

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148406.1 + phase: 0
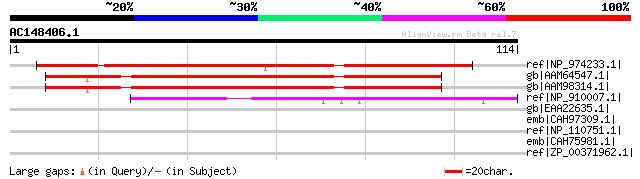
(114 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_974233.1| expressed protein [Arabidopsis thaliana] 81 5e-15
gb|AAM64547.1| unknown [Arabidopsis thaliana] 78 4e-14
gb|AAM98314.1| At5g26741/At5g26741 [Arabidopsis thaliana] gi|184... 78 4e-14
ref|NP_910007.1| hypothetical protein [Oryza sativa] gi|18855055... 45 3e-04
gb|EAA22635.1| hypothetical protein [Plasmodium yoelii yoelii] 35 0.33
emb|CAH97309.1| Pb-fam-2 protein [Plasmodium berghei] 33 1.6
ref|NP_110751.1| NADH:flavin oxidoreductase [Thermoplasma volcan... 32 3.7
emb|CAH75981.1| conserved hypothetical protein [Plasmodium chaba... 32 4.8
ref|ZP_00371962.1| conserved hypothetical protein [Campylobacter... 31 8.1
>ref|NP_974233.1| expressed protein [Arabidopsis thaliana]
Length = 205
Score = 81.3 bits (199), Expect = 5e-15
Identities = 48/99 (48%), Positives = 69/99 (69%), Gaps = 4/99 (4%)
Query: 7 NPLFRTESFGYYNFPDQNYTMIEKRQLFLRSYQFCRKKSLTERIKGSLVR-AKKVVWLKL 65
N ++++ S GYY++ +EKRQLFL+SYQF RK+S TE+IK S+ R KKVV ++L
Sbjct: 107 NGVWQSNSNGYYSY-GYGGGYVEKRQLFLKSYQFSRKQSFTEKIKISMKRVVKKVVCMRL 165
Query: 66 RYARGLRRKLVFFPRFKCGFYYRRRRFSQLLNNSGSSHH 104
+ AR L+R V + R K F+YRRRRF +LL+ + SS +
Sbjct: 166 KSARRLKR--VVWSRLKTAFFYRRRRFLRLLHPNKSSSY 202
>gb|AAM64547.1| unknown [Arabidopsis thaliana]
Length = 99
Score = 78.2 bits (191), Expect = 4e-14
Identities = 46/90 (51%), Positives = 60/90 (66%), Gaps = 5/90 (5%)
Query: 9 LFRTESFG-YYNFPDQNYTMIEKRQLFLRSYQFCRKKSLTERIKGSLVRAKKVVWLKLRY 67
+FR+ S G YY P Y +EKRQLFLRSYQF RK+S E++ S+ R K+VV +LR
Sbjct: 6 VFRSTSGGDYYGAPYGEY--MEKRQLFLRSYQFSRKQSFAEKVSRSVTRVKRVVLTRLRS 63
Query: 68 ARGLRRKLVFFPRFKCGFYYRRRRFSQLLN 97
A L+R V + R + FYYRRRRF +LL+
Sbjct: 64 APKLKR--VVWSRLRSAFYYRRRRFFRLLH 91
>gb|AAM98314.1| At5g26741/At5g26741 [Arabidopsis thaliana]
gi|18421036|ref|NP_568485.1| expressed protein
[Arabidopsis thaliana] gi|14488090|gb|AAK63865.1|
AT5g26741 [Arabidopsis thaliana]
Length = 99
Score = 78.2 bits (191), Expect = 4e-14
Identities = 46/90 (51%), Positives = 60/90 (66%), Gaps = 5/90 (5%)
Query: 9 LFRTESFG-YYNFPDQNYTMIEKRQLFLRSYQFCRKKSLTERIKGSLVRAKKVVWLKLRY 67
+FR+ S G YY P Y +EKRQLFLRSYQF RK+S E++ S+ R K+VV +LR
Sbjct: 6 VFRSTSGGDYYGAPYGEY--MEKRQLFLRSYQFSRKQSFAEKVSRSVTRVKRVVLTRLRS 63
Query: 68 ARGLRRKLVFFPRFKCGFYYRRRRFSQLLN 97
A L+R V + R + FYYRRRRF +LL+
Sbjct: 64 APKLKR--VVWSRLRSAFYYRRRRFFRLLH 91
>ref|NP_910007.1| hypothetical protein [Oryza sativa] gi|18855055|gb|AAL79747.1|
hypothetical protein [Oryza sativa]
Length = 125
Score = 45.4 bits (106), Expect = 3e-04
Identities = 37/101 (36%), Positives = 49/101 (47%), Gaps = 19/101 (18%)
Query: 28 IEKRQLFLRSYQFCRKKSLTERIKGSLVRAKKVVWLKLRYAR-----GLRR-----KLVF 77
+E+RQLFLRSY F R L+ R RA++VVW+ LR R GLRR +L F
Sbjct: 30 VERRQLFLRSYHFSRDVELSPR-----ARARRVVWVGLRRLRRAAATGLRRLRARLRLCF 84
