

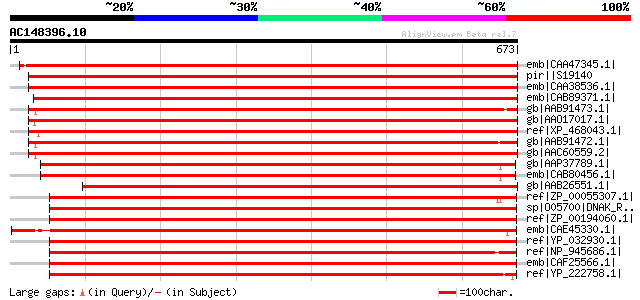
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148396.10 + phase: 0 /partial
(673 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAA47345.1| 70 kDa heat shock protein [Phaseolus vulgaris] g... 1164 0.0
pir||S19140 dnaK-type molecular chaperone PHSP1 precursor, mitoc... 1154 0.0
emb|CAA38536.1| HSP70 [Pisum sativum] gi|585272|sp|P37900|HSP7M_... 1152 0.0
emb|CAB89371.1| heat shock protein 70 (Hsc70-5) [Arabidopsis tha... 1087 0.0
gb|AAB91473.1| heat shock 70 protein [Spinacia oleracea] gi|2773... 1078 0.0
gb|AAO17017.1| Putative heat shock 70 KD protein, mitochondrial ... 1077 0.0
ref|XP_468043.1| putative dnaK-type molecular chaperone precurso... 1073 0.0
gb|AAB91472.1| heat shock 70 protein [Spinacia oleracea] gi|7441... 1073 0.0
gb|AAC60559.2| HSP68 [Solanum tuberosum] gi|7441872|pir||T07024 ... 1057 0.0
gb|AAP37789.1| At4g37910 [Arabidopsis thaliana] gi|27311569|gb|A... 1052 0.0
emb|CAB80456.1| heat shock protein 70 like protein [Arabidopsis ... 1052 0.0
gb|AAB26551.1| HSP68=68 kda heat-stress DnaK homolog [Lycopersic... 998 0.0
ref|ZP_00055307.1| COG0443: Molecular chaperone [Magnetospirillu... 846 0.0
sp|O05700|DNAK_RHOS7 Chaperone protein dnaK (Heat shock protein ... 842 0.0
ref|ZP_00194060.1| COG0443: Molecular chaperone [Mesorhizobium s... 842 0.0
emb|CAE45330.1| unnamed protein product [Magnetospirillum gryphi... 841 0.0
ref|YP_032930.1| Heat shock protein 70 DnaK [Bartonella henselae... 841 0.0
ref|NP_945686.1| heat shock protein DnaK (70) [Rhodopseudomonas ... 840 0.0
emb|CAF25566.1| Heat shock protein 70 DnaK [Bartonella quintana ... 837 0.0
ref|YP_222758.1| chaperone protein DnaK [Brucella abortus biovar... 837 0.0
>emb|CAA47345.1| 70 kDa heat shock protein [Phaseolus vulgaris]
gi|100004|pir||S25005 dnaK-type molecular chaperone
precursor, mitochondrial - kidney bean
gi|399940|sp|Q01899|HSP7M_PHAVU Heat shock 70 kDa
protein, mitochondrial precursor
Length = 675
Score = 1164 bits (3012), Expect = 0.0
Identities = 602/663 (90%), Positives = 630/663 (94%), Gaps = 3/663 (0%)
Query: 13 IVSQTYSDVVLQLTGNTKPAYVGHNWSSLSRPFSSRAAGNDVIGIDLGTTNSCVSVMEGK 72
+ S T+S LTG+TKPAYV WS L+RPFSSR AGNDVIGIDLGTTNSCVSVMEGK
Sbjct: 13 VASATFS-AYRSLTGSTKPAYVAQKWSCLARPFSSRPAGNDVIGIDLGTTNSCVSVMEGK 71
Query: 73 NPKVVENSEGARTTPSVVAFTQKGELLVGTPAKRQAVTNPENTISGAKRLIGRRFDDPQT 132
NPKV+ENSEGARTTPSVVAF QKGELLVGTPAKRQAVTNP NT+ G KRLIGRRFDDPQT
Sbjct: 72 NPKVIENSEGARTTPSVVAFNQKGELLVGTPAKRQAVTNPTNTVFGTKRLIGRRFDDPQT 131
Query: 133 QKEMKMVPYKIVKAPNGDAWVEAKGQQYSPSQIGAFVLTKMKETAEAYLGKTVPKAVITV 192
QKEMKMVP+KIVKAPNGDAWVEA GQQYSPSQIGAFVLTKMKETAEAYLGK+V KAVITV
Sbjct: 132 QKEMKMVPFKIVKAPNGDAWVEANGQQYSPSQIGAFVLTKMKETAEAYLGKSVSKAVITV 191
Query: 193 PAYFNDAQRQATKDAGRIAGLEVLRIINEPTAAALSYGMN-KEGLIAVFDLGGGTFDVSI 251
PAYFNDAQRQATKDAGRIAGL+V RIINEPTAAALSYGMN KEGLIAVFDLGGGTFDVSI
Sbjct: 192 PAYFNDAQRQATKDAGRIAGLDVQRIINEPTAAALSYGMNNKEGLIAVFDLGGGTFDVSI 251
Query: 252 LEISNGVFEVKATNGDTFLGGEDFDNALLDFLVNEFKRSESIDLSKDKLALQRLREAAEK 311
LEISNGVFEVKATNGDTFLGGEDFDNALLDFLVNEFKR+ESIDLSKD+LALQRLREAAEK
Sbjct: 252 LEISNGVFEVKATNGDTFLGGEDFDNALLDFLVNEFKRTESIDLSKDRLALQRLREAAEK 311
Query: 312 AKIELSSTSQTEINLPFITADASGAKHLNITLTRSKFEALVNNLIERTKAPCQSCLKDAN 371
AKIELSSTSQTEINLPFITADASGAKHLNITLTRSKFEALVN+LIERTKAPC+SCLKDAN
Sbjct: 312 AKIELSSTSQTEINLPFITADASGAKHLNITLTRSKFEALVNHLIERTKAPCKSCLKDAN 371
Query: 372 ISTKDIDEVLLVGGMTRVPKVQEVVSTIFGKSPCKGVNPDEAVAMGAALQGGILRGDVKE 431
+S KD+DEVLLVGGMTRVPKVQEVV IFGKSP KGVNPDEAVAMGAA+QGGILRGDVKE
Sbjct: 372 VSIKDVDEVLLVGGMTRVPKVQEVVLNIFGKSPSKGVNPDEAVAMGAAIQGGILRGDVKE 431
Query: 432 LLLLDVTPLSLGIETLGGIFTRLINRNTTIPTKKSQVFSTAADNQTQVGIKVLQGEREMA 491
LLLLDVTPLSLGIETLGGIFTRLINRNTTIPTKKSQVFSTAADNQTQVGIKVLQGEREMA
Sbjct: 432 LLLLDVTPLSLGIETLGGIFTRLINRNTTIPTKKSQVFSTAADNQTQVGIKVLQGEREMA 491
Query: 492 SDNKMLGEFELVGLPPAPRGMPQIEVTFDIDANGIVTVSAKDKSTGKEQQITIKSSGGLS 551
SDNKMLGEF+LVG+PPAPRG+PQIEVTFDIDANGIVTVSAKDKSTGKEQQITI+SSGGLS
Sbjct: 492 SDNKMLGEFDLVGIPPAPRGLPQIEVTFDIDANGIVTVSAKDKSTGKEQQITIRSSGGLS 551
Query: 552 DDEIQNMVKEAELHAQKDQERKSLIDIRNSADTSIYSIEKSLSEYREKIPAEVAKEIEDA 611
+DEI+ MVKEAELHAQKDQERK+LIDIRNSADT+IYSIEKSL EYREKIP+E AKEIEDA
Sbjct: 552 EDEIEKMVKEAELHAQKDQERKTLIDIRNSADTTIYSIEKSLGEYREKIPSETAKEIEDA 611
Query: 612 VSDLRSAMAGDSADEIKSKLDAANKAVSKIGEHMSGGSSSGPSSDGSQ-GGDQAPEAEYE 670
VSDLR AM+GD+ DEIKSKLDAANKAVSKIGEHMSGGSS G S+ GSQ GGDQAPEAEYE
Sbjct: 612 VSDLRKAMSGDNVDEIKSKLDAANKAVSKIGEHMSGGSSGGSSAGGSQGGGDQAPEAEYE 671
Query: 671 EVK 673
EVK
Sbjct: 672 EVK 674
>pir||S19140 dnaK-type molecular chaperone PHSP1 precursor, mitochondrial -
garden pea
Length = 675
Score = 1154 bits (2984), Expect = 0.0
Identities = 592/650 (91%), Positives = 625/650 (96%), Gaps = 1/650 (0%)
Query: 25 LTGNTKPAYVGHNWSSLSRPFSSRAAGNDVIGIDLGTTNSCVSVMEGKNPKVVENSEGAR 84
LTG+TK +Y H +SL+RPFSSR AGNDVIGIDLGTTNSCVSVMEGKNPKV+ENSEGAR
Sbjct: 25 LTGSTKTSYATHKLASLTRPFSSRPAGNDVIGIDLGTTNSCVSVMEGKNPKVIENSEGAR 84
Query: 85 TTPSVVAFTQKGELLVGTPAKRQAVTNPENTISGAKRLIGRRFDDPQTQKEMKMVPYKIV 144
TTPSVVAF QK ELLVGTPAKRQAVTNP NT+ G KRLIGRRFDD QTQKEMKMVPYKIV
Sbjct: 85 TTPSVVAFNQKSELLVGTPAKRQAVTNPTNTLFGTKRLIGRRFDDAQTQKEMKMVPYKIV 144
Query: 145 KAPNGDAWVEAKGQQYSPSQIGAFVLTKMKETAEAYLGKTVPKAVITVPAYFNDAQRQAT 204
+APNGDAWVEA GQQYSPSQIGAFVLTKMKETAEAYLGKT+ KAV+TVPAYFNDAQRQAT
Sbjct: 145 RAPNGDAWVEANGQQYSPSQIGAFVLTKMKETAEAYLGKTISKAVVTVPAYFNDAQRQAT 204
Query: 205 KDAGRIAGLEVLRIINEPTAAALSYGMN-KEGLIAVFDLGGGTFDVSILEISNGVFEVKA 263
KDAGRIAGL+V RIINEPTAAALSYGMN KEGLIAVFDLGGGTFDVSILEISNGVFEVKA
Sbjct: 205 KDAGRIAGLDVQRIINEPTAAALSYGMNNKEGLIAVFDLGGGTFDVSILEISNGVFEVKA 264
Query: 264 TNGDTFLGGEDFDNALLDFLVNEFKRSESIDLSKDKLALQRLREAAEKAKIELSSTSQTE 323
TNGDTFLGGEDFDNALLDFLV+EFKR+ESIDL+KDKLALQRLREAAEKAKIELSSTSQTE
Sbjct: 265 TNGDTFLGGEDFDNALLDFLVSEFKRTESIDLAKDKLALQRLREAAEKAKIELSSTSQTE 324
Query: 324 INLPFITADASGAKHLNITLTRSKFEALVNNLIERTKAPCQSCLKDANISTKDIDEVLLV 383
INLPFI+ADASGAKHLNITLTRSKFEALVNNLIERTKAPC+SCLKDANIS KD+DEVLLV
Sbjct: 325 INLPFISADASGAKHLNITLTRSKFEALVNNLIERTKAPCKSCLKDANISIKDVDEVLLV 384
Query: 384 GGMTRVPKVQEVVSTIFGKSPCKGVNPDEAVAMGAALQGGILRGDVKELLLLDVTPLSLG 443
GGMTRVPKVQ+VVS IFGKSP KGVNPDEAVAMGAALQGGILRGDVKELLLLDVTPLSLG
Sbjct: 385 GGMTRVPKVQQVVSEIFGKSPSKGVNPDEAVAMGAALQGGILRGDVKELLLLDVTPLSLG 444
Query: 444 IETLGGIFTRLINRNTTIPTKKSQVFSTAADNQTQVGIKVLQGEREMASDNKMLGEFELV 503
IETLGGIFTRLI+RNTTIPTKKSQVFSTAADNQTQVGIKVLQGEREMA+DNK LGEF+LV
Sbjct: 445 IETLGGIFTRLISRNTTIPTKKSQVFSTAADNQTQVGIKVLQGEREMAADNKSLGEFDLV 504
Query: 504 GLPPAPRGMPQIEVTFDIDANGIVTVSAKDKSTGKEQQITIKSSGGLSDDEIQNMVKEAE 563
G+PPAPRG+PQIEVTFDIDANGIVTVSAKDKSTGKEQQITI+SSGGLSDDEI MVKEAE
Sbjct: 505 GIPPAPRGLPQIEVTFDIDANGIVTVSAKDKSTGKEQQITIRSSGGLSDDEIDKMVKEAE 564
Query: 564 LHAQKDQERKSLIDIRNSADTSIYSIEKSLSEYREKIPAEVAKEIEDAVSDLRSAMAGDS 623
LHAQ+DQERK+LIDIRNSADTSIYSIEKSL+EYREKIPAEVAKEIEDAVSDLR+AMAG++
Sbjct: 565 LHAQRDQERKALIDIRNSADTSIYSIEKSLAEYREKIPAEVAKEIEDAVSDLRTAMAGEN 624
