

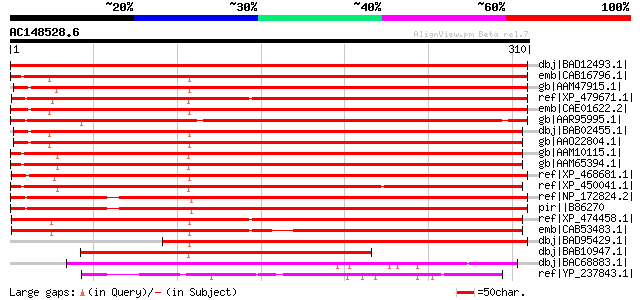
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148528.6 + phase: 0 /partial
(310 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAD12493.1| nodulin of unknown function [Lotus corniculatus ... 475 e-133
emb|CAB16796.1| MAP3K-like protein kinase [Arabidopsis thaliana]... 358 1e-97
gb|AAM47915.1| unknown protein [Arabidopsis thaliana] gi|2026021... 346 6e-94
ref|XP_479671.1| MAP3K-like protein kinase [Oryza sativa (japoni... 345 7e-94
emb|CAE01622.2| OSJNBa0042L16.16 [Oryza sativa (japonica cultiva... 335 1e-90
gb|AAR95995.1| hypothetical protein kinase [Musa acuminata] 329 6e-89
dbj|BAB02455.1| MAP3K protein kinase-like protein [Arabidopsis t... 325 8e-88
gb|AAO22804.1| unknown protein [Arabidopsis thaliana] 325 8e-88
gb|AAM10115.1| unknown protein [Arabidopsis thaliana] gi|1842515... 317 4e-85
gb|AAM65394.1| unknown [Arabidopsis thaliana] 315 1e-84
ref|XP_468681.1| expressed protein [Oryza sativa (japonica culti... 304 2e-81
ref|XP_450041.1| MAP3K protein kinase-like protein [Oryza sativa... 302 9e-81
ref|NP_172824.2| expressed protein [Arabidopsis thaliana] 291 2e-77
pir||B86270 hypothetical protein F21F23.12 - Arabidopsis thalian... 291 2e-77
ref|XP_474458.1| OSJNBa0039K24.18 [Oryza sativa (japonica cultiv... 288 2e-76
emb|CAB53483.1| CAA303710.1 protein [Oryza sativa] 266 7e-70
dbj|BAD95429.1| MAP3K-like protein kinase [Arabidopsis thaliana] 259 7e-68
dbj|BAB10947.1| unnamed protein product [Arabidopsis thaliana] 209 1e-52
dbj|BAC68883.1| hypothetical protein [Streptomyces avermitilis M... 96 1e-18
ref|YP_237843.1| hypothetical protein Psyr_4778 [Pseudomonas syr... 68 3e-10
>dbj|BAD12493.1| nodulin of unknown function [Lotus corniculatus var. japonicus]
Length = 337
Score = 475 bits (1223), Expect = e-133
Identities = 224/310 (72%), Positives = 258/310 (82%), Gaps = 1/310 (0%)
Query: 1 QIGETCSRDVNDCGTGLQCLECNSQSRCTRVRTSSPISKVMELPFNHYSWLTTHNSYASR 60
QI ETCSRD+NDC GLQCLEC+SQ+RCTR+RT SP SKVMELPFN YSWLTTHNS+A++
Sbjct: 25 QIAETCSRDINDCDLGLQCLECHSQNRCTRIRTISPTSKVMELPFNEYSWLTTHNSFAAK 84
Query: 61 AANLSIDSKISSVMNQEDSITDQLRNGVRGIMLDMHDYYGDIWLCRGPCTIFTAFQPAIN 120
N S S + + NQEDSITDQL+NGVRG+MLDM DY IWLCRGPCT +T FQPA+N
Sbjct: 85 GVNWSTGSPVLAFTNQEDSITDQLKNGVRGLMLDMWDYEDTIWLCRGPCTKYTTFQPALN 144
Query: 121 VLREINTFLTRHRTEIVTVFIKDRVTSPNGVNKVFNKAGLRKFWFPVYKMPKNGSDWLTV 180
VL+E+ FL H TEI+T+FI D VTS NGVNKVF+KA LRKFWFPV KMPKNGSDW TV
Sbjct: 145 VLKEVRVFLVTHPTEIITIFIDDHVTSGNGVNKVFDKARLRKFWFPVSKMPKNGSDWPTV 204
Query: 181 KKMLRMNHRLIVFTSNATKEASERIAYEWNYVVENKYGNDGMGRDHCLHRAESYPMNTTT 240
K M+R N+RLIVFTSNA++EASE IAYEWNYVVE+++GN G+ C +R ES PMN T
Sbjct: 205 KTMIRKNYRLIVFTSNASREASEGIAYEWNYVVESQFGNVGIKGGSCQNRPESLPMNNAT 264
Query: 241 KSLVLMNYYRNVLN-SNEACKDNSSPLIRKMHTCYKDAGNRWPNYIAVDFYKRGDGGGAP 299
KSLVLMNY+RNV N +EAC+DNSSPLI MH C++ AGNRWPN+IAVDFYKRGDGGGAP
Sbjct: 265 KSLVLMNYFRNVRNHDDEACRDNSSPLIAMMHVCFRAAGNRWPNFIAVDFYKRGDGGGAP 324
Query: 300 EALDVANRNL 309
EALD+ANRNL
Sbjct: 325 EALDLANRNL 334
>emb|CAB16796.1| MAP3K-like protein kinase [Arabidopsis thaliana]
gi|7270644|emb|CAB80361.1| MAP3K-like protein kinase
[Arabidopsis thaliana] gi|25407778|pir||D85436
MAP3K-like protein kinase [imported] - Arabidopsis
thaliana
Length = 799
Score = 358 bits (919), Expect = 1e-97
Identities = 177/316 (56%), Positives = 222/316 (70%), Gaps = 8/316 (2%)
Query: 1 QIGETCSRDVNDCGTGLQCLEC----NSQSRCTRVRTSSPISKVMELPFNHYSWLTTHNS 56
++GETCS ++C GL C C N+ S CTR++ +P SKV LPFN YSWLTTHNS
Sbjct: 338 EMGETCS-STSECDAGLSCQSCPANGNTGSTCTRIQPLNPTSKVNGLPFNKYSWLTTHNS 396
Query: 57 YASRAANLSIDSKISSVMNQEDSITDQLRNGVRGIMLDMHDYYGDIWLCR---GPCTIFT 113
