

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148360.13 + phase: 0
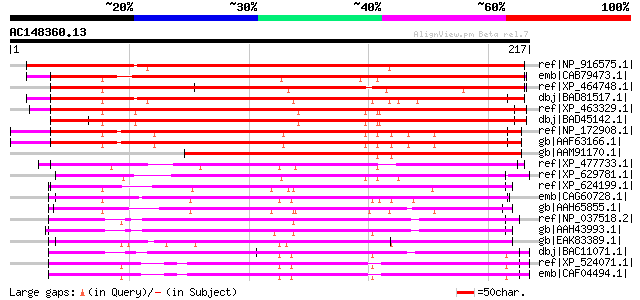
(217 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_916575.1| P0456F08.3 [Oryza sativa (japonica cultivar-gro... 249 3e-65
emb|CAB79473.1| putative mitochondrial carrier protein [Arabidop... 234 2e-60
ref|XP_464748.1| putative mitochondrial solute carrier protein [... 220 2e-56
dbj|BAD81517.1| Graves disease mitochondrial solute carrier prot... 215 8e-55
ref|XP_463329.1| putative mitochondrial carrier [Oryza sativa (j... 214 1e-54
dbj|BAD45142.1| mitochondrial carrier protein-like [Oryza sativa... 214 1e-54
ref|NP_172908.1| mitochondrial substrate carrier family protein ... 211 9e-54
gb|AAF63166.1| T5E21.6 [Arabidopsis thaliana] 211 9e-54
gb|AAM91170.1| putative mitochondrial carrier protein [Arabidops... 186 4e-46
ref|XP_477733.1| putative mitochondrial carrier protein [Oryza s... 116 5e-25
ref|XP_629781.1| hypothetical protein DDB0184176 [Dictyostelium ... 112 9e-24
ref|XP_624199.1| PREDICTED: similar to ENSANGP00000008222, parti... 111 2e-23
emb|CAG60728.1| unnamed protein product [Candida glabrata CBS138... 110 3e-23
gb|AAH65855.1| Unknown (protein for MGC:77742) [Danio rerio] gi|... 110 4e-23
ref|NP_037518.2| solute carrier family 25 member 24 isoform 1 [H... 110 4e-23
gb|AAH43993.1| LOC398474 protein [Xenopus laevis] 109 5e-23
gb|EAK83389.1| hypothetical protein UM02351.1 [Ustilago maydis 5... 109 6e-23
dbj|BAC11071.1| unnamed protein product [Homo sapiens] 108 1e-22
ref|XP_524071.1| PREDICTED: similar to solute carrier family 25 ... 108 1e-22
emb|CAF04494.1| small calcium-binding mitochondrial carrier 3 [H... 108 1e-22
>ref|NP_916575.1| P0456F08.3 [Oryza sativa (japonica cultivar-group)]
Length = 239
Score = 249 bits (637), Expect = 3e-65
Identities = 122/213 (57%), Positives = 161/213 (75%), Gaps = 6/213 (2%)
Query: 8 ILDHIPLFAKELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGM--VNNEQVYRG 65
+LD +P+FAKE++AGG+AG F+KT +APLERLKIL QV S +++ SG+ N + Y G
Sbjct: 20 VLDLVPVFAKEMIAGGVAGAFSKTAIAPLERLKILLQVNSSDQIS-SGLKRTNFQPKYGG 78
Query: 66 IRDCLSKTYKEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYKKSIMAKLTC 125
I+D Y EGG++ +YRGV PTL GI PYAGLKFY YE +K HVPEDYK S+ KL+C
Sbjct: 79 IKDVFRGVYSEGGVRALYRGVGPTLMGILPYAGLKFYIYEGLKAHVPEDYKNSVTLKLSC 138
Query: 126 GSVAGLLGQTFTYFLEVVRRQMQVQNLPASEE---AELKGTMRSMVLIAQKQGWKTLFSG 182
G+ AGL GQT TY L+VVRRQMQVQ+ ++ +++GT + +++I Q QGW+ LF+G
Sbjct: 139 GAAAGLFGQTLTYPLDVVRRQMQVQSQQYHDKFGGPQIRGTFQGLMIIKQTQGWRQLFAG 198
Query: 183 LSINYIKVVPSAAIGFTVYDTMKSYLRVPSRDE 215
LS+NYIKVVPS AIGFT YDTMKS L++P R++
Sbjct: 199 LSLNYIKVVPSVAIGFTAYDTMKSLLKIPPREK 231
>emb|CAB79473.1| putative mitochondrial carrier protein [Arabidopsis thaliana]
gi|4538947|emb|CAB39683.1| putative mitochondrial
carrier protein [Arabidopsis thaliana]
gi|15236140|ref|NP_194348.1| mitochondrial substrate
carrier family protein [Arabidopsis thaliana]
gi|7441723|pir||T04273 hypothetical protein F20B18.290 -
Arabidopsis thaliana
Length = 325
Score = 234 bits (596), Expect = 2e-60
Identities = 120/201 (59%), Positives = 150/201 (73%), Gaps = 9/201 (4%)
Query: 18 ELLAGGLAGGFAKTVVAPLE--RLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYK 75
+L+AG AGG A PL+ R K+ +Q V + + +YRGI DC S+TY+
Sbjct: 116 DLVAGSFAGGTAVLFTYPLDLVRTKLAYQT------QVKAIPVEQIIYRGIVDCFSRTYR 169
Query: 76 EGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYKKSIMAKLTCGSVAGLLGQT 135
E G +G+YRGVAP+L+GIFPYAGLKFYFYEEMKRHVP ++K+ I KL CGSVAGLLGQT
Sbjct: 170 ESGARGLYRGVAPSLYGIFPYAGLKFYFYEEMKRHVPPEHKQDISLKLVCGSVAGLLGQT 229
Query: 136 FTYFLEVVRRQMQVQNL-PASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSA 194
TY L+VVRRQMQV+ L A +E +GTM+++ IA+++GWK LFSGLSINY+KVVPS
Sbjct: 230 LTYPLDVVRRQMQVERLYSAVKEETRRGTMQTLFKIAREEGWKQLFSGLSINYLKVVPSV 289
Query: 195 AIGFTVYDTMKSYLRVPSRDE 215
AIGFTVYD MK +LRVP R+E
Sbjct: 290 AIGFTVYDIMKLHLRVPPREE 310
Score = 109 bits (273), Expect = 5e-23
Identities = 74/216 (34%), Positives = 112/216 (51%), Gaps = 19/216 (8%)
Query: 8 ILDHIPLFAKELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIR 67
I+D IPLFAKEL+AGG+ GG AKT VAPLER+KILFQ +E G+
Sbjct: 10 IIDSIPLFAKELIAGGVTGGIAKTAVAPLERIKILFQT-----------RRDEFKRIGLV 58
Query: 68 DCLSKTYKEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVP---EDYKKSIMAKLT 124
++K K G+ G YRG ++ I PYA L + YEE +R + D + + L
Sbjct: 59 GSINKIGKTEGLMGFYRGNGASVARIVPYAALHYMAYEEYRRWIIFGFPDTTRGPLLDLV 118
Query: 125 CGSVAGLLGQTFTYFLEVVRR----QMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLF 180
GS AG FTY L++VR Q QV+ +P E+ +G + ++ G + L+
Sbjct: 119 AGSFAGGTAVLFTYPLDLVRTKLAYQTQVKAIPV-EQIIYRGIVDCFSRTYRESGARGLY 177
Query: 181 SGLSINYIKVVPSAAIGFTVYDTMKSYLRVPSRDEV 216
G++ + + P A + F Y+ MK ++ + ++
Sbjct: 178 RGVAPSLYGIFPYAGLKFYFYEEMKRHVPPEHKQDI 213
Score = 36.6 bits (83), Expect = 0.50
Identities = 30/114 (26%), Positives = 50/114 (43%), Gaps = 17/114 (14%)
