

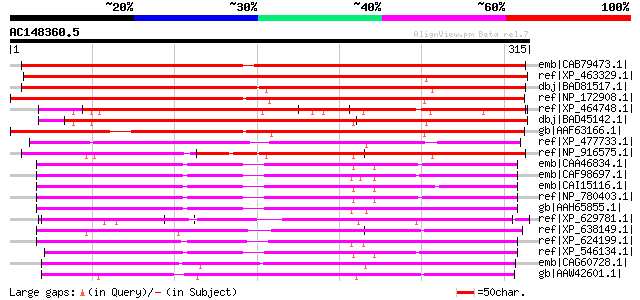
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148360.5 + phase: 0
(315 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAB79473.1| putative mitochondrial carrier protein [Arabidop... 425 e-117
ref|XP_463329.1| putative mitochondrial carrier [Oryza sativa (j... 371 e-101
dbj|BAD81517.1| Graves disease mitochondrial solute carrier prot... 371 e-101
ref|NP_172908.1| mitochondrial substrate carrier family protein ... 359 7e-98
ref|XP_464748.1| putative mitochondrial solute carrier protein [... 354 2e-96
dbj|BAD45142.1| mitochondrial carrier protein-like [Oryza sativa... 336 6e-91
gb|AAF63166.1| T5E21.6 [Arabidopsis thaliana] 323 3e-87
ref|XP_477733.1| putative mitochondrial carrier protein [Oryza s... 233 5e-60
ref|NP_916575.1| P0456F08.3 [Oryza sativa (japonica cultivar-gro... 209 6e-53
emb|CAA46834.1| Graves disease carrier protein from bovine heart... 206 7e-52
emb|CAF98697.1| unnamed protein product [Tetraodon nigroviridis] 203 6e-51
emb|CAI15116.1| solute carrier family 25 (mitochondrial carrier\... 202 1e-50
ref|NP_780403.1| solute carrier family 25 (mitochondrial carrier... 201 2e-50
gb|AAH65855.1| Unknown (protein for MGC:77742) [Danio rerio] gi|... 200 4e-50
ref|XP_629781.1| hypothetical protein DDB0184176 [Dictyostelium ... 196 1e-48
ref|XP_638149.1| hypothetical protein DDB0186597 [Dictyostelium ... 195 1e-48
ref|XP_624199.1| PREDICTED: similar to ENSANGP00000008222, parti... 195 2e-48
ref|XP_546134.1| PREDICTED: similar to solute carrier family 25,... 194 4e-48
emb|CAG60728.1| unnamed protein product [Candida glabrata CBS138... 191 3e-47
gb|AAW42601.1| conserved hypothetical protein [Cryptococcus neof... 190 5e-47
>emb|CAB79473.1| putative mitochondrial carrier protein [Arabidopsis thaliana]
gi|4538947|emb|CAB39683.1| putative mitochondrial
carrier protein [Arabidopsis thaliana]
gi|15236140|ref|NP_194348.1| mitochondrial substrate
carrier family protein [Arabidopsis thaliana]
gi|7441723|pir||T04273 hypothetical protein F20B18.290 -
Arabidopsis thaliana
Length = 325
Score = 425 bits (1092), Expect = e-117
Identities = 210/307 (68%), Positives = 251/307 (81%), Gaps = 7/307 (2%)
Query: 8 ILDHIPLFAKELLAGGLAGGFAKTVVAPLERLKILFQTRRTEFRSAGLSGSVRRIAKTEG 67
I+D IPLFAKEL+AGG+ GG AKT VAPLER+KILFQTRR EF+ GL GS+ +I KTEG
Sbjct: 10 IIDSIPLFAKELIAGGVTGGIAKTAVAPLERIKILFQTRRDEFKRIGLVGSINKIGKTEG 69
Query: 68 LLGFYRGNGASVARIIPYAGLHFMSYEEYRRLIMQAFPNVWKGPTLDLMAGSLSGGTAVL 127
L+GFYRGNGASVARI+PYA LH+M+YEEYRR I+ FP+ +GP LDL+AGS +GGTAVL
Sbjct: 70 LMGFYRGNGASVARIVPYAALHYMAYEEYRRWIIFGFPDTTRGPLLDLVAGSFAGGTAVL 129
Query: 128 FTYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIRGLYRGVAPT 187
FTYPLDL+RTKLAYQ V + + +YRGI DC S+TY+E G RGLYRGVAP+
Sbjct: 130 FTYPLDLVRTKLAYQ------TQVKAIPVEQIIYRGIVDCFSRTYRESGARGLYRGVAPS 183
Query: 188 LFGIFPYAGLKFYFYEEMKRRVPEDYKKSIMAKLTCGSVAGLLGQTFTYPLEVVRRQMQV 247
L+GIFPYAGLKFYFYEEMKR VP ++K+ I KL CGSVAGLLGQT TYPL+VVRRQMQV
Sbjct: 184 LYGIFPYAGLKFYFYEEMKRHVPPEHKQDISLKLVCGSVAGLLGQTLTYPLDVVRRQMQV 243
Query: 248 QNL-AASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSYL 306
+ L +A +E +GTM+++ IA+++GWK LFSGLSINY+KVVPS AIGFTVYD MK +L
Sbjct: 244 ERLYSAVKEETRRGTMQTLFKIAREEGWKQLFSGLSINYLKVVPSVAIGFTVYDIMKLHL 303
Query: 307 RVPSRDE 313
RVP R+E
Sbjct: 304 RVPPREE 310
>ref|XP_463329.1| putative mitochondrial carrier [Oryza sativa (japonica
cultivar-group)] gi|20161078|dbj|BAB90009.1|
mitochondrial carrier protein-like [Oryza sativa
(japonica cultivar-group)]
Length = 340
Score = 371 bits (953), Expect = e-101
Identities = 185/311 (59%), Positives = 235/311 (75%), Gaps = 5/311 (1%)
Query: 9 LDHIPLFAKELLAGGLAGGFAKTVVAPLERLKILFQTRRTEFRSAGLSGSVRRIAKTEGL 68
LD +P++AKEL+AGG AG FAKT VAPLER+KIL QTR F+S G+ S+R++ + EG+
Sbjct: 23 LDLLPVYAKELIAGGAAGAFAKTAVAPLERVKILLQTRTHGFQSLGILQSLRKLWQYEGI 82
Query: 69 LGFYRGNGASVARIIPYAGLHFMSYEEYRRLIMQAF-PNVWKGPTLDLMAGSLSGGTAVL 127
GFY+GNGASV RI+PYA LH+M+YE+YR I+ F P+V GP +DL+AGS +GGTAVL
Sbjct: 83 RGFYKGNGASVLRIVPYAALHYMTYEQYRCWILNNFAPSVGTGPVVDLLAGSAAGGTAVL 142
Query: 128 FTYPLDLIRTKLAYQIVSPTKL-NVSGMVNNEQVYRGIRDCLSKTYKEGGIRGLYRGVAP 186
TYPLDL RTKLAYQ+ + + N G + Y GI+D YKEGG R LYRGV P
Sbjct: 143 CTYPLDLARTKLAYQVSNVGQPGNALGNAGRQPAYGGIKDVFKTVYKEGGARALYRGVGP 202
Query: 187 TLFGIFPYAGLKFYFYEEMKRRVPEDYKKSIMAKLTCGSVAGLLGQTFTYPLEVVRRQMQ 246
TL GI PYAGLKFY YE++K RVPEDYK+S++ KL+CG++AGL GQT TYPL+VVRRQMQ
Sbjct: 203 TLIGILPYAGLKFYIYEDLKSRVPEDYKRSVVLKLSCGALAGLFGQTLTYPLDVVRRQMQ 262
Query: 247 VQNLA---ASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMK 303
VQN A++ ++GT + + LI + QGW+ LF+GLS+NY+KVVPS AIGFT YD MK
Sbjct: 263 VQNKQPHNANDAFRIRGTFQGLALIIRCQGWRQLFAGLSLNYVKVVPSVAIGFTTYDMMK 322