YA AN + S + S NQEDSIT+QL+NGVRGIMLD +D+ DIWLC G C FT
Sbjct: 397 YAITGANSATGSFLVSPKNQEDSITNQLKNGVRGIMLDTYDFQNDIWLCHSTGGTCFNFT 456
Query: 114 AFQPAINVLREINTFLTRHRTEIVTVFIKDRVTSPNGVNKVFNKAGLRKFWFPVYKMPKN 173
AFQPAIN L+EIN FL + +EIVT+ ++D V S G+ VFN +GL KF P+ +MPK+
Sbjct: 457 AFQPAINALKEINDFLESNLSEIVTIILEDYVKSQMGLTNVFNASGLSKFLLPISRMPKD 516
Query: 174 GSDWLTVKKMLRMNHRLIVFTSNATKEASERIAYEWNYVVENKYGNDGMGRDHCLHRAES 233
G+DW TV M++ N RL+VFTS KEASE +AY+WNY+VEN+YGNDGM C R+ES
Sbjct: 517 GTDWPTVDDMVKQNQRLVVFTSKKDKEASEGLAYQWNYMVENQYGNDGMKDGSCSSRSES 576
Query: 234 YPMNTTTKSLVLMNYYRNVLNSNEACKDNSSPLIRKMHTCYKDAGNRWPNYIAVDFYKRG 293
++T ++SLV NY+ NS +AC DNSSPLI M TC++ AG RWPN+IAVDFY+R
Sbjct: 577 SSLDTMSRSLVFQNYFETSPNSTQACADNSSPLIEMMRTCHEAAGKRWPNFIAVDFYQRS 636
Query: 294 DGGGAPEALDVANRNL 309
D GGA EA+D AN L
Sbjct: 637 DSGGAAEAVDEANGRL 652
>gb|AAM47915.1| unknown protein [Arabidopsis thaliana] gi|20260218|gb|AAM13007.1|
unknown protein [Arabidopsis thaliana]
gi|18402763|ref|NP_564553.1| expressed protein
[Arabidopsis thaliana] gi|25372963|pir||A96534
hypothetical protein F14J22.5 [imported] - Arabidopsis
thaliana gi|10120435|gb|AAG13060.1| Unknown protein
[Arabidopsis thaliana]
Length = 359
Score = 346 bits (887), Expect = 6e-94
Identities = 171/314 (54%), Positives = 220/314 (69%), Gaps = 8/314 (2%)
Query: 3 GETCSRDVNDCGTGLQCLECNSQS----RCTRVRTSSPISKVMELPFNHYSWLTTHNSYA 58
G+TC + N C GL C C + + RC+R + +PI+K LPFN YSWLTTHNS+A
Sbjct: 30 GKTCITNSN-CDAGLHCETCIANTDFRPRCSRTQPINPITKAKGLPFNKYSWLTTHNSFA 88
Query: 59 SRAANLSIDSKISSVMNQEDSITDQLRNGVRGIMLDMHDYYGDIWLCR---GPCTIFTAF 115
S I + NQ+DSIT QL NGVRG MLDM+D+ DIWLC G C FTAF
Sbjct: 89 RLGEVSRTGSAILAPTNQQDSITSQLNNGVRGFMLDMYDFQNDIWLCHSFDGTCFNFTAF 148
Query: 116 QPAINVLREINTFLTRHRTEIVTVFIKDRVTSPNGVNKVFNKAGLRKFWFPVYKMPKNGS 175
QPAIN+LRE FL +++ E+VT+ I+D V SP G+ KVF+ AGLRKF FPV +MPKNG
Sbjct: 149 QPAINILREFQVFLEKNKEEVVTIIIEDYVKSPKGLTKVFDAAGLRKFMFPVSRMPKNGG 208
Query: 176 DWLTVKKMLRMNHRLIVFTSNATKEASERIAYEWNYVVENKYGNDGMGRDHCLHRAESYP 235
DW + M+R N RL+VFTS++ KEA+E IAY+W Y+VEN+YGN G+ C +RA+S P
Sbjct: 209 DWPRLDDMVRKNQRLLVFTSDSHKEATEGIAYQWKYMVENQYGNGGLKVGVCPNRAQSAP 268
Query: 236 MNTTTKSLVLMNYYRNVLNSNEACKDNSSPLIRKMHTCYKDAGNRWPNYIAVDFYKRGDG 295
M+ +KSLVL+N++ + + ACK NS+ L+ + TCY+ AG RWPN+IAVDFYKR DG
Sbjct: 269 MSDKSKSLVLVNHFPDAADVIVACKQNSASLLESIKTCYQAAGQRWPNFIAVDFYKRSDG 328
Query: 296 GGAPEALDVANRNL 309
GGAP+A+DVAN NL
Sbjct: 329 GGAPQAVDVANGNL 342
>ref|XP_479671.1| MAP3K-like protein kinase [Oryza sativa (japonica cultivar-group)]
gi|51963696|ref|XP_506619.1| PREDICTED P0015C07.31 gene
product [Oryza sativa (japonica cultivar-group)]
gi|50725707|dbj|BAD33173.1| MAP3K-like protein [Oryza
sativa (japonica cultivar-group)]
Length = 411
Score = 345 bits (886), Expect = 7e-94
Identities = 166/315 (52%), Positives = 223/315 (70%), Gaps = 9/315 (2%)
Query: 2 IGETCSRDVNDCGTGLQCLECNS----QSRCTRVRTSSPISKVMELPFNHYSWLTTHNSY 57
+G+TCS + DCG GL C +C CTR + P++ +LPFN+YSWLTTHNSY
Sbjct: 23 VGDTCSSE-GDCGAGLHCSDCGGGGGGDKTCTRAKPIDPLTHGTDLPFNNYSWLTTHNSY 81
Query: 58 ASRAANLSIDSKISSVMNQEDSITDQLRNGVRGIMLDMHDYYGDIWLC---RGPCTIFTA 114
A ++ + S + + NQED+IT QL+NGVRG+MLD +D+ D+WLC +G C FTA
Sbjct: 82 ALAGSSSATGSALITQTNQEDTITAQLKNGVRGLMLDTYDFNNDVWLCHSFQGKCFNFTA 141
Query: 115 FQPAINVLREINTFLTRHRTEIVTVFIKDRVTSPNGVNKVFNKAGLRKFWFPVYKMPKNG 174
FQPAINVL+EI TFL + +E++T+F++D T+ + KVFN +GL K+WFPV KMPK+G
Sbjct: 142 FQPAINVLKEIRTFLDGNPSEVITIFLED-YTASGSLPKVFNASGLMKYWFPVAKMPKSG 200
Query: 175 SDWLTVKKMLRMNHRLIVFTSNATKEASERIAYEWNYVVENKYGNDGMGRDHCLHRAESY 234
DW +K M+ N RL+VFTS +KEASE IAYEW+YVVEN+YGN+GM C +RAES
Sbjct: 201 GDWPLLKDMISQNERLLVFTSKKSKEASEGIAYEWSYVVENQYGNEGMVEGKCPNRAESP 260
