

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148347.6 + phase: 0
(807 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
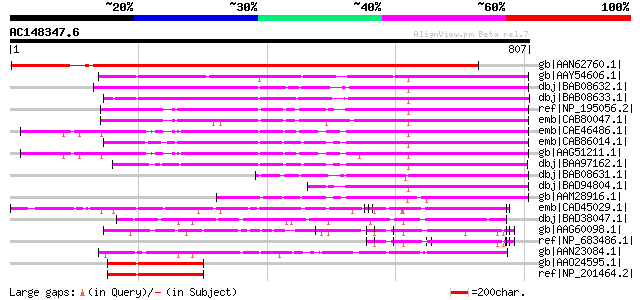
Score E
Sequences producing significant alignments: (bits) Value
gb|AAN62760.1| disease resistance protein-like protein MsR1 [Med... 1194 0.0
gb|AAY54606.1| NRG1 [Nicotiana benthamiana] 483 e-134
dbj|BAB08632.1| disease resistance protein-like [Arabidopsis tha... 444 e-123
dbj|BAB08633.1| disease resistance protein-like [Arabidopsis tha... 409 e-112
ref|NP_195056.2| disease resistance protein (CC-NBS-LRR class), ... 379 e-103
emb|CAB80047.1| putative protein [Arabidopsis thaliana] gi|44902... 359 2e-97
emb|CAE46486.1| CC-NBS-LRR disease resistance protein [Arabidops... 343 2e-92
emb|CAB86014.1| disease resistance-like protein [Arabidopsis tha... 337 7e-91
gb|AAG51211.1| disease resistance protein, putative; 92850-95636... 335 4e-90
dbj|BAA97162.1| disease resistance protein-like [Arabidopsis tha... 332 2e-89
dbj|BAB08631.1| unnamed protein product [Arabidopsis thaliana] g... 244 8e-63
dbj|BAD94804.1| putative protein [Arabidopsis thaliana] 238 6e-61
gb|AAM28916.1| NBS/LRR [Pinus taeda] 229 4e-58
emb|CAD45029.1| NBS-LRR disease resistance protein homologue [Ho... 165 5e-39
dbj|BAD38047.1| putative NBS-LRR resistance protein RGH2 [Oryza ... 153 2e-35
gb|AAG60098.1| disease resistance protein, putative [Arabidopsis... 132 5e-29
ref|NP_683486.1| leucine-rich repeat family protein [Arabidopsis... 132 5e-29
gb|AAN23084.1| putative rp3 protein [Zea mays] 121 1e-25
gb|AAO24595.1| At5g66630 [Arabidopsis thaliana] 119 3e-25
ref|NP_201464.2| LIM domain-containing protein [Arabidopsis thal... 119 3e-25
>gb|AAN62760.1| disease resistance protein-like protein MsR1 [Medicago sativa]
Length = 704
Score = 1194 bits (3090), Expect = 0.0
Identities = 612/728 (84%), Positives = 656/728 (90%), Gaps = 29/728 (3%)
Query: 3 SATTHIITLTTRF--HDVLNKISQIVETARKHQSARQLLRSILKDLTLVVQDIKQYNEHL 60
S+ THIITL TRF HDVLNKISQI E A+++QS+R+ LR IL++L+ VVQDIKQYN+HL
Sbjct: 2 SSATHIITLATRFQFHDVLNKISQIAEKAQENQSSRRSLRLILRNLSPVVQDIKQYNDHL 61
Query: 61 DHPREEIKTLLEENDAEESACKCSSENDSYVVDENQSLIVNDVEETLYKAREILELLNYE 120
DHPREEI +L+EENDAEESACKCSSENDS VV++ L E
Sbjct: 62 DHPREEINSLIEENDAEESACKCSSENDSNVVEK----------------------LKNE 99
Query: 121 TFEHKFNEAKPPFKRPFDVPENPKFTVGLDIPFSKLKMELIRDGSSTLVLTGLGGLGKTT 180
TFE A PPFK PFDVPENPKFTVGLDIPFSKLKMEL+RDGSSTLVLTGLGGLGKTT
Sbjct: 100 TFE-----AGPPFKCPFDVPENPKFTVGLDIPFSKLKMELLRDGSSTLVLTGLGGLGKTT 154
Query: 181 LATKLCWDQEVNGKFKENIIFVTFSKTPMLKTIVERIIEHCGYPVPEFQSDEDAVNKLEL 240
LATKLCWDQEVNGKF ENIIFVTFSKTPMLKTIVERI EHCGYPVPEFQ+DEDAVN+L L
Sbjct: 155 LATKLCWDQEVNGKFMENIIFVTFSKTPMLKTIVERIHEHCGYPVPEFQNDEDAVNRLGL 214
Query: 241 LLKKVEGSPLLLVLDDVWPTSESLVKKLQFEISDFKILVTSRVSFPRFRTTCILKPLAHE 300
LLKKVEGSPLLLVLDDVWP+SESLV+KLQF+ISDFKILVTSRV+FPRF TTCILKPLAHE
Sbjct: 215 LLKKVEGSPLLLVLDDVWPSSESLVEKLQFQISDFKILVTSRVAFPRFSTTCILKPLAHE 274
Query: 301 DAVTLFHHYAQMEKNSSDIIDKNLVEKVVRSCKGLPLTIKVIATSLRNRPDDLWRKIVME 360
DAVTLFHHYA MEKNSSDIIDKNLVEKVVRSC+GLPLTIKVIATSL+NRP DLWRKIV E
Sbjct: 275 DAVTLFHHYALMEKNSSDIIDKNLVEKVVRSCQGLPLTIKVIATSLKNRPHDLWRKIVKE 334
Query: 361 LSQGHSILDSNTELLTRLQKIFDVLEDNPTIMECFMDIALFPEDHRIPVAALVDMWAKLY 420
LSQGHSILDSNTELLTRLQKIFDVLEDNPTI+ECFMD+ALFPEDHRIPVAALVDMWA+LY
Sbjct: 335 LSQGHSILDSNTELLTRLQKIFDVLEDNPTIIECFMDLALFPEDHRIPVAALVDMWAELY 394
Query: 421 KLDDNGIQAMEIINKLGIMNLANVIIPRKNASDTDNNNYNNHFIILHDILRDLGIYQSTK 480
+LDD GIQAMEIINKLGIMNLANVIIPRK+ASDTDNNNYNNHFI+LHDILR+LGIY+STK
Sbjct: 395 RLDDTGIQAMEIINKLGIMNLANVIIPRKDASDTDNNNYNNHFIMLHDILRELGIYRSTK 454
Query: 481 QPFEQRKRLIIDINKNRSELAEKQQSLLTRILSKVMRLCIKQNPQQLAAHILSVSTDEAC 540
+PFEQRKRLIID++KN+S L EKQQ L+ RILSK MRLC+KQNPQQLAA ILSVSTDE C
Sbjct: 455 EPFEQRKRLIIDMHKNKSGLTEKQQGLMIRILSKFMRLCVKQNPQQLAARILSVSTDETC 514
Query: 541 ASDWSQMQPTQVEVLILNLHTKQYSFPESIKKMSKLKVLIITNYSFHPCELNNFELLGSL 600
A DWSQM+P QVEVLILNLHTKQYS PE I KMSKLKVLIITNY+ HP EL NFELL SL
Sbjct: 515 ALDWSQMEPAQVEVLILNLHTKQYSLPEWIGKMSKLKVLIITNYTVHPSELTNFELLSSL 574
Query: 601 QNLEKIRLERILVPSFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKD 660
QNLEKIRLERI VPSF T+KNLKKLSLYMCNT LAFEKGSILISDAFPNLEELNIDYCKD
Sbjct: 575 QNLEKIRLERISVPSFATVKNLKKLSLYMCNTRLAFEKGSILISDAFPNLEELNIDYCKD 634
Query: 661 LVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGKLL 720
L+VL TGICDIISLKKL+VTNCHK+SSLP+DIGKLENLELLS SSCTDLEAIPTSI KLL
Sbjct: 635 LLVLPTGICDIISLKKLSVTNCHKISSLPEDIGKLENLELLSLSSCTDLEAIPTSIEKLL 694
Query: 721 NLKHLDIS 728
NLKHLDIS
Sbjct: 695 NLKHLDIS 702
>gb|AAY54606.1| NRG1 [Nicotiana benthamiana]
Length = 850
Score = 483 bits (1242), Expect = e-134
Identities = 280/681 (41%), Positives = 405/681 (59%), Gaps = 35/681 (5%)
Query: 138 DVPENPKFTVGLDIPFSKLKMELIRDGSSTLVLTGLGGLGKTTLATKLCWDQEVNGKFKE 197
DVP+ VG D+P +LK++L+ + +VL+ G GKTTLA LC + ++ K+++
Sbjct: 187 DVPQFSDSVVGFDLPLQELKVKLLEEKEKVVVLSAPAGCGKTTLAAMLCQEDDIKDKYRD 246
Query: 198 NIIFVTFSKTPMLKTIVERIIEHCGYPVPEFQSDEDAVNKLELLLKKVEGSPLLLVLDDV 257
I FVT SK +K IV I E GY P+F S+ AV +L LL++ P+LLVLDDV
Sbjct: 247 -IFFVTVSKKANIKRIVGEIFEMKGYKGPDFASEHAAVCQLNNLLRRSTSQPVLLVLDDV 305
Query: 258 WPTSESLVKKLQFEISDFKILVTSRVSFPRFRTTCILKPLAHEDAVTLFHHYAQMEKNSS 317
W S+ +++ F+I FKILVTSR FP+F T L L+ +DA LF Y+ K+S
Sbjct: 306 WSESDFVIESFIFQIPGFKILVTSRSVFPKF-DTYKLNLLSEKDAKALF--YSSAFKDSI 362
Query: 318 DIIDKNLVEKVVRSCKGLPLTIKVIATSLRNRPDDLWRKIVMELSQGHSILDSNTELLTR 377
+ +LV K VRSC G PL +KV+ SL +P+ +W VM S+ + + +LL
Sbjct: 363 PYVQLDLVHKAVRSCCGFPLALKVVGRSLCGQPELIWFNRVMLQSKRQILFPTENDLLRT 422
Query: 378 LQKIFDVLED-------NPTIMECFMDIALFPEDHRIPVAALVDMWAKLYKLDDNGIQAM 430
L+ D L++ T+ +C++D+ FPEDHRI AA++DMW + Y LD++G++AM
Sbjct: 423 LRASIDALDEIDLYSSEATTLRDCYLDLGSFPEDHRIHAAAILDMWVERYNLDEDGMKAM 482
Query: 431 EIINKLGIMNLANVIIPRKNASDTDNNNYNNHFIILHDILRDLGIYQSTKQPFEQRKRLI 490
I +L NL N+ + RK+A +N H+I HD+LR+L I+Q ++ E+RKRL
Sbjct: 483 AIFFQLSSQNLVNLALARKDAPAV-LGLHNLHYIQQHDLLRELVIHQCDEKTVEERKRLY 541
Query: 491 IDINKNRSELAEKQQSLLTRILSKVMRLCIKQNPQQLAAHILSVSTDEACASDWSQMQPT 550
I+I N QQ L Q L A +LS+ TDE S W ++
Sbjct: 542 INIKGNDFPKWWSQQRL-----------------QPLQAEVLSIFTDEHFESVWYDVRFP 584
Query: 551 QVEVLILNLHTKQYSFPESIKKMSKLKVLIITNYSFHPCELNNFELLGSLQNLEKIRLER 610
+VEVL+LN TK Y+FP +++MS+LK LI+ N F P +LNNF+L SL NL++I LER
Sbjct: 585 KVEVLVLNFETKTYNFPPFVEQMSQLKTLIVANNYFFPTKLNNFQLC-SLLNLKRISLER 643
Query: 611 ILVPSFGT----LKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQT 666
I V S T L NL+K+S MC AFE + +S +P L E+NI+YC DLV +
Sbjct: 644 ISVTSIFTANLQLPNLRKISFIMCEIGEAFENYAANMSYMWPKLVEMNIEYCSDLVEVPA 703
Query: 667 GICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGKLLNLKHLD 726
CD++ LKKL++ CH+L +LP+++GKL NLE+L SCT++ +P S+ KL L LD
Sbjct: 704 ETCDLVGLKKLSICYCHELVALPEELGKLSNLEVLRLHSCTNVSKLPESVVKLNRLGFLD 763
Query: 727 ISNCISLSSLPEEFGNLCNLKNLDMAS-CASIELPFSVVNLQNLKTITCDEETAATWEDF 785
+ +C+ L LP E LC+L+ + M S ELP SV+ L L+ + CDEETA+ WE +
Sbjct: 764 VYDCVELDFLPREMDQLCSLRTICMGSRLGFTELPDSVLRLVKLEDVVCDEETASLWEYY 823
Query: 786 QHMLPNMKIEVLHVDVNLNWL 806
+ L N++I V+ D+NLN L
Sbjct: 824 KEHLRNLRITVIKEDINLNLL 844
>dbj|BAB08632.1| disease resistance protein-like [Arabidopsis thaliana]
gi|15240126|ref|NP_201491.1| disease resistance protein
(CC-NBS-LRR class), putative [Arabidopsis thaliana]
gi|46395984|sp|Q9FKZ1|DRL42_ARATH Probable disease
resistance protein At5g66900
Length = 809
Score = 444 bits (1143), Expect = e-123
Identities = 264/683 (38%), Positives = 392/683 (56%), Gaps = 36/683 (5%)
Query: 131 PPFKRPFDVPENPKFTVGLDIPFSKLKMELIRDGSSTLVLTGLGGLGKTTLATKLCWDQE 190
P F+ VP+ K VGLD P +LK L+ D TLV++ G GKTTL ++LC D +
Sbjct: 153 PVFRDLCSVPKLDKVIVGLDWPLGELKKRLLDDSVVTLVVSAPPGCGKTTLVSRLCDDPD 212
Query: 191 VNGKFKENIIFVTFSKTPMLKTIVERIIEHCGYPVPEFQSDEDAVNKLELLLKKV-EGSP 249
+ GKFK +I F S TP + IV+ +++H GY F++D A L LL+++ E P
Sbjct: 213 IKGKFK-HIFFNVVSNTPNFRVIVQNLLQHNGYNALTFENDSQAEVGLRKLLEELKENGP 271
Query: 250 LLLVLDDVWPTSESLVKKLQFEISDFKILVTSRVSFPRFRTTCILKPLAHEDAVTLFHHY 309
+LLVLDDVW ++S ++K Q ++ ++KILVTSR FP F + LKPL +DA L H+