Query: 304 SYLRVPSRDEV 314
+ LRVP R+ +
Sbjct: 323 NLLRVPPRERL 333
>dbj|BAD81517.1| Graves disease mitochondrial solute carrier protein-like [Oryza
sativa (japonica cultivar-group)]
Length = 337
Score = 371 bits (952), Expect = e-101
Identities = 181/311 (58%), Positives = 237/311 (76%), Gaps = 6/311 (1%)
Query: 8 ILDHIPLFAKELLAGGLAGGFAKTVVAPLERLKILFQTRRTEFRSAGLSGSVRRIAKTEG 67
+LD +P+FAKE++AGG+AG F+KT +APLERLKIL QTR EF S G+ S++++ + +G
Sbjct: 20 VLDLVPVFAKEMIAGGVAGAFSKTAIAPLERLKILLQTRTNEFSSLGVLKSLKKLKQHDG 79
Query: 68 LLGFYRGNGASVARIIPYAGLHFMSYEEYRRLIMQAFPNVWKGPTLDLMAGSLSGGTAVL 127
+LGFY+GNGASV RI+PYA LH+M+YE YR I+ P++ GP +DL+AGS SGGTAVL
Sbjct: 80 ILGFYKGNGASVLRIVPYAALHYMAYERYRCWILNNCPSLGTGPLVDLLAGSASGGTAVL 139
Query: 128 FTYPLDLIRTKLAYQIVSPTKLNVSGM--VNNEQVYRGIRDCLSKTYKEGGIRGLYRGVA 185
TYPLDL RTKLA+Q+ S +++ SG+ N + Y GI+D Y EGG+R LYRGV
Sbjct: 140 CTYPLDLARTKLAFQVNSSDQIS-SGLKRTNFQPKYGGIKDVFRGVYSEGGVRALYRGVG 198
Query: 186 PTLFGIFPYAGLKFYFYEEMKRRVPEDYKKSIMAKLTCGSVAGLLGQTFTYPLEVVRRQM 245
PTL GI PYAGLKFY YE +K VPEDYK S+ KL+CG+ AGL GQT TYPL+VVRRQM
Sbjct: 199 PTLMGILPYAGLKFYIYEGLKAHVPEDYKNSVTLKLSCGAAAGLFGQTLTYPLDVVRRQM 258
Query: 246 QVQNLAASEE---AELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTM 302
QVQ+ ++ +++GT + +++I Q QGW+ LF+GLS+NYIKVVPS AIGFT YDTM
Sbjct: 259 QVQSQQYHDKFGGPQIRGTFQGLMIIKQTQGWRQLFAGLSLNYIKVVPSVAIGFTAYDTM 318
Query: 303 KSYLRVPSRDE 313
KS L++P R++
Sbjct: 319 KSLLKIPPREK 329
>ref|NP_172908.1| mitochondrial substrate carrier family protein [Arabidopsis
thaliana]
Length = 331
Score = 359 bits (921), Expect = 7e-98
Identities = 180/319 (56%), Positives = 235/319 (73%), Gaps = 8/319 (2%)
Query: 1 MAASSESILDHIPLFAKELLAGGLAGGFAKTVVAPLERLKILFQTRRTEFRSAGLSGSVR 60
++A S++D +P+ AK L+AGG AG AKT VAPLER+KIL QTR +F++ G+S S++
Sbjct: 9 LSADVMSLVDTLPVLAKTLIAGGAAGAIAKTAVAPLERIKILLQTRTNDFKTLGVSQSLK 68
Query: 61 RIAKTEGLLGFYRGNGASVARIIPYAGLHFMSYEEYRRLIMQAFPNVWKGPTLDLMAGSL 120
++ + +G LGFY+GNGASV RIIPYA LH+M+YE YR I++ + GP +DL+AGS
Sbjct: 69 KVLQFDGPLGFYKGNGASVIRIIPYAALHYMTYEVYRDWILEKNLPLGSGPIVDLVAGSA 128
Query: 121 SGGTAVLFTYPLDLIRTKLAYQIVSPTKLNVSGMVN---NEQVYRGIRDCLSKTYKEGGI 177
+GGTAVL TYPLDL RTKLAYQ VS T+ ++ G N + Y GI++ L+ YKEGG
Sbjct: 129 AGGTAVLCTYPLDLARTKLAYQ-VSDTRQSLRGGANGFYRQPTYSGIKEVLAMAYKEGGP 187
Query: 178 RGLYRGVAPTLFGIFPYAGLKFYFYEEMKRRVPEDYKKSIMAKLTCGSVAGLLGQTFTYP 237
RGLYRG+ PTL GI PYAGLKFY YEE+KR VPE+++ S+ L CG++AGL GQT TYP
Sbjct: 188 RGLYRGIGPTLIGILPYAGLKFYIYEELKRHVPEEHQNSVRMHLPCGALAGLFGQTITYP 247
Query: 238 LEVVRRQMQVQNL----AASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAA 293
L+VVRRQMQV+NL + K T + I + QGWK LF+GLSINYIK+VPS A
Sbjct: 248 LDVVRRQMQVENLQPMTSEGNNKRYKNTFDGLNTIVRTQGWKQLFAGLSINYIKIVPSVA 307
Query: 294 IGFTVYDTMKSYLRVPSRD 312
IGFTVY++MKS++R+P R+
Sbjct: 308 IGFTVYESMKSWMRIPPRE 326
>ref|XP_464748.1| putative mitochondrial solute carrier protein [Oryza sativa
(japonica cultivar-group)] gi|49388534|dbj|BAD25656.1|
putative mitochondrial solute carrier protein [Oryza
sativa (japonica cultivar-group)]
Length = 426
Score = 354 bits (909), Expect = 2e-96
Identities = 177/289 (61%), Positives = 221/289 (76%), Gaps = 22/289 (7%)
Query: 45 TRRTEFRSAGLSGSVRRIAKTEGLLGFYRGNGASVARIIPYAGLHFMSYEEYRRLIMQAF 104
TRR EF +GL GS R I++TEGLLGFYRGNGASVARI+PYA LH+M+YEEYRR I+ F
Sbjct: 121 TRRAEFHGSGLIGSFRTISRTEGLLGFYRGNGASVARIVPYAALHYMAYEEYRRWIILGF 180
Query: 105 PNVWKGPTLDLMAGSLSGGTAVLFTYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGI 164
PNV +GP LDL+AGS++GGTAV+ TYPLDL+RTKLAYQ+ KL++ +EQVY+GI
Sbjct: 181 PNVEQGPILDLVAGSIAGGTAVICTYPLDLVRTKLAYQVKGAVKLSLREYKPSEQVYKGI 240
Query: 165 RDCLSKTYKEGGIRGLYRGVAPTLFGIFPYAGLKFYFYEEMKRRVPEDYKKSIMAKLTCG 224
DC+ Y++ G+RGLYRG+AP+L+GIFPY+GLKFYFYE MK VPE+++K I+AKL CG
Sbjct: 241 LDCVKTIYRQNGLRGLYRGMAPSLYGIFPYSGLKFYFYETMKTYVPEEHRKDIIAKLACG 300
Query: 225 SVAGLLGQTFTYPLEVVRRQMQVQNLAASEEAELKGTMRSMVLIAQKQGWKTLFSGLSIN 284
SVAGLLGQT TYPL+VVRRQMQ ++S + KGT S+ +IA+ QGW+ LFSGLSIN
Sbjct: 301 SVAGLLGQTITYPLDVVRRQMQA--FSSSNLEKGKGTFGSIAMIAKHQGWRQLFSGLSIN 358
Query: 285 YI--------------------KVVPSAAIGFTVYDTMKSYLRVPSRDE 313
Y+ KVVPS AIGFTVYD+MK +L+VPSR++
Sbjct: 359 YLKELYQLDTSVCAFIYVQCGEKVVPSVAIGFTVYDSMKVWLKVPSRED 407
Score = 65.5 bits (158), Expect = 2e-09
Identities = 42/151 (27%), Positives = 75/151 (48%), Gaps = 12/151 (7%)
Query: 176 GIRGLYRGVAPTLFGIFPYAGLKFYFYEEMKRRVPEDY---KKSIMAKLTCGSVAGLLGQ 232
G+ G YRG ++ I PYA L + YEE +R + + ++ + L GS+AG
Sbjct: 143 GLLGFYRGNGASVARIVPYAALHYMAYEEYRRWIILGFPNVEQGPILDLVAGSIAGGTAV 202
Query: 233 TFTYPLEVVRRQMQVQNLAASE---------EAELKGTMRSMVLIAQKQGWKTLFSGLSI 283
TYPL++VR ++ Q A + E KG + + I ++ G + L+ G++
Sbjct: 203 ICTYPLDLVRTKLAYQVKGAVKLSLREYKPSEQVYKGILDCVKTIYRQNGLRGLYRGMAP 262