Query: 235 PMNTTTKSLVLMNYYRNVLNSNEACKDNSSPLIRKMHTCYKDAGNRWPNYIAVDFYKRGD 294
M++ ++SLVLMN++ + C +NS+PL+ + TC+ +GNRWPNYIAVDFY R D
Sbjct: 261 AMDSKSQSLVLMNFFTTDPSQTGVCANNSAPLVSMLKTCHDLSGNRWPNYIAVDFYMRSD 320
Query: 295 GGGAPEALDVANRNL 309
GGGAP A D+AN +L
Sbjct: 321 GGGAPLATDIANGHL 335
>emb|CAE01622.2| OSJNBa0042L16.16 [Oryza sativa (japonica cultivar-group)]
gi|50924276|ref|XP_472498.1| OSJNBa0042L16.16 [Oryza
sativa (japonica cultivar-group)]
Length = 413
Score = 335 bits (858), Expect = 1e-90
Identities = 165/316 (52%), Positives = 217/316 (68%), Gaps = 8/316 (2%)
Query: 1 QIGETCSRDVNDCGTGLQCLEC----NSQSRCTRVRTSSPISKVMELPFNHYSWLTTHNS 56
++GETC+ D N C GL C C N + RCTRV P +K +LPFN Y+WLTTHNS
Sbjct: 33 KVGETCAADRN-CDAGLHCETCVADGNVRPRCTRVTPVDPQTKARDLPFNRYAWLTTHNS 91
Query: 57 YASRAANLSIDSKISSVMNQEDSITDQLRNGVRGIMLDMHDYYGDIWLCR---GPCTIFT 113
+A + I++ NQ+D+ITDQL NGVRG+MLDM+D+ DIWLC G C FT
Sbjct: 92 FARLGTRSRTGTAIATAWNQQDTITDQLNNGVRGLMLDMYDFRNDIWLCHSFGGACQNFT 151
Query: 114 AFQPAINVLREINTFLTRHRTEIVTVFIKDRVTSPNGVNKVFNKAGLRKFWFPVYKMPKN 173
AF PA+ VL EI FL R+ +E+VTVF++D V SP G+ +V N +GL K+ FP ++MPK+
Sbjct: 152 AFVPAVEVLGEIERFLARNPSEVVTVFVEDYVESPMGLTRVLNASGLTKYVFPAWRMPKS 211
Query: 174 GSDWLTVKKMLRMNHRLIVFTSNATKEASERIAYEWNYVVENKYGNDGMGRDHCLHRAES 233
G DW + M+R NHRL++FTS + KEA+E I YEW+YVVEN+YG GM + C +RAES
Sbjct: 212 GGDWPRLSDMVRDNHRLLLFTSKSAKEAAEGIPYEWHYVVENQYGTKGMIKGRCPNRAES 271
Query: 234 YPMNTTTKSLVLMNYYRNVLNSNEACKDNSSPLIRKMHTCYKDAGNRWPNYIAVDFYKRG 293
MN ++SLVL+NY+R++ N ACKDNS+ L+ + TC+ + +RW N+IAVDFYKR
Sbjct: 272 AAMNDLSRSLVLVNYFRDLPNFPVACKDNSAELLDMLTTCHDLSADRWANFIAVDFYKRS 331
Query: 294 DGGGAPEALDVANRNL 309
D GGA EA D AN L
Sbjct: 332 DRGGAAEATDRANGGL 347
>gb|AAR95995.1| hypothetical protein kinase [Musa acuminata]
Length = 376
Score = 329 bits (844), Expect = 6e-89
Identities = 171/342 (50%), Positives = 223/342 (65%), Gaps = 40/342 (11%)
Query: 1 QIGETCSRDVNDCGTGLQCLECNSQ-SRCTRVRTSSPISKVM------------------ 41
++GE CS + DC GL+C C+ C R+R P SKV
Sbjct: 26 KLGEGCSAN-QDCDAGLRCDGCDGDLGVCVRIRPYEPRSKVRIRHYPFSIRNLGLWVGWF 84
Query: 42 --------------ELPFNHYSWLTTHNSYASRAANLSIDSKISSVMNQEDSITDQLRNG 87
+LPFN YSWLTTHNS+A A+ + + + + NQ D+IT QL NG
Sbjct: 85 RFRANLGLECAQGKDLPFNKYSWLTTHNSFADAGAHSATGATLITFTNQHDNITSQLNNG 144
Query: 88 VRGIMLDMHDYYGDIWLCRGPCTIFTAFQPAINVLREINTFLTRHRTEIVTVFIKDRVTS 147
VRG+MLDM+D+ DIWLC QPAINVL+EI TFL + +E++T+FI+D V S
Sbjct: 145 VRGLMLDMYDFRNDIWLCHSTAVYQ---QPAINVLKEIETFLAANPSEVITIFIEDYVKS 201
Query: 148 PNGVNKVFNKAGLRKFWFPVYKMPKNGSDWLTVKKMLRMNHRLIVFTSNATKEASERIAY 207
P+G++KVFN +GL K+WFPV +MPKNGSDW + KM+ NHRL+VFTS A+KEASE IAY
Sbjct: 202 PSGLSKVFNASGLMKYWFPVDQMPKNGSDWPLLSKMIDQNHRLLVFTSVASKEASEGIAY 261
Query: 208 EWNYVVENKYGNDGMGRDHCLHRAESYPMNTTTKSLVLMNYYRNVLNSNEACKDNSSPLI 267
EWNYVVEN+YG++GM C RAES PM+TT KSLVLMNY+R +++ AC +NS+PL+
Sbjct: 262 EWNYVVENQYGDEGMTPGSCPSRAESSPMSTTLKSLVLMNYFRTNPSASSACHNNSAPLL 321
Query: 268 RKMHTCYKDAGNRWPNYIAVDFYKRGDGGGAPEALDVANRNL 309
+ TC+ + NRW N+IAVDFY +GD APEA DVAN ++
Sbjct: 322 DMLKTCHGLSANRWANFIAVDFYMKGD---APEAADVANGHM 360
>dbj|BAB02455.1| MAP3K protein kinase-like protein [Arabidopsis thaliana]
gi|15230348|ref|NP_188562.1| expressed protein
[Arabidopsis thaliana]
Length = 413
Score = 325 bits (834), Expect = 8e-88
Identities = 157/311 (50%), Positives = 212/311 (67%), Gaps = 8/311 (2%)
Query: 3 GETCSRDVNDCGTGLQCLEC----NSQSRCTRVRTSSPISKVMELPFNHYSWLTTHNSYA 58
GETC N C GL C C + + RC+R++ +P +KV LP+N YSWLTTHNS+A
Sbjct: 30 GETCIVSKN-CDRGLHCESCLASDSFRPRCSRMQPINPTTKVKGLPYNKYSWLTTHNSFA 88
Query: 59 SRAANLSIDSKISSVMNQEDSITDQLRNGVRGIMLDMHDYYGDIWLCR---GPCTIFTAF 115
A S I + NQ+DSIT QL NGVRG MLD++D+ DIWLC G C +TAF
