINSVWNALTDYEHLADFIPNLVWSGRIPCPFPGRIWLEQRGFQRAMYWHI 168
V +I I+A VWN LT+Y++L+ +P ++ S I G I LEQ G + +
Sbjct: 26 VTGKIFIDARPEEVWNTLTNYDNLSKTLPKVLESHLIENE-NGHIILEQTGRTGILIFEK 84
Query: 169 EARVVLDLQELLNSEWDRELHFSMVDGDFKKFEGKWSVKSGTRSSSTNLSYEVNVIPRFN 228
R L +QE E+ + F + GDF + G+W +++ T L Y + P F
Sbjct: 85 TVRFRLKIQE----EYLHRVTFEQISGDFHVYSGQWLLETLPDQRGTFLQYHALIKPLFF 140
Query: 229 FPAIFLERIVRSDLPVNLRALAYRVERNLLGNQKLP 264
P I + + R DLP L A + E L N+ P
Sbjct: 141 APPILVSFVQRQDLPGILSAHKQQAESAAL-NRNTP 175
>ref|ZP_00531162.1| Streptomyces cyclase/dehydrase [Chlorobium phaeobacteroides BS1]
gi|67915110|gb|EAM64436.1| Streptomyces
cyclase/dehydrase [Chlorobium phaeobacteroides BS1]
Length = 186
Score = 64.7 bits (156), Expect = 1e-08
Identities = 39/137 (28%), Positives = 68/137 (49%), Gaps = 5/137 (3%)
Query: 113 ISINADINSVWNALTDYEHLADFIPNLVWSGRIPCPFPGRIWLEQRGFQRAMYWHIEARV 172
+ + A ++W LTDY HL++ IP +V S + +I + Q G + +
Sbjct: 39 VFVAARPETIWAILTDYNHLSEKIPKVVESRLVEDNGDEKI-IAQTGRSGIFFIEKSVAI 97
Query: 173 VLDLQELLNSEWDRELHFSMVDGDFKKFEGKWSVKSGTRSSSTNLSYEVNVIPRFNFPAI 232
VL ++E + R L F +++G+F + G+W + S+T LS++ + PRF P
Sbjct: 98 VLSVKEF----FPRSLSFEILEGEFSVYRGEWRFEPSEDGSATFLSWQALLKPRFFAPPF 153
Query: 233 FLERIVRSDLPVNLRAL 249
+ + DLP LRA+
Sbjct: 154 LVSFVQHQDLPTILRAI 170
Score = 63.5 bits (153), Expect = 2e-08
Identities = 39/140 (27%), Positives = 70/140 (49%), Gaps = 3/140 (2%)
Query: 362 SITVKAPVRDVWNVMSSYETLPEIVPNLAISKILSRDNNKVRILQEGCKGLLYMVLHARV 421
++ V A +W +++ Y L E +P + S+++ + ++ I Q G G+ ++ +
Sbjct: 38 AVFVAARPETIWAILTDYNHLSEKIPKVVESRLVEDNGDEKIIAQTGRSGIFFIEKSVAI 97
Query: 422 VLDLCEQLEQEISFEQAEGDFDSFHGKWTFE-QLGNHHTLLKYSVDSKMRRDTFLSEAIM 480
VL + E + +SFE EG+F + G+W FE T L + K R F ++
Sbjct: 98 VLSVKEFFPRSLSFEILEGEFSVYRGEWRFEPSEDGSATFLSWQALLKPR--FFAPPFLV 155
Query: 481 EEVIYEDLPSNLCAIRDYVE 500
V ++DLP+ L AIR+ E
Sbjct: 156 SFVQHQDLPTILRAIRELAE 175
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.322 0.137 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,202,834,493
Number of Sequences: 2540612
Number of extensions: 50838943
Number of successful extensions: 127235
Number of sequences better than 10.0: 64
Number of HSP's better than 10.0 without gapping: 28
Number of HSP's successfully gapped in prelim test: 36
Number of HSP's that attempted gapping in prelim test: 127091
Number of HSP's gapped (non-prelim): 109
length of query: 719
length of database: 863,360,394
effective HSP length: 135
effective length of query: 584
effective length of database: 520,377,774
effective search space: 303900620016
effective search space used: 303900620016
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 79 (35.0 bits)
Medicago: description of AC148654.3