Query: 284 NYIKVVPSAAIGFTVYDTMKSYLRVPSRDEV 314
+ + P + + F Y+TMK+Y+ R ++
Sbjct: 263 SLYGIFPYSGLKFYFYETMKTYVPEEHRKDI 293
Score = 60.8 bits (146), Expect = 5e-08
Identities = 59/217 (27%), Positives = 95/217 (43%), Gaps = 37/217 (17%)
Query: 18 ELLAGGLAGGFAKTVVAPLE--RLKILFQTRRT------EFRSA-----GLSGSVRRIAK 64
+L+AG +AGG A PL+ R K+ +Q + E++ + G+ V+ I +
Sbjct: 190 DLVAGSIAGGTAVICTYPLDLVRTKLAYQVKGAVKLSLREYKPSEQVYKGILDCVKTIYR 249
Query: 65 TEGLLGFYRGNGASVARIIPYAGLHFMSYEEYRRLIMQAFPNVWKGPTLDLMAGSLSGGT 124
GL G YRG S+ I PY+GL F YE + + + K L GS++G
Sbjct: 250 QNGLRGLYRGMAPSLYGIFPYSGLKFYFYETMKTYVPEEHR---KDIIAKLACGSVAGLL 306
Query: 125 AVLFTYPLDLIRTKLAYQIVSPTKLNVS-------GMVNNEQVYRGIRDCLSKTYKEGGI 177
TYPLD++R ++ Q S + L M+ Q +R + LS Y +
Sbjct: 307 GQTITYPLDVVRRQM--QAFSSSNLEKGKGTFGSIAMIAKHQGWRQLFSGLSINY----L 360
Query: 178 RGLYR---GVAPTLF-----GIFPYAGLKFYFYEEMK 206
+ LY+ V ++ + P + F Y+ MK
Sbjct: 361 KELYQLDTSVCAFIYVQCGEKVVPSVAIGFTVYDSMK 397
>dbj|BAD45142.1| mitochondrial carrier protein-like [Oryza sativa (japonica
cultivar-group)]
Length = 330
Score = 336 bits (861), Expect = 6e-91
Identities = 167/286 (58%), Positives = 214/286 (74%), Gaps = 5/286 (1%)
Query: 34 APLERLKILFQTRRTEFRSAGLSGSVRRIAKTEGLLGFYRGNGASVARIIPYAGLHFMSY 93
+PLER+KIL QTR F+S G+ S+R++ + EG+ GFY+GNGASV RI+PYA LH+M+Y
Sbjct: 38 SPLERVKILLQTRTHGFQSLGILQSLRKLWQYEGIRGFYKGNGASVLRIVPYAALHYMTY 97
Query: 94 EEYRRLIMQAF-PNVWKGPTLDLMAGSLSGGTAVLFTYPLDLIRTKLAYQIVSPTKL-NV 151
E+YR I+ F P+V GP +DL+AGS +GGTAVL TYPLDL RTKLAYQ+ + + N
Sbjct: 98 EQYRCWILNNFAPSVGTGPVVDLLAGSAAGGTAVLCTYPLDLARTKLAYQVSNVGQPGNA 157
Query: 152 SGMVNNEQVYRGIRDCLSKTYKEGGIRGLYRGVAPTLFGIFPYAGLKFYFYEEMKRRVPE 211
G + Y GI+D YKEGG R LYRGV PTL GI PYAGLKFY YE++K RVPE
Sbjct: 158 LGNAGRQPAYGGIKDVFKTVYKEGGARALYRGVGPTLIGILPYAGLKFYIYEDLKSRVPE 217
Query: 212 DYKKSIMAKLTCGSVAGLLGQTFTYPLEVVRRQMQVQNLA---ASEEAELKGTMRSMVLI 268
DYK+S++ KL+CG++AGL GQT TYPL+VVRRQMQVQN A++ ++GT + + LI
Sbjct: 218 DYKRSVVLKLSCGALAGLFGQTLTYPLDVVRRQMQVQNKQPHNANDAFRIRGTFQGLALI 277
Query: 269 AQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSYLRVPSRDEV 314
+ QGW+ LF+GLS+NY+KVVPS AIGFT YD MK+ LRVP R+ +
Sbjct: 278 IRCQGWRQLFAGLSLNYVKVVPSVAIGFTTYDMMKNLLRVPPRERL 323
Score = 71.2 bits (173), Expect = 4e-11
Identities = 58/209 (27%), Positives = 91/209 (42%), Gaps = 25/209 (11%)
Query: 18 ELLAGGLAGGFAKTVVAPLE--RLKILFQTRRT------------EFRSAGLSGSVRRIA 63
+LLAG AGG A PL+ R K+ +Q + G+ + +
Sbjct: 119 DLLAGSAAGGTAVLCTYPLDLARTKLAYQVSNVGQPGNALGNAGRQPAYGGIKDVFKTVY 178
Query: 64 KTEGLLGFYRGNGASVARIIPYAGLHFMSYEEYRRLIMQAFPNVWKGPTLDLMAGSLSGG 123
K G YRG G ++ I+PYAGL F YE+ + + + + + L L G+L+G
Sbjct: 179 KEGGARALYRGVGPTLIGILPYAGLKFYIYEDLKSRVPEDYK---RSVVLKLSCGALAGL 235
Query: 124 TAVLFTYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIRGLYRG 183
TYPLD++R ++ Q P N + + RG L+ + G R L+ G
Sbjct: 236 FGQTLTYPLDVVRRQMQVQNKQPHNANDAFRI------RGTFQGLALIIRCQGWRQLFAG 289
Query: 184 VAPTLFGIFPYAGLKFYFYEEMKR--RVP 210
++ + P + F Y+ MK RVP
Sbjct: 290 LSLNYVKVVPSVAIGFTTYDMMKNLLRVP 318
>gb|AAF63166.1| T5E21.6 [Arabidopsis thaliana]
Length = 319
Score = 323 bits (829), Expect = 3e-87
Identities = 172/319 (53%), Positives = 219/319 (67%), Gaps = 20/319 (6%)
Query: 1 MAASSESILDHIPLFAKELLAGGLAGGFAKTVVAPLERLKILFQTRRTEFRSAGLSGSVR 60
++A S++D +P+ AK L+AGG AG AKT VAPLER+KIL Q T ++ + R
Sbjct: 9 LSADVMSLVDTLPVLAKTLIAGGAAGAIAKTAVAPLERIKILLQLSSTTLLNSCDVHNSR 68
Query: 61 RIAKTEGLLGFYRGNGASVARIIPYAGLHFMSYEEYRRLIMQAFPNVWKGPTLDLMAGSL 120
R GNGASV RIIPYA LH+M+YE YR I++ + GP +DL+AGS
Sbjct: 69 R------------GNGASVIRIIPYAALHYMTYEVYRDWILEKNLPLGSGPIVDLVAGSA 116
Query: 121 SGGTAVLFTYPLDLIRTKLAYQIVSPTKLNVSGMVNN---EQVYRGIRDCLSKTYKEGGI 177
+GGTAVL TYPLDL RTKLAYQ VS T+ ++ G N + Y GI++ L+ YKEGG
Sbjct: 117 AGGTAVLCTYPLDLARTKLAYQ-VSDTRQSLRGGANGFYRQPTYSGIKEVLAMAYKEGGP 175
Query: 178 RGLYRGVAPTLFGIFPYAGLKFYFYEEMKRRVPEDYKKSIMAKLTCGSVAGLLGQTFTYP 237
RGLYRG+ PTL GI PYAGLKFY YEE+KR VPE+++ S+ L CG++AGL GQT TYP
Sbjct: 176 RGLYRGIGPTLIGILPYAGLKFYIYEELKRHVPEEHQNSVRMHLPCGALAGLFGQTITYP 235
Query: 238 LEVVRRQMQVQNL----AASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAA 293
L+VVRRQMQV+NL + K T + I + QGWK LF+GLSINYIK+VPS A
Sbjct: 236 LDVVRRQMQVENLQPMTSEGNNKRYKNTFDGLNTIVRTQGWKQLFAGLSINYIKIVPSVA 295
Query: 294 IGFTVYDTMKSYLRVPSRD 312
IGFTVY++MKS++R+P R+
Sbjct: 296 IGFTVYESMKSWMRIPPRE 314
>ref|XP_477733.1| putative mitochondrial carrier protein [Oryza sativa (japonica
cultivar-group)] gi|34394749|dbj|BAC84113.1| putative
mitochondrial carrier protein [Oryza sativa (japonica
cultivar-group)]
Length = 333
Score = 233 bits (594), Expect = 5e-60
Identities = 129/305 (42%), Positives = 182/305 (59%), Gaps = 26/305 (8%)