Query: 624 ADEIKSKLDAANKAVSKIGEHMSGGSSSGPSSDGSQGGDQAPEAEYEEVK 673
AD+IK+KLDAANKAVSKIG+HMSGGSS GPS GSQGG+QAPEAEYEEVK
Sbjct: 625 ADDIKAKLDAANKAVSKIGQHMSGGSSGGPSEGGSQGGEQAPEAEYEEVK 674
>emb|CAA38536.1| HSP70 [Pisum sativum] gi|585272|sp|P37900|HSP7M_PEA Heat shock 70
kDa protein, mitochondrial precursor
Length = 675
Score = 1152 bits (2980), Expect = 0.0
Identities = 591/650 (90%), Positives = 625/650 (95%), Gaps = 1/650 (0%)
Query: 25 LTGNTKPAYVGHNWSSLSRPFSSRAAGNDVIGIDLGTTNSCVSVMEGKNPKVVENSEGAR 84
LTG+TK +Y H +SL+RPFSSR AGNDVIGIDLGTTNSCVSVMEGKNPKV+ENSEGAR
Sbjct: 25 LTGSTKTSYATHKLASLTRPFSSRPAGNDVIGIDLGTTNSCVSVMEGKNPKVIENSEGAR 84
Query: 85 TTPSVVAFTQKGELLVGTPAKRQAVTNPENTISGAKRLIGRRFDDPQTQKEMKMVPYKIV 144
TTPSVVAF QK ELLVGTPAKRQAVTNP NT+ G KRLIGRRFDD QTQKEMKMVPYKIV
Sbjct: 85 TTPSVVAFNQKSELLVGTPAKRQAVTNPTNTLFGTKRLIGRRFDDAQTQKEMKMVPYKIV 144
Query: 145 KAPNGDAWVEAKGQQYSPSQIGAFVLTKMKETAEAYLGKTVPKAVITVPAYFNDAQRQAT 204
+APNGDAWVEA GQQYSPSQIGAFVLTK+KETAEAYLGKT+ KAV+TVPAYFNDAQRQAT
Sbjct: 145 RAPNGDAWVEANGQQYSPSQIGAFVLTKIKETAEAYLGKTISKAVVTVPAYFNDAQRQAT 204
Query: 205 KDAGRIAGLEVLRIINEPTAAALSYGMN-KEGLIAVFDLGGGTFDVSILEISNGVFEVKA 263
KDAGRIAGL+V RIINEPTAAALSYGMN KEGLIAVFDLGGGTFDVSILEISNGVFEVKA
Sbjct: 205 KDAGRIAGLDVQRIINEPTAAALSYGMNNKEGLIAVFDLGGGTFDVSILEISNGVFEVKA 264
Query: 264 TNGDTFLGGEDFDNALLDFLVNEFKRSESIDLSKDKLALQRLREAAEKAKIELSSTSQTE 323
TNGDTFLGGEDFDNALLDFLV+EFKR+ESIDL+KDKLALQRLREAAEKAKIELSSTSQTE
Sbjct: 265 TNGDTFLGGEDFDNALLDFLVSEFKRTESIDLAKDKLALQRLREAAEKAKIELSSTSQTE 324
Query: 324 INLPFITADASGAKHLNITLTRSKFEALVNNLIERTKAPCQSCLKDANISTKDIDEVLLV 383
INLPFI+ADASGAKHLNITLTRSKFEALVNNLIERTKAPC+SCLKDANIS KD+DEVLLV
Sbjct: 325 INLPFISADASGAKHLNITLTRSKFEALVNNLIERTKAPCKSCLKDANISIKDVDEVLLV 384
Query: 384 GGMTRVPKVQEVVSTIFGKSPCKGVNPDEAVAMGAALQGGILRGDVKELLLLDVTPLSLG 443
GGMTRVPKVQ+VVS IFGKSP KGVNPDEAVAMGAALQGGILRGDVKELLLLDVTPLSLG
Sbjct: 385 GGMTRVPKVQQVVSEIFGKSPSKGVNPDEAVAMGAALQGGILRGDVKELLLLDVTPLSLG 444
Query: 444 IETLGGIFTRLINRNTTIPTKKSQVFSTAADNQTQVGIKVLQGEREMASDNKMLGEFELV 503
IETLGGIFTRLI+RNTTIPTKKSQVFSTAADNQTQVGIKVLQGEREMA+DNK LGEF+LV
Sbjct: 445 IETLGGIFTRLISRNTTIPTKKSQVFSTAADNQTQVGIKVLQGEREMAADNKSLGEFDLV 504
Query: 504 GLPPAPRGMPQIEVTFDIDANGIVTVSAKDKSTGKEQQITIKSSGGLSDDEIQNMVKEAE 563
G+PPAPRG+PQIEVTFDIDANGIVTVSAKDKSTGKEQQITI+SSGGLSDDEI MVKEAE
Sbjct: 505 GIPPAPRGLPQIEVTFDIDANGIVTVSAKDKSTGKEQQITIRSSGGLSDDEIDKMVKEAE 564
Query: 564 LHAQKDQERKSLIDIRNSADTSIYSIEKSLSEYREKIPAEVAKEIEDAVSDLRSAMAGDS 623
LHAQ+DQERK+LIDIRNSADTSIYSIEKSL+EYREKIPAEVAKEIEDAVSDLR+AMAG++
Sbjct: 565 LHAQRDQERKALIDIRNSADTSIYSIEKSLAEYREKIPAEVAKEIEDAVSDLRTAMAGEN 624
Query: 624 ADEIKSKLDAANKAVSKIGEHMSGGSSSGPSSDGSQGGDQAPEAEYEEVK 673
AD+IK+KLDAANKAVSKIG+HMSGGSS GPS GSQGG+QAPEAEYEEVK
Sbjct: 625 ADDIKAKLDAANKAVSKIGQHMSGGSSGGPSEGGSQGGEQAPEAEYEEVK 674
>emb|CAB89371.1| heat shock protein 70 (Hsc70-5) [Arabidopsis thaliana]
gi|15242459|ref|NP_196521.1| heat shock protein 70 /
HSP70 (HSC70-5) [Arabidopsis thaliana]
gi|6746590|gb|AAF27638.1| heat shock protein 70
[Arabidopsis thaliana] gi|11277110|pir||T49939 heat
shock protein 70 (Hsc70-5) - Arabidopsis thaliana
Length = 682
Score = 1087 bits (2812), Expect = 0.0
Identities = 552/645 (85%), Positives = 606/645 (93%), Gaps = 3/645 (0%)
Query: 32 AYVGHNWSSLSRPFSSRAAGNDVIGIDLGTTNSCVSVMEGKNPKVVENSEGARTTPSVVA 91
+Y+G N+ S SR FSS+ AGNDVIGIDLGTTNSCV+VMEGKNPKV+EN+EGARTTPSVVA
Sbjct: 37 SYLGQNFRSFSRAFSSKPAGNDVIGIDLGTTNSCVAVMEGKNPKVIENAEGARTTPSVVA 96
Query: 92 FTQKGELLVGTPAKRQAVTNPENTISGAKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDA 151
F KGELLVGTPAKRQAVTNP NT+SG KRLIGR+FDDPQTQKEMKMVPYKIV+APNGDA
Sbjct: 97 FNTKGELLVGTPAKRQAVTNPTNTVSGTKRLIGRKFDDPQTQKEMKMVPYKIVRAPNGDA 156
Query: 152 WVEAKGQQYSPSQIGAFVLTKMKETAEAYLGKTVPKAVITVPAYFNDAQRQATKDAGRIA 211
WVEA GQQYSPSQIGAF+LTKMKETAEAYLGK+V KAV+TVPAYFNDAQRQATKDAGRIA
Sbjct: 157 WVEANGQQYSPSQIGAFILTKMKETAEAYLGKSVTKAVVTVPAYFNDAQRQATKDAGRIA 216
Query: 212 GLEVLRIINEPTAAALSYGM-NKEGLIAVFDLGGGTFDVSILEISNGVFEVKATNGDTFL 270
GL+V RIINEPTAAALSYGM NKEGLIAVFDLGGGTFDVS+LEISNGVFEVKATNGDTFL
Sbjct: 217 GLDVERIINEPTAAALSYGMTNKEGLIAVFDLGGGTFDVSVLEISNGVFEVKATNGDTFL 276
Query: 271 GGEDFDNALLDFLVNEFKRSESIDLSKDKLALQRLREAAEKAKIELSSTSQTEINLPFIT 330
GGEDFDNALLDFLVNEFK +E IDL+KD+LALQRLREAAEKAKIELSSTSQTEINLPFIT
Sbjct: 277 GGEDFDNALLDFLVNEFKTTEGIDLAKDRLALQRLREAAEKAKIELSSTSQTEINLPFIT 336
Query: 331 ADASGAKHLNITLTRSKFEALVNNLIERTKAPCQSCLKDANISTKDIDEVLLVGGMTRVP 390
ADASGAKH NITLTRS+FE LVN+LIERT+ PC++CLKDA IS K++DEVLLVGGMTRVP
Sbjct: 337 ADASGAKHFNITLTRSRFETLVNHLIERTRDPCKNCLKDAGISAKEVDEVLLVGGMTRVP 396
Query: 391 KVQEVVSTIFGKSPCKGVNPDEAVAMGAALQGGILRGDVKELLLLDVTPLSLGIETLGGI 450
KVQ +V+ IFGKSP KGVNPDEAVAMGAALQGGILRGDVKELLLLDVTPLSLGIETLGG+
Sbjct: 397 KVQSIVAEIFGKSPSKGVNPDEAVAMGAALQGGILRGDVKELLLLDVTPLSLGIETLGGV 456
Query: 451 FTRLINRNTTIPTKKSQVFSTAADNQTQVGIKVLQGEREMASDNKMLGEFELVGLPPAPR 510
FTRLI RNTTIPTKKSQVFSTAADNQTQVGI+VLQGEREMA+DNK+LGEF+LVG+PP+PR
Sbjct: 457 FTRLITRNTTIPTKKSQVFSTAADNQTQVGIRVLQGEREMATDNKLLGEFDLVGIPPSPR 516
Query: 511 GMPQIEVTFDIDANGIVTVSAKDKSTGKEQQITIKSSGGLSDDEIQNMVKEAELHAQKDQ 570
G+PQIEVTFDIDANGIVTVSAKDK+TGK QQITI+SSGGLS+D+IQ MV+EAELHAQKD+
Sbjct: 517 GVPQIEVTFDIDANGIVTVSAKDKTTGKVQQITIRSSGGLSEDDIQKMVREAELHAQKDK 576
Query: 571 ERKSLIDIRNSADTSIYSIEKSLSEYREKIPAEVAKEIEDAVSDLRSAMAGDSADEIKSK 630
ERK LID +N+ADT+IYSIEKSL EYREKIP+E+AKEIEDAV+DLRSA +GD +EIK+K
Sbjct: 577 ERKELIDTKNTADTTIYSIEKSLGEYREKIPSEIAKEIEDAVADLRSASSGDDLNEIKAK 636
Query: 631 LDAANKAVSKIGEHMSGGSSSGPS-SDGSQGG-DQAPEAEYEEVK 673
++AANKAVSKIGEHMSGGS G + GS+GG DQAPEAEYEEVK
Sbjct: 637 IEAANKAVSKIGEHMSGGSGGGSAPGGGSEGGSDQAPEAEYEEVK 681
>gb|AAB91473.1| heat shock 70 protein [Spinacia oleracea] gi|2773052|gb|AAB96660.1|
heat shock 70 protein [Spinacia oleracea]
gi|7441879|pir||T08901 dnaK-type molecular chaperone
HSC70-11, mitochondrial - spinach
Length = 675
Score = 1078 bits (2787), Expect = 0.0
Identities = 553/654 (84%), Positives = 597/654 (90%), Gaps = 7/654 (1%)
Query: 25 LTGNTKPA----YVGHNWSSLSRPFSSRAAGNDVIGIDLGTTNSCVSVMEGKNPKVVENS 80
L GN P+ Y+ +SL RPFSSR AGNDVIGIDLGTTNSCVSVMEGK+ KV+EN+
Sbjct: 23 LAGNASPSLSSPYMAQRLASLVRPFSSRPAGNDVIGIDLGTTNSCVSVMEGKSAKVIENA 82
Query: 81 EGARTTPSVVAFTQKGELLVGTPAKRQAVTNPENTISGAKRLIGRRFDDPQTQKEMKMVP 140
EGARTTPSVVAF KGELLVGTPAKRQAVTNP NTI G KRLIGR F DPQTQKEMKMVP
Sbjct: 83 EGARTTPSVVAFNPKGELLVGTPAKRQAVTNPTNTIFGTKRLIGRVFSDPQTQKEMKMVP 142
Query: 141 YKIVKAPNGDAWVEAKGQQYSPSQIGAFVLTKMKETAEAYLGKTVPKAVITVPAYFNDAQ 200
YKIVKAPNGDAWVEA GQQYSPSQ+GAFVLTKMKETAEAYLGKTV KAV+TVPAYFNDAQ
Sbjct: 143 YKIVKAPNGDAWVEANGQQYSPSQVGAFVLTKMKETAEAYLGKTVSKAVVTVPAYFNDAQ 202
Query: 201 RQATKDAGRIAGLEVLRIINEPTAAALSYGM-NKEGLIAVFDLGGGTFDVSILEISNGVF 259
RQATKDAG+IAGL+V RIINEPTAAALSYGM NKEGLIAVFDLGGGTFDVSILEISNGVF
Sbjct: 203 RQATKDAGKIAGLDVQRIINEPTAAALSYGMTNKEGLIAVFDLGGGTFDVSILEISNGVF 262
Query: 260 EVKATNGDTFLGGEDFDNALLDFLVNEFKRSESIDLSKDKLALQRLREAAEKAKIELSST 319
EVKATNGDTFLGGEDFDN LL+FLV+EFK++E IDLS D+LALQRLREAAEKAKIELSST