Sbjct: 272 ILLVLDDVWRGADSFLQKFQIKLPNYKILVTSRFDFPSFDSNYRLKPLEDDDARALLIHW 331
Query: 310 AQMEKNSSDIIDKNLVEKVVRSCKGLPLTIKVIATSLRNRPDDLWRKIVMELSQGHSILD 369
A N+S ++L++K+++ C G P+ I+V+ SL+ R + W+ V S+G IL
Sbjct: 332 ASRPCNTSPDEYEDLLQKILKRCNGFPIVIEVVGVSLKGRSLNTWKGQVESWSEGEKILG 391
Query: 370 SN-TELLTRLQKIFDVLEDNPTIMECFMDIALFPEDHRIPVAALVDMWAKLYKLDDNGIQ 428
+L LQ FD L+ P + ECF+D+ F ED +I + ++DMW +LY + +
Sbjct: 392 KPYPTVLECLQPSFDALD--PNLKECFLDMGSFLEDQKIRASVIIDMWVELYGKGSSILY 449
Query: 429 AMEIINKLGIMNLANVIIPRKNASDTDNNNYNNHFIILHDILRDLGIYQSTKQPFEQRKR 488
+ L NL ++ N + ++ YN+ + HDILR+L I QS + +RKR
Sbjct: 450 MY--LEDLASQNLLKLVPLGTN--EHEDGFYNDFLVTQHDILRELAICQSEFKENLERKR 505
Query: 489 LIIDINKNRSELAEKQQSLLTRILSKVMRLCIKQNPQQLAAHILSVSTDEACASDWSQMQ 548
L ++I +N C+ + A +LS+STD+ +S W +M
Sbjct: 506 LNLEILENTFP-----------------DWCLNT----INASLLSISTDDLFSSKWLEMD 544
Query: 549 PTQVEVLILNLHTKQYSFPESIKKMSKLKVLIITNYSFHPCELNNFELLGSLQNLEKIRL 608
VE L+LNL + Y+ P I M KLKVL ITN+ F+P L+NF L SL NL++IRL
Sbjct: 545 CPNVEALVLNLSSSDYALPSFISGMKKLKVLTITNHGFYPARLSNFSCLSSLPNLKRIRL 604
Query: 609 ERILVPSFGT----LKNLKKLSLYMCNTILAF-EKGSILISDAFPNLEELNIDYCKDLVV 663
E++ + L +LKKLSL MC+ F + I++S+A L+E++IDYC DL
Sbjct: 605 EKVSITLLDIPQLQLSSLKKLSLVMCSFGEVFYDTEDIVVSNALSKLQEIDIDYCYDLDE 664
Query: 664 LQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGKLLNLK 723
L I +I+SLK L++TNC+KLS LP+ IG L LE+L S +L +P + L NL+
Sbjct: 665 LPYWISEIVSLKTLSITNCNKLSQLPEAIGNLSRLEVLRLCSSMNLSELPEATEGLSNLR 724
Query: 724 HLDISNCISLSSLPEEFGNLCNLKNLDMASCASIELPFSVVNLQNLKTITCDEETAATWE 783
LDIS+C+ L LP+E G L NLK + M C+ ELP SV NL+NL+ + CDEET WE
Sbjct: 725 FLDISHCLGLRKLPQEIGKLQNLKKISMRKCSGCELPESVTNLENLE-VKCDEETGLLWE 783
Query: 784 DFQHMLPNMKIEVLHVDVNLNWL 806
+ + N++++ ++ NLN L
Sbjct: 784 RLKPKMRNLRVQEEEIEHNLNLL 806
>dbj|BAB08633.1| disease resistance protein-like [Arabidopsis thaliana]
gi|18087526|gb|AAL58897.1| AT5g66910/MUD21_17
[Arabidopsis thaliana] gi|15240127|ref|NP_201492.1|
disease resistance protein (CC-NBS-LRR class), putative
[Arabidopsis thaliana] gi|46395983|sp|Q9FKZ0|DRL43_ARATH
Probable disease resistance protein At5g66910
Length = 815
Score = 409 bits (1052), Expect = e-112
Identities = 249/669 (37%), Positives = 381/669 (56%), Gaps = 36/669 (5%)
Query: 147 VGLDIPFSKLKMELIRDGSSTLVLTGLGGLGKTTLATKLCWDQEVNGKFKENIIFVTFSK 206
VGLD P +LK +L+ +S +V++G G GKTTL TKLC D E+ G+FK+ I + S
Sbjct: 173 VGLDWPLVELKKKLL--DNSVVVVSGPPGCGKTTLVTKLCDDPEIEGEFKK-IFYSVVSN 229
Query: 207 TPMLKTIVERIIEHCGYPVPEFQSDEDAVNKLELLLKKV-EGSPLLLVLDDVWPTSESLV 265
TP + IV+ +++ G F D A L LL+++ + +LLVLDDVW SE L+
Sbjct: 230 TPNFRAIVQNLLQDNGCGAITFDDDSQAETGLRDLLEELTKDGRILLVLDDVWQGSEFLL 289
Query: 266 KKLQFEISDFKILVTSRVSFPRFRTTCILKPLAHEDAVTLFHHYAQMEKNSSDIIDKNLV 325
+K Q ++ D+KILVTS+ F T L PL +E A +L +A ++S ++L+
Sbjct: 290 RKFQIDLPDYKILVTSQFDFTSLWPTYHLVPLKYEYARSLLIQWASPPLHTSPDEYEDLL 349
Query: 326 EKVVRSCKGLPLTIKVIATSLRNRPDDLWRKIVMELSQGHSIL-DSNTELLTRLQKIFDV 384
+K+++ C G PL I+V+ SL+ + LW+ V S+G +IL ++N + RLQ F+V
Sbjct: 350 QKILKRCNGFPLVIEVVGISLKGQALYLWKGQVESWSEGETILGNANPTVRQRLQPSFNV 409
Query: 385 LEDNPTIMECFMDIALFPEDHRIPVAALVDMWAKLYKLDDNGIQA-MEIINKLGIMNLAN 443
L+ P + ECFMD+ F +D +I + ++D+W +LY + M +N+L NL
Sbjct: 410 LK--PHLKECFMDMGSFLQDQKIRASLIIDIWMELYGRGSSSTNKFMLYLNELASQNLLK 467
Query: 444 VIIPRKNASDTDNNNYNNHFIILHDILRDLGIYQSTKQPFEQRKRLIIDINKNRSELAEK 503
++ + ++ YN + H+ILR+L I+QS +P QRK+L ++I ++
Sbjct: 468 LV--HLGTNKREDGFYNELLVTQHNILRELAIFQSELEPIMQRKKLNLEIREDNFPDE-- 523
Query: 504 QQSLLTRILSKVMRLCIKQNPQQLAAHILSVSTDEACASDWSQMQPTQVEVLILNLHTKQ 563
C+ Q + A +LS+ TD+ +S W +M VE L+LN+ +
Sbjct: 524 ---------------CLNQ---PINARLLSIYTDDLFSSKWLEMDCPNVEALVLNISSLD 565
Query: 564 YSFPESIKKMSKLKVLIITNYSFHPCELNNFELLGSLQNLEKIRLERILVPSFGT----L 619
Y+ P I +M KLKVL I N+ F+P L+NF L SL NL++IR E++ V L
Sbjct: 566 YALPSFIAEMKKLKVLTIANHGFYPARLSNFSCLSSLPNLKRIRFEKVSVTLLDIPQLQL 625
Query: 620 KNLKKLSLYMCNTILAF-EKGSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLN 678
+LKKLS +MC+ F + I +S A NL+E++IDYC DL L I +++SLK L+
Sbjct: 626 GSLKKLSFFMCSFGEVFYDTEDIDVSKALSNLQEIDIDYCYDLDELPYWIPEVVSLKTLS 685
Query: 679 VTNCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPE 738
+TNC+KLS LP+ IG L LE+L SC +L +P + +L NL+ LDIS+C+ L LP+
Sbjct: 686 ITNCNKLSQLPEAIGNLSRLEVLRMCSCMNLSELPEATERLSNLRSLDISHCLGLRKLPQ 745
Query: 739 EFGNLCNLKNLDMASCASIELPFSVVNLQNLKTITCDEETAATWEDFQHMLPNMKIEVLH 798
E G L L+N+ M C+ ELP SV L+NL+ + CDE T WE + N+++
Sbjct: 746 EIGKLQKLENISMRKCSGCELPDSVRYLENLE-VKCDEVTGLLWERLMPEMRNLRVHTEE 804
Query: 799 VDVNLNWLL 807
+ NL LL
Sbjct: 805 TEHNLKLLL 813
>ref|NP_195056.2| disease resistance protein (CC-NBS-LRR class), putative
[Arabidopsis thaliana]
Length = 816
Score = 379 bits (974), Expect = e-103
Identities = 230/679 (33%), Positives = 367/679 (53%), Gaps = 53/679 (7%)
Query: 141 ENPKFTVGLDIPFSKLKMELIRDGSSTLVLTGLGGLGKTTLATKLCWDQEVNGKFKENII 200
++ KF VGL++ K+K + ++G+GG+GKTTLA +L D EV F+ I+
Sbjct: 176 DSEKFGVGLELGKVKVKKMMFESQGGVFGISGMGGVGKTTLAKELQRDHEVQCHFENRIL 235
Query: 201 FVTFSKTPMLKTIVERI---IEHC--GYPVPEFQSDEDAVNKLELLLKKVEGSPLLLVLD 255
F+T S++P+L+ + E I + C G PVP+ D KL ++LD
Sbjct: 236 FLTVSQSPLLEELRELIWGFLSGCEAGNPVPDCNFPFDGARKL-------------VILD 282
Query: 256 DVWPTSESLVKKLQFEISDFKILVTSRVSFPRFRTTCILKPLAHEDAVTLFHHYAQMEKN 315
DVW T+++L + F+ LV SR + T ++ L+ ++A++LF A +K+
Sbjct: 283 DVW-TTQALDRLTSFKFPGCTTLVVSRSKLTEPKFTYDVEVLSEDEAISLFCLCAFGQKS 341
Query: 316 SSDIIDKNLVEKVVRSCKGLPLTIKVIATSLRNRPDDLWRKIVMELSQGHSILDSN-TEL 374
K+LV++V CKGLPL +KV SL +P+ W+ ++ LS+G DS+ + L
Sbjct: 342 IPLGFCKDLVKQVANECKGLPLALKVTGASLNGKPEMYWKGVLQRLSKGEPADDSHESRL 401
Query: 375 LTRLQKIFDVLEDNPTIMECFMDIALFPEDHRIPVAALVDMWAKLYKLDDNGIQAMEIIN 434
L +++ D L+ T +CF+D+ FPED +IP+ L+++W +L+ +D+ A I+
Sbjct: 402 LRQMEASLDNLDQ--TTKDCFLDLGAFPEDRKIPLDVLINIWIELHDIDEGN--AFAILV 457
Query: 435 KLGIMNLANVIIPRKNASDTDNNNYNNH---FIILHDILRDLGIYQSTKQPFEQRKRLII 491
L NL + + Y +H F+ HD+LRDL ++ S +RKRL++
Sbjct: 458 DLSHKNLLTL-----GKDPRLGSLYASHYDIFVTQHDVLRDLALHLSNAGKVNRRKRLLM 512
Query: 492 DINKNRSELAEKQQSLLTRILSKVMRLCIKQNPQQLAAHILSVSTDEACASDWSQMQPTQ 551
K +L + + N + A I+S+ T E W M+ +
Sbjct: 513 P--KRELDLPGDWE---------------RNNDEHYIAQIVSIHTGEMNEMQWFDMEFPK 555
Query: 552 VEVLILNLHTKQYSFPESIKKMSKLKVLIITNYSFHPCELNNFELLGSLQNLEKIRLERI 611
E+LILN + +Y P I KMS+LKVL+I N P L++F + L L + LER+
Sbjct: 556 AEILILNFSSDKYVLPPFISKMSRLKVLVIINNGMSPAVLHDFSIFAHLSKLRSLWLERV 615
Query: 612 LVPSFGT----LKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQTG 667
VP LKNL K+SL +C +F++ + ++D FP L +L ID+C DLV L +
Sbjct: 616 HVPQLSNSTTPLKNLHKMSLILCKINKSFDQTGLDVADIFPKLGDLTIDHCDDLVALPSS 675
Query: 668 ICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGKLLNLKHLDI 727
IC + SL L++TNC +L LP+++ KL+ LE+L +C +L+ +P I +L LK+LDI
Sbjct: 676 ICGLTSLSCLSITNCPRLGELPKNLSKLQALEILRLYACPELKTLPGEICELPGLKYLDI 735
Query: 728 SNCISLSSLPEEFGNLCNLKNLDMASCASIELPFSVVNLQNLKTITCDEETAATWEDFQH 787
S C+SLS LPEE G L L+ +DM C + P S V+L++L+ + CD + A WE+ +
Sbjct: 736 SQCVSLSCLPEEIGKLKKLEKIDMRECCFSDRPSSAVSLKSLRHVICDTDVAFMWEEVEK 795
Query: 788 MLPNMKIEVLHVDVNLNWL 806
+P +KIE +L+WL
Sbjct: 796 AVPGLKIEAAEKCFSLDWL 814
>emb|CAB80047.1| putative protein [Arabidopsis thaliana] gi|4490297|emb|CAB38788.1|
putative protein [Arabidopsis thaliana]
gi|46396028|sp|Q9SZA7|DRL29_ARATH Probable disease
resistance protein At4g33300
Length = 855
Score = 359 bits (921), Expect = 2e-97
Identities = 228/705 (32%), Positives = 371/705 (52%), Gaps = 66/705 (9%)
Query: 141 ENPKFTVGLDIPFSKLKMELIRDGSSTLVLTGLGGLGKTTLATKLCWDQEVNGKFKENII 200
++ KF VGL++ K+K + ++G+GG+GKTTLA +L D EV F+ I+
Sbjct: 176 DSEKFGVGLELGKVKVKKMMFESQGGVFGISGMGGVGKTTLAKELQRDHEVQCHFENRIL 235
Query: 201 FVTFSKTPMLKTIVERI---IEHC--GYPVPEFQSDEDAVNKLELLLKKVEGSPLLLVLD 255
F+T S++P+L+ + E I + C G PVP+ D KL ++LD
Sbjct: 236 FLTVSQSPLLEELRELIWGFLSGCEAGNPVPDCNFPFDGARKL-------------VILD 282
Query: 256 DVWPTSESLVKKLQFEISDFKILVTSRVSFPRFRTTCILKPLAHEDAVTLFHHYAQMEKN 315
DVW T+++L + F+ LV SR + T ++ L+ ++A++LF A +K+
Sbjct: 283 DVW-TTQALDRLTSFKFPGCTTLVVSRSKLTEPKFTYDVEVLSEDEAISLFCLCAFGQKS 341