Query: 6 ESILDHIPLFAKE-----LLAGGLAGGFAKTVVAPLERLKILFQV---VSPTKLNVSGMV 57
E + H+P K+ L+ G +AG +T+ PL+ ++ QV S K
Sbjct: 199 EEMKRHVPPEHKQDISLKLVCGSVAGLLGQTLTYPLDVVRRQMQVERLYSAVK------- 251
Query: 58 NNEQVYRGIRDCLSKTYKEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHV 111
E+ RG L K +E G K ++ G++ + P + F Y+ MK H+
Sbjct: 252 --EETRRGTMQTLFKIAREEGWKQLFSGLSINYLKVVPSVAIGFTVYDIMKLHL 303
>ref|XP_464748.1| putative mitochondrial solute carrier protein [Oryza sativa
(japonica cultivar-group)] gi|49388534|dbj|BAD25656.1|
putative mitochondrial solute carrier protein [Oryza
sativa (japonica cultivar-group)]
Length = 426
Score = 220 bits (561), Expect = 2e-56
Identities = 115/220 (52%), Positives = 153/220 (69%), Gaps = 24/220 (10%)
Query: 18 ELLAGGLAGGFAKTVVAPLE--RLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYK 75
+L+AG +AGG A PL+ R K+ +QV KL++ +EQVY+GI DC+ Y+
Sbjct: 190 DLVAGSIAGGTAVICTYPLDLVRTKLAYQVKGAVKLSLREYKPSEQVYKGILDCVKTIYR 249
Query: 76 EGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYKKSIMAKLTCGSVAGLLGQT 135
+ G++G+YRG+AP+L+GIFPY+GLKFYFYE MK +VPE+++K I+AKL CGSVAGLLGQT
Sbjct: 250 QNGLRGLYRGMAPSLYGIFPYSGLKFYFYETMKTYVPEEHRKDIIAKLACGSVAGLLGQT 309
Query: 136 FTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYI------- 188
TY L+VVRRQMQ +S + KGT S+ +IA+ QGW+ LFSGLSINY+
Sbjct: 310 ITYPLDVVRRQMQA--FSSSNLEKGKGTFGSIAMIAKHQGWRQLFSGLSINYLKELYQLD 367
Query: 189 -------------KVVPSAAIGFTVYDTMKSYLRVPSRDE 215
KVVPS AIGFTVYD+MK +L+VPSR++
Sbjct: 368 TSVCAFIYVQCGEKVVPSVAIGFTVYDSMKVWLKVPSRED 407
Score = 60.8 bits (146), Expect = 2e-08
Identities = 41/151 (27%), Positives = 74/151 (48%), Gaps = 12/151 (7%)
Query: 78 GIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDY---KKSIMAKLTCGSVAGLLGQ 134
G+ G YRG ++ I PYA L + YEE +R + + ++ + L GS+AG
Sbjct: 143 GLLGFYRGNGASVARIVPYAALHYMAYEEYRRWIILGFPNVEQGPILDLVAGSIAGGTAV 202
Query: 135 TFTYFLEVVRRQMQVQNLPASE---------EAELKGTMRSMVLIAQKQGWKTLFSGLSI 185
TY L++VR ++ Q A + E KG + + I ++ G + L+ G++
Sbjct: 203 ICTYPLDLVRTKLAYQVKGAVKLSLREYKPSEQVYKGILDCVKTIYRQNGLRGLYRGMAP 262
Query: 186 NYIKVVPSAAIGFTVYDTMKSYLRVPSRDEV 216
+ + P + + F Y+TMK+Y+ R ++
Sbjct: 263 SLYGIFPYSGLKFYFYETMKTYVPEEHRKDI 293
>dbj|BAD81517.1| Graves disease mitochondrial solute carrier protein-like [Oryza
sativa (japonica cultivar-group)]
Length = 337
Score = 215 bits (547), Expect = 8e-55
Identities = 110/205 (53%), Positives = 145/205 (70%), Gaps = 8/205 (3%)
Query: 18 ELLAGGLAGGFAKTVVAPLE--RLKILFQVVSPTKLNVSGM--VNNEQVYRGIRDCLSKT 73
+LLAG +GG A PL+ R K+ FQV S +++ SG+ N + Y GI+D
Sbjct: 126 DLLAGSASGGTAVLCTYPLDLARTKLAFQVNSSDQIS-SGLKRTNFQPKYGGIKDVFRGV 184
Query: 74 YKEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYKKSIMAKLTCGSVAGLLG 133
Y EGG++ +YRGV PTL GI PYAGLKFY YE +K HVPEDYK S+ KL+CG+ AGL G
Sbjct: 185 YSEGGVRALYRGVGPTLMGILPYAGLKFYIYEGLKAHVPEDYKNSVTLKLSCGAAAGLFG 244
Query: 134 QTFTYFLEVVRRQMQVQNLPASEE---AELKGTMRSMVLIAQKQGWKTLFSGLSINYIKV 190
QT TY L+VVRRQMQVQ+ ++ +++GT + +++I Q QGW+ LF+GLS+NYIKV
Sbjct: 245 QTLTYPLDVVRRQMQVQSQQYHDKFGGPQIRGTFQGLMIIKQTQGWRQLFAGLSLNYIKV 304
Query: 191 VPSAAIGFTVYDTMKSYLRVPSRDE 215
VPS AIGFT YDTMKS L++P R++
Sbjct: 305 VPSVAIGFTAYDTMKSLLKIPPREK 329
Score = 89.7 bits (221), Expect = 5e-17
Identities = 63/214 (29%), Positives = 106/214 (49%), Gaps = 24/214 (11%)
Query: 8 ILDHIPLFAKELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIR 67
+LD +P+FAKE++AGG+AG F+KT +APLERLKIL Q + NE G+
Sbjct: 20 VLDLVPVFAKEMIAGGVAGAFSKTAIAPLERLKILLQTRT-----------NEFSSLGVL 68
Query: 68 DCLSKTYKEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYKK---SIMAKLT 124
L K + GI G Y+G ++ I PYA L + YE + + + + L
Sbjct: 69 KSLKKLKQHDGILGFYKGNGASVLRIVPYAALHYMAYERYRCWILNNCPSLGTGPLVDLL 128
Query: 125 CGSVAGLLGQTFTYFLEVVRRQMQVQ-NLPASEEAELK--------GTMRSMVL-IAQKQ 174
GS +G TY L++ R ++ Q N + LK G ++ + + +
Sbjct: 129 AGSASGGTAVLCTYPLDLARTKLAFQVNSSDQISSGLKRTNFQPKYGGIKDVFRGVYSEG 188
Query: 175 GWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSYL 208
G + L+ G+ + ++P A + F +Y+ +K+++
Sbjct: 189 GVRALYRGVGPTLMGILPYAGLKFYIYEGLKAHV 222
>ref|XP_463329.1| putative mitochondrial carrier [Oryza sativa (japonica
cultivar-group)] gi|20161078|dbj|BAB90009.1|
mitochondrial carrier protein-like [Oryza sativa
(japonica cultivar-group)]
Length = 340
Score = 214 bits (546), Expect = 1e-54
Identities = 109/205 (53%), Positives = 142/205 (69%), Gaps = 6/205 (2%)
Query: 18 ELLAGGLAGGFAKTVVAPLE--RLKILFQVVSPTKL-NVSGMVNNEQVYRGIRDCLSKTY 74
+LLAG AGG A PL+ R K+ +QV + + N G + Y GI+D Y
Sbjct: 129 DLLAGSAAGGTAVLCTYPLDLARTKLAYQVSNVGQPGNALGNAGRQPAYGGIKDVFKTVY 188
Query: 75 KEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYKKSIMAKLTCGSVAGLLGQ 134
KEGG + +YRGV PTL GI PYAGLKFY YE++K VPEDYK+S++ KL+CG++AGL GQ
Sbjct: 189 KEGGARALYRGVGPTLIGILPYAGLKFYIYEDLKSRVPEDYKRSVVLKLSCGALAGLFGQ 248
Query: 135 TFTYFLEVVRRQMQVQNLP---ASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVV 191
T TY L+VVRRQMQVQN A++ ++GT + + LI + QGW+ LF+GLS+NY+KVV
