

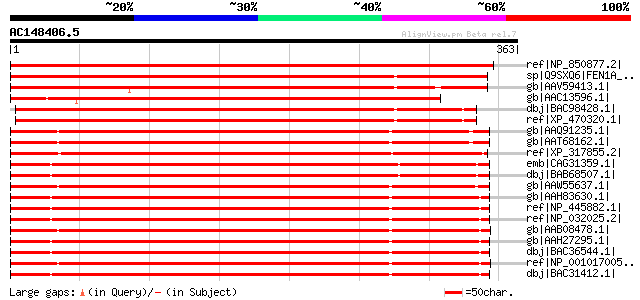
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148406.5 - phase: 0 /pseudo
(363 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_850877.2| endonuclease, putative [Arabidopsis thaliana] 600 e-170
sp|Q9SXQ6|FEN1A_ORYSA Flap endonuclease-1a (OsFEN-1a) gi|4587225... 563 e-159
gb|AAV59413.1| putative flap endonuclease 1 [Oryza sativa (japon... 547 e-154
gb|AAC13596.1| similar to FLAP endonuclease-1 (SW:P39748) [Arabi... 490 e-137
dbj|BAC98428.1| flap endonuclease-1b [Oryza sativa (japonica cul... 454 e-126
ref|XP_470320.1| flap endonuclease-1b [Oryza sativa (japonica cu... 452 e-126
gb|AAQ91235.1| flap structure-specific endonuclease 1 [Danio rer... 404 e-111
gb|AAT68162.1| flap structure specific endonuclease 1 [Danio rerio] 401 e-110
ref|XP_317855.2| ENSANGP00000014920 [Anopheles gambiae str. PEST... 395 e-108
emb|CAG31359.1| hypothetical protein [Gallus gallus] 394 e-108
dbj|BAB68507.1| FEN-1 nuclease [Gallus gallus] 394 e-108
gb|AAW55637.1| flap endonuclease-1 [Xiphophorus maculatus] gi|57... 394 e-108
gb|AAH83630.1| Fen1 protein [Rattus norvegicus] 392 e-108
ref|NP_445882.1| flap structure-specific endonuclease [Rattus no... 392 e-108
ref|NP_032025.2| flap structure specific endonuclease 1 [Mus mus... 392 e-107
gb|AAB08478.1| XFEN1b [Xenopus laevis] 392 e-107
gb|AAH27295.1| Fen1 protein [Mus musculus] 392 e-107
dbj|BAC36544.1| unnamed protein product [Mus musculus] 392 e-107
ref|NP_001017005.1| hypothetical protein LOC549759 [Xenopus trop... 391 e-107
dbj|BAC31412.1| unnamed protein product [Mus musculus] 389 e-107
>ref|NP_850877.2| endonuclease, putative [Arabidopsis thaliana]
Length = 453
Score = 600 bits (1547), Expect = e-170
Identities = 297/346 (85%), Positives = 321/346 (91%)
Query: 1 MKENKFESYFGRKIAVDASMSIYQFLIVVGRSGTEMLTNEAGEVTSHLQGMFARTIRLLE 60
MKE KFESYFGRKIAVDASMSIYQFLIVVGR+GTEMLTNEAGEVTSHLQGMF RTIRLLE
Sbjct: 18 MKEQKFESYFGRKIAVDASMSIYQFLIVVGRTGTEMLTNEAGEVTSHLQGMFNRTIRLLE 77
Query: 61 AGMKPVYVFDGKPPEMKNQELKKRLSKRAEATAGLTEALEADNKEDIEKFSKRTVKVTKQ 120
AG+KPVYVFDGKPPE+K QEL KR SKRA+ATA LT A+EA NKEDIEK+SKRTVKVTKQ
Sbjct: 78 AGIKPVYVFDGKPPELKRQELAKRYSKRADATADLTGAIEAGNKEDIEKYSKRTVKVTKQ 137
Query: 121 HNDDCKRLLRLMGVPVVEAPSEAEAQCAALCKAGKVYAVASEDMDSLTFGAPKFLRHLMD 180
HNDDCKRLLRLMGVPVVEA SEAEAQCAALCK+GKVY VASEDMDSLTFGAPKFLRHLMD
Sbjct: 138 HNDDCKRLLRLMGVPVVEATSEAEAQCAALCKSGKVYGVASEDMDSLTFGAPKFLRHLMD 197
Query: 181 PSSKKIPVMEFDVAKILEELDLTMDQFIDLCILSGCDYCDNIRGIGGMTALKLIRQHGSI 240
PSS+KIPVMEF+VAKILEEL LTMDQFIDLCILSGCDYCD+IRGIGG TALKLIRQHGSI
Sbjct: 198 PSSRKIPVMEFEVAKILEELQLTMDQFIDLCILSGCDYCDSIRGIGGQTALKLIRQHGSI 257
Query: 241 EKILENISKERYQVPDDWPYQEARRLFKEPEVSTDDEVLNLKWSPPDEEGLITFLVNENG 300
E ILEN++KERYQ+P++WPY EAR+LFKEP+V TD+E L++KW+ PDEEG++ FLVNENG
Sbjct: 258 ETILENLNKERYQIPEEWPYNEARKLFKEPDVITDEEQLDIKWTSPDEEGIVQFLVNENG 317
Query: 301 FNSDRVTKAIEKIKAAKNKSSQGRLESFFKPTANPSVPIKRKQLII 346
FN DRVTKAIEKIK AKNKSSQGRLESFFKP AN SVP KRK +I
Sbjct: 318 FNIDRVTKAIEKIKTAKNKSSQGRLESFFKPVANSSVPAKRKGNLI 363
>sp|Q9SXQ6|FEN1A_ORYSA Flap endonuclease-1a (OsFEN-1a) gi|4587225|dbj|BAA36171.1| FEN-1
[Oryza sativa (japonica cultivar-group)]
Length = 380
Score = 563 bits (1451), Expect = e-159
Identities = 280/342 (81%), Positives = 308/342 (89%), Gaps = 2/342 (0%)
Query: 1 MKENKFESYFGRKIAVDASMSIYQFLIVVGRSGTEMLTNEAGEVTSHLQGMFARTIRLLE 60
MKE KFESYFGR+IAVDASMSIYQFLIVVGR+G E LTNEAGEVTSHLQGMF RTIRLLE
Sbjct: 18 MKEQKFESYFGRRIAVDASMSIYQFLIVVGRTGMETLTNEAGEVTSHLQGMFNRTIRLLE 77
Query: 61 AGMKPVYVFDGKPPEMKNQELKKRLSKRAEATAGLTEALEADNKEDIEKFSKRTVKVTKQ 120
AG+KPVYVFDGKPP++K QEL KR SKR +AT LTEA+E +K+ IEKFSKRTVKVTKQ
Sbjct: 78 AGIKPVYVFDGKPPDLKKQELAKRYSKREDATKELTEAVEEGDKDAIEKFSKRTVKVTKQ 137
Query: 121 HNDDCKRLLRLMGVPVVEAPSEAEAQCAALCKAGKVYAVASEDMDSLTFGAPKFLRHLMD 180
HN++CKRLLRLMGVPVVEAP EAEA+CAALC VYAVASEDMDSLTFGAP+FLRHLMD
Sbjct: 138 HNEECKRLLRLMGVPVVEAPCEAEAECAALCINDMVYAVASEDMDSLTFGAPRFLRHLMD 197