Query: 316 -----SSDIIDKNLVE-----------KVVRSCKGLPLTIKVIATSLRNRPDDLWRKIVM 359
D++ ++ ++ +V CKGLPL +KV SL +P+ W+ ++
Sbjct: 342 IPLGFCKDLVKQHNIQSFSILRVLCLAQVANECKGLPLALKVTGASLNGKPEMYWKGVLQ 401
Query: 360 ELSQGHSILDSN-TELLTRLQKIFDVLEDNPTIMECFMDIALFPEDHRIPVAALVDMWAK 418
LS+G DS+ + LL +++ D L+ T +CF+D+ FPED +IP+ L+++W +
Sbjct: 402 RLSKGEPADDSHESRLLRQMEASLDNLDQ--TTKDCFLDLGAFPEDRKIPLDVLINIWIE 459
Query: 419 LYKLDDNGIQAMEIINKLGIMNLANVIIPRKNASDTDNNNYNNH---FIILHDILRDLGI 475
L+ +D+ A I+ L NL + + Y +H F+ HD+LRDL +
Sbjct: 460 LHDIDEGN--AFAILVDLSHKNLLTL-----GKDPRLGSLYASHYDIFVTQHDVLRDLAL 512
Query: 476 YQSTKQPFEQRKRLII---------DINKNRSELAEKQ-QSLLTRILSKVMRLCIKQNPQ 525
+ S +RKRL++ D +N E Q S+ T +++ + +
Sbjct: 513 HLSNAGKVNRRKRLLMPKRELDLPGDWERNNDEHYIAQIVSIHTGKCFSTLQIFLVLSCN 572
Query: 526 QLAAHILSVSTDEACASDWSQMQPTQVEVLILNLHTKQYSFPESIKKMSKLKVLIITNYS 585
H + E W M+ + E+LILN + +Y P I KMS+LKVL+I N
Sbjct: 573 NHQDH----NVGEMNEMQWFDMEFPKAEILILNFSSDKYVLPPFISKMSRLKVLVIINNG 628
Query: 586 FHPCELNNFELLGSLQNLEKIRLERILVPSFGT----LKNLKKLSLYMCNTILAFEKGSI 641
P L++F + L L + LER+ VP LKNL K+SL +C +F++ +
Sbjct: 629 MSPAVLHDFSIFAHLSKLRSLWLERVHVPQLSNSTTPLKNLHKMSLILCKINKSFDQTGL 688
Query: 642 LISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELL 701
++D FP L +L ID+C DLV L + IC + SL L++TNC +L LP+++ KL+ LE+L
Sbjct: 689 DVADIFPKLGDLTIDHCDDLVALPSSICGLTSLSCLSITNCPRLGELPKNLSKLQALEIL 748
Query: 702 SFSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMASCASIELPF 761
+C +L+ +P I +L LK+LDIS C+SLS LPEE G L L+ +DM C + P
Sbjct: 749 RLYACPELKTLPGEICELPGLKYLDISQCVSLSCLPEEIGKLKKLEKIDMRECCFSDRPS 808
Query: 762 SVVNLQNLKTITCDEETAATWEDFQHMLPNMKIEVLHVDVNLNWL 806
S V+L++L+ + CD + A WE+ + +P +KIE +L+WL
Sbjct: 809 SAVSLKSLRHVICDTDVAFMWEEVEKAVPGLKIEAAEKCFSLDWL 853
>emb|CAE46486.1| CC-NBS-LRR disease resistance protein [Arabidopsis thaliana]
gi|30692890|ref|NP_174620.2| disease resistance protein
(CC-NBS-LRR class), putative [Arabidopsis thaliana]
gi|46395988|sp|Q9FW44|ADR1_ARATH Disease resistance
protein ADR1 (Activated disease resistance protein 1)
Length = 787
Score = 343 bits (879), Expect = 2e-92
Identities = 242/821 (29%), Positives = 416/821 (50%), Gaps = 83/821 (10%)
Query: 18 VLNKISQIVETARKHQSARQLLRSILKDLTLVVQDIKQYNEHL-DHPREEIKTLLEENDA 76
+L ++ + T + + L ++++D+ +++I+ L +H + ++ E +
Sbjct: 16 LLKLLALVANTVYSCKGIAERLITMIRDVQPTIREIQYSGAELSNHHQTQLGVFYEILEK 75
Query: 77 EESAC----KCSSENDSYVVDENQSLIVNDVEET----------LYKAREILEL-LNYET 121
C +C+ N +V N+ + D+E+ L+ E+ L +N +
Sbjct: 76 ARKLCEKVLRCNRWNLKHVYHANK---MKDLEKQISRFLNSQILLFVLAEVCHLRVNGDR 132
Query: 122 FEHKFNEAKPPFKRPFDVPE---------NPKFTVGLDIPFSKLKMELIRDGSSTLV-LT 171
E + PE +P+ L++ K+K + + + L ++
Sbjct: 133 IERNMDRLLTERNDSLSFPETMMEIETVSDPEIQTVLELGKKKVKEMMFKFTDTHLFGIS 192
Query: 172 GLGGLGKTTLATKLCWDQEVNGKFKENIIFVTFSKTPMLKTIVERIIEHCGYPVPEFQSD 231
G+ G GKTTLA +L D +V G FK ++F+T S++P + + E C + EF D
Sbjct: 193 GMSGSGKTTLAIELSKDDDVRGLFKNKVLFLTVSRSPNFENL-----ESC---IREFLYD 244
Query: 232 EDAVNKLELLLKKVEGSPLLLVLDDVWPTSESLVKKLQFEISDFKILVTSRVSFPRFRTT 291
KL ++LDDVW T ESL +L +I LV SR RTT
Sbjct: 245 GVHQRKL-------------VILDDVW-TRESL-DRLMSKIRGSTTLVVSRSKLADPRTT 289
Query: 292 CILKPLAHEDAVTLFHHYAQMEKNSSDIIDKNLVEKVVRSCKGLPLTIKVIATSLRNRPD 351
++ L ++A++L A +K+ +K LV++VV CKGLPL++KV+ SL+N+P+
Sbjct: 290 YNVELLKKDEAMSLLCLCAFEQKSPPSPFNKYLVKQVVDECKGLPLSLKVLGASLKNKPE 349
Query: 352 DLWRKIVMELSQGHSILDSN-TELLTRLQKIFDVLEDNPTIMECFMDIALFPEDHRIPVA 410
W +V L +G + +++ + + +++ + L+ P I +CF+D+ FPED +IP+
Sbjct: 350 RYWEGVVKRLLRGEAADETHESRVFAHMEESLENLD--PKIRDCFLDMGAFPEDKKIPLD 407
Query: 411 ALVDMWAKLYKLDDNGIQAMEIINKLGIMNLANVIIPRKNASDTDNN-NYNNHFIILHDI 469
L +W + + +D+ A + +L NL ++ N D + Y + F+ HD+
Sbjct: 408 LLTSVWVERHDIDEE--TAFSFVLRLADKNLLTIV---NNPRFGDVHIGYYDVFVTQHDV 462
Query: 470 LRDLGIYQSTKQPFEQRKRLIIDINKNRSELAEKQQSLLTRILSKVMRLCIKQNPQQLAA 529
LRDL ++ S + +D+N+ L K + +L R K + A
Sbjct: 463 LRDLALHMSNR----------VDVNRRERLLMPKTEPVLPREWEK-------NKDEPFDA 505
Query: 530 HILSVSTDEACASDWSQMQPTQVEVLILNLHTKQYSFPESIKKMSKLKVLIITNYSFHPC 589
I+S+ T E +W M + EVLILN + Y P I KMS+L+VL+I N P
Sbjct: 506 KIVSLHTGEMDEMNWFDMDLPKAEVLILNFSSDNYVLPPFIGKMSRLRVLVIINNGMSPA 565
Query: 590 ELNNFELLGSLQNLEKIRLERILVPSFGT----LKNLKKLSLYMCNTILAFEKGSILISD 645
L+ F + +L L + L+R+ VP + LKNL K+ L C +F + S IS
Sbjct: 566 RLHGFSIFANLAKLRSLWLKRVHVPELTSCTIPLKNLHKIHLIFCKVKNSFVQTSFDISK 625
Query: 646 AFPNLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSFSS 705
FP+L +L ID+C DL+ L++ I I SL L++TNC ++ LP+++ +++LE L +
Sbjct: 626 IFPSLSDLTIDHCDDLLELKS-IFGITSLNSLSITNCPRILELPKNLSNVQSLERLRLYA 684
Query: 706 CTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMASCASIELPFSVVN 765
C +L ++P + +L LK++DIS C+SL SLPE+FG L +L+ +DM C+ + LP SV
Sbjct: 685 CPELISLPVEVCELPCLKYVDISQCVSLVSLPEKFGKLGSLEKIDMRECSLLGLPSSVAA 744
Query: 766 LQNLKTITCDEETAATWEDFQHMLPNMKIEVLHVDVNLNWL 806
L +L+ + CDEET++ WE + ++P + IEV ++WL
Sbjct: 745 LVSLRHVICDEETSSMWEMVKKVVPELCIEVAKKCFTVDWL 785
>emb|CAB86014.1| disease resistance-like protein [Arabidopsis thaliana]
gi|21281177|gb|AAM45000.1| putative disease resistance
[Arabidopsis thaliana] gi|15292721|gb|AAK92729.1|
putative disease resistance [Arabidopsis thaliana]
gi|9758447|dbj|BAB08976.1| unnamed protein product
[Arabidopsis thaliana] gi|15238281|ref|NP_196092.1|
disease resistance protein (CC-NBS-LRR class), putative
[Arabidopsis thaliana] gi|46396009|sp|Q9LZ25|DRL30_ARATH
Probable disease resistance protein At5g04720
Length = 811
Score = 337 bits (865), Expect = 7e-91
Identities = 216/668 (32%), Positives = 352/668 (52%), Gaps = 46/668 (6%)
Query: 147 VGLDIPFSKLKMELIR--DGSSTLVLTGLGGLGKTTLATKLCWDQEVNGKFKENIIFVTF 204
VGLD+ K+K L + DG + ++G+ G GKTTLA +L D+EV G F ++F+T
Sbjct: 180 VGLDLGKRKVKEMLFKSIDGERLIGISGMSGSGKTTLAKELARDEEVRGHFGNKVLFLTV 239
Query: 205 SKTPMLKTIVERIIEHCGYPVPEFQSDEDAVNKLELLLKKVEGSPLLLVLDDVWPTSESL 264
S++P L E + H + +++ A + S L++LDDVW T ESL
Sbjct: 240 SQSPNL----EELRAHIWGFLTSYEAGVGAT---------LPESRKLVILDDVW-TRESL 285
Query: 265 VKKLQFEISDFKILVTSRVSFPRFRTTCILKPLAHEDAVTLFHHYAQMEKNSSDIIDKNL 324
+ + I LV SR R T ++ L +A LF +K ++L
Sbjct: 286 DQLMFENIPGTTTLVVSRSKLADSRVTYDVELLNEHEATALFCLSVFNQKLVPSGFSQSL 345
Query: 325 VEKVVRSCKGLPLTIKVIATSLRNRPDDLWRKIVMELSQGHSILDSN-TELLTRLQKIFD 383
V++VV CKGLPL++KVI SL+ RP+ W V LS+G +++ + + +++ +
Sbjct: 346 VKQVVGECKGLPLSLKVIGASLKERPEKYWEGAVERLSRGEPADETHESRVFAQIEATLE 405
Query: 384 VLEDNPTIMECFMDIALFPEDHRIPVAALVDMWAKLYKLDDNGIQAMEIINKLGIMNLAN 443
L+ P +CF+ + FPED +IP+ L+++ +L+ L+D A +I L NL
Sbjct: 406 NLD--PKTRDCFLVLGAFPEDKKIPLDVLINVLVELHDLED--ATAFAVIVDLANRNLLT 461
Query: 444 VII-PRKNASDTDNNNYNNHFIILHDILRDLGIYQSTKQPFEQRKRLIIDINKNRSELAE 502
++ PR T +Y + F+ HD+LRD+ + S R+RL++
Sbjct: 462 LVKDPRFGHMYT---SYYDIFVTQHDVLRDVALRLSNHGKVNNRERLLMP---------- 508
Query: 503 KQQSLLTRILSKVMRLCIKQNPQQLAAHILSVSTDEACASDWSQMQPTQVEVLILNLHTK 562
K++S+L R + N + A ++S+ T E DW M+ + EVLIL+ +
Sbjct: 509 KRESMLPREWER-------NNDEPYKARVVSIHTGEMTQMDWFDMELPKAEVLILHFSSD 561
Query: 563 QYSFPESIKKMSKLKVLIITNYSFHPCELNNFELLGSLQNLEKIRLERILVPSFGT---- 618
+Y P I KM KL L+I N P L++F + +L L+ + L+R+ VP +
Sbjct: 562 KYVLPPFIAKMGKLTALVIINNGMSPARLHDFSIFTNLAKLKSLWLQRVHVPELSSSTVP 621
Query: 619 LKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLN 678
L+NL KLSL C + ++ + I+ FP L +L ID+C DL+ L + IC I SL ++
Sbjct: 622 LQNLHKLSLIFCKINTSLDQTELDIAQIFPKLSDLTIDHCDDLLELPSTICGITSLNSIS 681
Query: 679 VTNCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPE 738
+TNC ++ LP+++ KL+ L+LL +C +L ++P I +L LK++DIS C+SLSSLPE
Sbjct: 682 ITNCPRIKELPKNLSKLKALQLLRLYACHELNSLPVEICELPRLKYVDISQCVSLSSLPE 741
Query: 739 EFGNLCNLKNLDMASCASIELPFSVVNLQNLKTITCDEETAATWEDFQHMLPNMKIEVLH 798
+ G + L+ +D C+ +P SVV L +L+ + CD E WE Q + +++E
Sbjct: 742 KIGKVKTLEKIDTRECSLSSIPNSVVLLTSLRHVICDREALWMWEKVQKAVAGLRVEAAE 801
Query: 799 VDVNLNWL 806
+ +WL
Sbjct: 802 KSFSRDWL 809
>gb|AAG51211.1| disease resistance protein, putative; 92850-95636 [Arabidopsis
thaliana] gi|10998939|gb|AAG26078.1| disease resistance
protein, putative [Arabidopsis thaliana]
Length = 797
Score = 335 bits (859), Expect = 4e-90