Sbjct: 249 TLTYPLDVVRRQMQVQNKQPHNANDAFRIRGTFQGLALIIRCQGWRQLFAGLSLNYVKVV 308
Query: 192 PSAAIGFTVYDTMKSYLRVPSRDEV 216
PS AIGFT YD MK+ LRVP R+ +
Sbjct: 309 PSVAIGFTTYDMMKNLLRVPPRERL 333
Score = 99.4 bits (246), Expect = 6e-20
Identities = 71/217 (32%), Positives = 109/217 (49%), Gaps = 27/217 (12%)
Query: 9 LDHIPLFAKELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRD 68
LD +P++AKEL+AGG AG FAKT VAPLER+KIL Q G + GI
Sbjct: 23 LDLLPVYAKELIAGGAAGAFAKTAVAPLERVKILLQT------RTHGFQS-----LGILQ 71
Query: 69 CLSKTYKEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYKKSI----MAKLT 124
L K ++ GI+G Y+G ++ I PYA L + YE+ + + ++ S+ + L
Sbjct: 72 SLRKLWQYEGIRGFYKGNGASVLRIVPYAALHYMTYEQYRCWILNNFAPSVGTGPVVDLL 131
Query: 125 CGSVAGLLGQTFTYFLEVVRRQM--QVQNL--------PASEEAELKGTMRSMVLIAQKQ 174
GS AG TY L++ R ++ QV N+ A + G + ++
Sbjct: 132 AGSAAGGTAVLCTYPLDLARTKLAYQVSNVGQPGNALGNAGRQPAYGGIKDVFKTVYKEG 191
Query: 175 GWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSYLRVP 211
G + L+ G+ I ++P A + F +Y+ +KS RVP
Sbjct: 192 GARALYRGVGPTLIGILPYAGLKFYIYEDLKS--RVP 226
>dbj|BAD45142.1| mitochondrial carrier protein-like [Oryza sativa (japonica
cultivar-group)]
Length = 330
Score = 214 bits (546), Expect = 1e-54
Identities = 109/205 (53%), Positives = 142/205 (69%), Gaps = 6/205 (2%)
Query: 18 ELLAGGLAGGFAKTVVAPLE--RLKILFQVVSPTKL-NVSGMVNNEQVYRGIRDCLSKTY 74
+LLAG AGG A PL+ R K+ +QV + + N G + Y GI+D Y
Sbjct: 119 DLLAGSAAGGTAVLCTYPLDLARTKLAYQVSNVGQPGNALGNAGRQPAYGGIKDVFKTVY 178
Query: 75 KEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYKKSIMAKLTCGSVAGLLGQ 134
KEGG + +YRGV PTL GI PYAGLKFY YE++K VPEDYK+S++ KL+CG++AGL GQ
Sbjct: 179 KEGGARALYRGVGPTLIGILPYAGLKFYIYEDLKSRVPEDYKRSVVLKLSCGALAGLFGQ 238
Query: 135 TFTYFLEVVRRQMQVQNLP---ASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVV 191
T TY L+VVRRQMQVQN A++ ++GT + + LI + QGW+ LF+GLS+NY+KVV
Sbjct: 239 TLTYPLDVVRRQMQVQNKQPHNANDAFRIRGTFQGLALIIRCQGWRQLFAGLSLNYVKVV 298
Query: 192 PSAAIGFTVYDTMKSYLRVPSRDEV 216
PS AIGFT YD MK+ LRVP R+ +
Sbjct: 299 PSVAIGFTTYDMMKNLLRVPPRERL 323
Score = 63.9 bits (154), Expect = 3e-09
Identities = 52/192 (27%), Positives = 89/192 (46%), Gaps = 27/192 (14%)
Query: 34 APLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGIKGIYRGVAPTLFGI 93
+PLER+KIL Q + ++ GI L K ++ GI+G Y+G ++ I
Sbjct: 38 SPLERVKILLQTRTHGFQSL-----------GILQSLRKLWQYEGIRGFYKGNGASVLRI 86
Query: 94 FPYAGLKFYFYEEMKRHVPEDYKKSI----MAKLTCGSVAGLLGQTFTYFLEVVRRQM-- 147
PYA L + YE+ + + ++ S+ + L GS AG TY L++ R ++
Sbjct: 87 VPYAALHYMTYEQYRCWILNNFAPSVGTGPVVDLLAGSAAGGTAVLCTYPLDLARTKLAY 146
Query: 148 QVQNL--------PASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGFT 199
QV N+ A + G + ++ G + L+ G+ I ++P A + F
Sbjct: 147 QVSNVGQPGNALGNAGRQPAYGGIKDVFKTVYKEGGARALYRGVGPTLIGILPYAGLKFY 206
Query: 200 VYDTMKSYLRVP 211
+Y+ +KS RVP
Sbjct: 207 IYEDLKS--RVP 216
>ref|NP_172908.1| mitochondrial substrate carrier family protein [Arabidopsis
thaliana]
Length = 331
Score = 211 bits (538), Expect = 9e-54
Identities = 110/206 (53%), Positives = 145/206 (69%), Gaps = 10/206 (4%)
Query: 18 ELLAGGLAGGFAKTVVAPLE--RLKILFQVVSPTKLNVSGMVNN---EQVYRGIRDCLSK 72
+L+AG AGG A PL+ R K+ +QV S T+ ++ G N + Y GI++ L+
Sbjct: 122 DLVAGSAAGGTAVLCTYPLDLARTKLAYQV-SDTRQSLRGGANGFYRQPTYSGIKEVLAM 180
Query: 73 TYKEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYKKSIMAKLTCGSVAGLL 132
YKEGG +G+YRG+ PTL GI PYAGLKFY YEE+KRHVPE+++ S+ L CG++AGL
Sbjct: 181 AYKEGGPRGLYRGIGPTLIGILPYAGLKFYIYEELKRHVPEEHQNSVRMHLPCGALAGLF 240
Query: 133 GQTFTYFLEVVRRQMQVQNL-PASEEA---ELKGTMRSMVLIAQKQGWKTLFSGLSINYI 188
GQT TY L+VVRRQMQV+NL P + E K T + I + QGWK LF+GLSINYI
Sbjct: 241 GQTITYPLDVVRRQMQVENLQPMTSEGNNKRYKNTFDGLNTIVRTQGWKQLFAGLSINYI 300
Query: 189 KVVPSAAIGFTVYDTMKSYLRVPSRD 214
K+VPS AIGFTVY++MKS++R+P R+
Sbjct: 301 KIVPSVAIGFTVYESMKSWMRIPPRE 326
Score = 87.8 bits (216), Expect = 2e-16
Identities = 66/222 (29%), Positives = 105/222 (46%), Gaps = 25/222 (11%)
Query: 1 MAASSESILDHIPLFAKELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNE 60
++A S++D +P+ AK L+AGG AG AKT VAPLER+KIL Q + N+
Sbjct: 9 LSADVMSLVDTLPVLAKTLIAGGAAGAIAKTAVAPLERIKILLQTRT-----------ND 57
Query: 61 QVYRGIRDCLSKTYKEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPE---DYKK 117
G+ L K + G G Y+G ++ I PYA L + YE + + E
Sbjct: 58 FKTLGVSQSLKKVLQFDGPLGFYKGNGASVIRIIPYAALHYMTYEVYRDWILEKNLPLGS 117
Query: 118 SIMAKLTCGSVAGLLGQTFTYFLEVVRRQM--QVQNLPASEEAELKGTMR--------SM 167
+ L GS AG TY L++ R ++ QV + S G R +
Sbjct: 118 GPIVDLVAGSAAGGTAVLCTYPLDLARTKLAYQVSDTRQSLRGGANGFYRQPTYSGIKEV 177
Query: 168 VLIAQKQGW-KTLFSGLSINYIKVVPSAAIGFTVYDTMKSYL 208
+ +A K+G + L+ G+ I ++P A + F +Y+ +K ++
Sbjct: 178 LAMAYKEGGPRGLYRGIGPTLIGILPYAGLKFYIYEELKRHV 219
>gb|AAF63166.1| T5E21.6 [Arabidopsis thaliana]
Length = 319
Score = 211 bits (538), Expect = 9e-54
Identities = 110/206 (53%), Positives = 145/206 (69%), Gaps = 10/206 (4%)
Query: 18 ELLAGGLAGGFAKTVVAPLE--RLKILFQVVSPTKLNVSGMVNN---EQVYRGIRDCLSK 72
