

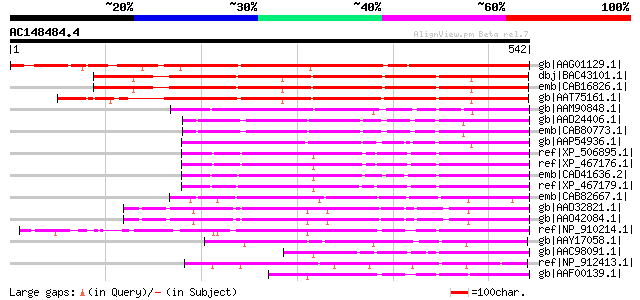
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148484.4 - phase: 0
(542 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAG01129.1| BAC19.14 [Lycopersicon esculentum] 469 e-131
dbj|BAC43101.1| ap2 SCARECROW-like protein [Arabidopsis thaliana] 462 e-128
emb|CAB16826.1| SCARECROW-like protein [Arabidopsis thaliana] gi... 459 e-128
gb|AAT75161.1| SCARECROW-like protein [Brassica napus] 424 e-117
gb|AAM90848.1| hairy meristem [Petunia x hybrida] 196 2e-48
gb|AAD24406.1| scarecrow-like 6 [Arabidopsis thaliana] gi|113588... 192 2e-47
emb|CAB80773.1| scarecrow-like 6 (SCL6) [Arabidopsis thaliana] g... 192 2e-47
gb|AAP54936.1| putative Scl1 protein [Oryza sativa (japonica cul... 188 4e-46
ref|XP_506895.1| PREDICTED P0516F12.29-2 gene product [Oryza sat... 187 6e-46
ref|XP_467176.1| Scl1 protein [Oryza sativa (japonica cultivar-g... 187 6e-46
emb|CAD41636.2| OSJNBb0012E24.1 [Oryza sativa (japonica cultivar... 187 6e-46
ref|XP_467179.1| putative Scl1 protein [Oryza sativa (japonica c... 185 3e-45
emb|CAB82667.1| scarecrow-like protein [Arabidopsis thaliana] gi... 184 5e-45
gb|AAD32821.1| putative SCARECROW gene regulator [Arabidopsis th... 175 3e-42
gb|AAO42084.1| putative SCARECROW gene regulator [Arabidopsis th... 173 1e-41
ref|NP_910214.1| Similar to Oryza sativa Scl1 protein (AF067401)... 163 1e-38
gb|AAY17058.1| f-171-1_1 [Ceratopteris thalictroides] 156 2e-36
gb|AAC98091.1| Scl1 protein [Oryza sativa] gi|11358961|pir||T512... 137 7e-31
ref|NP_912413.1| hypothetical protein [Oryza sativa (japonica cu... 112 3e-23
gb|AAF00139.1| Scl1 protein [Oryza sativa] 110 9e-23
>gb|AAG01129.1| BAC19.14 [Lycopersicon esculentum]
Length = 536
Score = 469 bits (1208), Expect = e-131
Identities = 277/569 (48%), Positives = 369/569 (64%), Gaps = 60/569 (10%)
Query: 1 MRVPPPAVSSSSSSPQPKLTTTQNIVVNVPTPNPTPTPTT-TTLCYEPRSVLELCRSPSP 59
M+VP + SS P +VN P T + LCYEP+SVLEL RSPSP
Sbjct: 1 MKVPFSTNDNVSSKP----------LVNSNNSFTFPAATNGSNLCYEPKSVLELRRSPSP 50
Query: 60 EKKPSQQEQEHNPEV----------EQEDHASLPNLDWWDSIMKDLGLQDDSSTPIIPLL 109
+Q NP++ + DH L N + WDS+M++LGL DDS++
Sbjct: 51 IV--DKQIITTNPDLSALCGGEDPLQLGDHV-LSNFEDWDSLMRELGLHDDSAS------ 101
Query: 110 KNTNNTSEIYPNPSQDQFDQTQDFTSLS------DIYSNNQNLAYNYPNTNTNLDHLVND 163
+ T+ + + S QF +F++ S D ++ N +P N +
Sbjct: 102 --LSKTNPLTHSESLTQFHNLSEFSAESNQFPSPDFSFSDTNFPQQFPTVNQ-----ASF 154
Query: 164 FNNNQPNNNTNSNW----DFIEELIRAADCFDNNHLQLAQAILERLNQRLRSPTGKPLHR 219
N + + + NW D+++ELIR A+CF+ N QLA IL RLNQRLRS GKPL R
Sbjct: 155 INALDLSGDIHQNWSVGFDYVDELIRFAECFETNAFQLAHVILARLNQRLRSAAGKPLQR 214
Query: 220 AAFHFKDALQSLLSGSNRTNPPRLSSMVEIVQTIRTFKAFSGISPIPMFSIFTTNQALLE 279
AAF+FK+ALQ+ L+GS R R SS +++QTI+++K S ISPIPMFS FT NQA+LE
Sbjct: 215 AAFYFKEALQAQLAGSARQT--RSSSSSDVIQTIKSYKILSNISPIPMFSSFTANQAVLE 272
Query: 280 ALHGSLYMHVVDFEIGLGIQYASLMKEIAEKAV---NGSPLLRITAVVPEEYAVESRLIR 336
A+ GS+ +HV+DF+IGLG +AS MKE+A+KA +P+LRITA+VPEEYAVESRLIR
Sbjct: 273 AVDGSMLVHVIDFDIGLGGHWASFMKELADKAECRKANAPILRITALVPEEYAVESRLIR 332
Query: 337 ENLNQFAHDLGIRVQVDFVPLRTFETVSFKAVRFVDGEKTAILLTPAIFCRLGSEGTAAF 396
ENL QFA +L I ++DFV +RTFE +SFKA++F++GEKTA+LL+PAIF R+GS F
Sbjct: 333 ENLTQFARELNIGFEIDFVLIRTFELLSFKAIKFMEGEKTAVLLSPAIFRRVGS----GF 388
Query: 397 LSDVRRITPGVVVFVDGEGWTEAAAAASFRRGVVNSLEFYSMMLESLDASVAAGGG-GEW 455
++++RRI+P VVV VD EG A SFR+ V++ LEFYS +LESL+A+ GG G+W
Sbjct: 389 VNELRRISPNVVVHVDSEG-LMGYGAMSFRQTVIDGLEFYSTLLESLEAANIGGGNCGDW 447
Query: 456 ARRIEMLLLRPKIIAAVEAAGRR--TTPWREAFYGAGMRPVQLSQFADFQAECLLAKVQI 513
R+IE +L PKI+ + A GRR WR+A AG RPV LSQFADFQA+CLL +VQ+
Sbjct: 448 MRKIENFVLFPKIVDMIGAVGRRGGGGSWRDAMVDAGFRPVGLSQFADFQADCLLGRVQV 507
Query: 514 RGFHVAKRQAELVLFWHERAMVATSAWRC 542
RGFHVAKRQAE++L WH+RA+VATSAWRC
Sbjct: 508 RGFHVAKRQAEMLLCWHDRALVATSAWRC 536
>dbj|BAC43101.1| ap2 SCARECROW-like protein [Arabidopsis thaliana]
Length = 486
Score = 462 bits (1188), Expect = e-128
Identities = 268/470 (57%), Positives = 330/470 (70%), Gaps = 38/470 (8%)
Query: 88 WDSIMKDLGLQDDSSTPIIPLLKNTNNT-SEIYPNPSQDQF-----DQTQ--DFTSLSDI 139
WDSIMK+L L DDS+ + T T S I P + D DQ Q DF S SD+
Sbjct: 38 WDSIMKELELDDDSAPNSLKTGFTTTTTDSTILPLYAVDSNLPGFPDQIQPSDFESSSDV 97
Query: 140 Y-SNNQNLAYNYPNTNTNLDHLVNDFNNNQPNNNTNSNWDFIEELIRAADCFDNNHLQLA 198
Y NQ Y + N ++ N +DFIE+LIR DC +++ LQLA
Sbjct: 98 YPGQNQTTGYGF----------------NSLDSVDNGGFDFIEDLIRVVDCVESDELQLA 141
Query: 199 QAILERLNQRLRSPTGKPLHRAAFHFKDALQSLLSGSNRTNPPRLSSMVEIVQTIRTFKA 258
Q +L RLNQRLRSP G+PL RAAF+FK+AL S L+GSNR NP RLSS EIVQ IR K
Sbjct: 142 QVVLSRLNQRLRSPAGRPLQRAAFYFKEALGSFLTGSNR-NPIRLSSWSEIVQRIRAIKE 200