Identities = 241/831 (29%), Positives = 417/831 (50%), Gaps = 93/831 (11%)
Query: 18 VLNKISQIVETARKHQSARQLLRSILKDLTLVVQDIKQYNEHL-DHPREEIKTLLEENDA 76
+L ++ + T + + L ++++D+ +++I+ L +H + ++ E +
Sbjct: 16 LLKLLALVANTVYSCKGIAERLITMIRDVQPTIREIQYSGAELSNHHQTQLGVFYEILEK 75
Query: 77 EESAC----KCSSENDSYVVDENQSLIVNDVEET----------LYKAREILEL-LNYET 121
C +C+ N +V N+ + D+E+ L+ E+ L +N +
Sbjct: 76 ARKLCEKVLRCNRWNLKHVYHANK---MKDLEKQISRFLNSQILLFVLAEVCHLRVNGDR 132
Query: 122 FEHKFNEAKPPFKRPFDVPE---------NPKFTVGLDIPFSKLKMELIRDGSSTLV-LT 171
E + PE +P+ L++ K+K + + + L ++
Sbjct: 133 IERNMDRLLTERNDSLSFPETMMEIETVSDPEIQTVLELGKKKVKEMMFKFTDTHLFGIS 192
Query: 172 GLGGLGKTTLATKLCWDQEVNGKFKENIIFVTFSKTPMLKTIVERIIEHCGYPVPEFQSD 231
G+ G GKTTLA +L D +V G FK ++F+T S++P + + E C + EF D
Sbjct: 193 GMSGSGKTTLAIELSKDDDVRGLFKNKVLFLTVSRSPNFENL-----ESC---IREFLYD 244
Query: 232 EDAVNKLELLLKKVEGSPLLLVLDDVWPTSESLVKKLQFEISDFKILVTSRVSFPRFRTT 291
KL ++LDDVW T ESL +L +I LV SR RTT
Sbjct: 245 GVHQRKL-------------VILDDVW-TRESL-DRLMSKIRGSTTLVVSRSKLADPRTT 289
Query: 292 CILKPLAHEDAVTLFHHYAQMEKNSSDIIDKNLVEKVVRSCKGLPLTIKVIATSLRNRPD 351
++ L ++A++L A +K+ +K LV++VV CKGLPL++KV+ SL+N+P+
Sbjct: 290 YNVELLKKDEAMSLLCLCAFEQKSPPSPFNKYLVKQVVDECKGLPLSLKVLGASLKNKPE 349
Query: 352 DLWRKIVMELSQGHSILDSN-TELLTRLQKIFDVLEDNPTIMECFMDIALFPEDHRIPVA 410
W +V L +G + +++ + + +++ + L+ P I +CF+D+ FPED +IP+
Sbjct: 350 RYWEGVVKRLLRGEAADETHESRVFAHMEESLENLD--PKIRDCFLDMGAFPEDKKIPLD 407
Query: 411 ALVDMWAKLYKLDDNGIQAMEIINKLGIMNLANVIIPRKNASDTDNN-NYNNHFIILHDI 469
L +W + + +D+ A + +L NL ++ N D + Y + F+ HD+
Sbjct: 408 LLTSVWVERHDIDEE--TAFSFVLRLADKNLLTIV---NNPRFGDVHIGYYDVFVTQHDV 462
Query: 470 LRDLGIYQSTKQPFEQRKRLIIDINKNRSELAEKQQSLLTRILSKVMRLCIKQNPQQLAA 529
LRDL ++ S + +D+N+ L K + +L R K + A
Sbjct: 463 LRDLALHMSNR----------VDVNRRERLLMPKTEPVLPREWEK-------NKDEPFDA 505
Query: 530 HILSVSTDEACAS----------DWSQMQPTQVEVLILNLHTKQYSFPESIKKMSKLKVL 579
I+S+ T + + +W M + EVLILN + Y P I KMS+L+VL
Sbjct: 506 KIVSLHTGKTSLTLNEFGEMDEMNWFDMDLPKAEVLILNFSSDNYVLPPFIGKMSRLRVL 565
Query: 580 IITNYSFHPCELNNFELLGSLQNLEKIRLERILVPSFGT----LKNLKKLSLYMCNTILA 635
+I N P L+ F + +L L + L+R+ VP + LKNL K+ L C +
Sbjct: 566 VIINNGMSPARLHGFSIFANLAKLRSLWLKRVHVPELTSCTIPLKNLHKIHLIFCKVKNS 625
Query: 636 FEKGSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKL 695
F + S IS FP+L +L ID+C DL+ L++ I I SL L++TNC ++ LP+++ +
Sbjct: 626 FVQTSFDISKIFPSLSDLTIDHCDDLLELKS-IFGITSLNSLSITNCPRILELPKNLSNV 684
Query: 696 ENLELLSFSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMASCA 755
++LE L +C +L ++P + +L LK++DIS C+SL SLPE+FG L +L+ +DM C+
Sbjct: 685 QSLERLRLYACPELISLPVEVCELPCLKYVDISQCVSLVSLPEKFGKLGSLEKIDMRECS 744
Query: 756 SIELPFSVVNLQNLKTITCDEETAATWEDFQHMLPNMKIEVLHVDVNLNWL 806
+ LP SV L +L+ + CDEET++ WE + ++P + IEV ++WL
Sbjct: 745 LLGLPSSVAALVSLRHVICDEETSSMWEMVKKVVPELCIEVAKKCFTVDWL 795
>dbj|BAA97162.1| disease resistance protein-like [Arabidopsis thaliana]
gi|15238054|ref|NP_199539.1| disease resistance protein
(NBS-LRR class), putative [Arabidopsis thaliana]
gi|46396005|sp|Q9LVT1|DRL39_ARATH Putative disease
resistance protein At5g47280
Length = 623
Score = 332 bits (852), Expect = 2e-89
Identities = 214/651 (32%), Positives = 352/651 (53%), Gaps = 41/651 (6%)
Query: 161 IRDGSSTLVLTGLGGLGKTTLATKLCWDQEVNGKFKENIIFVTFSKTPMLKTIVERIIEH 220
+ D + + ++G+ G GKT LA +L D+EV G F ++F+T S++P L+ + I
Sbjct: 5 LNDEARIIGISGMIGSGKTILAKELARDEEVRGHFANRVLFLTVSQSPNLEELRSLI--- 61
Query: 221 CGYPVPEFQSDEDAVNKLELLLKKVEGSPLLLVLDDVWPTSESLVKKLQFEISDFKILVT 280
+F + +A L + V + L++LDDV T ESL +L F I LV
Sbjct: 62 -----RDFLTGHEA-GFGTALPESVGHTRKLVILDDV-RTRESL-DQLMFNIPGTTTLVV 113
Query: 281 SRVSFPRFRTTCILKPLAHEDAVTLFHHYAQMEKNSSDIIDKNLVEKVVRSCKGLPLTIK 340
S+ RTT ++ L DA +LF A +K+ K+LV++VV KGLPL++K
Sbjct: 114 SQSKLVDPRTTYDVELLNEHDATSLFCLSAFNQKSVPSGFSKSLVKQVVGESKGLPLSLK 173
Query: 341 VIATSLRNRPDDLWRKIVMELSQGHSILDSN-TELLTRLQKIFDVLEDNPTIMECFMDIA 399
V+ SL +RP+ W V LS+G + +++ +++ +++ + L+ P ECF+D+
Sbjct: 174 VLGASLNDRPETYWAIAVERLSRGEPVDETHESKVFAQIEATLENLD--PKTKECFLDMG 231
Query: 400 LFPEDHRIPVAALVDMWAKLYKLDDNGIQAMEIINKLGIMNLANVII-PRKNASDTDNNN 458
FPE +IPV L++M K++ L+D A +++ L NL ++ P A T +
Sbjct: 232 AFPEGKKIPVDVLINMLVKIHDLEDAA--AFDVLVDLANRNLLTLVKDPTFVAMGT---S 286
Query: 459 YNNHFIILHDILRDLGIYQSTKQPFEQRKRLIIDINKNRSELAEKQQSLLTRILSKVMRL 518
Y + F+ HD+LRD+ ++ + + +R RL++ K++++L +
Sbjct: 287 YYDIFVTQHDVLRDVALHLTNRGKVSRRDRLLMP----------KRETMLPSEWER---- 332
Query: 519 CIKQNPQQLAAHILSVSTDEACASDWSQMQPTQVEVLILNLHTKQYSFPESIKKMSKLKV 578
N + A ++S+ T E DW M + EVLI+N + Y P I KM L+V
Sbjct: 333 ---SNDEPYNARVVSIHTGEMTEMDWFDMDFPKAEVLIVNFSSDNYVLPPFIAKMGMLRV 389
Query: 579 LIITNYSFHPCELNNFELLGSLQNLEKIRLERILVPSFGT----LKNLKKLSLYMCNTIL 634
+I N P L++F + SL NL + LER+ VP + LKNL KL L +C
Sbjct: 390 FVIINNGTSPAHLHDFPIPTSLTNLRSLWLERVHVPELSSSMIPLKNLHKLYLIICKINN 449
Query: 635 AFEKGSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGK 694
+F++ +I I+ FP L ++ IDYC DL L + IC I SL +++TNC + LP++I K
Sbjct: 450 SFDQTAIDIAQIFPKLTDITIDYCDDLAELPSTICGITSLNSISITNCPNIKELPKNISK 509
Query: 695 LENLELLSFSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMASC 754
L+ L+LL +C +L+++P I +L L ++DIS+C+SLSSLPE+ GN+ L+ +DM C
Sbjct: 510 LQALQLLRLYACPELKSLPVEICELPRLVYVDISHCLSLSSLPEKIGNVRTLEKIDMREC 569
Query: 755 ASIELPFSVVNLQNLKTITCDEETAATWEDFQHMLPNMKIEVLHVDVNLNW 805
+ +P S V+L +L +TC E W++ + +P ++IE N+ W
Sbjct: 570 SLSSIPSSAVSLTSLCYVTCYREALWMWKEVEKAVPGLRIEATEKWFNMTW 620
>dbj|BAB08631.1| unnamed protein product [Arabidopsis thaliana]
gi|15240125|ref|NP_201490.1| disease resistance protein
(CC-NBS-LRR class), putative [Arabidopsis thaliana]
gi|46395985|sp|Q9FKZ2|DRL41_ARATH Probable disease
resistance protein At5g66890
Length = 415
Score = 244 bits (623), Expect = 8e-63
Identities = 156/435 (35%), Positives = 237/435 (53%), Gaps = 41/435 (9%)
Query: 382 FDVLEDNPTIMECFMDIALFPEDHRIPVAALVDMWAKLYKLDDNGIQAMEIINKLGIMNL 441
FD L N + ECF+D+A F ED RI + ++D+W+ Y G + M + L NL
Sbjct: 9 FDALPHN--LRECFLDMASFLEDQRIIASTIIDLWSASY-----GKEGMNNLQDLASRNL 61
Query: 442 ANVIIPRKNASDTDNNNYNNHFIILHDILRDLGIYQSTKQPFE--QRKRLIIDINKNRSE 499
++ +N + ++ YN + ++LR+ I Q K+ +RKRL ++I N
Sbjct: 62 LKLLPIGRN--EYEDGFYNELLVKQDNVLREFAINQCLKESSSIFERKRLNLEIQDN--- 116
Query: 500 LAEKQQSLLTRILSKVMRLCIKQNPQQ---LAAHILSVSTDEACASDWSQMQPTQVEVLI 556
K C+ NP+Q + A + S+STD++ AS W +M VE L+
Sbjct: 117 --------------KFPNWCL--NPKQPIVINASLFSISTDDSFASSWFEMDCPNVEALV 160
Query: 557 LNLHTKQYSFPESIKKMSKLKVLIITNYSFHPCELNNFELLGSLQNLEKIRLERILV--- 613
LN+ + Y+ P I M +LKV+II N+ P +L N L SL NL++IR E++ +
Sbjct: 161 LNISSSNYALPNFIATMKELKVVIIINHGLEPAKLTNLSCLSSLPNLKRIRFEKVSISLL 220
Query: 614 --PSFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQTGICDI 671
P G LK+L+KLSL+ C+ + A + +S+ +L+E+ IDYC +L L I +
Sbjct: 221 DIPKLG-LKSLEKLSLWFCHVVDALNELED-VSETLQSLQEIEIDYCYNLDELPYWISQV 278
Query: 672 ISLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGKLLNLKHLDISNCI 731
+SLKKL+VTNC+KL + + IG L +LE L SSC L +P +I +L NL+ LD+S
Sbjct: 279 VSLKKLSVTNCNKLCRVIEAIGDLRDLETLRLSSCASLLELPETIDRLDNLRFLDVSGGF 338
Query: 732 SLSSLPEEFGNLCNLKNLDMASCASIELPFSVVNLQNLKTITCDEETAATWEDFQHMLPN 791
L +LP E G L L+ + M C ELP SV NL+NL+ + CDE+TA W+ + + N
Sbjct: 339 QLKNLPLEIGKLKKLEKISMKDCYRCELPDSVKNLENLE-VKCDEDTAFLWKILKPEMKN 397
Query: 792 MKIEVLHVDVNLNWL 806
+ I + NLN L
Sbjct: 398 LTITEEKTEHNLNLL 412
>dbj|BAD94804.1| putative protein [Arabidopsis thaliana]
Length = 366
Score = 238 bits (607), Expect = 6e-61
Identities = 128/348 (36%), Positives = 195/348 (55%), Gaps = 21/348 (6%)
Query: 463 FIILHDILRDLGIYQSTKQPFEQRKRLIIDINKNRSELAEKQQSLLTRILSKVMRLCIKQ 522
F+ HD+LRDL ++ S +RKRL++ K +L + +
Sbjct: 34 FVTQHDVLRDLALHLSNAGKVNRRKRLLMP--KRELDLPGDWE---------------RN 76
Query: 523 NPQQLAAHILSVSTDEACASDWSQMQPTQVEVLILNLHTKQYSFPESIKKMSKLKVLIIT 582
N + A I+S+ T E W M+ + E+LILN + +Y P I KMS+LKVL+I
Sbjct: 77 NDEHYIAQIVSIHTGEMNEMQWFDMEFPKAEILILNFSSDKYVLPPFISKMSRLKVLVII 136
Query: 583 NYSFHPCELNNFELLGSLQNLEKIRLERILVPSFGT----LKNLKKLSLYMCNTILAFEK 638
N P L++F + L L + LER+ VP LKNL K+SL +C +F++