Query: 13 PLFAKELLAGGLAGGFAKTVVAPLERLKILFQTRRTEFRSAGLSGSVRRIAKTEGLLGFY 72
P FA+E++AGG+AG AKT VAPLER+ ++ Q R AG +R I + EG+ G +
Sbjct: 39 PAFAREMVAGGVAGVVAKTAVAPLERVNLMRQVGAAP-RGAGAVQMLREIGRGEGVAGLF 97
Query: 73 RGNGASVARIIPYAGLHFMSYEEYRRLIMQAFPNVWKGPTLDLMAGSLSGGTAVLFTYPL 132
RGNGA+ R+ LHFM+YE Y+R ++ A P++ GP +DL+AGS +GGTAVL TYPL
Sbjct: 98 RGNGANALRVFHTKALHFMAYERYKRFLLGAAPSLGDGPVVDLLAGSAAGGTAVLATYPL 157
Query: 133 DLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKE-GGIRGLYRGVAPTLFGI 191
DL RT+LA P G+ L Y+E GG+RG+YRG+ P+L +
Sbjct: 158 DLARTRLACAAAPP-----------GAAAAGMSGVLRSAYREGGGVRGVYRGLCPSLARV 206
Query: 192 FPYAGLKFYFYEEMKRRVPEDYKK------SIMAKLTCGSVAGLLGQTFTYPLEVVRRQM 245
P +GL F YE +K ++P + ++ AK+ CG AGL+ T TYPL+VVRRQ+
Sbjct: 207 LPMSGLNFCVYEALKAQIPREEEEHGARGWRRAAKVACGVAAGLVASTATYPLDVVRRQI 266
Query: 246 QVQNLAASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSY 305
Q+ GT+++ I + QG + L++GL I Y+K VPS A+G YD MKS
Sbjct: 267 QLGGGGG-------GTLQAFRAIVRAQGARQLYAGLGITYVKKVPSTAVGLVAYDYMKSL 319
Query: 306 LRVPS 310
L +P+
Sbjct: 320 LMLPA 324
>ref|NP_916575.1| P0456F08.3 [Oryza sativa (japonica cultivar-group)]
Length = 239
Score = 209 bits (533), Expect = 6e-53
Identities = 106/205 (51%), Positives = 144/205 (69%), Gaps = 8/205 (3%)
Query: 114 DLMAGSLSGGTAVLFTYPLDLIRTKLAYQIVSPTKLNVSGM--VNNEQVYRGIRDCLSKT 171
+++AG ++G + PL+ R K+ Q+ S +++ SG+ N + Y GI+D
Sbjct: 30 EMIAGGVAGAFSKTAIAPLE--RLKILLQVNSSDQIS-SGLKRTNFQPKYGGIKDVFRGV 86
Query: 172 YKEGGIRGLYRGVAPTLFGIFPYAGLKFYFYEEMKRRVPEDYKKSIMAKLTCGSVAGLLG 231
Y EGG+R LYRGV PTL GI PYAGLKFY YE +K VPEDYK S+ KL+CG+ AGL G
Sbjct: 87 YSEGGVRALYRGVGPTLMGILPYAGLKFYIYEGLKAHVPEDYKNSVTLKLSCGAAAGLFG 146
Query: 232 QTFTYPLEVVRRQMQVQNLAASEE---AELKGTMRSMVLIAQKQGWKTLFSGLSINYIKV 288
QT TYPL+VVRRQMQVQ+ ++ +++GT + +++I Q QGW+ LF+GLS+NYIKV
Sbjct: 147 QTLTYPLDVVRRQMQVQSQQYHDKFGGPQIRGTFQGLMIIKQTQGWRQLFAGLSLNYIKV 206
Query: 289 VPSAAIGFTVYDTMKSYLRVPSRDE 313
VPS AIGFT YDTMKS L++P R++
Sbjct: 207 VPSVAIGFTAYDTMKSLLKIPPREK 231
Score = 105 bits (261), Expect = 2e-21
Identities = 73/222 (32%), Positives = 109/222 (48%), Gaps = 23/222 (10%)
Query: 8 ILDHIPLFAKELLAGGLAGGFAKTVVAPLERLKILFQT----------RRTEF--RSAGL 55
+LD +P+FAKE++AGG+AG F+KT +APLERLKIL Q +RT F + G+
Sbjct: 20 VLDLVPVFAKEMIAGGVAGAFSKTAIAPLERLKILLQVNSSDQISSGLKRTNFQPKYGGI 79
Query: 56 SGSVRRIAKTEGLLGFYRGNGASVARIIPYAGLHFMSYEEYRRLIMQAFPNVWKGPTLDL 115
R + G+ YRG G ++ I+PYAGL F YE + + + + N TL L
Sbjct: 80 KDVFRGVYSEGGVRALYRGVGPTLMGILPYAGLKFYIYEGLKAHVPEDYKN---SVTLKL 136
Query: 116 MAGSLSGGTAVLFTYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEG 175
G+ +G TYPLD++R ++ Q Q+ RG L +
Sbjct: 137 SCGAAAGLFGQTLTYPLDVVRRQMQVQ-----SQQYHDKFGGPQI-RGTFQGLMIIKQTQ 190
Query: 176 GIRGLYRGVAPTLFGIFPYAGLKFYFYEEMKR--RVPEDYKK 215
G R L+ G++ + P + F Y+ MK ++P KK
Sbjct: 191 GWRQLFAGLSLNYIKVVPSVAIGFTAYDTMKSLLKIPPREKK 232
>emb|CAA46834.1| Graves disease carrier protein from bovine heart mitochondria [Bos
taurus] gi|266574|sp|Q01888|GDC_BOVIN Grave's disease
carrier protein (GDC) (Mitochondrial solute carrier
protein homolog) gi|27807213|ref|NP_777097.1| solute
carrier family 25, member 16 [Bos taurus]
Length = 330
Score = 206 bits (524), Expect = 7e-52
Identities = 125/310 (40%), Positives = 171/310 (54%), Gaps = 34/310 (10%)
Query: 17 KELLAGGLAGGFAKTVVAPLERLKILFQTRRTEFRSAGLSGSVRRIAKTEGLLGFYRGNG 76
+ LAGG+AG AKT VAPL+R+K+L Q +R G+ ++R + K EG LG Y+GNG
Sbjct: 36 RSFLAGGIAGCCAKTTVAPLDRVKVLLQAHNHHYRHLGVFSTLRAVPKKEGYLGLYKGNG 95
Query: 77 ASVARIIPYAGLHFMSYEEYRRLIMQAFPNVWKGPTLDLMAGSLSGGTAVLFTYPLDLIR 136
A + RI PY + FM++E Y+ LI G LMAGS++G TAV+ TYPLD++R
Sbjct: 96 AMMIRIFPYGAIQFMAFEHYKTLITTKLG--VSGHVHRLMAGSMAGMTAVICTYPLDMVR 153
Query: 137 TKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTY-KEGGIRGLYRGVAPTLFGIFPYA 195
+LA+Q V E Y GI Y KEGG G YRG+ PT+ G+ PYA
Sbjct: 154 VRLAFQ------------VKGEHTYTGIIHAFKTIYAKEGGFLGFYRGLMPTILGMAPYA 201
Query: 196 GLKFYFYEEMKR-----------RVPEDYKKSIMAK----LTCGSVAGLLGQTFTYPLEV 240
G+ F+ + +K R D ++ K L CG VAG + QT +YP +V
Sbjct: 202 GVSFFTFGTLKSVGLSYAPTLLGRPSSDNPNVLVLKTHINLLCGGVAGAIAQTISYPFDV 261
Query: 241 VRRQMQVQNLAASEEAELKGTMR-SMVLIAQKQG-WKTLFSGLSINYIKVVPSAAIGFTV 298
RR+MQ+ A E E TMR +M + G K L+ GLS+NYI+ VPS A+ FT
Sbjct: 262 TRRRMQLG--AVLPEFEKCLTMRETMKYVYGHHGIRKGLYRGLSLNYIRCVPSQAVAFTT 319
Query: 299 YDTMKSYLRV 308
Y+ MK + +
Sbjct: 320 YELMKQFFHL 329
>emb|CAF98697.1| unnamed protein product [Tetraodon nigroviridis]
Length = 313
Score = 203 bits (516), Expect = 6e-51
Identities = 116/309 (37%), Positives = 176/309 (56%), Gaps = 32/309 (10%)
Query: 17 KELLAGGLAGGFAKTVVAPLERLKILFQTRRTEFRSAGLSGSVRRIAKTEGLLGFYRGNG 76
+ +AGG+AG AKT +APL+R+KIL Q + ++ G+ ++R + + EG LG Y+GNG
Sbjct: 19 RSFVAGGVAGCCAKTTIAPLDRIKILLQAQNPHYKHLGVFATLRAVPQKEGFLGLYKGNG 78