Query: 259 FSGISPIPMFSIFTTNQALLEALHG---SLYMHVVDFEIGLGIQYASLMKEIAEKAVNGS 315
+SGISPIP+FS FT NQA+L++L S ++HVVDFEIG G QYASLM+EI EK+V+G
Sbjct: 201 YSGISPIPLFSHFTANQAILDSLSSQSSSPFVHVVDFEIGFGGQYASLMREITEKSVSGG 260
Query: 316 PLLRITAVVPEEYAVESRLIRENLNQFAHDLGIRVQVDFVPLRTFETVSFKAVRFVDGEK 375
LR+TAVV EE AVE+RL++ENL QFA ++ IR Q++FV ++TFE +SFKA+RFV+GE+
Sbjct: 261 -FLRVTAVVAEECAVETRLVKENLTQFAAEMKIRFQIEFVLMKTFEMLSFKAIRFVEGER 319
Query: 376 TAILLTPAIFCRLGSEGTAAFLSDVRRITPGVVVFVDGEGWTEAAAAASFRRGVVNSLEF 435
T +L++PAIF RL G F++++RR++P VVVFVD EGWTE A + SFRR V++LEF
Sbjct: 320 TVVLISPAIFRRLS--GITDFVNNLRRVSPKVVVFVDSEGWTEIAGSGSFRREFVSALEF 377
Query: 436 YSMMLESLDASVAAGGGGEWARRIEMLLLRPKIIAAVE-AAGRRTT---PWREAFYGAGM 491
Y+M+LESLDA AA G + +E +LRPKI AAVE AA RR T WREAF AGM
Sbjct: 378 YTMVLESLDA--AAPPGDLVKKIVEAFVLRPKISAAVETAADRRHTGEMTWREAFCAAGM 435
Query: 492 RPVQLSQFADFQAECLLAKVQIRGFHVAKRQAELVLFWHERAMVATSAWR 541
RP+QLSQFADFQAECLL K Q+RGFHVAKRQ ELVL WH RA+VATSAWR
Sbjct: 436 RPIQLSQFADFQAECLLEKAQVRGFHVAKRQGELVLCWHGRALVATSAWR 485
>emb|CAB16826.1| SCARECROW-like protein [Arabidopsis thaliana]
gi|7270619|emb|CAB80337.1| SCARECROW-like protein
[Arabidopsis thaliana] gi|15234500|ref|NP_195389.1|
scarecrow transcription factor family protein
[Arabidopsis thaliana] gi|25407757|pir||E85433
SCARECROW-like protein [imported] - Arabidopsis thaliana
Length = 486
Score = 459 bits (1182), Expect = e-128
Identities = 267/470 (56%), Positives = 329/470 (69%), Gaps = 38/470 (8%)
Query: 88 WDSIMKDLGLQDDSSTPIIPLLKNTNNT-SEIYPNPSQDQF-----DQTQ--DFTSLSDI 139
WDSIMK+L L DDS+ + T T S I P + D DQ Q DF S SD+
Sbjct: 38 WDSIMKELELDDDSAPNSLKTGFTTTTTDSTILPLYAVDSNLPGFPDQIQPSDFESSSDV 97
Query: 140 Y-SNNQNLAYNYPNTNTNLDHLVNDFNNNQPNNNTNSNWDFIEELIRAADCFDNNHLQLA 198
Y NQ Y + N ++ N +DFIE+LIR DC +++ LQLA
Sbjct: 98 YPGQNQTTGYGF----------------NSLDSVDNGGFDFIEDLIRVVDCVESDELQLA 141
Query: 199 QAILERLNQRLRSPTGKPLHRAAFHFKDALQSLLSGSNRTNPPRLSSMVEIVQTIRTFKA 258
Q +L RLNQRLRSP G+PL RAAF+FK+AL S L+GSNR NP RLSS EIVQ IR K
Sbjct: 142 QVVLSRLNQRLRSPAGRPLQRAAFYFKEALGSFLTGSNR-NPIRLSSWSEIVQRIRAIKE 200
Query: 259 FSGISPIPMFSIFTTNQALLEALHG---SLYMHVVDFEIGLGIQYASLMKEIAEKAVNGS 315
+SGISPIP+FS FT NQA+L++L S ++HVVDFEIG G QYASLM+EI EK+V+G
Sbjct: 201 YSGISPIPLFSHFTANQAILDSLSSQSSSPFVHVVDFEIGFGGQYASLMREITEKSVSGG 260
Query: 316 PLLRITAVVPEEYAVESRLIRENLNQFAHDLGIRVQVDFVPLRTFETVSFKAVRFVDGEK 375
LR+TAVV EE AVE+RL++ENL QFA ++ IR Q++FV ++TFE +SFKA+RFV+GE+
Sbjct: 261 -FLRVTAVVAEECAVETRLVKENLTQFAAEMKIRFQIEFVLMKTFEMLSFKAIRFVEGER 319
Query: 376 TAILLTPAIFCRLGSEGTAAFLSDVRRITPGVVVFVDGEGWTEAAAAASFRRGVVNSLEF 435
T +L++PAIF RL G F++++RR++P VVVFVD EGWTE A + SFRR V++LEF
Sbjct: 320 TVVLISPAIFRRLS--GITDFVNNLRRVSPKVVVFVDSEGWTEIAGSGSFRREFVSALEF 377
Query: 436 YSMMLESLDASVAAGGGGEWARRIEMLLLRPKIIAAVE-AAGRRTT---PWREAFYGAGM 491
Y+M+LESLDA AA G + +E +LRPKI AAVE AA RR T WREAF AGM
Sbjct: 378 YTMVLESLDA--AAPPGDLVKKIVEAFVLRPKISAAVETAADRRHTGEMTWREAFCAAGM 435
Query: 492 RPVQLSQFADFQAECLLAKVQIRGFHVAKRQAELVLFWHERAMVATSAWR 541
RP+Q SQFADFQAECLL K Q+RGFHVAKRQ ELVL WH RA+VATSAWR
Sbjct: 436 RPIQQSQFADFQAECLLEKAQVRGFHVAKRQGELVLCWHGRALVATSAWR 485
>gb|AAT75161.1| SCARECROW-like protein [Brassica napus]
Length = 461
Score = 424 bits (1090), Expect = e-117
Identities = 247/510 (48%), Positives = 331/510 (64%), Gaps = 69/510 (13%)
Query: 51 LELCRSPSPEKKPSQ--QEQEHNPEVEQEDHASLPNLDWWDSIMKDLGLQDDSST----- 103
++L S + +PS + + ++HA + ++DW DSIMK+L + DDS+
Sbjct: 1 MKLQASSPQDNQPSNTTNNSTDSNHLSMDEHA-MRSMDW-DSIMKELEVDDDSAPYQLQP 58
Query: 104 -----PIIPLLKNTNNTSEIYPNPSQDQFDQTQDFTSLSDIYSNNQNLAYNYPNTNTNLD 158
P+ P + ++S++YP P+Q
Sbjct: 59 SSFNLPVFPDI----DSSDVYPGPNQ---------------------------------- 80
Query: 159 HLVNDFNNNQPNNNTNSNWDFIEELIRAADCFDNNHLQLAQAILERLNQRLRSPTGKPLH 218
+ + N ++ N +D+IE+LIR DC +++ L LA +L +LNQRL++ G+PL
Sbjct: 81 --ITGYGFNSLDSVDNGGFDYIEDLIRVVDCIESDELHLAHVVLSQLNQRLQTSAGRPLQ 138
Query: 219 RAAFHFKDALQSLLSGSNRTNPPRLSSMVEIVQTIRTFKAFSGISPIPMFSIFTTNQALL 278
RAAF+FK+AL SLL+G+NR +L S +IVQ IR K FSGISPIP+FS FT NQA+L
Sbjct: 139 RAAFYFKEALGSLLTGTNRN---QLFSWSDIVQKIRAIKEFSGISPIPLFSHFTANQAIL 195
Query: 279 EALHG---SLYMHVVDFEIGLGIQYASLMKEIAEKAVNGSPLLRITAVVPEEYAVESRLI 335
++L S ++HVVDFEIG G QYASLM+EIAEK+ NG LR+TAVV E+ AVE+RL+
Sbjct: 196 DSLSSQSSSPFVHVVDFEIGFGGQYASLMREIAEKSANGG-FLRVTAVVAEDCAVETRLV 254
Query: 336 RENLNQFAHDLGIRVQVDFVPLRTFETVSFKAVRFVDGEKTAILLTPAIFCRLGSEGTAA 395
+ENL QFA ++ IR Q++FV ++TFE +SFKA+RFVDGE+T +L++PAIF R+ G A
Sbjct: 255 KENLTQFAAEMKIRFQIEFVLMKTFEILSFKAIRFVDGERTVVLISPAIFRRV--IGIAE 312