Sbjct: 263 EVKATNGDTFLGGEDFDNTLLEFLVSEFKKTEGIDLSSDRLALQRLREAAEKAKIELSST 322
Query: 320 SQTEINLPFITADASGAKHLNITLTRSKFEALVNNLIERTKAPCQSCLKDANISTKDIDE 379
SQT+I+LPFI+AD+SGAKHLNITL+RSKFE+LV NLIERT+APC++CLKDA +S ++DE
Sbjct: 323 SQTDISLPFISADSSGAKHLNITLSRSKFESLVGNLIERTRAPCKNCLKDAGVSLNEVDE 382
Query: 380 VLLVGGMTRVPKVQEVVSTIFGKSPCKGVNPDEAVAMGAALQGGILRGDVKELLLLDVTP 439
VLLVGGMTRVPKVQE+V IFGKSP KGVNPDEAVAMGAA+QGGILRGDVK+LLLLDVTP
Sbjct: 383 VLLVGGMTRVPKVQEIVQEIFGKSPSKGVNPDEAVAMGAAIQGGILRGDVKDLLLLDVTP 442
Query: 440 LSLGIETLGGIFTRLINRNTTIPTKKSQVFSTAADNQTQVGIKVLQGEREMASDNKMLGE 499
LSLGIETLGG+FTRLINRNTTIPTKKSQVFSTAADNQTQVGIKVLQGEREMASDNKMLGE
Sbjct: 443 LSLGIETLGGVFTRLINRNTTIPTKKSQVFSTAADNQTQVGIKVLQGEREMASDNKMLGE 502
Query: 500 FELVGLPPAPRGMPQIEVTFDIDANGIVTVSAKDKSTGKEQQITIKSSGGLSDDEIQNMV 559
FEL G+PPAPRG+PQIEVTFDIDANGIVTVSAKDK+T KEQ ITIKSSGGLS+ EI+ MV
Sbjct: 503 FELQGIPPAPRGLPQIEVTFDIDANGIVTVSAKDKATNKEQNITIKSSGGLSEHEIEKMV 562
Query: 560 KEAELHAQKDQERKSLIDIRNSADTSIYSIEKSLSEYREKIPAEVAKEIEDAVSDLRSAM 619
KEAE+HAQKDQERKSLID RN+ADT+IYS+EKSL EYREKIP E+AKEIEDAVSDLRSAM
Sbjct: 563 KEAEIHAQKDQERKSLIDARNTADTTIYSVEKSLGEYREKIPTEIAKEIEDAVSDLRSAM 622
Query: 620 AGDSADEIKSKLDAANKAVSKIGEHMSGGSSSGPSSDGSQGGDQAPEAEYEEVK 673
D+ +EIK+K DAANKAVSKIGEHMSGG G SS G G DQ PEAEYEE K
Sbjct: 623 QNDNLEEIKAKTDAANKAVSKIGEHMSGGQGGGSSSSG--GADQTPEAEYEEAK 674
>gb|AAO17017.1| Putative heat shock 70 KD protein, mitochondrial precursor [Oryza
sativa (japonica cultivar-group)]
Length = 656
Score = 1077 bits (2785), Expect = 0.0
Identities = 553/654 (84%), Positives = 603/654 (91%), Gaps = 5/654 (0%)
Query: 25 LTGNTKP---AYVGHNWSSLSRPFSSRAAGNDVIGIDLGTTNSCVSVMEGKNPKVVENSE 81
LT N + A V W+ +R FS++A GN+VIGIDLGTTNSCVSVMEGKNPKV+ENSE
Sbjct: 2 LTANVQSKCAANVCSRWAGFARTFSAKATGNEVIGIDLGTTNSCVSVMEGKNPKVIENSE 61
Query: 82 GARTTPSVVAFTQKGELLVGTPAKRQAVTNPENTISGAKRLIGRRFDDPQTQKEMKMVPY 141
G RTTPSVVAF QKGELLVGTPAKRQAVTNP+NT G KRLIGRRFDDPQTQKEMKMVPY
Sbjct: 62 GTRTTPSVVAFNQKGELLVGTPAKRQAVTNPQNTFFGTKRLIGRRFDDPQTQKEMKMVPY 121
Query: 142 KIVKAPNGDAWVEAK-GQQYSPSQIGAFVLTKMKETAEAYLGKTVPKAVITVPAYFNDAQ 200
KIVKA NGDAW+E G+QYSPSQIGAFVLTKMKETAE+YLGK+V KAVITVPAYFNDAQ
Sbjct: 122 KIVKALNGDAWLETTDGKQYSPSQIGAFVLTKMKETAESYLGKSVSKAVITVPAYFNDAQ 181
Query: 201 RQATKDAGRIAGLEVLRIINEPTAAALSYGMN-KEGLIAVFDLGGGTFDVSILEISNGVF 259
RQATKDAGRIAGL+V RIINEPTAAALSYG N KEGLIAVFDLGGGTFDVSILEISNGVF
Sbjct: 182 RQATKDAGRIAGLDVQRIINEPTAAALSYGTNNKEGLIAVFDLGGGTFDVSILEISNGVF 241
Query: 260 EVKATNGDTFLGGEDFDNALLDFLVNEFKRSESIDLSKDKLALQRLREAAEKAKIELSST 319
EVKATNGDTFLGGEDFDN LL+FLV+EFKR+E IDLSKD+LALQRLREAAEKAKIELSST
Sbjct: 242 EVKATNGDTFLGGEDFDNTLLEFLVSEFKRTEGIDLSKDRLALQRLREAAEKAKIELSST 301
Query: 320 SQTEINLPFITADASGAKHLNITLTRSKFEALVNNLIERTKAPCQSCLKDANISTKDIDE 379
+QTEINLPFITAD+SGAKHLNITLTRSKFE+LVN+LIERT+ PC+SCLKDA I+TKD+DE
Sbjct: 302 AQTEINLPFITADSSGAKHLNITLTRSKFESLVNSLIERTRDPCKSCLKDAGITTKDVDE 361
Query: 380 VLLVGGMTRVPKVQEVVSTIFGKSPCKGVNPDEAVAMGAALQGGILRGDVKELLLLDVTP 439
VLLVGGMTRVPKVQEVVS IFGK+P KGVNPDEAVAMGAA+QGGILRGDVK+LLLLDVTP
Sbjct: 362 VLLVGGMTRVPKVQEVVSEIFGKAPSKGVNPDEAVAMGAAIQGGILRGDVKDLLLLDVTP 421
Query: 440 LSLGIETLGGIFTRLINRNTTIPTKKSQVFSTAADNQTQVGIKVLQGEREMASDNKMLGE 499
LSLGIETLGGIFTRLINRNTT+PTKKSQVFSTAADNQTQVGIKVLQGEREMA+DNK+LGE
Sbjct: 422 LSLGIETLGGIFTRLINRNTTVPTKKSQVFSTAADNQTQVGIKVLQGEREMAADNKLLGE 481
Query: 500 FELVGLPPAPRGMPQIEVTFDIDANGIVTVSAKDKSTGKEQQITIKSSGGLSDDEIQNMV 559
F+LVG+PPAPRGMPQIEVTFDIDANGIVTVSAKDK+TGKEQQITI+SSGGLS+ EIQ MV
Sbjct: 482 FDLVGIPPAPRGMPQIEVTFDIDANGIVTVSAKDKATGKEQQITIRSSGGLSEAEIQKMV 541
Query: 560 KEAELHAQKDQERKSLIDIRNSADTSIYSIEKSLSEYREKIPAEVAKEIEDAVSDLRSAM 619
EAELH+QKDQERK+LIDIRN+ADT+IYSIEKSL EYR+KIPAEVA EIE A++DLR+ M
Sbjct: 542 HEAELHSQKDQERKALIDIRNTADTTIYSIEKSLGEYRDKIPAEVASEIETAIADLRNEM 601
Query: 620 AGDSADEIKSKLDAANKAVSKIGEHMSGGSSSGPSSDGSQGGDQAPEAEYEEVK 673
A D ++IKSK++AANKAVSKIG+HMSGG S G + GGDQAPEAEYEEVK
Sbjct: 602 ASDDIEKIKSKIEAANKAVSKIGQHMSGGGSGGSQAGSQGGGDQAPEAEYEEVK 655
>ref|XP_468043.1| putative dnaK-type molecular chaperone precursor [Oryza sativa
(japonica cultivar-group)] gi|46805772|dbj|BAD17140.1|
putative dnaK-type molecular chaperone precursor [Oryza
sativa (japonica cultivar-group)]
Length = 679
Score = 1073 bits (2776), Expect = 0.0
Identities = 553/657 (84%), Positives = 605/657 (91%), Gaps = 8/657 (1%)
Query: 25 LTGNTKPAYVGH---NWSSLSRPFSSRAAGNDVIGIDLGTTNSCVSVMEGKNPKVVENSE 81
LT N + AY + W S +R FS + GN+VIGIDLGTTNSCVSVMEGKNPKV+ENSE
Sbjct: 22 LTANAQSAYSANICSQWGSFARAFSVKPTGNEVIGIDLGTTNSCVSVMEGKNPKVIENSE 81
Query: 82 GARTTPSVVAFTQKGELLVGTPAKRQAVTNPENTISGAKRLIGRRFDDPQTQKEMKMVPY 141
G RTTPSVVAF QKGE LVGTPAKRQAVTNP+NT G KRLIGRRF+DPQTQKEMKMVPY
Sbjct: 82 GTRTTPSVVAFNQKGERLVGTPAKRQAVTNPQNTFFGTKRLIGRRFEDPQTQKEMKMVPY 141
Query: 142 KIVKAPNGDAWVEAK-GQQYSPSQIGAFVLTKMKETAEAYLGKTVPKAVITVPAYFNDAQ 200
KIVKAPNGDAWVE G+QYSPSQIGAFVLTKMKETAE+YLGKTV KAVITVPAYFNDAQ
Sbjct: 142 KIVKAPNGDAWVETTDGKQYSPSQIGAFVLTKMKETAESYLGKTVSKAVITVPAYFNDAQ 201
Query: 201 RQATKDAGRIAGLEVLRIINEPTAAALSYGMN-KEGLIAVFDLGGGTFDVSILEISNGVF 259
RQATKDAGRIAGL+V RIINEPTAAALSYG N KEGLIAVFDLGGGTFDVSILEISNGVF
Sbjct: 202 RQATKDAGRIAGLDVQRIINEPTAAALSYGTNNKEGLIAVFDLGGGTFDVSILEISNGVF 261
Query: 260 EVKATNGDTFLGGEDFDNALLDFLVNEFKRSESIDLSKDKLALQRLREAAEKAKIELSST 319
EVKATNGDTFLGGEDFDN LL+FLV+EFKRSE+IDL+KD+LALQRLREAAEKAKIELSST
Sbjct: 262 EVKATNGDTFLGGEDFDNTLLEFLVSEFKRSEAIDLAKDRLALQRLREAAEKAKIELSST 321
Query: 320 SQTEINLPFITADASGAKHLNITLTRSKFEALVNNLIERTKAPCQSCLKDANISTKDIDE 379
+QTEINLPFITADASGAKHLNITLTRSKFE+LVN+LIERT+ PC++CLKDA I+TK++DE
Sbjct: 322 AQTEINLPFITADASGAKHLNITLTRSKFESLVNSLIERTREPCKNCLKDAGITTKEVDE 381
Query: 380 VLLVGGMTRVPKVQEVVSTIFGKSPCKGVNPDEAVAMGAALQGGILRGDVKELLLLDVTP 439
VLLVGGMTRVPKVQE+VS IFGKSP KGVNPDEAVAMGAA+QGGILRGDVKELLLLDVTP
Sbjct: 382 VLLVGGMTRVPKVQEIVSEIFGKSPSKGVNPDEAVAMGAAIQGGILRGDVKELLLLDVTP 441
Query: 440 LSLGIETLGGIFTRLINRNTTIPTKKSQVFSTAADNQTQVGIKVLQGEREMASDNKMLGE 499
LSLGIETLGGIFTRLINRNTTIPTKKSQVFSTAADNQTQVGI+VLQGEREMA+DNK+LGE
Sbjct: 442 LSLGIETLGGIFTRLINRNTTIPTKKSQVFSTAADNQTQVGIRVLQGEREMATDNKLLGE 501
Query: 500 FELVGLPPAPRGMPQIEVTFDIDANGIVTVSAKDKSTGKEQQITIKSSGGLSDDEIQNMV 559
F+LVG+PPAPRGMPQIEVTFDIDANGIVTVSAKDKSTGKEQQITI+SSGGLS+ EIQ MV
Sbjct: 502 FDLVGIPPAPRGMPQIEVTFDIDANGIVTVSAKDKSTGKEQQITIRSSGGLSEAEIQKMV 561
Query: 560 KEAELHAQKDQERKSLIDIRNSADTSIYSIEKSLSEYREKIPAEVAKEIEDAVSDLRSAM 619
+EAELH+QKDQERK+LIDIRN+ADT+IYS+EKSL EYR+KIPAEVA EIE A++DLRS M
Sbjct: 562 QEAELHSQKDQERKALIDIRNNADTTIYSVEKSLGEYRDKIPAEVATEIETAIADLRSVM 621
Query: 620 AGDSADEIKSKLDAANKAVSKIGEHMS--GGSSSGPSSDGSQ-GGDQAPEAEYEEVK 673
D ++IK+ ++AANKAVSKIG+HMS GG + G + GSQ GG+QAPEAEYEEVK
Sbjct: 622 TSDDIEKIKANIEAANKAVSKIGQHMSGGGGGAGGSETGGSQGGGEQAPEAEYEEVK 678
>gb|AAB91472.1| heat shock 70 protein [Spinacia oleracea] gi|7441881|pir||T08900
dnaK-type molecular chaperone HSC70-10, mitochondrial -
spinach
Length = 675
Score = 1073 bits (2775), Expect = 0.0