+L+AG AGG A PL+ R K+ +QV S T+ ++ G N + Y GI++ L+
Sbjct: 110 DLVAGSAAGGTAVLCTYPLDLARTKLAYQV-SDTRQSLRGGANGFYRQPTYSGIKEVLAM 168
Query: 73 TYKEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYKKSIMAKLTCGSVAGLL 132
YKEGG +G+YRG+ PTL GI PYAGLKFY YEE+KRHVPE+++ S+ L CG++AGL
Sbjct: 169 AYKEGGPRGLYRGIGPTLIGILPYAGLKFYIYEELKRHVPEEHQNSVRMHLPCGALAGLF 228
Query: 133 GQTFTYFLEVVRRQMQVQNL-PASEEA---ELKGTMRSMVLIAQKQGWKTLFSGLSINYI 188
GQT TY L+VVRRQMQV+NL P + E K T + I + QGWK LF+GLSINYI
Sbjct: 229 GQTITYPLDVVRRQMQVENLQPMTSEGNNKRYKNTFDGLNTIVRTQGWKQLFAGLSINYI 288
Query: 189 KVVPSAAIGFTVYDTMKSYLRVPSRD 214
K+VPS AIGFTVY++MKS++R+P R+
Sbjct: 289 KIVPSVAIGFTVYESMKSWMRIPPRE 314
Score = 81.6 bits (200), Expect = 1e-14
Identities = 65/222 (29%), Positives = 102/222 (45%), Gaps = 37/222 (16%)
Query: 1 MAASSESILDHIPLFAKELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNE 60
++A S++D +P+ AK L+AGG AG AKT VAPLER+KIL Q+ S T LN + N+
Sbjct: 9 LSADVMSLVDTLPVLAKTLIAGGAAGAIAKTAVAPLERIKILLQLSSTTLLNSCDVHNSR 68
Query: 61 QVYRGIRDCLSKTYKEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPE---DYKK 117
RG ++ I PYA L + YE + + E
Sbjct: 69 -----------------------RGNGASVIRIIPYAALHYMTYEVYRDWILEKNLPLGS 105
Query: 118 SIMAKLTCGSVAGLLGQTFTYFLEVVRRQM--QVQNLPASEEAELKGTMR--------SM 167
+ L GS AG TY L++ R ++ QV + S G R +
Sbjct: 106 GPIVDLVAGSAAGGTAVLCTYPLDLARTKLAYQVSDTRQSLRGGANGFYRQPTYSGIKEV 165
Query: 168 VLIAQKQGW-KTLFSGLSINYIKVVPSAAIGFTVYDTMKSYL 208
+ +A K+G + L+ G+ I ++P A + F +Y+ +K ++
Sbjct: 166 LAMAYKEGGPRGLYRGIGPTLIGILPYAGLKFYIYEELKRHV 207
>gb|AAM91170.1| putative mitochondrial carrier protein [Arabidopsis thaliana]
gi|20260324|gb|AAM13060.1| putative mitochondrial
carrier protein [Arabidopsis thaliana]
Length = 152
Score = 186 bits (472), Expect = 4e-46
Identities = 89/145 (61%), Positives = 112/145 (76%), Gaps = 4/145 (2%)
Query: 74 YKEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYKKSIMAKLTCGSVAGLLG 133
YKEGG +G+YRG+ PTL GI PYAGLKFY YEE+KRHVPE+++ S+ L CG++AGL G
Sbjct: 3 YKEGGPRGLYRGIGPTLIGILPYAGLKFYIYEELKRHVPEEHQNSVRMHLPCGALAGLFG 62
Query: 134 QTFTYFLEVVRRQMQVQNL-PASEEA---ELKGTMRSMVLIAQKQGWKTLFSGLSINYIK 189
QT TY L+VVRRQMQV+NL P + E K T + I + QGWK LF+GLSINYIK
Sbjct: 63 QTITYPLDVVRRQMQVENLQPMTSEGNNKRYKNTFDGLNTIVRTQGWKQLFAGLSINYIK 122
Query: 190 VVPSAAIGFTVYDTMKSYLRVPSRD 214
+VPS AIGFTVY++MKS++R+P R+
Sbjct: 123 IVPSVAIGFTVYESMKSWMRIPPRE 147
Score = 39.7 bits (91), Expect = 0.060
Identities = 26/90 (28%), Positives = 42/90 (45%), Gaps = 3/90 (3%)
Query: 19 LLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGG 78
L G LAG F +T+ PL+ ++ QV + + G N + Y+ D L+ + G
Sbjct: 52 LPCGALAGLFGQTITYPLDVVRRQMQVENLQPMTSEG---NNKRYKNTFDGLNTIVRTQG 108
Query: 79 IKGIYRGVAPTLFGIFPYAGLKFYFYEEMK 108
K ++ G++ I P + F YE MK
Sbjct: 109 WKQLFAGLSINYIKIVPSVAIGFTVYESMK 138
>ref|XP_477733.1| putative mitochondrial carrier protein [Oryza sativa (japonica
cultivar-group)] gi|34394749|dbj|BAC84113.1| putative
mitochondrial carrier protein [Oryza sativa (japonica
cultivar-group)]
Length = 333
Score = 116 bits (290), Expect = 5e-25
Identities = 73/203 (35%), Positives = 107/203 (51%), Gaps = 25/203 (12%)
Query: 18 ELLAGGLAGGFAKTVVAPLERLKI-LFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKE 76
+LLAG AGG A PL+ + L +P +GM L Y+E
Sbjct: 139 DLLAGSAAGGTAVLATYPLDLARTRLACAAAPPGAAAAGMSG----------VLRSAYRE 188
Query: 77 GG-IKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYKKS------IMAKLTCGSVA 129
GG ++G+YRG+ P+L + P +GL F YE +K +P + ++ AK+ CG A
Sbjct: 189 GGGVRGVYRGLCPSLARVLPMSGLNFCVYEALKAQIPREEEEHGARGWRRAAKVACGVAA 248
Query: 130 GLLGQTFTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIK 189
GL+ T TY L+VVRRQ+Q+ GT+++ I + QG + L++GL I Y+K
Sbjct: 249 GLVASTATYPLDVVRRQIQLGGGGG-------GTLQAFRAIVRAQGARQLYAGLGITYVK 301
Query: 190 VVPSAAIGFTVYDTMKSYLRVPS 212
VPS A+G YD MKS L +P+
Sbjct: 302 KVPSTAVGLVAYDYMKSLLMLPA 324
Score = 76.6 bits (187), Expect = 4e-13
Identities = 58/207 (28%), Positives = 97/207 (46%), Gaps = 20/207 (9%)
Query: 13 PLFAKELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSK 72
P FA+E++AGG+AG AKT VAPLER+ ++ QV + + G L +
Sbjct: 39 PAFAREMVAGGVAGVVAKTAVAPLERVNLMRQVGAAPR------------GAGAVQMLRE 86
Query: 73 TYKEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHV---PEDYKKSIMAKLTCGSVA 129
+ G+ G++RG +F L F YE KR + + L GS A
Sbjct: 87 IGRGEGVAGLFRGNGANALRVFHTKALHFMAYERYKRFLLGAAPSLGDGPVVDLLAGSAA 146
Query: 130 GLLGQTFTYFLEVVRRQMQVQNL-PASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYI 188
G TY L++ R ++ P + A + G +RS + G + ++ GL +
Sbjct: 147 GGTAVLATYPLDLARTRLACAAAPPGAAAAGMSGVLRS--AYREGGGVRGVYRGLCPSLA 204
Query: 189 KVVPSAAIGFTVYDTMKSYLRVPSRDE 215
+V+P + + F VY+ +K+ ++P +E
Sbjct: 205 RVLPMSGLNFCVYEALKA--QIPREEE 229
>ref|XP_629781.1| hypothetical protein DDB0184176 [Dictyostelium discoideum]
gi|60463180|gb|EAL61373.1| hypothetical protein
DDB0184176 [Dictyostelium discoideum]
Length = 297
Score = 112 bits (279), Expect = 9e-24