Query: 181 PSSKKIPVMEFDVAKILEELDLTMDQFIDLCILSGCDYCDNIRGIGGMTALKLIRQHGSI 240
PSSKKIPVMEF+VAK+LEEL+LTMDQFIDLCILSGCDYCD+I+GIGG TALKLIRQHGSI
Sbjct: 198 PSSKKIPVMEFEVAKVLEELELTMDQFIDLCILSGCDYCDSIKGIGGQTALKLIRQHGSI 257
Query: 241 EKILENISKERYQVPDDWPYQEARRLFKEPEVSTDDEVLNLKWSPPDEEGLITFLVNENG 300
E ILENI+K+RYQ+P+DWPYQEARRLFKEP V+ D + LKW+ PDEEGL+ FLV ENG
Sbjct: 258 ESILENINKDRYQIPEDWPYQEARRLFKEPNVTLD--IPELKWNAPDEEGLVEFLVKENG 315
Query: 301 FNSDRVTKAIEKIKAAKNKSSQGRLESFFKPTANPSVPIKRK 342
FN DRVTKAIEKIK AKNKSSQGRLESFFKP + SVP+KRK
Sbjct: 316 FNQDRVTKAIEKIKFAKNKSSQGRLESFFKPVVSTSVPLKRK 357
>gb|AAV59413.1| putative flap endonuclease 1 [Oryza sativa (japonica
cultivar-group)] gi|50932545|ref|XP_475800.1| putative
flap endonuclease 1 [Oryza sativa (japonica
cultivar-group)]
Length = 380
Score = 547 bits (1409), Expect = e-154
Identities = 277/345 (80%), Positives = 305/345 (88%), Gaps = 8/345 (2%)
Query: 1 MKENKFESYFGRKIAVDASMSIYQFLIVVGRSGTEMLTNEAGEVTSHLQGMFARTIRLLE 60
MKE KFESYFGR+IAVDASMSIYQFLIVVGR+G E LTNEAGEVTSHLQGMF RTIRLLE
Sbjct: 18 MKEQKFESYFGRRIAVDASMSIYQFLIVVGRTGMETLTNEAGEVTSHLQGMFNRTIRLLE 77
Query: 61 AGMKPVYVFDGKPPEMKNQELKKR---LSKRAEATAGLTEALEADNKEDIEKFSKRTVKV 117
AG+KPVYVFDGKPP++K QEL KR SKR +AT LTEA+E +K+ IEKFSKRTVKV
Sbjct: 78 AGIKPVYVFDGKPPDLKKQELAKRHDMYSKREDATKELTEAVEEGDKDAIEKFSKRTVKV 137
Query: 118 TKQHNDDCKRLLRLMGVPVVEAPSEAEAQCAALCKAGKVYAVASEDMDSLTFGAPKFLRH 177
TKQHN++CKRLLRLMGVPVVEAP EAEA+CAALC VYAVASEDMDSLTFGAP+FLRH
Sbjct: 138 TKQHNEECKRLLRLMGVPVVEAPCEAEAECAALCINDMVYAVASEDMDSLTFGAPRFLRH 197
Query: 178 LMDPSSKKIPVMEFDVAKILEELDLTMDQFIDLCILSGCDYCDNIRGIGGMTALKLIRQH 237
LMDPSSKKIPVMEF+VAK+LEEL+LTMDQFIDLCILSGCDYCD+I+GIGG TALKLIRQH
Sbjct: 198 LMDPSSKKIPVMEFEVAKVLEELELTMDQFIDLCILSGCDYCDSIKGIGGQTALKLIRQH 257
Query: 238 GSIEKILENISKERYQVPDDWPYQEARRLFKEPEVSTDDEVLNLKWSPPDEEGLITFLVN 297
GSIE ILENI+K+RYQ+P+DWPYQEARRLFKEP V+ D + LKW+ PDEEGL+ FLV
Sbjct: 258 GSIESILENINKDRYQIPEDWPYQEARRLFKEPNVTLD--IPELKWNAPDEEGLVEFLVK 315
Query: 298 ENGFNSDRVTKAIEKIKAAKNKSSQGRLESFFKPTANPSVPIKRK 342
ENGFN DR AIEKIK AKNKSSQGRLESFFKP + SVP+KRK
Sbjct: 316 ENGFNQDR---AIEKIKFAKNKSSQGRLESFFKPVVSTSVPLKRK 357
>gb|AAC13596.1| similar to FLAP endonuclease-1 (SW:P39748) [Arabidopsis thaliana]
gi|7446863|pir||T01198 endonuclease homolog F21E10.3 -
Arabidopsis thaliana
Length = 362
Score = 490 bits (1262), Expect = e-137
Identities = 253/342 (73%), Positives = 276/342 (79%), Gaps = 35/342 (10%)
Query: 1 MKENKFESYFGRKIAVDASMSIYQFLIVVGRSGTEMLTNEAGEVTS-------------- 46
MKE KFESYFGRKIAVDASMSIYQFL V S +L E G S
Sbjct: 18 MKEQKFESYFGRKIAVDASMSIYQFL-VRAPSSPLLLLEELGLKCSLMKLVKSLVWIFLC 76
Query: 47 --------------------HLQGMFARTIRLLEAGMKPVYVFDGKPPEMKNQELKKRLS 86
HLQGMF RTIRLLEAG+KPVYVFDGKPPE+K QEL KR S
Sbjct: 77 DVLCLYVFFVYVVIYMMFNRHLQGMFNRTIRLLEAGIKPVYVFDGKPPELKRQELAKRYS 136
Query: 87 KRAEATAGLTEALEADNKEDIEKFSKRTVKVTKQHNDDCKRLLRLMGVPVVEAPSEAEAQ 146
KRA+ATA LT A+EA NKEDIEK+SKRTVKVTKQHNDDCKRLLRLMGVPVVEA SEAEAQ
Sbjct: 137 KRADATADLTGAIEAGNKEDIEKYSKRTVKVTKQHNDDCKRLLRLMGVPVVEATSEAEAQ 196
Query: 147 CAALCKAGKVYAVASEDMDSLTFGAPKFLRHLMDPSSKKIPVMEFDVAKILEELDLTMDQ 206
CAALCK+GKVY VASEDMDSLTFGAPKFLRHLMDPSS+KIPVMEF+VAKILEEL LTMDQ
Sbjct: 197 CAALCKSGKVYGVASEDMDSLTFGAPKFLRHLMDPSSRKIPVMEFEVAKILEELQLTMDQ 256
Query: 207 FIDLCILSGCDYCDNIRGIGGMTALKLIRQHGSIEKILENISKERYQVPDDWPYQEARRL 266
FIDLCILSGCDYCD+IRGIGG TALKLIRQHGSIE ILEN++KERYQ+P++WPY EAR+L
Sbjct: 257 FIDLCILSGCDYCDSIRGIGGQTALKLIRQHGSIETILENLNKERYQIPEEWPYNEARKL 316
Query: 267 FKEPEVSTDDEVLNLKWSPPDEEGLITFLVNENGFNSDRVTK 308
FKEP+V TD+E L++KW+ PDEEG++ FLVNENGFN DRVTK
Sbjct: 317 FKEPDVITDEEQLDIKWTSPDEEGIVQFLVNENGFNIDRVTK 358
>dbj|BAC98428.1| flap endonuclease-1b [Oryza sativa (japonica cultivar-group)]
gi|29467052|dbj|BAC66965.1| flap endonuclease-1b [Oryza
sativa (japonica cultivar-group)]
Length = 412
Score = 454 bits (1167), Expect = e-126
Identities = 225/330 (68%), Positives = 269/330 (81%), Gaps = 3/330 (0%)