Query: 396 FLSDVRRITPGVVVFVDGEGWTEAAAAASFRRGVVNSLEFYSMMLESLDASVAAGGGGEW 455
F++++ R++P VVVFVD EG TE A + SFRR V++ EFY+M+LESLDA AA G
Sbjct: 313 FVNNLGRVSPNVVVFVDSEGCTETAGSGSFRREFVSAFEFYTMVLESLDA--AAPPGDLV 370
Query: 456 ARRIEMLLLRPKIIAAVEAAGRRTT----PWREAFYGAGMRPVQLSQFADFQAECLLAKV 511
+ +E LLRPKI AAVE A R + WRE AGMRPVQLSQFADFQAECLL K
Sbjct: 371 KKIVETFLLRPKISAAVETAANRRSAGQMTWREMLCAAGMRPVQLSQFADFQAECLLEKA 430
Query: 512 QIRGFHVAKRQAELVLFWHERAMVATSAWR 541
Q+RGFHVAKRQ ELVL WH RA+VATSAWR
Sbjct: 431 QVRGFHVAKRQGELVLCWHGRALVATSAWR 460
>gb|AAM90848.1| hairy meristem [Petunia x hybrida]
Length = 721
Score = 196 bits (498), Expect = 2e-48
Identities = 133/391 (34%), Positives = 215/391 (54%), Gaps = 34/391 (8%)
Query: 169 PNNNTNSNWDFIEELIRAADCFDNNHLQLAQAILERLNQRLRSPTGKPLHRAAFHFKDAL 228
P + +++ +A++ H AQ IL RLNQ+L SP GKP RAAF+FK+AL
Sbjct: 348 PRHQQQEQQFIYDQIFQASELLLAGHFSNAQMILARLNQQL-SPIGKPFKRAAFYFKEAL 406
Query: 229 QS--LLSGSNRTNPPRLSSMVEIVQTIRTFKAFSGISPIPMFSIFTTNQALLEALHGSLY 286
Q LL ++ PPR + + V + +K+FS +SP+ F FT+NQA+LEAL
Sbjct: 407 QLPFLLPCTSTFLPPRSPTPFDCVLKMDAYKSFSEVSPLIQFMNFTSNQAILEALGDVER 466
Query: 287 MHVVDFEIGLGIQYASLMKEIAEKAVNGSPLLRITAVVPEE--YAVESRLIRENLNQFAH 344
+H++DF+IG G Q++S M+E+ + L +ITA ++VE ++ E+L QFA+
Sbjct: 467 IHIIDFDIGFGAQWSSFMQELPSSNRKATSL-KITAFASPSTHHSVEIAIMHESLTQFAN 525
Query: 345 DLGIRVQVDFVPLRTFETVSF--KAVRFVDGEKTAI-----LLTPAIFCRLGSEGTAAFL 397
D GIR +++ + L TF+ S+ ++R D E AI ++ +F + L
Sbjct: 526 DAGIRFELEVINLDTFDPKSYPLSSLRSSDCEAIAINFPIWSISSCLFA------FPSLL 579
Query: 398 SDVRRITPGVVVFVD-GEGWTEAAAAASFRRGVVNSLEFYSMMLESLDASVAAGGGGEWA 456
+++++P ++V ++ G TE + ++++L++Y ++L S+DA A E
Sbjct: 580 HYMKQLSPKIIVSLERGCERTELP----LKHHLLHALQYYEILLASIDA---ANVTPEIG 632
Query: 457 RRIEMLLLRPKIIAAVEAAGRRTTP-----WREAFYGAGMRPVQLSQFADFQAECLLAKV 511
++IE LL P I V GR +P WR F AG PV S + QAEC++ +
Sbjct: 633 KKIEKSLLLPSIENMV--LGRLRSPDRMPQWRNLFASAGFSPVAFSNLTEIQAECVVKRT 690
Query: 512 QIRGFHVAKRQAELVLFWHERAMVATSAWRC 542
Q+ GFHV KRQ+ LVL W ++ +++ WRC
Sbjct: 691 QVGGFHVEKRQSSLVLCWKQQELLSALTWRC 721
>gb|AAD24406.1| scarecrow-like 6 [Arabidopsis thaliana] gi|11358828|pir||T51237
scarecrow-like protein 6 [imported] - Arabidopsis
thaliana (fragment)
Length = 378
Score = 192 bits (488), Expect = 2e-47
Identities = 131/370 (35%), Positives = 203/370 (54%), Gaps = 29/370 (7%)
Query: 181 EELIRAADCFDNNHLQLAQAILERLNQRLRSPTGKPLHRAAFHFKDALQSLLSGSNRTNP 240
E+L++AA+ +++ LAQ IL RLNQ+L SP GKPL RAAF+FK+AL +LL ++T
Sbjct: 27 EQLVKAAEVIESDTC-LAQGILARLNQQLSSPVGKPLERAAFYFKEALNNLLHNVSQTLN 85
Query: 241 PRLSSMVEIVQTIRTFKAFSGISPIPMFSIFTTNQALLEALHGSLYMHVVDFEIGLGIQY 300
P ++ I +K+FS ISP+ F+ FT+NQALLE+ HG +H++DF+IG G Q+
Sbjct: 86 P-----YSLIFKIAAYKSFSEISPVLQFANFTSNQALLESFHGFHRLHIIDFDIGYGGQW 140
Query: 301 ASLMKEIAEKAVNGSPL-LRIT--AVVPEEYAVESRLIRENLNQFAHDLGIRVQVDFVPL 357
ASLM+E+ + N +PL L+IT A +E ++NL FA ++ I + + + L
Sbjct: 141 ASLMQELVLRD-NAAPLSLKITVFASPANHDQLELGFTQDNLKHFASEINISLDIQVLSL 199
Query: 358 RTFETVSFKAVRFVDGEKTAILLTPAIFCRLGSEGTAAFLSDVRRITPGVVVFVD-GEGW 416
++S+ + E A+ ++ A F L L V+ ++P ++V D G
Sbjct: 200 DLLGSISWP--NSSEKEAVAVNISAASFSHL-----PLVLRFVKHLSPTIIVCSDRGCER 252
Query: 417 TEAAAAASFRRGVVNSLEFYSMMLESLDASVAAGGGGEWARRIEMLLLRPKIIAAV---- 472
T+ F + + +SL ++ + ESLD A + ++IE L++P+I V
Sbjct: 253 TD----LPFSQQLAHSLHSHTALFESLD---AVNANLDAMQKIERFLIQPEIEKLVLDRS 305
Query: 473 EAAGRRTTPWREAFYGAGMRPVQLSQFADFQAECLLAKVQIRGFHVAKRQAELVLFWHER 532
R W+ F G PV S F + QAECL+ + +RGFHV K+ L+L W
Sbjct: 306 RPIERPMMTWQAMFLQMGFSPVTHSNFTESQAECLVQRTPVRGFHVEKKHNSLLLCWQRT 365
Query: 533 AMVATSAWRC 542
+V SAWRC
Sbjct: 366 ELVGVSAWRC 375
>emb|CAB80773.1| scarecrow-like 6 (SCL6) [Arabidopsis thaliana]
gi|22137044|gb|AAM91367.1| At4g00150/F6N15_20
[Arabidopsis thaliana] gi|18087577|gb|AAL58919.1|
AT4g00150/F6N15_20 [Arabidopsis thaliana]
gi|15236725|ref|NP_191926.1| scarecrow-like
transcription factor 6 (SCL6) [Arabidopsis thaliana]
gi|3193314|gb|AAC19296.1| contains similarity to
Arabidopsis scarecrow (GB:U62798) [Arabidopsis thaliana]
gi|7486541|pir||T01343 hypothetical protein F6N15.20 -
Arabidopsis thaliana
Length = 558
Score = 192 bits (488), Expect = 2e-47
Identities = 131/370 (35%), Positives = 203/370 (54%), Gaps = 29/370 (7%)
Query: 181 EELIRAADCFDNNHLQLAQAILERLNQRLRSPTGKPLHRAAFHFKDALQSLLSGSNRTNP 240
E+L++AA+ +++ LAQ IL RLNQ+L SP GKPL RAAF+FK+AL +LL ++T
Sbjct: 207 EQLVKAAEVIESDTC-LAQGILARLNQQLSSPVGKPLERAAFYFKEALNNLLHNVSQTLN 265
Query: 241 PRLSSMVEIVQTIRTFKAFSGISPIPMFSIFTTNQALLEALHGSLYMHVVDFEIGLGIQY 300
P ++ I +K+FS ISP+ F+ FT+NQALLE+ HG +H++DF+IG G Q+
Sbjct: 266 P-----YSLIFKIAAYKSFSEISPVLQFANFTSNQALLESFHGFHRLHIIDFDIGYGGQW 320