Identities = 71/198 (35%), Positives = 105/198 (52%), Gaps = 25/198 (12%)
Query: 20 LAGGLAGGFAKTVVAPLERLKILFQVV---SPTKLNVSGMVNNEQVYRGIRDCLSKTYKE 76
LAG AGG A PL+ L+ + PTK + L T+ +
Sbjct: 111 LAGSAAGGIAVCATYPLDLLRARLAIEIHKKPTKPH---------------HLLKSTFTK 155
Query: 77 GGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVP-----EDYKKSIMAKLTCGSVAGL 131
G+KGIYRG+ PTL GI PY G+ F +E +KR P E+ + S KL G +AG
Sbjct: 156 DGVKGIYRGIQPTLIGILPYGGISFSTFEFLKRIAPLNEIDENGQISGTYKLIAGGIAGG 215
Query: 132 LGQTFTYFLEVVRRQMQVQNL-PASEEAELK-GTMRSMVLIAQKQGWKTLFSGLSINYIK 189
+ QT Y +VVRR++Q A L+ GT+R++ I +++G L+ GLSINY+K
Sbjct: 216 VAQTVAYPFDVVRRRVQTHGFGDAKAVVNLEHGTLRTIAHILKEEGILALYKGLSINYVK 275
Query: 190 VVPSAAIGFTVYDTMKSY 207
V+P+A+I F Y+ + ++
Sbjct: 276 VIPTASIAFYTYEYLSNF 293
Score = 95.1 bits (235), Expect = 1e-18
Identities = 65/200 (32%), Positives = 103/200 (51%), Gaps = 24/200 (12%)
Query: 20 LAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGI 79
L+GGLAG AK+ VAPLER+KIL+Q+ S + + VY + K + GI
Sbjct: 19 LSGGLAGVTAKSAVAPLERVKILYQIKSE-------LYSLNSVYGS----MLKIVENEGI 67
Query: 80 KGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYKKSIMAKLTCGSVAGLLGQTFTYF 139
KG++RG + T+ +FPYA ++F YE +K H+ D K S GS AG + TY
Sbjct: 68 KGLWRGNSATILRVFPYAAVQFLSYETIKNHLVAD-KSSSFQIFLAGSAAGGIAVCATYP 126
Query: 140 LEVVRRQM--QVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIG 197
L+++R ++ ++ P LK T K G K ++ G+ I ++P I
Sbjct: 127 LDLLRARLAIEIHKKPTKPHHLLKSTF-------TKDGVKGIYRGIQPTLIGILPYGGIS 179
Query: 198 FTVYDTMKSYLRVPSRDEVD 217
F+ ++ +K R+ +E+D
Sbjct: 180 FSTFEFLK---RIAPLNEID 196
Score = 43.9 bits (102), Expect = 0.003
Identities = 28/88 (31%), Positives = 47/88 (52%), Gaps = 5/88 (5%)
Query: 18 ELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEG 77
+L+AGG+AGG A+TV P + ++ +V + + +VN E G ++ KE
Sbjct: 206 KLIAGGIAGGVAQTVAYPFDVVR--RRVQTHGFGDAKAVVNLE---HGTLRTIAHILKEE 260
Query: 78 GIKGIYRGVAPTLFGIFPYAGLKFYFYE 105
GI +Y+G++ + P A + FY YE
Sbjct: 261 GILALYKGLSINYVKVIPTASIAFYTYE 288
>ref|XP_624199.1| PREDICTED: similar to ENSANGP00000008222, partial [Apis mellifera]
Length = 315
Score = 111 bits (277), Expect = 2e-23
Identities = 70/212 (33%), Positives = 114/212 (53%), Gaps = 31/212 (14%)
Query: 18 ELLAGGLAGGFAKTVVAPLE--RLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYK 75
+ LAG AG A T+ PL+ R ++ FQV E +Y GI +K
Sbjct: 111 KFLAGSAAGVTAVTLTYPLDIIRARLAFQVAG------------EHIYIGIVHAGITIFK 158
Query: 76 -EGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMK--------RHVPEDYKK-------SI 119
EGGI+ +YRG PT+FG+ PYAG FY +E++K + E+ + +I
Sbjct: 159 NEGGIRALYRGFWPTIFGMIPYAGFSFYSFEKLKYFCMKYASNYFCENCDRNTGGLVLTI 218
Query: 120 MAKLTCGSVAGLLGQTFTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQG-WKT 178
A+L CG +AG + Q+F+Y L+V RR MQ+ + + ++++ +I ++ G K
Sbjct: 219 PARLLCGGIAGAVAQSFSYPLDVTRRHMQLGIMHHANHKYSSSMLQTIKMIYKENGIIKG 278
Query: 179 LFSGLSINYIKVVPSAAIGFTVYDTMKSYLRV 210
L+ G+SINY++ +P ++ FT Y+ MK L++
Sbjct: 279 LYRGMSINYLRAIPMVSVSFTTYEIMKQILQL 310
Score = 74.7 bits (182), Expect = 2e-12
Identities = 52/192 (27%), Positives = 90/192 (46%), Gaps = 14/192 (7%)
Query: 17 KELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKE 76
K L+AGG+AG +KT VAPL+R+KIL Q N + G+ L + +
Sbjct: 16 KSLIAGGVAGMCSKTTVAPLDRIKILLQA-----------HNKYYKHLGVLSGLREVIQR 64
Query: 77 GGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDY-KKSIMAKLTCGSVAGLLGQT 135
+Y+G + IFPYA +F +E K+++ + K + + K GS AG+ T
Sbjct: 65 ERFFALYKGNFAQMIRIFPYAATQFTTFELYKKYLGGLFGKHTHIDKFLAGSAAGVTAVT 124
Query: 136 FTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAA 195
TY L+++R ++ Q A E + + + + G + L+ G ++P A
Sbjct: 125 LTYPLDIIRARLAFQ--VAGEHIYIGIVHAGITIFKNEGGIRALYRGFWPTIFGMIPYAG 182
Query: 196 IGFTVYDTMKSY 207
F ++ +K +
Sbjct: 183 FSFYSFEKLKYF 194
Score = 37.0 bits (84), Expect = 0.39
Identities = 25/96 (26%), Positives = 44/96 (45%), Gaps = 5/96 (5%)
Query: 113 EDYKKSIMAKLTCGSVAGLLGQTFTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQ 172
+DY+ ++ L G VAG+ +T L+ ++ +Q N L G + Q
Sbjct: 9 KDYE-FLLKSLIAGGVAGMCSKTTVAPLDRIKILLQAHNKYYKHLGVLSGLRE----VIQ 63
Query: 173 KQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSYL 208
++ + L+ G I++ P AA FT ++ K YL
Sbjct: 64 RERFFALYKGNFAQMIRIFPYAATQFTTFELYKKYL 99
>emb|CAG60728.1| unnamed protein product [Candida glabrata CBS138]
gi|50290697|ref|XP_447781.1| unnamed protein product
[Candida glabrata]
Length = 327
Score = 110 bits (275), Expect = 3e-23
Identities = 68/202 (33%), Positives = 110/202 (53%), Gaps = 10/202 (4%)
Query: 17 KELLAGGLAGGFAKTVVAPLE--RLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTY 74
+ L +G L GG + PL+ R ++ Q + +KL+ S ++ G+ LSK Y
Sbjct: 125 QRLFSGALCGGCSVVATYPLDLVRTRLSVQTANLSKLSKS-RASDIAKPPGVWKLLSKAY 183
Query: 75 -KEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYK-----KSIMAKLTCGSV 128
+EGGI G+YRGV PT GI PY L F YE++K +P D + + KL+ G++
Sbjct: 184 AEEGGIMGLYRGVWPTSLGIVPYVALNFAVYEQLKEFMPSDENGNSSMRDSLYKLSMGAI 243