Identities = 552/654 (84%), Positives = 598/654 (91%), Gaps = 7/654 (1%)
Query: 25 LTGNTKPA----YVGHNWSSLSRPFSSRAAGNDVIGIDLGTTNSCVSVMEGKNPKVVENS 80
L GN P+ Y+ +SL RPFSSR AGNDVIGIDLGTTNSCVSVMEGK+ KV+EN+
Sbjct: 23 LAGNASPSLSSPYMAQRLASLVRPFSSRPAGNDVIGIDLGTTNSCVSVMEGKSAKVIENA 82
Query: 81 EGARTTPSVVAFTQKGELLVGTPAKRQAVTNPENTISGAKRLIGRRFDDPQTQKEMKMVP 140
EGARTTPSVVAF KGELLVGTPAKRQAVTNP NTI G KRLIGR F DPQTQKEMKMVP
Sbjct: 83 EGARTTPSVVAFNPKGELLVGTPAKRQAVTNPTNTIFGTKRLIGRVFSDPQTQKEMKMVP 142
Query: 141 YKIVKAPNGDAWVEAKGQQYSPSQIGAFVLTKMKETAEAYLGKTVPKAVITVPAYFNDAQ 200
YKIVKAPNGDAWVEA GQQYSPSQ+GAFVLTKMKETAEAYLGKTV KAV+TVPAYFNDAQ
Sbjct: 143 YKIVKAPNGDAWVEANGQQYSPSQVGAFVLTKMKETAEAYLGKTVSKAVVTVPAYFNDAQ 202
Query: 201 RQATKDAGRIAGLEVLRIINEPTAAALSYGM-NKEGLIAVFDLGGGTFDVSILEISNGVF 259
RQATKDAG+IAGL+V RIINEPTAAALSYGM NKEGLIAVFDLGGGTFDVSILEISNGVF
Sbjct: 203 RQATKDAGKIAGLDVQRIINEPTAAALSYGMTNKEGLIAVFDLGGGTFDVSILEISNGVF 262
Query: 260 EVKATNGDTFLGGEDFDNALLDFLVNEFKRSESIDLSKDKLALQRLREAAEKAKIELSST 319
EVKATNGDTFLGGEDFDN LL+FLV+EFK++E IDLS D+LALQRLREAAEKAKIELSST
Sbjct: 263 EVKATNGDTFLGGEDFDNTLLEFLVSEFKKTEGIDLSSDRLALQRLREAAEKAKIELSST 322
Query: 320 SQTEINLPFITADASGAKHLNITLTRSKFEALVNNLIERTKAPCQSCLKDANISTKDIDE 379
SQT+I+LPFI+AD+SGAKHLNITL+RSKFE+LV NLIERT+APC++CLKDA +S ++DE
Sbjct: 323 SQTDISLPFISADSSGAKHLNITLSRSKFESLVGNLIERTRAPCKNCLKDAGVSLNEVDE 382
Query: 380 VLLVGGMTRVPKVQEVVSTIFGKSPCKGVNPDEAVAMGAALQGGILRGDVKELLLLDVTP 439
VLLVGGMTRVPKVQE+V IFGKSP KGVNPDEAVAMGAA+QGGILRGDVK+LLLLDVTP
Sbjct: 383 VLLVGGMTRVPKVQEIVQEIFGKSPSKGVNPDEAVAMGAAIQGGILRGDVKDLLLLDVTP 442
Query: 440 LSLGIETLGGIFTRLINRNTTIPTKKSQVFSTAADNQTQVGIKVLQGEREMASDNKMLGE 499
LSLGIETLGG+FTRLINRNTTIPTKKSQVFSTAADNQTQVGIKVLQGEREMASDNKMLGE
Sbjct: 443 LSLGIETLGGVFTRLINRNTTIPTKKSQVFSTAADNQTQVGIKVLQGEREMASDNKMLGE 502
Query: 500 FELVGLPPAPRGMPQIEVTFDIDANGIVTVSAKDKSTGKEQQITIKSSGGLSDDEIQNMV 559
FEL G+PPAPRG+PQIEVTFDIDANGIVTVSAKDK+T KEQ ITIKSSGGLS+ EI+ MV
Sbjct: 503 FELQGIPPAPRGLPQIEVTFDIDANGIVTVSAKDKATNKEQNITIKSSGGLSEHEIEKMV 562
Query: 560 KEAELHAQKDQERKSLIDIRNSADTSIYSIEKSLSEYREKIPAEVAKEIEDAVSDLRSAM 619
KEAE+HAQKDQERKSLID RN+ADT+IYS+EKSL EYREKI +E+AKEIEDAVSDLRSAM
Sbjct: 563 KEAEIHAQKDQERKSLIDARNTADTTIYSVEKSLGEYREKITSEIAKEIEDAVSDLRSAM 622
Query: 620 AGDSADEIKSKLDAANKAVSKIGEHMSGGSSSGPSSDGSQGGDQAPEAEYEEVK 673
D+ +EIK+K DAANKAVSKIGEHMSGG G SS G+ GDQ PEAEYEE K
Sbjct: 623 QNDNLEEIKAKTDAANKAVSKIGEHMSGG--QGGSSGGTGTGDQTPEAEYEEAK 674
>gb|AAC60559.2| HSP68 [Solanum tuberosum] gi|7441872|pir||T07024 dnaK-type
molecular chaperone HSP68, mitochondrial - potato
gi|585273|sp|Q08276|HSP7M_SOLTU Heat shock 70 kDa
protein, mitochondrial precursor
Length = 682
Score = 1057 bits (2733), Expect = 0.0
Identities = 543/656 (82%), Positives = 597/656 (90%), Gaps = 7/656 (1%)
Query: 25 LTGNTKPAY----VGHNWSSLSRPFSSRAAGNDVIGIDLGTTNSCVSVMEGKNPKVVENS 80
L NTKP++ VG W+ L+RPFSS+ AGN++IGIDLGTTNSCV+VMEGKNPKV+ENS
Sbjct: 26 LASNTKPSWCPSLVGAKWAGLARPFSSKPAGNEIIGIDLGTTNSCVAVMEGKNPKVIENS 85
Query: 81 EGARTTPSVVAFTQKGELLVGTPAKRQAVTNPENTISGAKRLIGRRFDDPQTQKEMKMVP 140
EGARTTPSVVAF QKGELLVGTPAKRQAVTNP NT+SG KRLIGRRFDDPQTQKEMKMVP
Sbjct: 86 EGARTTPSVVAFNQKGELLVGTPAKRQAVTNPTNTLSGTKRLIGRRFDDPQTQKEMKMVP 145
Query: 141 YKIVKAPNGDAWVEAKGQQYSPSQIGAFVLTKMKETAEAYLGKTVPKAVITVPAYFNDAQ 200
YKIV+ NGDAWVEA GQQYSP+QIGAF+LTKMKETAEAYLGK++ KAVITVPAYFNDAQ
Sbjct: 146 YKIVRGSNGDAWVEANGQQYSPTQIGAFILTKMKETAEAYLGKSINKAVITVPAYFNDAQ 205
Query: 201 RQATKDAGRIAGLEVLRIINEPTAAALSYGMN-KEGLIAVFDLGGGTFDVSILEISNGVF 259
RQA KDAG IAGL+V RIINEPTAAALSYGMN KEGL+AVFDLGGGTFDVSILEISNGVF
Sbjct: 206 RQAIKDAGAIAGLDVQRIINEPTAAALSYGMNSKEGLVAVFDLGGGTFDVSILEISNGVF 265
Query: 260 EVKATNGDTFLGGEDFDNALLDFLVNEFKRSESIDLSKDKLALQRLREAAEKAKIELSST 319
EVKATNGDTFLGGEDFDNALL+FLV+EFKR+E IDLSKDKLALQRLREAAEKAKIELSST
Sbjct: 266 EVKATNGDTFLGGEDFDNALLEFLVSEFKRTEGIDLSKDKLALQRLREAAEKAKIELSST 325
Query: 320 SQTEINLPFITADASGAKHLNITLTRSKFEALVNNLIERTKAPCQSCLKDANISTKDIDE 379
SQT+INLPFITADASGAKHLNITLTRSKFE LVN+LIERT+ PC++CLKDA +S KD+DE
Sbjct: 326 SQTDINLPFITADASGAKHLNITLTRSKFETLVNHLIERTRNPCKNCLKDAGVSLKDVDE 385
Query: 380 VLLVGGMTRVPKVQEVVSTIFGKSPCKGVNPDEAVAMGAALQGGILRGDVKELLLLDVTP 439
VLLVGGMTRVPKVQE+VS IFGKSP KGVNPDEAVAMGAALQGGILRGDVKELLLLDVTP
Sbjct: 386 VLLVGGMTRVPKVQEIVSEIFGKSPSKGVNPDEAVAMGAALQGGILRGDVKELLLLDVTP 445
Query: 440 LSLGIETLGGIFTRLINRNTTIPTKKSQVFSTAADNQTQVGIKVLQGEREMASDNKMLGE 499
L+ GIETLGGIFTRLINRNTTIPTKKSQVFSTAADNQTQVGIKVLQGEREMASDNK+LGE
Sbjct: 446 LARGIETLGGIFTRLINRNTTIPTKKSQVFSTAADNQTQVGIKVLQGEREMASDNKLLGE 505
Query: 500 FELVGLPPAPRG-MPQIEVTFDIDANGIVTVSAKDKSTGKEQQITIKSSGGLSDDEIQNM 558
F+LVG+PPAP+G PQIEV FDIDANG+VTVSAKDK+T KEQQITI+SSGGLS+DEI M
Sbjct: 506 FDLVGIPPAPKGYCPQIEVIFDIDANGMVTVSAKDKATSKEQQITIRSSGGLSEDEIDKM 565
Query: 559 VKEAELHAQKDQERKSLIDIRNSADTSIYSIEKSLSEYREKIPAEVAKEIEDAVSDLRSA 618
V+EAE+HAQ+ + + L+ T+IYSIEKSLSEY+EK+P EV EIE A+SDLR+A
Sbjct: 566 VREAEMHAQRIKNARHLLISGIVQSTTIYSIEKSLSEYKEKVPKEVVTEIETAISDLRAA 625
Query: 619 MAGDSADEIKSKLDAANKAVSKIGEHMSGGSSSGPS-SDGSQGGDQAPEAEYEEVK 673
M ++ D+IK+KLDAANKAVSKIGEHM+GGSS G S G+QGGDQ PEAEYEEVK
Sbjct: 626 MGTENIDDIKAKLDAANKAVSKIGEHMAGGSSGGASGGGGAQGGDQPPEAEYEEVK 681
>gb|AAP37789.1| At4g37910 [Arabidopsis thaliana] gi|27311569|gb|AAO00750.1| heat
shock protein 70 like protein [Arabidopsis thaliana]
gi|30691626|ref|NP_195504.2| heat shock protein 70,
mitochondrial, putative / HSP70, mitochondrial, putative
[Arabidopsis thaliana]
Length = 682
Score = 1052 bits (2721), Expect = 0.0
Identities = 534/636 (83%), Positives = 590/636 (91%), Gaps = 5/636 (0%)
Query: 41 LSRPFSSRAAGNDVIGIDLGTTNSCVSVMEGKNPKVVENSEGARTTPSVVAFTQKGELLV 100
L+RPF SR GNDVIGIDLGTTNSCVSVMEGK +V+EN+EG+RTTPSVVA QKGELLV
Sbjct: 41 LARPFCSRPVGNDVIGIDLGTTNSCVSVMEGKTARVIENAEGSRTTPSVVAMNQKGELLV 100
Query: 101 GTPAKRQAVTNPENTISGAKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWVEAKGQQY 160
GTPAKRQAVTNP NTI G+KRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWVEA GQ++
Sbjct: 101 GTPAKRQAVTNPTNTIFGSKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWVEANGQKF 160
Query: 161 SPSQIGAFVLTKMKETAEAYLGKTVPKAVITVPAYFNDAQRQATKDAGRIAGLEVLRIIN 220
SPSQIGA VLTKMKETAEAYLGK++ KAV+TVPAYFNDAQRQATKDAG+IAGL+V RIIN
Sbjct: 161 SPSQIGANVLTKMKETAEAYLGKSINKAVVTVPAYFNDAQRQATKDAGKIAGLDVQRIIN 220
Query: 221 EPTAAALSYGMN-KEGLIAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNAL 279
EPTAAALSYGMN KEG+IAVFDLGGGTFDVSILEIS+GVFEVKATNGDTFLGGEDFDN L
Sbjct: 221 EPTAAALSYGMNNKEGVIAVFDLGGGTFDVSILEISSGVFEVKATNGDTFLGGEDFDNTL 280
Query: 280 LDFLVNEFKRSESIDLSKDKLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHL 339
L++LVNEFKRS++IDL+KD LALQRLREAAEKAKIELSST+QTEINLPFITADASGAKHL
Sbjct: 281 LEYLVNEFKRSDNIDLTKDNLALQRLREAAEKAKIELSSTTQTEINLPFITADASGAKHL 340
Query: 340 NITLTRSKFEALVNNLIERTKAPCQSCLKDANISTKDIDEVLLVGGMTRVPKVQEVVSTI 399
NITLTRSKFE LV LIERT++PCQ+CLKDA ++ K++DEVLLVGGMTRVPKVQE+VS I
Sbjct: 341 NITLTRSKFEGLVGKLIERTRSPCQNCLKDAGVTIKEVDEVLLVGGMTRVPKVQEIVSEI 400