Query: 301 ASLMKEIAEKAVNGSPL-LRIT--AVVPEEYAVESRLIRENLNQFAHDLGIRVQVDFVPL 357
ASLM+E+ + N +PL L+IT A +E ++NL FA ++ I + + + L
Sbjct: 321 ASLMQELVLRD-NAAPLSLKITVFASPANHDQLELGFTQDNLKHFASEINISLDIQVLSL 379
Query: 358 RTFETVSFKAVRFVDGEKTAILLTPAIFCRLGSEGTAAFLSDVRRITPGVVVFVD-GEGW 416
++S+ + E A+ ++ A F L L V+ ++P ++V D G
Sbjct: 380 DLLGSISWP--NSSEKEAVAVNISAASFSHL-----PLVLRFVKHLSPTIIVCSDRGCER 432
Query: 417 TEAAAAASFRRGVVNSLEFYSMMLESLDASVAAGGGGEWARRIEMLLLRPKIIAAV---- 472
T+ F + + +SL ++ + ESLD A + ++IE L++P+I V
Sbjct: 433 TD----LPFSQQLAHSLHSHTALFESLD---AVNANLDAMQKIERFLIQPEIEKLVLDRS 485
Query: 473 EAAGRRTTPWREAFYGAGMRPVQLSQFADFQAECLLAKVQIRGFHVAKRQAELVLFWHER 532
R W+ F G PV S F + QAECL+ + +RGFHV K+ L+L W
Sbjct: 486 RPIERPMMTWQAMFLQMGFSPVTHSNFTESQAECLVQRTPVRGFHVEKKHNSLLLCWQRT 545
Query: 533 AMVATSAWRC 542
+V SAWRC
Sbjct: 546 ELVGVSAWRC 555
>gb|AAP54936.1| putative Scl1 protein [Oryza sativa (japonica cultivar-group)]
gi|37536694|ref|NP_922649.1| putative Scl1 protein
[Oryza sativa (japonica cultivar-group)]
gi|10140637|gb|AAG13473.1| putative Scl1 protein [Oryza
sativa (japonica cultivar-group)]
Length = 582
Score = 188 bits (477), Expect = 4e-46
Identities = 139/377 (36%), Positives = 203/377 (52%), Gaps = 28/377 (7%)
Query: 180 IEELIRAADCFDNNHLQLAQAILERLNQRL-RSPT-GKPLHRAAFHFKDALQSLLSGSNR 237
+E+L AA + A+ IL RLN RL +PT G PL R+AF+FK+AL+ LS +
Sbjct: 220 VEQLAEAAKLAEAGDAFGAREILARLNYRLPAAPTAGTPLLRSAFYFKEALRLTLSPTGD 279
Query: 238 TNPPRLSSMVEIVQTIRTFKAFSGISPIPMFSIFTTNQALLEALHGSLYMHVVDFEIGLG 297
P S+ ++V + +KAFS +SP+ F+ T QA+L+ L G+ +HV+DF+IG+G
Sbjct: 280 APAPSASTPYDVVVKLGAYKAFSEVSPVLQFAHLTCVQAVLDELGGAGCIHVLDFDIGMG 339
Query: 298 IQYASLMKEIAEKAVNGSPLLRITAVV--PEEYAVESRLIRENLNQFAHDLGIRVQVDFV 355
Q+ASLM+E+A+ + + L++TA+V + +E +LI ENL+ FA +LG+
Sbjct: 340 EQWASLMQELAQ--LRPAAALKVTALVSPASHHPLELQLIHENLSGFAAELGVFFHFTVF 397
Query: 356 PLRTFETVSFKAVRFVDGEKTAILLT--PAIFCRLGSEGTAAFLSDVRRITPGVVVFVDG 413
+ T + A G+ A+ L PA + T A L V+R+ VVV VD
Sbjct: 398 NIDTLDPAELLA-NATAGDAVAVHLPVGPA-----HAAATPAVLRLVKRLGAKVVVSVD- 450
Query: 414 EGWTEAAAAASFRRGVVNSLEFYSMMLESLDASVAAGGGGEWARRIEMLLLRPKIIAAVE 473
G + F + +S +LES+D A G + A +IE L+ P I V
Sbjct: 451 RGCDR--SDLPFAAHLFHSFHSAVYLLESID---AVGTDPDTASKIERYLIHPAIEQCVV 505
Query: 474 AAGRRTT--------PWREAFYGAGMRPVQLSQFADFQAECLLAKVQIRGFHVAKRQAEL 525
A+ R + PWR AF AG PVQ + FA+ QAE LL+KV +RGF V KR L
Sbjct: 506 ASHRAASAMDKAPPPPWRAAFAAAGFAPVQATTFAESQAESLLSKVHVRGFRVEKRAGSL 565
Query: 526 VLFWHERAMVATSAWRC 542
L+W +V+ SAWRC
Sbjct: 566 CLYWQRGELVSVSAWRC 582
>ref|XP_506895.1| PREDICTED P0516F12.29-2 gene product [Oryza sativa (japonica
cultivar-group)] gi|50911539|ref|XP_467177.1| Scl1
protein [Oryza sativa (japonica cultivar-group)]
gi|50251747|dbj|BAD27680.1| Scl1 protein [Oryza sativa
(japonica cultivar-group)] gi|49388513|dbj|BAD25637.1|
Scl1 protein [Oryza sativa (japonica cultivar-group)]
Length = 531
Score = 187 bits (476), Expect = 6e-46
Identities = 127/372 (34%), Positives = 206/372 (55%), Gaps = 21/372 (5%)
Query: 180 IEELIRAADCFDNNHLQLAQAILERLNQRLRSPTGKPLHRAAFHFKDALQSLLSGSNRTN 239
++EL AA + + A+ IL RLNQ+L GKP R+A + K+AL L+ S+ +
Sbjct: 172 LDELAAAAKATEAGNSVGAREILARLNQQLPQ-LGKPFLRSASYLKEALLLALADSHHGS 230
Query: 240 PPRLSSMVEIVQTIRTFKAFSGISPIPMFSIFTTNQALLEALHG--SLYMHVVDFEIGLG 297
++S +++ + +K+FS +SP+ F+ FT QALL+ + G + +HV+DF++G+G
Sbjct: 231 SG-VTSPLDVALKLAAYKSFSDLSPVLQFTNFTATQALLDEIGGMATSCIHVIDFDLGVG 289
Query: 298 IQYASLMKEIAEKAVNGS---PLLRITAVVP--EEYAVESRLIRENLNQFAHDLGIRVQV 352
Q+AS ++E+A + G PLL++TA + + +E L ++NL+QFA +L I +
Sbjct: 290 GQWASFLQELAHRRGAGGMALPLLKLTAFMSTASHHPLELHLTQDNLSQFAAELRIPFEF 349
Query: 353 DFVPLRTFETVSFKAVRFVDGEKTAILLTPAIFCRLGSEGTAAFLSDVRRITPGVVVFVD 412
+ V L F + G++ + P + C + A L V+++ P VVV +D
Sbjct: 350 NAVSLDAFNPAELISS---SGDEVVAVSLP-VGCSARAPPLPAILRLVKQLCPKVVVAID 405
Query: 413 GEGWTEAAAAASFRRGVVNSLEFYSMMLESLDASVAAGGGGEWARRIEMLLLRPKIIAAV 472
G A F + +N + +L+SLDA AG + A +IE L++P++ AV
Sbjct: 406 HGG---DRADLPFSQHFLNCFQSCVFLLDSLDA---AGIDADSACKIERFLIQPRVEDAV 459
Query: 473 --EAAGRRTTPWREAFYGAGMRPVQLSQFADFQAECLLAKVQIRGFHVAKRQAELVLFWH 530
++ WR F G +PVQLS A+ QA+CLL +VQ+RGFHV KR A L L+W
Sbjct: 460 IGRHKAQKAIAWRSVFAATGFKPVQLSNLAEAQADCLLKRVQVRGFHVEKRGAALTLYWQ 519
Query: 531 ERAMVATSAWRC 542
+V+ S+WRC
Sbjct: 520 RGELVSISSWRC 531
>ref|XP_467176.1| Scl1 protein [Oryza sativa (japonica cultivar-group)]
gi|50251746|dbj|BAD27679.1| Scl1 protein [Oryza sativa
(japonica cultivar-group)] gi|49388512|dbj|BAD25636.1|
Scl1 protein [Oryza sativa (japonica cultivar-group)]
Length = 709