Sbjct: 89 RMGAKSGTGSMILAPSNQQDSITSQLLNGVRGFMLDLYDFQNDIWLCHSYGGNCFNYTAF 148
Query: 116 QPAINVLREINTFLTRHRTEIVTVFIKDRVTSPNGVNKVFNKAGLRKFWFPVYKMPKNGS 175
QPA+N+L+E FL +++ +VT+ ++D V SPNG+ +VF+ +GLR F FPV +MPKNG
Sbjct: 149 QPAVNILKEFQVFLDKNKDVVVTLILEDYVKSPNGLTRVFDASGLRNFMFPVSRMPKNGE 208
Query: 176 DWLTVKKMLRMNHRLIVFTSNATKEASERIAYEWNYVVENKYGNDGMGRDHCLHRAESYP 235
DW T+ M+ N RL+VFTSN KEASE IA+ W Y++EN+YG+ GM C +R ES
Sbjct: 209 DWPTLDDMICQNQRLLVFTSNPQKEASEGIAFMWRYMIENQYGDGGMKAGVCTNRPESVA 268
Query: 236 MNTTTKSLVLMNYYRNVLNSNEACKDNSSPLIRKMHTCYKDAGNRWPNYIAVDFYKRGDG 295
M ++SL+L+NY+ + + +CK NS+PL+ + C + +G RWPN+IAVDFYKR DG
Sbjct: 269 MGDRSRSLILVNYFPDTADVIGSCKQNSAPLLDTVKNCQEASGKRWPNFIAVDFYKRSDG 328
Query: 296 GGAPEALDVAN 306
GGAP+A+DVAN
Sbjct: 329 GGAPKAVDVAN 339
>gb|AAO22804.1| unknown protein [Arabidopsis thaliana]
Length = 397
Score = 325 bits (834), Expect = 8e-88
Identities = 157/311 (50%), Positives = 212/311 (67%), Gaps = 8/311 (2%)
Query: 3 GETCSRDVNDCGTGLQCLEC----NSQSRCTRVRTSSPISKVMELPFNHYSWLTTHNSYA 58
GETC N C GL C C + + RC+R++ +P +KV LP+N YSWLTTHNS+A
Sbjct: 14 GETCIVSKN-CDRGLHCESCLASDSFRPRCSRMQPINPTTKVKGLPYNKYSWLTTHNSFA 72
Query: 59 SRAANLSIDSKISSVMNQEDSITDQLRNGVRGIMLDMHDYYGDIWLCR---GPCTIFTAF 115
A S I + NQ+DSIT QL NGVRG MLD++D+ DIWLC G C +TAF
Sbjct: 73 RMGAKSGTGSMILAPSNQQDSITSQLLNGVRGFMLDLYDFQNDIWLCHSYGGNCFNYTAF 132
Query: 116 QPAINVLREINTFLTRHRTEIVTVFIKDRVTSPNGVNKVFNKAGLRKFWFPVYKMPKNGS 175
QPA+N+L+E FL +++ +VT+ ++D V SPNG+ +VF+ +GLR F FPV +MPKNG
Sbjct: 133 QPAVNILKEFQVFLDKNKDVVVTLILEDYVKSPNGLTRVFDASGLRNFMFPVSRMPKNGE 192
Query: 176 DWLTVKKMLRMNHRLIVFTSNATKEASERIAYEWNYVVENKYGNDGMGRDHCLHRAESYP 235
DW T+ M+ N RL+VFTSN KEASE IA+ W Y++EN+YG+ GM C +R ES
Sbjct: 193 DWPTLDDMICQNQRLLVFTSNPQKEASEGIAFMWRYMIENQYGDGGMKAGVCTNRPESVA 252
Query: 236 MNTTTKSLVLMNYYRNVLNSNEACKDNSSPLIRKMHTCYKDAGNRWPNYIAVDFYKRGDG 295
M ++SL+L+NY+ + + +CK NS+PL+ + C + +G RWPN+IAVDFYKR DG
Sbjct: 253 MGDRSRSLILVNYFPDTADVIGSCKQNSAPLLDTVKNCQEASGKRWPNFIAVDFYKRSDG 312
Query: 296 GGAPEALDVAN 306
GGAP+A+DVAN
Sbjct: 313 GGAPKAVDVAN 323
>gb|AAM10115.1| unknown protein [Arabidopsis thaliana] gi|18425155|ref|NP_569045.1|
expressed protein [Arabidopsis thaliana]
gi|15451188|gb|AAK96865.1| Unknown protein [Arabidopsis
thaliana]
Length = 426
Score = 317 bits (811), Expect = 4e-85
Identities = 157/313 (50%), Positives = 213/313 (67%), Gaps = 8/313 (2%)
Query: 1 QIGETCSRDVNDCGTGLQCLECNSQSR----CTRVRTSSPISKVMELPFNHYSWLTTHNS 56
Q+ ++CS DC +GL C +C + R CTR + +SP S + LPFN Y+WL THN+
Sbjct: 35 QLLDSCS-SATDCVSGLYCGDCPAVGRSKPVCTRGQATSPTSIINGLPFNKYTWLMTHNA 93
Query: 57 YASRAANLSIDSKISSVMNQEDSITDQLRNGVRGIMLDMHDYYGDIWLC---RGPCTIFT 113
+++ A L + + NQED+IT+QL+NGVRG+MLDM+D+ DIWLC RG C FT
Sbjct: 94 FSNANAPLLPGVERITFYNQEDTITNQLQNGVRGLMLDMYDFNNDIWLCHSLRGQCFNFT 153
Query: 114 AFQPAINVLREINTFLTRHRTEIVTVFIKDRVTSPNGVNKVFNKAGLRKFWFPVYKMPKN 173
AFQPAIN+LRE+ FL+++ TEIVT+ I+D V P G++ +F AGL K+WFPV KMP+
Sbjct: 154 AFQPAINILREVEAFLSQNPTEIVTIIIEDYVHRPKGLSTLFANAGLDKYWFPVSKMPRK 213
Query: 174 GSDWLTVKKMLRMNHRLIVFTSNATKEASERIAYEWNYVVENKYGNDGMGRDHCLHRAES 233
G DW TV M++ NHRL+VFTS A KE E +AY+W Y+VEN+ G+ G+ R C +R ES
Sbjct: 214 GEDWPTVTDMVQENHRLLVFTSVAAKEDEEGVAYQWRYMVENESGDPGVKRGSCPNRKES 273
Query: 234 YPMNTTTKSLVLMNYYRNVLNSNEACKDNSSPLIRKMHTCYKDAGNRWPNYIAVDFYKRG 293
P+N+ + SL LMNY+ +ACK++S+PL + TC K GNR PN++AV+FY R
Sbjct: 274 QPLNSKSSSLFLMNYFPTYPVEKDACKEHSAPLAEMVGTCLKSGGNRMPNFLAVNFYMRS 333
Query: 294 DGGGAPEALDVAN 306
DGGG E LD N
Sbjct: 334 DGGGVFEILDRMN 346
>gb|AAM65394.1| unknown [Arabidopsis thaliana]
Length = 426
Score = 315 bits (807), Expect = 1e-84
Identities = 156/313 (49%), Positives = 212/313 (66%), Gaps = 8/313 (2%)