Query: 77 ASVARIIPYAGLHFMSYEEYRRLIMQAFPNVWKGPTLDLMAGSLSGGTAVLFTYPLDLIR 136
A + RI PY + FM+++ Y++L+ G LMAGS++G TAV+ TYPLD++R
Sbjct: 79 AMMVRIFPYGAIQFMAFDNYKKLLSTQIG--ISGHIHRLMAGSMAGMTAVICTYPLDVVR 136
Query: 137 TKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTY-KEGGIRGLYRGVAPTLFGIFPYA 195
+LA+Q V E Y GI + Y KEGG+ G YRG+ PTL G+ PYA
Sbjct: 137 ARLAFQ------------VTGEHRYTGIANAFHTIYLKEGGVLGFYRGLTPTLIGMAPYA 184
Query: 196 GLKFYFYEEMK----RRVPE-------DYKKSIMAK----LTCGSVAGLLGQTFTYPLEV 240
G F+ + +K + PE D ++ K L CG +AG + QT +YPL+V
Sbjct: 185 GFSFFTFGTLKSLGLKHFPELLGRPSSDNPNVLVLKPQVNLLCGGMAGAVAQTISYPLDV 244
Query: 241 VRRQMQVQNLAASEEAELKGTMRSMVLIAQKQGWKT-LFSGLSINYIKVVPSAAIGFTVY 299
RR+MQ+ + + +++ + ++ G K L+ GLS+NYI+ VPS A+ FT Y
Sbjct: 245 ARRRMQL-GAVLPDSDKCVSLSKTLTYVYKQYGIKKGLYRGLSLNYIRCVPSQAMAFTTY 303
Query: 300 DTMKSYLRV 308
+ MK L +
Sbjct: 304 EFMKQVLHL 312
Score = 40.0 bits (92), Expect = 0.089
Identities = 23/89 (25%), Positives = 40/89 (44%), Gaps = 4/89 (4%)
Query: 218 MAKLTCGSVAGLLGQTFTYPLEVVRRQMQVQNLAASEEAELKGTMRSMVLIAQKQGWKTL 277
+ G VAG +T PL+ ++ +Q QN + G ++ + QK+G+ L
Sbjct: 18 LRSFVAGGVAGCCAKTTIAPLDRIKILLQAQN----PHYKHLGVFATLRAVPQKEGFLGL 73
Query: 278 FSGLSINYIKVVPSAAIGFTVYDTMKSYL 306
+ G +++ P AI F +D K L
Sbjct: 74 YKGNGAMMVRIFPYGAIQFMAFDNYKKLL 102
>emb|CAI15116.1| solute carrier family 25 (mitochondrial carrier\; Graves disease
autoantigen), member 16 [Homo sapiens]
gi|20988432|gb|AAH30266.1| Solute carrier family 25,
member 16 [Homo sapiens] gi|27544933|ref|NP_689920.1|
solute carrier family 25, member 16 [Homo sapiens]
Length = 332
Score = 202 bits (514), Expect = 1e-50
Identities = 121/310 (39%), Positives = 171/310 (55%), Gaps = 34/310 (10%)
Query: 17 KELLAGGLAGGFAKTVVAPLERLKILFQTRRTEFRSAGLSGSVRRIAKTEGLLGFYRGNG 76
+ LAGG+AG AKT VAPL+R+K+L Q ++ G+ ++R + + EG LG Y+GNG
Sbjct: 38 RSFLAGGIAGCCAKTTVAPLDRVKVLLQAHNHHYKHLGVFSALRAVPQKEGFLGLYKGNG 97
Query: 77 ASVARIIPYAGLHFMSYEEYRRLIMQAFPNVWKGPTLDLMAGSLSGGTAVLFTYPLDLIR 136
A + RI PY + FM++E Y+ LI G LMAGS++G TAV+ TYPLD++R
Sbjct: 98 AMMIRIFPYGAIQFMAFEHYKTLITTKLG--ISGHVHRLMAGSMAGMTAVICTYPLDMVR 155
Query: 137 TKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTY-KEGGIRGLYRGVAPTLFGIFPYA 195
+LA+Q V E Y GI Y KEGG G YRG+ PT+ G+ PYA
Sbjct: 156 VRLAFQ------------VKGEHSYTGIIHAFKTIYAKEGGFFGFYRGLMPTILGMAPYA 203
Query: 196 GLKFYFYEEMKR-----------RVPEDYKKSIMAK----LTCGSVAGLLGQTFTYPLEV 240
G+ F+ + +K R D ++ K L CG VAG + QT +YP +V
Sbjct: 204 GVSFFTFGTLKSVGLSHAPTLLGRPSSDNPNVLVLKTHVNLLCGGVAGAIAQTISYPFDV 263
Query: 241 VRRQMQVQNLAASEEAELKGTMR-SMVLIAQKQG-WKTLFSGLSINYIKVVPSAAIGFTV 298
RR+MQ+ + E L TMR +M + G K L+ GLS+NYI+ +PS A+ FT
Sbjct: 264 TRRRMQLGTVLPEFEKCL--TMRDTMKYVYGHHGIRKGLYRGLSLNYIRCIPSQAVAFTT 321
Query: 299 YDTMKSYLRV 308
Y+ MK + +
Sbjct: 322 YELMKQFFHL 331
>ref|NP_780403.1| solute carrier family 25 (mitochondrial carrier, Graves disease
autoantigen), member 16 [Mus musculus]
gi|26326839|dbj|BAC27163.1| unnamed protein product [Mus
musculus]
Length = 332
Score = 201 bits (512), Expect = 2e-50
Identities = 120/310 (38%), Positives = 171/310 (54%), Gaps = 34/310 (10%)
Query: 17 KELLAGGLAGGFAKTVVAPLERLKILFQTRRTEFRSAGLSGSVRRIAKTEGLLGFYRGNG 76
+ LAGG+AG AKT VAPL+R+K+L Q ++ G+ ++R + + EG LG Y+GNG
Sbjct: 38 RSFLAGGIAGCCAKTTVAPLDRVKVLLQAHNRHYKHLGVLSTLRAVPQKEGYLGLYKGNG 97
Query: 77 ASVARIIPYAGLHFMSYEEYRRLIMQAFPNVWKGPTLDLMAGSLSGGTAVLFTYPLDLIR 136
A + RI PY + FM++E Y+ I G LMAGS++G TAV+ TYPLD++R
Sbjct: 98 AMMIRIFPYGAIQFMAFEHYKTFITTKLG--VSGHVHRLMAGSMAGMTAVICTYPLDVVR 155
Query: 137 TKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTY-KEGGIRGLYRGVAPTLFGIFPYA 195
+LA+Q V E Y GI Y KEGG G YRG+ PT+ G+ PYA
Sbjct: 156 VRLAFQ------------VKGEHTYSGIIHAFKTIYAKEGGFLGFYRGLMPTILGMAPYA 203
Query: 196 GLKFYFYEEMKR-----------RVPEDYKKSIMAK----LTCGSVAGLLGQTFTYPLEV 240
G+ F+ + +K R D ++ K L CG VAG + QT +YP +V
Sbjct: 204 GVSFFTFGTLKSVGLSYAPALLGRPSSDNPNVLVLKTHINLLCGGVAGAIAQTISYPFDV 263
Query: 241 VRRQMQVQNLAASEEAELKGTMR-SMVLIAQKQGWKT-LFSGLSINYIKVVPSAAIGFTV 298
RR+MQ+ A E E TMR +M + + G + L+ GLS+NYI+ +PS A+ FT
Sbjct: 264 TRRRMQLG--AVLPEFEKCLTMRETMKYVYGQHGIRRGLYRGLSLNYIRCIPSQAVAFTT 321
Query: 299 YDTMKSYLRV 308
Y+ MK + +
Sbjct: 322 YELMKQFFHL 331
>gb|AAH65855.1| Unknown (protein for MGC:77742) [Danio rerio]
gi|28277902|gb|AAH45977.1| Unknown (protein for
MGC:77742) [Danio rerio] gi|45387539|ref|NP_991112.1|
solute carrier family 25, member 16 [Danio rerio]
Length = 321
Score = 200 bits (509), Expect = 4e-50
Identities = 116/308 (37%), Positives = 165/308 (52%), Gaps = 30/308 (9%)
Query: 17 KELLAGGLAGGFAKTVVAPLERLKILFQTRRTEFRSAGLSGSVRRIAKTEGLLGFYRGNG 76
+ AGG+AG AK+ +APL+R+KIL Q + ++ G+ +++ + K EG LG Y+GNG