Query: 400 FGKSPCKGVNPDEAVAMGAALQGGILRGDVKELLLLDVTPLSLGIETLGGIFTRLINRNT 459
FGKSPCKGVNPDEAVAMGAA+QGGILRGDVK+LLLLDV PLSLGIETLG +FT+LI RNT
Sbjct: 401 FGKSPCKGVNPDEAVAMGAAIQGGILRGDVKDLLLLDVVPLSLGIETLGAVFTKLIPRNT 460
Query: 460 TIPTKKSQVFSTAADNQTQVGIKVLQGEREMASDNKMLGEFELVGLPPAPRGMPQIEVTF 519
TIPTKKSQVFSTAADNQ QVGIKVLQGEREMA+DNK+LGEF+LVG+PPAPRGMPQIEVTF
Sbjct: 461 TIPTKKSQVFSTAADNQMQVGIKVLQGEREMAADNKVLGEFDLVGIPPAPRGMPQIEVTF 520
Query: 520 DIDANGIVTVSAKDKSTGKEQQITIKSSGGLSDDEIQNMVKEAELHAQKDQERKSLIDIR 579
DIDANGI TVSAKDK+TGKEQ ITI+SSGGLSDDEI MVKEAEL+AQKDQE+K LID+R
Sbjct: 521 DIDANGITTVSAKDKATGKEQNITIRSSGGLSDDEINRMVKEAELNAQKDQEKKQLIDLR 580
Query: 580 NSADTSIYSIEKSLSEYREKIPAEVAKEIEDAVSDLRSAMAGDSADEIKSKLDAANKAVS 639
NSADT+IYS+EKSLSEYREKIPAE+A EIE AVSDLR+AMAG+ ++IK+K++AANKAVS
Sbjct: 581 NSADTTIYSVEKSLSEYREKIPAEIASEIETAVSDLRTAMAGEDVEDIKAKVEAANKAVS 640
Query: 640 KIGEHMSGGS----SSGPSSDGSQGGDQAPEAEYEE 671
KIGEHMS GS S G S +G+ G +Q PEAE+EE
Sbjct: 641 KIGEHMSKGSGSSGSDGSSGEGTSGTEQTPEAEFEE 676
>emb|CAB80456.1| heat shock protein 70 like protein [Arabidopsis thaliana]
gi|4467097|emb|CAB37531.1| heat shock protein 70 like
protein [Arabidopsis thaliana] gi|7441874|pir||T05618
dnaK-type molecular chaperone F20D10.30 - Arabidopsis
thaliana
Length = 666
Score = 1052 bits (2721), Expect = 0.0
Identities = 534/636 (83%), Positives = 590/636 (91%), Gaps = 5/636 (0%)
Query: 41 LSRPFSSRAAGNDVIGIDLGTTNSCVSVMEGKNPKVVENSEGARTTPSVVAFTQKGELLV 100
L+RPF SR GNDVIGIDLGTTNSCVSVMEGK +V+EN+EG+RTTPSVVA QKGELLV
Sbjct: 25 LARPFCSRPVGNDVIGIDLGTTNSCVSVMEGKTARVIENAEGSRTTPSVVAMNQKGELLV 84
Query: 101 GTPAKRQAVTNPENTISGAKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWVEAKGQQY 160
GTPAKRQAVTNP NTI G+KRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWVEA GQ++
Sbjct: 85 GTPAKRQAVTNPTNTIFGSKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWVEANGQKF 144
Query: 161 SPSQIGAFVLTKMKETAEAYLGKTVPKAVITVPAYFNDAQRQATKDAGRIAGLEVLRIIN 220
SPSQIGA VLTKMKETAEAYLGK++ KAV+TVPAYFNDAQRQATKDAG+IAGL+V RIIN
Sbjct: 145 SPSQIGANVLTKMKETAEAYLGKSINKAVVTVPAYFNDAQRQATKDAGKIAGLDVQRIIN 204
Query: 221 EPTAAALSYGMN-KEGLIAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNAL 279
EPTAAALSYGMN KEG+IAVFDLGGGTFDVSILEIS+GVFEVKATNGDTFLGGEDFDN L
Sbjct: 205 EPTAAALSYGMNNKEGVIAVFDLGGGTFDVSILEISSGVFEVKATNGDTFLGGEDFDNTL 264
Query: 280 LDFLVNEFKRSESIDLSKDKLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHL 339
L++LVNEFKRS++IDL+KD LALQRLREAAEKAKIELSST+QTEINLPFITADASGAKHL
Sbjct: 265 LEYLVNEFKRSDNIDLTKDNLALQRLREAAEKAKIELSSTTQTEINLPFITADASGAKHL 324
Query: 340 NITLTRSKFEALVNNLIERTKAPCQSCLKDANISTKDIDEVLLVGGMTRVPKVQEVVSTI 399
NITLTRSKFE LV LIERT++PCQ+CLKDA ++ K++DEVLLVGGMTRVPKVQE+VS I
Sbjct: 325 NITLTRSKFEGLVGKLIERTRSPCQNCLKDAGVTIKEVDEVLLVGGMTRVPKVQEIVSEI 384
Query: 400 FGKSPCKGVNPDEAVAMGAALQGGILRGDVKELLLLDVTPLSLGIETLGGIFTRLINRNT 459
FGKSPCKGVNPDEAVAMGAA+QGGILRGDVK+LLLLDV PLSLGIETLG +FT+LI RNT
Sbjct: 385 FGKSPCKGVNPDEAVAMGAAIQGGILRGDVKDLLLLDVVPLSLGIETLGAVFTKLIPRNT 444
Query: 460 TIPTKKSQVFSTAADNQTQVGIKVLQGEREMASDNKMLGEFELVGLPPAPRGMPQIEVTF 519
TIPTKKSQVFSTAADNQ QVGIKVLQGEREMA+DNK+LGEF+LVG+PPAPRGMPQIEVTF
Sbjct: 445 TIPTKKSQVFSTAADNQMQVGIKVLQGEREMAADNKVLGEFDLVGIPPAPRGMPQIEVTF 504
Query: 520 DIDANGIVTVSAKDKSTGKEQQITIKSSGGLSDDEIQNMVKEAELHAQKDQERKSLIDIR 579
DIDANGI TVSAKDK+TGKEQ ITI+SSGGLSDDEI MVKEAEL+AQKDQE+K LID+R
Sbjct: 505 DIDANGITTVSAKDKATGKEQNITIRSSGGLSDDEINRMVKEAELNAQKDQEKKQLIDLR 564
Query: 580 NSADTSIYSIEKSLSEYREKIPAEVAKEIEDAVSDLRSAMAGDSADEIKSKLDAANKAVS 639
NSADT+IYS+EKSLSEYREKIPAE+A EIE AVSDLR+AMAG+ ++IK+K++AANKAVS
Sbjct: 565 NSADTTIYSVEKSLSEYREKIPAEIASEIETAVSDLRTAMAGEDVEDIKAKVEAANKAVS 624
Query: 640 KIGEHMSGGS----SSGPSSDGSQGGDQAPEAEYEE 671
KIGEHMS GS S G S +G+ G +Q PEAE+EE
Sbjct: 625 KIGEHMSKGSGSSGSDGSSGEGTSGTEQTPEAEFEE 660
>gb|AAB26551.1| HSP68=68 kda heat-stress DnaK homolog [Lycopersicon
peruvianum=tomatoes, Peptide Mitochondrial Partial, 580
aa]
Length = 580
Score = 998 bits (2580), Expect = 0.0
Identities = 511/579 (88%), Positives = 547/579 (94%), Gaps = 2/579 (0%)
Query: 97 ELLVGTPAKRQAVTNPENTISGAKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWVEAK 156
ELLVGTPAKRQAVTNP NT+SG KRLIGRRFDDPQTQKEMKMVPYKIVK NGDAWVEA
Sbjct: 1 ELLVGTPAKRQAVTNPTNTLSGTKRLIGRRFDDPQTQKEMKMVPYKIVKGSNGDAWVEAN 60
Query: 157 GQQYSPSQIGAFVLTKMKETAEAYLGKTVPKAVITVPAYFNDAQRQATKDAGRIAGLEVL 216
GQQYSPSQIGAF+LTKMKETAEAYLGK++ KAVITVPAYFNDAQRQATKDAGRIAGL+V
Sbjct: 61 GQQYSPSQIGAFILTKMKETAEAYLGKSINKAVITVPAYFNDAQRQATKDAGRIAGLDVQ 120
Query: 217 RIINEPTAAALSYGMN-KEGLIAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDF 275
RIINEPTAAALSYGMN KEGL+AVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDF
Sbjct: 121 RIINEPTAAALSYGMNSKEGLVAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDF 180
Query: 276 DNALLDFLVNEFKRSESIDLSKDKLALQRLREAAEKAKIELSSTSQTEINLPFITADASG 335
DNALL+FLV+EFKR+E IDLSKDKLALQRLREAAEKAKIELSSTSQT+INLPFITADASG
Sbjct: 181 DNALLEFLVSEFKRTEGIDLSKDKLALQRLREAAEKAKIELSSTSQTDINLPFITADASG 240
Query: 336 AKHLNITLTRSKFEALVNNLIERTKAPCQSCLKDANISTKDIDEVLLVGGMTRVPKVQEV 395
AKHLNITLTRSKFE LVNNLIERT+ PC++CLKDA +S KD+DEVLLVGGMTRVPKVQE+
Sbjct: 241 AKHLNITLTRSKFETLVNNLIERTRNPCKNCLKDAGVSLKDVDEVLLVGGMTRVPKVQEI 300
Query: 396 VSTIFGKSPCKGVNPDEAVAMGAALQGGILRGDVKELLLLDVTPLSLGIETLGGIFTRLI 455
VS IFGKSP KGVNPDEAVAMGAALQGGILRGDVKELLLLDVTPLSLGIETLGGIFTRLI
Sbjct: 301 VSEIFGKSPSKGVNPDEAVAMGAALQGGILRGDVKELLLLDVTPLSLGIETLGGIFTRLI 360
Query: 456 NRNTTIPTKKSQVFSTAADNQTQVGIKVLQGEREMASDNKMLGEFELVGLPPAPRGMPQI 515
NRNTTIPTKKSQVFSTAADNQTQVGIKVLQGEREMASDNK+LGEFELVG+PPAPRGMPQI
Sbjct: 361 NRNTTIPTKKSQVFSTAADNQTQVGIKVLQGEREMASDNKLLGEFELVGIPPAPRGMPQI 420
Query: 516 EVTFDIDANGIVTVSAKDKSTGKEQQITIKSSGGLSDDEIQNMVKEAELHAQKDQERKSL 575
EVTFDIDANG+VTVSAKDK+T KEQQITI+SSGGLS+DEI MV+EAE+HAQKDQERK+L
Sbjct: 421 EVTFDIDANGMVTVSAKDKATSKEQQITIRSSGGLSEDEIDKMVREAEMHAQKDQERKAL 480
Query: 576 IDIRNSADTSIYSIEKSLSEYREKIPAEVAKEIEDAVSDLRSAMAGDSADEIKSKLDAAN 635
IDIRNSADT+IYSIEKSLSEY++K+P EV EIE A+SDLR+AM ++ D+IK KLDAAN
Sbjct: 481 IDIRNSADTTIYSIEKSLSEYKDKVPKEVVTEIETAISDLRAAMGTENIDDIKPKLDAAN 540
Query: 636 KAVSKIGEHMSGGSSSGPS-SDGSQGGDQAPEAEYEEVK 673
KAVSKIGEHM+GGSS G S G+QGGDQ PEAEYEEVK
Sbjct: 541 KAVSKIGEHMAGGSSGGASGGGGAQGGDQPPEAEYEEVK 579
>ref|ZP_00055307.1| COG0443: Molecular chaperone [Magnetospirillum magnetotacticum
MS-1]
Length = 644
Score = 846 bits (2185), Expect = 0.0
Identities = 429/634 (67%), Positives = 523/634 (81%), Gaps = 15/634 (2%)
Query: 54 VIGIDLGTTNSCVSVMEGKNPKVVENSEGARTTPSVVAFTQKGELLVGTPAKRQAVTNPE 113
VIGIDLGTTNSCV+VMEGKN KV+EN+EG RTTPS+ AFT+ GE LVG PAKRQAVTNP
Sbjct: 4 VIGIDLGTTNSCVAVMEGKNAKVIENAEGMRTTPSMTAFTESGERLVGQPAKRQAVTNPT 63
Query: 114 NTISGAKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWVEAKGQQYSPSQIGAFVLTKM 173
+T+ KRLIGRRFDDP T+K+M +VPY IV NGDAWVEA+ +YSPSQ+ AF+L KM
Sbjct: 64 STLFAIKRLIGRRFDDPITKKDMNLVPYHIVAGDNGDAWVEARDAKYSPSQVSAFILQKM 123