Sbjct: 137 NNGMSPAVLHDFSIFAHLSKLRSLWLERVHVPQLSNSTTPLKNLHKMSLILCKINKSFDQ 196
Query: 639 GSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENL 698
+ ++D FP L +L ID+C DLV L + IC + SL L++TNC +L LP+++ KL+ L
Sbjct: 197 TGLDVADIFPKLGDLTIDHCDDLVALPSSICGLTSLSCLSITNCPRLGELPKNLSKLQAL 256
Query: 699 ELLSFSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMASCASIE 758
E+L +C +L+ +P I +L LK+LDIS C+SLS LPEE G L L+ +DM C +
Sbjct: 257 EILRLYACPELKTLPGEICELPGLKYLDISQCVSLSCLPEEIGKLKKLEKIDMRECCFSD 316
Query: 759 LPFSVVNLQNLKTITCDEETAATWEDFQHMLPNMKIEVLHVDVNLNWL 806
P S V+L++L+ + CD + A WE+ + +P +KIE +L+WL
Sbjct: 317 RPSSAVSLKSLRHVICDTDVAFMWEEVEKAVPGLKIEAAEKCFSLDWL 364
>gb|AAM28916.1| NBS/LRR [Pinus taeda]
Length = 479
Score = 229 bits (583), Expect = 4e-58
Identities = 158/500 (31%), Positives = 253/500 (50%), Gaps = 42/500 (8%)
Query: 322 KNLVEKVVRSCKGLPLTIKVIATSLRNRPDDLWRKIVMELSQGHSILDSNTE-LLTRLQK 380
++LV++V CKGLPL +KVI +SLR + +W +LS+ SI + E LL RL+
Sbjct: 5 EDLVKQVAAECKGLPLALKVIGSSLRGKRRPIWINAERKLSKSESISEYYKESLLKRLET 64
Query: 381 IFDVLEDNPTIMECFMDIALFPEDHRIPVAALVDMWAKLYKLDDNGIQAMEIINKLGIMN 440
DVL+D +CF+D+ FP+ + V L+D+W + +++ A E++ +L N
Sbjct: 65 SIDVLDDKHK--QCFLDLGAFPKGRKFSVETLLDIWVYVRQME--WTDAFEVLLELASRN 120
Query: 441 LANVI-IPRKNASDTDNNNYNNHFIILHDILRDLGIYQSTKQPFEQRKRLIIDINKNRSE 499
L N+ P A D + + HD++RDL +Y +++ KRL
Sbjct: 121 LLNLTGYPGSGA--IDYSCASELTFSQHDVMRDLALYLASQDNIISPKRLFTP------- 171
Query: 500 LAEKQQSLLTRILSKVMRLCIKQNPQQLAAHILSVSTDEACASDWSQMQPTQVEVLILNL 559
K+ + T LS + Q A +S+ T DW Q+ +VE L L
Sbjct: 172 --SKEDKIPTEWLSTL-------KDQASRAQFVSIYTGAMQEQDWCQIDFPEVEALALFF 222
Query: 560 HTKQYSFPESIKKMSKLKVLIITNYSFHPCELNNFELLGSLQNLEKIRLERILVPSF--- 616
QY P + + KLKVLI+ NYS + S + + L +++VP+
Sbjct: 223 SANQYCLPTFLHRTPKLKVLIVYNYSSMRANIIGLPRFSSPIQIRSVFLHKLIVPASLYE 282
Query: 617 --GTLKNLKKLSLYMCNTILAFEKGSILISDA------FPNLEELNIDYCKDLVVLQTGI 668
+ + L+KLS+ +C + G+ + D FPN+ E+NID+C DL L +
Sbjct: 283 NCRSWERLEKLSVCLCEGL-----GNSSLVDMELEPLNFPNITEINIDHCSDLGELPLKL 337
Query: 669 CDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGKLLNLKHLDIS 728
C++ SL++L+VTNCH + +LP D+G+L++L +L S+C L +P SI KL L++LDIS
Sbjct: 338 CNLTSLQRLSVTNCHLIQNLPDDMGRLKSLRVLRLSACPSLSRLPPSICKLGQLEYLDIS 397
Query: 729 NCISLSSLPEEFGNLCNLKNLDMASCASIELPFSVV--NLQNLKTITCDEETAATWEDFQ 786
C L LP EF L NL+ LDM C+ ++ +V+ +L+ + D+E A
Sbjct: 398 LCRCLQDLPSEFDQLSNLETLDMRECSGLKKVPTVIQSSLKRVVISDSDKEYEAWXSIKA 457
Query: 787 HMLPNMKIEVLHVDVNLNWL 806
L + I+V+ +L WL
Sbjct: 458 STLHTLTIDVVPEIFSLAWL 477
>emb|CAD45029.1| NBS-LRR disease resistance protein homologue [Hordeum vulgare]
Length = 1262
Score = 165 bits (418), Expect = 5e-39
Identities = 203/837 (24%), Positives = 364/837 (43%), Gaps = 106/837 (12%)
Query: 1 MSSATTHIITLTTRFHDVLNKISQIVETARKHQSARQLLRSILKDL---TLVVQDIKQYN 57
+ SA +T+ F D L + +E+ + ++LKD ++ + ++ +
Sbjct: 20 LGSAIGDEVTMLWSFKDDLKDMKDTLES----------MEAVLKDAERRSVKEELVRLWL 69
Query: 58 EHLDHPREEIKTLLEENDA--EESACKCSSENDSYVVDENQSLIVNDVEETLYKAREILE 115
L H +I +L+E A E ++ K + D + + +L +++ + R+I E
Sbjct: 70 NRLKHAAYDISYMLDEFQANSEPASRKMIGKLDCFAIAPKITLAYK-MKKMRGQLRKIKE 128
Query: 116 LLNYETFEHKFNEAKPPFKRPFDVPE----NPKFTVGLDIPFSKLKMEL---------IR 162
++E+F KF A +P+ + L I K +M + I+
Sbjct: 129 --DHESF--KFTHANSSLINVHQLPDPRETSSNVVESLIIGREKDRMNVLSLLSTSNNIK 184
Query: 163 DGSSTLVLTGLGGLGKTTLATKLCWDQEVNGKFKENIIFVTFSKTPMLKTIVERIIEHCG 222
+ + L + GLGG+GKTTLA + D + N + ++V S+ L I II
Sbjct: 185 EDFTVLPICGLGGIGKTTLAQLVFNDAQFNDYHR---VWVYVSQVFDLNKIGNSIISQVS 241
Query: 223 YPVPEFQSDEDAVNKLELLLKKVEGSPLLLVLDDVWPTSESLVKKLQFEIS---DFKILV 279
E ++K L ++ L+VLDD+W T + +L+ ++ K+LV
Sbjct: 242 GKGSEHSHTLQHISKQ--LKDLLQDKKTLIVLDDLWETGYFQLDQLKLMLNVSTKMKVLV 299
Query: 280 TSR-VSFPRFRTTC-----ILKPLAHEDAVTLFHHYAQMEKNSSDIIDKNLVE----KVV 329
T+R + R +L PL ++ + ++ + DK +E K+
Sbjct: 300 TTRSIDIARKMGNVGVEPYMLDPLDNDMCWRIIKQSSRFQSRP----DKEQLEPNGQKIA 355
Query: 330 RSCKGLPLTIKVIATSLRNRPDDLWRKIVMELSQGHSILDSNTELLTRLQKIFDVLEDNP 389
R C GLPL + + L W I + DS +L L+ ++ L P
Sbjct: 356 RKCGGLPLAAQALGFLLSGMDLSEWEAICISDIWDEPFSDST--VLPSLKLSYNTL--TP 411
Query: 390 TIMECFMDIALFPEDHRIPVAALVDMWAKLYKLD-DNGIQAMEIINKLGIMNLANVIIPR 448
+ CF +FP+ H I L+ W L ++ N A+++ K L +
Sbjct: 412 YMRLCFAYCGIFPKGHNISKDYLIHQWIALGFIEPSNKFSAIQLGGKYVRQFLGMSFLHH 471
Query: 449 KNASDTDNNNYNNHFIILHDILRDLGIYQSTKQPFEQRKRLIIDINKNRSELAEKQQSLL 508
+T + N +HD++ DL T++ ++ D NR + SL
Sbjct: 472 SKLPET----FGNAMFTMHDLVHDLARSVITEELVVFDAEIVSD---NRIKEYCIYASLT 524
Query: 509 TRILS---KVMRLCIKQNPQQLAAHI--------------------LSVSTDEACASDWS 545
+S KV ++ P+ H LS + + AS
Sbjct: 525 NCNISDHNKVRKMTTIFPPKLRVMHFSDCKLHGSAFSFQKCLRVLDLSGCSIKDFASALG 584
Query: 546 QMQPTQVEVLILNLHTKQYSFPESIKKMSKLKVLIITN---YSFHPCELNNFELLGSLQN 602
Q++ Q+EVLI + FPESI ++SKL L ++ S P + L L +
Sbjct: 585 QLK--QLEVLIAQ-KLQDRQFPESITRLSKLHYLNLSGSRGISEIPSSVGKLVSLVHL-D 640
Query: 603 LEKIRLERILVPSFGTLKNLKKLSLYMCNTILAFEK--GSILISDAFPNLEELNIDYCKD 660
L +++ + G L+NL+ L L C + + + GS+ NL+ LN+ C +
Sbjct: 641 LSYCTNVKVIPKALGILRNLQTLDLSWCEKLESLPESLGSV------QNLQRLNLSNCFE 694
Query: 661 LVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGKLL 720
L L + + ++ L++++C+KL SLP+ +G L+N++ L S C L ++P ++G+L
Sbjct: 695 LEALPESLGSLKDVQTLDLSSCYKLESLPESLGSLKNVQTLDLSRCYKLVSLPKNLGRLK 754
Query: 721 NLKHLDISNCISLSSLPEEFGNLCNLKNLDMASCASIE-LPFSVVNLQNLKTITCDE 776
NL+ +D+S C L + PE FG+L NL+ L++++C +E LP S +L+NL+T+ E
Sbjct: 755 NLRTIDLSGCKKLETFPESFGSLENLQILNLSNCFELESLPESFGSLKNLQTLNLVE 811
Score = 127 bits (318), Expect = 2e-27
Identities = 84/227 (37%), Positives = 129/227 (56%), Gaps = 13/227 (5%)
Query: 552 VEVLILNLHTKQYSFPESIKKMSKLKVLIITNYSFHPCELNNFELLGSLQNLEKIRLE-- 609
+E L L+ K S PES+ + L+ L + H E + E LG L+NL+ ++L
Sbjct: 972 LETLNLSKCFKLESLPESLGGLQNLQTLDLL--VCHKLE-SLPESLGGLKNLQTLQLSFC 1028
Query: 610 ---RILVPSFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQT 666
L S G LKNL+ L+L +C+ + + + + NL L + C L L
Sbjct: 1029 HKLESLPESLGGLKNLQTLTLSVCDKLESLPESL----GSLKNLHTLKLQVCYKLKSLPE 1084
Query: 667 GICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGKLLNLKHLD 726
+ I +L LN++ CH L S+P+ +G LENL++L+ S+C LE+IP S+G L NL+ L
Sbjct: 1085 SLGSIKNLHTLNLSVCHNLESIPESVGSLENLQILNLSNCFKLESIPKSLGSLKNLQTLI 1144
Query: 727 ISNCISLSSLPEEFGNLCNLKNLDMASCASIE-LPFSVVNLQNLKTI 772
+S C L SLP+ GNL NL+ LD++ C +E LP S+ +L+NL+T+
Sbjct: 1145 LSWCTRLVSLPKNLGNLKNLQTLDLSGCKKLESLPDSLGSLENLQTL 1191
Score = 126 bits (316), Expect = 3e-27
Identities = 84/236 (35%), Positives = 135/236 (56%), Gaps = 20/236 (8%)
Query: 552 VEVLILNLHTKQYSFPESIKKMSKLKVLIITNYSFHPCE--LNNFELLGSLQNLEKIRLE 609
++ L ++ K S PES+ ++ L+ L ++ C+ ++ + LGSL+NL+ + L
Sbjct: 828 LQTLDFSVCHKLESVPESLGGLNNLQTLKLS-----VCDNLVSLLKSLGSLKNLQTLDLS 882
Query: 610 -----RILVPSFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVL 664
L S G+L+NL+ L+L C + + + NL+ LNI +C +LV L
Sbjct: 883 GCKKLESLPESLGSLENLQILNLSNCFKLESLPESL----GRLKNLQTLNISWCTELVFL 938
Query: 665 QTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGKLLNLKH 724
+ ++ +L +L+++ C KL SLP +G LENLE L+ S C LE++P S+G L NL+
Sbjct: 939 PKNLGNLKNLPRLDLSGCMKLESLPDSLGSLENLETLNLSKCFKLESLPESLGGLQNLQT 998
Query: 725 LDISNCISLSSLPEEFGNLCNLKNLDMASCASIE-LPFSVVNLQNLKTIT---CDE 776
LD+ C L SLPE G L NL+ L ++ C +E LP S+ L+NL+T+T CD+
Sbjct: 999 LDLLVCHKLESLPESLGGLKNLQTLQLSFCHKLESLPESLGGLKNLQTLTLSVCDK 1054
Score = 117 bits (294), Expect = 1e-24
Identities = 80/229 (34%), Positives = 130/229 (55%), Gaps = 17/229 (7%)
Query: 552 VEVLILNLHTKQYSFPESIKKMSKLKVLIITNYSFHPCELNNFELLGSLQNLEKIRLE-- 609
++ L L + K S PES+ + L+ L ++ H E + E LG L+NL+ + L
Sbjct: 996 LQTLDLLVCHKLESLPESLGGLKNLQTLQLS--FCHKLE-SLPESLGGLKNLQTLTLSVC 1052
Query: 610 ---RILVPSFGTLKNLKKLSLYMCNTILAFEK--GSILISDAFPNLEELNIDYCKDLVVL 664
L S G+LKNL L L +C + + + GSI NL LN+ C +L +
Sbjct: 1053 DKLESLPESLGSLKNLHTLKLQVCYKLKSLPESLGSI------KNLHTLNLSVCHNLESI 1106
Query: 665 QTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGKLLNLKH 724
+ + +L+ LN++NC KL S+P+ +G L+NL+ L S CT L ++P ++G L NL+
Sbjct: 1107 PESVGSLENLQILNLSNCFKLESIPKSLGSLKNLQTLILSWCTRLVSLPKNLGNLKNLQT 1166
Query: 725 LDISNCISLSSLPEEFGNLCNLKNLDMASCASIE-LPFSVVNLQNLKTI 772