Score = 187 bits (476), Expect = 6e-46
Identities = 127/372 (34%), Positives = 206/372 (55%), Gaps = 21/372 (5%)
Query: 180 IEELIRAADCFDNNHLQLAQAILERLNQRLRSPTGKPLHRAAFHFKDALQSLLSGSNRTN 239
++EL AA + + A+ IL RLNQ+L GKP R+A + K+AL L+ S+ +
Sbjct: 350 LDELAAAAKATEAGNSVGAREILARLNQQLPQ-LGKPFLRSASYLKEALLLALADSHHGS 408
Query: 240 PPRLSSMVEIVQTIRTFKAFSGISPIPMFSIFTTNQALLEALHG--SLYMHVVDFEIGLG 297
++S +++ + +K+FS +SP+ F+ FT QALL+ + G + +HV+DF++G+G
Sbjct: 409 SG-VTSPLDVALKLAAYKSFSDLSPVLQFTNFTATQALLDEIGGMATSCIHVIDFDLGVG 467
Query: 298 IQYASLMKEIAEKAVNGS---PLLRITAVVP--EEYAVESRLIRENLNQFAHDLGIRVQV 352
Q+AS ++E+A + G PLL++TA + + +E L ++NL+QFA +L I +
Sbjct: 468 GQWASFLQELAHRRGAGGMALPLLKLTAFMSTASHHPLELHLTQDNLSQFAAELRIPFEF 527
Query: 353 DFVPLRTFETVSFKAVRFVDGEKTAILLTPAIFCRLGSEGTAAFLSDVRRITPGVVVFVD 412
+ V L F + G++ + P + C + A L V+++ P VVV +D
Sbjct: 528 NAVSLDAFNPAELISS---SGDEVVAVSLP-VGCSARAPPLPAILRLVKQLCPKVVVAID 583
Query: 413 GEGWTEAAAAASFRRGVVNSLEFYSMMLESLDASVAAGGGGEWARRIEMLLLRPKIIAAV 472
G A F + +N + +L+SLDA AG + A +IE L++P++ AV
Sbjct: 584 HGG---DRADLPFSQHFLNCFQSCVFLLDSLDA---AGIDADSACKIERFLIQPRVEDAV 637
Query: 473 --EAAGRRTTPWREAFYGAGMRPVQLSQFADFQAECLLAKVQIRGFHVAKRQAELVLFWH 530
++ WR F G +PVQLS A+ QA+CLL +VQ+RGFHV KR A L L+W
Sbjct: 638 IGRHKAQKAIAWRSVFAATGFKPVQLSNLAEAQADCLLKRVQVRGFHVEKRGAALTLYWQ 697
Query: 531 ERAMVATSAWRC 542
+V+ S+WRC
Sbjct: 698 RGELVSISSWRC 709
>emb|CAD41636.2| OSJNBb0012E24.1 [Oryza sativa (japonica cultivar-group)]
gi|38345487|emb|CAE01700.2| OSJNBa0010H02.25 [Oryza
sativa (japonica cultivar-group)]
gi|50927847|ref|XP_473451.1| OSJNBa0010H02.25 [Oryza
sativa (japonica cultivar-group)]
Length = 711
Score = 187 bits (476), Expect = 6e-46
Identities = 129/372 (34%), Positives = 202/372 (53%), Gaps = 22/372 (5%)
Query: 180 IEELIRAADCFDNNHLQLAQAILERLNQRLRSPTGKPLHRAAFHFKDALQSLLSGSNRTN 239
++EL AA + + A+ IL RLNQ+L P GKP R+A + KDAL L+ +
Sbjct: 353 LDELAAAAKATEVGNSIGAREILARLNQQL-PPIGKPFLRSASYLKDALLLALADGHHA- 410
Query: 240 PPRLSSMVEIVQTIRTFKAFSGISPIPMFSIFTTNQALLEALHGSLY--MHVVDFEIGLG 297
RL+S +++ + +K+FS +SP+ F+ FT QALL+ + + + V+DF++G+G
Sbjct: 411 ATRLTSPLDVALKLTAYKSFSDLSPVLQFANFTVTQALLDEIASTTASCIRVIDFDLGVG 470
Query: 298 IQYASLMKEIAEKAVNGS---PLLRITAVVP--EEYAVESRLIRENLNQFAHDLGIRVQV 352
Q+AS ++E+A + +G P+L++TA V + +E L ++NL+QFA DLGI +
Sbjct: 471 GQWASFLQELAHRCGSGGVSLPMLKLTAFVSAASHHPLELHLTQDNLSQFAADLGIPFEF 530
Query: 353 DFVPLRTFETVSFKAVRFVDGEKTAILLTPAIFCRLGSEGTAAFLSDVRRITPGVVVFVD 412
+ + L F+ + A E A+ L R A L V+++ P +VV +D
Sbjct: 531 NAINLDAFDPMELIAP--TADEVVAVSLPVGCSARTP---LPAMLQLVKQLAPKIVVAID 585
Query: 413 GEGWTEAAAAASFRRGVVNSLEFYSMMLESLDASVAAGGGGEWARRIEMLLLRPKIIAAV 472
+ + F + +N L+ +LESLDA AG + +IE L++P++ AV
Sbjct: 586 ---YGSDRSDLPFSQHFLNCLQSCLCLLESLDA---AGTDADAVSKIERFLIQPRVEDAV 639
Query: 473 EAAGR--RTTPWREAFYGAGMRPVQLSQFADFQAECLLAKVQIRGFHVAKRQAELVLFWH 530
R + WR AG P LS A+ QA+CLL +VQ+RGFHV KR A L L+W
Sbjct: 640 LGRRRADKAIAWRTVLTSAGFAPQPLSNLAEAQADCLLKRVQVRGFHVEKRGAGLALYWQ 699
Query: 531 ERAMVATSAWRC 542
+V+ SAWRC
Sbjct: 700 RGELVSVSAWRC 711
>ref|XP_467179.1| putative Scl1 protein [Oryza sativa (japonica cultivar-group)]
gi|50251749|dbj|BAD27682.1| putative Scl1 protein [Oryza
sativa (japonica cultivar-group)]
gi|49388515|dbj|BAD25639.1| putative Scl1 protein [Oryza
sativa (japonica cultivar-group)]
Length = 715
Score = 185 bits (470), Expect = 3e-45
Identities = 126/372 (33%), Positives = 204/372 (53%), Gaps = 21/372 (5%)
Query: 180 IEELIRAADCFDNNHLQLAQAILERLNQRLRSPTGKPLHRAAFHFKDALQSLLSGSNRTN 239
++EL AA + + A+ IL RLNQ+L P GKP R+A + ++AL L+ S+
Sbjct: 356 LDELAAAAKATEAGNSVGAREILARLNQQL-PPLGKPFLRSASYLREALLLALADSHH-G 413
Query: 240 PPRLSSMVEIVQTIRTFKAFSGISPIPMFSIFTTNQALLEALHGSLY--MHVVDFEIGLG 297
+++ +++ + +K+FS +SP+ F+ FT QALL+ + G+ +HV+DF++G+G
Sbjct: 414 VSSVTTPLDVALKLAAYKSFSDLSPVLQFANFTATQALLDEIGGTATSCIHVIDFDLGVG 473
Query: 298 IQYASLMKEIAEKAVNGS---PLLRITAVVP--EEYAVESRLIRENLNQFAHDLGIRVQV 352
Q+AS ++E+A + G PLL++TA V + +E L ++NL+QFA DLGI +
Sbjct: 474 GQWASFLQELAHRRAAGGVTLPLLKLTAFVSTASHHPLELHLTQDNLSQFAADLGIPFEF 533
Query: 353 DFVPLRTFETVSFKAVRFVDGEKTAILLTPAIFCRLGSEGTAAFLSDVRRITPGVVVFVD 412
+ V L F + E A+ L + C + A L V++++P +VV +D
Sbjct: 534 NAVSLDAFNPGEL--ISSTGDEVVAVSLP--VGCSARAPPLPAILRLVKQLSPKIVVAID 589
Query: 413 GEGWTEAAAAASFRRGVVNSLEFYSMMLESLDASVAAGGGGEWARRIEMLLLRPKIIAAV 472
A SF + +N + +L+SLDA AG + A +IE L++P++ V
Sbjct: 590 HGA---DRADLSFSQHFLNCFQSCVFLLDSLDA---AGIDADSACKIERFLIQPRVHDMV 643
Query: 473 EAAGR--RTTPWREAFYGAGMRPVQLSQFADFQAECLLAKVQIRGFHVAKRQAELVLFWH 530
+ + WR F AG +PV S A+ QA+CLL +VQ+RGFHV K A L L+W