Query: 174 KETAEAYLGKTVPKAVITVPAYFNDAQRQATKDAGRIAGLEVLRIINEPTAAALSYGMNK 233
KETAE YLG+ V +AVITVPAYFNDAQRQATKDAGRIAGLEVLRIINEPTAAAL+YG+ K
Sbjct: 124 KETAEGYLGEKVTQAVITVPAYFNDAQRQATKDAGRIAGLEVLRIINEPTAAALAYGLEK 183
Query: 234 E--GLIAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDFLVNEFKRSE 291
+ G IAV+DLGGGTFDVS+LEI +GVFEVK+TNGDTFLGGEDFD ++D+L +EFK+ +
Sbjct: 184 KSAGTIAVYDLGGGTFDVSVLEIGDGVFEVKSTNGDTFLGGEDFDARIMDYLADEFKKEQ 243
Query: 292 SIDLSKDKLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNITLTRSKFEAL 351
IDL KD+LALQRL+EAAEKAKIELSS+ QTE+NLPFITADASG KHLNI LTRSK EAL
Sbjct: 244 GIDLRKDRLALQRLKEAAEKAKIELSSSMQTEVNLPFITADASGPKHLNIKLTRSKLEAL 303
Query: 352 VNNLIERTKAPCQSCLKDANISTKDIDEVLLVGGMTRVPKVQEVVSTIFGKSPCKGVNPD 411
V +L+ RT PC++ LKDA + +IDEV+LVGGMTR+PK+QEVV FG+ P KGVNPD
Sbjct: 304 VEDLVARTVEPCKAALKDAGVKASEIDEVILVGGMTRMPKIQEVVKEFFGREPHKGVNPD 363
Query: 412 EAVAMGAALQGGILRGDVKELLLLDVTPLSLGIETLGGIFTRLINRNTTIPTKKSQVFST 471
E VA+GAA+QGG+L+G+VK++LLLDVTPLSLGIETLGG+FTRLI+RNTTIPT+KSQVFST
Sbjct: 364 EVVAIGAAIQGGVLKGEVKDVLLLDVTPLSLGIETLGGVFTRLIDRNTTIPTRKSQVFST 423
Query: 472 AADNQTQVGIKVLQGEREMASDNKMLGEFELVGLPPAPRGMPQIEVTFDIDANGIVTVSA 531
A DNQT V I+V QGEREMA+DNK+LG+F+LVG+PPAPRG+PQ+EVTFDIDANG+V VSA
Sbjct: 424 AEDNQTAVTIRVFQGEREMAADNKVLGQFDLVGIPPAPRGVPQVEVTFDIDANGLVNVSA 483
Query: 532 KDKSTGKEQQITIKSSGGLSDDEIQNMVKEAELHAQKDQERKSLIDIRNSADTSIYSIEK 591
KDK+TGKEQQI I++SGGLSD +I+ MVKEAE+HA +D++RK LI+ RN AD I++ EK
Sbjct: 484 KDKATGKEQQIRIQASGGLSDTDIEKMVKEAEIHAAEDKKRKELIEARNHADGLIHTTEK 543
Query: 592 SLSEYREKIPAEVAKEIEDAVSDLRSAMAGDSADEIKSKLDAANKAVSKIGEHM------ 645
SL E+ +K AE+ IE ++ L++ M D + IK+K + +A K+GE M
Sbjct: 544 SLKEFGDKAGAELTGAIEKEITALKAVMEKDDVEAIKAKTEDLMQASMKLGEAMYKAQEA 603
Query: 646 SGGS-----SSGPSSDGSQGG--DQAPEAEYEEV 672
+GGS + G + G+ GG D+ +A++EEV
Sbjct: 604 AGGSEAEAAAGGGAHGGASGGHDDKVVDADFEEV 637
>sp|O05700|DNAK_RHOS7 Chaperone protein dnaK (Heat shock protein 70) (Heat shock 70 kDa
protein) (HSP70) gi|2058265|dbj|BAA19796.1| DnaK protein
[Rhodopseudomonas sp.]
Length = 631
Score = 842 bits (2176), Expect = 0.0
Identities = 426/621 (68%), Positives = 517/621 (82%), Gaps = 3/621 (0%)
Query: 54 VIGIDLGTTNSCVSVMEGKNPKVVENSEGARTTPSVVAFTQKGELLVGTPAKRQAVTNPE 113
VIGIDLGTTNSCV+VM+GK+ KV+EN+EG RTTPS+VA T GE LVG PAKRQAVTNPE
Sbjct: 4 VIGIDLGTTNSCVAVMDGKSAKVIENAEGMRTTPSIVAITDDGERLVGQPAKRQAVTNPE 63
Query: 114 NTISGAKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWVEAKGQQYSPSQIGAFVLTKM 173
T KRLIGRR+DDP +K+ +VPYKIVKA NGDAWVEA G+ YSPSQ+ AF+L KM
Sbjct: 64 RTFFAVKRLIGRRYDDPMVEKDKGLVPYKIVKASNGDAWVEADGKTYSPSQVSAFILQKM 123
Query: 174 KETAEAYLGKTVPKAVITVPAYFNDAQRQATKDAGRIAGLEVLRIINEPTAAALSYGMNK 233
KETAEA+LG+ V +AVITVPAYFNDAQRQATKDAG+IAGLEVLRIINEPTAAAL+YG++K
Sbjct: 124 KETAEAHLGQKVDQAVITVPAYFNDAQRQATKDAGKIAGLEVLRIINEPTAAALAYGLDK 183
Query: 234 E--GLIAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDFLVNEFKRSE 291
G IAV+DLGGGTFDVSILEI +GVFEVK+TNGDTFLGGEDFD L+++L +EF++ +
Sbjct: 184 AKTGTIAVYDLGGGTFDVSILEIGDGVFEVKSTNGDTFLGGEDFDMRLVNYLADEFQKEQ 243
Query: 292 SIDLSKDKLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNITLTRSKFEAL 351
IDL KDKLALQRL+EAAEKAKIELSST+QTEINLPFITAD SG KHL + LTR+KFEAL
Sbjct: 244 GIDLRKDKLALQRLKEAAEKAKIELSSTTQTEINLPFITADQSGPKHLTMKLTRAKFEAL 303
Query: 352 VNNLIERTKAPCQSCLKDANISTKDIDEVLLVGGMTRVPKVQEVVSTIFGKSPCKGVNPD 411
V++L+++T PC+ LKDA ++ +I EV+LVGGMTR+PKVQEVV +FGK P KGVNPD
Sbjct: 304 VDDLVQKTIEPCRKALKDAGLTAGEISEVVLVGGMTRMPKVQEVVKQLFGKEPHKGVNPD 363
Query: 412 EAVAMGAALQGGILRGDVKELLLLDVTPLSLGIETLGGIFTRLINRNTTIPTKKSQVFST 471
E VA+GAA+Q G+L+GDVK++LLLDVTPLSLGIETLGG+FTR+I+RNTTIPTKKSQVFST
Sbjct: 364 EVVAIGAAIQAGVLQGDVKDVLLLDVTPLSLGIETLGGVFTRIIDRNTTIPTKKSQVFST 423
Query: 472 AADNQTQVGIKVLQGEREMASDNKMLGEFELVGLPPAPRGMPQIEVTFDIDANGIVTVSA 531
A DNQ V I+V QGEREMA+DNKMLG+F+L+G+PPAPRGMPQIEVTFDIDANGIV VSA
Sbjct: 424 AEDNQNAVTIRVFQGEREMAADNKMLGQFDLMGIPPAPRGMPQIEVTFDIDANGIVNVSA 483
Query: 532 KDKSTGKEQQITIKSSGGLSDDEIQNMVKEAELHAQKDQERKSLIDIRNSADTSIYSIEK 591
KDK+TGKEQQI I++SGGLSD EI MVK+AE +A +D++R+ +D +N AD ++S EK
Sbjct: 484 KDKATGKEQQIRIQASGGLSDSEIDKMVKDAEANAAEDKKRREAVDAKNHADALVHSTEK 543
Query: 592 SLSEYREKIPAEVAKEIEDAVSDLRSAMAGDSADEIKSKLDAANKAVSKIGEHMSGGSSS 651
+L+E+ KI + IEDA+SDLR A+ GD A+ IK+K + +A K+GE M + +
Sbjct: 544 ALAEHGSKIDEGERRSIEDALSDLREALKGDDAEAIKTKSNTLAQASMKLGEAMYKQAEA 603
Query: 652 GPSSDGSQGGDQAPEAEYEEV 672
G ++ G D +AE+ EV
Sbjct: 604 GGAAQ-QAGKDDVVDAEFTEV 623
>ref|ZP_00194060.1| COG0443: Molecular chaperone [Mesorhizobium sp. BNC1]
Length = 636
Score = 842 bits (2176), Expect = 0.0
Identities = 427/625 (68%), Positives = 523/625 (83%), Gaps = 6/625 (0%)
Query: 54 VIGIDLGTTNSCVSVMEGKNPKVVENSEGARTTPSVVAFTQKGELLVGTPAKRQAVTNPE 113
VIGIDLGTTNSCV+VM+GK+ KV+EN+EGARTTPS+VAFT E LVG PAKRQAVTNPE
Sbjct: 4 VIGIDLGTTNSCVAVMDGKDAKVIENAEGARTTPSIVAFTDSDERLVGQPAKRQAVTNPE 63
Query: 114 NTISGAKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWVEAKGQQYSPSQIGAFVLTKM 173
NT KRLIGRRF+DP +K+ K+VPYKIVKA NGDAWVE+ G +YSPSQI A +L KM
Sbjct: 64 NTFFAIKRLIGRRFEDPMVEKDKKLVPYKIVKADNGDAWVESHGTKYSPSQISAMILQKM 123
Query: 174 KETAEAYLGKTVPKAVITVPAYFNDAQRQATKDAGRIAGLEVLRIINEPTAAALSYGMNK 233
KETAE+YLG+ V KAVITVPAYFNDAQRQATKDAG+IAGLEVLRIINEPTAAAL+YG++K
Sbjct: 124 KETAESYLGEKVEKAVITVPAYFNDAQRQATKDAGKIAGLEVLRIINEPTAAALAYGLDK 183
Query: 234 -EG-LIAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDFLVNEFKRSE 291
EG IAV+DLGGGTFD+SILEI +GVFEVK+TNGDTFLGGEDFD L+++L +EFK+ +
Sbjct: 184 KEGKTIAVYDLGGGTFDISILEIGDGVFEVKSTNGDTFLGGEDFDMRLVNYLADEFKKEQ 243
Query: 292 SIDLSKDKLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNITLTRSKFEAL 351
IDL DKLALQRL+EAAEKAKIELSS+SQTEINLPFITAD +G KHL I LTR+KFE+L
Sbjct: 244 GIDLKNDKLALQRLKEAAEKAKIELSSSSQTEINLPFITADQTGPKHLAIKLTRAKFESL 303
Query: 352 VNNLIERTKAPCQSCLKDANISTKDIDEVLLVGGMTRVPKVQEVVSTIFGKSPCKGVNPD 411
V +L+ RT PC++ LKDA +S +IDEV+LVGGMTR+PKVQE V FGK P KGVNPD
Sbjct: 304 VEDLVTRTIEPCRAALKDAGLSAGEIDEVVLVGGMTRMPKVQETVKNFFGKEPHKGVNPD 363
Query: 412 EAVAMGAALQGGILRGDVKELLLLDVTPLSLGIETLGGIFTRLINRNTTIPTKKSQVFST 471
E VAMGAA+Q G+L+GDVK++LLLDVTPLSLGIETLGG+FTRLI+RNTTIPTKKSQVFST
Sbjct: 364 EVVAMGAAIQAGVLQGDVKDVLLLDVTPLSLGIETLGGVFTRLIDRNTTIPTKKSQVFST 423
Query: 472 AADNQTQVGIKVLQGEREMASDNKMLGEFELVGLPPAPRGMPQIEVTFDIDANGIVTVSA 531
A DNQ V I+V QGEREMA+DNK+LG+F+LVG+PPAPRGMPQIEVTFDIDANGIV VSA
Sbjct: 424 AEDNQNAVTIRVFQGEREMAADNKLLGQFDLVGIPPAPRGMPQIEVTFDIDANGIVNVSA 483
Query: 532 KDKSTGKEQQITIKSSGGLSDDEIQNMVKEAELHAQKDQERKSLIDIRNSADTSIYSIEK 591
KDK TGKEQQI I++SGGLSD EI+ MVK+AE +A+ D++R+ ++++N A+ I+S EK
Sbjct: 484 KDKGTGKEQQIRIQASGGLSDAEIEKMVKDAEANAEADKKRRETVEVKNQAEALIHSTEK 543
Query: 592 SLSEYREKIPAEVAKEIEDAVSDLRSAMAGDSADEIKSKLDAANKAVSKIGEHMSGGSSS 651
SL +Y +K+ + K IE+A+++L++A G+ AD I++K A +A K+G+ + S +
Sbjct: 544 SLKDYGDKVSEDDRKAIENAIAELKTATEGEDADAIRAKTTALAEASMKLGQAVYEASQA 603
Query: 652 GPSSDGSQ--GG--DQAPEAEYEEV 672
++ G++ GG D +A++EE+
Sbjct: 604 ENAAGGTEETGGAKDDVVDADFEEI 628