Query: 78 -FPRFKCGFYYRRRRFSQLLNNSGSSHHR---KVDSSSCLW 114
+ + + R RF + SG + H SS C W
Sbjct: 85 AWVSRRRNIHRRGARFGRYGRLSGGAAHAPAPAASSSVCFW 125
>gb|EAA22635.1| hypothetical protein [Plasmodium yoelii yoelii]
Length = 610
Score = 35.4 bits (80), Expect = 0.33
Identities = 16/39 (41%), Positives = 28/39 (71%), Gaps = 1/39 (2%)
Query: 49 RIKGSLVRAKKVVWLKLRYARGLRRKLVFFPRFKCGFYY 87
RIK +R KK +++K+ + RG+R+KL+ F + K GF++
Sbjct: 44 RIKSKDIRFKKKIFVKVPF-RGIRQKLLLFDKLKNGFFF 81
>emb|CAH97309.1| Pb-fam-2 protein [Plasmodium berghei]
Length = 162
Score = 33.1 bits (74), Expect = 1.6
Identities = 14/39 (35%), Positives = 27/39 (68%), Gaps = 1/39 (2%)
Query: 49 RIKGSLVRAKKVVWLKLRYARGLRRKLVFFPRFKCGFYY 87
R+K +R KK +++K+ + RG+R+KL+ F + K G ++
Sbjct: 44 RVKSKDIRFKKNIYVKIPF-RGIRQKLLLFDKLKNGLFF 81
>ref|NP_110751.1| NADH:flavin oxidoreductase [Thermoplasma volcanium GSS1]
gi|14324447|dbj|BAB59375.1| NADH oxidase [Thermoplasma
volcanium GSS1]
Length = 547
Score = 32.0 bits (71), Expect = 3.7
Identities = 22/69 (31%), Positives = 34/69 (48%), Gaps = 12/69 (17%)
Query: 16 GYYNFPDQNYTMI-EKRQLFLRS-----------YQFCRKKSLTERIKGSLVRAKKVVWL 63
G Y PD+ Y + EK ++F+ S Y K +T R SL RA+K +L
Sbjct: 436 GIYCLPDKTYPDLQEKPEIFIDSNVYMYHDIALKYSSTSKVFVTYRSLSSLDRARKAAFL 495
Query: 64 KLRYARGLR 72
K+ +RG++
Sbjct: 496 KIAESRGIK 504
>emb|CAH75981.1| conserved hypothetical protein [Plasmodium chabaudi]
Length = 251
Score = 31.6 bits (70), Expect = 4.8
Identities = 14/39 (35%), Positives = 27/39 (68%), Gaps = 1/39 (2%)
Query: 49 RIKGSLVRAKKVVWLKLRYARGLRRKLVFFPRFKCGFYY 87
RI+ + +R KK +++K+ RG+R+KL+ F + K G ++
Sbjct: 15 RIRNNNIRFKKKIFVKVP-VRGIRQKLLLFDKLKNGLFF 52
>ref|ZP_00371962.1| conserved hypothetical protein [Campylobacter upsaliensis RM3195]
gi|57015647|gb|EAL52438.1| conserved hypothetical
protein [Campylobacter upsaliensis RM3195]
Length = 365
Score = 30.8 bits (68), Expect = 8.1
Identities = 23/64 (35%), Positives = 31/64 (47%), Gaps = 13/64 (20%)
Query: 32 QLFLRSYQFCRKKSLTERIKGSLVRAKKVVWLKLRYARGLRRKLVFFPRFKCGFYYRRRR 91
+LFLR +F + AK+ V+LK+ AR LR KL+ F RF F Y + R
Sbjct: 259 ELFLREGEFYKS-------------AKEEVFLKVFVARILRLKLIEFKRFYENFSYEKFR 305
Query: 92 FSQL 95
L
Sbjct: 306 LKCL 309
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.329 0.140 0.442
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 186,309,110
Number of Sequences: 2540612
Number of extensions: 6193179
Number of successful extensions: 13477
Number of sequences better than 10.0: 9
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 13465
Number of HSP's gapped (non-prelim): 9
length of query: 114
length of database: 863,360,394
effective HSP length: 90
effective length of query: 24
effective length of database: 634,705,314
effective search space: 15232927536
effective search space used: 15232927536
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 68 (30.8 bits)
Medicago: description of AC148406.1