Query: 5 KFESYFGRKIAVDASMSIYQFLIVVGRSGTEMLTNEAGEVTSHLQGMFARTIRLLEAGMK 64
+ E Y GR +A+DAS+SIYQFLIVVGR GTE+LTNEAGEVTSHLQGM RT+R+LEAG+K
Sbjct: 22 RVEDYRGRVVAIDASLSIYQFLIVVGRKGTEVLTNEAGEVTSHLQGMLNRTVRILEAGIK 81
Query: 65 PVYVFDGKPPEMKNQELKKRLSKRAEATAGLTEALEADNKEDIEKFSKRTVKVTKQHNDD 124
PV+VFDG+PP+MK +EL KR KR ++ L A+E +++ IEKFSKRTVKVTK+HN+D
Sbjct: 82 PVFVFDGEPPDMKKKELAKRSLKRDGSSEDLNRAIEVGDEDLIEKFSKRTVKVTKKHNED 141
Query: 125 CKRLLRLMGVPVVEAPSEAEAQCAALCKAGKVYAVASEDMDSLTFGAPKFLRHLMDPSSK 184
CKRLL LMGVPVV+AP EAEAQCAALC+ KV+A+ASEDMDSLTFGA +FLRHL D S K
Sbjct: 142 CKRLLSLMGVPVVQAPGEAEAQCAALCENHKVFAIASEDMDSLTFGARRFLRHLTDLSFK 201
Query: 185 KIPVMEFDVAKILEELDLTMDQFIDLCILSGCDYCDNIRGIGGMTALKLIRQHGSIEKIL 244
+ PV EF+V+K+LEEL LTMDQFIDLCILSGCDYC+NIRGIGG ALKLIRQHG IE+++
Sbjct: 202 RSPVTEFEVSKVLEELGLTMDQFIDLCILSGCDYCENIRGIGGQRALKLIRQHGYIEEVV 261
Query: 245 ENISKERYQVPDDWPYQEARRLFKEPEVSTDDEVLNLKWSPPDEEGLITFLVNENGFNSD 304
+N+S+ RY VP+DWPYQE R LFKEP V TD + + W+PPDEEGLI FL EN F+ D
Sbjct: 262 QNLSQTRYSVPEDWPYQEVRALFKEPNVCTD--IPDFLWTPPDEEGLINFLAAENNFSPD 319
Query: 305 RVTKAIEKIKAAKNKSSQGRLESFFKPTAN 334
RV K++EKIKAA +K S GR P AN
Sbjct: 320 RVVKSVEKIKAANDKFSLGR-GKLLAPVAN 348
>ref|XP_470320.1| flap endonuclease-1b [Oryza sativa (japonica cultivar-group)]
gi|62510683|sp|Q75LI2|FEN1B_ORYSA Flap endonuclease-1b
(OsFEN-1b) gi|40714676|gb|AAR88582.1| flap
endonuclease-1b [Oryza sativa (japonica cultivar-group)]
Length = 412
Score = 452 bits (1163), Expect = e-126
Identities = 224/330 (67%), Positives = 268/330 (80%), Gaps = 3/330 (0%)
Query: 5 KFESYFGRKIAVDASMSIYQFLIVVGRSGTEMLTNEAGEVTSHLQGMFARTIRLLEAGMK 64
+ E Y GR +A+D S+SIYQFLIVVGR GTE+LTNEAGEVTSHLQGM RT+R+LEAG+K
Sbjct: 22 RVEDYRGRVVAIDTSLSIYQFLIVVGRKGTEVLTNEAGEVTSHLQGMLNRTVRILEAGIK 81
Query: 65 PVYVFDGKPPEMKNQELKKRLSKRAEATAGLTEALEADNKEDIEKFSKRTVKVTKQHNDD 124
PV+VFDG+PP+MK +EL KR KR ++ L A+E +++ IEKFSKRTVKVTK+HN+D
Sbjct: 82 PVFVFDGEPPDMKKKELAKRSLKRDGSSEDLNRAIEVGDEDLIEKFSKRTVKVTKKHNED 141
Query: 125 CKRLLRLMGVPVVEAPSEAEAQCAALCKAGKVYAVASEDMDSLTFGAPKFLRHLMDPSSK 184
CKRLL LMGVPVV+AP EAEAQCAALC+ KV+A+ASEDMDSLTFGA +FLRHL D S K
Sbjct: 142 CKRLLSLMGVPVVQAPGEAEAQCAALCENHKVFAIASEDMDSLTFGARRFLRHLTDLSFK 201
Query: 185 KIPVMEFDVAKILEELDLTMDQFIDLCILSGCDYCDNIRGIGGMTALKLIRQHGSIEKIL 244
+ PV EF+V+K+LEEL LTMDQFIDLCILSGCDYC+NIRGIGG ALKLIRQHG IE+++
Sbjct: 202 RSPVTEFEVSKVLEELGLTMDQFIDLCILSGCDYCENIRGIGGQRALKLIRQHGYIEEVV 261
Query: 245 ENISKERYQVPDDWPYQEARRLFKEPEVSTDDEVLNLKWSPPDEEGLITFLVNENGFNSD 304
+N+S+ RY VP+DWPYQE R LFKEP V TD + + W+PPDEEGLI FL EN F+ D
Sbjct: 262 QNLSQTRYSVPEDWPYQEVRALFKEPNVCTD--IPDFLWTPPDEEGLINFLAAENNFSPD 319
Query: 305 RVTKAIEKIKAAKNKSSQGRLESFFKPTAN 334
RV K++EKIKAA +K S GR P AN
Sbjct: 320 RVVKSVEKIKAANDKFSLGR-GKLLAPVAN 348
>gb|AAQ91235.1| flap structure-specific endonuclease 1 [Danio rerio]
gi|47940409|gb|AAH71488.1| Fen1 protein [Danio rerio]
Length = 380
Score = 404 bits (1037), Expect = e-111
Identities = 202/343 (58%), Positives = 259/343 (74%), Gaps = 4/343 (1%)
Query: 1 MKENKFESYFGRKIAVDASMSIYQFLIVVGRSGTEMLTNEAGEVTSHLQGMFARTIRLLE 60
+KE++ +SYFGRKIA+DASM IYQFLI V + G +L NE GE TSHL GMF RTIR+LE
Sbjct: 18 IKEHEIKSYFGRKIAIDASMCIYQFLIAVRQDGN-VLQNEDGETTSHLMGMFYRTIRMLE 76
Query: 61 AGMKPVYVFDGKPPEMKNQELKKRLSKRAEATAGLTEALEADNKEDIEKFSKRTVKVTKQ 120
+G+KPVYVFDGKPP++K+ EL+KR+ +RAEA L +A EA +E+I+KFSKR VKVTKQ
Sbjct: 77 SGIKPVYVFDGKPPQLKSGELEKRVERRAEAEKLLAQAQEAGEQENIDKFSKRLVKVTKQ 136
Query: 121 HNDDCKRLLRLMGVPVVEAPSEAEAQCAALCKAGKVYAVASEDMDSLTFGAPKFLRHLMD 180
HN++CK+LL LMGVP +EAP EAEA CAAL KAGKVYA A+EDMD LTFG LRHL
Sbjct: 137 HNEECKKLLSLMGVPYIEAPCEAEASCAALVKAGKVYATATEDMDGLTFGTTVLLRHLTA 196
Query: 181 PSSKKIPVMEFDVAKILEELDLTMDQFIDLCILSGCDYCDNIRGIGGMTALKLIRQHGSI 240
+KK+P+ EF ++IL++++LT QFIDLCIL GCDYC I+GIG A+ LI+QHGSI
Sbjct: 197 SEAKKLPIQEFHFSRILQDMELTHQQFIDLCILLGCDYCGTIKGIGPKRAIDLIKQHGSI 256
Query: 241 EKILENISKERYQVPDDWPYQEARRLFKEPEVSTDDEVLNLKWSPPDEEGLITFLVNENG 300
E+ILENI ++ P+DW Y+EAR LF EPEV D ++LKW+ PDE+GLI F+ E