>emb|CAE45330.1| unnamed protein product [Magnetospirillum gryphiswaldense]
Length = 686
Score = 841 bits (2173), Expect = 0.0
Identities = 427/676 (63%), Positives = 536/676 (79%), Gaps = 18/676 (2%)
Query: 3 NPTQACCSILIVSQTYSDVVLQLTGNTKPAYVGHNWSSLSRPFSSRAAGNDVIGIDLGTT 62
NP + C+ +S+ + G+ PA + N +S+ VIGIDLGTT
Sbjct: 16 NPAEGVCAPSGLSKGVPAIAALSQGSPPPAKL--NEDDMSK----------VIGIDLGTT 63
Query: 63 NSCVSVMEGKNPKVVENSEGARTTPSVVAFTQKGELLVGTPAKRQAVTNPENTISGAKRL 122
NSCV+VM+GK +V+EN+EG RTTPS+VAFT GE LVG PAKRQAVTNP NT+ KRL
Sbjct: 64 NSCVAVMDGKTARVIENAEGVRTTPSMVAFTDSGERLVGQPAKRQAVTNPTNTLFAIKRL 123
Query: 123 IGRRFDDPQTQKEMKMVPYKIVKAPNGDAWVEAKGQQYSPSQIGAFVLTKMKETAEAYLG 182
IGRRFDDP T+K+M +VPY IV NGDAWVE+ +++SPSQ+ A++L KMKETAE+YLG
Sbjct: 124 IGRRFDDPITKKDMNLVPYHIVNGENGDAWVESGDKKHSPSQVSAYILMKMKETAESYLG 183
Query: 183 KTVPKAVITVPAYFNDAQRQATKDAGRIAGLEVLRIINEPTAAALSYGMNKEG--LIAVF 240
+ V +AVITVPAYFNDAQRQATKDAGRIAGLEVLRIINEPTAAAL+YGM K+G ++AV+
Sbjct: 184 EKVTQAVITVPAYFNDAQRQATKDAGRIAGLEVLRIINEPTAAALAYGMEKKGSGIVAVY 243
Query: 241 DLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDFLVNEFKRSESIDLSKDKL 300
DLGGGTFDVS+LEI +GVFEVK+TNGDTFLGGEDFD ++D+L +EFK+ + IDL D+L
Sbjct: 244 DLGGGTFDVSVLEIGDGVFEVKSTNGDTFLGGEDFDARIMDYLADEFKKEQGIDLRADRL 303
Query: 301 ALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNITLTRSKFEALVNNLIERTK 360
ALQRL+EAAEKAK ELSS+ QTE+NLPFITADASG KHLNI LTRSK EALV +L+ RT
Sbjct: 304 ALQRLKEAAEKAKCELSSSMQTEVNLPFITADASGPKHLNIKLTRSKLEALVEDLVARTV 363
Query: 361 APCQSCLKDANISTKDIDEVLLVGGMTRVPKVQEVVSTIFGKSPCKGVNPDEAVAMGAAL 420
PC++ L+DA + +IDEV+LVGGMTR+PK+QEVV FG+ P KGVNPDE VA+GAA+
Sbjct: 364 EPCKAALRDAGLKASEIDEVILVGGMTRMPKIQEVVKEFFGREPHKGVNPDEVVAIGAAI 423
Query: 421 QGGILRGDVKELLLLDVTPLSLGIETLGGIFTRLINRNTTIPTKKSQVFSTAADNQTQVG 480
QGG+L+G+VK++LLLDVTPLSLGIETLGG+FTRLI+RNTTIPT+KSQVFSTA DNQT V
Sbjct: 424 QGGVLKGEVKDVLLLDVTPLSLGIETLGGVFTRLIDRNTTIPTRKSQVFSTAEDNQTAVT 483
Query: 481 IKVLQGEREMASDNKMLGEFELVGLPPAPRGMPQIEVTFDIDANGIVTVSAKDKSTGKEQ 540
I+V QGEREMA+DNK+LG+F+L+G+PPAPRG+PQ+EVTFDIDANGIV VSAKDK+T KEQ
Sbjct: 484 IRVFQGEREMAADNKILGQFDLMGIPPAPRGVPQVEVTFDIDANGIVNVSAKDKATNKEQ 543
Query: 541 QITIKSSGGLSDDEIQNMVKEAELHAQKDQERKSLIDIRNSADTSIYSIEKSLSEYREKI 600
QI I++SGGL+D +I+ MV+EAE HA +D++RK ++ RN AD ++S EKSL E+ +K
Sbjct: 544 QIRIQASGGLADADIERMVREAEAHAAEDKKRKETVEARNHADGLVHSTEKSLKEFGDKA 603
Query: 601 PAEVAKEIEDAVSDLRSAMAGDSADEIKSKLDAANKAVSKIGEHMSGGSSSGPSSDGSQ- 659
A + EIE A++ LR+ M GD A+ IK+K +A ++ K+GE M +G +D S
Sbjct: 604 DASLKAEIEGAITALRAVMDGDDAEAIKTKTEALMQSSMKLGEAMYKAQEAGGGADDSAP 663
Query: 660 ---GGDQAPEAEYEEV 672
G D+ +A++EEV
Sbjct: 664 QAGGDDKVVDADFEEV 679
>ref|YP_032930.1| Heat shock protein 70 DnaK [Bartonella henselae str. Houston-1]
gi|49237694|emb|CAF26881.1| Heat shock protein 70 DnaK
[Bartonella henselae str. Houston-1]
Length = 630
Score = 841 bits (2172), Expect = 0.0
Identities = 421/622 (67%), Positives = 514/622 (81%), Gaps = 3/622 (0%)
Query: 54 VIGIDLGTTNSCVSVMEGKNPKVVENSEGARTTPSVVAFTQKGELLVGTPAKRQAVTNPE 113
VIGIDLGTTNSCV+VM+GKN KV+ENSEGARTTPSVVAFT GE LVG PAKRQAVTNPE
Sbjct: 4 VIGIDLGTTNSCVAVMDGKNAKVIENSEGARTTPSVVAFTDGGERLVGQPAKRQAVTNPE 63
Query: 114 NTISGAKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWVEAKGQQYSPSQIGAFVLTKM 173
TI KRLIGRRFDDP +K+ +VPYKIVK NGDAWVE G++YSPSQI A +L KM
Sbjct: 64 GTIFAVKRLIGRRFDDPMVEKDKALVPYKIVKGDNGDAWVEEAGKKYSPSQISAMILQKM 123
Query: 174 KETAEAYLGKTVPKAVITVPAYFNDAQRQATKDAGRIAGLEVLRIINEPTAAALSYGMNK 233
KETAE+YLG+ V +AVITVPAYFNDAQRQATKDAG+IAGLEVLRIINEPTAAAL+YG++K
Sbjct: 124 KETAESYLGEKVEQAVITVPAYFNDAQRQATKDAGKIAGLEVLRIINEPTAAALAYGLDK 183
Query: 234 EG--LIAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDFLVNEFKRSE 291
+ IAV+DLGGGTFD+S+LEI +GVFEVK+TNGDTFLGGEDFD L+ + +EFK+ +
Sbjct: 184 KDGKTIAVYDLGGGTFDISVLEIGDGVFEVKSTNGDTFLGGEDFDMRLVGYFADEFKKEQ 243
Query: 292 SIDLSKDKLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNITLTRSKFEAL 351
IDL DKLALQRL+EAAEKAKIELSS+ QTEINLPFITAD SG KHL + LTR+KFE+L
Sbjct: 244 GIDLKNDKLALQRLKEAAEKAKIELSSSQQTEINLPFITADQSGPKHLTMKLTRAKFESL 303
Query: 352 VNNLIERTKAPCQSCLKDANISTKDIDEVLLVGGMTRVPKVQEVVSTIFGKSPCKGVNPD 411
V++L++RT PC++ LKDA + +IDEV+LVGGMTR+PK+QEVV + FGK P KGVNPD
Sbjct: 304 VDDLVKRTVEPCKAALKDAGLKAGEIDEVVLVGGMTRMPKIQEVVQSFFGKDPHKGVNPD 363
Query: 412 EAVAMGAALQGGILRGDVKELLLLDVTPLSLGIETLGGIFTRLINRNTTIPTKKSQVFST 471
E VAMGAA+QGG+L+GDVK++LLLDVTPLSLGIETLGG+FTRLI RNTTIPTKKSQVFST
Sbjct: 364 EVVAMGAAIQGGVLQGDVKDVLLLDVTPLSLGIETLGGVFTRLIERNTTIPTKKSQVFST 423
Query: 472 AADNQTQVGIKVLQGEREMASDNKMLGEFELVGLPPAPRGMPQIEVTFDIDANGIVTVSA 531
A DNQ V I+V QGEREMA+DNK+LG+F+LVG+PPAPRG+PQIEVTFDIDANGIV VSA
Sbjct: 424 ADDNQNAVTIRVFQGEREMANDNKLLGQFDLVGIPPAPRGVPQIEVTFDIDANGIVNVSA 483
Query: 532 KDKSTGKEQQITIKSSGGLSDDEIQNMVKEAELHAQKDQERKSLIDIRNSADTSIYSIEK 591
KDK TGKE QI I++SGGLSD +I+ MVK+AE HA +D++R+ ++ RN A+ I+S EK
Sbjct: 484 KDKGTGKEHQIRIQASGGLSDADIEKMVKDAEEHAAEDKKRREGVEARNQAEALIHSTEK 543
Query: 592 SLSEYREKIPAEVAKEIEDAVSDLRSAMAGDSADEIKSKLDAANKAVSKIGEHMSGGSSS 651
SL+EY +K+ E ++IE A+SDL+S + +E+K+K+ + K+G+ M S +
Sbjct: 544 SLTEYGDKVSTEEKEQIETAISDLKSVLDSTDTEEVKAKMQKLAEVSMKLGQAMYEASQA 603
Query: 652 G-PSSDGSQGGDQAPEAEYEEV 672
P+++ D +A++EE+
Sbjct: 604 ATPNTETDTKSDDVVDADFEEI 625
>ref|NP_945686.1| heat shock protein DnaK (70) [Rhodopseudomonas palustris CGA009]
gi|39647256|emb|CAE25777.1| heat shock protein DnaK (70)
[Rhodopseudomonas palustris CGA009]
Length = 631
Score = 840 bits (2171), Expect = 0.0
Identities = 426/623 (68%), Positives = 517/623 (82%), Gaps = 7/623 (1%)
Query: 54 VIGIDLGTTNSCVSVMEGKNPKVVENSEGARTTPSVVAFTQKGELLVGTPAKRQAVTNPE 113
VIGIDLGTTNSCV+VM+GK+ KV+EN+EG RTTPS+VA T GE LVG PAKRQAVTNPE
Sbjct: 4 VIGIDLGTTNSCVAVMDGKSAKVIENAEGMRTTPSIVAITDDGERLVGQPAKRQAVTNPE 63
Query: 114 NTISGAKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWVEAKGQQYSPSQIGAFVLTKM 173
T KRLIGRR+DDP +K+ +VPYKIVKA NGDAWVEA G+ YSPSQ+ AF+L KM
Sbjct: 64 RTFFAVKRLIGRRYDDPMVEKDKGLVPYKIVKASNGDAWVEADGKTYSPSQVSAFILQKM 123
Query: 174 KETAEAYLGKTVPKAVITVPAYFNDAQRQATKDAGRIAGLEVLRIINEPTAAALSYGMNK 233
KETAEA+LG+ V +AVITVPAYFNDAQRQATKDAG+IAGLEVLRIINEPTAAAL+YG++K
Sbjct: 124 KETAEAHLGQKVDQAVITVPAYFNDAQRQATKDAGKIAGLEVLRIINEPTAAALAYGLDK 183
Query: 234 E--GLIAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDFLVNEFKRSE 291
G IAV+DLGGGTFDVSILEI +GVFEVK+TNGDTFLGGEDFD L+++L +EF++ +
Sbjct: 184 AKTGTIAVYDLGGGTFDVSILEIGDGVFEVKSTNGDTFLGGEDFDMRLVNYLADEFQKEQ 243
Query: 292 SIDLSKDKLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNITLTRSKFEAL 351
IDL KDKLALQRL+EAAEKAKIELSST+QTEINLPFITAD SG KHL + LTR+KFEAL
Sbjct: 244 GIDLRKDKLALQRLKEAAEKAKIELSSTTQTEINLPFITADQSGPKHLTMKLTRAKFEAL 303
Query: 352 VNNLIERTKAPCQSCLKDANISTKDIDEVLLVGGMTRVPKVQEVVSTIFGKSPCKGVNPD 411
V++L+++T PC+ LKDA ++ +I EV+LVGGMTR+PKVQEVV +FGK P KGVNPD
Sbjct: 304 VDDLVQKTIEPCRKALKDAGLTAGEISEVVLVGGMTRMPKVQEVVKQLFGKEPHKGVNPD 363
Query: 412 EAVAMGAALQGGILRGDVKELLLLDVTPLSLGIETLGGIFTRLINRNTTIPTKKSQVFST 471