Sbjct: 644 LGRHKVHKAIAWRSVFAAAGFKPVPPSNLAEAQADCLLKRVQVRGFHVEKCGAALTLYWQ 703
Query: 531 ERAMVATSAWRC 542
+V+ S+WRC
Sbjct: 704 RGELVSISSWRC 715
>emb|CAB82667.1| scarecrow-like protein [Arabidopsis thaliana]
gi|20465817|gb|AAM20013.1| putative scarecrow protein
[Arabidopsis thaliana] gi|17381212|gb|AAL36418.1|
putative scarecrow protein [Arabidopsis thaliana]
gi|15232385|ref|NP_191622.1| scarecrow transcription
factor family protein [Arabidopsis thaliana]
gi|11358821|pir||T47874 scarecrow-like protein -
Arabidopsis thaliana
Length = 623
Score = 184 bits (468), Expect = 5e-45
Identities = 142/401 (35%), Positives = 207/401 (51%), Gaps = 35/401 (8%)
Query: 168 QPNNNTNSNWDFIEELIRAA-----DCFDNNHLQLAQAILERLNQRLRSPTGK------- 215
+ N+ + + I++L AA + DNN + LAQ IL RLN L +
Sbjct: 232 EDQNDQDQSAVIIDQLFSAAAELTTNGGDNNPV-LAQGILARLNHNLNNNNDDTNNNPKP 290
Query: 216 PLHRAAFHFKDALQSLLSGSNRTNPPRLSSMVEIVQTIRTFKAFSGISPIPMFSIFTTNQ 275
P HRAA + +AL SLL S+ +PP LS ++ I ++AFS SP F FT NQ
Sbjct: 291 PFHRAASYITEALHSLLQDSS-LSPPSLSPPQNLIFRIAAYRAFSETSPFLQFVNFTANQ 349
Query: 276 ALLEALHGSLYMHVVDFEIGLGIQYASLMKEIAEKAVNGS--PLLRITA-----VVPEEY 328
+LE+ G +H+VDF+IG G Q+ASL++E+A K S P L+ITA V +E+
Sbjct: 350 TILESFEGFDRIHIVDFDIGYGGQWASLIQELAGKRNRSSSAPSLKITAFASPSTVSDEF 409
Query: 329 AVESRLIRENLNQFAHDLGIRVQVDFVPLRTFETVSFKAVR-FVDGEKTAILLTPAIFCR 387
E R ENL FA + G+ +++ + + ++ + F EK AI + I
Sbjct: 410 --ELRFTEENLRSFAGETGVSFEIELLNMEILLNPTYWPLSLFRSSEKEAIAVNLPI-SS 466
Query: 388 LGSEGTAAFLSDVRRITPGVVVFVDGEGWTEAAAAASFRRGVVNSLEFYSMMLESLDASV 447
+ S L +++I+P VVV D + A F GV+N+L++Y+ +LESLD+
Sbjct: 467 MVSGYLPLILRFLKQISPNVVVCSDRS--CDRNNDAPFPNGVINALQYYTSLLESLDSGN 524
Query: 448 AAGGGGEWARRIEMLLLRPKIIAAVEAAGR---RTTPWREAFYGAGMRPVQLSQFADFQA 504
E A IE ++P I + R R+ PWR F G PV LSQ A+ QA
Sbjct: 525 L--NNAEAATSIERFCVQPSIQKLLTNRYRWMERSPPWRSLFGQCGFTPVTLSQTAETQA 582
Query: 505 ECLLAKVQIRGFHVAKRQA---ELVLFWHERAMVATSAWRC 542
E LL + +RGFH+ KRQ+ LVL W + +V SAW+C
Sbjct: 583 EYLLQRNPMRGFHLEKRQSSSPSLVLCWQRKELVTVSAWKC 623
>gb|AAD32821.1| putative SCARECROW gene regulator [Arabidopsis thaliana]
gi|58652078|gb|AAW80864.1| At2g45160 [Arabidopsis
thaliana] gi|25408931|pir||B84887 probable SCARECROW
gene regulator [imported] - Arabidopsis thaliana
gi|15225417|ref|NP_182041.1| scarecrow transcription
factor family protein [Arabidopsis thaliana]
Length = 640
Score = 175 bits (444), Expect = 3e-42
Identities = 135/444 (30%), Positives = 217/444 (48%), Gaps = 39/444 (8%)
Query: 120 PNPSQDQFDQTQDFTSLSDIYSNNQNLAYNYPNTNTNLDHLVNDFNNNQPNNNTNSNWDF 179
P+P D + F Y NNQ + +++T + + P +
Sbjct: 215 PDPGHDPVRRQHQFQF--PFYHNNQQQQFPSSSSSTAVAMVPVP----SPGMAGDDQSVI 268
Query: 180 IEELIRAADCF------DNNHLQLAQAILERLNQRLRSPTG--KPLHRAAFHFKDALQSL 231
IE+L AA+ + +H LAQ IL RLN L + + P RAA H +AL SL
Sbjct: 269 IEQLFNAAELIGTTGNNNGDHTVLAQGILARLNHHLNTSSNHKSPFQRAASHIAEALLSL 328
Query: 232 LSGSNRTNPPRLSSMVEIVQTIRTFKAFSGISPIPMFSIFTTNQALLEALHGSLY--MHV 289
+ N ++PP L + ++ I +++FS SP F FT NQ++LE+ + S + +H+
Sbjct: 329 IH--NESSPP-LITPENLILRIAAYRSFSETSPFLQFVNFTANQSILESCNESGFDRIHI 385
Query: 290 VDFEIGLGIQYASLMKEIAE----KAVNGSPLLRITAVVPEEYAV----ESRLIRENLNQ 341
+DF++G G Q++SLM+E+A + N + L++T P V E R ENL
Sbjct: 386 IDFDVGYGGQWSSLMQELASGVGGRRRNRASSLKLTVFAPPPSTVSDEFELRFTEENLKT 445
Query: 342 FAHDLGIRVQVDFVPLRTFETVSFKAVRFVDGEKTAILLTPAIFCRLGSEGTAAFLSDVR 401
FA ++ I +++ + + ++ + EK AI + + + S L ++
Sbjct: 446 FAGEVKIPFEIELLSVELLLNPAYWPLSLRSSEKEAIAVNLPVNS-VASGYLPLILRFLK 504
Query: 402 RITPGVVVFVDGEGWTEAAAAASFRRGVVNSLEFYSMMLESLDASVAAGGGGEWARRIEM 461
+++P +VV D G A F V++SL++++ +LESLDA+ IE
Sbjct: 505 QLSPNIVVCSD-RGCDRNDAP--FPNAVIHSLQYHTSLLESLDANQNQDDSS-----IER 556
Query: 462 LLLRPKIIAAVEAAGR---RTTPWREAFYGAGMRPVQLSQFADFQAECLLAKVQIRGFHV 518
++P I + R R+ PWR F G P LSQ A+ QAECLL + +RGFHV
Sbjct: 557 FWVQPSIEKLLMKRHRWIERSPPWRILFTQCGFSPASLSQMAEAQAECLLQRNPVRGFHV 616
Query: 519 AKRQAELVLFWHERAMVATSAWRC 542
KRQ+ LV+ W + +V SAW+C
Sbjct: 617 EKRQSSLVMCWQRKELVTVSAWKC 640
>gb|AAO42084.1| putative SCARECROW gene regulator [Arabidopsis thaliana]
Length = 640
Score = 173 bits (438), Expect = 1e-41
Identities = 137/444 (30%), Positives = 218/444 (48%), Gaps = 39/444 (8%)
Query: 120 PNPSQDQFDQTQDFTSLSDIYSNNQNLAYNYPNTNTNLDHLVNDFNNNQPNNNTNSNWDF 179
P+P D + F Y NNQ + +++T + + P +
Sbjct: 215 PDPGHDPVRRQHQFQF--PFYHNNQQQQFPSSSSSTAVAMVPVP----SPGMAGDDQSVI 268
Query: 180 IEELIRAADCF----DNNHLQ--LAQAILERLNQRLRSPTG--KPLHRAAFHFKDALQSL 231
IE+L AA+ +NN Q LAQ IL RLN L + + P RAA H +AL SL
Sbjct: 269 IEQLFNAAELIGTTGNNNGDQTVLAQGILARLNHHLNTSSNHKSPFQRAASHIAEALLSL 328