Sbjct: 257 EEILENIDPNKHPAPEDWLYKEARGLFLEPEV-VDGTSVDLKWNEPDEDGLIQFMCAEKQ 315
Query: 301 FNSDRVTKAIEKIKAAKNKSSQGRLESFFKPTANPSVPIKRKQ 343
F+ DR+ +KI ++ S+QGRL++FF T S+ KRK+
Sbjct: 316 FSEDRIRNGCKKITKSRQGSTQGRLDTFF--TVTGSISSKRKE 356
>gb|AAT68162.1| flap structure specific endonuclease 1 [Danio rerio]
Length = 380
Score = 401 bits (1031), Expect = e-110
Identities = 201/343 (58%), Positives = 259/343 (74%), Gaps = 4/343 (1%)
Query: 1 MKENKFESYFGRKIAVDASMSIYQFLIVVGRSGTEMLTNEAGEVTSHLQGMFARTIRLLE 60
+KE++ +SYFGRKIA+DASM IYQFLI V + G +L NE GE TSHL GMF RTIR+LE
Sbjct: 18 IKEHEIKSYFGRKIAIDASMCIYQFLIAVRQDGN-VLQNEDGETTSHLMGMFYRTIRMLE 76
Query: 61 AGMKPVYVFDGKPPEMKNQELKKRLSKRAEATAGLTEALEADNKEDIEKFSKRTVKVTKQ 120
+G+KPVYVFDGKPP++K+ EL+KR+ +RAEA L +A EA +E+I+KFSKR VKVTKQ
Sbjct: 77 SGIKPVYVFDGKPPQLKSGELEKRVERRAEAEKLLAQAQEAGEQENIDKFSKRLVKVTKQ 136
Query: 121 HNDDCKRLLRLMGVPVVEAPSEAEAQCAALCKAGKVYAVASEDMDSLTFGAPKFLRHLMD 180
HN++CK+LL LMGVP +EAP EAEA CAAL KAGKVYA A+EDM LTFG LRHL
Sbjct: 137 HNEECKKLLSLMGVPYIEAPCEAEASCAALVKAGKVYATATEDMAGLTFGTTVLLRHLTA 196
Query: 181 PSSKKIPVMEFDVAKILEELDLTMDQFIDLCILSGCDYCDNIRGIGGMTALKLIRQHGSI 240
+KK+P+ EF ++IL++++LT QFIDLCIL GCDYC I+GIG A+ LI+QHGSI
Sbjct: 197 SEAKKLPIQEFHFSRILQDMELTHQQFIDLCILLGCDYCGTIKGIGPKRAIDLIKQHGSI 256
Query: 241 EKILENISKERYQVPDDWPYQEARRLFKEPEVSTDDEVLNLKWSPPDEEGLITFLVNENG 300
E+ILENI +++ P+DW Y+EAR LF EPEV D ++LKW+ PDE+GLI F+ E
Sbjct: 257 EEILENIDPNKHRAPEDWLYKEARGLFLEPEV-VDGTSVDLKWNEPDEDGLIQFMCAEKQ 315
Query: 301 FNSDRVTKAIEKIKAAKNKSSQGRLESFFKPTANPSVPIKRKQ 343
F+ DR+ +KI ++ S+QGRL++FF T S+ KRK+
Sbjct: 316 FSEDRIRNGCKKITKSRQGSTQGRLDTFF--TVTGSISSKRKE 356
>ref|XP_317855.2| ENSANGP00000014920 [Anopheles gambiae str. PEST]
gi|55237026|gb|EAA13029.2| ENSANGP00000014920 [Anopheles
gambiae str. PEST]
Length = 383
Score = 395 bits (1014), Expect = e-108
Identities = 199/342 (58%), Positives = 251/342 (73%), Gaps = 3/342 (0%)
Query: 1 MKENKFESYFGRKIAVDASMSIYQFLIVVGRSGTEMLTNEAGEVTSHLQGMFARTIRLLE 60
+KE + + +FGRK+A+DASM +YQFLI V G + LT+ GE TSHL G F RTIRLLE
Sbjct: 18 VKEGEIKQFFGRKVAIDASMCLYQFLIAVRAEGAQ-LTSVDGETTSHLMGTFYRTIRLLE 76
Query: 61 AGMKPVYVFDGKPPEMKNQELKKRLSKRAEATAGLTEALEADNKEDIEKFSKRTVKVTKQ 120
G+KPVYVFDGKPP++K+ EL KR +R EA L +A EA EDIEKF++R VKVTK
Sbjct: 77 NGIKPVYVFDGKPPDLKSGELNKRAERREEAQKALDKATEAGATEDIEKFNRRLVKVTKH 136
Query: 121 HNDDCKRLLRLMGVPVVEAPSEAEAQCAALCKAGKVYAVASEDMDSLTFGAPKFLRHLMD 180
H ++ K LLRLMGVP VEAP EAEAQCAAL +AGKVYA A+EDMD+LTFG+ LRHL
Sbjct: 137 HANEAKELLRLMGVPYVEAPCEAEAQCAALVRAGKVYATATEDMDALTFGSNILLRHLTF 196
Query: 181 PSSKKIPVMEFDVAKILEELDLTMDQFIDLCILSGCDYCDNIRGIGGMTALKLIRQHGSI 240
++K+PV EF K+L+ +LT D+FIDLCIL GCDYCD IRGIG A++LI +H SI
Sbjct: 197 SEARKMPVQEFAYEKVLKGFELTQDEFIDLCILLGCDYCDTIRGIGPKKAIELINKHRSI 256
Query: 241 EKILENISKERYQVPDDWPYQEARRLFKEPEVSTDDEVLNLKWSPPDEEGLITFLVNENG 300
EKILE++ +++Y VP+ W Y++AR+LFKEPEV D + + LKWS PDEEGL+ FL +
Sbjct: 257 EKILEHLDRQKYIVPEGWNYEQARKLFKEPEVQ-DADTIELKWSEPDEEGLVKFLCGDRQ 315
Query: 301 FNSDRVTKAIEKIKAAKNKSSQGRLESFFKPTANPSVPIKRK 342
FN DR+ +KI KN ++QGRL+SFFK + P KRK
Sbjct: 316 FNEDRIRSGAKKILKTKNTATQGRLDSFFKVLPSTGTP-KRK 356
>emb|CAG31359.1| hypothetical protein [Gallus gallus]
Length = 381
Score = 394 bits (1013), Expect = e-108
Identities = 190/343 (55%), Positives = 260/343 (75%), Gaps = 3/343 (0%)
Query: 1 MKENKFESYFGRKIAVDASMSIYQFLIVVGRSGTEMLTNEAGEVTSHLQGMFARTIRLLE 60
++EN +SYFGRK+A+DASMSIYQFLI V R G E+L NE GE TSHL GMF RTIR++E
Sbjct: 18 IRENDIKSYFGRKVAIDASMSIYQFLIAV-RQGAEVLQNEEGETTSHLMGMFYRTIRMVE 76
Query: 61 AGMKPVYVFDGKPPEMKNQELKKRLSKRAEATAGLTEALEADNKEDIEKFSKRTVKVTKQ 120
G+KPVYVFDGKPP++K+ EL KR +R+EA L EA EA + +IEKFSKR VKVT+Q
Sbjct: 77 NGIKPVYVFDGKPPQLKSGELAKRTERRSEAEKHLQEAQEAGEEANIEKFSKRLVKVTQQ 136
Query: 121 HNDDCKRLLRLMGVPVVEAPSEAEAQCAALCKAGKVYAVASEDMDSLTFGAPKFLRHLMD 180
H D+CK+LL LMG+P VEAP EAEA CA L KAGKVYA A+EDMD LTFG+P +RHL
Sbjct: 137 HTDECKKLLMLMGIPYVEAPGEAEASCATLVKAGKVYAAATEDMDCLTFGSPVLMRHLTA 196