Query: 129 AGLLGQTFTYFLEVVRRQMQVQNLPASEEA-ELKGTMRSMVLIAQKQGWKTLFSGLSINY 187
+G + QT TY +++RR+ QV + +E ++V I + +G+K + GL+ N
Sbjct: 244 SGGVAQTITYPFDLLRRRFQVLAMGGNELGFHYNSVWDALVTIGKTEGFKGYYKGLTANL 303
Query: 188 IKVVPSAAIGFTVYDTMKSYLR 209
KVVPS A+ + VY+ Y++
Sbjct: 304 FKVVPSTAVSWLVYELTWDYMK 325
Score = 92.4 bits (228), Expect = 8e-18
Identities = 63/204 (30%), Positives = 106/204 (51%), Gaps = 25/204 (12%)
Query: 20 LAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKEGGI 79
LAGG+AG ++TVV+P ER+KIL QV S T +G+ D + + YKE I
Sbjct: 28 LAGGIAGAISRTVVSPFERVKILLQVQSSTTA----------YNKGLFDAIGQVYKEENI 77
Query: 80 KGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHV------PEDYKKSIMAKLTCGSVAGLLG 133
KG++RG +FPY+ ++F +E K+H+ + + + +L G++ G
Sbjct: 78 KGLFRGNGLNCIRVFPYSAVQFVVFEGCKKHIFHVDTKGKGEQLNNWQRLFSGALCGGCS 137
Query: 134 QTFTYFLEVVRRQMQVQ--NLP------ASEEAELKGTMRSM-VLIAQKQGWKTLFSGLS 184
TY L++VR ++ VQ NL AS+ A+ G + + A++ G L+ G+
Sbjct: 138 VVATYPLDLVRTRLSVQTANLSKLSKSRASDIAKPPGVWKLLSKAYAEEGGIMGLYRGVW 197
Query: 185 INYIKVVPSAAIGFTVYDTMKSYL 208
+ +VP A+ F VY+ +K ++
Sbjct: 198 PTSLGIVPYVALNFAVYEQLKEFM 221
>gb|AAH65855.1| Unknown (protein for MGC:77742) [Danio rerio]
gi|28277902|gb|AAH45977.1| Unknown (protein for
MGC:77742) [Danio rerio] gi|45387539|ref|NP_991112.1|
solute carrier family 25, member 16 [Danio rerio]
Length = 321
Score = 110 bits (274), Expect = 4e-23
Identities = 77/212 (36%), Positives = 107/212 (50%), Gaps = 34/212 (16%)
Query: 19 LLAGGLAGGFAKTVVAPLE--RLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTY-K 75
L+AG +AG A PL+ R ++ FQV + Y GIR Y K
Sbjct: 123 LMAGSMAGMTAVICTYPLDVIRARLAFQVTGHHR------------YSGIRHAFQTIYHK 170
Query: 76 EGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMK----RHVPEDYKK-----------SIM 120
EGGI G YRG+ PT+ G+ PYAG F+ + +K H PE K
Sbjct: 171 EGGISGFYRGLIPTIIGMAPYAGFSFFTFGTLKTLGLTHFPEQLGKPSLDNPDVLVLKTQ 230
Query: 121 AKLTCGSVAGLLGQTFTYFLEVVRRQMQV-QNLPASEE-AELKGTMRSMVLIAQKQGWKT 178
L CG VAG + QT +Y L+V RR+MQ+ +LP ++ L T++ + +Q K
Sbjct: 231 VNLLCGGVAGAIAQTISYPLDVARRRMQLGASLPDHDKCCSLTKTLKH--VYSQYGVKKG 288
Query: 179 LFSGLSINYIKVVPSAAIGFTVYDTMKSYLRV 210
L+ GLS+NYI+ VPS A+ FT Y+ MK L +
Sbjct: 289 LYRGLSLNYIRCVPSQAVAFTTYEFMKQVLHL 320
Score = 73.6 bits (179), Expect = 4e-12
Identities = 53/192 (27%), Positives = 88/192 (45%), Gaps = 16/192 (8%)
Query: 17 KELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKE 76
+ AGG+AG AK+ +APL+R+KIL Q +P + G+ L K+
Sbjct: 27 RSFTAGGVAGCCAKSTIAPLDRVKILLQAQNP-----------HYKHLGVFATLKAVPKK 75
Query: 77 GGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYKKS-IMAKLTCGSVAGLLGQT 135
G G+Y+G + IFPY ++F ++ K+ + S + +L GS+AG+
Sbjct: 76 EGFLGLYKGNGAMMIRIFPYGAIQFMAFDNYKKFLHTKVGISGHVHRLMAGSMAGMTAVI 135
Query: 136 FTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGW-KTLFSGLSINYIKVVPSA 194
TY L+V+R ++ Q + G + I K+G + GL I + P A
Sbjct: 136 CTYPLDVIRARLAFQ---VTGHHRYSGIRHAFQTIYHKEGGISGFYRGLIPTIIGMAPYA 192
Query: 195 AIGFTVYDTMKS 206
F + T+K+
Sbjct: 193 GFSFFTFGTLKT 204
>ref|NP_037518.2| solute carrier family 25 member 24 isoform 1 [Homo sapiens]
Length = 476
Score = 110 bits (274), Expect = 4e-23
Identities = 65/202 (32%), Positives = 104/202 (51%), Gaps = 20/202 (9%)
Query: 17 KELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKE 76
+ ++G +AG A+T + P+E +K T+L V Y GI DC K K
Sbjct: 290 ERFISGSMAGATAQTFIYPMEVMK--------TRL----AVGKTGQYSGIYDCAKKILKH 337
Query: 77 GGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYKK-----SIMAKLTCGSVAGL 131
G+ Y+G P L GI PYAG+ YE +K + +++ K +M L CG+++
Sbjct: 338 EGLGAFYKGYVPNLLGIIPYAGIDLAVYELLKSYWLDNFAKDSVNPGVMVLLGCGALSST 397
Query: 132 LGQTFTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVV 191
GQ +Y L +VR +MQ Q L S + + G R ++ K+G L+ G++ N++KV+
Sbjct: 398 CGQLASYPLALVRTRMQAQVLEGSPQLNMVGLFRRII---SKEGIPGLYRGITPNFMKVL 454
Query: 192 PSAAIGFTVYDTMKSYLRVPSR 213
P+ I + VY+ MK L V +
Sbjct: 455 PAVGISYVVYENMKQTLGVTQK 476
Score = 97.1 bits (240), Expect = 3e-19
Identities = 62/194 (31%), Positives = 100/194 (50%), Gaps = 21/194 (10%)
Query: 17 KELLAGGLAGGFAKTVVAPLERLKILFQV--VSPTKLNVSGMVNNEQVYRGIRDCLSKTY 74
++LLAGG+AG ++T APL+RLKI+ QV K+N+ G G R +
Sbjct: 196 RQLLAGGIAGAVSRTSTAPLDRLKIMMQVHGSKSDKMNIFG---------GFRQMV---- 242
Query: 75 KEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYKK-SIMAKLTCGSVAGLLG 133
KEGGI+ ++RG + I P +KF+ YE+ K+ + E+ +K + GS+AG
Sbjct: 243 KEGGIRSLWRGNGTNVIKIAPETAVKFWAYEQYKKLLTEEGQKIGTFERFISGSMAGATA 302
Query: 134 QTFTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPS 193
QTF Y +EV++ ++ V + + G I + +G + G N + ++P
Sbjct: 303 QTFIYPMEVMKTRLAV-----GKTGQYSGIYDCAKKILKHEGLGAFYKGYVPNLLGIIPY 357
Query: 194 AAIGFTVYDTMKSY 207
A I VY+ +KSY
Sbjct: 358 AGIDLAVYELLKSY 371
>gb|AAH43993.1| LOC398474 protein [Xenopus laevis]
Length = 535
Score = 109 bits (273), Expect = 5e-23
Identities = 65/201 (32%), Positives = 109/201 (53%), Gaps = 21/201 (10%)
Query: 16 AKELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYK 75