Query: 232 LSGSNRTNPPRLSSMVEIVQTIRTFKAFSGISPIPMFSIFTTNQALLEALHGSLY--MHV 289
+ N ++PP L + ++ I +++FS SP F FT NQ++LE+ + S + +H+
Sbjct: 329 IH--NESSPP-LITPENLILRIAAYRSFSETSPFLQFVNFTANQSILESCNESGFDRIHI 385
Query: 290 VDFEIGLGIQYASLMKEIAE----KAVNGSPLLRITAVVPEEYAV----ESRLIRENLNQ 341
+DF++G G Q++SLM+E+A + N + L++T P V E R ENL
Sbjct: 386 IDFDVGYGGQWSSLMQELASGVGGRRRNRASSLKLTVFAPPPSTVSDEFELRFTEENLKT 445
Query: 342 FAHDLGIRVQVDFVPLRTFETVSFKAVRFVDGEKTAILLTPAIFCRLGSEGTAAFLSDVR 401
FA ++ I +++ + + ++ + EK AI + + + S L ++
Sbjct: 446 FAGEVKIPFEIELLSVELLLNPAYWPLSLRSSEKEAIAVNLPVNS-VASGYLPLILRFLK 504
Query: 402 RITPGVVVFVDGEGWTEAAAAASFRRGVVNSLEFYSMMLESLDASVAAGGGGEWARRIEM 461
+++P +VV D G A F V++SL++++ +LESLDA+ IE
Sbjct: 505 QLSPNIVVCSD-RGCDRNDAP--FPNAVIHSLQYHTSLLESLDANQNQDDSS-----IER 556
Query: 462 LLLRPKIIAAVEAAGR---RTTPWREAFYGAGMRPVQLSQFADFQAECLLAKVQIRGFHV 518
++P I + R R+ PWR F G P LSQ A+ QAECLL + +RGFHV
Sbjct: 557 FWVQPSIEKLLMKRHRWIERSPPWRILFTQCGFSPASLSQMAEAQAECLLQRNPVRGFHV 616
Query: 519 AKRQAELVLFWHERAMVATSAWRC 542
KRQ+ LV+ W + +V SAW+C
Sbjct: 617 EKRQSSLVMCWQRKELVTVSAWKC 640
>ref|NP_910214.1| Similar to Oryza sativa Scl1 protein (AF067401) [Oryza sativa
(japonica cultivar-group)]
Length = 602
Score = 163 bits (413), Expect = 1e-38
Identities = 157/549 (28%), Positives = 246/549 (44%), Gaps = 87/549 (15%)
Query: 11 SSSSPQPKLTTTQNIVVNVPTPNPTPTPTTTTLCYE---PRSVLELCRSPSPEKKPSQQE 67
SSSS P TT + +P P+ + +P T++ Y P + L C S PE P
Sbjct: 124 SSSSCHPHNPTT----LPLPEPDSSKSPEPTSVLYNRSSPSTSLGSCSSKPPEDPP---- 175
Query: 68 QEHNPEVEQEDHASLPNLDWWDSIMKDLGLQDDSSTPIIPLLKNTNNTSEIYPNPSQDQF 127
P + +D WD+++ D+ + + P P+ D
Sbjct: 176 ----PPIAADDDCD------WDAVV-DMHMH------------------MLAPAPAPD-- 204
Query: 128 DQTQDFTSLSDIYSNNQNLAYNYPNTNTNLDHLVNDFNNNQPNNNTNSNWDFIEELIRAA 187
+ + D + + ++P+ +++L L ++ P +++L+ AA
Sbjct: 205 --SSFLRWIMDTGYADADTFPDHPSFDSDLLQLPMPMPSDHPPQ------PLVDDLLDAA 256
Query: 188 DCFDNNHLQLAQAILERLNQRLRS---PTGK---PLHRAAFHFKDALQSLLSGSNRTNPP 241
D A+ IL RLN RL S P G PL RAA +DAL T P
Sbjct: 257 RLLDAGDSTSAREILARLNHRLPSLPSPPGHAHPPLLRAAALLRDALLP------PTALP 310
Query: 242 RLSSMVEIVQTIRTFKAFSGISPIPMFSIFTTNQALLEALHGSLYMHVVDFEIGLGIQYA 301
S+ +++ + KA + SP F+ FT+ QA L+AL + +H++DF++G G +
Sbjct: 311 VSSTPLDVPLKLAAHKALADASPTVQFTTFTSTQAFLDALGSARRLHLLDFDVGFGAHWP 370
Query: 302 SLMKEIAE---KAVNGSPLLRITAVVP--EEYAVESRLIRENLNQFAHDLGIRVQVDFVP 356
LM+E+A +A P L++TA+V + +E L E+L +FA +LGI + +
Sbjct: 371 PLMQELAHHWRRAAGPPPNLKVTALVSPGSSHPLELHLTNESLTRFAAELGIPFEFTALV 430
Query: 357 LRTFETVSFK-AVRFVDGEKTAILLTPAIFCRLGSEGTA-AFLSDVRRITPGVVVFVDGE 414
+ S + E A+ LT G+ A A L V+ + P VVV VD
Sbjct: 431 FDPLSSASPPLGLSAAPDEAVAVHLTAGS----GAFSPAPAHLRVVKELRPAVVVCVDH- 485
Query: 415 GWTEAAAAASFRRGVVNSLEFYSMMLESLDASVAAGGGGEWARRIEMLLLRPKIIA-AVE 473
RG +N L+ + +LESLDA AG + ++E +LRP++ AV
Sbjct: 486 ---------GCERGALNLLQSCAALLESLDA---AGASPDVVSKVEQFVLRPRVERLAVG 533
Query: 474 AAGRRTTPWREAFYGAGMRPVQLSQFADFQAECLLAKVQIRGFHVAKRQAELVLFWHERA 533
+ P + AG +Q+S A+ QAECLL + GFHV KRQA L L+W
Sbjct: 534 GGDKLPPPLQSMLASAGFAALQVSNAAEAQAECLLRRTASHGFHVEKRQAALALWWQRSE 593
Query: 534 MVATSAWRC 542
+V+ SAWRC
Sbjct: 594 LVSVSAWRC 602
>gb|AAY17058.1| f-171-1_1 [Ceratopteris thalictroides]
Length = 348
Score = 156 bits (394), Expect = 2e-36
Identities = 114/357 (31%), Positives = 176/357 (48%), Gaps = 31/357 (8%)
Query: 204 RLNQRLRSPTGK-PLHRAAFHFKDALQSLLSGSNRTNPPRL-------SSMVEIVQTIRT 255
RLNQ++ + P R F DAL +S S+ + P R +S +++++ I
Sbjct: 2 RLNQQVPLKGQQLPSQRVIPLFVDALAKWVSASSPSAPSRFQQHSSLANSPLDLIKKISA 61
Query: 256 FKAFSGISPIPMFSIFTTNQALLEALHGSLYMHVVDFEIGLGIQYASLMKEIAEKAVNGS 315
+K F +SPI F+ FT NQA+L+A+ G ++H++DFE+G G Q+AS M+E++++ G
Sbjct: 62 YKKFCEVSPISQFAHFTANQAILDAIEGEDHVHLIDFELGFGGQWASFMQELSQRC-RGP 120
Query: 316 PLLRITAVVPEEYAVESRLIRENLNQFAHDLGIRVQVDFVPLRTFETVSFKAVRFVDGEK 375
P L+IT + + +E++L +ENL QFA ++GI+++V+ VPL E V E
Sbjct: 121 PELKITTMGTD--TLETKLAKENLLQFATEMGIKLEVNVVPLEKLELVKSVVANKASKEA 178
Query: 376 TAI----LLTPAIFCRLGSEGTAAFLSDVRRITPGVVVFVDGEGWTEAAAAASFRRGVVN 431
A+ + I E +FL V+ + P V+ +D E + + G+
Sbjct: 179 VAVNFGFAMNRMISDFASMEEVLSFLQLVKTLCPKVIAVMDSECEFDGPS------GLSE 232
Query: 432 SLEFYSMMLESLDASVAAGGGGEWARRIEMLLLRPKIIAAVEAA--------GRRTTPWR 483
+L+FYS LESLDAS E IE L+L PKI V A G WR
Sbjct: 233 ALQFYSCNLESLDASTKL--SAEVVSNIEGLVLGPKIAELVNARLTNXSSTDGGALPQWR 290
Query: 484 EAFYGAGMRPVQLSQFADFQAECLLAKVQIRGFHVAKRQAELVLFWHERAMVATSAW 540
G P S A+ A + GF K+QA L L W+ + +V+ AW
Sbjct: 291 ILLQKVGFSPCPFSSAAEXXASWXVNNPLNLGFTYXKQQATLFLGWYNKTLVSAXAW 347
>gb|AAC98091.1| Scl1 protein [Oryza sativa] gi|11358961|pir||T51243 Scl1 protein