Query: 181 PSSKKIPVMEFDVAKILEELDLTMDQFIDLCILSGCDYCDNIRGIGGMTALKLIRQHGSI 240
+KK+P+ EF + ++L++L LT +QF+DLCIL GCDYC++IRGIG A++LI+QH +I
Sbjct: 197 SEAKKLPIQEFHLNRVLQDLGLTWEQFVDLCILLGCDYCESIRGIGPKRAVELIKQHKTI 256
Query: 241 EKILENISKERYQVPDDWPYQEARRLFKEPEVSTDDEVLNLKWSPPDEEGLITFLVNENG 300
E+I+++I ++Y +P++W ++EA++LF EP+V D+V LKW+ P+EE L+ F+ E
Sbjct: 257 EEIIQHIDTKKYPLPENWLHKEAQKLFLEPDVINPDDV-ELKWTEPNEEELVQFMCGEKQ 315
Query: 301 FNSDRVTKAIEKIKAAKNKSSQGRLESFFKPTANPSVPIKRKQ 343
FN +R+ ++++ ++ S+QGRL+ FFK T + KRK+
Sbjct: 316 FNEERIRNGVKRLSKSRQGSTQGRLDDFFKVTGS-ITSAKRKE 357
>dbj|BAB68507.1| FEN-1 nuclease [Gallus gallus]
Length = 381
Score = 394 bits (1013), Expect = e-108
Identities = 190/343 (55%), Positives = 260/343 (75%), Gaps = 3/343 (0%)
Query: 1 MKENKFESYFGRKIAVDASMSIYQFLIVVGRSGTEMLTNEAGEVTSHLQGMFARTIRLLE 60
++EN +SYFGRK+A+DASMSIYQFLI V R G E+L NE GE TSHL GMF RTIR++E
Sbjct: 18 IRENDIKSYFGRKVAIDASMSIYQFLIAV-RQGAEVLQNEEGETTSHLMGMFYRTIRMVE 76
Query: 61 AGMKPVYVFDGKPPEMKNQELKKRLSKRAEATAGLTEALEADNKEDIEKFSKRTVKVTKQ 120
G+KPVYVFDGKPP++K+ EL KR +R+EA L EA EA + +IEKFSKR VKVT+Q
Sbjct: 77 NGIKPVYVFDGKPPQLKSGELAKRTERRSEAEKHLQEAQEAGEEANIEKFSKRLVKVTQQ 136
Query: 121 HNDDCKRLLRLMGVPVVEAPSEAEAQCAALCKAGKVYAVASEDMDSLTFGAPKFLRHLMD 180
H D+CK+LL LMG+P VEAP EAEA CA L KAGKVYA A+EDMD LTFG+P +RHL
Sbjct: 137 HTDECKKLLMLMGIPYVEAPGEAEASCATLVKAGKVYAAATEDMDCLTFGSPVLMRHLTA 196
Query: 181 PSSKKIPVMEFDVAKILEELDLTMDQFIDLCILSGCDYCDNIRGIGGMTALKLIRQHGSI 240
+KK+P+ EF + ++L++L LT +QF+DLCIL GCDYC++IRGIG A++LI+QH +I
Sbjct: 197 SEAKKLPIQEFHLNRVLQDLGLTWEQFVDLCILLGCDYCESIRGIGPKRAVELIKQHKTI 256
Query: 241 EKILENISKERYQVPDDWPYQEARRLFKEPEVSTDDEVLNLKWSPPDEEGLITFLVNENG 300
E+I+++I ++Y +P++W ++EA++LF EP+V D+V LKW+ P+EE L+ F+ E
Sbjct: 257 EEIIQHIDTKKYPLPENWLHKEAQKLFLEPDVINPDDV-ELKWTEPNEEELVQFMCGEKQ 315
Query: 301 FNSDRVTKAIEKIKAAKNKSSQGRLESFFKPTANPSVPIKRKQ 343
FN +R+ ++++ ++ S+QGRL+ FFK T + KRK+
Sbjct: 316 FNEERIRNGVKRLSKSRQGSTQGRLDDFFKVTGS-ITSAKRKE 357
>gb|AAW55637.1| flap endonuclease-1 [Xiphophorus maculatus]
gi|57639537|gb|AAW55636.1| flap endonuclease-1
[Xiphophorus maculatus]
Length = 380
Score = 394 bits (1011), Expect = e-108
Identities = 200/343 (58%), Positives = 251/343 (72%), Gaps = 4/343 (1%)
Query: 1 MKENKFESYFGRKIAVDASMSIYQFLIVVGRSGTEMLTNEAGEVTSHLQGMFARTIRLLE 60
+KE ++YFGRKIA+DASM IYQFLI V + G +L +E GE TSHL GMF RTIR+LE
Sbjct: 18 IKEQDIKNYFGRKIAIDASMCIYQFLIAVRQDGN-VLQSEDGETTSHLMGMFYRTIRMLE 76
Query: 61 AGMKPVYVFDGKPPEMKNQELKKRLSKRAEATAGLTEALEADNKEDIEKFSKRTVKVTKQ 120
G+KPVYVFDGKPP++K+ EL+KR +RAEA L +A E +E+I+KFSKR VKVTKQ
Sbjct: 77 NGIKPVYVFDGKPPQLKSAELEKRGERRAEAEKMLAKAQELGEQENIDKFSKRLVKVTKQ 136
Query: 121 HNDDCKRLLRLMGVPVVEAPSEAEAQCAALCKAGKVYAVASEDMDSLTFGAPKFLRHLMD 180
HNDDCK+LL LMGVP +EAP EAEA CAAL K GKV+A A+EDMD LTFG LRHL
Sbjct: 137 HNDDCKKLLTLMGVPYIEAPCEAEASCAALVKEGKVFATATEDMDGLTFGTNVLLRHLTA 196
Query: 181 PSSKKIPVMEFDVAKILEELDLTMDQFIDLCILSGCDYCDNIRGIGGMTALKLIRQHGSI 240
+KK+PV EF +IL+++ LT +QFIDLCIL GCDYC I+GIG A+ LIRQHGSI
Sbjct: 197 SEAKKLPVQEFHFNRILQDIGLTSEQFIDLCILLGCDYCGTIKGIGPKRAIDLIRQHGSI 256
Query: 241 EKILENISKERYQVPDDWPYQEARRLFKEPEVSTDDEVLNLKWSPPDEEGLITFLVNENG 300
E+ILENI ++ P+DW Y+EAR LF +PEV D ++LKW PDEE LI F+ +E
Sbjct: 257 EEILENIDTSKHPAPEDWLYKEARNLFLKPEV-VDSSTVDLKWREPDEEALIQFMCSEKQ 315
Query: 301 FNSDRVTKAIEKIKAAKNKSSQGRLESFFKPTANPSVPIKRKQ 343
F+ DR+ +K+ ++ S+QGRL+SFF T S+ KRK+
Sbjct: 316 FSEDRIRNGCKKMMKSRQGSTQGRLDSFFSVTG--SLSSKRKE 356
>gb|AAH83630.1| Fen1 protein [Rattus norvegicus]
Length = 380
Score = 392 bits (1008), Expect = e-108
Identities = 190/343 (55%), Positives = 259/343 (75%), Gaps = 3/343 (0%)
Query: 1 MKENKFESYFGRKIAVDASMSIYQFLIVVGRSGTEMLTNEAGEVTSHLQGMFARTIRLLE 60
++EN +SYFGRK+A+DASMSIYQFLI V R G ++L NE GE TSHL GMF RTIR++E
Sbjct: 18 IRENDIKSYFGRKVAIDASMSIYQFLIAV-RQGGDVLQNEEGETTSHLMGMFYRTIRMME 76
Query: 61 AGMKPVYVFDGKPPEMKNQELKKRLSKRAEATAGLTEALEADNKEDIEKFSKRTVKVTKQ 120
G+KPVY+FDGKPP++K+ EL KR +RAEA L +A EA +E++EKF+KR VKVTKQ