A+ +AG LAG A+T + P+E LK T+L V Y G+ DC K +
Sbjct: 350 AERFIAGSLAGATAQTSIYPMEVLK--------TRL----AVGKTGQYSGMFDCAKKIMQ 397
Query: 76 EGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYKK-----SIMAKLTCGSVAG 130
+ GI Y+G P + GI PYAG+ YE +K + ++Y K ++ L CG+V+
Sbjct: 398 KEGILAFYKGYIPNILGIIPYAGIDLAIYETLKNYWLQNYAKDSANPGVLVLLGCGTVSS 457
Query: 131 LLGQTFTYFLEVVRRQMQVQ-NLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIK 189
GQ +Y L ++R +MQ Q ++ + + + G R +V K+G+ L++G++ N++K
Sbjct: 458 TCGQLASYPLALIRTRMQAQASIEGAPQLNMGGLFRKIV---AKEGFFGLYTGIAPNFLK 514
Query: 190 VVPSAAIGFTVYDTMKSYLRV 210
V+P+ +I + VY+ MK L +
Sbjct: 515 VLPAVSISYVVYEKMKIQLGI 535
Score = 98.6 bits (244), Expect = 1e-19
Identities = 59/192 (30%), Positives = 96/192 (49%), Gaps = 18/192 (9%)
Query: 17 KELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKE 76
K LLAGG+AG ++T APL+RLK++ Q V G N + G++ + KE
Sbjct: 258 KHLLAGGMAGAVSRTGTAPLDRLKVMMQ--------VHGTKGNSNIITGLKQMV----KE 305
Query: 77 GGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKR-HVPEDYKKSIMAKLTCGSVAGLLGQT 135
GG++ ++RG + I P +KF+ YE+ K+ E K + GS+AG QT
Sbjct: 306 GGVRSLWRGNGVNVIKIAPETAMKFWAYEQYKKLFTSESGKLGTAERFIAGSLAGATAQT 365
Query: 136 FTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAA 195
Y +EV++ ++ V + + G I QK+G + G N + ++P A
Sbjct: 366 SIYPMEVLKTRLAV-----GKTGQYSGMFDCAKKIMQKEGILAFYKGYIPNILGIIPYAG 420
Query: 196 IGFTVYDTMKSY 207
I +Y+T+K+Y
Sbjct: 421 IDLAIYETLKNY 432
>gb|EAK83389.1| hypothetical protein UM02351.1 [Ustilago maydis 521]
gi|49071354|ref|XP_399966.1| hypothetical protein
UM02351.1 [Ustilago maydis 521]
Length = 495
Score = 109 bits (272), Expect = 6e-23
Identities = 76/225 (33%), Positives = 115/225 (50%), Gaps = 32/225 (14%)
Query: 17 KELLAGGLAGGFAKTVVAPLERLKILFQVVSP--------------TKLNVSGMVNNEQV 62
++L AG +AG + PL+ ++ + S TK+ V+ V EQV
Sbjct: 257 RKLTAGAIAGIASVVSTYPLDLVRSRISIASANMYNEAKSEAISASTKMAVAERVP-EQV 315
Query: 63 YR-----------GIRDCLSKTYKE-GGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRH 110
R GI +K Y+E GG++G+YRG PT G+ PY L FYFYE ++
Sbjct: 316 LRTQIAARQKAVPGIWAMTTKVYREEGGLRGLYRGCVPTSVGVAPYVALNFYFYEAARKR 375
Query: 111 V-PED-YKKSIMAKLTCGSVAGLLGQTFTYFLEVVRRQMQVQNLPASEEA---ELKGTMR 165
+ P D S + KL CG++AG + QT TY L+V+RR+MQV + S+E + K +
Sbjct: 376 ISPADGSDPSALLKLACGALAGSISQTLTYPLDVLRRRMQVAGMKDSQEKLGYKDKNAIN 435
Query: 166 SMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSYLRV 210
++ I + +G L+ GL N +KV PS F Y+ +K +L V
Sbjct: 436 AIQNIIKAEGVTGLYRGLLPNLLKVAPSIGTSFLTYEAVKGFLEV 480
Score = 77.0 bits (188), Expect = 3e-13
Identities = 51/154 (33%), Positives = 81/154 (52%), Gaps = 16/154 (10%)
Query: 20 LAGGLAGGFAKTVVAPLERLKILFQV------------VSPTKLNVSGMVNNEQVYRGIR 67
+AGG AG ++TVV+PLERLKI+ QV S T + S + N + Y G+
Sbjct: 144 VAGGAAGATSRTVVSPLERLKIIMQVQPQSSQSSSSGAASTTAKSRSAVKN--RAYNGVW 201
Query: 68 DCLSKTYKEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPED--YKKSIMAKLTC 125
L K ++E G G RG I PY+ ++F YE K ++ ++ + +M KLT
Sbjct: 202 TGLVKMWQEEGFAGFMRGNGINCLRIAPYSAVQFTTYEMCKTYLRQEGSDELDVMRKLTA 261
Query: 126 GSVAGLLGQTFTYFLEVVRRQMQVQNLPASEEAE 159
G++AG+ TY L++VR ++ + + EA+
Sbjct: 262 GAIAGIASVVSTYPLDLVRSRISIASANMYNEAK 295
Score = 45.4 bits (106), Expect = 0.001
Identities = 30/116 (25%), Positives = 50/116 (42%), Gaps = 17/116 (14%)
Query: 119 IMAKLTCGSVAGLLGQTFTYFLEVVRRQMQVQ-----------------NLPASEEAELK 161
++ G AG +T LE ++ MQVQ + A +
Sbjct: 139 LITYFVAGGAAGATSRTVVSPLERLKIIMQVQPQSSQSSSSGAASTTAKSRSAVKNRAYN 198
Query: 162 GTMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMKSYLRVPSRDEVD 217
G +V + Q++G+ G IN +++ P +A+ FT Y+ K+YLR DE+D
Sbjct: 199 GVWTGLVKMWQEEGFAGFMRGNGINCLRIAPYSAVQFTTYEMCKTYLRQEGSDELD 254
>dbj|BAC11071.1| unnamed protein product [Homo sapiens]
Length = 208
Score = 108 bits (270), Expect = 1e-22
Identities = 64/203 (31%), Positives = 106/203 (51%), Gaps = 21/203 (10%)
Query: 17 KELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKE 76
+ +AG LAG A+T++ P+E LK T+L + Y+G+ DC + +
Sbjct: 21 ERFVAGSLAGATAQTIIYPMEVLK--------TRLTL----RRTGQYKGLLDCARRILER 68
Query: 77 GGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYK-----KSIMAKLTCGSVAGL 131
G + YRG P + GI PYAG+ YE +K + Y I+ L CG+++
Sbjct: 69 EGPRAFYRGYLPNVLGIIPYAGIDLAVYETLKNWWLQQYSHDSADPGILVLLACGTISST 128
Query: 132 LGQTFTYFLEVVRRQMQVQ-NLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKV 190
GQ +Y L +VR +MQ Q ++ + + G +R I ++G + L+ G++ N++KV
Sbjct: 129 CGQIASYPLALVRTRMQAQASIEGGPQLSMLGLLRH---ILSQEGMRGLYRGIAPNFMKV 185
Query: 191 VPSAAIGFTVYDTMKSYLRVPSR 213
+P+ +I + VY+ MK L V SR
Sbjct: 186 IPAVSISYVVYENMKQALGVTSR 208
Score = 49.7 bits (117), Expect = 6e-05
Identities = 30/116 (25%), Positives = 59/116 (50%), Gaps = 7/116 (6%)
Query: 104 YEEMKRHV-PEDYKKSIMAKLTCGSVAGLLGQTFTYFLEVVRRQMQVQNLPASEEAELKG 162
YE++KR + + + + GS+AG QT Y +EV++ ++ ++ + KG
Sbjct: 3 YEQIKRAILGQQETLHVQERFVAGSLAGATAQTIIYPMEVLKTRLTLR-----RTGQYKG 57
Query: 163 TMRSMVLIAQKQGWKTLFSGLSINYIKVVPSAAIGFTVYDTMKS-YLRVPSRDEVD 217