[imported] - rice (fragment)
Length = 261
Score = 137 bits (346), Expect = 7e-31
Identities = 92/263 (34%), Positives = 145/263 (54%), Gaps = 18/263 (6%)
Query: 287 MHVVDFEIGLGIQYASLMKEIAEKAVNGS---PLLRITAVVP--EEYAVESRLIRENLNQ 341
+HV+DF++G+G Q+AS ++E+A + G PLL++TA + + +E L ++NL+Q
Sbjct: 10 IHVIDFDLGVGGQWASFLQELAHRRGAGGMALPLLKLTAFMSTASHHPLELHLTQDNLSQ 69
Query: 342 FAHDLGIRVQVDFVPLRTFETVSFKAVRFVDGEKTAILLTPAIFCRLGSEGTAAFLSDVR 401
FA +L I + + V L F +++ E A+ L + C + A L V+
Sbjct: 70 FAAELRIPFEFNAVSLDAFNPA--ESISSSGDEVVAVSLP--VGCSARAPPLPAILRLVK 125
Query: 402 RITPGVVVFVDGEGWTEAAAAASFRRGVVNSLEFYSMMLESLDASVAAGGGGEWARRIEM 461
++ P VVV +D G A F + +N + + L+SLDA AG + A +IE
Sbjct: 126 QLCPKVVVAIDHGG---DRADLPFSQHFLNCFQS-CVFLDSLDA---AGIDADSACKIER 178
Query: 462 LLLRPKIIAAV--EAAGRRTTPWREAFYGAGMRPVQLSQFADFQAECLLAKVQIRGFHVA 519
L++P++ AV ++ WR F G +PVQLS A+ QA+CLL +VQ+RGFHV
Sbjct: 179 FLIQPRVEDAVIGRHKAQKAIAWRSVFAATGFKPVQLSNLAEAQADCLLKRVQVRGFHVE 238
Query: 520 KRQAELVLFWHERAMVATSAWRC 542
KR A L L+W +V+ S+WRC
Sbjct: 239 KRGAALTLYWQRGELVSISSWRC 261
>ref|NP_912413.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|29893602|gb|AAP06856.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 575
Score = 112 bits (280), Expect = 3e-23
Identities = 118/394 (29%), Positives = 171/394 (42%), Gaps = 40/394 (10%)
Query: 183 LIRAADCFDNNHL--QLAQAILERLNQRLR-----SPTGKPLHRAAFHFKDALQSLLSGS 235
L+ AA+ H +LA+ IL RL + + + + R A HF DALQ LL GS
Sbjct: 136 LMAAAEALSGPHKSRELARVILVRLKEMVSHTASANAAASNMERLAAHFTDALQGLLDGS 195
Query: 236 NRTN-----PPRLSSMVEIVQTIRTFKAFSGISPIPMFSIFTTNQALLEALHGSLYMHVV 290
+ +S + F+ +SP F FT NQA+LEA+ G +H+V
Sbjct: 196 HPVGGSGRQAAAAASHHHAGDVLTAFQMLQDMSPYMKFGHFTANQAILEAVSGDRRVHIV 255
Query: 291 DFEIGLGIQYASLMKEIAEKAVN-GSPLLRITAVVPEEYAVESRLIRE---NLNQFAHDL 346
D++I GIQ+ASLM+ + +A +P LRITA V +R ++E L+ FA +
Sbjct: 256 DYDIAEGIQWASLMQAMTSRADGVPAPHLRITA-VSRSGGGGARAVQEAGRRLSAFAASI 314
Query: 347 GIRVQVDFVPLRTFETVSFKAVRFVDGE---KTAILLTPA--IFCRLGSEGTAAFLSDVR 401
G L + E VR V GE +L A R + A+FLS +
Sbjct: 315 GQPFSFGQCRLDSDERFRPATVRMVKGEALVANCVLHQAAATTTIRRPTGSVASFLSGMA 374
Query: 402 RITPGVVVFVDGEGWTEA---------AAAASFRRGVVNSLEFYSMMLESLDASVAAGGG 452
+ +V V+ EG E AAA F R + L YS + +SL+A
Sbjct: 375 ALGAKLVTVVEEEGEAEKDDDGDSAGDAAAGGFVRQFMEELHRYSAVWDSLEAGFPT--Q 432
Query: 453 GEWARRIEMLLLRPKIIAAVEAA-----GRRTTPWREAFYGAGMRPVQLSQFADFQAECL 507
+E ++L P I AV A G W + G+G V LS F QA L
Sbjct: 433 SRVRGLVERVILAPNIAGAVSRAYRGVDGEGRCGWGQWMRGSGFTAVPLSCFNHSQARLL 492
Query: 508 LAKVQIRGFHVAKR-QAELVLFWHERAMVATSAW 540
L G+ V + ++VL W R +++ S W
Sbjct: 493 LGLFN-DGYTVEETGPNKIVLGWKARRLMSASVW 525
>gb|AAF00139.1| Scl1 protein [Oryza sativa]
Length = 360
Score = 110 bits (276), Expect = 9e-23
Identities = 86/279 (30%), Positives = 130/279 (45%), Gaps = 25/279 (8%)
Query: 271 FTTNQALLEALHGSLYMHVVDFEIGLGIQYASLMKEIAE---KAVNGSPLLRITAVVP-- 325
FT+ QA L+A + +H++ ++G G + LM+E+A +A P L++TA+V
Sbjct: 90 FTSTQAFLDAXGSARRLHLLXXDVGFGAHWPPLMQELAHHWRRAXGPPPNLKVTALVSPG 149
Query: 326 EEYAVESRLIRENLNQFAHDLGIRVQVDFVPLRTFETVSFK-AVRFVDGEKTAILLTPAI 384
+ E L E+L +FA +LGI + + + S + E A+ LT
Sbjct: 150 SSHPXELHLTNESLTRFAAELGIPFEFTALVFDPLSSASPPLGLSAAPDEAVAVHLTAGS 209
Query: 385 FCRLGSEGTA-AFLSDVRRITPGVVVFVDGEGWTEAAAAASFRRGVVNSLEFYSMMLESL 443
G+ A A L V+ + P VVV VD RG +N L + +LESL
Sbjct: 210 ----GAFSPAPAHLRVVKELRPAVVVCVDH----------GCERGALNLLRSCAALLESL 255
Query: 444 DASVAAGGGGEWARRIEMLLLRPKIIAAVEAAGRRTTPWREAFYGAGMRPVQLSQFADFQ 503
DA AG + ++E +LRP++ G P + AG +Q++ A Q
Sbjct: 256 DA---AGASPDVVSKVEQFVLRPRVERLAVGVGGGP-PLQSMLASAGFAALQVNNAAXAQ 311
Query: 504 AECLLAKVQIRGFHVAKRQAELVLFWHERAMVATSAWRC 542
AECLL + GFHV R A L L+W +V+ S WRC
Sbjct: 312 AECLLRRTAXHGFHVEXRPAALALWWXRSELVSVSXWRC 350
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.318 0.133 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 953,380,533
Number of Sequences: 2540612
Number of extensions: 45119595
Number of successful extensions: 512281
Number of sequences better than 10.0: 3378
Number of HSP's better than 10.0 without gapping: 2029
Number of HSP's successfully gapped in prelim test: 1524
Number of HSP's that attempted gapping in prelim test: 292059
Number of HSP's gapped (non-prelim): 56750
length of query: 542
length of database: 863,360,394
effective HSP length: 133
effective length of query: 409
effective length of database: 525,458,998
effective search space: 214912730182
effective search space used: 214912730182
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 78 (34.7 bits)
Medicago: description of AC148484.4