Query: 1 QIGETCSRDVNDCGTGLQCLECNSQSR----CTRVRTSSPISKVMELPFNHYSWLTTHNS 56
Q+ ++CS DC +GL C +C + R CTR + +SP S + LPFN Y+WL THN+
Sbjct: 35 QLLDSCS-SATDCVSGLYCGDCPAVGRSKPVCTRGQATSPTSIINGLPFNKYTWLMTHNA 93
Query: 57 YASRAANLSIDSKISSVMNQEDSITDQLRNGVRGIMLDMHDYYGDIWLC---RGPCTIFT 113
+++ A L + + NQED+IT+QL+NGVRG+MLDM+D+ DIWLC RG C FT
Sbjct: 94 FSNANAPLLPGVERITFYNQEDTITNQLQNGVRGLMLDMYDFNNDIWLCHSLRGQCFNFT 153
Query: 114 AFQPAINVLREINTFLTRHRTEIVTVFIKDRVTSPNGVNKVFNKAGLRKFWFPVYKMPKN 173
FQPAIN+LRE+ FL+++ TEIVT+ I+D V P G++ +F AGL K+WFPV KMP+
Sbjct: 154 XFQPAINILREVEAFLSQNPTEIVTIIIEDYVHRPKGLSTLFANAGLDKYWFPVSKMPRK 213
Query: 174 GSDWLTVKKMLRMNHRLIVFTSNATKEASERIAYEWNYVVENKYGNDGMGRDHCLHRAES 233
G DW TV M++ NHRL+VFTS A KE E +AY+W Y+VEN+ G+ G+ R C +R ES
Sbjct: 214 GEDWPTVTDMVQENHRLLVFTSVAAKEDEEGVAYQWRYMVENESGDPGVKRGSCPNRKES 273
Query: 234 YPMNTTTKSLVLMNYYRNVLNSNEACKDNSSPLIRKMHTCYKDAGNRWPNYIAVDFYKRG 293
P+N+ + SL LMNY+ +ACK++S+PL + TC K GNR PN++AV+FY R
Sbjct: 274 QPLNSKSSSLFLMNYFPTYPVEKDACKEHSAPLAEMVGTCLKSGGNRMPNFLAVNFYMRS 333
Query: 294 DGGGAPEALDVAN 306
DGGG E LD N
Sbjct: 334 DGGGVFEILDRMN 346
>ref|XP_468681.1| expressed protein [Oryza sativa (japonica cultivar-group)]
gi|41469135|gb|AAS07086.1| expressed protein [Oryza
sativa (japonica cultivar-group)]
Length = 360
Score = 304 bits (779), Expect = 2e-81
Identities = 153/315 (48%), Positives = 204/315 (64%), Gaps = 8/315 (2%)
Query: 2 IGETCSRDVNDCGTGLQCLECNSQ---SRCTRVRTSSPISKVME-LPFNHYSWLTTHNSY 57
+G++CS V DCG G C +C + S C R ++P LPFN Y++LTTHNS+
Sbjct: 27 VGDSCSTAV-DCGGGQWCFDCQPEFAGSSCVRSAATNPFQLTNNSLPFNKYAYLTTHNSF 85
Query: 58 ASRAANLSIDSKISSVMNQEDSITDQLRNGVRGIMLDMHDYYGDIWLCR---GPCTIFTA 114
A + NQED++TDQL NGVR +MLD +D+ GD+WLC G C FTA
Sbjct: 86 AIVGEPSHTGVPRITFDNQEDTVTDQLNNGVRALMLDTYDFKGDVWLCHSNGGKCNDFTA 145
Query: 115 FQPAINVLREINTFLTRHRTEIVTVFIKDRVTSPNGVNKVFNKAGLRKFWFPVYKMPKNG 174
F+PA++ +EI FL + +EIVT+ ++D V +PNG+ VF +GL K+WFPV KMP+ G
Sbjct: 146 FEPALDTFKEIEAFLGANPSEIVTLILEDYVHAPNGLTNVFKASGLMKYWFPVSKMPQKG 205
Query: 175 SDWLTVKKMLRMNHRLIVFTSNATKEASERIAYEWNYVVENKYGNDGMGRDHCLHRAESY 234
DW V M+ N RL+VFTS +K+A+E IAY+WNY+VEN YG+DGM C +RAES
Sbjct: 206 KDWPLVSDMVASNQRLLVFTSIRSKQATEGIAYQWNYMVENNYGDDGMDAGKCSNRAESA 265
Query: 235 PMNTTTKSLVLMNYYRNVLNSNEACKDNSSPLIRKMHTCYKDAGNRWPNYIAVDFYKRGD 294
P+N TKSLVL+NY+ +V AC +S L ++TCY AGNRW N +AVD+YKR D
Sbjct: 266 PLNDKTKSLVLVNYFPSVPVKVTACLQHSKSLTDMVNTCYGAAGNRWANLLAVDYYKRSD 325
Query: 295 GGGAPEALDVANRNL 309
GGGA +A D+ N L
Sbjct: 326 GGGAFQATDLLNGRL 340
>ref|XP_450041.1| MAP3K protein kinase-like protein [Oryza sativa (japonica
cultivar-group)] gi|51963708|ref|XP_506634.1| PREDICTED
OJ1310_F05.17 gene product [Oryza sativa (japonica
cultivar-group)] gi|46806453|dbj|BAD17589.1| MAP3K
protein kinase-like protein [Oryza sativa (japonica
cultivar-group)] gi|46389989|dbj|BAD16231.1| MAP3K
protein kinase-like protein [Oryza sativa (japonica
cultivar-group)]
Length = 412
Score = 302 bits (773), Expect = 9e-81
Identities = 150/313 (47%), Positives = 208/313 (65%), Gaps = 9/313 (2%)
Query: 1 QIGETCSRDVNDCGTGLQCLECNSQSR----CTRVRTSSPISKVMELPFNHYSWLTTHNS 56
Q+G++CS DCG GL C C + + C R P S V LPFN YSWL THNS
Sbjct: 23 QVGDSCS-SARDCGAGLYCGNCAATGKTRPSCIRDLAIQPTSIVKGLPFNRYSWLVTHNS 81
Query: 57 YASRAANLSIDSKISSVMNQEDSITDQLRNGVRGIMLDMHDYYGDIWLC---RGPCTIFT 113
++ + + NQED++T+QLRNGVRG+MLDM+D+ DIWLC +G C FT
Sbjct: 82 FSIIGEPSHTGVERVTFYNQEDTVTNQLRNGVRGLMLDMYDFNDDIWLCHSLQGQCYNFT 141
Query: 114 AFQPAINVLREINTFLTRHRTEIVTVFIKDRVTSPNGVNKVFNKAGLRKFWFPVYKMPKN 173
AFQPAI+ L+E+ FL+ + TEI+T+FI+D V S G++K+F A L K+W+P+ +MP N
Sbjct: 142 AFQPAIDTLKEVEAFLSENPTEIITIFIEDYVHSTMGLSKLFTAADLTKYWYPISEMPTN 201