LD+S C L SLP+ G+L NL+ L++++C +E LP + +L+ L+T+
Sbjct: 1167 LDLSGCKKLESLPDSLGSLENLQTLNLSNCFKLESLPEILGSLKKLQTL 1215
Score = 117 bits (292), Expect = 2e-24
Identities = 78/229 (34%), Positives = 131/229 (57%), Gaps = 22/229 (9%)
Query: 558 NLHTKQYSF-------PESIKKMSKLKVLIITNYSFHPCELNNF-ELLGSLQNLEKIRLE 609
NL T Q SF PES+ + L+ L ++ +L + E LGSL+NL ++L+
Sbjct: 1019 NLQTLQLSFCHKLESLPESLGGLKNLQTLTLSVCD----KLESLPESLGSLKNLHTLKLQ 1074
Query: 610 -----RILVPSFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVL 664
+ L S G++KNL L+L +C+ + + + + NL+ LN+ C L +
Sbjct: 1075 VCYKLKSLPESLGSIKNLHTLNLSVCHNLESIPESV----GSLENLQILNLSNCFKLESI 1130
Query: 665 QTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGKLLNLKH 724
+ + +L+ L ++ C +L SLP+++G L+NL+ L S C LE++P S+G L NL+
Sbjct: 1131 PKSLGSLKNLQTLILSWCTRLVSLPKNLGNLKNLQTLDLSGCKKLESLPDSLGSLENLQT 1190
Query: 725 LDISNCISLSSLPEEFGNLCNLKNLDMASCASIE-LPFSVVNLQNLKTI 772
L++SNC L SLPE G+L L+ L++ C +E LP S+ +L++L+T+
Sbjct: 1191 LNLSNCFKLESLPEILGSLKKLQTLNLFRCGKLESLPESLGSLKHLQTL 1239
Score = 115 bits (289), Expect = 4e-24
Identities = 83/229 (36%), Positives = 127/229 (55%), Gaps = 17/229 (7%)
Query: 552 VEVLILNLHTKQYSFPESIKKMSKLKVLIITN-YSFHPCELNNFELLGSLQNLEKIRLE- 609
V+ L L+ K S PES+ + ++ L ++ Y N LG L+NL I L
Sbjct: 708 VQTLDLSSCYKLESLPESLGSLKNVQTLDLSRCYKLVSLPKN----LGRLKNLRTIDLSG 763
Query: 610 ----RILVPSFGTLKNLKKLSLYMCNTILAFEKGSILIS-DAFPNLEELNIDYCKDLVVL 664
SFG+L+NL+ L+L C FE S+ S + NL+ LN+ CK L L
Sbjct: 764 CKKLETFPESFGSLENLQILNLSNC-----FELESLPESFGSLKNLQTLNLVECKKLESL 818
Query: 665 QTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGKLLNLKH 724
+ + +L+ L+ + CHKL S+P+ +G L NL+ L S C +L ++ S+G L NL+
Sbjct: 819 PESLGGLKNLQTLDFSVCHKLESVPESLGGLNNLQTLKLSVCDNLVSLLKSLGSLKNLQT 878
Query: 725 LDISNCISLSSLPEEFGNLCNLKNLDMASCASIE-LPFSVVNLQNLKTI 772
LD+S C L SLPE G+L NL+ L++++C +E LP S+ L+NL+T+
Sbjct: 879 LDLSGCKKLESLPESLGSLENLQILNLSNCFKLESLPESLGRLKNLQTL 927
Score = 115 bits (287), Expect = 7e-24
Identities = 82/230 (35%), Positives = 128/230 (55%), Gaps = 19/230 (8%)
Query: 552 VEVLILNLHTKQYSFPESIKKMSKLKVLIITNYSFHPCELNNF-ELLGSLQNLEKIRL-- 608
++ L L+ K S PES+ + L++L ++N +L + E LG L+NL+ + +
Sbjct: 876 LQTLDLSGCKKLESLPESLGSLENLQILNLSNCF----KLESLPESLGRLKNLQTLNISW 931
Query: 609 --ERILVP-SFGTLKNLKKLSLYMCNTI--LAFEKGSILISDAFPNLEELNIDYCKDLVV 663
E + +P + G LKNL +L L C + L GS+ NLE LN+ C L
Sbjct: 932 CTELVFLPKNLGNLKNLPRLDLSGCMKLESLPDSLGSL------ENLETLNLSKCFKLES 985
Query: 664 LQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGKLLNLK 723
L + + +L+ L++ CHKL SLP+ +G L+NL+ L S C LE++P S+G L NL+
Sbjct: 986 LPESLGGLQNLQTLDLLVCHKLESLPESLGGLKNLQTLQLSFCHKLESLPESLGGLKNLQ 1045
Query: 724 HLDISNCISLSSLPEEFGNLCNLKNLDMASCASIE-LPFSVVNLQNLKTI 772
L +S C L SLPE G+L NL L + C ++ LP S+ +++NL T+
Sbjct: 1046 TLTLSVCDKLESLPESLGSLKNLHTLKLQVCYKLKSLPESLGSIKNLHTL 1095
Score = 114 bits (286), Expect = 1e-23
Identities = 74/215 (34%), Positives = 120/215 (55%), Gaps = 15/215 (6%)
Query: 565 SFPESIKKMSKLKVLIITN-YSFHPCELNNFELLGSLQNLEKIRLERI-----LVPSFGT 618
+ PES+ + ++ L +++ Y E LGSL+N++ + L R L + G
Sbjct: 697 ALPESLGSLKDVQTLDLSSCYKLESLP----ESLGSLKNVQTLDLSRCYKLVSLPKNLGR 752
Query: 619 LKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLN 678
LKNL+ + L C + F + + NL+ LN+ C +L L + +L+ LN
Sbjct: 753 LKNLRTIDLSGCKKLETFPESF----GSLENLQILNLSNCFELESLPESFGSLKNLQTLN 808
Query: 679 VTNCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPE 738
+ C KL SLP+ +G L+NL+ L FS C LE++P S+G L NL+ L +S C +L SL +
Sbjct: 809 LVECKKLESLPESLGGLKNLQTLDFSVCHKLESVPESLGGLNNLQTLKLSVCDNLVSLLK 868
Query: 739 EFGNLCNLKNLDMASCASIE-LPFSVVNLQNLKTI 772
G+L NL+ LD++ C +E LP S+ +L+NL+ +
Sbjct: 869 SLGSLKNLQTLDLSGCKKLESLPESLGSLENLQIL 903
Score = 113 bits (283), Expect = 2e-23
Identities = 76/227 (33%), Positives = 121/227 (52%), Gaps = 16/227 (7%)
Query: 552 VEVLILNLHTKQYSFPESIKKMSKLKVLIITNYSFHPCELNNF-ELLGSLQNLEKIRLER 610
V+ L L+ K S P+++ ++ L+ + ++ +L F E GSL+NL+ + L
Sbjct: 732 VQTLDLSRCYKLVSLPKNLGRLKNLRTIDLSGCK----KLETFPESFGSLENLQILNLSN 787
Query: 611 I-----LVPSFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVVLQ 665
L SFG+LKNL+ L+L C + + + NL+ L+ C L +
Sbjct: 788 CFELESLPESFGSLKNLQTLNLVECKKLESLPESL----GGLKNLQTLDFSVCHKLESVP 843
Query: 666 TGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGKLLNLKHL 725
+ + +L+ L ++ C L SL + +G L+NL+ L S C LE++P S+G L NL+ L
Sbjct: 844 ESLGGLNNLQTLKLSVCDNLVSLLKSLGSLKNLQTLDLSGCKKLESLPESLGSLENLQIL 903
Query: 726 DISNCISLSSLPEEFGNLCNLKNLDMASCASIELPFSVVNLQNLKTI 772
++SNC L SLPE G L NL+ L+++ C EL F NL NLK +
Sbjct: 904 NLSNCFKLESLPESLGRLKNLQTLNISWCT--ELVFLPKNLGNLKNL 948
>dbj|BAD38047.1| putative NBS-LRR resistance protein RGH2 [Oryza sativa (japonica
cultivar-group)]
Length = 1216
Score = 153 bits (386), Expect = 2e-35
Identities = 174/658 (26%), Positives = 296/658 (44%), Gaps = 78/658 (11%)
Query: 166 STLVLTGLGGLGKTTLATKLCWDQEVNGKFKENIIFVTFSKTPMLKTIVERIIEHCGYPV 225
S + + GLGG+GKTTLA + D+E + + +V S L IV II H
Sbjct: 195 SIIPVVGLGGMGKTTLAKAVYTDKETH--MFDVKAWVHVSMEFQLNKIVSGIISHVEGST 252
Query: 226 PEFQSDEDAVNKLELLLKKVEGSPL-LLVLDDVWPTS----ESLVKKLQFEISDFKILVT 280
P +D + L+ L ++ + L L++LDD+W E L++ LQ KI+VT
Sbjct: 253 PANIAD---LQYLKSQLDRILCNKLYLIILDDLWEEGWSKLEKLMEMLQSGKKGSKIIVT 309
Query: 281 SR----------VSFPRFRTTCILKPLAHE-DAVTLFHHYAQMEK-NSSDIIDKNLVEKV 328
+R + F T +K + D ME SD++D + +++
Sbjct: 310 TRSEKVVNTLSTIRLSYFHTVDPIKLVGMSIDECWFIMKPRNMENCEFSDLVD--IGKEI 367
Query: 329 VRSCKGLPLTIKVIATSL-RNRPDDLWRKIVMELSQGHSILDSNTELLTRLQKIF-DVLE 386
+ C G+PL K + + ++R + W +I + +ILD+ + L+ +
Sbjct: 368 AQRCSGVPLVAKALGYVMQKHRTREEWMEI-----KNSNILDTKDDEEGILKGLLLSYYH 422
Query: 387 DNPTIMECFMDIALFPEDHRIPVAALVDMWAKLYKLDDNGIQ-----AMEIINKL----- 436
P + CFM ++FP H I L+ W L + D Q AME +N+L
Sbjct: 423 MPPQLKLCFMYCSMFPMSHVIDHDCLIQQWIALGFIQDTDGQPLQKVAMEYVNELLGMSF 482
Query: 437 -GIMNLANVIIPRKNASDTDNNNYNNHFIILHDILRDL-GIYQSTKQPFEQRKRLIIDIN 494
I V+ R T + ++ ++H++ R + G S E R ++N
Sbjct: 483 LTIFTSPTVLASRMLFKPTLKLHMHD---MVHELARHVAGNEFSHTNGAENRNTKRDNLN 539
Query: 495 ------KNRSELAEKQQSLLTRILSKVMRLCIKQN-PQQLAAHILSVSTDEACASDWSQM 547
N++E + +SL T++ + R C K + P+Q +H L + + S++
Sbjct: 540 FHYHLLLNQNETSSAYKSLATKVRALHFRGCDKMHLPKQAFSHTLCLRVLDLGGRQVSEL 599
Query: 548 QPTQVEVLILNL----HTKQYSFPESIKKMSKLKVLIITNYSFHPCELNNFELLGSLQNL 603
+ ++ +L + SF +S + L+ LI++N N +G LQ L
Sbjct: 600 PSSVYKLKLLRYLDASSLRISSFSKSFNHLLNLQALILSNTYLKTLPTN----IGCLQKL 655
Query: 604 EKIRLERI-----LVPSFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEEL---NI 655
+ L L SFG L +L L+L C+ + A +F NL L ++
Sbjct: 656 QYFDLSGCANLNELPTSFGDLSSLLFLNLASCHELEALPM-------SFGNLNRLQFLSL 708
Query: 656 DYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTS 715
C L L C + L L++++C+ L LP I +L LE L+ +SC+ ++A+P S
Sbjct: 709 SDCYKLNSLPESCCQLHDLAHLDLSDCYNLGKLPDCIDQLSKLEYLNMTSCSKVQALPES 768
Query: 716 IGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDM-ASCASIELPFSVVNLQNLKTI 772
+ KL L+HL++S C+ L +LP G+L L++LD+ S +LP S+ N+ LKT+
Sbjct: 769 LCKLTMLRHLNLSYCLRLENLPSCIGDL-QLQSLDIQGSFLLRDLPNSIFNMSTLKTV 825
Score = 47.0 bits (110), Expect = 0.003
Identities = 28/83 (33%), Positives = 42/83 (49%), Gaps = 1/83 (1%)
Query: 673 SLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGKLLN-LKHLDISNCI 731
S++ L + + L +LP+ I +L LS C +LE +P +G L+ + I C
Sbjct: 1110 SIESLTLMSIAGLRALPEAIQCFTSLWRLSILGCGELETLPEWLGDYFTCLEEISIDTCP 1169
Query: 732 SLSSLPEEFGNLCNLKNLDMASC 754
LSSLPE L LK L + +C
Sbjct: 1170 MLSSLPESIRRLTKLKKLRITNC 1192
Score = 39.3 bits (90), Expect = 0.52
Identities = 29/92 (31%), Positives = 45/92 (48%), Gaps = 5/92 (5%)
Query: 597 LGSLQNLEKIRLE--RILVPSFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELN 654
L S+++L + + R L + +L +LS+ C + + + D F LEE++
Sbjct: 1108 LSSIESLTLMSIAGLRALPEAIQCFTSLWRLSILGCGELETLPEW---LGDYFTCLEEIS 1164
Query: 655 IDYCKDLVVLQTGICDIISLKKLNVTNCHKLS 686
ID C L L I + LKKL +TNC LS
Sbjct: 1165 IDTCPMLSSLPESIRRLTKLKKLRITNCPVLS 1196
Score = 36.6 bits (83), Expect = 3.4
Identities = 25/80 (31%), Positives = 40/80 (49%), Gaps = 2/80 (2%)