Sbjct: 77 NGIKPVYIFDGKPPQLKSGELAKRSERRAEAEKQLQQAQEAGAEEEVEKFTKRLVKVTKQ 136
Query: 121 HNDDCKRLLRLMGVPVVEAPSEAEAQCAALCKAGKVYAVASEDMDSLTFGAPKFLRHLMD 180
HND+CK LL LMG+P ++APSEAEA CAAL KAGKVYA A+EDMD LTFG+P +RHL
Sbjct: 137 HNDECKHLLSLMGIPYLDAPSEAEASCAALAKAGKVYAAATEDMDCLTFGSPVLMRHLTA 196
Query: 181 PSSKKIPVMEFDVAKILEELDLTMDQFIDLCILSGCDYCDNIRGIGGMTALKLIRQHGSI 240
+KK+P+ EF ++++L+EL L +QF+DLCIL G DYC+++RGIG A+ LI++H SI
Sbjct: 197 SEAKKLPIQEFHLSRVLQELGLNQEQFVDLCILLGSDYCESVRGIGPKRAVDLIQKHKSI 256
Query: 241 EKILENISKERYQVPDDWPYQEARRLFKEPEVSTDDEVLNLKWSPPDEEGLITFLVNENG 300
E+I+ + +Y VP++W ++EAR+LF EPEV D E + LKWS P+EE L+ F+ E
Sbjct: 257 EEIVRRLDPSKYPVPENWLHKEARQLFLEPEV-LDPESVELKWSEPNEEELVKFMCGEKQ 315
Query: 301 FNSDRVTKAIEKIKAAKNKSSQGRLESFFKPTANPSVPIKRKQ 343
F+ +R+ ++++ ++ S+QGRL+ FFK T + S KRK+
Sbjct: 316 FSEERIRSGVKRLNKSRQGSTQGRLDDFFKVTGSLS-SAKRKE 357
>ref|NP_445882.1| flap structure-specific endonuclease [Rattus norvegicus]
gi|8896169|gb|AAF81265.1| flag structure-specific
endonuclease [Rattus norvegicus]
Length = 380
Score = 392 bits (1007), Expect = e-108
Identities = 189/343 (55%), Positives = 259/343 (75%), Gaps = 3/343 (0%)
Query: 1 MKENKFESYFGRKIAVDASMSIYQFLIVVGRSGTEMLTNEAGEVTSHLQGMFARTIRLLE 60
++EN +SYFGRK+A+DASMSIYQFLI V R G ++L NE GE TSHL GMF RTIR++E
Sbjct: 18 IRENDIKSYFGRKVAIDASMSIYQFLIAV-RQGGDVLQNEEGETTSHLMGMFYRTIRMME 76
Query: 61 AGMKPVYVFDGKPPEMKNQELKKRLSKRAEATAGLTEALEADNKEDIEKFSKRTVKVTKQ 120
G+KPVY+FDGKPP++K+ +L KR +RAEA L +A EA +E++EKF+KR VKVTKQ
Sbjct: 77 NGIKPVYIFDGKPPQLKSAQLAKRSERRAEAEKQLQQAQEAGAEEEVEKFTKRLVKVTKQ 136
Query: 121 HNDDCKRLLRLMGVPVVEAPSEAEAQCAALCKAGKVYAVASEDMDSLTFGAPKFLRHLMD 180
HND+CK LL LMG+P ++APSEAEA CAAL KAGKVYA A+EDMD LTFG+P +RHL
Sbjct: 137 HNDECKHLLSLMGIPYLDAPSEAEASCAALAKAGKVYAAATEDMDCLTFGSPVLMRHLTA 196
Query: 181 PSSKKIPVMEFDVAKILEELDLTMDQFIDLCILSGCDYCDNIRGIGGMTALKLIRQHGSI 240
+KK+P+ EF ++++L+EL L +QF+DLCIL G DYC+++RGIG A+ LI++H SI
Sbjct: 197 SEAKKLPIQEFHLSRVLQELGLNQEQFVDLCILLGSDYCESVRGIGPKRAVDLIQKHKSI 256
Query: 241 EKILENISKERYQVPDDWPYQEARRLFKEPEVSTDDEVLNLKWSPPDEEGLITFLVNENG 300
E+I+ + +Y VP++W ++EAR+LF EPEV D E + LKWS P+EE L+ F+ E
Sbjct: 257 EEIVRRLDPSKYPVPENWLHKEARQLFLEPEV-VDPESVELKWSEPNEEELVKFMCGEKQ 315
Query: 301 FNSDRVTKAIEKIKAAKNKSSQGRLESFFKPTANPSVPIKRKQ 343
F+ +R+ ++++ ++ S+QGRL+ FFK T + S KRK+
Sbjct: 316 FSEERIRSGVKRLNKSRQGSTQGRLDDFFKVTGSLS-SAKRKE 357
>ref|NP_032025.2| flap structure specific endonuclease 1 [Mus musculus]
gi|16307328|gb|AAH10203.1| Flap structure specific
endonuclease 1 [Mus musculus]
gi|53791234|dbj|BAD52443.1| flap endonuclease 1 [Mus
musculus] gi|26347483|dbj|BAC37390.1| unnamed protein
product [Mus musculus] gi|26337949|dbj|BAC32660.1|
unnamed protein product [Mus musculus]
Length = 380
Score = 392 bits (1006), Expect = e-107
Identities = 191/343 (55%), Positives = 259/343 (74%), Gaps = 3/343 (0%)
Query: 1 MKENKFESYFGRKIAVDASMSIYQFLIVVGRSGTEMLTNEAGEVTSHLQGMFARTIRLLE 60
++EN +SYFGRK+A+DASMSIYQFLI V R G ++L NE GE TSHL GMF RTIR++E
Sbjct: 18 IRENDIKSYFGRKVAIDASMSIYQFLIAV-RQGGDVLQNEEGETTSHLMGMFYRTIRMME 76
Query: 61 AGMKPVYVFDGKPPEMKNQELKKRLSKRAEATAGLTEALEADNKEDIEKFSKRTVKVTKQ 120
G+KPVYVFDGKPP++K+ EL KR +RAEA L +A EA +E++EKF+KR VKVTKQ
Sbjct: 77 NGIKPVYVFDGKPPQLKSGELAKRSERRAEAEKQLQQAQEAGMEEEVEKFTKRLVKVTKQ 136
Query: 121 HNDDCKRLLRLMGVPVVEAPSEAEAQCAALCKAGKVYAVASEDMDSLTFGAPKFLRHLMD 180
HND+CK LL LMG+P ++APSEAEA CAAL KAGKVYA A+EDMD LTFG+P +RHL
Sbjct: 137 HNDECKHLLSLMGIPYLDAPSEAEASCAALAKAGKVYAAATEDMDCLTFGSPVLMRHLTA 196
Query: 181 PSSKKIPVMEFDVAKILEELDLTMDQFIDLCILSGCDYCDNIRGIGGMTALKLIRQHGSI 240
+KK+P+ EF ++++L+EL L +QF+DLCIL G DYC++IRGIG A+ LI++H SI
Sbjct: 197 SEAKKLPIQEFHLSRVLQELGLNQEQFVDLCILLGSDYCESIRGIGPKRAVDLIQKHKSI 256
Query: 241 EKILENISKERYQVPDDWPYQEARRLFKEPEVSTDDEVLNLKWSPPDEEGLITFLVNENG 300
E+I+ + +Y VP++W ++EA++LF EPEV D E + LKWS P+EE L+ F+ E
Sbjct: 257 EEIVRRLDPSKYPVPENWLHKEAQQLFLEPEV-LDPESVELKWSEPNEEELVKFMCGEKQ 315
Query: 301 FNSDRVTKAIEKIKAAKNKSSQGRLESFFKPTANPSVPIKRKQ 343