Query: 174 GSDWLTVKKMLRMNHRLIVFTSNATKEASERIAYEWNYVVENKYGNDGMGRDHCLHRAES 233
G DW +V M+ NHRL+VFTS+++KEASE IAY+W+Y++EN+ G+ G+ C +R ES
Sbjct: 202 GKDWPSVTDMVAKNHRLLVFTSDSSKEASEGIAYQWSYLLENESGDPGI-TGSCPNRKES 260
Query: 234 YPMNTTTKSLVLMNYYRNVLNSNEACKDNSSPLIRKMHTCYKDAGNRWPNYIAVDFYKRG 293
P+N+ + SL + NY+ + NEACK+NS L + + TCY AGNR PN+IAV++Y R
Sbjct: 261 QPLNSRSASLFMQNYFPTIPVENEACKENSVGLPQMVQTCYTAAGNRIPNFIAVNYYMRS 320
Query: 294 DGGGAPEALDVAN 306
DGGG + D N
Sbjct: 321 DGGGVFDVQDRIN 333
>ref|NP_172824.2| expressed protein [Arabidopsis thaliana]
Length = 317
Score = 291 bits (745), Expect = 2e-77
Identities = 149/314 (47%), Positives = 200/314 (63%), Gaps = 13/314 (4%)
Query: 1 QIGETCSRDVNDCGTGLQCLECNSQ-SRCTRVRTSSPISKVME-LPFNHYSWLTTHNSYA 58
Q+G+ CS D DC GL C +C +RC R + S V +PFN Y++LTTHNSYA
Sbjct: 2 QLGDQCSSD-EDCNVGLGCFKCGIDVARCVRSNITDQFSIVNNSMPFNKYAFLTTHNSYA 60
Query: 59 SRAANLSIDSKISSVMNQEDSITDQLRNGVRGIMLDMHDYYGDIWLCRG---PCTIFTAF 115
I+ K V QED+I QL +GVR +MLD +DY GD+W C C FT F
Sbjct: 61 -------IEGKALHVATQEDTIVQQLNSGVRALMLDTYDYEGDVWFCHSFDEQCFEFTKF 113
Query: 116 QPAINVLREINTFLTRHRTEIVTVFIKDRVTSPNGVNKVFNKAGLRKFWFPVYKMPKNGS 175
AI+ +EI FLT + +EIVT+ ++D V S NG+ KVF +GL+KFWFPV MP G
Sbjct: 114 NRAIDTFKEIFAFLTANPSEIVTLILEDYVKSQNGLTKVFTDSGLKKFWFPVQNMPIGGQ 173
Query: 176 DWLTVKKMLRMNHRLIVFTSNATKEASERIAYEWNYVVENKYGNDGMGRDHCLHRAESYP 235
DW VK M+ NHRLIVFTS +K+ +E IAY+WNY+VEN+YG+DG+ D C +RA+S
Sbjct: 174 DWPLVKDMVANNHRLIVFTSAKSKQETEGIAYQWNYMVENQYGDDGVKPDECSNRADSAL 233
Query: 236 MNTTTKSLVLMNYYRNVLNSNEACKDNSSPLIRKMHTCYKDAGNRWPNYIAVDFYKRGDG 295
+ TK+LV +N+++ V C++NS L+ + TCY AGNRW N++AV+FYKR +G
Sbjct: 234 LTDKTKALVSVNHFKTVPVKILTCEENSEQLLDMIKTCYVAAGNRWANFVAVNFYKRSNG 293
Query: 296 GGAPEALDVANRNL 309
GG +A+D N L
Sbjct: 294 GGTFQAIDKLNGEL 307
>pir||B86270 hypothetical protein F21F23.12 - Arabidopsis thaliana
gi|8920573|gb|AAF81295.1| Contains similarity to
MAP3K-like protein kinase from Arabidopsis thaliana
gb|Z99707
Length = 346
Score = 291 bits (745), Expect = 2e-77
Identities = 149/314 (47%), Positives = 200/314 (63%), Gaps = 13/314 (4%)
Query: 1 QIGETCSRDVNDCGTGLQCLECNSQ-SRCTRVRTSSPISKVME-LPFNHYSWLTTHNSYA 58
Q+G+ CS D DC GL C +C +RC R + S V +PFN Y++LTTHNSYA
Sbjct: 31 QLGDQCSSD-EDCNVGLGCFKCGIDVARCVRSNITDQFSIVNNSMPFNKYAFLTTHNSYA 89
Query: 59 SRAANLSIDSKISSVMNQEDSITDQLRNGVRGIMLDMHDYYGDIWLCRG---PCTIFTAF 115
I+ K V QED+I QL +GVR +MLD +DY GD+W C C FT F
Sbjct: 90 -------IEGKALHVATQEDTIVQQLNSGVRALMLDTYDYEGDVWFCHSFDEQCFEFTKF 142
Query: 116 QPAINVLREINTFLTRHRTEIVTVFIKDRVTSPNGVNKVFNKAGLRKFWFPVYKMPKNGS 175
AI+ +EI FLT + +EIVT+ ++D V S NG+ KVF +GL+KFWFPV MP G
Sbjct: 143 NRAIDTFKEIFAFLTANPSEIVTLILEDYVKSQNGLTKVFTDSGLKKFWFPVQNMPIGGQ 202
Query: 176 DWLTVKKMLRMNHRLIVFTSNATKEASERIAYEWNYVVENKYGNDGMGRDHCLHRAESYP 235
DW VK M+ NHRLIVFTS +K+ +E IAY+WNY+VEN+YG+DG+ D C +RA+S
Sbjct: 203 DWPLVKDMVANNHRLIVFTSAKSKQETEGIAYQWNYMVENQYGDDGVKPDECSNRADSAL 262
Query: 236 MNTTTKSLVLMNYYRNVLNSNEACKDNSSPLIRKMHTCYKDAGNRWPNYIAVDFYKRGDG 295
+ TK+LV +N+++ V C++NS L+ + TCY AGNRW N++AV+FYKR +G
Sbjct: 263 LTDKTKALVSVNHFKTVPVKILTCEENSEQLLDMIKTCYVAAGNRWANFVAVNFYKRSNG 322
Query: 296 GGAPEALDVANRNL 309
GG +A+D N L
Sbjct: 323 GGTFQAIDKLNGEL 336
>ref|XP_474458.1| OSJNBa0039K24.18 [Oryza sativa (japonica cultivar-group)]
gi|38345515|emb|CAE01799.2| OSJNBa0039K24.18 [Oryza
sativa (japonica cultivar-group)]
Length = 468
Score = 288 bits (736), Expect = 2e-76
Identities = 139/314 (44%), Positives = 201/314 (63%), Gaps = 10/314 (3%)
Query: 2 IGETCSRD-VNDCGTGLQCLECN-----SQSRCTRVRTSSPISKVMELPFNHYSWLTTHN 55
+G+TC+ + CG G++C C+ C+R P + +L FN Y+WLTTHN
Sbjct: 30 VGDTCTASSASSCGAGMRCATCSPLPGMGPPVCSRTTPLDPKAHGTDLAFNRYTWLTTHN 89