Sbjct: 27 RSFTAGGVAGCCAKSTIAPLDRVKILLQAQNPHYKHLGVFATLKAVPKKEGFLGLYKGNG 86
Query: 77 ASVARIIPYAGLHFMSYEEYRRLIMQAFPNVWKGPTLDLMAGSLSGGTAVLFTYPLDLIR 136
A + RI PY + FM+++ Y++ + G LMAGS++G TAV+ TYPLD+IR
Sbjct: 87 AMMIRIFPYGAIQFMAFDNYKKFLHTKVG--ISGHVHRLMAGSMAGMTAVICTYPLDVIR 144
Query: 137 TKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTY-KEGGIRGLYRGVAPTLFGIFPYA 195
+LA+Q V Y GIR Y KEGGI G YRG+ PT+ G+ PYA
Sbjct: 145 ARLAFQ------------VTGHHRYSGIRHAFQTIYHKEGGISGFYRGLIPTIIGMAPYA 192
Query: 196 GLKFYFYEEMK----RRVPEDYKK-----------SIMAKLTCGSVAGLLGQTFTYPLEV 240
G F+ + +K PE K L CG VAG + QT +YPL+V
Sbjct: 193 GFSFFTFGTLKTLGLTHFPEQLGKPSLDNPDVLVLKTQVNLLCGGVAGAIAQTISYPLDV 252
Query: 241 VRRQMQVQNLAASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYD 300
RR+MQ+ + T + +Q K L+ GLS+NYI+ VPS A+ FT Y+
Sbjct: 253 ARRRMQLGASLPDHDKCCSLTKTLKHVYSQYGVKKGLYRGLSLNYIRCVPSQAVAFTTYE 312
Query: 301 TMKSYLRV 308
MK L +
Sbjct: 313 FMKQVLHL 320
Score = 41.6 bits (96), Expect = 0.030
Identities = 24/89 (26%), Positives = 42/89 (46%), Gaps = 4/89 (4%)
Query: 218 MAKLTCGSVAGLLGQTFTYPLEVVRRQMQVQNLAASEEAELKGTMRSMVLIAQKQGWKTL 277
+ T G VAG ++ PL+ V+ +Q QN + G ++ + +K+G+ L
Sbjct: 26 LRSFTAGGVAGCCAKSTIAPLDRVKILLQAQN----PHYKHLGVFATLKAVPKKEGFLGL 81
Query: 278 FSGLSINYIKVVPSAAIGFTVYDTMKSYL 306
+ G I++ P AI F +D K +L
Sbjct: 82 YKGNGAMMIRIFPYGAIQFMAFDNYKKFL 110
>ref|XP_629781.1| hypothetical protein DDB0184176 [Dictyostelium discoideum]
gi|60463180|gb|EAL61373.1| hypothetical protein
DDB0184176 [Dictyostelium discoideum]
Length = 297
Score = 196 bits (497), Expect = 1e-48
Identities = 113/294 (38%), Positives = 174/294 (58%), Gaps = 27/294 (9%)
Query: 20 LAGGLAGGFAKTVVAPLERLKILFQTRRTEFRSAGLSGSVRRIAKTEGLLGFYRGNGASV 79
L+GGLAG AK+ VAPLER+KIL+Q + + + GS+ +I + EG+ G +RGN A++
Sbjct: 19 LSGGLAGVTAKSAVAPLERVKILYQIKSELYSLNSVYGSMLKIVENEGIKGLWRGNSATI 78
Query: 80 ARIIPYAGLHFMSYEEYRRLIMQAFPNVWKGPTLDLMAGSLSGGTAVLFTYPLDLIRTKL 139
R+ PYA + F+SYE + ++ + ++ +AGS +GG AV TYPLDL+R +L
Sbjct: 79 LRVFPYAAVQFLSYETIKNHLVADKSSSFQ----IFLAGSAAGGIAVCATYPLDLLRARL 134
Query: 140 AYQI-VSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIRGLYRGVAPTLFGIFPYAGLK 198
A +I PTK + L T+ + G++G+YRG+ PTL GI PY G+
Sbjct: 135 AIEIHKKPTKPH---------------HLLKSTFTKDGVKGIYRGIQPTLIGILPYGGIS 179
Query: 199 FYFYEEMKRRVP-----EDYKKSIMAKLTCGSVAGLLGQTFTYPLEVVRRQMQVQNLA-A 252
F +E +KR P E+ + S KL G +AG + QT YP +VVRR++Q A
Sbjct: 180 FSTFEFLKRIAPLNEIDENGQISGTYKLIAGGIAGGVAQTVAYPFDVVRRRVQTHGFGDA 239
Query: 253 SEEAELK-GTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSY 305
L+ GT+R++ I +++G L+ GLSINY+KV+P+A+I F Y+ + ++
Sbjct: 240 KAVVNLEHGTLRTIAHILKEEGILALYKGLSINYVKVIPTASIAFYTYEYLSNF 293
Score = 75.9 bits (185), Expect = 1e-12
Identities = 58/205 (28%), Positives = 97/205 (47%), Gaps = 26/205 (12%)
Query: 113 LDLMAGSLSGGTAVLFTYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTY 172
+ ++G L+G TA PL+ R K+ YQI S + + VY + K
Sbjct: 16 VSFLSGGLAGVTAKSAVAPLE--RVKILYQIKSE-------LYSLNSVYGS----MLKIV 62
Query: 173 KEGGIRGLYRGVAPTLFGIFPYAGLKFYFYEEMKRRVPEDYKKSIMAKLTCGSVAGLLGQ 232
+ GI+GL+RG + T+ +FPYA ++F YE +K + D S L GS AG +
Sbjct: 63 ENEGIKGLWRGNSATILRVFPYAAVQFLSYETIKNHLVADKSSSFQIFLA-GSAAGGIAV 121
Query: 233 TFTYPLEVVRRQM--QVQNLAASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVP 290
TYPL+++R ++ ++ LK T K G K ++ G+ I ++P
Sbjct: 122 CATYPLDLLRARLAIEIHKKPTKPHHLLKSTFT-------KDGVKGIYRGIQPTLIGILP 174
Query: 291 SAAIGFTVYDTMKSYLRVPSRDEVD 315
I F+ ++ +K R+ +E+D
Sbjct: 175 YGGISFSTFEFLK---RIAPLNEID 196
Score = 50.1 bits (118), Expect = 9e-05
Identities = 30/83 (36%), Positives = 48/83 (57%), Gaps = 6/83 (7%)
Query: 18 ELLAGGLAGGFAKTVVAPLERLKILFQTRRTEFRSAGLS---GSVRRIA---KTEGLLGF 71
+L+AGG+AGG A+TV P + ++ QT A ++ G++R IA K EG+L
Sbjct: 206 KLIAGGIAGGVAQTVAYPFDVVRRRVQTHGFGDAKAVVNLEHGTLRTIAHILKEEGILAL 265
Query: 72 YRGNGASVARIIPYAGLHFMSYE 94
Y+G + ++IP A + F +YE
Sbjct: 266 YKGLSINYVKVIPTASIAFYTYE 288
>ref|XP_638149.1| hypothetical protein DDB0186597 [Dictyostelium discoideum]
gi|60466585|gb|EAL64637.1| hypothetical protein
DDB0186597 [Dictyostelium discoideum]
Length = 434
Score = 195 bits (496), Expect = 1e-48
Identities = 103/303 (33%), Positives = 173/303 (56%), Gaps = 22/303 (7%)
Query: 17 KELLAGGLAGGFAKTVVAPLERLKILFQT-------RRTEFRSAGLSGSVRRIAKTEGLL 69
K LL+GG+AG ++T +PLERLKIL Q +++ G+ S++ + TEG +
Sbjct: 140 KLLLSGGVAGAVSRTCTSPLERLKILNQVGHMNLEQNAPKYKGRGIIQSLKTMYTTEGFI 199
Query: 70 GFYRGNGASVARIIPYAGLHFMSYEEYRRLIMQAFPNVWKGPTLDLMAGSLSGGTAVLFT 129
GF++GNG +V RI PY+ + F+SYE+Y+ ++ +L G +G T++L T
Sbjct: 200 GFFKGNGTNVIRIAPYSAIQFLSYEKYKNFLLNNNDQTHLTTYENLFVGGAAGVTSLLCT 259
Query: 130 YPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIRGLYRGVAPTLF 189
YPLDLIR++L Q+ Y GI D +E G+ GLY+G+ +
Sbjct: 260 YPLDLIRSRLTVQVFG-------------NKYNGIADTCKMIIREEGVAGLYKGLFASAL 306
Query: 190 GIFPYAGLKFYFYEEMKRR-VPEDYKKSIMAKLTCGSVAGLLGQTFTYPLEVVRRQMQVQ 248
G+ PY + F YE +K+ +P+D +++ LT G+++G QT TYP++++RR++QVQ
Sbjct: 307 GVAPYVAINFTTYENLKKTFIPKDTTPTVVQSLTFGAISGATAQTLTYPIDLIRRRLQVQ 366
Query: 249 NLAASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSYLRV 308
+ ++ GT + I + +G L++G+ Y+KV+P+ +I F VY+ MK L++
Sbjct: 367 GI-GGKDILYNGTFDAFRKIIRDEGVLGLYNGMIPCYLKVIPAISISFCVYEVMKKILKI 425
Query: 309 PSR 311
S+
Sbjct: 426 DSK 428
Score = 93.6 bits (231), Expect = 7e-18
Identities = 58/201 (28%), Positives = 95/201 (46%), Gaps = 16/201 (7%)
Query: 17 KELLAGGLAGGFAKTVVAPLERLKILFQTRRTEFRSAGLSGSVRRIAKTEGLLGFYRGNG 76
+ L GG AG + PL+ ++ + + G++ + + I + EG+ G Y+G
Sbjct: 243 ENLFVGGAAGVTSLLCTYPLDLIRSRLTVQVFGNKYNGIADTCKMIIREEGVAGLYKGLF 302
Query: 77 ASVARIIPYAGLHFMSYEEYRRLIM--QAFPNVWKGPTLDLMAGSLSGGTAVLFTYPLDL 134
AS + PY ++F +YE ++ + P V + T G++SG TA TYP+DL
Sbjct: 303 ASALGVAPYVAINFTTYENLKKTFIPKDTTPTVVQSLTF----GAISGATAQTLTYPIDL 358
Query: 135 IRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIRGLYRGVAPTLFGIFPY 194
IR +L V G+ + +Y G D K ++ G+ GLY G+ P + P
Sbjct: 359 IRRRL----------QVQGIGGKDILYNGTFDAFRKIIRDEGVLGLYNGMIPCYLKVIPA 408
Query: 195 AGLKFYFYEEMKRRVPEDYKK 215
+ F YE MK+ + D KK
Sbjct: 409 ISISFCVYEVMKKILKIDSKK 429
>ref|XP_624199.1| PREDICTED: similar to ENSANGP00000008222, partial [Apis mellifera]
Length = 315
Score = 195 bits (495), Expect = 2e-48
Identities = 113/310 (36%), Positives = 174/310 (55%), Gaps = 33/310 (10%)
Query: 17 KELLAGGLAGGFAKTVVAPLERLKILFQTRRTEFRSAGLSGSVRRIAKTEGLLGFYRGNG 76
K L+AGG+AG +KT VAPL+R+KIL Q ++ G+ +R + + E Y+GN
Sbjct: 16 KSLIAGGVAGMCSKTTVAPLDRIKILLQAHNKYYKHLGVLSGLREVIQRERFFALYKGNF 75
Query: 77 ASVARIIPYAGLHFMSYEEYRRLIMQAFPNVWKGPTLD-LMAGSLSGGTAVLFTYPLDLI 135
A + RI PYA F ++E Y++ + F K +D +AGS +G TAV TYPLD+I
Sbjct: 76 AQMIRIFPYAATQFTTFELYKKYLGGLF---GKHTHIDKFLAGSAAGVTAVTLTYPLDII 132
Query: 136 RTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYK-EGGIRGLYRGVAPTLFGIFPY 194
R +LA+Q+ E +Y GI +K EGGIR LYRG PT+FG+ PY
Sbjct: 133 RARLAFQVA------------GEHIYIGIVHAGITIFKNEGGIRALYRGFWPTIFGMIPY 180
Query: 195 AGLKFYFYEEMK---RRVPEDY------------KKSIMAKLTCGSVAGLLGQTFTYPLE 239
AG FY +E++K + +Y +I A+L CG +AG + Q+F+YPL+
Sbjct: 181 AGFSFYSFEKLKYFCMKYASNYFCENCDRNTGGLVLTIPARLLCGGIAGAVAQSFSYPLD 240
Query: 240 VVRRQMQVQNLAASEEAELKGTMRSMVLIAQKQG-WKTLFSGLSINYIKVVPSAAIGFTV 298
V RR MQ+ + + ++++ +I ++ G K L+ G+SINY++ +P ++ FT
Sbjct: 241 VTRRHMQLGIMHHANHKYSSSMLQTIKMIYKENGIIKGLYRGMSINYLRAIPMVSVSFTT 300
Query: 299 YDTMKSYLRV 308
Y+ MK L++
Sbjct: 301 YEIMKQILQL 310
Score = 41.6 bits (96), Expect = 0.030
Identities = 26/96 (27%), Positives = 45/96 (46%), Gaps = 5/96 (5%)
Query: 211 EDYKKSIMAKLTCGSVAGLLGQTFTYPLEVVRRQMQVQNLAASEEAELKGTMRSMVLIAQ 270
+DY+ ++ L G VAG+ +T PL+ ++ +Q N L G + Q
Sbjct: 9 KDYE-FLLKSLIAGGVAGMCSKTTVAPLDRIKILLQAHNKYYKHLGVLSGLRE----VIQ 63
Query: 271 KQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSYL 306
++ + L+ G I++ P AA FT ++ K YL
Sbjct: 64 RERFFALYKGNFAQMIRIFPYAATQFTTFELYKKYL 99
>ref|XP_546134.1| PREDICTED: similar to solute carrier family 25, member 16 [Canis
familiaris]
Length = 515
Score = 194 bits (492), Expect = 4e-48
Identities = 115/304 (37%), Positives = 164/304 (53%), Gaps = 32/304 (10%)
Query: 22 GGLAGGFAKTVVAPLERLKILFQTRRTEFRSAGLSGSVRRIAKTEGLLGFYRGNGASVAR 81
G +AG AKT VAPL+R+K+L Q ++ G+ ++R + + EG LG Y+GNGA + R
Sbjct: 226 GCIAGCCAKTTVAPLDRVKVLLQAHNHHYKHLGVFSALRAVPQKEGYLGLYKGNGAMMIR 285
Query: 82 IIPYAGLHFMSYEEYRRLIMQAFPNVWKGPTLDLMAGSLSGGTAVLFTYPLDLIRTKLAY 141
I PY + FM++E Y+ LI G LMAGS++G TAV+ TYPLD++R +LA+
Sbjct: 286 IFPYGAIQFMAFEHYKTLITTKLG--VSGHVHRLMAGSMAGMTAVICTYPLDMVRVRLAF 343
Query: 142 QIVSPTKLNVSGMVNNEQVYRGIRDCLSKTY-KEGGIRGLYRGVAPTLFGIFPYAGLKFY 200
Q V E Y GI Y KEGG G YRG+ PT+ G+ PYAG+ F+
Sbjct: 344 Q------------VKGEHTYTGIIHAFKTIYAKEGGFLGFYRGLMPTILGMAPYAGVSFF 391
Query: 201 FYEEMKR-----------RVPEDYKKSIMAK----LTCGSVAGLLGQTFTYPLEVVRRQM 245
+ +K R D ++ K L CG VAG + QT +YP +V RR+M
Sbjct: 392 TFGTLKSVGLSHAPTLLGRPSSDNPNVLVLKTHINLLCGGVAGAIAQTISYPFDVTRRRM 451
Query: 246 QVQNLAASEEAELKGTMRSMVLIAQKQGWKT-LFSGLSINYIKVVPSAAIGFTVYDTMKS 304
Q+ A E + +M + G + L+ GLS+NYI+ VPS A+ FT Y+ MK
Sbjct: 452 QL-GTALPEFEKCLTMWETMKYVYGHHGIRRGLYRGLSLNYIRCVPSQAVAFTTYELMKQ 510
Query: 305 YLRV 308
+ +
Sbjct: 511 FFHL 514
>emb|CAG60728.1| unnamed protein product [Candida glabrata CBS138]
gi|50290697|ref|XP_447781.1| unnamed protein product
[Candida glabrata]
Length = 327
Score = 191 bits (484), Expect = 3e-47
Identities = 112/300 (37%), Positives = 176/300 (58%), Gaps = 14/300 (4%)