F+ +R+ ++++ ++ S+QGRL+ FFK T + S KRK+
Sbjct: 316 FSEERIRSGVKRLSKSRQGSTQGRLDDFFKVTGSLS-SAKRKE 357
>gb|AAB08478.1| XFEN1b [Xenopus laevis]
Length = 382
Score = 392 bits (1006), Expect = e-107
Identities = 195/344 (56%), Positives = 255/344 (73%), Gaps = 3/344 (0%)
Query: 1 MKENKFESYFGRKIAVDASMSIYQFLIVVGRSGTEMLTNEAGEVTSHLQGMFARTIRLLE 60
+KE+ +SYFGRK+AVDASM IYQFLI V + G ML NE GE TSHL GMF RTIR+LE
Sbjct: 18 IKEHDIKSYFGRKVAVDASMCIYQFLIAVRQDGN-MLQNEEGETTSHLMGMFYRTIRMLE 76
Query: 61 AGMKPVYVFDGKPPEMKNQELKKRLSKRAEATAGLTEALEADNKEDIEKFSKRTVKVTKQ 120
G+KPVYVFDGKPP+MK+ EL KR +RAEA L A EA E+IEKF+KR VKVTKQ
Sbjct: 77 HGIKPVYVFDGKPPQMKSGELAKRSERRAEAEKLLEAAEEAGEVENIEKFNKRLVKVTKQ 136
Query: 121 HNDDCKRLLRLMGVPVVEAPSEAEAQCAALCKAGKVYAVASEDMDSLTFGAPKFLRHLMD 180
HN++CK+LL LMG+P V+AP EAEA CAAL KAGKVYA A+EDMD+LTFG P LRHL
Sbjct: 137 HNEECKKLLSLMGIPYVDAPCEAEATCAALVKAGKVYAAATEDMDALTFGTPVLLRHLTA 196
Query: 181 PSSKKIPVMEFDVAKILEELDLTMDQFIDLCILSGCDYCDNIRGIGGMTALKLIRQHGSI 240
+KK+P+ EF + ++ +++ + +QF+DLCIL G DYC+ IRGIG A+ LIRQH +I
Sbjct: 197 SEAKKLPIQEFHLNRVFQDIGINHEQFVDLCILLGSDYCETIRGIGPKRAIDLIRQHKTI 256
Query: 241 EKILENISKERYQVPDDWPYQEARRLFKEPEVSTDDEVLNLKWSPPDEEGLITFLVNENG 300
E+I++NI ++Y +P++W ++EAR+LF EPEV D ++ LKW+ PDEEGL+ F+ E
Sbjct: 257 EEIIDNIDLKKYPIPENWLHKEARQLFLEPEV-IDADITELKWTEPDEEGLVAFMCGEKQ 315
Query: 301 FNSDRVTKAIEKIKAAKNKSSQGRLESFFKPTANPSVPIKRKQL 344
F+ DR+ +K+ + S+QGRL+ FFK T + S KRK++
Sbjct: 316 FSEDRIRNGAKKLAKNRQGSTQGRLDDFFKVTGSIS-STKRKEV 358
>gb|AAH27295.1| Fen1 protein [Mus musculus]
Length = 380
Score = 392 bits (1006), Expect = e-107
Identities = 191/343 (55%), Positives = 259/343 (74%), Gaps = 3/343 (0%)
Query: 1 MKENKFESYFGRKIAVDASMSIYQFLIVVGRSGTEMLTNEAGEVTSHLQGMFARTIRLLE 60
++EN +SYFGRK+A+DASMSIYQFLI V R G ++L NE GE TSHL GMF RTIR++E
Sbjct: 18 IRENDIKSYFGRKVAIDASMSIYQFLIAV-RQGGDVLQNEEGETTSHLMGMFYRTIRMME 76
Query: 61 AGMKPVYVFDGKPPEMKNQELKKRLSKRAEATAGLTEALEADNKEDIEKFSKRTVKVTKQ 120
G+KPVYVFDGKPP++K+ EL KR +RAEA L +A EA +E++EKF+KR VKVTKQ
Sbjct: 77 NGVKPVYVFDGKPPQLKSGELAKRSERRAEAEKQLQQAQEAGMEEEVEKFTKRLVKVTKQ 136
Query: 121 HNDDCKRLLRLMGVPVVEAPSEAEAQCAALCKAGKVYAVASEDMDSLTFGAPKFLRHLMD 180
HND+CK LL LMG+P ++APSEAEA CAAL KAGKVYA A+EDMD LTFG+P +RHL
Sbjct: 137 HNDECKHLLSLMGIPYLDAPSEAEASCAALAKAGKVYAAATEDMDCLTFGSPVLMRHLTA 196
Query: 181 PSSKKIPVMEFDVAKILEELDLTMDQFIDLCILSGCDYCDNIRGIGGMTALKLIRQHGSI 240
+KK+P+ EF ++++L+EL L +QF+DLCIL G DYC++IRGIG A+ LI++H SI
Sbjct: 197 SEAKKLPIQEFHLSRVLQELGLNQEQFVDLCILLGSDYCESIRGIGPKRAVDLIQKHKSI 256
Query: 241 EKILENISKERYQVPDDWPYQEARRLFKEPEVSTDDEVLNLKWSPPDEEGLITFLVNENG 300
E+I+ + +Y VP++W ++EA++LF EPEV D E + LKWS P+EE L+ F+ E
Sbjct: 257 EEIVRRLDPSKYPVPENWLHKEAQQLFLEPEV-LDPESVELKWSEPNEEELVKFMCGEKQ 315
Query: 301 FNSDRVTKAIEKIKAAKNKSSQGRLESFFKPTANPSVPIKRKQ 343
F+ +R+ ++++ ++ S+QGRL+ FFK T + S KRK+
Sbjct: 316 FSEERIRSGVKRLSKSRQGSTQGRLDDFFKVTGSLS-SAKRKE 357
>dbj|BAC36544.1| unnamed protein product [Mus musculus]
Length = 411
Score = 392 bits (1006), Expect = e-107
Identities = 191/343 (55%), Positives = 259/343 (74%), Gaps = 3/343 (0%)
Query: 1 MKENKFESYFGRKIAVDASMSIYQFLIVVGRSGTEMLTNEAGEVTSHLQGMFARTIRLLE 60
++EN +SYFGRK+A+DASMSIYQFLI V R G ++L NE GE TSHL GMF RTIR++E
Sbjct: 18 IRENDIKSYFGRKVAIDASMSIYQFLIAV-RQGGDVLQNEEGETTSHLMGMFYRTIRMME 76
Query: 61 AGMKPVYVFDGKPPEMKNQELKKRLSKRAEATAGLTEALEADNKEDIEKFSKRTVKVTKQ 120
G+KPVYVFDGKPP++K+ EL KR +RAEA L +A EA +E++EKF+KR VKVTKQ
Sbjct: 77 NGIKPVYVFDGKPPQLKSGELAKRSERRAEAEKQLQQAQEAGMEEEVEKFTKRLVKVTKQ 136
Query: 121 HNDDCKRLLRLMGVPVVEAPSEAEAQCAALCKAGKVYAVASEDMDSLTFGAPKFLRHLMD 180
HND+CK LL LMG+P ++APSEAEA CAAL KAGKVYA A+EDMD LTFG+P +RHL
Sbjct: 137 HNDECKHLLSLMGIPYLDAPSEAEASCAALAKAGKVYAAATEDMDCLTFGSPVLMRHLTA 196
Query: 181 PSSKKIPVMEFDVAKILEELDLTMDQFIDLCILSGCDYCDNIRGIGGMTALKLIRQHGSI 240
+KK+P+ EF ++++L+EL L +QF+DLCIL G DYC++IRGIG A+ LI++H SI
Sbjct: 197 SEAKKLPIQEFHLSRVLQELGLNQEQFVDLCILLGSDYCESIRGIGPKRAVDLIQKHKSI 256
Query: 241 EKILENISKERYQVPDDWPYQEARRLFKEPEVSTDDEVLNLKWSPPDEEGLITFLVNENG 300