Query: 56 SYASRAANLSIDSKISSVMNQEDSITDQLRNGVRGIMLDMHDYYGDIWLCR---GPCTIF 112
S+A + + I + NQED++T QL+NGVRG+MLD +D+ ++WLC G C F
Sbjct: 90 SFAIVGSPSRTGTPIIAPPNQEDTVTAQLKNGVRGLMLDAYDFQNEVWLCHSFGGKCYNF 149
Query: 113 TAFQPAINVLREINTFLTRHRTEIVTVFIKDRVTSPNGVNKVFNKAGLRKFWFPVYKMPK 172
A+Q A++VL+EI FL + +E++TVF++D P + KV +GL K+ FP KMPK
Sbjct: 150 AAYQRAMDVLKEIGAFLDANPSEVITVFVED-YAGPGSLGKVVGGSGLSKYLFPPAKMPK 208
Query: 173 NGSDWLTVKKMLRMNHRLIVFTSNATKEASERIAYEWNYVVENKYGNDGMGRDHCLHRAE 232
G DW +K M+ NHRL++FTS K+ S+ +AYEW+YV+E +YGNDG+ C RAE
Sbjct: 209 GGGDWPLLKDMIAQNHRLLMFTSKRGKDGSDGLAYEWDYVLETQYGNDGLVGGSCPKRAE 268
Query: 233 SYPMNTTTKSLVLMNYYRNVLNSNEACKDNSSPLIRKMHTCYKDAGNRWPNYIAVDFYKR 292
S M++T +SL+LMN++ + + AC +NS+PL+ K+ CY + RWPN+IAVD+Y R
Sbjct: 269 SMAMDSTKQSLILMNFFSTNPSQSWACGNNSAPLVAKLKACYDASAKRWPNFIAVDYYMR 328
Query: 293 GDGGGAPEALDVAN 306
GGGAP A DVAN
Sbjct: 329 SKGGGAPLATDVAN 342
>emb|CAB53483.1| CAA303710.1 protein [Oryza sativa]
Length = 416
Score = 266 bits (679), Expect = 7e-70
Identities = 132/314 (42%), Positives = 193/314 (61%), Gaps = 22/314 (7%)
Query: 2 IGETCSRD-VNDCGTGLQCLECN-----SQSRCTRVRTSSPISKVMELPFNHYSWLTTHN 55
+G+TC+ + CG G++C C+ C+R P + +L FN Y+WLTTHN
Sbjct: 30 VGDTCTASSASSCGAGMRCATCSPLPGMGPPVCSRTTPLDPKAHGTDLAFNRYTWLTTHN 89
Query: 56 SYASRAANLSIDSKISSVMNQEDSITDQLRNGVRGIMLDMHDYYGDIWLCR---GPCTIF 112
S+A + + I + NQED++T QL+NGVRG+MLD +D+ ++WLC G C F
Sbjct: 90 SFAIVGSPSRTGTPIIAPPNQEDTVTAQLKNGVRGLMLDAYDFQNEVWLCHSFGGKCYNF 149
Query: 113 TAFQPAINVLREINTFLTRHRTEIVTVFIKDRVTSPNGVNKVFNKAGLRKFWFPVYKMPK 172
A+Q A++VL+EI FL + +E++TVF++D P + K + MPK
Sbjct: 150 AAYQRAMDVLKEIGAFLDANPSEVITVFVED-YAGPGSLGKSGGR------------MPK 196
Query: 173 NGSDWLTVKKMLRMNHRLIVFTSNATKEASERIAYEWNYVVENKYGNDGMGRDHCLHRAE 232
G DW +K M+ NHRL++FTS K+ S+ +AYEW+YV+E +YGNDG+ C RAE
Sbjct: 197 GGGDWPLLKDMIAQNHRLLMFTSKRGKDGSDGLAYEWDYVLETQYGNDGLVGGSCPKRAE 256
Query: 233 SYPMNTTTKSLVLMNYYRNVLNSNEACKDNSSPLIRKMHTCYKDAGNRWPNYIAVDFYKR 292
S M++T +SL+LMN++ + + AC +NS+PL+ K+ CY + RWPN+IAVD+Y R
Sbjct: 257 SMAMDSTKQSLILMNFFSTNPSQSWACGNNSAPLVAKLKACYDASAKRWPNFIAVDYYMR 316
Query: 293 GDGGGAPEALDVAN 306
GGGAP A DVAN
Sbjct: 317 SKGGGAPLATDVAN 330
>dbj|BAD95429.1| MAP3K-like protein kinase [Arabidopsis thaliana]
Length = 291
Score = 259 bits (662), Expect = 7e-68
Identities = 124/221 (56%), Positives = 157/221 (70%), Gaps = 3/221 (1%)
Query: 92 MLDMHDYYGDIWLCR---GPCTIFTAFQPAINVLREINTFLTRHRTEIVTVFIKDRVTSP 148
MLD +D+ DIWLC G C FTAFQPAIN L+EIN FL + +EIVT+ ++D V S
Sbjct: 1 MLDTYDFQNDIWLCHSTGGTCFNFTAFQPAINALKEINDFLESNLSEIVTIILEDYVKSQ 60
Query: 149 NGVNKVFNKAGLRKFWFPVYKMPKNGSDWLTVKKMLRMNHRLIVFTSNATKEASERIAYE 208
G+ VFN +GL KF P+ +MPK+G+DW TV M++ N RL+VFTS KEASE +AY+
Sbjct: 61 MGLTNVFNASGLSKFLLPISRMPKDGTDWPTVDDMVKQNQRLVVFTSKKDKEASEGLAYQ 120
Query: 209 WNYVVENKYGNDGMGRDHCLHRAESYPMNTTTKSLVLMNYYRNVLNSNEACKDNSSPLIR 268
WNY+VEN+YGNDGM C R+ES ++T ++SLV NY+ NS +AC DNSSPLI
Sbjct: 121 WNYMVENQYGNDGMKDGSCSSRSESSSLDTMSRSLVFQNYFETSPNSTQACADNSSPLIE 180
Query: 269 KMHTCYKDAGNRWPNYIAVDFYKRGDGGGAPEALDVANRNL 309
M TC++ AG RWPN+IAVDFY+R D GGA EA+D AN L
Sbjct: 181 MMRTCHEAAGKRWPNFIAVDFYQRSDSGGAAEAVDEANGRL 221
>dbj|BAB10947.1| unnamed protein product [Arabidopsis thaliana]
Length = 365
Score = 209 bits (531), Expect = 1e-52
Identities = 99/177 (55%), Positives = 131/177 (73%), Gaps = 3/177 (1%)
Query: 43 LPFNHYSWLTTHNSYASRAANLSIDSKISSVMNQEDSITDQLRNGVRGIMLDMHDYYGDI 102
LPFN Y+WL THN++++ A L + + NQED+IT+QL+NGVRG+MLDM+D+ DI
Sbjct: 60 LPFNKYTWLMTHNAFSNANAPLLPGVERITFYNQEDTITNQLQNGVRGLMLDMYDFNNDI 119
Query: 103 WLC---RGPCTIFTAFQPAINVLREINTFLTRHRTEIVTVFIKDRVTSPNGVNKVFNKAG 159