Query: 695 LENLELLSFSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCN-LKNLDMAS 753
L ++E L+ S L A+P +I +L L I C L +LPE G+ L+ + + +
Sbjct: 1108 LSSIESLTLMSIAGLRALPEAIQCFTSLWRLSILGCGELETLPEWLGDYFTCLEEISIDT 1167
Query: 754 CASI-ELPFSVVNLQNLKTI 772
C + LP S+ L LK +
Sbjct: 1168 CPMLSSLPESIRRLTKLKKL 1187
>gb|AAG60098.1| disease resistance protein, putative [Arabidopsis thaliana]
Length = 1398
Score = 132 bits (332), Expect = 5e-29
Identities = 86/221 (38%), Positives = 123/221 (54%), Gaps = 23/221 (10%)
Query: 567 PESIKKMSKLKVLIITNYSFHPCELNNFELLGSLQNLEKIRLERI--------LVPSFGT 618
P SI + LK+L + S + E+ S+ NL ++L + L S G
Sbjct: 827 PSSIGNLISLKILYLKRIS------SLVEIPSSIGNLINLKLLNLSGCSSLVELPSSIGN 880
Query: 619 LKNLKKLSLYMCNTI--LAFEKGSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKK 676
L NLKKL L C+++ L G+++ NL+EL + C LV L + I ++I+LK
Sbjct: 881 LINLKKLDLSGCSSLVELPLSIGNLI------NLQELYLSECSSLVELPSSIGNLINLKT 934
Query: 677 LNVTNCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSL 736
LN++ C L LP IG L NL+ L S C+ L +P+SIG L+NLK LD+S C SL L
Sbjct: 935 LNLSECSSLVELPSSIGNLINLQELYLSECSSLVELPSSIGNLINLKKLDLSGCSSLVEL 994
Query: 737 PEEFGNLCNLKNLDMASCAS-IELPFSVVNLQNLKTITCDE 776
P GNL NLK L+++ C+S +ELP S+ NL NL+ + E
Sbjct: 995 PLSIGNLINLKTLNLSECSSLVELPSSIGNLINLQELYLSE 1035
Score = 121 bits (304), Expect = 8e-26
Identities = 75/184 (40%), Positives = 110/184 (59%), Gaps = 15/184 (8%)
Query: 597 LGSLQNLEKIRLERI-----LVPSFGTLKNLKKLSLYMCNTILAFEK--GSILISDAFPN 649
+G+L NL+ + L L S G L NL++L L C++++ G+++ N
Sbjct: 926 IGNLINLKTLNLSECSSLVELPSSIGNLINLQELYLSECSSLVELPSSIGNLI------N 979
Query: 650 LEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTDL 709
L++L++ C LV L I ++I+LK LN++ C L LP IG L NL+ L S C+ L
Sbjct: 980 LKKLDLSGCSSLVELPLSIGNLINLKTLNLSECSSLVELPSSIGNLINLQELYLSECSSL 1039
Query: 710 EAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMASCAS-IELPFSVVNLQN 768
+P+SIG L+NLK LD+S C SL LP GNL NLK L+++ C+S +ELP S+ NL N
Sbjct: 1040 VELPSSIGNLINLKKLDLSGCSSLVELPLSIGNLINLKTLNLSGCSSLVELPSSIGNL-N 1098
Query: 769 LKTI 772
LK +
Sbjct: 1099 LKKL 1102
Score = 121 bits (303), Expect = 1e-25
Identities = 97/307 (31%), Positives = 144/307 (46%), Gaps = 29/307 (9%)
Query: 476 YQSTKQPFEQRKRLIIDINKNRSELAEKQQSLLTRILSKVMRLCIKQNPQQLAAHILSVS 535
Y T P + + ++ I SEL + + + + KVM L + ++L +++
Sbjct: 656 YPMTSLPSKFNLKFLVKIILKHSELEKLWEGIQPLVNLKVMDLRYSSHLKELPNLSTAIN 715
Query: 536 TDEACASDWSQMQPTQVEVLILNLHTKQYSFPESIKKMSKLKVLIITNYSFH---PCELN 592
E SD S + P SI + +K L I S P +
Sbjct: 716 LLEMVLSDCSSL----------------IELPSSIGNATNIKSLDIQGCSSLLKLPSSIG 759
Query: 593 NFELLGSLQNLEKIRLERILVPSFGTLKNLKKLSLYMCNTILAFEK--GSILISDAFPNL 650
N L L + L L S G L NL +L L C++++ G+++ NL
Sbjct: 760 NLITLPRLDLMGCSSLVE-LPSSIGNLINLPRLDLMGCSSLVELPSSIGNLI------NL 812
Query: 651 EELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTDLE 710
E C L+ L + I ++ISLK L + L +P IG L NL+LL+ S C+ L
Sbjct: 813 EAFYFHGCSSLLELPSSIGNLISLKILYLKRISSLVEIPSSIGNLINLKLLNLSGCSSLV 872
Query: 711 AIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMASCAS-IELPFSVVNLQNL 769
+P+SIG L+NLK LD+S C SL LP GNL NL+ L ++ C+S +ELP S+ NL NL
Sbjct: 873 ELPSSIGNLINLKKLDLSGCSSLVELPLSIGNLINLQELYLSECSSLVELPSSIGNLINL 932
Query: 770 KTITCDE 776
KT+ E
Sbjct: 933 KTLNLSE 939
Score = 120 bits (301), Expect = 2e-25
Identities = 181/708 (25%), Positives = 293/708 (40%), Gaps = 123/708 (17%)
Query: 147 VGLDIPFSKLKMELIRDGSS---TLVLTGLGGLGKTTLATKLC------WDQEVNGKFKE 197
+G+ K+K L D + T+ ++G G+GK+T+A L + V KFK
Sbjct: 253 IGMKAHIEKMKQLLCLDSTDERRTVGISGPSGIGKSTIARVLHNQISDGFQMSVFMKFKP 312
Query: 198 NIIFVTFSKTPMLKTIVERIIEHCGYPVPEFQSDEDA-VNKLELLLKKVEGSPLLLVLDD 256
+ S +K +E+ + + + ED +++L V G +L+VLD
Sbjct: 313 SYTRPICSDDHDVKLQLEQQF------LAQLINQEDIKIHQLGTAQNFVMGKKVLIVLDG 366
Query: 257 VWPTSESLVKKLQFEIS-----DFKILVTSRVS--FPRFRTTCILK---PLAHEDAVTLF 306
V + LV+ L + +I++T++ F+ I P HE A+ +F
Sbjct: 367 V----DQLVQLLAMPKAVCLGPGSRIIITTQDQQLLKAFQIKHIYNVDFPPDHE-ALQIF 421
Query: 307 HHYAQMEKNSSDIIDKNLVEKVVRSCKGLPLTIKVIATSLRNRPDDLWRKIVMELSQGHS 366
+A + D +K L KV R LPL ++V+ + R + W+ EL +
Sbjct: 422 CIHAFGHDSPDDGFEK-LATKVTRLAGNLPLGLRVMGSHFRGMSKEDWKG---ELPRLRI 477
Query: 367 ILDSNTELLTRLQKIFDVLEDNPTIMECFMDIALFPEDHRIPVAALVDMWAKLYKLDDNG 426
LD E+ + L+ +DVL+D + F+ IA F D I D + G
Sbjct: 478 RLDG--EIGSILKFSYDVLDDED--KDLFLHIACFFNDEGID-HTFEDTLRHKFSNVQRG 532
Query: 427 IQAMEIINKLGIMNLANVIIPRKNASDTDNNNYNNHFIIL-HDILRDLGIYQSTKQPF-- 483
+Q V++ R S+ +N + L +I+R+ +Y+ K+ F
Sbjct: 533 LQ---------------VLVQRSLISEDLTQPMHNLLVQLGREIVRNQSVYEPGKRQFLV 577
Query: 484 -----------EQRKRLIIDIN---------KNRSELAEKQQSLLTRIL---SKVMRLCI 520
+I IN N S+ + S L + RL +
Sbjct: 578 DGKEICEVLTSHTGSESVIGINFEVYWSMDELNISDRVFEGMSNLQFFRFDENSYGRLHL 637
Query: 521 KQNPQQLAAHILSVSTDEACASDWSQMQPTQVEVLILNLHTKQYSFPESIKKMSKLKVLI 580
Q L + + D + + V I+ H++ E I+ + LKV+
Sbjct: 638 PQGLNYLPPKLRILHWDYYPMTSLPSKFNLKFLVKIILKHSELEKLWEGIQPLVNLKVMD 697
Query: 581 ITNYSFHPCELNNFELLGSLQNLEKIRLERI-----LVPSFGTLKNLKKLSLYMCNTILA 635
+ YS H EL N L + NL ++ L L S G N+K L + C+++L
Sbjct: 698 L-RYSSHLKELPN---LSTAINLLEMVLSDCSSLIELPSSIGNATNIKSLDIQGCSSLLK 753
Query: 636 FEK--GSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIG 693
G+++ L L++ C LV L + I ++I+L +L++ C L LP IG
Sbjct: 754 LPSSIGNLI------TLPRLDLMGCSSLVELPSSIGNLINLPRLDLMGCSSLVELPSSIG 807
Query: 694 KLENLELLSFSSC------------------------TDLEAIPTSIGKLLNLKHLDISN 729
L NLE F C + L IP+SIG L+NLK L++S
Sbjct: 808 NLINLEAFYFHGCSSLLELPSSIGNLISLKILYLKRISSLVEIPSSIGNLINLKLLNLSG 867
Query: 730 CISLSSLPEEFGNLCNLKNLDMASCAS-IELPFSVVNLQNLKTITCDE 776
C SL LP GNL NLK LD++ C+S +ELP S+ NL NL+ + E
Sbjct: 868 CSSLVELPSSIGNLINLKKLDLSGCSSLVELPLSIGNLINLQELYLSE 915
Score = 115 bits (289), Expect = 4e-24
Identities = 87/251 (34%), Positives = 129/251 (50%), Gaps = 29/251 (11%)
Query: 556 ILNLHTKQYS-------FPESIKKMSKLKVLIITNYSFHPCELNNFELLGSLQNLEKIRL 608
++NL T S P SI + L+ L ++ S EL + +G+L NL+K+ L
Sbjct: 1001 LINLKTLNLSECSSLVELPSSIGNLINLQELYLSECS-SLVELPSS--IGNLINLKKLDL 1057
Query: 609 ERI-----LVPSFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVV 663
L S G L NLK L+L C++++ S NL++L++ C LV
Sbjct: 1058 SGCSSLVELPLSIGNLINLKTLNLSGCSSLVELPS-----SIGNLNLKKLDLSGCSSLVE 1112
Query: 664 LQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGKLLNLK 723
L + I ++I+LKKL+++ C L LP IG L NL+ L S C+ L +P+SIG L+NL+
Sbjct: 1113 LPSSIGNLINLKKLDLSGCSSLVELPLSIGNLINLQELYLSECSSLVELPSSIGNLINLQ 1172
Query: 724 HLDISNCISLSSLPEEFGNLCNLKNLDMASCASI----ELPFSVVNL-----QNLKTITC 774
L +S C SL LP GNL NLK LD+ C + +LP S+ L ++L+T+ C
Sbjct: 1173 ELYLSECSSLVELPSSIGNLINLKKLDLNKCTKLVSLPQLPDSLSVLVAESCESLETLAC 1232
Query: 775 DEETAATWEDF 785
W F
Sbjct: 1233 SFPNPQVWLKF 1243
>ref|NP_683486.1| leucine-rich repeat family protein [Arabidopsis thaliana]
Length = 703
Score = 132 bits (332), Expect = 5e-29
Identities = 86/221 (38%), Positives = 123/221 (54%), Gaps = 23/221 (10%)
Query: 567 PESIKKMSKLKVLIITNYSFHPCELNNFELLGSLQNLEKIRLERI--------LVPSFGT 618
P SI + LK+L + S + E+ S+ NL ++L + L S G
Sbjct: 132 PSSIGNLISLKILYLKRIS------SLVEIPSSIGNLINLKLLNLSGCSSLVELPSSIGN 185
Query: 619 LKNLKKLSLYMCNTI--LAFEKGSILISDAFPNLEELNIDYCKDLVVLQTGICDIISLKK 676
L NLKKL L C+++ L G+++ NL+EL + C LV L + I ++I+LK
Sbjct: 186 LINLKKLDLSGCSSLVELPLSIGNLI------NLQELYLSECSSLVELPSSIGNLINLKT 239
Query: 677 LNVTNCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSL 736
LN++ C L LP IG L NL+ L S C+ L +P+SIG L+NLK LD+S C SL L
Sbjct: 240 LNLSECSSLVELPSSIGNLINLQELYLSECSSLVELPSSIGNLINLKKLDLSGCSSLVEL 299
Query: 737 PEEFGNLCNLKNLDMASCAS-IELPFSVVNLQNLKTITCDE 776
P GNL NLK L+++ C+S +ELP S+ NL NL+ + E
Sbjct: 300 PLSIGNLINLKTLNLSECSSLVELPSSIGNLINLQELYLSE 340
Score = 121 bits (304), Expect = 8e-26
Identities = 75/184 (40%), Positives = 110/184 (59%), Gaps = 15/184 (8%)
Query: 597 LGSLQNLEKIRLERI-----LVPSFGTLKNLKKLSLYMCNTILAFEK--GSILISDAFPN 649
+G+L NL+ + L L S G L NL++L L C++++ G+++ N
Sbjct: 231 IGNLINLKTLNLSECSSLVELPSSIGNLINLQELYLSECSSLVELPSSIGNLI------N 284
Query: 650 LEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTDL 709
L++L++ C LV L I ++I+LK LN++ C L LP IG L NL+ L S C+ L