Query: 20 LAGGLAGGFAKTVVAPLERLKILFQTRR-TEFRSAGLSGSVRRIAKTEGLLGFYRGNGAS 78
LAGG+AG ++TVV+P ER+KIL Q + T + GL ++ ++ K E + G +RGNG +
Sbjct: 28 LAGGIAGAISRTVVSPFERVKILLQVQSSTTAYNKGLFDAIGQVYKEENIKGLFRGNGLN 87
Query: 79 VARIIPYAGLHFMSYEEYRRLIMQAFPNVWKGPTLD----LMAGSLSGGTAVLFTYPLDL 134
R+ PY+ + F+ +E ++ I KG L+ L +G+L GG +V+ TYPLDL
Sbjct: 88 CIRVFPYSAVQFVVFEGCKKHIFHV-DTKGKGEQLNNWQRLFSGALCGGCSVVATYPLDL 146
Query: 135 IRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTY-KEGGIRGLYRGVAPTLFGIFP 193
+RT+L+ Q + +KL+ S ++ G+ LSK Y +EGGI GLYRGV PT GI P
Sbjct: 147 VRTRLSVQTANLSKLSKS-RASDIAKPPGVWKLLSKAYAEEGGIMGLYRGVWPTSLGIVP 205
Query: 194 YAGLKFYFYEEMKRRVPEDYK-----KSIMAKLTCGSVAGLLGQTFTYPLEVVRRQMQVQ 248
Y L F YE++K +P D + + KL+ G+++G + QT TYP +++RR+ QV
Sbjct: 206 YVALNFAVYEQLKEFMPSDENGNSSMRDSLYKLSMGAISGGVAQTITYPFDLLRRRFQVL 265
Query: 249 NLAASEEA-ELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSYLR 307
+ +E ++V I + +G+K + GL+ N KVVPS A+ + VY+ Y++
Sbjct: 266 AMGGNELGFHYNSVWDALVTIGKTEGFKGYYKGLTANLFKVVPSTAVSWLVYELTWDYMK 325
Score = 46.2 bits (108), Expect = 0.001
Identities = 29/103 (28%), Positives = 55/103 (53%), Gaps = 7/103 (6%)
Query: 204 EMKRRVPEDYKKSIMAKLTCGSVAGLLGQTFTYPLEVVRRQMQVQNLAASEEAELKGTMR 263
++K + +D + +A G +AG + +T P E V+ +QVQ +S A KG
Sbjct: 14 QLKNFLKQDTNVAFLA----GGIAGAISRTVVSPFERVKILLQVQ---SSTTAYNKGLFD 66
Query: 264 SMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSYL 306
++ + +++ K LF G +N I+V P +A+ F V++ K ++
Sbjct: 67 AIGQVYKEENIKGLFRGNGLNCIRVFPYSAVQFVVFEGCKKHI 109
Score = 43.9 bits (102), Expect = 0.006
Identities = 23/82 (28%), Positives = 43/82 (52%), Gaps = 5/82 (6%)
Query: 18 ELLAGGLAGGFAKTVVAPLERLKILFQT-----RRTEFRSAGLSGSVRRIAKTEGLLGFY 72
+L G ++GG A+T+ P + L+ FQ F + ++ I KTEG G+Y
Sbjct: 237 KLSMGAISGGVAQTITYPFDLLRRRFQVLAMGGNELGFHYNSVWDALVTIGKTEGFKGYY 296
Query: 73 RGNGASVARIIPYAGLHFMSYE 94
+G A++ +++P + ++ YE
Sbjct: 297 KGLTANLFKVVPSTAVSWLVYE 318
>gb|AAW42601.1| conserved hypothetical protein [Cryptococcus neoformans var.
neoformans JEC21] gi|50259254|gb|EAL21927.1|
hypothetical protein CNBC0670 [Cryptococcus neoformans
var. neoformans B-3501A] gi|58265504|ref|XP_569908.1|
conserved hypothetical protein [Cryptococcus neoformans
var. neoformans JEC21]
Length = 378
Score = 190 bits (482), Expect = 5e-47
Identities = 118/307 (38%), Positives = 170/307 (54%), Gaps = 26/307 (8%)
Query: 20 LAGGLAGGFAKTVVAPLERLKILFQTRRTEFRS------AGLSGSVRRIAKTEGLLGFYR 73
+AGGLAG ++TVV+PLERLKI+ Q + + +S AG+ S+ R+ K EG GF R
Sbjct: 75 IAGGLAGAASRTVVSPLERLKIILQVQASGSKSGVGQAYAGVWESLGRMWKDEGWRGFMR 134
Query: 74 GNGASVARIIPYAGLHFMSYEEYRRLIMQAFPNVWKGPT-----LDLMAGSLSGGTAVLF 128
GNG +V RI+PY+ L F SY ++ ++ + W G L L AG+ +G AV+
Sbjct: 135 GNGINVVRILPYSALQFTSYGAFKGVL-----STWSGQEALSTPLRLTAGAGAGVVAVVA 189
Query: 129 TYPLDLIRTKLAYQIVSPTKLNVSGMVNNEQVYRGIRDCLSKTYK-EGGIRGLYRGVAPT 187
TYPLDL+R +L+ + NE GI K YK EGG+RGLYRG T
Sbjct: 190 TYPLDLVRARLSIATANMAVRQPGAAFTNEDARLGIVGMTKKVYKAEGGLRGLYRGCWAT 249
Query: 188 LFGIFPYAGLKFYFYEEMKRRV-------PEDYKKSIMAKLTCGSVAGLLGQTFTYPLEV 240
G+ PY L F+FYE +K V P + KL CG+V+G FT+P +V
Sbjct: 250 ALGVAPYVSLNFFFYESVKTHVLPDPPSPPLSETDLALRKLFCGAVSGASSLIFTHPFDV 309
Query: 241 VRRQMQVQNLAASEEAELKGTMRSMVLIAQKQG-WKTLFSGLSINYIKVVPSAAIGFTVY 299
+RR++QV L ++ G + +M I + +G WK ++ GL+ N IKV PS A+ F V+
Sbjct: 310 LRRKLQVAGL-STLTPHYDGAIDAMRQIIRNEGFWKGMYRGLTPNLIKVTPSIAVSFYVF 368
Query: 300 DTMKSYL 306
+ ++ L
Sbjct: 369 ELVRDSL 375
Score = 43.5 bits (101), Expect = 0.008
Identities = 26/92 (28%), Positives = 43/92 (46%), Gaps = 2/92 (2%)
Query: 224 GSVAGLLGQTFTYPLEVVRRQMQVQNLAASEEA--ELKGTMRSMVLIAQKQGWKTLFSGL 281
G +AG +T PLE ++ +QVQ + G S+ + + +GW+ G
Sbjct: 77 GGLAGAASRTVVSPLERLKIILQVQASGSKSGVGQAYAGVWESLGRMWKDEGWRGFMRGN 136
Query: 282 SINYIKVVPSAAIGFTVYDTMKSYLRVPSRDE 313
IN ++++P +A+ FT Y K L S E
Sbjct: 137 GINVVRILPYSALQFTSYGAFKGVLSTWSGQE 168
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.322 0.139 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 502,438,044
Number of Sequences: 2540612
Number of extensions: 20153000
Number of successful extensions: 62279
Number of sequences better than 10.0: 2069
Number of HSP's better than 10.0 without gapping: 1452
Number of HSP's successfully gapped in prelim test: 618
Number of HSP's that attempted gapping in prelim test: 45211
Number of HSP's gapped (non-prelim): 6085
length of query: 315
length of database: 863,360,394
effective HSP length: 128
effective length of query: 187
effective length of database: 538,162,058
effective search space: 100636304846
effective search space used: 100636304846
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 75 (33.5 bits)
Medicago: description of AC148360.5