WLC RG C FTAFQPAIN+LRE+ FL+++ TEIVT+ I+D V P G++ +F AG
Sbjct: 120 WLCHSLRGQCFNFTAFQPAINILREVEAFLSQNPTEIVTIIIEDYVHRPKGLSTLFANAG 179
Query: 160 LRKFWFPVYKMPKNGSDWLTVKKMLRMNHRLIVFTSNATKEASERIAYEWNYVVENK 216
L K+WFPV KMP+ G DW TV M++ NHRL+VFTS A KE E +AY+W Y+VEN+
Sbjct: 180 LDKYWFPVSKMPRKGEDWPTVTDMVQENHRLLVFTSVAAKEDEEGVAYQWRYMVENE 236
>dbj|BAC68883.1| hypothetical protein [Streptomyces avermitilis MA-4680]
gi|29827714|ref|NP_822348.1| hypothetical protein
SAV1173 [Streptomyces avermitilis MA-4680]
Length = 464
Score = 96.3 bits (238), Expect = 1e-18
Identities = 77/287 (26%), Positives = 136/287 (46%), Gaps = 19/287 (6%)
Query: 35 SPISKVMELPFNHYSWLTTHNSYASRAANLSIDSKISSVMNQEDSITDQLRNGVRGIMLD 94
+P+ + + ++LT HN+YA+ ++ V NQ I QL +GVRG M+D
Sbjct: 174 NPMPSPDQRTLDQVTFLTAHNAYANGVDGGFAPPFVNLVPNQTRGINQQLTDGVRGFMMD 233
Query: 95 MHDYYGDIWLCRGPCTIFTAFQPAINVLREINTFLTRHRTEIVTVFIKDRVTSPNGVNKV 154
+H LC CT+ + ++ + FL +H ++VTVF++D V +++
Sbjct: 234 IHQTSDGAILCHNSCTLVSKPVALWVDIQRMVDFLKQHPDQVVTVFLEDYVDPGVLRSEL 293
Query: 155 FNKAGLRKFWFPVYKMPKNGSDWLTVKKMLRMNHRLIVFT--SNATKEA------SERIA 206
+GL + + S W + ++ NHRL++FT S ++ E+ S +
Sbjct: 294 ARVSGLSDVLYRPDRTGVRQSGWPRMADLIAANHRLLIFTDHSRSSDESAGLTRDSFGVM 353
Query: 207 YEWNYVVENKYG-NDGMGRD--HCLHR---AESYPMNTTTKS----LVLMNYYRNVLNSN 256
Y+ + VEN + G+G C R A++ T T+S L +MN++R+ ++
Sbjct: 354 YQREWTVENYWSMGSGLGSSDWSCYSRWYGADTNIPLTYTESAFRPLFVMNHFRDAAIAS 413
Query: 257 EACKDNSSPLIRKMHTCYKDAGNRWPNYIAVDFYKRGDGGGAPEALD 303
A DN+ R C + A + PN++AVD Y G+ A + L+
Sbjct: 414 TATTDNTKLADRAQRFC-RPAARKKPNFLAVDRYDLGNPTSAVDTLN 459
>ref|YP_237843.1| hypothetical protein Psyr_4778 [Pseudomonas syringae pv. syringae
B728a] gi|63258709|gb|AAY39805.1| hypothetical protein
Psyr_4778 [Pseudomonas syringae pv. syringae B728a]
Length = 2378
Score = 68.2 bits (165), Expect = 3e-10
Identities = 69/281 (24%), Positives = 122/281 (42%), Gaps = 58/281 (20%)
Query: 44 PFNHYSWLTTHNSYASRAANLSIDSKISSVMNQEDSITDQLRNGVRGIMLDMHDYYGDIW 103
PF+ Y+W+T HN+Y D+IT QL G+RG MLD+H GD
Sbjct: 1845 PFDQYTWVTAHNAYL-------------------DAITPQLERGIRGFMLDIHMDVGDY- 1884
Query: 104 LCRGPCTIFTAFQPAI-----------NVLREINTFLTRHRTEIVTVFIKDRVTSPNGVN 152
G + PAI +VLRE ++ + R ++++ + ++S + +
Sbjct: 1885 --NGQKRVRVCHLPAIGACWADAPLLSDVLREFVAYMQKDRNAVISLLFESTLSS-DELR 1941
Query: 153 KVFNKAGLRKFWFPVYKMPKNGSDWLTVKKMLRMNHRLIVFTSNATKE----ASERIAYE 208
V + Y NG W T+++M+ N RL++ ++ + A ++
Sbjct: 1942 PVLEQVP----ELADYSHVSNGYSWPTLREMIDSNKRLVMLSNGEVAKTYTLAGKQAEVL 1997
Query: 209 W--NYVVENKY--GNDGMGRD-HCLHRAESYPMNTTTKS-----LVLMNYYRN----VLN 254
W + VEN Y G + D C R ++ T+ L ++N + + L+
Sbjct: 1998 WAPDTEVENTYNLGITSLVHDWQCKSRYGYMDLSLRTRDGGLPRLFVLNQFHSWGSTTLH 2057
Query: 255 SNEACKDNSSPLIRKMHT-CYKDAGNRWPNYIAVDFYKRGD 294
+ + +N + L R++ C + G R PNY+ +DF + GD
Sbjct: 2058 AGDV-DNNLTWLQRRVENHCGEATGWRKPNYLGIDFNQVGD 2097
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.320 0.134 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 544,389,676
Number of Sequences: 2540612
Number of extensions: 21966057
Number of successful extensions: 43891
Number of sequences better than 10.0: 63
Number of HSP's better than 10.0 without gapping: 20
Number of HSP's successfully gapped in prelim test: 43
Number of HSP's that attempted gapping in prelim test: 43803
Number of HSP's gapped (non-prelim): 63
length of query: 310
length of database: 863,360,394
effective HSP length: 127
effective length of query: 183
effective length of database: 540,702,670
effective search space: 98948588610
effective search space used: 98948588610
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 75 (33.5 bits)
Medicago: description of AC148528.6