Sbjct: 285 LKKLDLSGCSSLVELPLSIGNLINLKTLNLSECSSLVELPSSIGNLINLQELYLSECSSL 344
Query: 710 EAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMASCAS-IELPFSVVNLQN 768
+P+SIG L+NLK LD+S C SL LP GNL NLK L+++ C+S +ELP S+ NL N
Sbjct: 345 VELPSSIGNLINLKKLDLSGCSSLVELPLSIGNLINLKTLNLSGCSSLVELPSSIGNL-N 403
Query: 769 LKTI 772
LK +
Sbjct: 404 LKKL 407
Score = 115 bits (289), Expect = 4e-24
Identities = 87/251 (34%), Positives = 129/251 (50%), Gaps = 29/251 (11%)
Query: 556 ILNLHTKQYS-------FPESIKKMSKLKVLIITNYSFHPCELNNFELLGSLQNLEKIRL 608
++NL T S P SI + L+ L ++ S EL + +G+L NL+K+ L
Sbjct: 306 LINLKTLNLSECSSLVELPSSIGNLINLQELYLSECS-SLVELPSS--IGNLINLKKLDL 362
Query: 609 ERI-----LVPSFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDLVV 663
L S G L NLK L+L C++++ S NL++L++ C LV
Sbjct: 363 SGCSSLVELPLSIGNLINLKTLNLSGCSSLVELPS-----SIGNLNLKKLDLSGCSSLVE 417
Query: 664 LQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGKLLNLK 723
L + I ++I+LKKL+++ C L LP IG L NL+ L S C+ L +P+SIG L+NL+
Sbjct: 418 LPSSIGNLINLKKLDLSGCSSLVELPLSIGNLINLQELYLSECSSLVELPSSIGNLINLQ 477
Query: 724 HLDISNCISLSSLPEEFGNLCNLKNLDMASCASI----ELPFSVVNL-----QNLKTITC 774
L +S C SL LP GNL NLK LD+ C + +LP S+ L ++L+T+ C
Sbjct: 478 ELYLSECSSLVELPSSIGNLINLKKLDLNKCTKLVSLPQLPDSLSVLVAESCESLETLAC 537
Query: 775 DEETAATWEDF 785
W F
Sbjct: 538 SFPNPQVWLKF 548
Score = 115 bits (288), Expect = 6e-24
Identities = 82/228 (35%), Positives = 118/228 (50%), Gaps = 13/228 (5%)
Query: 555 LILNLHTKQYSFPESIKKMSKLKVLIITNYSFH---PCELNNFELLGSLQNLEKIRLERI 611
++L+ + P SI + +K L I S P + N L L + L
Sbjct: 24 MVLSDCSSLIELPSSIGNATNIKSLDIQGCSSLLKLPSSIGNLITLPRLDLMGCSSLVE- 82
Query: 612 LVPSFGTLKNLKKLSLYMCNTILAFEK--GSILISDAFPNLEELNIDYCKDLVVLQTGIC 669
L S G L NL +L L C++++ G+++ NLE C L+ L + I
Sbjct: 83 LPSSIGNLINLPRLDLMGCSSLVELPSSIGNLI------NLEAFYFHGCSSLLELPSSIG 136
Query: 670 DIISLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSIGKLLNLKHLDISN 729
++ISLK L + L +P IG L NL+LL+ S C+ L +P+SIG L+NLK LD+S
Sbjct: 137 NLISLKILYLKRISSLVEIPSSIGNLINLKLLNLSGCSSLVELPSSIGNLINLKKLDLSG 196
Query: 730 CISLSSLPEEFGNLCNLKNLDMASCAS-IELPFSVVNLQNLKTITCDE 776
C SL LP GNL NL+ L ++ C+S +ELP S+ NL NLKT+ E
Sbjct: 197 CSSLVELPLSIGNLINLQELYLSECSSLVELPSSIGNLINLKTLNLSE 244
Score = 90.5 bits (223), Expect = 2e-16
Identities = 49/125 (39%), Positives = 69/125 (55%), Gaps = 1/125 (0%)
Query: 649 NLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTD 708
NL E+ + C L+ L + I + ++K L++ C L LP IG L L L C+
Sbjct: 20 NLLEMVLSDCSSLIELPSSIGNATNIKSLDIQGCSSLLKLPSSIGNLITLPRLDLMGCSS 79
Query: 709 LEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMASCAS-IELPFSVVNLQ 767
L +P+SIG L+NL LD+ C SL LP GNL NL+ C+S +ELP S+ NL
Sbjct: 80 LVELPSSIGNLINLPRLDLMGCSSLVELPSSIGNLINLEAFYFHGCSSLLELPSSIGNLI 139
Query: 768 NLKTI 772
+LK +
Sbjct: 140 SLKIL 144
Score = 83.6 bits (205), Expect = 2e-14
Identities = 46/115 (40%), Positives = 66/115 (57%), Gaps = 5/115 (4%)
Query: 657 YCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSFSSCTDLEAIPTSI 716
+ K+L L T I +L ++ +++C L LP IG N++ L C+ L +P+SI
Sbjct: 8 HLKELPNLSTAI----NLLEMVLSDCSSLIELPSSIGNATNIKSLDIQGCSSLLKLPSSI 63
Query: 717 GKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMASCAS-IELPFSVVNLQNLK 770
G L+ L LD+ C SL LP GNL NL LD+ C+S +ELP S+ NL NL+
Sbjct: 64 GNLITLPRLDLMGCSSLVELPSSIGNLINLPRLDLMGCSSLVELPSSIGNLINLE 118
>gb|AAN23084.1| putative rp3 protein [Zea mays]
Length = 944
Score = 121 bits (303), Expect = 1e-25
Identities = 166/669 (24%), Positives = 278/669 (40%), Gaps = 76/669 (11%)
Query: 139 VPENPKFTV-------GLDIPFSKLKMELIRDGSSTLVLT--GLGGLGKTTLATKLCWDQ 189
+ E P +T+ G D +++ ELI S +++ GLGG GKTTLA + D
Sbjct: 155 IGEVPLYTIVDETTIFGRDQAKNQIISELIETDSQQKIVSVIGLGGSGKTTLAKLVFNDG 214
Query: 190 EVNGKFKENIIFVTFSKTPMLKTIVERIIEHCGYPVPEFQSDEDAVNKLELLLK-KVEGS 248
+ F E +++V S+ + VE+++E + SD + + + K+ G
Sbjct: 215 NIIKHF-EVVLWVHVSR----EFAVEKLVEKLFKAIAGDMSDHPPLQHVSRTISDKLVGK 269
Query: 249 PLLLVLDDVWPTS----ESLVKKLQFEISDFKILVTSR----VSFPRFRTTCILKPLAHE 300
L VLDDVW E + L+ IL+T+R L L+ E
Sbjct: 270 RFLAVLDDVWTEDRVEWEQFMVHLKSGAPGSSILLTTRSRKVAEAVDSSYAYNLPFLSKE 329
Query: 301 DAVTLFHHYAQMEKNSSDIIDKNLVEKVVRSCKGLPLTIKVIATSLRN-RPDDLWRKIVM 359
D+ +F + + D +++V C G+PL IKVIA L + + WR I
Sbjct: 330 DSWKVFQQCFGIALKALDPEFLQTGKEIVEKCGGVPLAIKVIAGVLHGIKGIEEWRSICD 389
Query: 360 ELSQGHSILDSNTELLTRLQKIFDVLEDNPTIMECFMDIALFPEDHRIPVAALVDMWA-- 417
S + D + L F L D+ + CF+ ++FP + I L+ W
Sbjct: 390 --SNLLDVQDDEHRVFACLSLSFVHLPDH--LKPCFLHCSIFPRGYVINRRHLISQWIAH 445
Query: 418 ------KLYKLDDNGIQAMEIINKLGIMNLANVIIPRKNASDTDNNNYNNHFIILHDILR 471
+ + +D GI + + K+G + +V I T +HD++
Sbjct: 446 GFVPTNQARQAEDVGIGYFDSLLKVGFLQ-DHVQIWSTRGEVTCK---------MHDLVH 495
Query: 472 DLGIYQSTKQPFEQRKRLIIDINKNRSELAEKQQSLLTRILSKVMRLCIKQNPQQLAAHI 531
DL +Q R + +I N+ + SL + +LC K +
Sbjct: 496 DLA-----RQIL--RDEFVSEIETNKQIKRCRYLSLTSCTGKLDNKLCGKVRALYVCGRE 548
Query: 532 LSVSTDEACASDWSQMQPTQVEVLILNLHTKQYSFPESIKKMSKLKVLIITNYSFHPCEL 591
L D + + V +IL T S P + K L L I++ + CE
Sbjct: 549 LEF--------DKTMNKQCCVRTIILKYITDD-SLPLFVSKFEYLGYLEISDVN---CEA 596
Query: 592 --NNFELLGSLQNLEKIRLERILV--PSFGTLKNLKKLSLYMCNTILAFEKGSILISDAF 647
+LQ L + R+ V S G LK L+ L L ++I + + I D
Sbjct: 597 LPEALSRCWNLQALHVLNCSRLAVVPESIGKLKKLRTLELNGVSSIKSLPQS---IGDC- 652
Query: 648 PNLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLP--QDIGKLENLELLSFSS 705
NL L ++ C+ + + + + +L+ L++ +C L LP GKL NL+ ++F+
Sbjct: 653 DNLRRLYLEECRGIEDIPNSLGKLENLRILSIVDCVSLQKLPPSDSFGKLLNLQTITFNL 712
Query: 706 CTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMASCASIE-LPFSVV 764
C +L +P + L++L+ +D+ C L LPE GNL NLK L++ C + LP
Sbjct: 713 CYNLRNLPQCMTSLIHLESVDLGYCFQLVELPEGMGNLRNLKVLNLKKCKKLRGLPAGCG 772
Query: 765 NLQNLKTIT 773
L L+ ++
Sbjct: 773 KLTRLQQLS 781
>gb|AAO24595.1| At5g66630 [Arabidopsis thaliana]
Length = 702
Score = 119 bits (299), Expect = 3e-25
Identities = 66/151 (43%), Positives = 95/151 (62%), Gaps = 2/151 (1%)
Query: 152 PFSKLKMELIRDGSSTLVLTGLGGLGKTTLATKLCWDQEVNGKFKENIIFVTFSKTPMLK 211
P +LK L DG T+V++ LGKTTL TKLC D +V KFK+ I F++ SK P ++
Sbjct: 175 PLMELKKMLFEDGVVTVVVSAPYALGKTTLVTKLCHDADVKEKFKQ-IFFISVSKFPNVR 233
Query: 212 TIVERIIEHCGYPVPEFQSDEDAVNKLELLLKKV-EGSPLLLVLDDVWPTSESLVKKLQF 270
I +++EH G E+++D DA+ ++ LLK++ +LLVLDDVW ESL++K
Sbjct: 234 LIGHKLLEHIGCKANEYENDLDAMLYIQQLLKQLGRNGSILLVLDDVWAEEESLLQKFLI 293
Query: 271 EISDFKILVTSRVSFPRFRTTCILKPLAHED 301
++ D+KILVTSR F F T LKPL ++
Sbjct: 294 QLPDYKILVTSRFEFTSFGPTFHLKPLIDDE 324
>ref|NP_201464.2| LIM domain-containing protein [Arabidopsis thaliana]
Length = 702
Score = 119 bits (299), Expect = 3e-25
Identities = 66/151 (43%), Positives = 95/151 (62%), Gaps = 2/151 (1%)
Query: 152 PFSKLKMELIRDGSSTLVLTGLGGLGKTTLATKLCWDQEVNGKFKENIIFVTFSKTPMLK 211
P +LK L DG T+V++ LGKTTL TKLC D +V KFK+ I F++ SK P ++
Sbjct: 175 PLMELKKMLFEDGVVTVVVSAPYALGKTTLVTKLCHDADVKEKFKQ-IFFISVSKFPNVR 233
Query: 212 TIVERIIEHCGYPVPEFQSDEDAVNKLELLLKKV-EGSPLLLVLDDVWPTSESLVKKLQF 270
I +++EH G E+++D DA+ ++ LLK++ +LLVLDDVW ESL++K
Sbjct: 234 LIGHKLLEHIGCKANEYENDLDAMLYIQQLLKQLGRNGSILLVLDDVWAEEESLLQKFLI 293
Query: 271 EISDFKILVTSRVSFPRFRTTCILKPLAHED 301
++ D+KILVTSR F F T LKPL ++
Sbjct: 294 QLPDYKILVTSRFEFTSFGPTFHLKPLIDDE 324
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.319 0.136 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,281,578,580
Number of Sequences: 2540612
Number of extensions: 53192101
Number of successful extensions: 218692
Number of sequences better than 10.0: 4171
Number of HSP's better than 10.0 without gapping: 1459
Number of HSP's successfully gapped in prelim test: 2747
Number of HSP's that attempted gapping in prelim test: 192463
Number of HSP's gapped (non-prelim): 15335
length of query: 807
length of database: 863,360,394
effective HSP length: 136
effective length of query: 671
effective length of database: 517,837,162
effective search space: 347468735702
effective search space used: 347468735702
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 79 (35.0 bits)
Medicago: description of AC148347.6