+ I +++G + + G N + ++P A I VY+T+K+ +L+ S D D
Sbjct: 58 LLDCARRILEREGPRAFYRGYLPNVLGIIPYAGIDLAVYETLKNWWLQQYSHDSAD 113
>ref|XP_524071.1| PREDICTED: similar to solute carrier family 25 (mitochondrial
carrier; phosphate carrier), member 23; short
calcium-binding mitochondrial carrier 3; mitochondrial
Ca2+-dependent solute carrier protein 2 [Pan
troglodytes]
Length = 562
Score = 108 bits (270), Expect = 1e-22
Identities = 64/203 (31%), Positives = 106/203 (51%), Gaps = 21/203 (10%)
Query: 17 KELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKE 76
+ +AG LAG A+T++ P+E LK T+L + Y+G+ DC + +
Sbjct: 375 ERFVAGSLAGATAQTIIYPMEVLK--------TRLTL----RQTGQYKGLLDCARRILER 422
Query: 77 GGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYK-----KSIMAKLTCGSVAGL 131
G + YRG P + GI PYAG+ YE +K + Y I+ L CG+++
Sbjct: 423 EGPRAFYRGYLPNVLGIIPYAGIDLAVYETLKNWWLQQYSHDSADPGILVLLACGTISST 482
Query: 132 LGQTFTYFLEVVRRQMQVQ-NLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKV 190
GQ +Y L +VR +MQ Q ++ + + G +R I ++G + L+ G++ N++KV
Sbjct: 483 CGQIASYPLALVRTRMQAQASIEGGPQLSMLGLLRH---ILSQEGMRGLYRGIAPNFMKV 539
Query: 191 VPSAAIGFTVYDTMKSYLRVPSR 213
+P+ +I + VY+ MK L V SR
Sbjct: 540 IPAVSISYVVYENMKQALGVTSR 562
Score = 92.0 bits (227), Expect = 1e-17
Identities = 60/205 (29%), Positives = 107/205 (51%), Gaps = 22/205 (10%)
Query: 17 KELLAGGLAGGFAKTVVAPLERLKILFQV--VSPTKLNVSGMVNNEQVYRGIRDCLSKTY 74
K+L+AG +AG ++T APL+RLK+ QV +LN+ G G+R +
Sbjct: 281 KQLVAGAVAGAVSRTGTAPLDRLKVFMQVHASKTNRLNILG---------GLRSMV---- 327
Query: 75 KEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHV-PEDYKKSIMAKLTCGSVAGLLG 133
EGGI+ ++RG + I P + +KF YE++KR + + + + GS+AG
Sbjct: 328 LEGGIRSLWRGNGINVLKIAPESAIKFMAYEQIKRAILGQQETLHVQERFVAGSLAGATA 387
Query: 134 QTFTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPS 193
QT Y +EV++ ++ ++ + + KG + I +++G + + G N + ++P
Sbjct: 388 QTIIYPMEVLKTRLTLR-----QTGQYKGLLDCARRILEREGPRAFYRGYLPNVLGIIPY 442
Query: 194 AAIGFTVYDTMKS-YLRVPSRDEVD 217
A I VY+T+K+ +L+ S D D
Sbjct: 443 AGIDLAVYETLKNWWLQQYSHDSAD 467
>emb|CAF04494.1| small calcium-binding mitochondrial carrier 3 [Homo sapiens]
gi|47109342|emb|CAF04059.1| mitochondrial ATP-Mg/Pi
carrier [Homo sapiens] gi|53830367|gb|AAU95077.1|
mitochondrial Ca2+-dependent solute carrier protein 2
[Homo sapiens] gi|48476342|ref|NP_077008.2| solute
carrier family 25 (mitochondrial carrier; phosphate
carrier), member 23 [Homo sapiens]
Length = 468
Score = 108 bits (270), Expect = 1e-22
Identities = 64/203 (31%), Positives = 106/203 (51%), Gaps = 21/203 (10%)
Query: 17 KELLAGGLAGGFAKTVVAPLERLKILFQVVSPTKLNVSGMVNNEQVYRGIRDCLSKTYKE 76
+ +AG LAG A+T++ P+E LK T+L + Y+G+ DC + +
Sbjct: 281 ERFVAGSLAGATAQTIIYPMEVLK--------TRLTL----RRTGQYKGLLDCARRILER 328
Query: 77 GGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHVPEDYK-----KSIMAKLTCGSVAGL 131
G + YRG P + GI PYAG+ YE +K + Y I+ L CG+++
Sbjct: 329 EGPRAFYRGYLPNVLGIIPYAGIDLAVYETLKNWWLQQYSHDSADPGILVLLACGTISST 388
Query: 132 LGQTFTYFLEVVRRQMQVQ-NLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKV 190
GQ +Y L +VR +MQ Q ++ + + G +R I ++G + L+ G++ N++KV
Sbjct: 389 CGQIASYPLALVRTRMQAQASIEGGPQLSMLGLLRH---ILSQEGMRGLYRGIAPNFMKV 445
Query: 191 VPSAAIGFTVYDTMKSYLRVPSR 213
+P+ +I + VY+ MK L V SR
Sbjct: 446 IPAVSISYVVYENMKQALGVTSR 468
Score = 91.3 bits (225), Expect = 2e-17
Identities = 60/205 (29%), Positives = 106/205 (51%), Gaps = 22/205 (10%)
Query: 17 KELLAGGLAGGFAKTVVAPLERLKILFQV--VSPTKLNVSGMVNNEQVYRGIRDCLSKTY 74
K+L+AG +AG ++T APL+RLK+ QV +LN+ G G+R +
Sbjct: 187 KQLVAGAVAGAVSRTGTAPLDRLKVFMQVHASKTNRLNILG---------GLRSMV---- 233
Query: 75 KEGGIKGIYRGVAPTLFGIFPYAGLKFYFYEEMKRHV-PEDYKKSIMAKLTCGSVAGLLG 133
EGGI+ ++RG + I P + +KF YE++KR + + + + GS+AG
Sbjct: 234 LEGGIRSLWRGNGINVLKIAPESAIKFMAYEQIKRAILGQQETLHVQERFVAGSLAGATA 293
Query: 134 QTFTYFLEVVRRQMQVQNLPASEEAELKGTMRSMVLIAQKQGWKTLFSGLSINYIKVVPS 193
QT Y +EV++ ++ ++ + KG + I +++G + + G N + ++P
Sbjct: 294 QTIIYPMEVLKTRLTLR-----RTGQYKGLLDCARRILEREGPRAFYRGYLPNVLGIIPY 348
Query: 194 AAIGFTVYDTMKS-YLRVPSRDEVD 217
A I VY+T+K+ +L+ S D D
Sbjct: 349 AGIDLAVYETLKNWWLQQYSHDSAD 373
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.321 0.138 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 346,788,854
Number of Sequences: 2540612
Number of extensions: 13659300
Number of successful extensions: 40952
Number of sequences better than 10.0: 1960
Number of HSP's better than 10.0 without gapping: 1375
Number of HSP's successfully gapped in prelim test: 586
Number of HSP's that attempted gapping in prelim test: 32224
Number of HSP's gapped (non-prelim): 4940
length of query: 217
length of database: 863,360,394
effective HSP length: 123
effective length of query: 94
effective length of database: 550,865,118
effective search space: 51781321092
effective search space used: 51781321092
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 72 (32.3 bits)
Medicago: description of AC148360.13