E VA+GAA+Q G+L+GDVK++LLLDVTPLSLGIETLGG+FTR+I+RNTTIPTKKSQVFST
Sbjct: 364 EVVAIGAAIQAGVLQGDVKDVLLLDVTPLSLGIETLGGVFTRIIDRNTTIPTKKSQVFST 423
Query: 472 AADNQTQVGIKVLQGEREMASDNKMLGEFELVGLPPAPRGMPQIEVTFDIDANGIVTVSA 531
A DNQ V I+V QGEREMA+DNKMLG+F+L+G+PPAPRGMPQIEVTFDIDANGIV VSA
Sbjct: 424 AEDNQNAVTIRVFQGEREMAADNKMLGQFDLMGIPPAPRGMPQIEVTFDIDANGIVNVSA 483
Query: 532 KDKSTGKEQQITIKSSGGLSDDEIQNMVKEAELHAQKDQERKSLIDIRNSADTSIYSIEK 591
KDK+TGKEQQI I++SGGLSD EI MVK+AE +A +D++R+ +D +N AD ++S EK
Sbjct: 484 KDKATGKEQQIRIQASGGLSDSEIDKMVKDAEANAAEDKKRREAVDAKNHADALVHSTEK 543
Query: 592 SLSEYREKIPAEVAKEIEDAVSDLRSAMAGDSADEIKSKLDAANKAVSKIGEHMSGGSSS 651
+L+E+ K+ + IEDA+SDLR A+ GD A+ IK+K + +A K+GE M
Sbjct: 544 ALAEHGSKVDESERRSIEDALSDLREALKGDDAEAIKAKSNTLAQASMKLGEAM---YKQ 600
Query: 652 GPSSDGSQ--GGDQAPEAEYEEV 672
++ G+Q G D +AE+ EV
Sbjct: 601 AEAAGGAQQAGKDDVVDAEFTEV 623
>emb|CAF25566.1| Heat shock protein 70 DnaK [Bartonella quintana str. Toulouse]
gi|49473743|ref|YP_031785.1| Heat shock protein 70 DnaK
[Bartonella quintana str. Toulouse]
Length = 630
Score = 837 bits (2162), Expect = 0.0
Identities = 421/622 (67%), Positives = 513/622 (81%), Gaps = 3/622 (0%)
Query: 54 VIGIDLGTTNSCVSVMEGKNPKVVENSEGARTTPSVVAFTQKGELLVGTPAKRQAVTNPE 113
VIGIDLGTTNSCV+VM+GKN KV+ENSEGARTTPSVVAFT GE LVG PAKRQAVTNPE
Sbjct: 4 VIGIDLGTTNSCVAVMDGKNAKVIENSEGARTTPSVVAFTDGGERLVGQPAKRQAVTNPE 63
Query: 114 NTISGAKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWVEAKGQQYSPSQIGAFVLTKM 173
TI KRLIGRRFDDP +K+ +VPYKIVK NGDAWVE G++YSPSQI A +L KM
Sbjct: 64 GTIFAVKRLIGRRFDDPMVEKDKALVPYKIVKGDNGDAWVEEAGKKYSPSQISAMILQKM 123
Query: 174 KETAEAYLGKTVPKAVITVPAYFNDAQRQATKDAGRIAGLEVLRIINEPTAAALSYGMNK 233
KETAE+YLG+ V +AVITVPAYFNDAQRQATKDAG+IAGLEVLRIINEPTAAAL+YG++K
Sbjct: 124 KETAESYLGEKVEQAVITVPAYFNDAQRQATKDAGKIAGLEVLRIINEPTAAALAYGLDK 183
Query: 234 EG--LIAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDFLVNEFKRSE 291
+ IAV+DLGGGTFD+S+LEI +GVFEVK+TNGDTFLGGEDFD L+ + +EFK+ +
Sbjct: 184 KDGKTIAVYDLGGGTFDISVLEIGDGVFEVKSTNGDTFLGGEDFDMRLVGYFADEFKKEQ 243
Query: 292 SIDLSKDKLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNITLTRSKFEAL 351
IDL DKLALQRL+EAAEKAKIELSS+ QTEINLPFITAD SG KHL + LTR+KFE+L
Sbjct: 244 GIDLKNDKLALQRLKEAAEKAKIELSSSQQTEINLPFITADQSGPKHLTMKLTRAKFESL 303
Query: 352 VNNLIERTKAPCQSCLKDANISTKDIDEVLLVGGMTRVPKVQEVVSTIFGKSPCKGVNPD 411
V++L++RT PC++ LKDA + +IDEV+LVGGMTR+PK+QEVV + FGK P KGVNPD
Sbjct: 304 VDDLVQRTIEPCKAALKDAGLKAGEIDEVVLVGGMTRMPKIQEVVQSFFGKDPHKGVNPD 363
Query: 412 EAVAMGAALQGGILRGDVKELLLLDVTPLSLGIETLGGIFTRLINRNTTIPTKKSQVFST 471
E VAMGAA+QGG+L+GDVK++LLLDVTPLSLGIETLGG+FTRLI RNTTIPTKKSQVFST
Sbjct: 364 EVVAMGAAIQGGVLQGDVKDVLLLDVTPLSLGIETLGGVFTRLIERNTTIPTKKSQVFST 423
Query: 472 AADNQTQVGIKVLQGEREMASDNKMLGEFELVGLPPAPRGMPQIEVTFDIDANGIVTVSA 531
A DNQ V I+V QGEREMA+DNK+L +F+LVG+PPAPRG+PQIEVTFDIDANGIV VSA
Sbjct: 424 ADDNQNAVTIRVFQGEREMANDNKLLAQFDLVGIPPAPRGVPQIEVTFDIDANGIVNVSA 483
Query: 532 KDKSTGKEQQITIKSSGGLSDDEIQNMVKEAELHAQKDQERKSLIDIRNSADTSIYSIEK 591
KDK TGKE QI I++SGGLSD +I+ MVK+AE HA +D++R+ ++ RN A+ I+S EK
Sbjct: 484 KDKGTGKEHQIRIQASGGLSDADIEKMVKDAEEHAAEDKKRREGVEARNQAEALIHSTEK 543
Query: 592 SLSEYREKIPAEVAKEIEDAVSDLRSAMAGDSADEIKSKLDAANKAVSKIGEHMSGGSSS 651
SL+EY +KI E ++IE A+SDL+SA+ G +E+ +K+ + K+G+ M S +
Sbjct: 544 SLTEYGDKISTEEKEQIETAISDLKSALDGTDPEEVTAKMQKLAEVSMKLGQAMYEASQA 603
Query: 652 G-PSSDGSQGGDQAPEAEYEEV 672
+++ D +A++EE+
Sbjct: 604 ATANTETDTKSDDVVDADFEEI 625
>ref|YP_222758.1| chaperone protein DnaK [Brucella abortus biovar 1 str. 9-941]
gi|62197097|gb|AAX75397.1| chaperone protein DnaK
[Brucella abortus biovar 1 str. 9-941]
Length = 637
Score = 837 bits (2161), Expect = 0.0
Identities = 420/628 (66%), Positives = 522/628 (82%), Gaps = 10/628 (1%)
Query: 54 VIGIDLGTTNSCVSVMEGKNPKVVENSEGARTTPSVVAFTQKGELLVGTPAKRQAVTNPE 113
VIGIDLGTTNSCV+VM+GKN KV+EN+EGARTTPS++AFT E L G PAKRQAVTNPE
Sbjct: 4 VIGIDLGTTNSCVAVMDGKNAKVIENAEGARTTPSIIAFTDGDERLAGQPAKRQAVTNPE 63
Query: 114 NTISGAKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWVEAKGQQYSPSQIGAFVLTKM 173
T+ KRLIGRR+DDP K+ +VPYKIVK NGDAWVE G++YSPSQI A +L KM
Sbjct: 64 GTLFAVKRLIGRRYDDPMVTKDKDLVPYKIVKGDNGDAWVEVHGKKYSPSQISAMILQKM 123
Query: 174 KETAEAYLGKTVPKAVITVPAYFNDAQRQATKDAGRIAGLEVLRIINEPTAAALSYGMNK 233
KETAE+YLG+TV +AVITVPAYFNDAQRQATKDAG+IAGLEVLRIINEPTAAAL+YG++K
Sbjct: 124 KETAESYLGETVTQAVITVPAYFNDAQRQATKDAGKIAGLEVLRIINEPTAAALAYGLDK 183
Query: 234 -EG-LIAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDFLVNEFKRSE 291
EG IAV+DLGGGTFDVS+LEI +GVFEVK+TNGDTFLGGEDFD L+++LV EFK+
Sbjct: 184 SEGKTIAVYDLGGGTFDVSVLEIGDGVFEVKSTNGDTFLGGEDFDIRLVEYLVAEFKKES 243
Query: 292 SIDLSKDKLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNITLTRSKFEAL 351
IDL DKLALQRL+EAAEKAKIELSS+ QTEINLPFITAD +G KHL I L+R+KFE+L
Sbjct: 244 GIDLKNDKLALQRLKEAAEKAKIELSSSQQTEINLPFITADQTGPKHLAIKLSRAKFESL 303
Query: 352 VNNLIERTKAPCQSCLKDANISTKDIDEVLLVGGMTRVPKVQEVVSTIFGKSPCKGVNPD 411
V++L++RT PC++ LKDA + +IDEV+LVGGMTR+PK+QEVV FGK P KGVNPD
Sbjct: 304 VDDLVQRTVEPCKAALKDAGLKAGEIDEVVLVGGMTRMPKIQEVVKAFFGKEPHKGVNPD 363
Query: 412 EAVAMGAALQGGILRGDVKELLLLDVTPLSLGIETLGGIFTRLINRNTTIPTKKSQVFST 471
E VAMGAA+QGG+L+GDVK++LLLDVTPLSLGIETLGG+FTRLI RNTTIPTKKSQ FST
Sbjct: 364 EVVAMGAAIQGGVLQGDVKDVLLLDVTPLSLGIETLGGVFTRLIERNTTIPTKKSQTFST 423
Query: 472 AADNQTQVGIKVLQGEREMASDNKMLGEFELVGLPPAPRGMPQIEVTFDIDANGIVTVSA 531
A DNQ+ V I+V QGEREMA+DNK+LG+F+LVG+PPAPRG+PQIEVTFDIDANGIV VSA
Sbjct: 424 AEDNQSAVTIRVFQGEREMAADNKLLGQFDLVGIPPAPRGVPQIEVTFDIDANGIVNVSA 483
Query: 532 KDKSTGKEQQITIKSSGGLSDDEIQNMVKEAELHAQKDQERKSLIDIRNSADTSIYSIEK 591
KDK TGKE QI I++SGGLSD +I+ MVK+AE +A+ D++R+ ++ +N A++ ++S EK
Sbjct: 484 KDKGTGKEHQIRIQASGGLSDADIEKMVKDAEANAEADKKRRESVEAKNQAESLVHSTEK 543
Query: 592 SLSEYREKIPAEVAKEIEDAVSDLRSAMAGDSADEIKSKLDAANKAVSKIGEHMSGGSSS 651
SL+EY +K+ A+ K IEDA++ L++++ G+ A++IK+K A + K+G+ M + +
Sbjct: 544 SLAEYGDKVSADDKKAIEDAIAALKTSLEGEDAEDIKAKTQALAEVSMKLGQAMYEAAQA 603
Query: 652 GPSSDGSQGGDQAP-------EAEYEEV 672
+ G++GG+QA +A+YEE+
Sbjct: 604 AEGA-GAEGGEQASSSKDDVVDADYEEI 630
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.312 0.131 0.355
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,071,880,873
Number of Sequences: 2540612
Number of extensions: 44320159
Number of successful extensions: 136808
Number of sequences better than 10.0: 3294
Number of HSP's better than 10.0 without gapping: 2750
Number of HSP's successfully gapped in prelim test: 546
Number of HSP's that attempted gapping in prelim test: 122398
Number of HSP's gapped (non-prelim): 4017
length of query: 673
length of database: 863,360,394
effective HSP length: 135
effective length of query: 538
effective length of database: 520,377,774
effective search space: 279963242412
effective search space used: 279963242412
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 79 (35.0 bits)
Medicago: description of AC148396.10