E+I+ + +Y VP++W ++EA++LF EPEV D E + LKWS P+EE L+ F+ E
Sbjct: 257 EEIVRRLDPSKYPVPENWLHKEAQQLFLEPEV-LDPESVELKWSEPNEEELVKFMCGEKQ 315
Query: 301 FNSDRVTKAIEKIKAAKNKSSQGRLESFFKPTANPSVPIKRKQ 343
F+ +R+ ++++ ++ S+QGRL+ FFK T + S KRK+
Sbjct: 316 FSEERIRSGVKRLSKSRQGSTQGRLDDFFKVTGSLS-SAKRKE 357
>ref|NP_001017005.1| hypothetical protein LOC549759 [Xenopus tropicalis]
Length = 382
Score = 391 bits (1004), Expect = e-107
Identities = 197/344 (57%), Positives = 255/344 (73%), Gaps = 3/344 (0%)
Query: 1 MKENKFESYFGRKIAVDASMSIYQFLIVVGRSGTEMLTNEAGEVTSHLQGMFARTIRLLE 60
+KE+ +SYFGRK+AVDASM IYQFLI V + G ML NE GE TSHL GMF RTIR++E
Sbjct: 18 IKEHDIKSYFGRKVAVDASMCIYQFLIAVRQDGN-MLQNEDGETTSHLMGMFYRTIRMIE 76
Query: 61 AGMKPVYVFDGKPPEMKNQELKKRLSKRAEATAGLTEALEADNKEDIEKFSKRTVKVTKQ 120
G+KPVYVFDGKPP+MK+ EL KR +RAEA L A EA E+IEKF+KR VKVTKQ
Sbjct: 77 HGIKPVYVFDGKPPQMKSGELAKRSERRAEAEKLLEAAEEAGEVENIEKFNKRLVKVTKQ 136
Query: 121 HNDDCKRLLRLMGVPVVEAPSEAEAQCAALCKAGKVYAVASEDMDSLTFGAPKFLRHLMD 180
HN++CK+LL LMGVP V+AP EAEA CAAL KAGKVYA A+EDMD+LTFG P LRHL
Sbjct: 137 HNEECKKLLTLMGVPYVDAPCEAEATCAALVKAGKVYAAATEDMDALTFGTPVLLRHLTA 196
Query: 181 PSSKKIPVMEFDVAKILEELDLTMDQFIDLCILSGCDYCDNIRGIGGMTALKLIRQHGSI 240
+KK+P+ EF + ++++++ ++ +QF+DLCIL G DYC+ IRGIG A+ LIRQH SI
Sbjct: 197 SEAKKLPIQEFHLNRVMQDMGVSHEQFVDLCILLGSDYCETIRGIGPKRAIDLIRQHKSI 256
Query: 241 EKILENISKERYQVPDDWPYQEARRLFKEPEVSTDDEVLNLKWSPPDEEGLITFLVNENG 300
E+I++NI ++Y +P++W ++EAR+LF EPEV D E LKW PDEEGL+ F+ E
Sbjct: 257 EEIVDNIDLKKYPIPENWLHKEARQLFLEPEV-VDTESTELKWIEPDEEGLVAFMCAEKQ 315
Query: 301 FNSDRVTKAIEKIKAAKNKSSQGRLESFFKPTANPSVPIKRKQL 344
F+ DR+ +K+ + S+QGRL+ FFK T + S KRK++
Sbjct: 316 FSEDRIRNGAKKLSKNRQGSTQGRLDDFFKVTGSIS-STKRKEV 358
>dbj|BAC31412.1| unnamed protein product [Mus musculus]
Length = 380
Score = 389 bits (1000), Expect = e-107
Identities = 190/343 (55%), Positives = 258/343 (74%), Gaps = 3/343 (0%)
Query: 1 MKENKFESYFGRKIAVDASMSIYQFLIVVGRSGTEMLTNEAGEVTSHLQGMFARTIRLLE 60
++EN +SYFGRK+A+DASMSIYQFLI V R G ++L NE GE TSHL GMF RTIR++E
Sbjct: 18 IRENDIKSYFGRKVAIDASMSIYQFLIAV-RQGGDVLQNEEGETTSHLMGMFYRTIRMME 76
Query: 61 AGMKPVYVFDGKPPEMKNQELKKRLSKRAEATAGLTEALEADNKEDIEKFSKRTVKVTKQ 120
G+KPVYV DGKPP++K+ EL KR +RAEA L +A EA +E++EKF+KR VKVTKQ
Sbjct: 77 NGIKPVYVLDGKPPQLKSGELAKRSERRAEAEKQLQQAQEAGMEEEVEKFTKRLVKVTKQ 136
Query: 121 HNDDCKRLLRLMGVPVVEAPSEAEAQCAALCKAGKVYAVASEDMDSLTFGAPKFLRHLMD 180
HND+CK LL LMG+P ++APSEAEA CAAL KAGKVYA A+EDMD LTFG+P +RHL
Sbjct: 137 HNDECKHLLSLMGIPYLDAPSEAEASCAALAKAGKVYAAATEDMDCLTFGSPVLMRHLTA 196
Query: 181 PSSKKIPVMEFDVAKILEELDLTMDQFIDLCILSGCDYCDNIRGIGGMTALKLIRQHGSI 240
+KK+P+ EF ++++L+EL L +QF+DLCIL G DYC++IRGIG A+ LI++H SI
Sbjct: 197 SEAKKLPIQEFHLSRVLQELGLNQEQFVDLCILLGSDYCESIRGIGPKRAVDLIQKHKSI 256
Query: 241 EKILENISKERYQVPDDWPYQEARRLFKEPEVSTDDEVLNLKWSPPDEEGLITFLVNENG 300
E+I+ + +Y VP++W ++EA++LF EPEV D E + LKWS P+EE L+ F+ E
Sbjct: 257 EEIVRRLDPSKYPVPENWLHKEAQQLFLEPEV-LDPESVELKWSEPNEEELVKFMCGEKQ 315
Query: 301 FNSDRVTKAIEKIKAAKNKSSQGRLESFFKPTANPSVPIKRKQ 343
F+ +R+ ++++ ++ S+QGRL+ FFK T + S KRK+
Sbjct: 316 FSEERIRSGVKRLSKSRQGSTQGRLDDFFKVTGSLS-SAKRKE 357
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.318 0.135 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 597,431,520
Number of Sequences: 2540612
Number of extensions: 24567548
Number of successful extensions: 73884
Number of sequences better than 10.0: 556
Number of HSP's better than 10.0 without gapping: 333
Number of HSP's successfully gapped in prelim test: 223
Number of HSP's that attempted gapping in prelim test: 72907
Number of HSP's gapped (non-prelim): 689
length of query: 363
length of database: 863,360,394
effective HSP length: 129
effective length of query: 234
effective length of database: 535,621,446
effective search space: 125335418364
effective search space used: 125335418364
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 76 (33.9 bits)
Medicago: description of AC148406.5


